Abstract
To the best of our knowledge, the microRNA (miR/miRNA) expression profile of plasma exosomes in ovarian cancer has not been previously studied. The aim of the present study was to investigate the practicality of using plasma exosomal miRNAs as novel serological biomarkers of ovarian cancer. In the study, exosome-like vesicles were purified from the plasma of patients with ovarian cancer and healthy women using differential centrifugation. The purified vesicles, ranging from 50–100 nm in size, were identified as exosomes by transmission electron microscopy and western blotting. High-throughput sequencing demonstrated that 65 known miRNAs, 34 of which were upregulated and 31 downregulated, were differentially expressed between patients with ovarian cancer and healthy women (P<0.05; fold change ≥2). The miRNA expression levels of hsa-miR-106a-5p, hsa-let-7d-5p and hsa-miR-93-5p were significantly increased, whereas hsa-miR-122-5p, hsa-miR-185-5p and hsa-miR-99b-5p expression levels were significantly decreased in the exosomes of patients with ovarian cancer compared with those in the healthy controls. Additionally, the miRNA expression levels of plasma hsa-miR-93-5p were significantly increased in patients with ovarian cancer compared with those in the healthy controls, while the plasma expression levels of hsa-miR-122-5p and hsa-miR-99b-5p were significantly decreased in patients with ovarian cancer compared with those in the healthy controls. Overall, the present study identified plasma and exosomal miRNAs with dysregulated expression in patients with ovarian cancer compared with that in healthy controls, and the differentially expressed miRNAs may have potential as diagnostic and prognostic targets for the treatment of patients with ovarian cancer.
Keywords: ovarian cancer, exosome, microRNA, plasma
Introduction
Ovarian cancer is the seventh most common cancer and the fifth leading cause of cancer-associated mortality among women worldwide (1), with metastasis being the principal cause of mortality in patients with this disease (2). Although progress has been made in the treatment of ovarian cancer by improved debulking surgery followed by platinum/taxane-based chemotherapy, the overall 5-year survival rate is at present 15–30% (3). Therefore, a novel biomarker to improve the early detection of ovarian cancer is required.
MicroRNAs (miRs/miRNAs) are a class of small non-coding RNA molecules that regulate post-transcriptional gene regulation (4,5). miRNAs suppress the protein expression of specific mRNAs through either repressing translation or initiating degradation of a targeted mRNA (6). Increasing evidence has demonstrated that miRNAs serve key roles in the development and pathogenesis of a range of cancer types due to abnormal expression of miRNAs (7). miRNAs have been used as biomarkers for clinical diagnosis and prognosis in human cancer (8). In particular, circulating miRNAs have been demonstrated to be candidates for biomarkers of cancer (9). Due to the ease and repeatability of sample collection, serum and plasma miRNAs are frequently used as non-invasive biomarkers for cancer detection (9).
Exosomes are small (30- to 150-nm) membrane vesicles originating from the endosomal membrane compartment (10). Exosomes secreted by donor cells mediate cellular communication to recipient cells (11). Exosomes are present in the body fluids, including the plasma, urine, saliva, breast milk and synovial fluid (8,12). A previous study demonstrated that exosomal miRNAs are stable in the blood, as exosomes are able to protect them from degradation by enzymes, including RNase (9). Cancer cells additionally release exosomes into the tumour microenvironment and peripheral blood of patients with cancer (13). Exosomes contain numerous cell-specific proteins [such as CD63 antigen (CD63) and heat shock protein 70 kDa (HSP70)], mRNAs and miRNAs (14). Exosomal miRNA has a potential role as a diagnostic biomarker in human cancer (9). However, the functional role of plasma exosomal miRNA in patients with ovarian cancer remains largely unknown. In the present study, the miRNA profile of plasma exosomes in healthy individuals and patients with ovarian cancer was examined.
Materials and methods
Patients and plasma collection
The present study was approved by The Human Investigation and Ethical Committees of the Gansu Provincial Maternity and Child Care Hospital (Gansu, China) and conducted according to their guidelines and regulations. All participants provided written informed consent. Plasma samples were collected from 30 patients with ovarian cancer at The Gansu Provincial Maternity and Child Care Hospital between January 2014 and December 2017. The age of the patients ranged between 42 and 72 years [mean age ± standard deviation (SD), 53.34±2.34 years]. All patients were diagnosed with ovarian cancer according to the Tumor-Node-Metastasis (TNM) staging system (15), and none of the patients had received any surgery, radiotherapy, chemotherapy or other treatments. Patients with systemic diseases or other ocular diseases were excluded from the present study. The plasma samples of 30 healthy women (the control group) were additionally obtained during the same period. The age range of the control group was 45–62 years, with a mean age ± SD of 52.67±3.21 years.
Fasting blood (10 ml) was collected in ethylenediaminetetraacetic acid tubes, thoroughly mixed and centrifuged at 2,000 × g (5430R; Eppendorf, Germany) for 10 min at 4°C. The plasma was carefully removed using a pipette, collected in 750-µl aliquots and frozen at −80°C until required.
Cell culture
Human ovarian carcinoma cells (OVCa-3), purchased from Cells Resource Center Shanghai Institutes Biological Sciences, Chinese Academy Sciences (Shanghai, China), were cultured in RPMI-1640 medium containing 10% fetal bovine serum, with 5% penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in 5% CO2 at 37°C.
Exosome preparation and size determination
Exosomes were prepared using differential ultra-centrifugation as follows: Plasma (5 ml) was centrifuged at 1,600 × g for 20 min at 4°C and at 10,000 × g for 30 min at 4°C. The supernatant was filtered through 0.22-µm disposable filter units (Merck KGaA, Darmstadt, Germany) and centrifuged at 100,000 × g for 2 h at 4°C. Subsequent to washing with 10 ml PBS (Invitrogen; Thermo Fisher Scientific, Inc.), the exosomes were suspended in 0.1 ml PBS and stored at −80°C until required. The diameter of the exosomes was determined using a Zetasizer Nano (Malvern Instruments China, Shanghai, China) according to the manufacturer's protocol. The purified exosomes were observed using an ×250,000 magnification EM-2010 transmission electron microscope (JEOL, Ltd., Tokyo, Japan).
Protein preparation and western blotting
The total protein was extracted from exosomes using RIPA lysis buffer (Sigma-Aldrich; Merck KGaA) containing a protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA). Samples were also quantified using the BCA protein assay kit (Thermo Fisher Scientific, Inc.) and 20 µg proteins were separated on a 10% SDS-PAGE gel and transferred to nitrocellulose membranes. After the membranes were blocked with 5% skimmed milk in PBS-Tween (0.5% Tween-20) (Invitrogen; Thermo Fisher Scientific, Inc.) for 2 h at room temperature, they were incubated overnight at 4°C with either mouse anti-CD63 (1/1,000; cat. no. ab193349; Abcam, Cambridge, UK) or mouse anti-HSP70 (1/1,000; cat. no. SAB4200714; Sigma-Aldrich; Merck KGaA). Subsequent to incubating with the anti-mouse (cat. no. PA1-74421; Invitrogen; Thermo Fisher Scientific, Inc.) secondary antibody at 1/10,000 dilution for 1 h at room temperature, the membranes were visualized by exposing X-ray films using Pierce enhanced chemiluminescent visualization reagent (Thermo Fisher Scientific, Inc.).
Isolation of exosomal RNAs and small RNA sequencing
The total RNA was extracted from plasma exosomes using an RNeasy Mini Spin kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocol. The RNA concentration and integrity were determined using an Agilent 2100 bioanalyzer system (Agilent Technologies GmbH, Waldbronn, Germany). Small RNA sequencing was performed using the BGISEQ-500 sequencer (BGI Park, Shenzhen, China). Low-quality reads were removed, and adaptor sequences and small RNAs, which mapped perfectly to the human genome (ftp://ncbi.nlm.nih.gov/genomes/Homo_sapiens) were used for further analysis. According to the seed sequences, target mRNAs were predicted and obtained from TargetScan (release 7.1; http://www.targetscan.org/vert_71/). The miRNA expression levels were estimated by the number of reads per million and the relative miRNA expression levels were analyzed using the DEGseq method (16).
Target gene prediction and pathway analysis
Target genes of differentially expressed miRNAs were predicted by three databases, including TargetScan (17), miRanda (18) and RNAhybrid (19). The target genes were analyzed in terms of Gene Ontology (GO) functional annotation (20) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (21) using KOBAS v3.0 (http://kobas.cbi.pku.edu.cn/anno_iden.php).
Plasma RNA extraction
Total RNA was extracted from plasma using an miRNeasy Serum/Plasma kit (cat. no. 217184, Qiagen GmbH). A total of 200 µl plasma was mixed with QIAzol Lysis Reagent (cat. no. 79306; Qiagen GmbH) according to the manufacturer's protocols and the homogenate was incubated at room temperature for 5 min. Subsequently, 25 fmol synthetic Caenorhabditis elegans miR-39 (cel-miR-39; Ambion; Thermo Fisher Scientific, Inc.) was spiked into the mixture. Subsequently, the RNA was extracted according to the manufacturer's protocol. The total RNA was eluted into 14 µl nuclease-free water and the RNA concentration was measured using a NanoDrop ND-2000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA (100 ng) was used for reverse transcription to cDNA using the miScript II RT kit (Qiagen GmbH). The reverse transcription reaction contained 2 µl total RNA (~100 ng), 2.2 µl plasma spike-in control, 5 µl 5X miScript HiFlex Buffer, 2 µl 10X miScript Nucleics mix, 2 µl miScript Reverse Transcriptase mix and 7.8 µl RNase-free water. The reverse transcription temperature protocol used was as follows: 37°C for 60 min, 95°C for 5 min and then held at 4°C until required. qPCR was performed using miScript SYBR Green PCR kit (Qiagen GmbH). The reactions were incubated in a 96-well plate at 95°C for 15 min, followed by 40 cycles of 94°C for 15 sec and 60°C for 60 sec. The miScript primers are listed in Table SI. The Cq values were median-normalized to synthetic spike-in cel-miR-39 as described previously (9). Relative fold-changes were calculated as using the 2−ΔΔCq method (22).
Statistical analysis
All statistical analyses were performed using GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA), and a one-tailed unpaired t-test was used for comparing differences between two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Clinical characteristics
Plasma samples from 30 patients with epithelial ovarian cancer were obtained. According to the TNM staging, 2 cases were classed as stage I, 4 cases were classed as stage II, and 24 cases were classed as stage III–IV (Table SII). Patients with ovarian cancer were diagnosed by postoperative histopathology, and none of the patients had received any surgery, radiotherapy, chemotherapy or any other form of treatment. Any patients with systemic diseases or ocular diseases were excluded from the present study. The plasma samples of 30 healthy women (the control group) were additionally obtained during the same time period (Table SII). There was no significant difference between the age distributions of the two groups (P>0.05; Table SII).
Identification of plasma exosomes from patients with ovarian cancer
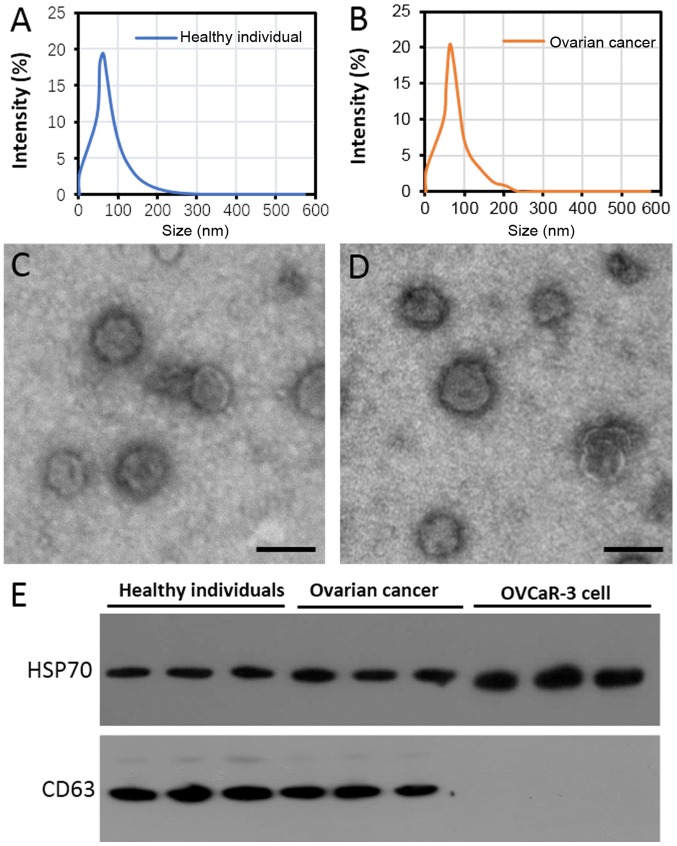
The diameter of extracellular vesicles (EVs) isolated from the plasma of healthy individuals and patients with ovarian cancer primarily ranged between 50 and 100 nm in diameter (Fig. 1A and B). Transmission electron microscopy demonstrated that the EVs were spherical vesicles with a similar diameter to the exosomes (Fig. 1C and D). Furthermore, two known exosome markers, CD63 and HSP70 (23), were observed in the purified EVs of the patients with ovarian cancer and the healthy individuals (Fig. 1E). However, CD63 was not detected in the lysates from the OVCar-3 cells (Fig. 1E), which indicates that the exosomes had been adequately purified.
Figure 1.
Identification of exosome-like vesicles purified from the plasma of healthy women and patients with ovarian cancer. Diameter distribution of purified exosome-like vesicles from (A) healthy individuals and (B) patients with ovarian cancer. Transmission electron microscopy of purified exosome-like vesicles from (C) healthy individuals and (D) patients with ovarian cancer. Scale bar, 100 nm. (E) Western blotting of CD63 and HSP70 protein expression in exosome-like vesicles purified from the plasma of patients with ovarian cancer and healthy women. Proteins from OVCaR-3 cells were used as a control. CD63, granulophysin; HSP70, heat-shock-protein 70 KDa.
Differentially expressed miRNAs in plasma exosomes
Using a two-fold change and P<0.05 as a threshold cut-off, 65 miRNAs were identified to be differentially expressed in the exosomes in the healthy individuals compared with those in the patients with ovarian cancer. Among the differentially expressed miRNAs, 34 were upregulated (Table SIII), and 31 were downregulated (Table SIV) in the exosomes of patients with ovarian cancer compared with the exosomes of healthy individuals. The top 10 differentially expressed miRNAs were hsa-miR-106a-5p, hsa-miR-185-5p, hsa-miR-99b-5p, hsa-miR-122-5p, hsa-miR-92b-3p, hsa-miR-584-3p, hsa-miR-744-5p, hsa-miR-92a-3p, hsa-miR-93-5p and hsa-miR-150-5p. A volcano plot of the differentially expressed miRNAs, where red and green indicate significantly upregulated and downregulated miRNAs, respectively is presented in Fig. 2.
Figure 2.
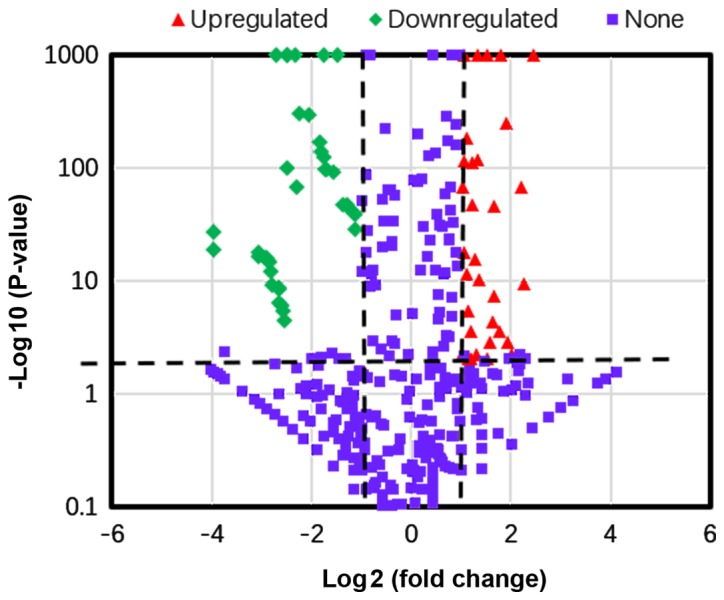
Volcano plot of differentially expressed miRNAs in patients with ovarian cancer compared with healthy women. Plotted along the x-axis is the mean of log2 fold-change and the y-axis is the negative logarithm of the -log P-values. Red points represent significantly upregulated miRNAs and green points represent significantly downregulated miRNAs with a >2.0-fold change.
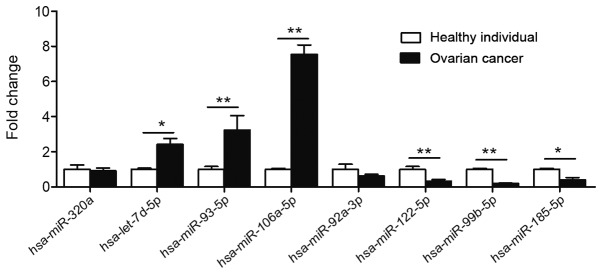
Validation of differentially expressed miRNAs by RT-qPCR analysis
The exosomes were isolated from the plasma of 30 patients with ovarian cancer and from the 30 healthy individuals. Relative expression levels of hsa-miR-106a-5p, hsa-let-7d-5p, hsa-miR-93-5p, hsa-miR-320a, hsa-miR-185-5p, hsa-miR-99b-5p, hsa-miR-122-5p and hsa-miR-92a-3p were analyzed. Compared with those in healthy individuals, the expression levels of hsa-let-7d-5p, hsa-miR-93-5p, hsa-miR-106a-5p were significantly upregulated, whereas the expression levels of hsa-miR-122-5p, hsa-miR-99b-5p and hsa-miR-185-5p were significantly downregulated in the plasma exosomes of ovarian cancer (Fig. 3). The expression levels of hsa-miR-320a and hsa-miR-92a-3p were not significantly changed (Fig. 3).
Figure 3.
Validation of the differential expression of 8 miRNAs identified in the microarray using RT-qPCR. The expression levels of plasma exosomal miRNA were analysed in patients with ovarian cancer (n=30) and healthy women (n=30) using RT-qPCR. A total of 8 miRNAs were randomly selected, including 4 upregulated miRNAs (hsa-miR-320a, hsa-let-7d-5p, hsa-miR-93-5p and hsa-miR-106a-5p) and 4 downregulated miRNAs (hsa-miR-92a-3p, hsa-miR-122-5p, hsa-miR-99b-5p and hsa-miR-185-5p). *P<0.05 and **P<0.01. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR/miRNA, microRNA.
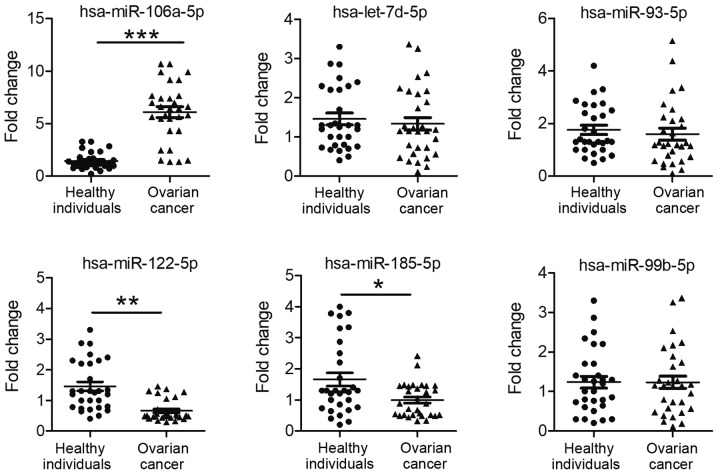
Expression levels of exosomal miRNAs in plasma
The expression levels of six selected miRNAs in the exosome (including hsa-miR-106a-5p, hsa-miR-122-5p, hsa-miR-185-5p, hsa-let-7d-5p, hsa-miR-93-5p, and hsa-miR-99b-5p) were consistent with the sequencing data (Tables SIII and SIV). Subsequently, the plasma expression levels of these 6 miRNAs were examined in healthy individuals and patients with ovarian cancer. Compared with the expression levels in healthy individuals, the expression levels of exosomal hsa-miR-106a-5p were significantly increased (P<0.001), and hsa-miR-122-5p and hsa-miR-185-5p levels were significantly decreased, in the plasma of patients with ovarian cancer (P=0.007 and P=0.018, respectively; Fig. 4). There was no significant difference in exosomal hsa-let-7d-5p, hsa-miR-93-5p and hsa-miR-99b-5p expression in the plasma between healthy individuals and patients with ovarian cancer (P=0.562, P=0.569 and P=0.997, respectively; Fig. 4).
Figure 4.
Expression of 6 exosomal miRNAs in the plasma of patients with ovarian cancer and healthy women. Expression levels of 6 miRNAs (hsa-let-7d-5p, hsa-miR-93-5p, hsa-miR-106a-5p, hsa-miR-122-5p, hsa-miR-99b-5p and hsa-miR-185-5) were analysed in the plasma of patients with ovarian cancer (n=30) and healthy women (n=30). The values of the relative gene expression for target miRNA were normalized to Caenorhabditis elegans miR-39 expression. *P<0.05, **P<0.01 and ***P<0.001. miR/miRNA, microRNA.
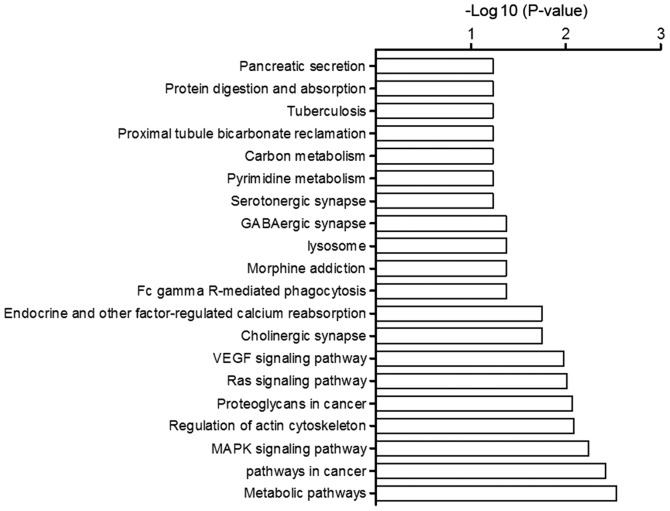
Enriched biological processes and molecular functions of miRNA targeted genes
GO analysis of the target genes of the differentially expressed miRNAs suggested that they were highly enriched in signaling and metabolism of biological process, transition metal ion binding, adenyl ribonucleotide binding, and signal transducer activity of molecular function. KEGG pathway analysis suggested that the target genes of those miRNAs were primarily involved in metabolic pathways, pathways in cancer, and MAPK signaling pathways (Fig. 5).
Figure 5.
Top Kyoto Encyclopedia of Genes and Genomes pathways regulated by predicted target genes of differentially expressed miRNAs. GABA, γ-aminobutyric acid; VEGF, vascular endothelial growth factor; MAPK, mitogen-activated protein kinase.
Discussion
Ovarian cancer is the leading cause of mortality among gynecological malignancies in developed countries (24). Due to the asymptomatic development of ovarian cancer, it the diagnosis frequently occurs at an advanced and incurable stage (25). At present, there is a dearth of reliable diagnostic markers and methods for the early detection and screening of the disease. Previous studies have demonstrated that circulating miRNAs may be valuable biomarkers for a variety of cancer types using blood plasma or serum (26,27). Additionally, various studies demonstrated the diagnostic potential of circulating miRNAs by analyzing the body fluids of patients with ovarian cancer, including the serum and plasma (28,29). Previous studies demonstrated that miRNAs were bound to miRNA-binding proteins or packaged into exosomes to avoid degradation (11,30). Specific miRNAs are packaged into exosomes secreted by the tumour cells that promote tumour spreading and growth in the surrounding microenvironment (31). However, to the best of our knowledge, there are no studies examining plasma exosomal miRNAs in patients with ovarian cancer.
Exosomes are membrane vesicles ~40–100 nm in size, and are released from a variety of cells (32); they are present in a number of body fluids, including blood, amniotic fluid, urine, saliva, breast milk and cerebrospinal fluid (33). Exosomes contain numerous types of molecules, including proteins, lipids, metabolites, mRNAs and miRNAs (34). In the present study, EVs were purified from the plasma of patients with ovarian cancer and healthy women. The mean diameters of the vesicles from the two groups ranged between 100 and 150 nm, which is similar to those previously observed in patients with ovarian cancer (35). These EVs were demonstrated to enrich CD63 and HSP70, which are markers that have been used for exosome identification (36), suggesting that the EVs were most likely exosome-like EVs. Taylor and Gercel-Taylor (37) first demonstrated that 8 exosomal miRNAs (miR-21, miR-141, miR-200a, miR-200b, miR-200c, miR-203, miR-205 and miR-214) from the serum of patients with ovarian cancer were upregulated compared with those from the benign controls. The aforementioned miRNA signatures were similar between ovarian tumour cells and exosomes from patients with ovarian cancer (37). Recent studies demonstrated that serum exosomal miRNAs, including miR-99a-5p and miR-145-5p, may serve as important biomarkers for the diagnosis and treatment of ovarian cancer (38,39). The miRNA profile in plasma exosomes derived from patients with ovarian cancer and healthy women was examined using high-throughput sequencing in the present study. A total of 65 plasma exosomal miRNAs were deregulated in patients with ovarian cancer compared with those in the healthy women. A total of 8 differentially expressed miRNAs were selected at random. The expression levels in the plasma samples of the 8 miRNAs were evaluated in the patients with ovarian cancer and healthy controls using RT-qPCR. The expression levels of plasma hsa-miR-106a-5p, hsa-let-7d-5p and hsa-miR-93-5p were increased, and the expression levels of hsa-miR-122-5p, hsa-miR-185-5p and hsa-miR-99b-5p were decreased in the patients with ovarian cancer compared with those in the healthy controls. A recent study by Pan et al (40) demonstrated that 8 miRNAs (miR-16, miR-21, miR-93, miR-100, miR-126, miR-200b, miR-223 and miR-320) were deregulated in the plasma exosomes of patients with ovarian cancer compared with those in healthy women. Similar to these results, the expression levels of hsa-miR-320 were increased in the patients with ovarian cancer, whereas the expression levels of hsa-miR-92a-3p were decreased. High expression levels of hsa-miR-320 are associated with the invasion and metastasis of ovarian cancer (41). The present data suggest that plasma exosomal miRNA may be used as a novel biomarker for ovarian cancer. The exosomal miRNA signatures were similar to those from the originating tumour cells, suggesting that the tumour profiles were accurately reflected by the circulating miRNA profiles (37). Emerging evidence has indicated that circulating miRNAs may serve as potential biomarkers for the early detection of ovarian cancer, the associated prognosis and the sensitivity of the disease to chemotherapy (28,42). In the present study, the plasma expression levels of exosomal miRNAs in patients with ovarian cancer and healthy female individuals were examined. The expression levels of plasma hsa-miR-93-5p were significantly increased in patients with ovarian cancer compared with those in the healthy controls. Furthermore, the expression levels of hsa-miR-122-5p and hsa-miR-99b-5p were decreased in patients with ovarian cancer compared with those in healthy women. The management strategies used at present for ovarian cancer rely on clinicopathological factors, including tumour histology and cancer stage. A recent study demonstrated that alterations in the expression levels of circulating miRNAs were associated with the prognosis of patients with ovarian cancer (28).
To examine the regulatory roles of differentially expressed miRNAs, the target genes of those miRNAs were predicted. Among the target genes of differentially expressed miRNAs, certain miRNAs are involved in pathways associated with cancer, MAPK signaling, regulation of the actin cytoskeleton or proteoglycans in cancer. For example, hsa-miR-122 inhibits the migration, invasion and epithelial-mesenchymal transition of ovarian cancer cells by regulating prolyl 4-hydroxylase subunit α-1 (43).
In conclusion, there was a significant difference in the expression levels of plasma exosomal miRNAs in patients with ovarian cancer compared with those in healthy women. The role of circulating exosomal miRNAs and the possibility of their diagnostic use in early detection requires further investigation.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by a grant from The Gansu Provincial Health Industry Research Project (Gansu, China; grant no. GSWSKY2017-14).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
HZ designed the study and wrote the manuscript. HZ, SX and XL conducted the experiments and analyzed the data. All authors approved the final manuscript.
Ethics approval and consent to participate
The study was approved by The Human Investigation and Ethical Committees of The Gansu Provincial Maternity and Child Care Hospital (Gansu, China) and conducted according to their guidelines and regulations.
Patient consent for publication
All participants provided informed consent.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Reid BM, Permuth JB, Sellers TA. Epidemiology of ovarian cancer: A review. Cancer Biol Med. 2017;14:9–32. doi: 10.20892/j.issn.2095-3941.2016.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decio A, Giavazzi R. Orthotopic model of ovarian cancer. Methods Mol Biol. 2016;1464:139–149. doi: 10.1007/978-1-4939-3999-2_13. [DOI] [PubMed] [Google Scholar]
- 3.Despierre E, Lambrechts D, Neven P, Amant F, Lambrechts S, Vergote I. The molecular genetic basis of ovarian cancer and its roadmap towards a better treatment. Gynecol Oncol. 2010;117:358–365. doi: 10.1016/j.ygyno.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 7.Nana-Sinkam SP, Croce CM. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther. 2013;93:98–104. doi: 10.1038/clpt.2012.192. [DOI] [PubMed] [Google Scholar]
- 8.Lässer C. Exosomal RNA as biomarkers and the therapeutic potential of exosome vectors. Expert Opin Biol Ther. 2012;12(Suppl 1):S189–S197. doi: 10.1517/14712598.2012.680018. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrovčíková E, Vičíková K, Leksa V. Extracellular vesicles-biogenesis, composition, function, uptake and therapeutic applications. Biologia. 2018;73:437–448. doi: 10.2478/s11756-018-0047-0. [DOI] [Google Scholar]
- 11.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 12.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suetsugu A, Honma K, Saji S, Moriwaki H, Ochiya T, Hoffman RM. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv Drug Deliv Rev. 2013;65:383–390. doi: 10.1016/j.addr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Subra C, Grand D, Laulagnier K, Stella A, Lambeau G, Paillasse M, De Medina P, Monsarrat B, Perret B, Silvente-Poirot S, et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J Lipid Res. 2010;51:2105–2120. doi: 10.1194/jlr.M003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuo K, Machida H, Mandelbaum RS, Konishi I, Mikami M. Validation of the 2018 FIGO cervical cancer staging system. Gynecol Oncol. 2019;152:87–93. doi: 10.1016/j.ygyno.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4. 2015 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;3:e264. doi: 10.1371/journal.pbio.0030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:451–454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Research 36 (Database Issue) 2008:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Pant S, Hilton H, Burczynski ME. The multifaceted exosome: Biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83:1484–1494. doi: 10.1016/j.bcp.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koshiyama M, Matsumura N, Konishi I. Subtypes of ovarian cancer and ovarian cancer screening. Diagnostics (Basel) 2017;7:E12. doi: 10.3390/diagnostics7010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandai M, Yamaguchi K, Matsumura N, Baba T, Konishi I. Ovarian cancer in endometriosis: Molecular biology, pathology, and clinical management. Int J Clin Oncol. 2009;14:383–391. doi: 10.1007/s10147-009-0935-y. [DOI] [PubMed] [Google Scholar]
- 26.Weiland M, Gao XH, Zhou L, Mi QS. Small RNAs have a large impact: Circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012;9:850–859. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 27.He Y, Lin J, Kong D, Huang M, Xu C, Kim TK, Etheridge A, Luo Y, Ding Y, Wang K. Current state of circulating MicroRNAs as cancer biomarkers. Clin Chem. 2015;61:1138–1155. doi: 10.1373/clinchem.2015.241190. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura K, Sawada K, Yoshimura A, Kinose Y, Nakatsuka E, Kimura T. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol Cancer. 2016;15:48. doi: 10.1186/s12943-016-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Wang T, Shi W, Yuan L, Chen L, Li Z. Comprehensive analysis on diagnostic value of circulating miRNAs for patients with ovarian cancer. Oncotarget. 2017;8:66620–66628. doi: 10.18632/oncotarget.18129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaksman O, Tropé C, Davidson B, Reich R. Exosome-derived miRNAs and ovarian carcinoma progression. Carcinogenesis. 2014;35:2113–2120. doi: 10.1093/carcin/bgu130. [DOI] [PubMed] [Google Scholar]
- 32.EL Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 33.van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64:676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 34.Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: From biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Szajnik M, Derbis M, Lach M, Patalas P, Michalak M, Drzewiecka H, Szpurek D, Nowakowski A, Spaczynski M, Baranowski W, Whiteside TL. Exosomes in plasma of patients with ovarian carcinoma: Potential biomarkers of tumor progression and response to therapy. Gynecol Obstet (Sunnyvale) 2013;(Suppl 4):S3. doi: 10.4172/2161-0932.S4-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 38.Yoshimura A, Sawada K, Nakamura K, Kinose Y, Nakatsuka E, Kobayashi M, Miyamoto M, Ishida K, Matsumoto Y, Kodama M, et al. Exosomal miR-99a-5p is elevated in sera of ovarian cancer patients and promotes cancer cell invasion by increasing fibronectin and vitronectin expression in neighboring peritoneal mesothelial cells. BMC Cancer. 2018;18:1065. doi: 10.1186/s12885-018-4974-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hang W, Feng Y, Sang Z, Yang Y, Zhu Y, Huang Q, Xi X. Downregulation of miR-145-5p in cancer cells and their derived exosomes may contribute to the development of ovarian cancer by targeting CT. Int J Mol Med. 2019;43:256–266. doi: 10.3892/ijmm.2018.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan C, Stevic I, Müller V, Ni Q, Oliveira-Ferrer L, Pantel K, Schwarzenbach H. Exosomal microRNAs as tumor markers in epithelial ovarian cancer. Mol Oncol. 2018;12:1935–1948. doi: 10.1002/1878-0261.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W, Yang J, Xiang YY, Pi J, Bian J. Overexpression of Hsa-miR-320 is associated with invasion and metastasis of ovarian cancer. J Cell Biochem. 2017;118:3654–3661. doi: 10.1002/jcb.26009. [DOI] [PubMed] [Google Scholar]
- 42.Zheng H, Liu JY, Song FJ, Chen KX. Advances in circulating microRNAs as diagnostic and prognostic markers for ovarian cancer. Cancer Biol Med10. 2013:123–130. doi: 10.7497/j.issn.2095-3941.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan Y, Dong Y, Dang R, Hu Z, Yang Y, Hu Y, Cheng J. MiR-122 inhibits epithelial mesenchymal transition by regulating P4HA1 in ovarian cancer cells. Cell Biol Int. 2018;42:1564–1574. doi: 10.1002/cbin.11052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.