Abstract
Nervous system polycomb 1 (NSPc1) is a member of the polycomb group (PcG) family of proteins and has been demonstrated to maintain the differentiation and pluripotency of stem cells. Long non-coding RNAs (lncRNAs) have been demonstrated to be involved in the control of pluripotency and differentiation in embryonic and pluripotent cells. In the present study, the expression levels of NSPc1 were associated with the malignant potential of various glioma cell lines. Additionally, lncRNAs were differentially expressed in glioblastoma cell lines. Following induced differentiation of U87 glioblastoma cells with all-trans retinoic acid, the expression levels of NSPc1 decreased initially, reaching its lowest point on day 6, but then subsequently increased until day 10. The expression of lncRNA candidates decreased in the cell differentiation stage. Additionally, the expression of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), sex-determining region of the Y chromosome-box 2 overlapping transcript (SOX2OT) and antisense non-coding RNA in the INK4 locus (ANRIL) was significantly altered relative to the expression levels of NSPc1. RNA immunoprecipitation (RIP) assays demonstrated that MALAT1, SOX2OT and ANRIL bind to NSPc1 in U87 glioblastoma cells and the enrichment of ANRIL in anti-NSPc1 antibody group was associated with the expression levels of NSPc1 during U87 cell differentiation. Small interfering RNA mediated downregulation of NSPc1 expression with MALAT1, SOX2OT and ANRIL, inhibited the proliferation, and promoted apoptosis in U87 cells. The results of the present study demonstrate that MALAT1, SOX2OT and ANRIL combine and crosstalk with NSPc1 in U87 cells to affect proliferation and apoptosis.
Keywords: polycomb protein, long-noncoding RNA, glioma, epigenetics
Introduction
Glioblastoma (GBM) is one of most fatal types of primary brain tumors with high tumorigenesis, rapid growth and high invasiveness. GBM is defined as a grade III or IV malignant tumor by the World Health Organization (WHO) that most frequently involves the frontal, parietal and temporal lobes (1). Complete resection of tumors is made difficult as a result of its location and its resistance to chemical and radical therapies (2). Therefore, the two-year survival rate of patients with high-grade malignancy of glioblastoma is <9% (3). Due to the limits of standard therapy, a novel and effective therapy is needed to improve prognosis. Targeted molecular therapies have made marked progress in treatment, with less severe adverse effects and a significant increase in the cure rate (4). Therefore, seeking out potential molecular targets for glioblastoma is becoming an increasing area of interest.
Polycomb group (PcG) proteins, originally discovered in the Drosophila melanogaster, are abundantly present in cells, suppress gene transcription through epigenetic remodeling, and serve as determinants of cell fate, thus serving important roles in embryonic development, embryonic stem cells (ESC) and adult stem cells maintenance and tumor formation (5–9). The PcG family functions as complexes, which are divided into polycomb repressive complex (PRC) 1 and PRC2 (10). Furthermore, PRC2 regulates histone H3K27 methyltransferase activity, resulting in the recruitment of PRC1, transcriptional repression and gene silencing (11,12). Nervous system polycomb 1 (NSPc1), a novel member of PRC1, was originally identified in 2001 (13). The expression of mouse NSPc1 is relatively high and localized to the nervous system during early embryonic development. Additionally, NSPc1 serves an important role in stemness maintenance of octamer (oct)-sex determining region of the Y chromosome-box 2 (Sox2)-homeobox protein nanog axis by directly activating the promoter of Oct4 in P19 embryonic carcinoma stem cells (14) and overexpression of Nspc1 induces ESC to self-renew independent of leukemia inhibitory factor in the culture media in vitro (15). Furthermore, Yan et al (16) recently demonstrated that although the Nspc1 mutant cells were viable and retained normal self-renewal capabilities, they presented with severe defects in differentiation. Additionally, NSPc1 regulates retinoic acid response element to downregulate p21Waf1/COP1 interacting protin-1, which is associated with the differentiation and metastasis of tumor cells (17). As NSPc1 possesses a marked amount of homology with the proto-oncogene and polycomb protein BMI-1 (13), it may additionally serve a role in tumorigenesis as a proto-oncogene, as has been demonstrated in malignant glioma (18).
Long non-coding RNA (lncRNAs) is a variant of non-coding RNA of 200 bp in length with a lack of an open reading frame (19). LncRNA was firstly identified by Okazaki et al (20) in mouse full-length cDNA library. Recent studies have demonstrated that lncRNAs are involved in X-chromosome silencing, gene imprinting, chromosome modification, transcription activation and the occurrence and progression of a number of diseases (21). Aside from forming triplets by specifically combining with DNA and RNA, lncRNA binds to protein to regulate gene expression and possibly cancer progression. It has been indicated that lncRNA recruits chromosome to certain loci for gene silence in epigenetic regulation (19).
At present, a number of lncRNAs have been identified as directing epigenetic silencing through interactions with PcG proteins during the initiation and progression of cancer (22). It has been demonstrated that the 5′ end of lncRNA, homeobox protein transcript antisense RNA (HOTAIR) combined with PRC2 drives target gene epigenetically silencing (23). Furthermore, lncRNA antisense non-coding RNA in the INK4 locus (ANRIL) in conjunction with PRC1 and PRC2 may regulate H3K27 methylation of the INK4b-ARF-INK4a locus (24).
The crosstalk between PRC members and lncRNAs may serve a crucial role in epigenetically silencing gene expression, resulting in the formation and development of tumors. In the present study, eight lncRNAs were selected via bioinformatics (25) as potential NSPc1 interacting and crosstalking molecules in various types of nervous system tumor cell lines. The present study aimed to determine among these eight candidates the potential lncRNAs binding to PRC1 member NSPc1, and to examine the possible crosstalks between NSPc1 and lncRNAs in glioma cells. The results demonstrated that metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), Sox2 overlapping transcript (SOX2OT) and ANRIL may interact with NSPc1 in U87 glioma cells, and ANRIL may combine with NSPc1 to affect the proliferation and apoptosis of U87 glioma cells.
Materials and methods
Cell culture and induction of differentiation
U87 cells (derived from malignant glioma of unknown origin; WHO grade IV) and H4 cells (derived from low-grade malignant glioma; WHO grade II) were acquired from American Type Culture Collection (Manassas, VA, USA). U251 cells (derived from glioblastoma astrocytoma, additionally known as U-373; WHO grade III) were obtained from the Type Culture Collection of the Chinese Academy of Medical Science (Shanghai, China). All the above cell lines used have been authenticated using STR profiling. The cell lines were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific Inc.) at 37°C in an incubator containing 5% CO2. Differentiation of U87 cells was induced by adding 1 µM of all-trans retinoic acid (ATRA) (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) to the medium.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using TRIzol® Reagent (Invitrogen; Thermo Fisher Scientific Inc.). RT-qPCR was performed with the primer pair sequences for lncRNAs and NSPc1 designed as follows: MALAT1, forward 5′-AAAGCAAGGTCTCCCCACAAG-3′, reverse 5′-GGTCTGTGCTAGATCAAAAGGCA-3′; SOX2OT, forward 5′-AGGCATAGCATCCACCCTAA-3′, reverse 5′-AGCCCTCACACCTCCTTACTT-3′; HOTAIR, forward 5′-GGTAGAAAAAGCAACCACGAAGC-3′, reverse 5′-ACATAAACCTCTGTCTGTGAGTGCC-3′; H19, forward 5′-TTCAAAGCCTCCACGACTCT-3′, reverse 5′-GCTCACACTCACGCACACTC-3′; ANRIL forward 5′-GAAGAAGCAAAAGCGGAAAC-3′, reverse 5′-GGACCCAGAGGGAGGTAAAT-3′; alu-mediated CDKN1A/P21 transcriptional regulator (APTR), forward 5′-GTGGGTATCAGGGAAGAACC-3′, reverse 5-′CAGGAAAAATGGAAGGGAAA-3′; maternally expressed 3 (MEG3), forward 5′-GGGAGTGGAAAGAAACA-3′, reverse 5′-TGTGGATGGCTTGTGCTAAA-3; ADAMTS9 antisense RNA 2 (ADAMTS9-AS2), forward 5′-AACACAAACACCTTGGGTA-3, reverse 5′-GGGTCTCTTGGCTTCGTAT-3′; NSPc1, forward 5′-GAGACACAGCCACTGCTCAA-3′, reverse 5′-TGCACATGCTGAGGGTTTAG-3′; and GAPDH, forward, 5′-GAAGGTCGGAGTCAACGGATT-3′ and reverse 5′-CGCTCCTGGAAGATGGTGAT-3′. RT-qPCR was achieved on Step one Plus™ System (Applied Biosystems; Thermo Fisher Scientific Inc.) using SYBR Premix EX Taq™ (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocol. The amplification conditions were 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 1 min. Relative gene expression was normalized to the housekeeping gene GAPDH. The relative expression fold change was calculated using the 2−ΔΔCq method (26). The RT-qPCR products of NSPc1 and GAPDH were also removed for electrophoresis in 2% agarose gel, and were further visualized using a UV Transilluminator Gel Docu 2000 (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All quantifications are the averages of at least three biological replicates.
Western blot analysis
Western blot analysis was performed as described previously (14). Cells were lysed using radioimmunoprecipitation assay buffer (cat. no. P0013B; Beyotime Institute of Biotechnology, Nanjing, China) and the protein concentration was determined using a bicinchonininc acid assay kit (cat. no. P0009; Beyotime Institute of Biotechnology). Proteins (20 µg) were separated by 10% SDS-PAGE, and then transferred onto polyvinylidene difluoride membranes (ΕΜD Millipore, Billerica, MA, USA). Membranes were blocked with 5% skimmed milk dissolved inTris-buffered saline supplemented with 1% Tween-20 at room temperature for 1 h, and incubated with primary antibodies against NSPc1 [1:5,000, prepared in our laboratory (27)] and GAPDH (1:6,000; cat. no. AP0063; Bioworld, Inc., St. Louis Park, MN, USA) at 4°C overnight. Membranes were then incubated with the horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin (Ig) G secondary antibody (1:4,000; cat. no. sc-2004; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) for 30 min at room temperature. Bands were visualized with enhanced chemiluminescence reagent (Merck KGaA). Data were analysed via densitometry using ImageJ (version 1.47v; National Institute of Health, Bethesda, MD, USA) and normalized to expression of GAPDH. All the densitometry analyses are based on four-five repeats and the blots presented are representative of the technical repeats.
RNA binding protein immunoprecipitation (RIP) assay
RIP assays were performed using U87 cells using the Magna RIP kit (Merck KGaA), according to the manufacturer's protocol. lncRNAs-NSPc1 immunoprecipitation was performed using anti-NSPc1 monoclonal antibody (m-NSPc1) (2 µg) developed in our laboratory (14), anti-enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) antibody (1 µg; cat. no. 07689; EMD Millipore) as a positive control and IgG beads (Merck KGaA) as a negative control. In addition, purified RNAs isolated by RIP were detected by RT-qPCR.
Cell transfection
U87 cells were cultured to 60–70% confluence and transfected with either pCDEF-NSPc1 (in order to overexpress NSPc1; pCDEF-empty as control vector), pSE-NSPc1 (in order to knockdown the NSPc1 gene; pSE-empty as control vector) (27) or small interfering (si)RNAs against MALAT1, SOX2OT and ANRIL. A universal siRNA with no homology with mammalian gene sequence was used as negative control. Sequences were designed as follows: MALAT1, forward 5′-GGGCUUCUCUUAACAUUUAUU-3′, reverse 5′-UAAAUGUUAAGAGAAGCCCUU)-3′ (50 nM) (27,28); SOX2OT, forward 5′-GGAGAUUGUGACCUGGCUUTT-3′, reverse 5′-AAGCCAGGUCACAAUCUCCTT)-3′ (50 nM) (29); ANRIL, forward 5′-GGUCAUCUCAUUGCUCUAUTT-3′, reverse 5′-CCAGUAGAGUAACGAGAUATT)-3′ (25 nM) (24,30); and universal siRNA negative control, forward 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse 5′-ACGUGACACGUUCGGAGAATT-3′ (50 nM). Transfections were performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature for 25 min and at 37°C in an incubator containing 5% CO2 for 6 h. Plasmids were constructed and extracted in our laboratory and siRNAs were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China).
Flow cytometry
Fluorescein isothiocyanate-AnnexinV (BD Biosciences, Franklin Lakes, NJ, USA) and propidium iodide (Sigma-Aldrich; Merck KGaA) were used to assess cell apoptosis according to the manufacturer's protocol. U87 cells were cultured to 60–70% confluence and transfected with siMALAT1, siSOX2OT, siANRIL and the negative control in 6-well plate. Apoptosis of transfected U87 cells were detected using a flow cytometer (BD FACS Calibur™, BD Biosciences) equipped with CellQuest software version 5.1 (BD Biosciences).
MTT assay
U87 cells were cultured to 30–50% confluence and transfected with siMALAT1, siSOX2OT, siANRIL, pSE-NSPc1 and the negative control in a 24-well plate. MTT assay (Sigma-Aldrich; Merck KGaA) was used to measure the proliferation of the transfected U87 cells. Following the manufacturer's protocol, the absorbance values were detected at optical density 490 nm, 24, 48, 72 and 96 h subsequent to transfection (independently).
Statistical methods
All results were expressed as the means ± standard deviation. Statistical analyses were performed using SPSS software (version 13.0; SPSS, Inc., Chicago, IL, USA). Comparisons between two groups were analyzed by a Student's t-test. One-way analysis of variance was applied for multiple comparisons with a post-hoc Student-Newman-Keuls test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of endogenous lncRNAs and NSPc1 in glioma cells
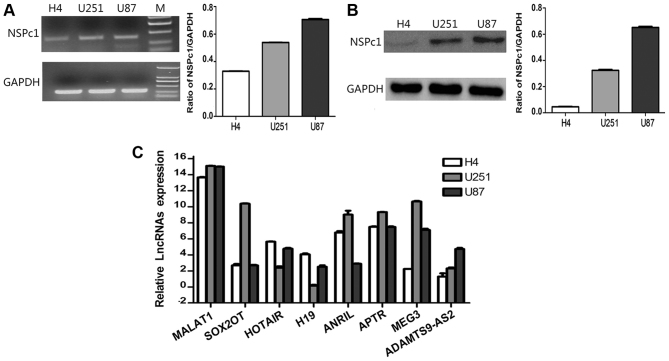
mRNA and protein expression levels of NSPc1 were associated with the degree of malignancy of glioma cell lines (Fig. 1A and B). Furthermore, the expression levels of MALAT1 and APTR were higher in glioma U87 cells, H4 cells and U251 cells, whereas HOTAIR, H19 and ADAMTS9-AS2 expression was lower in glioma cells. ANRIL expression levels were relatively high in H4 cells and U251 cells compared with U87 cells. However, the expression levels of MEG3 were relatively higher in U251 and U87 cells compared with H4 cells (Fig. 1C).
Figure 1.
Expression levels of NSPc1 and lncRNAs in H4, U251 and U87 glioma cell lines. (A) mRNA expression levels of NSPc1 as detected by RT-qPCR in glioma cells normalized to GAPDH and quantified using the 2−ΔΔCq method (right panel). RT-qPCR products were also removed for electrophoresis and visualized using a UV Transilluminator (left panel). (B) Protein expression levels of NSPc1 in glioma cells as detected by western blotting with GAPDH as an internal control. (C) Expression levels of lncRNAs in the glioma cell lines. lncRNA candidates' expression analysis was performed using RT-qPCR. Expression of each lncRNA candidate was quantified using the 2−ΔΔCq method with the expression of GAPDH used as the internal control. NSPc1, nervous system polycomb-1; lncRNAs, long non-coding RNAs; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; MALAT1, metastasis associated lung adenocarcinoma transcript 1; ANRIL, antisense non-coding RNA in the INK4 locus; SOX2OT, sex-determining region of the Y chromosome-box 2 overlapping transcript; HOTAIR, homeobox protein transcript antisense RNA; APTR, alu-mediated CDKN1A/P21 transcriptional regulator; MEG3, maternally expressed 3; ADAMTS9-AS2, ADAMTS9 antisense RNA 2.
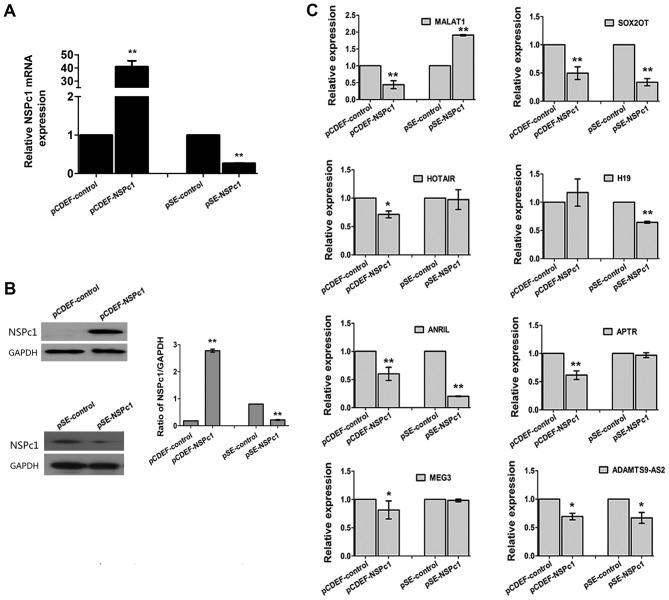
MALAT1 expression is inversely associated with NSPc1 expression in U87 cells
To investigate whether the variability of NSPc1 expression influenced the expression of any of the eight lncRNAs, the overexpression vector, pCDEF-NSPc1, and the knockdown vector, pSE-NSPc1, were used. The expression of MALAT1 decreased following upregulation of NSPc1, and it increased when expression of NSPc1 was knocked down (Fig. 2). The expression of SOX2OT and ANRIL decreased, irrespective of NSPc1 expression levels in U87 cells. The remaining lncRNA candidates exhibited a significant difference in expression (P<0.01) only following NSPc1 overexpression or knockdown, whereas SOX2OT and ANRIL expressions were both significantly decreased (P<0.01) following NSPc1 overexpression and knockdown (Fig. 2C).
Figure 2.
The expression of NSPc1 and lncRNAs in U87 cells subsequent to increasing or decreasing the expression of NSPc1. U87 cells were transfected with pCDEF-NSPc1 (overexpression plasmid) or pSE-NSPc1 (knockdown plasmid). (A) NSPc1 expression levels were determined using reverse transcription-quantitative polymerase chain reaction. GAPDH was used as the internal control. Results were analyzed using the 2−ΔΔCq method. (B) Protein expression levels of NSPc1 following transfection or knockdown of NSPc1 in U87 cells. GAPDH was used as an internal control. (C) lncRNA expression levels subsequent to regulation of NSPc1 expression with GAPDH as an internal control. Results were analyzed using the 2−ΔΔCq method. *P<0.05, **P<0.01 vs. control group. NSPc1, nervous system polycomb-1; lncRNAs, long non-coding RNAs; lncRNAs, long non-coding RNAs; MALAT1, metastasis associated lung adenocarcinoma transcript 1; ANRIL, antisense non-coding RNA in the INK4 locus; SOX2OT, sex-determining region of the Y chromosome-box 2 overlapping transcript; HOTAIR, homeobox protein transcript antisense RNA; APTR, alu-mediated CDKN1A/P21 transcriptional regulator; MEG3, maternally expressed 3; ADAMTS9-AS2, ADAMTS9 antisense RNA 2.
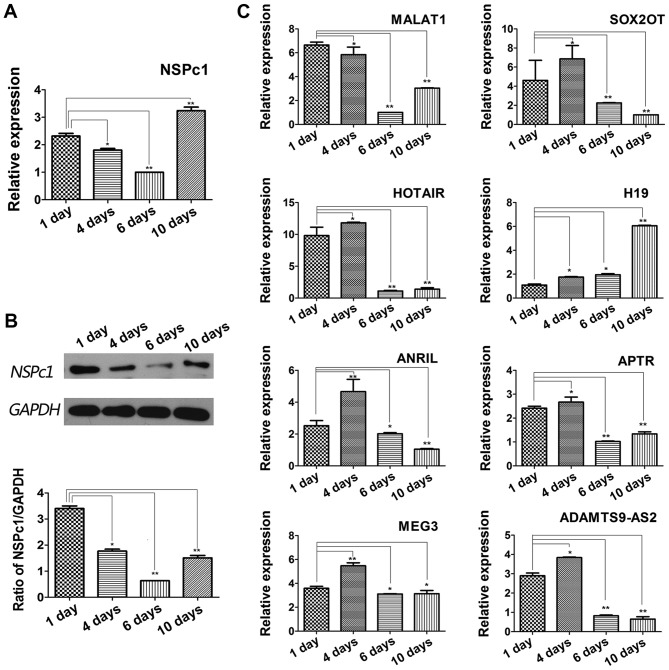
lncRNA candidate expression levels are variable following ATRA mediated induction of differentiation in U87 glioblastoma cells
Following induction of differentiation using ATRA, alterations in expression of NSPc1 and lncRNAs during the differentiation process were examined. mRNA and protein expression levels of NSPc1 decreased at the beginning and reached the lowest expression levels measured by the 6th day of differentiation. However, the expression levels increased subsequent to the 6th day till the 10th day of differentiation. By the 10th day, the expression levels of NSPc1 were higher compared with the 1st day (Fig. 3A and B). MALAT1 expression decreased initially, but increased by the 10th day, H19 increased for the duration of the experiment, and MEG3 decreased significantly by the 10th day (Fig. 3C). The expression changes of the remaining lncRNA candidates largely followed a similar pattern; a slight increase in expression was seen on day 1 after which expression decreased (Fig. 3C).
Figure 3.
Expression of NSPc1 and lncRNAs in U87 cells following ATRA-induced differentiation. (A) Measurement of the expression levels of NSPc1 by reverse transcription-quantitative polymerase chain reaction following ATRA-induced differentiation of U87 cells. GAPDH was used as an internal control. Results were analyzed using the 2−ΔΔCq method. (B) Protein expression levels of NSPc1 following ATRA-induced differentiation of U87 cell by western blotting. GAPDH was used as an internal control. (C) Expression of lncRNAs following ATRA-induced differentiation of U87 cells. GAPDH was used as the internal control. Results were analyzed using the 2−ΔΔCq method. *P<0.05, **P<0.01. NSPc1, nervous system polycomb-1; lncRNAs, long non-coding RNAs; ATRA, all-trans retinoic acid; MALAT1, metastasis associated lung adenocarcinoma transcript 1; ANRIL, antisense non-coding RNA in the INK4 locus; SOX2OT, sex-determining region of the Y chromosome-box 2 overlapping transcript; HOTAIR, homeobox protein transcript antisense RNA; APTR, alu-mediated CDKN1A/P21 transcriptional regulator; MEG3, maternally expressed 3; ADAMTS9-AS2, ADAMTS9 antisense RNA 2.
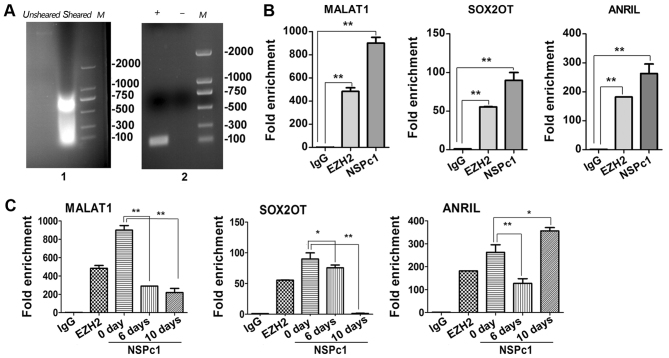
Evaluation of lncRNA candidates interacting with NSPc1 during ATRA-induced U87 differentiation
To investigate a potential interaction between endogenous NSPc1 and lncRNA candidates, RIP assays were used. According to the aforementioned data, MALAT1, SOX2OT and ANRIL were selected for further studies (as presented in Fig. 2). From the RIP assays on U87 cells (Fig. 4A-C), the enrichment of immunoprecipitation of MALAT1 and SOX2OT decreased throughout the whole differentiation process and the enrichment of SOX2OT was almost absent by the 10th day (Fig. 4C; left and middle panels). The enrichment of immunoprecipitation of ANRIL decreased until the 6th day of differentiation, following which the expression levels increased until the 10th day of differentiation (Fig. 4C; right panel).
Figure 4.
Identification of lncRNAs interactions with NSPc1 in U87 cells. (A) Identification of nucleic acid shear fragments by 2% agarose gel electrophoresis. The nucleic acid clip was between ~200 and 1,000 bp (left panel). Efficiency test of immunoprecipitation enrichment of positive and negative control analysed by 2% agarose gel electrophoresis (right panel). (B) Purified RNA immunoprecipitated by antibodies and analysed by RT-qPCR in U87 cells. Verification of RIP enrichment was performed by applying the comparative 2−ΔΔCq method with anti-NSPc1 binding lncRNA compared with positive control (anti-EZH2) binding lncRNA and negative control (IgG) binding lncRNA. (C) The relative enrichment folds of three lncRNA candidates in the all-trans retinoic assay differentiated cells were calculated using the 2−ΔΔCq methodfor each group. All data are presented as the mean ± standard deviation of three independent repeats. *P<0.05, **P<0.01. EZH2, enhancer of zeste 2 polycomb repressive complex 2 subunit; NSPc1, nervous system polycom-1; RIP, RNA immunoprecipitation; MALAT1, metastasis associated lung adenocarcinoma transcript 1; ANRIL, antisense non-coding RNA in the INK4 locus; SOX2OT, sex-determining region of the Y chromosome-box 2 overlapping transcript; IgG, immunoglobulin G; M, ladder; +, positive control; -, negative control.
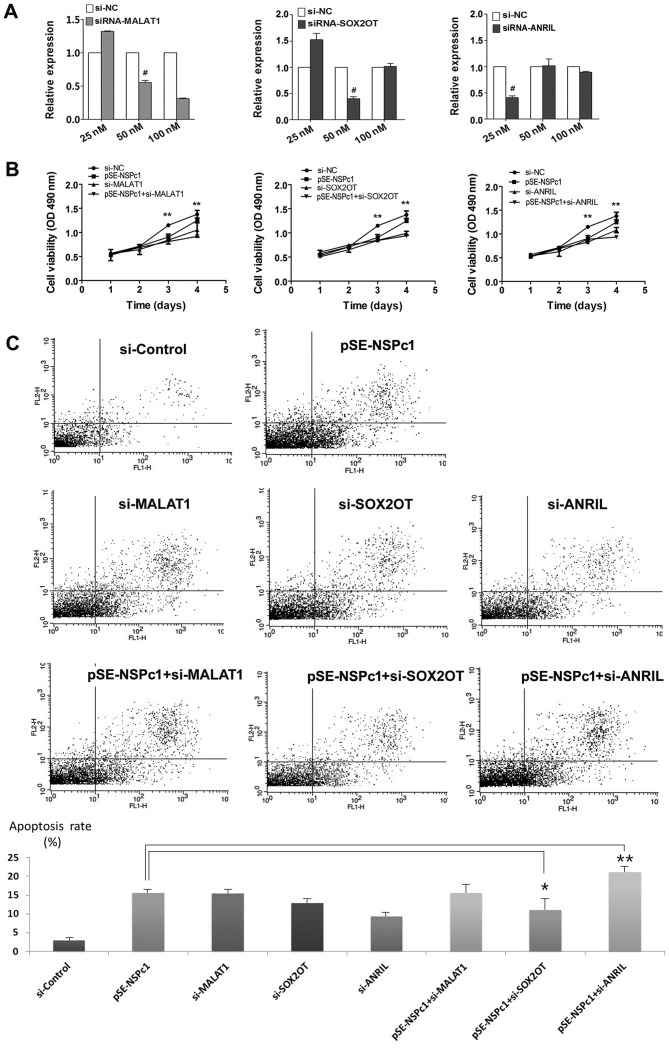
Knocking down expression of lncRNA candidates reduces proliferation and apoptosis in U87 cells
The proliferation and apoptosis of U87 cells following siRNA transfection are presented in Fig. 5. The knockdown efficiency of MALAT1, SOX2OT and ANRIL in U87 glioma cells was examined with increasing concentrations of siRNAs using RT-qPCR (Fig. 5A). The MTT assay demonstrated that knocking down the expression of MALAT1, SOX2OT and ANRIL using their respective siRNAs, reduced proliferation of U87 cells, and the reduction in proliferation was more prominent when NSPc1 and ANRIL were knocked down together (Fig. 5B). Furthermore, using flow cytometry, it was determined that the rates of apoptosis was significantly increased following downregulation of MALAT1, SOX2OT and ANRIL expression compared with the control group. Similarly, the apoptotic rate was further reduced when NSPc1 and ANRIL expression were knocked down together (Fig. 5C).
Figure 5.
Proliferative and apoptotic rates of U87 glioma cells under different NSPc1 and lncRNAs expression conditions. (A) siRNA knockdown efficiency of MALAT1, SOX2OT and ANRIL in U87 glioma cells. The knockdown efficiency was determined using a concentration gradient of each siRNA and reverse transcription-quantitative polymerase chain reaction with GAPDH as the loading control. (B) MTT assay was used to determine the viability of U87 cells following transfection with si-NC, pSE-NSPc1, si-MALAT1, si-SOX2OT, si-ANRIL, pSE-NSPc1+si-MALAT1, pSE-NSPc1+ si-SOX2OT, pSE-NSPc1+ si-ANRIL. (C) Flow cytometry analysis was performed to evaluate the apoptotic rate of U87 cells following knockdown of MALAT1, SOX2OT and ANRIL, and following the knockdown of the three candidate lncRNAs combined with knockdown of NSPc1. *P<0.05, **P<0.01; #siRNA concentration used for subsequent experiments. si, small-interfering; lncRNAs, long non-coding RNAs, NSPc1, nervous system polycomb-1; pSE-NSPc1, NSPc1 knockdown plasmid; MALAT1, metastasis associated lung adenocarcinoma transcript 1; ANRIL, antisense non-coding RNA in the INK4 locus; SOX2OT, sex-determining region of the Y chromosome-box 2 overlapping transcript.
Discussion
Emerging evidence has demonstrated that glioma cells recruited undifferentiated and differentiated cells, providing heterogeneity for tumorigenesis (31). NSPc1, as a key member of PRC1, was associated with the differentiation of tumor cells or cancer stem cells in (17,18). Certain lncRNAs were identified as novel epigenetic regulators (32). Previous studies have hypothesized a link between PcG complexes and lncRNAs (24). There are a number of established glioma cell line models, including U251, H4 and U87. In the present study, only the U87 cell line was used as: i) U251 cells are difficult to transiently transfected; ii) H4 neuroglioma cells are derived from low-grade malignant glioma and has a relatively low NSPc1 expression level, thus making it unsuitable for RIP or RNAi assays; and iii) of the three cell lines, only the U87 cells have been demonstrated to respond to differentiation following treatment with ATRA (33), thus making it a suitable model for the present studies. Therefore, the association between NSPc1 and a number of lncRNAs in differentiated U87 glioma cells was examined.
A total of eight lncRNAs were selected, which were most commonly associated with glioma, and exhibit high correlation scores with NSPc1 based on bioinformatics analysis (32). The expression of NSPc1 and lncRNA candidates is associated with the degree of malignancy of different glioma cells lines. This agrees with a recent study by Hu et al (18), where it was demonstrated that NSPc1 was highly expressed in stem cell-like glioma cells (SLCs).
The expression levels of HOTAIR, APTR and ADAMTS9-AS2 were decreased following NSPc1 overexpression, and the levels of MALAT1, SOX2OT and ANRIL were significantly reduced (P<0.001); therefore, these were investigated further. SOX2OT and ANRIL were downregulated upon either overexpression or silencing of NSPc1. NSPc1 is typically considered to be a transcription repressor (17); however, a previous study has demonstrated that it may additionally be an activator of the SOX2 gene (14). The present data raise the possibility of an, as yet, unidentified indirect regulatory mechanism between the expression levels of NSPc1 and those of SOX2OT and ANRIL under certain circumstances.
Previously, Hu et al (18) demonstrated that ATRA partly reversed the NSPc1-induced stemness enhancement in SLCs through mechanisms associated with an ATRA-dependent decrease in the expression of NSPc1. It was demonstrated that there was a significant change in MALAT1, SOX2OT and ANRIL with variation of NSPc1 expression levels in the ATRA mediated differentiation process of U87 malignant glioma cells. Hu et al (18) additionally demonstrated that NSPc1 promotes self-renewal of stem cell-like glioma cells by repressing the synthesis of ATRA by targeting RDH16, suggesting that NSPc1 expression levels in U87 cells may be crucial for determining whether a cell should differentiate or proliferate.
In the ATRA-induced differentiation of U87 cells, results demonstrated that NSPc1 and MALAT1 expression levels were decreased in a similar manner. Furthermore, the lncRNAs that did not demonstrate a decrease in expression upon NSPc1 silencing were downregulated following ATRA-induced differentiation (for example, HOTAIR and APTR expression), suggesting that the lncRNAs respond to ATRA-induced differentiation independently of NSPc1 expression levels. An experiment overexpressing NSPc1 following ATRA-induced differentiation may help to determine which effects are dependent on NSPc1 expression levels and which effects are dependent on the differentiation process itself. However, it is very difficult to efficiently transfect the cells following induced differentiation of U87 cells. At present, it is difficult to ascertain which effects are the result of NSPc1 expression, and which effects are the results of the differentiation process.
MALAT1 was demonstrated to interact with NSPc1 in U87 cells using an RIP assay. It was previously demonstrated that MALAT1 was able to maintain self-renewing ability and increase the proportion of pancreatic cancer stem cells (CSCs) (34). In U87 cells, ATRA-induced differentiation activates the same upstream as retinoic acid signaling pathway of NSPc1 and MALAT1. Additionally, SOX2OT and ANRIL were also demonstrated to interact with NSPc1 in U87 glioblastoma cells by RIP assay. The expression of SOX2OT is associated with that of regulators of pluripotency, including SOX2 and OCT4, in the pluripotent cell line NT2 (35). ANRIL binds to PRC2 to promote proliferation in gastric cancer, resulting in epigenetic suppression in trans (as transcription factor) (36). Importantly, it was demonstrated that ANRIL interacted with the PRC2 member at the INK4b/ARF/INL4a locus (24). Since MALAT1, SOX2OT and ANRIL interact with NSPc1 in the U87 cells during the differentiation process, it is hypothesized that these three lncRNAs are closely associated with NSPc1 in the canonical PRC1 complex in glioma cells.
The downregulated expression of MALAT1, SOX2OT and ANRIL inhibited the proliferation, and promoted apoptosis of U87 cells. Furthermore, downregulation of NSPc1 combined with ANRIL but not with MALAT1 or SOX2OT was determined to exhibit a more significant effect on proliferation and apoptosis of U87 glioma cells compared with downregulating NSPc1 or ANRIL alone. Therefore, an in-depth study on the interaction mechanism between ANRIL and NSPc1 is required to better understand the formation and development of malignant glioma.
In conclusion, the present data demonstrated that MALAT1, SOX2OT and ANRIL interacted with NSPc1. In addition, ANRIL may interact with NSPc1 during ATRA-induced differentiation of U87 cells, and may be involved in regulating proliferation and apoptosis of U87 cells. Further studies will be required to determine the role of the ANRIL-NSPc1-PRC1 complex in tumorigenesis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Sciences Foundation of China (grant no. 31070929).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YW, ZL and HL performed experiments. HL, YS and YG analysed and interpreted the data. ZL, YW and YG were major contributors in writing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Sathornsumetee S, Rich JN, Reardon DA. Diagnosis and treatment of high-grade astrocytoma. Neurol Clin. 2007;25(x):1111–1139. doi: 10.1016/j.ncl.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Filippini G, Falcone C, Boiardi A, Broggi G, Bruzzone MG, Caldiroli D, Farina R, Farinotti M, Fariselli L, Finocchiaro G, et al. Prognostic factors for survival in 676 consecutive patients with newly diagnosed primary glioblastoma. Neuro Oncol. 2008;10:79–87. doi: 10.1215/15228517-2007-038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes MN, Deshane JS, Rosenfeld M, Siegal GP, Curiel DT, Alvarez RD. Gene therapy and ovarian cancer: A review. Obstet Gynecol. 1997;89:145–155. doi: 10.1016/S0029-7844(96)00296-7. [DOI] [PubMed] [Google Scholar]
- 5.Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, Koellhoffer EC, Pomeroy SL, Orkin SH, Roberts CW. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su Y, Deng B, Xi R. Polycomb group genes in stem cell self-renewal: A double-edged sword. Epigenetics. 2011;6:16–19. doi: 10.4161/epi.6.1.13298. [DOI] [PubMed] [Google Scholar]
- 7.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35:323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Sauvageau M, Sauvageau G. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prezioso C, Orlando V. Polycomb proteins in mammalian cell differentiation and plasticity. FEBS Lett. 2011;585:2067–2077. doi: 10.1016/j.febslet.2011.04.062. [DOI] [PubMed] [Google Scholar]
- 10.Pasini D, Bracken AP, Helin K. Polycomb group proteins in cell cycle progression and cancer. Cell Cycle. 2004;3:396–400. doi: 10.4161/cc.3.4.773. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs JJ, van Lohuizen M. Polycomb repression: From cellular memory to cellular proliferation and cancer. Biochim Biophys Acta. 2002;1602:151–161. doi: 10.1016/s0304-419x(02)00052-5. [DOI] [PubMed] [Google Scholar]
- 12.Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2012;106:243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nunes M, Blanc I, Maes J, Fellous M, Robert B, McElreavey K. NSPc1, a novel mammalian Polycomb gene, is expressed in neural crest-derived structures of the peripheral nervous system. Mech Dev. 2001;102:219–222. doi: 10.1016/S0925-4773(01)00288-X. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Fan R, Sun M, Jiang T, Gong Y. Nspc1 regulates the key pluripotent Oct4-Nanog-Sox2 axis in P19 embryonal carcinoma cells via directly activating Oct4. Biochem Biophys Res Commun. 2013;440:527–532. doi: 10.1016/j.bbrc.2013.09.095. [DOI] [PubMed] [Google Scholar]
- 15.Pritsker M, Ford NR, Jenq HT, Lemischka IR. Genomewide gain-of-function genetic screen identifies functionally active genes in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2006;103:6946–6951. doi: 10.1073/pnas.0509861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan Y, Zhao W, Huang Y, Tong H, Xia Y, Jiang Q, Qin J. Loss of polycomb group protein Pcgf1 severely compromises proper differentiation of embryonic stem cells. Sci Rep. 2017;7:46276. doi: 10.1038/srep46276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong Y, Yue J, Wu X, Wang X, Wen J, Lu L, Peng X, Qiang B, Yuan J. NSPc1 is a cell growth regulator that acts as a transcriptional repressor of p21Waf1/Cip1 via the RARE element. Nucleic Acids Res. 2006;34:6158–6169. doi: 10.1093/nar/gkl834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu PS, Xia QS, Wu F, Li DK, Qi YJ, Hu Y, Wei ZZ, Li SS, Tian NY, Wei QF, et al. NSPc1 promotes cancer stem cell self-renewal by repressing the synthesis of all-trans retinoic acid via targeting RDH16 in malignant glioma. Oncogene. 2017;36:4706–4718. doi: 10.1038/onc.2017.34. [DOI] [PubMed] [Google Scholar]
- 19.Huang Y, Liu N, Wang JP, Wang YQ, Yu XL, Wang ZB, Cheng XC, Zou Q. Regulatory long non-coding RNA and its functions. J Physiol Biochem. 2012;68:611–618. doi: 10.1007/s13105-012-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 21.Wilusz JE. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2016;1859:128–138. doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: New links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Q, Ren S, Lu M, Zhang Y, Zhu D, Zhang X, Li T. Computational prediction of associations between long non-coding RNAs and proteins. BMC Genomics. 2013;14:651. doi: 10.1186/1471-2164-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Wu X, Gong Y, Yue J, Qiang B, Yuan J, Peng X. Cooperation between EZH2, NSPc1-mediated histone H2A ubiquitination and Dnmt1 in HOX gene silencing. Nucleic Acids Res. 2008;36:3590–3599. doi: 10.1093/nar/gkn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang J, Guo S, Jiang S, Xu Y, Li J, Li L, Xiang J. Silencing of long non-coding RNA MALAT1 promotes apoptosis of glioma cells. J Korean Med Sci. 2016;31:688–694. doi: 10.3346/jkms.2016.31.5.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shahryari A, Rafiee MR, Fouani Y, Oliae NA, Samaei NM, Shafiee M, Semnani S, Vasei M, Mowla SJ. Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cells. 2014;32:126–134. doi: 10.1002/stem.1542. [DOI] [PubMed] [Google Scholar]
- 30.Zhang D, Ding L, Li Y, Ren J, Shi G, Wang Y, Zhao S, Ni Y, Hou Y. Midkine derived from cancer-associated fibroblasts promotes cisplatin-resistance via up-regulation of the expression of lncRNA ANRIL in tumour cells. Sci Rep. 2017;7:16231. doi: 10.1038/s41598-017-13431-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rolle K. miRNA multiplayers in glioma. From bench to bedside. Acta Biochim Pol. 2015;62:353–365. doi: 10.18388/abp.2015_1072. [DOI] [PubMed] [Google Scholar]
- 32.Caley DP, Pink RC, Trujillano D, Carter DR. Long noncoding RNAs, chromatin, and development. ScientificWorldJournal. 2010;10:90–102. doi: 10.1100/tsw.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling GQ, Liu YJ, Ke YQ, Chen L, Jiang XD, Jiang CL, Ye W. All-trans retinoic acid impairs the vasculogenic mimicry formation ability of U87 stem-like cells through promoting differentiation. Mol Med Rep. 2015;12:165–172. doi: 10.3892/mmr.2015.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiao F, Hu H, Han T, Yuan C, Wang L, Jin Z, Guo Z, Wang L. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int J Mol Sci. 2015;16:6677–6693. doi: 10.3390/ijms16046677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shahryari A, Rafiee MR, Fouani Y, Oliae NA, Samaei NM, Shafiee M, Semnani S, Vasei M, Mowla SJ. Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cells. 2014;32:126–134. doi: 10.1002/stem.1542. [DOI] [PubMed] [Google Scholar]
- 36.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Chen JF. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.