Abstract
The recombinant HIV-1 CRF02_AG is prevalent in West-Central Africa but its effects on the blood-brain barrier (BBB) and HIV-associated neurocognitive disorders (HAND) are not known. We analyzed the effects of Tat from HIV-1 subtype-B (Tat.B) and CRF02_AG(Tat.AG) on primary human brain microvascular endothelial cells (HBMEC), the major BBB component. Exposure of HBMEC to Tat.B increased IL-6 expression and transcription by 9-fold (p<0.001) and 113-fold (p<0.001) respectively, whereas Tat.AG increased IL-6 expression and transcription by 2.7–3.8-fold and 35.7-fold (p<0.001) respectively. Tat.B induced IL-6 through the interleukin-1 receptor associated kinase(IRAK)-1/4 / mitogen-activated protein kinase kinase(MKK) / C-jun N-terminal kinase(JNK) pathways, in an activator protein-1(AP1)- and nuclear factor-kappaB(NFκB)-independent manner, whereas Tat.AG effects occurred via MKK/JNK/AP1/NFκB pathways. Tat-induced effects were associated with activation of c-jun (serine-63) and SAPK/JNK (Thr183/Tyr185). We demonstrated increased expression of transcription factors associated with these pathways (Jun, RELB, CEBPA), with higher levels in Tat.B-treated cells compared to Tat.AG. Functional studies showed that Tat.B and Tat.AG decreased the expression of tight junction proteins claudin-5 and ZO-1, and decreased the trans-endothelial electric resistance (TEER); Tat.B induced greater reduction in TEER, claudin-5 and ZO-1, compared to Tat.AG. Overall, our data showed increased inflammation and BBB dysfunction with Tat.B, compared to Tat.AG. This suggests these two HIV-1 subtypes differentially affect the BBB and central nervous system; our data provides novel insights into the molecular basis of these differential Tat-mediated effects.
Keywords: HIV-1 Tat subtypes, CRF02_AG, blood-brain barrier, IL-6, JNK/NFκB signaling
Introduction
The human immunodeficiency virus (HIV) infection of the central nervous system (CNS) commonly results in behavioral, motor, and cognitive impairments termed HIV-1-associated neurocognitive disorders (HAND) [1]. This CNS infection occurs as a result of the blood-brain barrier (BBB) breakdown, which enables HIV-1 and infected mononuclear phagocytes to enter the brain and spread infection to resident brain cells [2,3]. This BBB breakdown can be caused by HIV and secreted viral factors such as HIV proteins. In fact, there is in vitro and in vivo evidence that HIV-1 virions and secreted viral proteins alter the BBB integrity and function, and increase monocyte entry into the CNS.[3] One such viral protein is the HIV-1 transactivator of transcription (Tat). Tat is essential for viral replication and immune response [4,5]; it is expressed early in the viral life cycle, is released by HIV-infected cells and can be detected in the serum of infected patients [6]. Tat can cross the BBB [7] and is present in the CNS of HIV- infected humans [8]. However most of these studies showing the effects of Tat on the BBB and CNS were performed using Tat proteins from HIV-1 subtype-B, the predominant clade circulating in the US and Europe; whereas, over two-thirds of the 37 million people living with HIV/AIDS are in sub-Saharan Africa and are infected with non-B HIV subtypes, including the circulating recombinant form (CRF)02_AG [9].
HIV-1 accounts for over 95% of all infections [9,10]. and includes four groups: M (major), O (outlier), N (non-M non-O), and P [11,12]. HIV-1 group M accounts for the vast majority of infection globally and includes 9 pure subtypes (A-D, F-H, J and K), sub-subtypes (A1 and A2, F1 and F2), about 70 CRFs and several unique (unclassified) recombinant forms (URFs) [11,13]. HIV-1 CRF02_AG is a recombinant of subtypes A and G, circulating in West and Central Africa; with 52 to 84% of HIV-infected humans in that region infected with HIV-1 CRF02_AG [14,15]. This Central and West Africa region includes 26 countries with over 456 millions inhabitants [16]. The effects of this recombinant virus on the BBB and HAND are not known. In the current study, we show differential inflammation with Tat from HIV-1 subtype B (Tat.B) and Tat from HIV-1 CRF02_AG (Tat.AG) on primary human brain microvascular endothelial cells (HBMEC), the major component of the BBB. We show significantly increased transcription, expression and secretion of the pro-inflammatory cytokine interleukin (IL)-6 by primary HBMEC exposed to Tat.B, compared to cells exposed to Tat.AG. We demonstrate that Tat.B induced BBB inflammation through the IRAK1/4,MKK/JNK pathways, in an AP1- and nuclear factor kappa-B (NFκB)-independent manner, whereas Tat.AG-induced inflammation occurred via the MKK/JNK/AP1/NFκB pathways. We further demonstrated increased expression of transcription factors associated with these pathways (Jun, RELB, CEBPA), with higher levels in cells exposed to Tat.B compared Tat.AG. Functional studies showed that both Tat.B and Tat.AG decreased the expression of endothelial tight junction proteins claudin-5 and ZO-1, and decreased brain trans-endothelial electric resistance (TEER), but greater reduction in TEER, claudin-5 and ZO-1 levels was observed with Tat.B compared to Tat.AG. Compared to Tat.AG, Tat.B significantly increased monocytes adhesion and migration through in vitro BBB models.
Materials and Methods
Brain Endothelial Cell Culture
Primary HBMEC were isolated from brain tissue obtained during surgical removal of epileptogenic cerebral cortex in adult patients, under an Institutional Review Board-approved protocol at the University of Arizona as described previously [17]. Routine evaluation by immunostaining for von-Willebrand factor, Ulex europaeus lectin and CD31 showed that cells were >99% pure. Freshly isolated cells were cultured in collagen-coated flasks or 6-well culture plates using Dulbecco’s Modified Eagle Medium (DMEM)/F12 (Life Technologies, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS, Atlanta Biologicals, Flowery Branch, GA, USA), supplemented with 10 mmol/l L-glutamine (Life Technologies), 1% heparin (Thermo Fisher Scientific, Pittsburgh, PA, USA), 1% endothelial cell growth supplement (ECGS; BD Bioscience, San Jose, CA, USA), 1% penicillin-streptomycin (Life Technologies), 1% fungizone (MP Biomedicals, Solon, OH, USA). Cells at passage 2 to 4 were used in this study.
Recombinant HIV Tat.AG and Tat .B
Recombinant Tat proteins from a subtype-B HIV-1 isolate (Tat.B) (amino acids 1 to 86; accession number: P69697) were purchased from Diatheva (Viale Piceno, Fano, Italy). Recombinant Tat proteins from HIV-1 CRF02_AG (Tat.AG) (amino acids 1 to 86; accession number: AY371128) were made by Diatheva under a custom-order agreement with our laboratory, using similar procedures as for Tat.B. For controls, HIV Tat proteins were heat inactivated at 100°C for 30 minutes, cooled down and centrifuged at 1000 rpm for 5 minutes to recover all of the solution. Protein aliquots were stored at −80°C.
Interleukin-6 ELISA
Interleukin (IL)-6 expression and secretion was quantified using the BD OptEIA Human IL-6 ELISA kit II (BD Biosciences, CA, USA) according to manufacturer’s protocol. Briefly, confluent HBMEC were treated with Tat.AG or Tat.B at 100 or 1000 ng/mL and/or pharmacological inhibitors (Table 1) for 48 hours. Controls consisted of untreated cells and cells treated with similar concentrations of heat-inactivated Tat proteins (HI-Tat.B and HI-Tat.AG). Following cell treatment, culture supernatants were collected and further centrifuged for 5 min at 2350 g to remove any cellular debris, and 100 μl of each sample used for IL-6 ELISA according to manufacturer’s instructions. For each experiment, the IL-6 standards provided with kit were used to generate standard curve and determine IL-6 concentrations in each samples. Each experimental condition was tested in triplicate.
Table 1:
Pharmacological inhibitors used in the study
| Inhibitor name | Catalog# / Abbreviation | Mechanism of action | Manufacturer |
|---|---|---|---|
| NFκB Activation Inhibitor | 481406 | Inhibitor of NFκB transcriptional activation | Calbiochem- EMD Millipore |
| SR 11302 | SR11302 | Inhibitor of AP-1 transcriptional activity | Tocris Biosciences |
| JNK Inhibitor II | 420119 | Selective and reversible inhibitor of JNK1, JNK2, and JNK3. | Calbiochem- EMD Millipore |
| JNK Inhibitor V | 420129 | Reversible and ATP-competitive inhibitor of JNK1, JNK2, and JNK3. | Calbiochem- EMD Millipore |
| JNK Inhibitor III | 420130 | Specifically disrupts c-Jun/JNK complex formation and subsequent phosphorylation and activation of c-Jun by JNK | Calbiochem- EMD Millipore |
| IRAK-1/4 inhibitor | IRAK-1/4 | Inhibitor of IRAK-1 and IRAK-4 | Sigma-Aldrich |
| 5Z-7-Oxozeaenol | 5ZO | ATP-competitive irreversible inhibitor of TAK1 (MEKK7), MKK7, MEK1, and ERK2. Has no activity against other MAPKs | Sigma-Aldrich |
Abbreviations: JNK: c-Jun N-terminal kinase; AP-1: activator protein-1; IRAK: Interleukin-1 Receptor-Associated Kinase; NFκB: nuclear factor kappaB; TAK1: Transforming growth factor β activated kinase-1; MEKK7: MAP Kinase Kinase Kinase-7; MKK7: MAP Kinase Kinase-7; MEK1: MAP Kinase Kinase-1; ERK2: extracellular-signal-regulated kinase-2 / MAP Kinase-1
RNA extraction and real-time PCR
Confluent HBMEC in six-well plates were treated for 48 hours with 100 ng/ml Tat.AG or Tat.B. Controls consisted of untreated cells, and cells treated with similar concentrations of heat inactivated Tat proteins. Following Tat treatment, cells were harvested and total RNA was extracted using the Trizol reagent (Life Technologies-Ambion, Austin, TX) according to the manufacturer’s protocol. RNA was further cleaned using Total RNA cleanup kit (Qiagen, Valencia, CA). RNA yield and quality were checked using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) and for all samples, absorbance ratio of 260/280 was ≥ 2. For each sample, cDNA was generated from 1 μg RNA, using the Verso cDNA kit (Thermo Fisher, Waltham, MA USA), according to the manufacturer’s instructions. Reverse transcription was carried out for 30 min at 42°C. The cDNA obtained was used for quantitative real-time PCR using Roche Real-Time PCR Systems (Roche diagnostics, Indianapolis, IN, USA). Gene quantification was performed using the Ct method as described in the software user manual. All PCR primers probes were obtained from Applied Biosystems (Grand Island, NY, USA); and primer IDs were as follows: CEBGA (Hs00269972_s1), CEBPG (Hs01922818_s1), Jun (Hs99999141_s1), RELB (Hs00232399_m1) and IL-6 (Hs00985639). For endogenous control, each gene expression was normalized to the sample GAPDH (Hs99999905_m1).
Protein Extraction and Western Blot Analyses
Protein extraction, quantification, and western blot analyses were performed as previously described [18,19].Briefly, for the analysis of JNK and c-Jun phosphorylation, HBMEC were treated with Tat.AG and Tat.B at 100 ng/ml for various time points (0, 7.5, 15, 30, 60, and 120 minutes). For tight junction protein analysis, HBMEC were treated with Tat.AG and Tat.B at 100ng/ml for 48 hours. Untreated cells or cells treated with HI-Tat.AG and HI-Tat.B served as controls. Cells were lysed using the mammalian cell lysis buffer CelLytic M (Sigma, St Louis, MO, USA), and protein quantified using the bicinchoninic acid assay as we previously described [18,19]. Protein (35 μg) was fractionated in a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. Membranes were blocked for 1 hour with SuperBlock T-20 (Pierce, Rockford, IL, USA) and blotted for 2 hours or overnight with monoclonal antibodies to JNK, phopho-JNK, c-Jun, phospho-c-Jun (Cell Signaling Technology, Danvers, MA, USA), Claudin-5, or ZO-1 (Abcam, Cambridge, MA, USA), at 1:1,000 dilution. Membranes were then washed, blotted for 1 hour with horseradish peroxidase- conjugated secondary antibody, washed again, and visualized using the enhanced chemiluminescence (Pierce, Rockford, IL, USA) and gel doc system (Syngene, Frederick, MD, USA). After each western blot experiment, the membranes were stripped using the Restore Western Blot Stripping Buffer (Pierce, Rockford, IL, USA) and re-blotted with β-actin antibody (Abcam, Cambridge, MA) to confirm equal loading.
Transendothelial Electrical Resistance (TEER)
Electrical resistance across the endothelial cell monolayer was measured using the Electric Cell-substrate Impedance Sensing (ECIS) instrument (Applied Biophysics, Troy, NY, USA) and the ECIS V1.2.215.msi software (Applied Biophysics), according to the manufacturer’s instructions. The BBB model was constructed by seeding 2 × 104 HBMECs in ECIS- 8W10E+PET array chambers containing gold film electrodes delineated with an insulating film (Applied Biophysics). The ECIS electrode array containing cells were placed in an array holder located in the CO2 incubator. Cells were cultured for 7 to 10 days to enable confluence, and before TEER measurements, electrodes were calibrated and stabilized according to the manufacturer’s recommendations. Confluent cells were treated with Tat.AG or Tat.B (100 or 1000 ng/ml), and TEER recorded for up to 72 hours. The ohmic resistance of a blank (culture insert without cells) was measured in parallel and was subtracted from the TEER readings obtained from inserts with confluent endothelial cell monolayers. The resulting TEER values represented the resistance (‘tightness’) of the endothelial cell monolayers. HBMEC treated with saponin (0.1%) served as positive control.
Monocyte isolation and labeling
Human monocytes were obtained from HIV-1, HIV-2 and hepatitis-B seronegative donor leukopaks, and separated by countercurrent centrifugal elutriation as previously described [20,18]. Cells were identified as >98% pure monocytes by Wright staining and CD68 immunostaining (at 1:50 dilution, Dako, Carpentaria, CA). Monocytes were used for adhesion and transendothelial migration assays within 24 hours of elutriation. Freshly elutriated monocytes were washed twice with pre-warmed (37°C) PBS, re-suspended in pre-warmed PBS containing 5μM of 5-carboxyfluorescein diacetate, succinimidyl ester (CFDA-SE) (Life Technologies); 1 × 106 cells / ml, and incubated at 37°C, 5% CO2, for 15 min. Monocytes were then pelleted by centrifugation at 300 g for 10 minutes, resuspended in pre-warmed media and incubated at 37°C, 5% CO2, for 30 min. CFDA-SE-labeled monocytes were washed three times with pre-warmed serum-free media before use for adhesion and migration assays.
Monocyte adhesion and migration through in vitro BBB models
Adhesion and migration experiments were performed as we previously described [20,18]. Briefly, for adhesion assays, HBMEC were plated on 96-well collagen-coated black plates, and for migration assay, HBMEC were plated on collagen-coated FluoroBlok-tinted tissue culture inserts (3-μm pore size; BD Biosciences). Endothelial cells were cultured to confluence, and were treated with Tat.AG or Tat.B proteins (100 or 1000 ng/ml) for 48 hours. Controls consisted of untreated cells and cells treated with similar concentrations of heat-inactivated proteins for 48 hours. For adhesion assays, following Tat treatment, HBMEC were rinsed to remove all Tat proteins, exposed to 2.5 × 105 CFDA-SE-labeled monocytes, and incubated at 37°C, 5% CO2, for 15 min. After the 15 min adhesion, HBMEC were washed 3 times with PBS, and the number of adherent monocytes quantified by spectrophotometry (top readings, absorbance 494 nm; emission, 517 nm), with a standard curve derived from a serial dilution of a known number of CFDA-SE-labeled monocytes. For migration assays, following Tat treatment, 2.5 × 105 CFDA- SE-labeled monocytes were added to the upper chamber of the FluoroBlok inserts and allowed to migrate for 2 hours (37°C, 5% CO2). The numbers of migrated monocytes were quantified by spectrophotometry (bottom readings, absorbance 494 nm; emission, 517 nm), with a standard curve derived from a serial dilution of a known number of CFDA SE-labeled monocytes.
Statistical analyses
Data were analyzed by t-test (two-tailed) for two-group comparisons and one- or two-way ANOVA followed by Tukey’s multiple-comparisons tests using GraphPad Prism 5.0b. (GraphPad Software, La Jolla, CA). Threshold of significance level was 0.05. Data are presented as means ± standard error of the mean.
Results
Increased IL-6 expression and secretion in HBMEC exposed to Tat.B, compared to cells exposed to Tat.AG
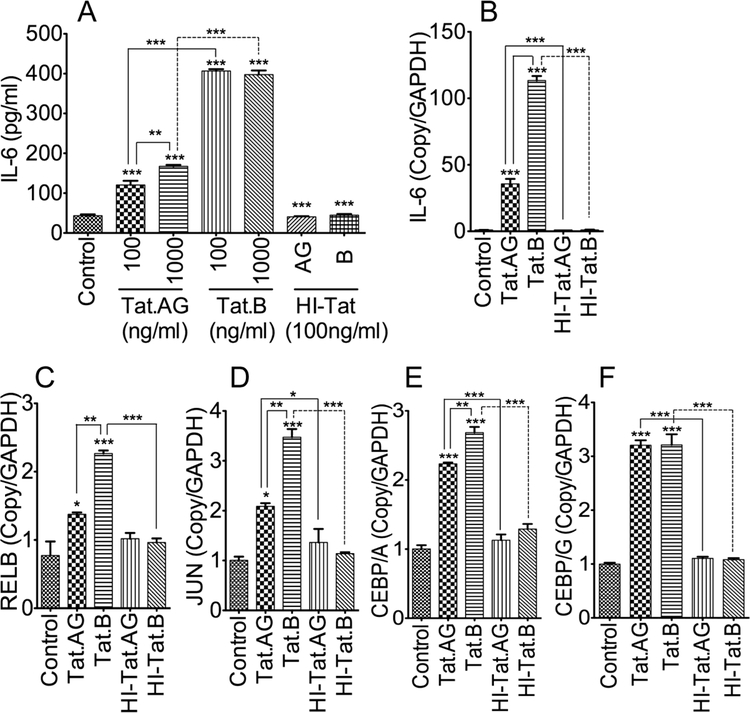
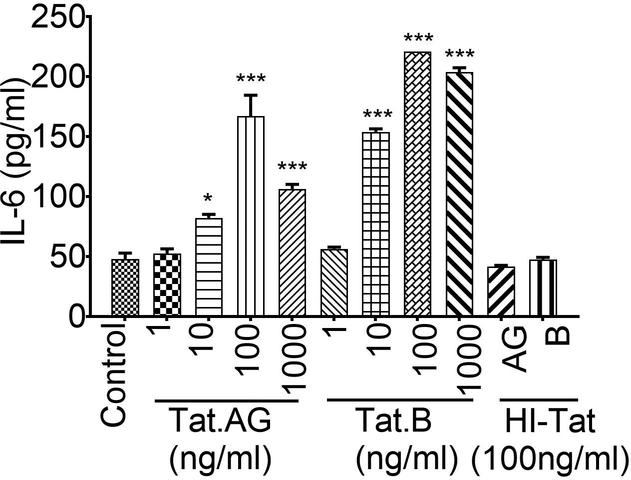
To determine the effect of Tat proteins derived from HIV-1 subtypes B and CRF02_AG on BBB inflammation, we tested the effects of these two Tat proteins on IL-6 expression in HBMEC. Both Tat.AG and Tat.B (100 or 1000ng/ml) induced IL-6 expression and secretion in primary HBMEC but much larger increase was observed with Tat.B compared to Tat.AG (Fig. 1A). Compared to untreated controls and HBMEC treated with heat-inactivated Tat proteins, Tat.AG increased IL-6 levels in HBMEC by 2.7 to 3.8-fold, whereas Tat.B increased IL-6 expression in HBMEC by over 9-fold (Fig. 1A, P<0.001). Overall, IL-6 expression in Tat.B-treated HBMEC were 2.37- to 3.37-times higher than the levels in Tat.AG-treated cells (Fig. 1A, P<0.001). No increased in IL-6 was observed in HBMEC treated with heat inactivated Tat proteins (HI-Tat.AG and HI-Tat.B), with IL-6 levels comparable to levels in untreated controls (Fig. 1A). Additional dose-dependent experiments showed that at similar doses, Tat.B induced higher IL-6 expression and secretion in primary HBMEC than Tat.AG (Appendix).
Fig. 1: Increased expression and transcription of IL-6 and transcription factors associated with NFκB and JNK pathways in HBMEC exposed to Tat.B, compared to cells exposed to Tat.AG.
Exposure of primary HBMEC to Tat proteins (100 or 1000 ng/ml) increased IL-6 levels, and higher IL-6 expression (A) and transcription (B) were observed in Tat.B-treated cells, compared to cells exposed to Tat.AG. Quantitative real-time PCR also showed higher transcriptional upregulation of RELB (C), JUN (D), and CEBPA (E), in Tat.B-treated cells, compared to cells exposed to Tat.AG. CEBPG mRNA levels were similar in cells exposed to Tat.B or Tat.AG (F). No increased expression or transcription of IL-6, RELB, JUN, CEBP/A, or CEBP/G was observed in cells exposed to of 100 ng/ml heat-inactivated Tat.B (HI-Tat.B) or Tat.AG (HI- Tat.AG). *P<0.05, **P<0.01, ***P<0.001, compared to cells treated with heat-inactivated proteins and untreated controls. For all experiments, each experimental condition was performed in triplicate. Fig. shows representative data from two to three independent experiments.
Tat.AG and Tat.B induced transcriptional upregulation of IL-6 and transcription factors associated with NFκB and JNK pathways in HBMEC
To determine whether differential Tat-induced IL-6 expression correlated with IL-6 transcription, we used real-time PCR to quantify IL-6 mRNA in HBMEC exposed to Tat.B and Tat.AG. Both Tat.B and Tat.AG significantly increased IL-6 mRNA in HBMEC (Fig. 1B, P<0.001), but the levels of IL-6 mRNA in Tat.B treated cells were over 3-times higher than the levels in Tat.AG- treated cells (Fig. 1B, P<0.001). The IL-6 promoter contains several cis-acting response elements that regulate its transcription and expression. These included the activator protein-1 (AP-1), NFκB, and the cAMP response element binding protein (CREB/CEBP). Therefore, we analyzed the effects of Tat.B and Tat.AG on the expression of these transcription factors. Compared to untreated controls or cells treated with heat-inactivated Tat proteins, exposure of HBMEC to Tat.B and Tat.AG increased JUN transcription by 3.47-fold (Fig. 1C, P<0.001) and 2- fold (Fig. 1C, P<0.01) respectively, and JUN mRNA levels in Tat.B-treated cells were significantly higher than the levels in Tat.AG-treated cells (Fig. 1C, P<0.01). Compared to untreated controls or cells treated with heat-inactivated Tat proteins, exposure of HBMEC to Tat.B and Tat.AG increased RELB transcription by 2.93-fold (Fig. 1D, P<0.001) and 1.78-fold (Fig. 1D, P<0.05) respectively, and RELB mRNA levels in Tat.B-treated cells were significantly higher than the levels in Tat.AG-treated cells (Fig. 1D, P<0.01). Compared to untreated controls or cells treated with heat-inactivated Tat proteins, exposure of HBMEC to Tat.B and Tat.AG increased CEBP/A by 2.7-fold (Fig. 1E, P<0.001) and 2.2-fold (Fig. 1E, P<0.001) respectively, and CEBP/A mRNA levels in Tat.B-treated cells were significantly higher than the levels in Tat.AG-treated cells (Fig. 1E, P<0.01). Similarly, Tat.B and Tat.AG increased CEPB/G in HBMEC by 3.2-fold, compared to untreated controls or cells treated with heat-inactivated Tat proteins (Fig. 1F, P<0.001).
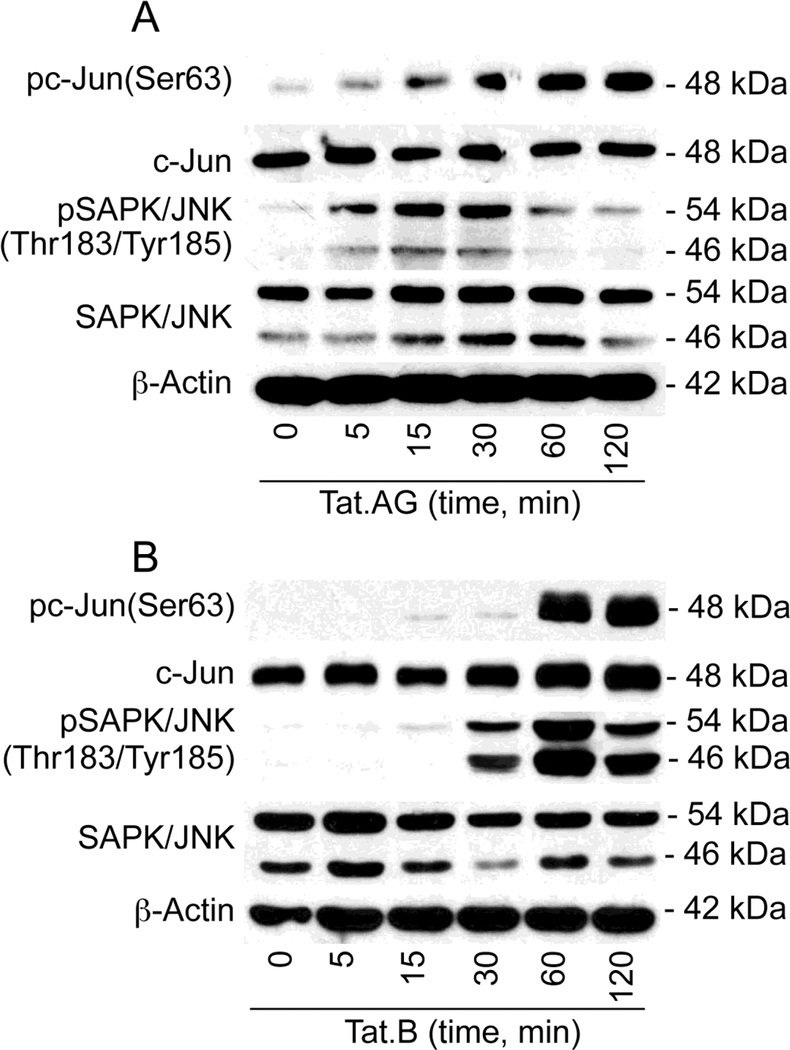
Tat.AG and Tat.B phosphorylate c-Jun and JNK in HBMEC
Because Tat induced upregulation of transcription factors associated with the JNK pathways (Fig. 1), we analyzed the effects of Tat.B and Tat.AG on c-Jun and JNK activation. Exposure of HBMEC to both Tat.AG (Fig. 2A) and Tat.B (Fig. 2B) induced the phosphorylation of c-Jun at serine-63 (Ser63). Tat.AG- and Tat.B-induced c-Jun phosphorylation occurred as early as 5 min and 15 min respectively, and continued for up to 2 hours, with maximal phosphorylation observed at 1 and 2 hours (Fig. 2). Both Tat.AG (Fig. 2A) and Tat.B (Fig. 2B) induced the phosphorylation of SAPK/JNK at Thr183/Tyr185. Tat.AG-induced SAPK/JNK phosphorylation occurred as early as 5 min following Tat exposure, with maximal phosphorylation at 15 and 30 min, and gradual decrease in phosphorylation thereafter (Fig. 2A). Tat.B-induced SAPK/JNK phosphorylation started at 15 min, with maximal phosphorylation observed at 1 hour (Fig. 2B).
Fig. 2: HIV-1 Tat.AG and Tat.B phosphorylate c-Jun and JNK in HBMEC.
Exposure of primary HBMEC to both Tat.AG (A) and Tat.B (B) (100 ng/ml) induced the phosphorylation of c-Jun at Ser63, with maximal phosphorylation at 1 and 2 hours. Tat.AG and Tat.B also induced the phosphorylation of SAPK/JNK at Thr183/Tyr185, with maximal phosphorylation at 15 to 30 min with Tat.AG (A), and 1 hour with Tat.B (B). Tat treatment did not induce major change in total c-Jun and SAPK/JNK, or actin levels. Fig. shows representative data from three independent experiments.
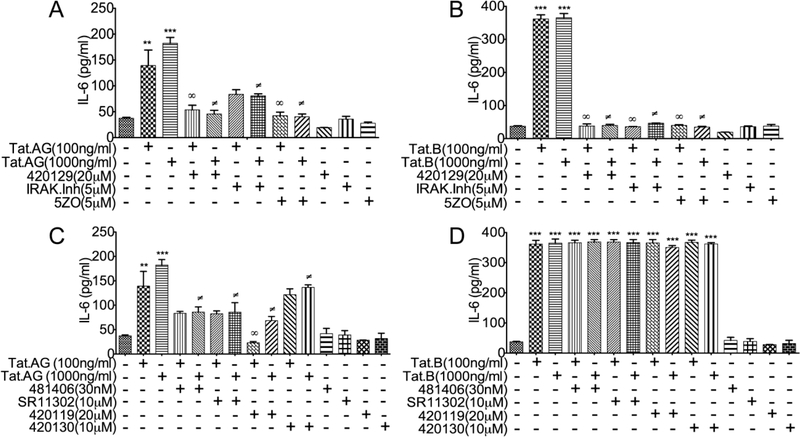
JNK and NFκB pathways mediates Tat.B- and Tat.AG-induced IL-6 expression
To determine whether blocking the pathways associated with transcription factors upregulated by Tat (Fig. 1) can alter Tat-induced IL-6 expression, we quantified IL-6 expression and secretion in HBMEC exposed to Tat.B or Tat.AG, in the presence or absence of specific inhibitors of effectors associated with those transcription factors. The ATP-competitive JNK inhibitor (420129), the IRAK1/4 inhibitor and the MEKK7/MKK7 inhibitor (5ZO) significantly diminished Tat.AG induced IL-6 expression (Fig. 3A), and totally blocked Tat.B-induced IL-6 expression and secretion (Fig. 3B). The JNK inhibitor 420119, the inhibitor of c-Jun/JNK complex (420130), the inhibitor of AP-1 transcription (SR11302), and the inhibitor of NFκB transcriptional activation (481406), partially blocked Tat.AG-induced IL-6 expression (Fig. 3C), but had no effect on Tat.B induced IL-6 expression and secretion (Fig. 3D).
Fig. 3: JNK and NFκB pathways mediate Tat.B- and Tat.AG-induced IL-6 expression.
Exposure of HBMEC to Tat.AG (A, C) and Tat.B (B, D) increased IL-6 expression. The ATP- competitive JNK inhibitor (420129), the IRAK1/4 inhibitor and the MEKK7/MKK7 inhibitor (5ZO) significantly diminished Tat.AG induced IL-6 expression (A), and blocked Tat.B-induced IL-6 expression (B). The JNK inhibitor (420119), the inhibitor of c-Jun/JNK complex (420130), the inhibitor of AP-1 transcription (SR11302), and the inhibitor of NFκB transcriptional activation (481406) partially blocked Tat.AG-induced IL-6 expression (C), but had no effect on Tat.B induced IL-6 expression (D). For all experiments, each experimental condition was performed in triplicate. Fig. shows representative data from two to three independent experiments.
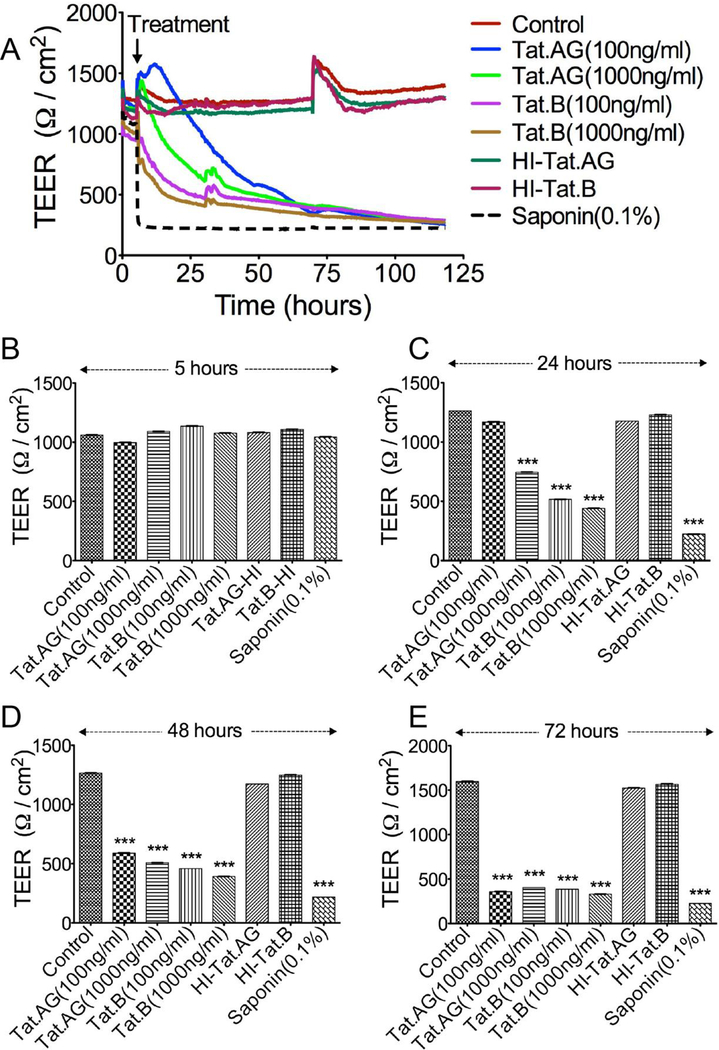
Effects of Tat.AG and Tat.B on the endothelial barrier properties
We performed experiments to determine whether Tat.AG and Tat.B alter the endothelial barrier property. For this purpose, we assessed the effects of Tat.B and Tat.AG on the brain TEER. Exposure of HBMEC to both Tat proteins decreased TEER, compared to TEER values of confluent cells before Tat treatment (Fig. 4A). No major effect was observed within 5 hours of Tat treatment (Fig. 4B), but TEER gradually decreased thereafter and Tat.B induced larger decreases in TEER, compared to Tat.AG. At 24 hours, 100 ng/ml Tat.AG did not affect TEER, whereas similar concentrations of Tat.B decreased the TEER by 54.5% (Fig. 4A and 4C, P<0.001). At 24 hours, 1000 ng/ml Tat.AG decreased TEER by 32% whereas similar concentrations of Tat.B decreased the TEER by 59% (Fig. 4A and 4C, P<0.001). At 48 hours, 100 ng/ml Tat.AG decreased the TEER by 41%, whereas similar concentrations of Tat.B decreased the TEER by 59.5%; 1000 ng/ml Tat.AG decreased the TEER by 53%, whereas similar concentrations of Tat.B decreased the TEER by 63.5% (Fig. 4A and 4D, P<0.001). Longer exposure to Tat further decreased TEER; at 72h Tat.AG and Tat.B decreased the TEER by 63 to 64% and by 66 to 69% respectively (Fig. 4A and 4E). No decrease in TEER was observed in cells treated with similar concentrations (100 ng/ml) of heat-inactivated Tat proteins (Fig. 4). Positive controls consisted of cells exposed to 0.1% saponin (Fig. 4).
Fig. 4: Effects of Tat.B and Tat.AG on the BBB integrity.
Confluent HBMEC were exposed Tat.B or Tat.AG (100 or 1000 ng/ml) and TEER measured in real-time for up to 120 hours (A) as described in the Method section. No major change in TEER was observed at 5 hours post-Tat exposure, compared to TEER values before Tat treatment (B). TEER gradually decreased thereafter, with larger decrease observed at 24 hours (C), 48 hours (D), and 72 hours (E). Tat.B induced larger decrease in TEER than Tat.AG. At 24h (C), 100 ng/ml Tat.B decrease TEER by 54.4% whereas 100ng/ml Tat.AG did not affect TEER; 1000 ng/ml Tat.B decrease TEER by 59% compared to 32% with 1000 ng/ml Tat.G. At 48h (D), 100 ng/ml Tat.B decreased the TEER by 59.5% compared to 41% with 100 ng/ml Tat.AG; 1000 ng/ml Tat.B decreased the TEER by 63.5% compared to 53% with 1000 ng/ml Tat.AG. Further decreases in TEER were observed at 72h (E). ***P<0.001, compared to untreated controls or cells treated with heat-inactivated proteins. Data shown are representative of two to three independent experiments. Saponin (0.1%) was used as a positive control for lowering TEER.
Effects of Tat.AG and Tat.B on the BBB integrity and function
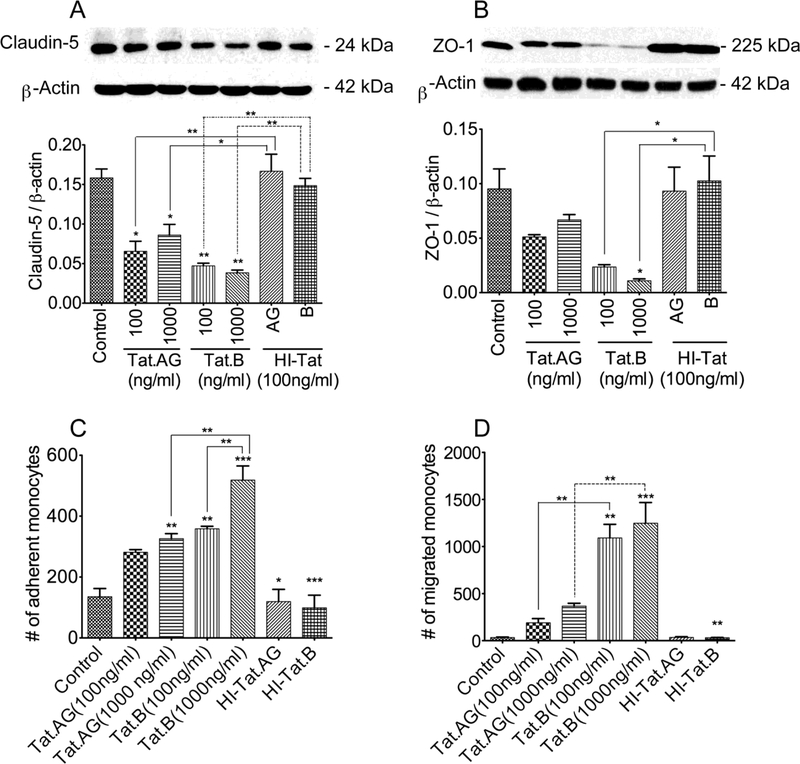
To determine the effects of Tat.B and Tat.AG on the BBB integrity, we analyzed the effects of both Tat proteins on the expression of tight junction proteins claudin-5 and ZO-1 in HBMEC. Both Tat proteins significantly decreased claudin-5 expression, but larger decrease in claudin-5 levels was observed in HBMEC treated with Tat.B, compared to cells treated with Tat.AG (Fig. 5A). Similarly, both Tat proteins decreased ZO-1 expression, but larger decrease in ZO-1 was observed in HBMEC treated with Tat.B, compared to cells treated with Tat.AG (Fig. 5B). Cells treated with heat-inactivated Tat.B or heat-inactivated Tat.AG showed no decrease in claudin-5 (Fig. 5A) or ZO-1 (Fig. 5B) expression.
Fig. 5: Effects of Tat.B and Tat.AG on endothelial tight junction proteins and BBB function.
Exposure of HBMEC to Tat.B or Tat.AG (100 and 1000 ng/ml) for 48 hours decreased claudin-5 (A) and ZO-1 (B) expression, but larger decreases were observed with Tat.B, compared to Tat.AG. Similarly, Tat.B increased monocytes adhesion to HBMEC by 2.6 to 3.8-fold compared 2 to 2.4-fold increase with Tat.AG (C). Tat.B increased monocyte BBB transmigration by 32 to 36-fold, compared 5.6 to 10.8-fold increase with Tat.AG (D). *P<0.05, **P<0.01, ***P<0.001. HI- Tat: heat-inactivated Tat (AG or B). For panels C and D, p-values of HI-Tat.AG and HI-Tat.B are in comparison to HBMEC treated with similar concentrations of Tat proteins. Data shown are representative of three independent experiments.
Additional functional assays demonstrated that both HIV-1 Tat proteins increased the adhesion (Fig. 5C) and migration (Fig. 5D) of monocytes through in vitro BBB models, with significantly larger increase in adhesion and migration when HBMEC were exposed to Tat.B. Exposure of HBMEC to Tat.B (100 or 1000 ng/ml) increased monocytes adhesion by 2.6 to 3.8- fold (Fig. 5C), and increased monocyte BBB transmigration by 32 to 36-fold (Fig. 5D), compare to untreated controls or cells exposed to similar concentrations of heat-inactivated proteins. Exposure of HBMEC to Tat.AG (100 or 1000 ng/ml) increased monocytes adhesion by 2 to 2.4- fold (Fig. 5C), and increased monocyte BBB transmigration by 5.6 to 10.8-fold (Fig. 5D), compare to untreated controls or cells exposed to similar concentrations of heat-inactivated proteins. No increase in monocyte adhesion or migration was observed in HBMEC exposed to heat-inactivated Tat proteins (Fig. 5C, D).
Discussion
HIV-1 Tat proteins are released by infected cells, can be detected in the serum and CNS of infected patients [6,8,7], and is involved in HAND. Tat can induce injury to many cells, including neurons [21–23], astrocytes [24,23], monocytes and brain endothelial cells [25,3]. Most of the studies investigating these Tat-mediated effects were performed using Tat from HIV-1 subtype B [6,8,7,21–25,3], with few others studies using subtype-C Tat [26,27]. Over two-thirds of individuals currently living with HIV/AIDS reside in sub-Saharan Africa and are infected with non-B HIV subtypes, including HIV-1 CRF02_AG, the predominant subtype circulating in West and Central Africa [28,14,15]. In the present study, we show that viral genotype can influence Tat-induced BBB inflammation; we demonstrate significantly higher IL-6 transcription, expression, and secretion in HBMEC exposed to Tat.B, compared to Tat.AG.
IL-6 plays a major role in inflammation and anti-IL-6 monoclonal antibodies or antibodies to IL-6 receptors are clinically used for the treatment of inflammatory diseases such as rheumatoid arthritis [29–31]. IL-6 is synthesized following infection or tissue injury and triggers acute-phase responses and subsequent immune responses [29]. In normal conditions, the production of IL-6 and IL-6-induced immune reactions stop once the infection or injury-inducing agent is cleared. However, a dysregulated immune response often leads to continuous IL-6 synthesis and chronic inflammation. Therefore, dysregulation and continued IL-6 production play a major role in the pathogenesis of several diseases associated with chronic inflammation, including HIV/AIDS. Human studies showed that increased IL-6 expression in HIV-infected subjects positively correlate with viral RNA and is associated with worse clinical outcomes [32]. In fact, increased IL-6 levels in the serum or plasma of HIV-infected humans at seroconversion independently predicts faster progression to AIDS [33,34]. Increased IL-6 levels before antiretroviral therapy (ART) initiation is associated with faster disease progression [35], and predicts early mortality [36–38]. In chronic HIV infection, increased IL-6 levels predicts the development of opportunistic infections and mortality [39], and is associated with increased risks of cardiovascular disease [40,41]. ART use diminished IL-6 levels in vivo and discontinuation of ART is associated with significant rebound in IL-6 levels [39]. Even for subjects on ART, persistent increase in serum IL-6 levels is a strong predictor of mortality [42].
Studies using primary HBMEC also showed that HIV-1 [18] and viral gp120 proteins [19] induced IL-6 expression and dysfunction of the vascular endothelium; and brain microvessels from HIV-infected humans showed increased IL-6 transcription [18]. Our current study show that although both Tat.B and Tat.AG increased the transcription and expression of IL-6 in HBMEC, IL-6 levels in Tat.B-treated cells were three times higher than the levels in Tat.AG-treated cells. This suggests that viral genotype influence HIV-induced BBB inflammation. These findings are in agreement with our previous studies showing significantly increased levels of chemokines, matrix metalloproteinases, and acute-phase response factors such as complement factors-B and C3 in HBMEC exposed to Tat.B, compared to cells exposed to Tat.AG [17]. Studies in human monocytes also showed increased IL-6 expression in monocytes exposed to Tat.B, compared to monocytes exposed to Tat from HIV-1 subtype-C [43], confirming the effects of viral genotype on Tat-induced IL-6 expression in other cell types.
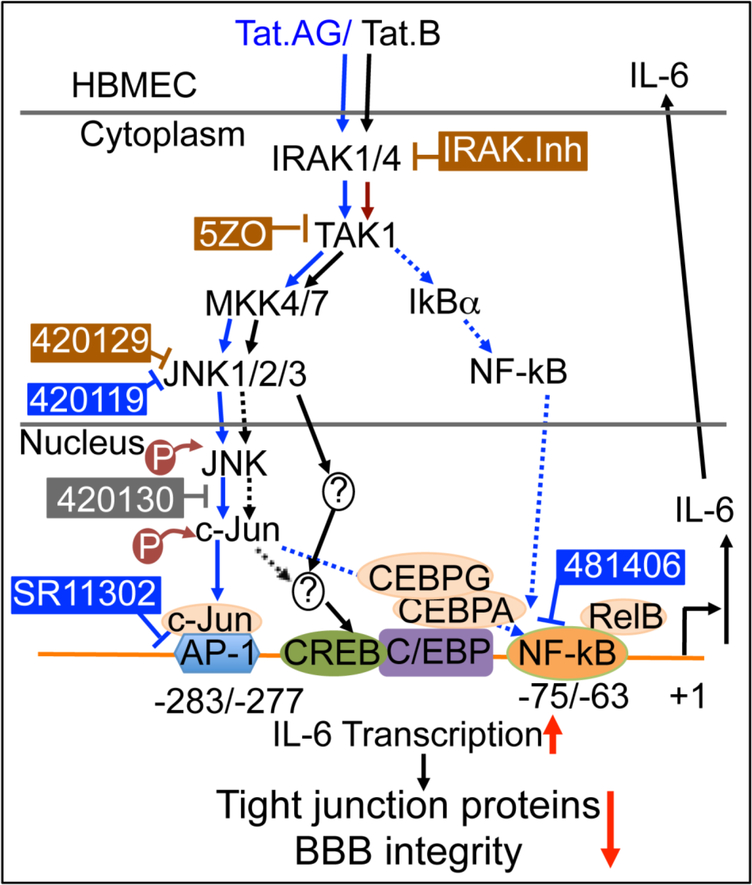
The pharmacological inhibitors of IRAK1/4 and TAK1 (5ZO) blocked both Tat.B- and Tat.AG- induced IL-6 expression and secretion. 5ZO is an ATP-dependent and competitive inhibitor of TAK1 and the MAP kinases MEKK7, MKK7, MEK1, and ERK2 (Table 1); suggesting that both Tat proteins use similar IRAK1/4- and TAK1-dependent pathways upstream to induce IL-6 expression in HBMEC (Fig. 6). It has also been shown that microRNA-146 target IRAK1 to block IL-6 transcription [30]. The ATP-competitive JNK inhibitor 420129 blocked both Tat.B- and Tat.AG-induced IL-6 expression, while the JNK inhibitor 420119 only blocked Tat.AG and had no effect on Tat.B-induced IL-6 expression. This suggests that both Tat.B- and Tat.AG-induced IL-6 expression occurs via ATP-dependent activation of JNK. This was confirmed by our Western blot data showing that both Tat.B and Tat.AG induced phosphorylation of c-Jun at Ser36, and phosphorylation of SAPK/JNK at Thr183 and Tyr185. The fact that the JNK inhibitor 420119 only blocked Tat.AG and had no effect on Tat.B-induced IL-6 expression suggests that Tat.AG-mediated JNK activation can be ATP-dependent or independent of ATP, and that Tat.B- mediated JNK activation may be strictly ATP-dependent. c-Jun is activated through phosphorylation by the JNK pathways [44,45], and c-Jun dimerizes with c-Fos to form AP-1 [44]. The inhibitor of c-Jun/JNK complex formation had no major effect on Tat.B- or Tat.AG-induced IL-6 expression, while the inhibitor of AP-1 transcriptional activity, as well as the inhibitor of NFκB transcriptional activation, blocked Tat.AG-mediated IL-6 expression and had no effect on Tat.B-induced IL-6 expression. This would suggest that following activation of JNK and c-Jun upstream, and translocation of the c- Jun/JNK complex into the nucleus, Tat.B-induced IL-6 expression occurs principally via the JNK pathways, whereas Tat.AG-mediated IL-6 expression occurs predominantly via JNK/c-Jun crosstalk with NFκB and the alternative NFκB pathway (Fig. 6).
Fig. 6: Model illustrating the signaling pathways in Tat.B- and Tat.AG-induced endothelial inflammation and BBB injury.
Arrows with solid blue lines indicates activation and direct signaling of Tat.AG. Dotted blue lines show other pathways involved in Tat.AG-mediated effects, and cross-talk of JNK and NFκB pathways. Arrows with solid black lines indicates activation and direct signaling of Tat.B. Dotted black lines indicated potential / alternative pathways. The ⊥ symbol indicates pharmacological inhibitors; brown symbol and brown rectangular boxes indicates compounds that inhibited both Tat.B- and Tat.AG-mediated effects. Blue symbol and blue rectangular boxes indicates compounds that inhibited only Tat.AG- mediated effects. The grey symbol and grey rectangular box indicates compound that had no effect on Tat.B- or Tat.AG-mediated effects. Red arrows ↓ indicates a decrease; ↑ indicates an increase.
The IL-6 gene has functional cis-regulatory elements that contain binding sites for NFκB, CEBP, and AP-1 [29,46], and it has been shown that other viral proteins such as the human T- lymphotropic virus-1 Tax, the HIV-1 Tat, and the human hepatitis-B X proteins increased IL-6 transcription by enhancing NFκB and CEBP-B DNA-binding activity [47,48,30]. It has also been shown that HIV-1 gp120-, Vpr-, and Tat.B-induced IL-6 expression in astrocytes and astrocytic cell lines are regulated by NFκB, AP-1, and CEBP-delta [49–51]. Our study confirmed these previous findings and showed that different mechanisms regulate Tat.B- and Tat.AG-induced IL- 6 expression on the brain endothelium. This suggests that viral genotype may influence Tat- induced endothelial inflammation, BBB injury and HAND.
The main feature of the brain endothelium is the presence of tight junctions between endothelial cells that restrict BBB permeability [52,3]. In HIV and SIV encephalitis there is evidence that BBB dysfunction is associated with disruption of TJ and increased trafficking of HIV-infected mononuclear phagocytes into the CNS [53,54]. We now provide new insights into the role of viral genotype into this seemingly complex process. We show that both Tat.B and Tat.AG decreased the TEER and endothelial tight junction proteins claudin-5 and ZO-1, but larger decrease in TEER and tight junction proteins were seen with Tat.B, compared to Tat.AG. Additional functional studies further showed significantly more increase in adhesion and migration of monocytes through in vitro BBB models when HBMEC were exposed to Tat.B, compared to Tat.AG. BBB dysfunction is common among HIV-infected patients and plays an important role in the pathogenesis of HAND; it enables viral particles, viral factors, and infected mononuclear phagocytes to infiltrate the CNS where they spread infection to resident macrophages and glial cells, cause neuronal injury and HAND [52,3]. Our current study shows that viral genotype influence this process and the mechanism and magnitude of Tat-induced BBB inflammation and injury can differ based on viral subtype.
Acknowledgements:
This work was supported by grant from the National Institute of Health, National Institute of Mental Health R01 MH094160.
Appendix. Figure 7
Fig. 7.
Exposure of primary HBMEC to Tat proteins (1, 10, 100, or 1000 ng/ml) increased IL-6 levels. At similar doses, higher IL-6 expression was observed in Tat.B-treated cells, compared to cells exposed to Tat.AG. *P < 0.05; ***P < 0.001, compared to cells treated with heat-inactivated proteins and untreated controls. For all experiments, each experimental condition was performed in triplicate.
Footnotes
Conflicting interests: The authors declare that they have no conflict of interest.
References:
- 1.Antinori A, Arendt G, Becker JT, Brew BJ, Byrd DA, Cherner M, Clifford DB, Cinque P, Epstein LG, Goodkin K, Gisslen M, Grant I, Heaton RK, Joseph J, Marder K, Marra CM, McArthur JC, Nunn M, Price RW, Pulliam L, Robertson KR, Sacktor N, Valcour V, Wojna VE (2007) Updated research nosology for HIV-associated neurocognitive disorders. Neurology 69 (18):1789–1799. doi: 10.1212/01.WNL.0000287431.88658.8b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banks WA, Ercal N, Price TO (2006) The blood-brain barrier in neuroAIDS. Curr HIV Res 4 (3):259–266 [DOI] [PubMed] [Google Scholar]
- 3.Persidsky Y, Ramirez SH, Haorah J, Kanmogne GD (2006) Blood-brain barrier: structural components and function under physiologic and pathologic conditions. J Neuroimmune Pharmacol 1 (3):223–236. doi: 10.1007/s11481-006-9025-3 [DOI] [PubMed] [Google Scholar]
- 4.Bannwarth S, Gatignol A (2005) HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Curr HIV Res 3 (1):61–71 [DOI] [PubMed] [Google Scholar]
- 5.Gupta S, Mitra D (2007) Human immunodeficiency virus-1 Tat protein: immunological facets of a transcriptional activator. Indian J Biochem Biophys 44 (5):269–275 [PubMed] [Google Scholar]
- 6.Westendorp MO, Frank R, Ochsenbauer C, Stricker K, Dhein J, Walczak H, Debatin KM, Krammer PH (1995) Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature 375 (6531):497–500. doi: 10.1038/375497a0 [DOI] [PubMed] [Google Scholar]
- 7.Banks WA, Robinson SM, Nath A (2005) Permeability of the blood-brain barrier to HIV-1 Tat. Exp Neurol 193 (1):218–227. doi:S0014–4886(04)00461–3 [pii] 10.1016/j.expneurol.2004.11.019 [DOI] [PubMed] [Google Scholar]
- 8.Hudson L, Liu J, Nath A, Jones M, Raghavan R, Narayan O, Male D, Everall I (2000) Detection of the human immunodeficiency virus regulatory protein tat in CNS tissues. J Neurovirol 6 (2):145–155 [DOI] [PubMed] [Google Scholar]
- 9.UNAIDS (2015) Fact Sheet 2015: Global Statistics. http://wwwunaidsorg/sites/default/files/media_asset/20150901_FactSheet_2015_en.pdf
- 10.WHO/UNAIDS (2015) AIDS by the numbers 2015 http://wwwunaidsorg/en/resources/documents/2015/AIDS_by_the_numbers_2015
- 11.Anastassopoulou CG, Kostrikis LG (2006) Global genetic variation of HIV-1 infection. Curr HIV Res 4 (3):365–373 [DOI] [PubMed] [Google Scholar]
- 12.Robertson DLAJ, Bradac JA, Carr JK, Foley B, et al. (2000) HIV-1 nomenclature proposal. Science 288:55–56 [DOI] [PubMed] [Google Scholar]
- 13.LANL (2016) HIV Circulating Recombinant Forms (CRFs). HIV sequence database http://wwwhivlanlgov/content/sequence/HIV/CRFs/CRFshtml
- 14.Montavon C, Toure-Kane C, Liegeois F, Mpoudi E, Bourgeois A, Vergne L, Perret JL, Boumah A, Saman E, Mboup S, Delaporte E, Peeters M (2000) Most env and gag subtype A HIV-1 viruses circulating in West and West Central Africa are similar to the prototype AG recombinant virus IBNG. J Acquir Immune Defic Syndr 23 (5):363–374 [DOI] [PubMed] [Google Scholar]
- 15.Brennan CA, Bodelle P, Coffey R, Devare SG, Golden A, Hackett J Jr., Harris B, Holzmayer V, Luk KC, Schochetman G, Swanson P, Yamaguchi J, Vallari A, Ndembi N, Ngansop C, Makamche F, Mbanya D, Gurtler LG, Zekeng L, Kaptue L (2008) The prevalence of diverse HIV-1 strains was stable in Cameroonian blood donors from 1996 to 2004. J Acquir Immune Defic Syndr 49 (4):432–439. doi: 10.1097/QAI.0b013e31818a6561 [DOI] [PubMed] [Google Scholar]
- 16.PRB (2010) 2010 World Population Data Sheet. http://www.prb.org. The Population Reference Bureau, Washington DC [Google Scholar]
- 17.Woollard SM, Bhargavan B, Yu F, Kanmogne GD (2014) Differential effects of Tat proteins derived from HIV-1 subtypes B and recombinant CRF02_AG on human brain microvascular endothelial cells: implications for blood-brain barrier dysfunction. J Cereb Blood Flow Metab 34 (6):1047–1059. doi: 10.1038/jcbfm.2014.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaudhuri A, Yang B, Gendelman HE, Persidsky Y, Kanmogne GD (2008) STAT1 signaling modulates HIV-1-induced inflammatory responses and leukocyte transmigration across the blood-brain barrier. Blood 111 (4):2062–2072. doi:blood-2007-05-091207 [pii] 10.1182/blood-2007-05-091207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang B, Akhter S, Chaudhuri A, Kanmogne GD (2009) HIV-1 gp120 induces cytokine expression, leukocyte adhesion, and transmigration across the blood-brain barrier: modulatory effects of STAT1 signaling. Microvascular research 77 (2):212–219. doi: 10.1016/j.mvr.2008.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanmogne GD, Schall K, Leibhart J, Knipe B, Gendelman HE, Persidsky Y (2007) HIV-1 gp120 compromises blood-brain barrier integrity and enhances monocyte migration across blood-brain barrier: implication for viral neuropathogenesis. J Cereb Blood Flow Metab 27 (1):123–134. doi:9600330 [pii] 10.1038/sj.jcbfm.9600330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maggirwar SB, Tong N, Ramirez S, Gelbard HA, Dewhurst S (1999) HIV-1 Tat- mediated activation of glycogen synthase kinase-3beta contributes to Tat-mediated neurotoxicity. Journal of neurochemistry 73 (2):578–586 [DOI] [PubMed] [Google Scholar]
- 22.Silvers JM, Aksenova MV, Aksenov MY, Mactutus CF, Booze RM (2007) Neurotoxicity of HIV-1 Tat protein: involvement of D1 dopamine receptor. Neurotoxicology 28 (6):1184–1190. doi: 10.1016/j.neuro.2007.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eugenin EA, King JE, Nath A, Calderon TM, Zukin RS, Bennett MV, Berman JW (2007) HIV-tat induces formation of an LRP-PSD-95- NMDAR-nNOS complex that promotes apoptosis in neurons and astrocytes. Proc Natl Acad Sci U S A 104 (9):3438–3443. doi:0611699104 [pii] 10.1073/pnas.0611699104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou BY, Liu Y, Kim B, Xiao Y, He JJ (2004) Astrocyte activation and dysfunction and neuron death by HIV-1 Tat expression in astrocytes. Molecular and cellular neurosciences 27 (3):296–305. doi: 10.1016/j.mcn.2004.07.003 [DOI] [PubMed] [Google Scholar]
- 25.Toborek M, Lee YW, Pu H, Malecki A, Flora G, Garrido R, Hennig B, Bauer HC, Nath A (2003) HIV-Tat protein induces oxidative and inflammatory pathways in brain endothelium. J Neurochem 84. doi: 10.1046/j.1471-4159.2003.01543.x [DOI] [PubMed] [Google Scholar]
- 26.Ranga U, Shankarappa R, Siddappa NB, Ramakrishna L, Nagendran R, Mahalingam M, Mahadevan A, Jayasuryan N, Satishchandra P, Shankar SK, Prasad VR (2004) Tat protein of human immunodeficiency virus type 1 subtype C strains is a defective chemokine. J Virol 78 (5):2586–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell GR, Watkins JD, Singh KK, Loret EP, Spector SA (2007) Human immunodeficiency virus type 1 subtype C Tat fails to induce intracellular calcium flux and induces reduced tumor necrosis factor production from monocytes. J Virol 81 (11):5919–5928. doi:JVI.01938–06 [pii] 10.1128/JVI.01938-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carr JK, Salminen MO, Albert J, Sanders-Buell E, Gotte D, Birx DL, McCutchan FE (1998) Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology 247 (1):22–31. doi: 10.1006/viro.1998.9211 [DOI] [PubMed] [Google Scholar]
- 29.Tanaka T, Narazaki M, Kishimoto T (2014) IL-6 in Inflammation, Immunity, and Disease. Cold Spring Harbor Perspectives in Biology 6 (10). doi: 10.1101/cshperspect.a016295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka T, Narazaki M, Ogata A, Kishimoto T (2014) A new era for the treatment of inflammatory autoimmune diseases by interleukin-6 blockade strategy. Seminars in Immunology 26 (1):88–96. doi: 10.1016/j.smim.2014.01.009 [DOI] [PubMed] [Google Scholar]
- 31.Rossi J-F, Lu Z-Y, Jourdan M, Klein B (2015) Interleukin-6 as a Therapeutic Target. Clinical Cancer Research 21 (6):1248–1257. doi: 10.1158/1078-0432.ccr-14-2291 [DOI] [PubMed] [Google Scholar]
- 32.Bastard JP, Soulie C, Fellahi S, Haim-Boukobza S, Simon A, Katlama C, Calvez V, Marcelin AG, Capeau J (2012) Circulating interleukin-6 levels correlate with residual HIV viraemia and markers of immune dysfunction in treatment-controlled HIV- infected patients. Antivir Ther 17 (5):915–919. doi: 10.3851/IMP2093 [DOI] [PubMed] [Google Scholar]
- 33.Hamlyn E, Fidler S, Stohr W, Cooper DA, Tambussi G, Schechter M, Miro JM, McClure M, Weber J, Babiker A, Porter K (2014) Interleukin-6 and D-dimer levels at seroconversion as predictors of HIV-1 disease progression. AIDS 28 (6):869–874. doi: 10.1097/QAD.0000000000000155 [DOI] [PubMed] [Google Scholar]
- 34.Olwenyi OA, Naluyima P, Cham F, Quinn TC, Serwadda D, Sewankambo NK, Gray RH, Sandberg JK, Michael NL, Wabwire-Mangen F, Robb ML, Eller MA (2016) Brief Report: Differential Associations of Interleukin 6 and Intestinal Fatty Acid-Binding Protein With Progressive Untreated HIV-1 Infection in Rakai, Uganda. J Acquir Immune Defic Syndr 72 (1):15–20. doi: 10.1097/QAI.0000000000000915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalayjian RC, Machekano RN, Rizk N, Robbins GK, Gandhi RT, Rodriguez BA, Pollard RB, Lederman MM, Landay A (2010) Pretreatment Levels of Soluble Cellular Receptors and Interleukin-6 Are Associated with HIV Disease Progression in Subjects Treated with Highly Active Antiretroviral Therapy. Journal of Infectious Diseases 201 (12):1796–1805. doi: 10.1086/652750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boulware DR, Hullsiek KH, Puronen CE, Rupert A, Baker JV, French MA, Bohjanen PR, Novak RM, Neaton JD, Sereti I, Group IS (2011) Higher levels of CRP, D- dimer, IL-6, and hyaluronic acid before initiation of antiretroviral therapy (ART) are associated with increased risk of AIDS or death. The Journal of infectious diseases 203 (11):1637–1646. doi: 10.1093/infdis/jir134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.French MA, Cozzi-Lepri A, Arduino RC, Johnson M, Achhra AC, Landay A (2015) Plasma levels of cytokines and chemokines and the risk of mortality in HIV-infected individuals: a case-control analysis nested in a large clinical trial. AIDS 29 (7):847–851. doi: 10.1097/QAD.0000000000000618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borges ÁH, O’Connor JL, Phillips AN, Neaton JD, Grund B, Neuhaus J, Vjecha MJ, Calmy A, Koelsch KK, Lundgren JD (2016) Interleukin 6 Is a Stronger Predictor of Clinical Events Than High-Sensitivity C-Reactive Protein or D-Dimer During HIV Infection. Journal of Infectious Diseases 214 (3):408–416. doi: 10.1093/infdis/jiw173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, Ledergerber B, Lundgren J, Neuhaus J, Nixon D, Paton NI, Neaton JD, Group ISS (2008) Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med 5 (10):e203. doi: 10.1371/journal.pmed.0050203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nordell AD, McKenna M, Borges ÁH, Duprez D, Neuhaus J, Neaton JD, the Insight Smart ESG, Committee SS (2014) Severity of Cardiovascular Disease Outcomes Among Patients With HIV Is Related to Markers of Inflammation and Coagulation. Journal of the American Heart Association: Cardiovascular and Cerebrovascular Disease 3 (3):e000844. doi: 10.1161/jaha.114.000844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu DC, Ma YF, Hur S, Li D, Rupert A, Scherzer R, Kalapus SC, Deeks S, Sereti I, Hsue PY (2016) Plasma IL-6 levels are independently associated with atherosclerosis and mortality in HIV-infected individuals on suppressive antiretroviral therapy. AIDS 30 (13):2065–2074. doi: 10.1097/QAD.0000000000001149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald B, Moyo S, Gabaitiri L, Gaseitsiwe S, Bussmann H, Koethe JR, Musonda R, Makhema J, Novitsky V, Marlink RG, Wester CW, Essex M (2013) Persistently elevated serum interleukin-6 predicts mortality among adults receiving combination antiretroviral therapy in Botswana: results from a clinical trial. AIDS research and human retroviruses 29 (7):993–999. doi: 10.1089/AID.2012.0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gandhi N, Saiyed Z, Thangavel S, Rodriguez J, Rao KV, Nair MP (2009) Differential effects of HIV type 1 clade B and clade C Tat protein on expression of proinflammatory and antiinflammatory cytokines by primary monocytes. AIDS research and human retroviruses 25 (7):691–699. doi: 10.1089/aid.2008.0299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kallunki T, Deng T, Hibi M, Karin M (1996) c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell 87 (5):929–939 [DOI] [PubMed] [Google Scholar]
- 45.Gupta S, Barrett T, Whitmarsh AJ, Cavanagh J, Sluss HK, Derijard B, Davis RJ (1996) Selective interaction of JNK protein kinase isoforms with transcription factors. The EMBO journal 15 (11):2760–2770 [PMC free article] [PubMed] [Google Scholar]
- 46.Akira S, Kishimoto T (1992) IL-6 and NF-IL6 in acute-phase response and viral infection. Immunol Rev 127:25–50 [DOI] [PubMed] [Google Scholar]
- 47.Ambrosino C, Ruocco MR, Chen X, Mallardo M, Baudi F, Trematerra S, Quinto I, Venuta S, Scala G (1997) HIV-1 Tat induces the expression of the interleukin-6 (IL6) gene by binding to the IL6 leader RNA and by interacting with CAAT enhancer-binding protein beta (NF-IL6) transcription factors. The Journal of biological chemistry 272 (23):14883–14892 [DOI] [PubMed] [Google Scholar]
- 48.Ohno H, Kaneko S, Lin Y, Kobayashi K, Murakami S (1999) Human hepatitis B virus X protein augments the DNA binding of nuclear factor for IL-6 through its basic- leucine zipper domain. J Med Virol 58 (1):11–18 [DOI] [PubMed] [Google Scholar]
- 49.Nookala AR, Kumar A (2014) Molecular mechanisms involved in HIV-1 Tat- mediated induction of IL-6 and IL-8 in astrocytes. Journal of Neuroinflammation 11 (1):1–18. doi: 10.1186/s12974-014-0214-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah A, Verma AS, Patel KH, Noel R, Rivera-Amill V, Silverstein PS, Chaudhary S, Bhat HK, Stamatatos L, Singh DP, Buch S, Kumar A (2011) HIV-1 gp120 induces expression of IL-6 through a nuclear factor-kappa B-dependent mechanism: suppression by gp120 specific small interfering RNA. PloS one 6. doi: 10.1371/journal.pone.0021261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gangwani MR, Kumar A (2015) Multiple Protein Kinases via Activation of Transcription Factors NF-κB, AP-1 and C/EBP-δ Regulate the IL-6/IL-8 Production by HIV-1 Vpr in Astrocytes. PloS one 10 (8):e0135633. doi: 10.1371/journal.pone.0135633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toborek M, Lee YW, Flora G, Pu H, Andras IE, Wylegala E, Hennig B, Nath A (2005) Mechanisms of the blood-brain barrier disruption in HIV-1 infection. Cell Mol Neurobiol 25 (1):181–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dallasta LM, Pisarov LA, Esplen JE, Werley JV, Moses AV, Nelson JA, Achim CL (1999) Blood-brain barrier tight junction disruption in human immunodeficiency virus-1 encephalitis. Am J Pathol 155 (6):1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boven LA, Middel J, Verhoef J, De Groot CJ, Nottet HS (2000) Monocyte infiltration is highly associated with loss of the tight junction protein zonula occludens in HIV-1- associated dementia. Neuropathology and applied neurobiology 26 (4):356–360 [DOI] [PubMed] [Google Scholar]