Abstract
Increasing evidence has revealed the importance of microRNA (miRNA/miR) in cancer genesis and progression. The aim of the current study was to identify the key miRNAs involved in the onset and development of nasopharyngeal carcinoma (NPC) and to further evaluate their diagnostic and prognostic values. Microarray data were obtained and analyzed to screen differentially expressed miRNAs (DEMs) between patients with NPC and healthy controls. The target genes of the DEMs were predicted and their possible functions were evaluated. The diagnostic and prognostic values of the DEMs were subsequently investigated. A total of 4 DEMs, including miR-18a, miR-135b, miR-204 and miR-497, were identified. Gene Ontology (GO) and pathway enrichment analysis revealed that the target genes were enriched in a number of GO terms and signaling pathways. The results demonstrated that the selected DEMs may present potential diagnostic factors for NPC. In addition, miR-18a [Hazard ratio (HR), 3.405; 95% confidence interval (CI), 1.334–8.693] and miR-135b (HR, 2.482; 95% CI, 1.014–6.076) may serve prognostic roles for patients with NPC. In summary, the present study identified 4 miRNAs that may be involved in the genesis and development of NPC. In addition, miR-18a and miR-135b may present useful prognostic markers for patients with NPC. Future in vitro and in vivo investigations are warranted to substantiate the results obtained in the current study.
Keywords: nasopharyngeal carcinoma, microarray, microRNA, diagnosis, prognosis
Introduction
Nasopharyngeal carcinoma (NPC) is a squamous cell malignant tumor arising from the nasopharynx. The incidence of NPC in Southeast Asia reached 20 cases per 100,000 people in 2003 (1). Currently, the standard treatment strategy for patients with NPC is chemotherapy and radiotherapy (2). Despite advances in the treatment of NPC, the rate of disease recurrence remains high, with a 5-year recurrence rate of 10.8%. Disease recurrence typically occurs within a few years of treatment completion and is associated with poor prognosis (3).
The molecular mechanisms underlying the development of NPC are complex have not yet been fully elucidated. Previous studies have revealed that a number of genes and signaling pathways may be involved in the genesis and progression of NPC. For example, high expression levels of Janus kinase 2, signal transducer and activator of transcription 3 and vascular endothelial growth factor may be associated with the clinicopathological characteristics and prognosis of patients with NPC (4). Overexpression of the downstream mediator of protein kinase B (AKT), NUAK family kinase 1, may also be a prognostic factor for NPC (5). Furthermore, increased expression of the transcription factor, SRY-box 2 (SOX2) has been observed in NPC, and may be associated with the prognosis of patients with NPC; in vitro assays have revealed that SOX2 recruits the nuclear transcription factor, Kruppel like factor 4, to bind to the phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α promoter and upregulate its expression, thus enhancing phosphoinositide 3-kinase (PI3K)/AKT signaling and NPC tumorigenesis (6).
microRNAs (miRNAs/miRs) are a class of short non-coding RNA sequences that bind to the 3′-untranslated region of mRNAs and inhibit their translation (7). As such, miRNAs may modulate various biological processes by regulating gene expression. miRNAs have been implicated in the genesis and/or development of a variety of disorders, particularly in cancer (7). Previous studies have revealed that miRNAs may serve roles in the development of NPC, and may provide novel therapeutic targets. miR-185 negatively targets homeobox C6 and promotes cell apoptosis by inhibiting the transforming growth factor β1/mechanistic target of rapamycin axis in NPC (8). Similarly, miR-379 negatively regulates the growth and migration of NPC cells by suppressing tumor protein D52 expression (9). Modulating the expression levels of the aforementioned miRNAs may be a potential therapeutic strategy for NPC. However, studies investigating the roles of miRNAs in NPC remains limited. Therefore, the identification of miRNAs that serve important roles in the genesis and development of NPC is required.
The present study aimed to screen miRNAs which may serve important roles in the development of NPC, and to further assess their diagnostic and prognostic values. Differentially expressed miRNAs (DEMs) between NPC tissues and benign nasopharyngeal tissues were identified by analyzing public microarray-based data. Their target genes were subsequently predicted and their potential functions were annotated. The diagnostic and prognostic values of the miRNAs in NPC were then further assessed.
Materials and methods
Data source
To obtain DEMs between NPC tissues and benign controls, microarray-based datasets were retrieved from the Gene Expression Omnibus database (ncbi.nlm.nih.gov/geo). Datasets that met the following criteria were selected for inclusion in the study: i) Datasets obtained from Homo sapiens; ii) datasets that contained normalized data regarding healthy adjacent tissues from the patients with NPC or from healthy controls; iii) datasets amenable to analysis by online tools or/and other software including dCHIP (version no. 2011.01; www.hsph.harvard.edu) and R.
Screening of DEMs
The selected miRNA expression profiles were analyzed using the GEO2R web tool (ncbi.nlm.nih.gov/geo/geo2r/), based on the limma R package from the Bioconductor project. The results were downloaded in text format, in which the miRNAs that met the cut-off criteria of P<0.05 and a |log fold-change|>1.0 were considered as DEMs. dCHIP was used for datasets that could not be analyzed by GEO2R as the Matrix data required manual filtering. A paired Student's t-test was used to compare the two groups (NPC vs. controls).
Determining the intersection of DEGs
If there were ≥2 datasets that met the aforementioned cut-off criteria, the overall range of the DEMs may be large as different datasets may generate different ranges of DEMs. To narrow the scope of the DEMs, the intersecting DEMs from each dataset were identified using Venn diagram analysis (10).
miRNA target gene prediction and functional annotation
Several computational miRNA-target prediction tools have been developed to predict target mRNAs of DEMs. The mirDIP database integrates a number of tools that can reduce the weaknesses of individual tools (11). Therefore, target genes of the DEMs in the current study were predicted using the mirDIP database. An integrative score, which was statistically inferred from the obtained predictions and assigned to each unique miRNA-target interaction to provide a unified measure of confidence, was introduced. Candidate mRNAs with integrated scores of >0.8 were selected as target mRNAs in this database. To annotate the functions of the predicted target genes, Gene Ontology (GO) (12) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (13) was performed using the Kobas tool (version 3.0) (14). P<0.05 was considered to indicate a statistically significant difference.
Association between DEMs and clinical features, diagnosis and prognosis of NPC
To evaluate the association between the expression levels of DEMs and the clinical features of patients, and to further determine their diagnostic and prognostic values, datasets containing a cohort of patients with NPC with sufficient information were retrieved and selected for analysis. The expression levels of the screened DEMs and the associated clinical data were extracted from the datasets. The diagnostic and prognostic values of the DEMs were subsequently assessed as mentioned below.
Statistical analysis
Chi-squared values were calculated to evaluate the association between the expression of the DEMs and clinicopathological features. The diagnostic accuracy of miRNAs was measured using receiver operating characteristic (ROC) curves and the area under the ROC curve (AUC). The optimal diagnostic point of the signature was assessed at cut-off values with the largest Youden's index (sensitivity + specificity −1). The probability of survival and its significance were evaluated by using the Kaplan-Meier method and Cox proportional hazard models, respectively. Log-rank test was used to compare the survival rates between low and high expression groups. Two-tailed P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using MedCalc software (version 11.1.1.0; MedCalc Software, Mariakerke, Belgium).
Results
Identification of DEMs from the miRNA expression profiles
Two miRNA expression profiles, GSE22587 (no available reference) and GSE46172 (15), met the aforementioned inclusion criteria and were selected for the purposes of the current study. These datasets comprised non-coding RNA profiling data generated by array analysis. GSE22587 was deposited by Li et al. The dataset was generated using the Illumina Human Beta-version microRNA expression BeadChip (Illumina Inc., San Diego, CA, USA) and contained 8 NPC tissue specimens and 4 benign nasopharyngeal tissue specimens. GSE46172 was based on the Agilent-031181 Unrestricted_Human_miRNA_V16.0_Microarray (miRBase release 16.0 miRNA ID version; Agilent Technologies, Inc., Santa Clara, CA, USA) and contained 4 NPC specimens and 4 controls.
GEO2R analysis identified 18 upregulated and 20 downregulated miRNAs from the GSE22587 dataset, and 30 upregulated and 36 downregulated miRNAs from the GSE46172 dataset. In this context, upregulated miRNAs were downregulated in NPC tissues relative to the controls, while downregulated miRNAs were upregulated in NPC specimens compared with the controls.
Intersection of DEMs
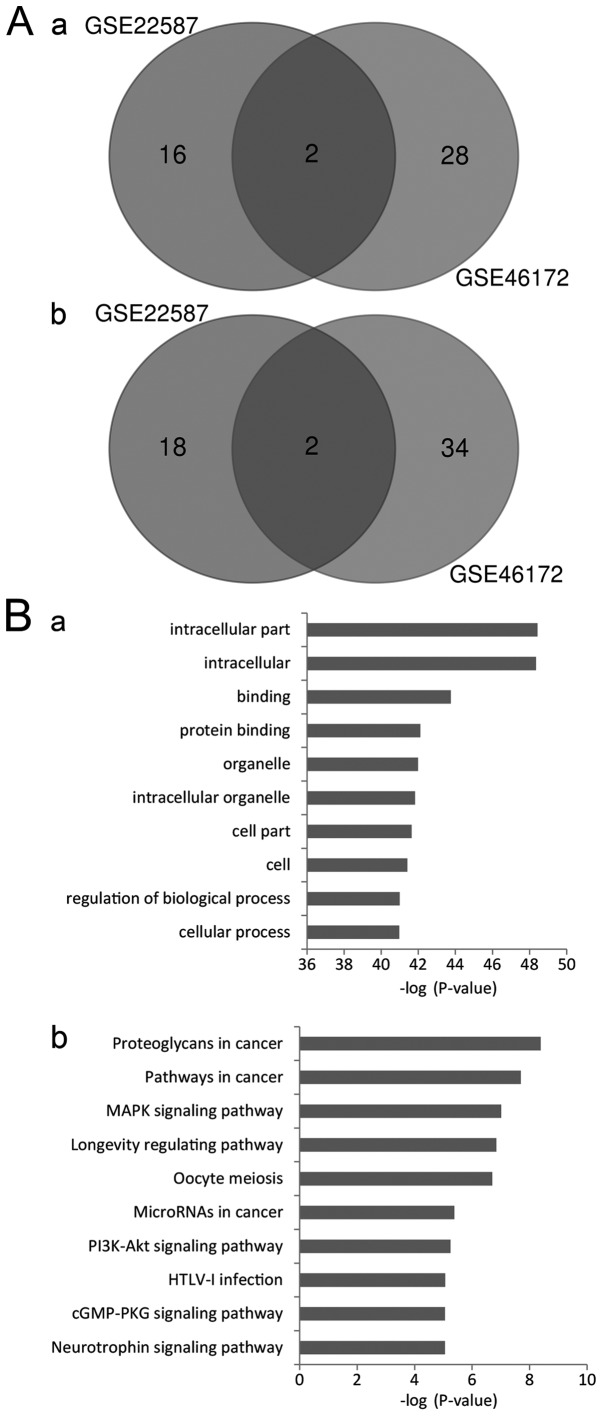
To narrow the range of the DEMs identified, Venn diagram analysis was used to identify the intersecting DEM profiles. A total of 2 upregulated and 2 downregulated DEMs were identified in the intersection (Fig. 1A). The upregulated DEMs were miR-204, and miR-497, and the downregulated DEMs were miR-135b and miR-18a.
Figure 1.
Screening and annotation of the DEMs. (A) Intersection of the DEMs from the GSE22587 and GSE46172 datasets. (A-a) Upregulated DEMs. (A-b) Downregulated DEMs. Upregulated DEMs refer to DEMs that were downregulated in NPC tissues compared with the controls, and vice versa. (B) Pathway enrichment analysis of the target genes. (B-a) Gene Ontology analysis. (B-b) Kyoto Encyclopedia of Genes and Genomes analysis. DEMs, differentially expressed microRNAs.
Prediction of miRNA target genes and annotation of their functions
A total of 417 genes with integrated scores of >0.8 were considered as predicted target genes. The Kobas tool was used to annotate the putative functions of these target genes. GO analysis revealed that the targeted genes were enriched in 2,340 GO items. The results revealed that these genes may be associated with a number of biological processes, including regulation of gene expression, cell metabolism, structure development and compound binding. The top significant items were ‘intracellular part’, ‘protein binding’, ‘intracellular organelle’, ‘regulation of biological process’ and ‘cellular process’ (Fig. 1B-a). The results indicated that various aspects of biological processes may be involved in the genesis and development of NPC.
KEGG analysis revealed that the target genes may be enriched in 108 signaling pathways. The most significant pathways were ‘proteoglycans in cancer’, ‘pathways in cancer’, ‘MAPK signaling pathway’, ‘longevity regulating pathway’, ‘oocyte meiosis’, ‘microRNAs in cancer’ and ‘PI3K-AKT signaling pathway’ (Fig. 1B-b). These results suggest that multiple signaling pathways may mediate the onset and progression of NPC.
Diagnostic value of DEMs in NPC
To further assess the roles of the DEMs in NPC, the GSE36682 dataset was downloaded for analysis. The GSE36682 dataset includes non-coding RNA profiling results generated using the Human miRNA 1K array. The data were deposited by Wei et al (no available reference). This dataset contained 62 NPC tissue specimens and 6 nasopharyngitis tissue specimens.
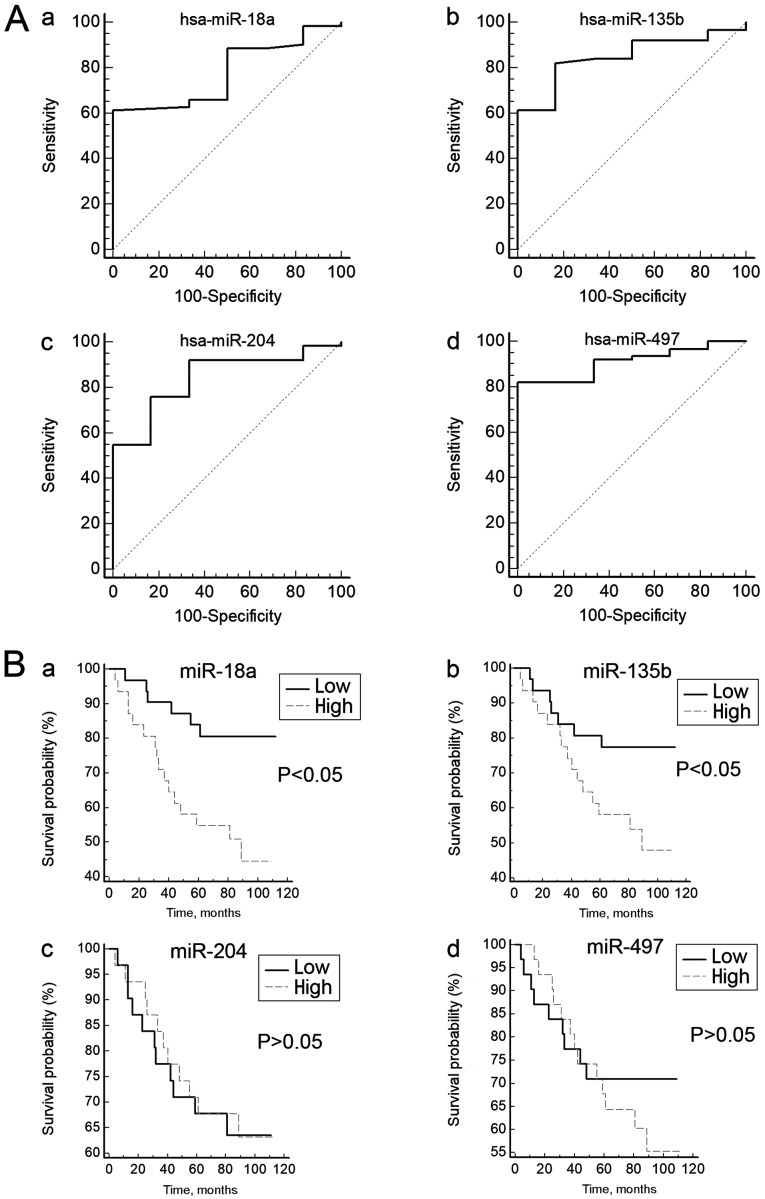
To test whether the expression of these DEMs can distinguish patients with NPC from the controls, ROC curves were constructed. The range of the AUCs of these 4 miRNAs (miR-204, miR-497, miR-18a and miR-135b) was 0.778–0.911 (Table I; Fig. 2A), suggesting that any of these miRNAs may be used as a potential biomarker to distinguish patients with NPC from healthy controls.
Table I.
Diagnostic accuracy of the selected differentially expressed miRNAs in patients with nasopharyngeal carcinoma.
| miRNA | Sensitivity, % | Specificity, % | AUC | Standard error | 95% confidence interval of AUC |
|---|---|---|---|---|---|
| miR-18a | 61.29 | 100.00 | 0.778 | 0.0767 | 0.661–0.870 |
| miR-135b | 82.26 | 83.33 | 0.848 | 0.0618 | 0.740–0.924 |
| miR-204 | 75.81 | 83.33 | 0.841 | 0.0732 | 0.733–0.919 |
| miR-497 | 82.26 | 100.00 | 0.911 | 0.0404 | 0.733–0.919 |
miRNA/miR, microRNA; AUC, area under the curve.
Figure 2.
Diagnostic and prognostic values of the candidate miRNAs. (A) Candidate differentially expressed microRNAs as potential biomarkers for the diagnosis of nasopharyngeal carcinoma. (A-a) miR-18a, (A-b) miR-135b, (A-c) miR-204 and (A-d) miR-497. (B) Kaplan Meier curves showing the overall survival rates of patients with nasopharyngeal carcinoma grouped into high and low expression of (B-a) miR-18a, (B-b) miR-135b, (B-c) miR-204 and (B-d) miR-497. miR, microRNA.
Association between DEM expression levels and clinical features
The expression levels of the DEMs from the GSE36682 dataset were classified as high or low according to their median expression levels. The association between the expression levels of these miRNAs and the clinicopathologic parameters are presented in Table II. Details of four confounding factors, including age, gender, metastasis status and maximum diameters of lymph nodes, were available in the dataset.
Table II.
Association of miR-18a, miR-135b, miR-204 and miR-497 expression with clinicopathological factors.
| miR-18a | miR-135b | miR-204 | miR-497 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | Total no. of patients | High | Low | P-value | High | Low | P-value | High | Low | P-value | High | Low | P-value |
| Sex | |||||||||||||
| Male | 11 | 6 | 5 | 0.740 | 6 | 5 | 0.740 | 4 | 7 | 0.319 | 3 | 8 | 0.965 |
| Female | 51 | 25 | 26 | 25 | 26 | 27 | 24 | 28 | 23 | ||||
| Age, years | |||||||||||||
| ≥50 | 26 | 14 | 12 | 0.607 | 13 | 13 | 0.797 | 14 | 12 | 0.607 | 15 | 11 | 0.303 |
| <50 | 36 | 17 | 19 | 18 | 18 | 17 | 19 | 16 | 20 | ||||
| Metastasis status | |||||||||||||
| Yes | 17 | 15 | 2 | <0.001 | 14 | 3 | 0.002 | 8 | 9 | 0.776 | 9 | 8 | 0.776 |
| No | 45 | 16 | 29 | 17 | 28 | 23 | 22 | 22 | 23 | ||||
| Maximum diameter of lymph nodes, mm | |||||||||||||
| ≥10 | 43 | 25 | 18 | 0.054 | 24 | 19 | 0.168 | 19 | 24 | 0.168 | 22 | 21 | 0.783 |
| <10 | 19 | 6 | 13 | 7 | 12 | 12 | 7 | 9 | 10 | ||||
miR, microRNA.
No association between the expression levels of the 4 miRNAs and 3 factors, including age, gender and maximum diameters of lymph nodes, was observed (P>0.05). However, for miR-18a and miR-135b, a significant association with metastasis status was observed (P<0.05), indicating that high expression of miR-18a and miR-135b expression may be associated with metastasis.
Prognostic value of the DEMs in NPC
Survival curves were generated to assess the association between the expression levels of the DEMs and the prognosis of patients with NPC. The log-rank test did not reveal any association between the expression levels of miR-204 and miR-497 and the overall survival rate of patients with NPC (Fig. 2B). However, the survival curves revealed that the low miR-18a-expression group had a significantly longer survival rate compared with the high miR-18a-expression group (P<0.05). Similar results were observed for miR-135b (P<0.05; Fig. 2B). These results indicated that high expression of miR-18a or miR-135b may be associated with a poor prognosis for patients with NPC.
Cox proportional hazard regression analysis was used to analyze the effects of the confounding factors on the survival time of patients with NPC. Univariate analysis revealed that metastasis [hazard ratio (HR), 13.343; 95% confidence interval (CI), 5.138–34.648], miR-18a (HR, 3.405; 95% CI, 1.334–8.693), and miR-135b (HR, 2.482; 95% CI, 1.014–6.076) may be prognostic indicators for patients with NPC (Table III). However, multivariate analysis revealed that only metastasis (HR, 12.140; 95% CI, 4.355–33.839) may be an independent prognostic indicator for patients with NPC. Thus, although miR-18a and miR-135b may be prognostic factors, they are not independent prognostic indicators for NPC patients.
Table III.
Cox proportional hazard regression.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (≥50 vs. <50 years) | 2.146 (0.921–5.002) | 0.0785 | ||
| Sex (male vs. female) | 0.183 (0.025–1.344) | 0.0967 | ||
| Metastasis (yes vs. no) | 13.343 (5.138–34.648) | <0.001 | 12.140 (4.355–33.839) | <0.001 |
| Max diameter of lymph nodes (≥10 vs. <10 mm) | 2.340 (0.795–6.889) | 0.1246 | ||
| miR-18a | 3.405 (1.334–8.693) | 0.0108 | 2.203 (0.430–11.298) | 0.3460 |
| miR-135b | 2.482 (1.014–6.076) | 0.0477 | 0.616 (0.126–3.013) | 0.5517 |
| miR-204 | 0.945 (0.411–2.171) | 0.8938 | ||
| miR-497 | 1.424 (0.611–3.321) | 0.4151 | ||
HR, hazard ratio; CI, confidence interval; miR, microRNA.
Discussion
The incidence rate of NPC is high in South Asia (1). A previous study investigated the roles of miRNAs in the development of NPC (16). However, the molecular mechanisms underlying the development of this disease have not yet been fully elucidated. The current study identified several DEMs that may be involved in NPC by analyzing microarray-based data. The predicted target genes of the identified DEMs were enriched in a number of GO items and signaling pathways. The results identified 4 DEMs that may be used as potential biomarkers to distinguish patients with NPC from healthy controls. In addition, miR-18a and miR135b were identified as potential prognostic indicators for patients with NPC.
miRNAs exert their influence on biological processes by targeting specific mRNAs. Thus, the functions of the target genes may reflect the roles of miRNAs in the biological processes involved. In the present study, pathway enrichment analysis suggested that the mechanisms by which the 4 DEMs contribute to NPC progression are complex. A pathway crosstalk network may be used to determine how different biological processes coordinate with each other. However, the primary aim of the present study was to annotate the possible functions of the target genes. Therefore, bar charts instead of a crosstalk network were used to present the pathway items.
A previous study revealed that miR-18a may be highly expressed in lung cancer and may function as an oncogene in the development of the disease (17). Increased circulating miR-18a may also be a prognostic factor for lung cancer (18). A meta-analysis revealed that miR-18a may be a promising biomarker for the diagnosis of patients with gastric carcinoma (19). The results obtained in the current study are in accordance with the aforementioned studies. However, an additional study demonstrated that miR-18a may serve a role as a tumor suppressor by inhibiting cell proliferation and inducing apoptosis in ovarian cancer (20). Thus, miRNAs may serve different roles in different types of cancer or in the same type of cancer under different conditions. In vitro overexpression of miR-135b promoted the progression and migration of gastric cancer and by targeting CKLF like MARVEL transmembrane domain containing 3 (21). miR-135b promoted the migration, invasion and epithelial-mesenchymal transition of pancreatic cancer cells by targeting nuclear receptor subfamily 3 group C member 2 (22). The results obtained in the aforementioned studies are consistent with the results of the present study, suggesting that upregulation of miR-135b may contribute to NPC progression. In the current study, miR-204 expression was downregulated in NPC tissues when compared with controls. A previous study revealed that miR-204 may exert antitumor effects on melanoma cells (23). miR-204 may decrease the growth, migration and invasion of colon cancer cells by deactivating the PI3K/AKT/mTOR signaling pathway and targeting C-X-C motif chemokine ligand 8 expression (24). Therefore, miR-204 may function as tumor suppressor in NPC.
In the present study, miR-497 was observed to be downregulated in NPC tissues compared with healthy controls. miR-497 may inhibit renal cancer cell proliferation and migration by targeting the CD274 molecule (25). Furthermore, miR-497 may suppress the proliferation and invasion of thyroid cancer cells by targeting Yes associated protein 1 (26). Therefore, miR-497 may also function as a tumor suppressor in NPC. Taken together, the results suggest that miR-18a and miR135b may function as oncogenes, while miR-204 and miR497 may function as tumor suppressors in NPC.
The occurrence of metastasis affects the prognosis of patients with NPC (27). In the present study, univariate analysis revealed that metastasis, miR-18a and miR-135b were prognostic indicators for NPC. Multivariate analysis revealed that only metastasis was an independent prognostic factor for NPC. Since miR-18a and miR-135b may be associated with metastasis, these miRNAs may target genes that serve roles in the invasion and migration of NPC cells. Although miR-18a and miR-135b were not independent prognostic factors for NPC, they may be useful biomarkers for monitoring NPC prognosis. However, the current study did not investigate how these miRNAs may affect the prognosis of patients with NPC. Future laboratory experiments investigating the association of these miRNAs and their target genes are required to elucidate the underlying molecular mechanisms of NPC.
The present study had a number of limitations. Firstly, confounding factors only included age, gender, metastasis status and maximum diameter of the lymph nodes. Other confounding factors that may influence the results, including Epstein-Barr virus infection status, clinical stage of the tumor, and smoking and drinking status, were not evaluated in the present study, as this information was not available from the datasets. Secondly, only microarray-based data were analyzed in this study. Further experiments using cell models and clinical specimens are required to substantiate the results obtained in the current study. Thirdly, imaging modalities, including computed tomography and magnetic resonance imaging, may be used for the detection of cancer metastasis in patients. However, comparing the expression levels of the identified miRNAs with the results of imaging for predicting metastasis of NPC could not be assessed because the relevant data was not included in the datasets.
Despite these limitations, the present study revealed 4 miRNAs (miR-18a, miR-135b, miR-204 and miR-497) as key miRNAs that may serve roles in the genesis and development of NPC. The target genes of these miRNAs were enriched in multiple biological processes and signaling pathways. Among these miRNAs, miR-18a and miR-135b, may be associated with metastasis and may be useful prognostic indicators for NPC. Future studies using cell and animal models and clinical specimens are required to elucidate the molecular mechanisms in NPC.
Acknowledgements
Not applicable.
Funding
This study was partly supported by the Fund of Chongqing Health and Family Planning Commission (grant no. 20141020) and funding from the China Science Foundation for Postdoctoral Researchers (grant no. 2015T80962).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/gds/).
Authors' contributions
XZ and QZ designed the experiments. WZ, HY, DL, and AC performed the experiments. XZ, WZ, and YW analyzed the data. XZ and QZ wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Perri F, Della Vittoria Scarpati G, Caponigro F, Ionna F, Longo F, Buonopane S, Muto P, Di Marzo M, Pisconti S, Solla R. Management of recurrent nasopharyngeal carcinoma: Current perspectives. Onco Targets Ther. 2019;12:1583–1591. doi: 10.2147/OTT.S188148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He Y, Guo T, Guan H, Wang J, Sun Y, Peng X. Concurrent chemoradiotherapy versus radiotherapy alone for locoregionally advanced nasopharyngeal carcinoma in the era of intensity-modulated radiotherapy: A meta-analysis. Cancer Manag Res. 2018;10:1419–1428. doi: 10.2147/CMAR.S160469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Guo Y, Xu J, Chen Z, Jiang X, Zhang L, Huang S, He X, Zhang Y. Clinical analysis of recurrence patterns in patients with nasopharyngeal carcinoma treated with intensity-modulated radiotherapy. Ann Otol Rhinol Laryngol. 2017;126:789–797. doi: 10.1177/0003489417734229. [DOI] [PubMed] [Google Scholar]
- 4.Cheng JZ, Chen JJ, Xue K, Wang ZG, Yu D. Clinicopathologic and prognostic significance of VEGF, JAK2 and STAT3 in patients with nasopharyngeal carcinoma. Cancer Cell Int. 2018;18:110. doi: 10.1186/s12935-018-0605-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Tang G, Huang H, Li H, Zhang P, Xu L. Expression level of NUAK1 in human nasopharyngeal carcinoma and its prognostic significance. Eur Arch Otorhinolaryngol. 2018;275:2563–2573. doi: 10.1007/s00405-018-5095-0. [DOI] [PubMed] [Google Scholar]
- 6.Tang J, Zhong G, Wu J, Chen H, Jia Y. SOX2 recruits KLF4 to regulate nasopharyngeal carcinoma proliferation via PI3K/AKT signaling. Oncogenesis. 2018;7:61. doi: 10.1038/s41389-018-0074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dragomir M, Mafra ACP, Dias SMG, Vasilescu C, Calin GA. Using microRNA networks to understand cancer. Int J Mol Sci. 2018;19:E1871. doi: 10.3390/ijms19071871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng JZ, Chen JJ, Wang ZG, Yu D. MicroRNA-185 inhibits cell proliferation while promoting apoptosis and autophagy through negative regulation of TGF-β1/mTOR axis and HOXC6 in nasopharyngeal carcinoma. Cancer Biomark. 2018;23:107–123. doi: 10.3233/CBM-181459. [DOI] [PubMed] [Google Scholar]
- 9.Zhao X, Chu J. MicroRNA-379 suppresses cell proliferation, migration and invasion in nasopharyngeal carcinoma by targeting tumor protein D52. Exp Ther Med. 2018;16:1232–1240. doi: 10.3892/etm.2018.6302. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Yu JW, Mai W, Cui YL, Kong LY. Genes and pathways identified in thyroid carcinoma based on bioinformatics analysis. Neoplasma. 2016;63:559–568. doi: 10.4149/neo_2016_409. [DOI] [PubMed] [Google Scholar]
- 11.Tokar T, Pastrello C, Rossos AEM, Abovsky M, Hauschild AC, Tsay M, Lu R, Jurisica I. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46:D360–D370. doi: 10.1093/nar/gkx1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Mubaid H. Gene multifunctionality scoring using gene ontology. J Bioinform Comput Biol. 2018;16:1840018. doi: 10.1142/S0219720018400188. [DOI] [PubMed] [Google Scholar]
- 13.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plieskatt JL, Rinaldi G, Feng Y, Levine PH, Easley S, Martinez E, Hashmi S, Sadeghi N, Brindley PJ, Bethony JM, Mulvenna JP. Methods and matrices: Approaches to identifying miRNAs for nasopharyngeal carcinoma. J Transl Med. 2014;12:3. doi: 10.1186/1479-5876-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee KT, Tan JK, Lam AK, Gan SY. MicroRNAs serving as potential biomarkers and therapeutic targets in nasopharyngeal carcinoma: A critical review. Crit Rev Oncol Hematol. 2016;103:1–9. doi: 10.1016/j.critrevonc.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Liang C, Zhang X, Wang HM, Liu XM, Zhang XJ, Zheng B, Qian GR, Ma ZL. MicroRNA-18a-5p functions as an oncogene by directly targeting IRF2 in lung cancer. Cell Death Dis. 2017;8:e2764. doi: 10.1038/cddis.2017.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, Zhu S, Tao Z, Ye S. High circulating miR-18a, miR-20a, and miR-92a expression correlates with poor prognosis in patients with non-small cell lung cancer. Cancer Med. 2018;7:21–31. doi: 10.1002/cam4.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang Q, Zhang G, Wang J, Sheng S. Diagnostic value of MicroRNA-18a for gastric cancer: A meta-analysis. Clin Lab. 2018;64:177–184. doi: 10.7754/Clin.Lab.2017.170827. [DOI] [PubMed] [Google Scholar]
- 20.Liu P, Qi X, Bian C, Yang F, Lin X, Zhou S, Xie C, Zhao X, Yi T. MicroRNA-18a inhibits ovarian cancer growth via directly targeting TRIAP1 and IPMK. Oncol Lett. 2017;13:4039–4046. doi: 10.3892/ol.2017.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu M, Huang Y, Sun W, Li P, Li L. miR-135b-5p promotes gastric cancer progression by targeting CMTM3. Int J Oncol. 2018;52:589–598. doi: 10.3892/ijo.2017.4222. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Che X, Yang N, Bai Z, Wu Y, Zhao L, Pei H. miR-135b-5p promotes migration, invasion and EMT of pancreatic cancer cells by targeting NR3C2. Biomed Pharmacother. 2017;96:1341–1348. doi: 10.1016/j.biopha.2017.11.074. [DOI] [PubMed] [Google Scholar]
- 23.Palkina N, Komina A, Aksenenko M, Moshev A, Savchenko A, Ruksha T. miR-204-5p and miR-3065-5p exert antitumor effects on melanoma cells. Oncol Lett. 2018;15:8269–8280. doi: 10.3892/ol.2018.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shuai F, Wang B, Dong S. microRNA-204 inhibits the growth and motility of colorectal cancer cells by downregulation of CXCL8. Oncol Res. 2018;15:8269–8280. doi: 10.3727/096504018X15172747209020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Qu F, Ye J, Pan X, Wang J, Gan S, Chu C, Chu J, Zhang X, Liu M, He H, Cui X. MicroRNA-497-5p down-regulation increases PD-L1 expression in clear cell renal cell carcinoma. J Drug Target. 2018;27:1–15. doi: 10.1080/1061186X.2018.1479755. [DOI] [PubMed] [Google Scholar]
- 26.Cheng H, Dong H, Feng J, Tian H, Zhang H, Xu L. miR-497 inhibited proliferation, migration and invasion of thyroid papillary carcinoma cells by negatively regulating YAP1 expression. Onco Targets Ther. 2018;11:4711–4721. doi: 10.2147/OTT.S164052. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Feng Y, Cao C, Hu Q, Chen X. Prognostic value and staging classification of lymph nodal necrosis in nasopharyngeal carcinoma after intensity-modulated radiotherapy. Cancer Res Treat. 2018 doi: 10.4143/crt.2018.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/gds/).