Abstract
Lung cancer causes severe health problems worldwide and, in China, besides being the principal cause of mortality among urbanites, it is the second leading cause of mortality in the rural population, preceded by hepatocellular carcinoma. Between 80 and 85% of lung cancer is classified as non-small cell lung cancer (NSCLC). The high mobility group box 1 (HMGB1) protein serves an important function in the tumor microenvironment and antitumor response, and may be targeted by microRNA (miR). In the present study, miR-129-5p was identified to target HMGB1 and miR-129-5p exhibited low expression levels in NSCLC tissues. Overexpression of miR-129-5p inhibited cell proliferation and promoted cell apoptosis. In conclusion, the results of the present study suggested the inhibitory function of miR-129-5p and revealed a novel therapeutic target for further investigation.
Keywords: microRNA-129-5p, lung cancer, cell proliferation, apoptosis
Introduction
Lung cancer is the leading cause of cancer-associated mortality in China (1). Two major types of lung cancer exist: Non-small cell lung cancer (NSCLC), which accounts for between 80 and 85% of lung cancer cases, and small cell lung cancer (SCLC), which accounts for between 10 and 15% of lung cancer cases (2). NSCLCs include adenocarcinoma, large cell carcinoma, bronchioloalveolar carcinoma and squamous cell carcinoma (3,4).
It has been reported that the high mobility group box 1 (HMGB1) protein is involved in the immune response of tumors (5). The HMGB1 protein was first identified as a highly conserved nuclear protein that serves a pivotal function in chromatin organization and transcriptional regulation (6,7). In addition, HMGB1 also serves important functions in the immune response (8). Preclinical data revealed that dying tumor cells emit certain danger molecular signals, known as damage-associated molecular patterns (DAMPs) (9–14). In brief, DAMPs are intracellular sequestered biomolecules that evade recognition by the immune system under normal physiological conditions (15). However, under conditions of cellular stress or injury, these molecules are actively secreted by stressed immune cells or released from dying cells, and trigger a non-infectious inflammatory response (16). HMGB1 is a typical DAMP biomolecule, as HMGB1 is released from dying cells and can trigger an immune response (17). A previous study demonstrated that, during chemotherapy or radiotherapy, HMGB1 was released from dying cells and stimulated dendritic cells (DCs) via Toll-like receptor 4 (TLR4) (5). DCs require signaling through TLR4 and its adaptor myeloid differentiation primary response 88 (MyD88) for efficient processing and cross-presentation of antigens from dying tumor cells (5).
MicroRNAs (miRNAs/miRs) serve multiple functions, including promoting cellular proliferation and apoptosis, in the pathogenesis of lung cancer (18–24). HMGB1 may be a target for miRNAs. A previous study demonstrated that miR-218 is able to suppress cell proliferation and invasion, and promotes the apoptosis of pancreatic cancer cells by targeting HMGB1 (25).
In the present study, the function of miR-129-5p in lung cancer was examined and it was demonstrated that miR-129-5p targets HMGB1. The results of the present study may provide novel insights into the molecular mechanism for lung cancer progression.
Patients and methods
Tissue samples
A total of 10 NSCLC tissue samples were collected from The Pulmonary Department at Sichuan Cancer Center (Chengdu, China). Of the ten patients included in the study, 4 were male and 6 were female, with a mean age of 60 years (range, 43–77 years). The 10 NSCLC tissue samples were collected between January 2011 and May 2012. The exclusion criteria was any patient who had been diagnosed with another type of cancer. The protocol for the collection and use of human tissues in the present study was evaluated and approved by the Ethics Committee of Sichuan Cancer Hospital (Chengdu, China). All patients enrolled in the present study provided written informed consent, and all specimens were handled and blinded as required by the legal standards of China. All NSCLC tissue samples were evaluated and confirmed by a senior pathologist at Sichuan Cancer Center. The clinical information are presented in Table I.
Table I.
Characteristics of the 10 patients with non-small cell lung cancer.
| Patient number | Age, years | Sex | TNM stage |
|---|---|---|---|
| 1 | 65 | Female | I |
| 2 | 55 | Female | II |
| 3 | 56 | Female | II |
| 4 | 43 | Female | III |
| 5 | 59 | Male | III |
| 6 | 50 | Male | IV |
| 7 | 56 | Male | III |
| 8 | 77 | Female | IV |
| 9 | 66 | Male | II |
| 10 | 73 | Female | II |
Cell culture
The NSCLC cell lines A549 and SPC-A-1 were obtained from the Cell Bank of Sichuan University (Sichuan, China). All cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified incubator with 5% CO2.
Detection of miR-129-5p in tissues and cells
The levels of miR-129-5p in tissues and cells were determined using the reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Briefly, the total RNA from the tissue samples or A549 and SPC-A-1 cells was extracted using TRIzol® reagent, according to the manufacturer's protocol (Invitrogen; Thermo Fisher Scientific, Inc.). The corresponding cDNA was obtained using the reverse transcription kit (cat. no. 12574018, Thermo Fisher Scientific. Inc.). The expression of miR-129-5p was determined using TaqMan miRNA assay (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 95°C for 5 min, 40 cycles of 95°C for 15 sec, 58°C for 30 sec and 74°C for 30 sec. U6 small nuclear RNA was used as an internal loading control. RT-qPCR analysis was performed on an ABI 7900HT instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). The relative expression levels were calculated with the 2−ΔΔCq method (26). The primers were synthesized and tested by the Sangon Biotech Co., Ltd. (Shanghai, China) (27–30). The following primer sequences were used: miR-129-5p forward, 5′-GGGGGCTTTTTGCGGTCTGG-3′ and reverse, 5′-AGTGCGTGTCGTGGAGTC-3; and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Overexpression and downregulation of miR-129-5p in NSCLC cells
The miR-129-5p levels in the A549 and SPC-A-1 cells were increased and decreased by miR-129-5p mimic and miR-129-5p antisense oligonucleotide (ASO), respectively. The miR-129-5p mimic and were purchased from Sangon Biotech Co., Ltd. Prior to transfection with 500 ng mimic or ASO, the cells were cultured overnight (1×106 cells/well) at 37°C. The cell transfection was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The sequences of miR-129-5p mimic and miR-129-5p ASO were as follows: miR-129-5p mimic, 5′-GGAUCUUUUUGCGGUCUGGGCUUGCUGUUCCUCUCAACAGUAGUCAGGAAGCCCUUACCCCAAAAAGUAUCU-3′, and miR-129-5p ASO, 5′-CCUAGAAAAACGCCAGACCCGAACGACAAGGAGAGUUGUCAUCAGUCCUUCGGGAAUGGGGUUUUUCAUAGAAA-3′.
Cell proliferation assay
Cell proliferation was assessed using the MTT assay. Briefly, A549 and SPC-A-1 cells were seeded into 96-well plates at a density of 5×105 cells/well. The MTT reagent was added to the medium at a final concentration of 0.1 mg/ml. A total of 100 µl DMSO was added to each well to dissolve the formazan. The optical density was determined using a microplate reader at 570 nm.
Cell apoptosis analysis
A549 or SPC-A-1 cell suspensions (5×105 cells/ml) and Annexin V-FITC (Abcam, Shanghai, China) binding buffer was prepared. An Annexin V-FITC Apoptosis Detection kit (cat. no. APOAF-20TST; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used according the manufacturer's protocol. Annexin V-fluorescein isothiocyanate (FITC; 0.5 mg/ml) was added to the cell mixture and incubated at room temperature for 15 min. Subsequently, propidium iodide (PI; 0.1 µg/ml; Abcam) was added for 5 min at room temperature, and samples were analyzed on a fluorescence-activated cell sorting analyzer instrument using the 488 nm excitation wavelength (argon-ion laser or solid state laser) and emission detected was at 530 nm (green, FITC) and 575–610 nm (orange, PI) (28). Data were analyzed with BD FACSuite™ software (23–12943; BD Biosciences, San Jose, CA, USA).
Prediction of the possible targets of miR-129-5p
TargetScan software (www.targetscan.org) was used to predict the possible targets of miR-129-5p.
Dual-luciferase reporter assays
A549 cells were seeded at 1×105 cells/well and serum-starved for 6 h pre-transfection. The 3′-untranslated region (3′-UTR) of HMGB1 and mutated controls were cloned and inserted into a reporter plasmid (500 ng) and the pGL3-control (100 ng) (Promega Corporation, Madison, WI, USA). miR-129-5p mimics were then transfected into A549 cells containing wild-type or mutant 3′-UTR plasmids with Lipofectamine 2000. Cells were harvested and luciferase activity was analyzed after 24 h using the Dual-Luciferase Reporter assay system (Promega Corporation). Luciferase activity was normalized to Renilla luciferase activity. Mutants of HMGB1 3′-UTR were generated using a Site-Directed Mutagenesis kit (Promega Corporation).
Western blotting
A549 cells were frozen and lysed in lysis buffer (150 mM NaCl, 50 mM Tris/HCl, 1% Triton X-100 and 0.1% SDS) with protease inhibitors cocktail (cat. no. ab65621; Abcam). Protein concentration was determined using a BCA protein assay (cat. no. ab146331; Abcam). Subsequently, 20 µg total protein from the cell lysate was loaded onto a 10% SDS-PAGE gel. The protein was then transferred to PVDF membrane and the membrane was blocked with 5% milk for 1 h at room temperature. For the HMGB1 western blot analysis, anti-HMGB1 antibodies (cat. no. ab79823; Abcam) were used at a dilution of 1:1,000 at 4°C overnight. β-actin was used as an internal reference (cat. no. ab179467; 1:1,000; Abcam). The membrane was then incubated with horseradish peroxidase-conjugated anti-rabbit IgG (1:2,000; cat. no. ab150077; Abcam) at room temperature for 2 h. Proteins were detected by Enhanced Chemiluminescence Western Blotting Detection Reagents (GE Healthcare, Little Chalfont, UK). Images were analyzed using ImageJ 1.8.0 software (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
All experiments were performed three times. Results are presented as the mean ± standard deviation. A two-tailed Student's t-test was used to analyze the mean value between two groups; One-way analysis of variance was used to test the mean value among three or more groups with post hoc contrasts by Student-Newman-Keuls test. P<0.05 was considered to indicate a statistically significant difference. All calculations were performed using SPSS software (version 16.0; SPSS, Inc., Chicago, IL, USA).
Results
miR-129-5p is expressed at low levels in lung cancer tissues
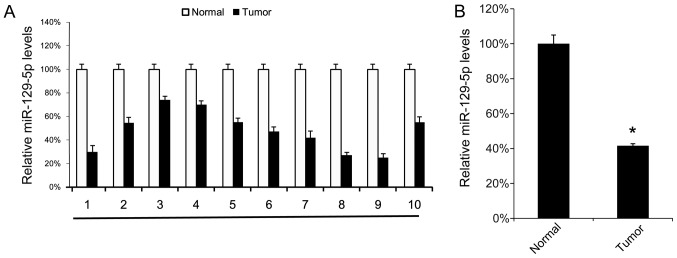
First, the miR-129-5p expression levels in 10 NSCLC tissue samples and their corresponding normal adjacent tissue samples were analyzed using RT-qPCR. Tumor tissue samples exhibited lower miR-129-5p levels compared with their corresponding normal adjacent tissues (Fig. 1A). The mean expression levels of miR-129-5p in all 10 tumor tissue samples and normal tissue samples were determined, and were revealed to be significantly lower in tumor tissue samples compared with in normal tissue samples (Fig. 1B). These results suggest that miR-129-5p may exhibit a tumor suppressor function in NSCLC.
Figure 1.
NSCLC tissues express low levels of miR-129-5p. A total of 10 NSCLC tissue samples were collected. (A) The miR-129-5p levels in the 10 NSCLC tissue samples were determined using the reverse transcription-quantitative polymerase chain reaction. (B) The mean expression level of the miR-129-5p in the 10 NSCLC tissue samples and their corresponding adjacent normal tissues were calculated (B). Results are presented as the mean ± standard deviation. The experiment was performed at least three times. *P<0.05 vs. normal tissues. NSCLC, non-small cell lung cancer; miR, microRNA.
Overexpression of miR-129-5p inhibits cell proliferation and promotes cell apoptosis
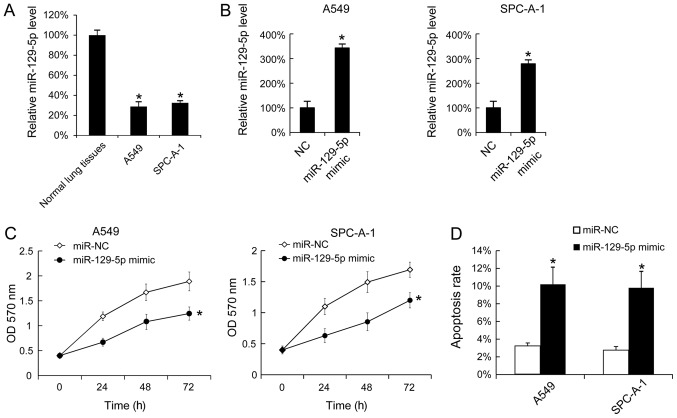
To investigate the function of miR-129-5p in vitro, the levels of miR-129-5p in A549 and SPC-A-1 cells were analyzed using RT-qPCR, normal lung tissues were used as control. A549 and SPC-A-1 cells expressed significantly lower levels of miR-129-5p compared with normal tissues (Fig. 2A). Additionally, miR-129-5p mimic was transfected into A549 and SPC-A-1 cells, and it was identified that the miR-129-5p mimic effectively upregulated the miR-129-5p levels in A549 and SPC-A-1 cells (Fig. 2B). As the miR-129-5p mimic was able to increase the miR-129-5p level in vitro, cell proliferation following transfection with the miR-129-5p mimic was determined, and it was identified that the upregulation of miR-129-5p inhibited cell proliferation in A549 and SPC-A-1 cells (Fig. 2C). The effect of the miR-129-5p mimic on cell apoptosis was investigated and transfection of the miR-129-5p mimic was identified to increase the rate of apoptosis (Fig. 2D).
Figure 2.
Overexpression of miR-129-5p inhibited the proliferation of A549 and SPC-A-1 cells and promoted cell apoptosis. (A) The miR-129-5p levels in normal lung tissues, A549 and SPC-A-1 cells were determined using RT-qPCR. The miR-129-5p levels in the tumor adjacent normal lung tissues were arbitrarily defined as 100%. (B) A549 and SPC-A-1 cells were seeded and then were transfected with miR-129-5p mimic. After 24 h, the miR-129-5p expression was determined using RT-qPCR. (C) Following miR-129-5p mimic transfection, cell proliferation was determined using an MTT assay. (D) At 48 h after miR-129-5p mimic transfection, SPC-A-1 and A549 cells were stained with Annexin V-fluorescein isothiocyanate and propidium iodide, and the cell apoptosis rate was evaluated using fluorescence-activated cell sorting analysis. Results are presented as the mean ± standard deviation. The experiments were performed at least three times. *P<0.05 vs. normal lung tissues, NC or miR-NC. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NC, negative control; miR, microRNA; OD, optical density.
Downregulation of miR-129-5p promotes cell proliferation
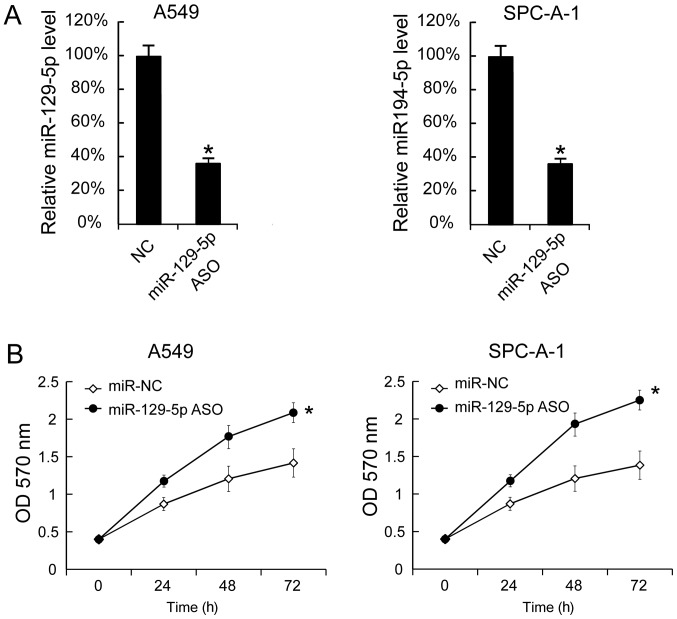
miR-129-5p ASO was transfected into A549 and SPC-A-1 cells to decrease the miR-129-5p levels. The miR-129-5p levels in A549 and SPC-A-1 cells 24 h after miR-129-5p ASO transfection revealed that miR-129-5p were significantly inhibited by miR-129-5p ASO (Fig. 3A). Additionally, cell proliferation was determined using the MTT assay and it was identified that miR-129-5p ASO transfection significantly increased cell proliferation (Fig. 3B).
Figure 3.
Suppression of miR-129-5p levels promotes A549 and SPC-A-1 cell proliferation. A549 and SPC-A-1 cells were seeded and then transfected with miR-129-5p ASO. (A) At 24 h after transfection, miR-129-5p expression was determined using the reverse transcription-quantitative polymerase chain reaction. (B) Following miR-129-5p ASO transfection, cell proliferation was determined. Results are presented as the mean ± standard deviation. The experiment was performed at least three times. *P<0.05 vs. NC, or miR-NC. OD, optical density, NC, negative control; ASO, antisense oligonucleotide; miR, microRNA.
miR-129-5p inhibits HMGB1 expression in lung cancer cells
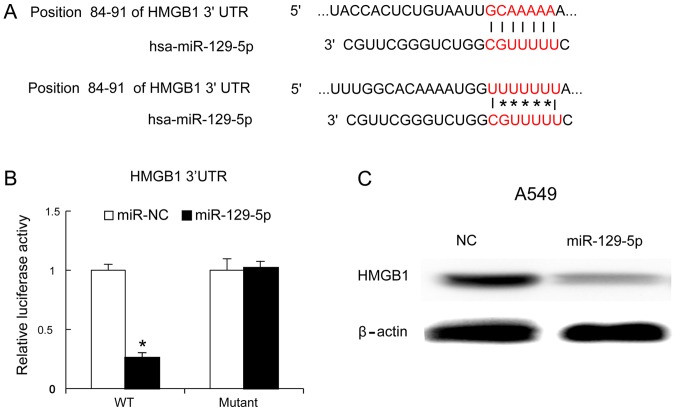
HMGB1 is an important immune factor and one of the genes targeted by miR-129-5p (1,6–8). The present study identified the binding site of miR-129-5p on the HMGB1 gene using TargetScan software (Fig. 4A). To confirm whether miR-129-5p targeted HMGB1, a mutant of the 3′-UTR of HMGB1 was generated and the mutant and wild-type versions were cloned into luciferase reporter plasmids. The miR-129-5p mimic and mutant version were co-transfected into A549 cells. At 24 h later, it was identified that the miR-129-5p mimic decreased the luciferase values of the 3′UTR of HMGB1 (wild-type version), but not that of the mutated version of the 3′UTR of HMGB1 (Fig. 4B). Furthermore, the miR-129-5p mimic was transfected into A549 cells and, 48 h later, the HMGB1 protein levels were evaluated using western blot analysis. The results identified that the miR-129-5p mimic inhibited HMGB1 protein expression (Fig. 4C).
Figure 4.
HMGB1 is targeted by miR-129-5p in A549 cells. (A) The binding sites and its mutated version of HMGB1 and miR-129-5p. (B) miR-129-5p mimic and plasmid containing WT or mutated 3′-UTR of HMGB1 were transfected into A549 cells. After 48 h, the luciferase activity was measured. (C) The miR-129-5p mimic was transfected into A549 cells, and the abundance of the HMGB1 protein was analyzed by western blotting. The experiment was performed at least three times. *P<0.05 vs. WT. HMGB1, high mobility group box 1; UTR, untranslated region; WT, wild-type; NC, negative control; miR, microRNA.
Discussion
The present study investigated the function of miR-129-5p in NSCLC and identified that miR-129-5p exhibited tumor suppressor activity against NSCLC via HMGB1. The function of miR-129-5p has previously been investigated in various types of cancer. In gastric cancer, miR-129-5p was identified to be downregulated and involved in the migration and invasion of gastric cancer cells by targeting interleukin-8 (31). In laryngeal cancer, the upregulation of miR-129-5p inhibited laryngeal cancer cell proliferation, invasiveness and migration by affecting signal transducer and activator of transcription 3 expression (32), and in breast cancer, miR-129-5p attenuates irradiation-induced autophagy and decreases radioresistance of breast cancer cells by targeting HMGB1 (33). The results also indicated that miR-129-5p serves a function as an inhibitor of tumor growth.
HMGB1 may activate and induce the maturation of DCs (34). Accordingly, HMGB1 is one of the links between tumor cells and the immune system of the host (35). Recent studies have identified that, in NSCLC, DCs are in contact with tumor cells (36), and tumor-infiltrating lymphocytes (TILs) are observed in the peritumoral zones (37). HMGB1 produced by tumor cells recruits DCs, which associate with an increased number of TILs (38). Furthermore, HMGB1 may be released from dying cells and stimulate DCs (5). DCs required signaling through TLR4 and its adaptor MyD88 for the efficient processing and cross-presentation of antigens from dying tumor cells (5). We hypothesize that HMGB1 may serve an important function in the immune response against tumors and therefore represents a potential immune therapy target in cancer. Accordingly, miR-129-5p may serve a function in the NSCLC immune therapy via HMGB1.
A limitation of the present study was that only ten NSCLC tissue samples were analyzed. The number of patients involved in the study limited the patient survival analysis. Therefore, a larger number of patients should be included in further investigations.
In conclusion, the results of the present study revealed the suppressive function of miR-129-5p in NSCLC and offer a novel therapeutic target for the treatment of NSCLC that is worthy of further investigation.
Acknowledgements
The authors wish to thank Dr Tao Huang (Department of Cancer, West China Hospital, Chengdu, China) for discussion
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
GL collected patient data and performed the cell experiments. JX performed reverse transcription-quantitative polymerase chain reaction analysis, western blotting and other molecular experiments. JW contributed to the study design and manuscript writing.
Ethics approval and consent to participate
The protocol for the collection and use of human tissues in the present study was evaluated and approved by the Ethics Committee of Sichuan Cancer Hospital. All patients enrolled in the present study provided written informed consent, and all specimens were handled and blinded as required by the legal standards of China.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wen C, Dehnel T. China wrestles with lung cancer. Lancet Oncol. 2011;12:15. doi: 10.1016/S1470-2045(10)70303-X. [DOI] [PubMed] [Google Scholar]
- 2.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wakelee HA, Chang ET, Gomez SL, Keegan TH, Feskanich D, Clarke CA, Holmberg L, Yong LC, Kolonel LN, Gould MK, West DW. Lung cancer incidence in never-smokers. J Clin Oncol. 2007;25:472–478. doi: 10.1200/JCO.2006.07.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujii T, Dracheva T, Player A, Chacko S, Clifford R, Strausberg RL, Buetow K, Azumi N, Travis WD, Jen J. A preliminary transcriptome map of non-small cell lung cancer. Cancer Res. 2002;62:3340–3346. [PubMed] [Google Scholar]
- 5.Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050–1059. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi ME. HMGB1 loves company. J Leukoc Biol. 2009;86:573–576. doi: 10.1189/jlb.1008585. [DOI] [PubMed] [Google Scholar]
- 7.Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr Opin Genet Dev. 2003;13:170–178. doi: 10.1016/S0959-437X(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 8.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): Nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 9.Seong SY, Matzinger P. Hydrophobicity: An ancient damage-associated molecular pattern that initiates innate immune responses. Nat Rev Immunol. 2004;4:469–478. doi: 10.1038/nri1372. [DOI] [PubMed] [Google Scholar]
- 10.Foell D, Wittkowski H, Vogl T, Roth J. S100 proteins expressed in phagocytes: A novel group of damage-associated molecular pattern molecules. J Leukoc Biol. 2007;81:28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- 11.Kaczmarek A, Vandenabeele P, Krysko DV. Necroptosis: The release of damage-associated molecular patterns and its physiological relevance. Immunity. 2013;38:209–223. doi: 10.1016/j.immuni.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Krysko DV, Agostinis P, Krysko O, Garg AD, Bachert C, Lambrecht BN, Vandenabeele P. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 2011;32:157–164. doi: 10.1016/j.it.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Ahrens S, Zelenay S, Sancho D, Hanč P, Kjær S, Feest C, Fletcher G, Durkin C, Postigo A, Skehel M, et al. F-actin is an evolutionarily conserved damage-associated molecular pattern recognized by DNGR-1, a receptor for dead cells. Immunity. 2012;36:635–645. doi: 10.1016/j.immuni.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Rubartelli A, Lotze MT. Inside, outside, upside down: Damage-associated molecular-pattern molecules (DAMPs) and redox. Trends Immunol. 2007;28:429–436. doi: 10.1016/j.it.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez K, Vasquez G. Damage-associated molecular patterns and their role as initiators of inflammatory and auto-immune signals in systemic lupus erythematosus. Int Rev Immunol. 2017;36:259–270. doi: 10.1080/08830185.2017.1365146. [DOI] [PubMed] [Google Scholar]
- 16.Land WG. The role of damage-associated molecular patterns (DAMPs) in human diseases: Part II: DAMPs as diagnostics, prognostics and therapeutics in clinical medicine. Sultan Qaboos Univ Med J. 2015;15:e157–e170. [PMC free article] [PubMed] [Google Scholar]
- 17.Laursen TL, Stoy S, Deleuran B, Vilstrup H, Gronbaek H, Sandahl TD. The damage-associated molecular pattern HMGB1 is elevated in human alcoholic hepatitis, but does not seem to be a primary driver of inflammation. APMIS. 2016;124:741–747. doi: 10.1111/apm.12568. [DOI] [PubMed] [Google Scholar]
- 18.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 20.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, Bader AG. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–1704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 2010;411:846–852. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 24.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Xu Y, Long J, Guo K, Ge C, Du R. microRNA-218 suppresses the proliferation, invasion and promotes apoptosis of pancreatic cancer cells by targeting HMGB1. Chin J Cancer Res. 2015;27:247–257. doi: 10.3978/j.issn.1000-9604.2015.04.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P, Li J, Song Y, Wang X. MiR-129-5p inhibits proliferation and invasion of chondrosarcoma cells by regulating SOX4/Wnt/β-catenin signaling pathway. Cell Physiol Biochem. 2017;42:242–253. doi: 10.1159/000477323. [DOI] [PubMed] [Google Scholar]
- 27.Li D, Liu X, Lin L, Hou J, Li N, Wang C, Wang P, Zhang Q, Zhang P, Zhou W, et al. MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J Biol Chem. 2011;286:36677–36685. doi: 10.1074/jbc.M111.270561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song B, Zhang C, Li G, Jin G, Liu C. MiR-940 inhibited pancreatic ductal adenocarcinoma growth by targeting MyD88. Cell Physiol Biochem. 2015;35:1167–1177. doi: 10.1159/000369686. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Q, Yu Y. Upregulated CDK16 expression in serous epithelial ovarian cancer cells. Med Sci Monit. 2015;21:3409–3414. doi: 10.12659/MSM.894990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Xu Y, Qiu W, Zhao D, Zhang Y. Tissue miR-193b as a novel biomarker for patients with ovarian cancer. Med Sci Monit. 2015;21:3929–3934. doi: 10.12659/MSM.895407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang Z, Wang H, Li Y, Hou Z, Ma N, Chen W, Zong Z, Chen S. MiR-129-5p is down-regulated and involved in migration and invasion of gastric cancer cells by targeting interleukin-8. Neoplasma. 2016;63:673–680. doi: 10.4149/neo_2016_503. [DOI] [PubMed] [Google Scholar]
- 32.Shen N, Huang X, Li J. Upregulation of miR-129-5p affects laryngeal cancer cell proliferation, invasiveness, and migration by affecting STAT3 expression. Tumor Biol. 2016;37:1789–1796. doi: 10.1007/s13277-015-3969-y. [DOI] [PubMed] [Google Scholar]
- 33.Luo J, Chen J, He L. mir-129-5p attenuates irradiation-induced autophagy and decreases radioresistance of breast cancer cells by targeting HMGB1. Med Sci Monit. 2015;21:4122–4129. doi: 10.12659/MSM.896661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dumitriu IE, Bianchi ME, Bacci M, Manfredi AA, Rovere-Querini P. The secretion of HMGB1 is required for the migration of maturing dendritic cells. J Leukoc Biol. 2007;81:84–91. doi: 10.1189/jlb.0306171. [DOI] [PubMed] [Google Scholar]
- 35.Kang R, Xie Y, Zhang Q, Hou W, Jiang Q, Zhu S, Liu J, Zeng D, Wang H, Bartlett DL, et al. Intracellular HMGB1 as a novel tumor suppressor of pancreatic cancer. Cell Res. 2017;27:916–932. doi: 10.1038/cr.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perrot I, Blanchard D, Freymond N, Isaac S, Guibert B, Pachéco Y, Lebecque S. Dendritic cells infiltrating human non-small cell lung cancer are blocked at immature stage. J Immunol. 2007;178:2763–2769. doi: 10.4049/jimmunol.178.5.2763. [DOI] [PubMed] [Google Scholar]
- 37.Konishi J, Yamazaki K, Azuma M, Kinoshita I, Dosaka-Akita H, Nishimura M. B7-H1 expression on non-small cell lung cancer cells and its relationship with tumor-infiltrating lymphocytes and their PD-1 expression. Clin Cancer Res. 2004;10:5094–5100. doi: 10.1158/1078-0432.CCR-04-0428. [DOI] [PubMed] [Google Scholar]
- 38.Kusume A, Sasahira T, Luo Y, Isobe M, Nakagawa N, Tatsumoto N, Fujii K, Ohmori H, Kuniyasu H. Suppression of dendritic cells by HMGB1 is associated with lymph node metastasis of human colon cancer. Pathobiology. 2009;76:155–162. doi: 10.1159/000218331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.