Abstract
Cluster of differentiation 151 (CD151) is a potent therapeutic target for regulating tumor metastasis. In the present study, the role of CD151 in clear cell sarcoma of soft tissue was examined using a xenoplanted tumor model, which had high rates of metastasis. A clear cell sarcoma cell line, HS-MM, which was transplanted to the aponeuroses of the thighs, the most affected sites of human clear cell sarcoma, exhibited robust lymphatic invasion and nodal metastasis in SCID-beige mice. Serial in vivo passaging of peritoneally disseminated tumor cells accelerated the metastatic activity, which was accompanied by increased CD151 expression, and were designated as HS-MMhigh. Notably, inoculation of anti-CD151 antibody significantly suppressed the lymphatic invasion, peritoneal dissemination and distant metastasis of the present clear cell sarcoma model without affecting local tumor growth at the transplantation site. Small interfering RNA (siRNA)-mediated downregulation of CD151 did not alter cell proliferation, but significantly inhibited Matrigel invasion activity of HS-MMhigh cells. Downregulation of CD151 impaired matrix metalloproteinase-9 activity and phosphorylation of SMAD3 protein in HS-MMhigh cells. The present results suggest that CD151 may contribute to invasion and metastasis of clear cell sarcoma of soft tissue. Therefore, CD151 may serve as a potent target to regulate metastasis of clear cell sarcoma.
Keywords: clear cell sarcoma, cluster of differentiation 151, matrix metalloproteinase-9, SMAD3, xenoplant assay
Introduction
Cluster of differentiation 151 (CD151) is a member of the tetraspanins superfamily, and it is composed of four-transmembrane-spanning proteins containing two extracellular regions of unequal size and three short intracellular regions (1). In various malignant tumors, CD151 is overexpressed and characterized as a ‘facilitator’ for tumor metastasis (2). Currently, CD151 appears to assist metastasis of renal cell carcinoma (3), osteosarcoma (4), hepatocellular carcinoma (5), and prostate cancer (6). CD151 is a positive regulator of TGF-β-induced signaling in cancer metastasis (3). Therefore, CD151 is thought to be a suitable target molecule for various malignant tumors (7).
Clear cell sarcoma (CCS) of soft tissue primarily affects young adults (8,9). Radical surgical resection is the first choice for treatment of CCS. However, in contrast to other malignant soft tissue tumors, CCS often possesses a robust lymphatic metastatic activity (8–11). Therefore, the rate of local recurrence is as high as 84%, while the rate of late metastases is as high as 63%, which is associated with the 5 to 20-year survival rate of 67–10% (12). Unfortunately, CCS is relatively insensitive to conventional soft tissue sarcoma chemotherapy regimens. These unfavourable clinicopathological features encouraged us for new targeted therapies development of CCS.
Unfortunately, the rare instances of CCS cases not only pose an impediment to know the molecular mechanisms that are responsible for metastatic activity of CCS, but also create obstacles to develop novel therapies. To overcome this limitation, we have recently generated an orthotropic metastatic model of CCS through xenoplanting human clear sarcoma cells, HS-MM, to SCID-beige mice (13). This tumor model very well reflects clinicopathobiological features of CCS, i.e., high lymphatic metastatic activity and distant metastasis.
Furthermore, by serial in vivo passaging of HS-MM from peritoneally disseminated tumor, we established a HS-MM cell clone, designated as HS-MMhigh, which harbored the prominent lymphatic invasion and metastatic activity. In this study, we found that CD151, which is recently designed to be a target molecule to regulate cancer progression (7), is related to metastatic activity of CCS in the animal models.
Materials and methods
Antibodies
For intraperitoneal injection, anti-CD151 antibody was obtained from ascites of nude mice (BLAB/c nu/nu, female, 8 weeks old) after intraperitoneal injection of hybridoma cells. Hybridoma cells, which produce anti-CD151 IgG1κ antibody [clone 50-6 (14), CRL-2696] were purchased from American Type Culture Collection (Manassas, VA, USA).
For immunoblotting, a murine antibody specific to CD151 (clone 11G5a), and rabbit GAPDH antibody were purchased from Abcam Inc. (Cambridge, MA, USA) and Sigma-Aldrich; Merck KGaA (Darmstadt, Germany), respectively. Rabbit monoclonal SMAD3 antibodies (clone C67H9) and phospho-SMAD3 (Ser423/425) (clone C25A9) were purchased from Cell Signaling Technology (CST, Inc., Danvers, MA, USA).
Alexa Fluor 555-conjugated anti-rabbit and Alexa Fluor 488-conjugated anti-mouse antibodies were purchased from Invitrogen; Thermo Fisher Scientific, Inc. (Waltham, MA, USA). Control murine antibody was isolated by Protein A-affinity chromatography from normal mouse sera (Caltag Lab, Burlingame, CA, USA).
Xenografts
The experimental protocol was approved by the Animal Care Committee of Gifu Graduate School of Gifu, Japan (approval no. H30-32). Detailed procedure of orthotropic metastatic model of CCS has been previously described (13). Briefly, SCID-Beige (CB17.Cg-PrkdcscidLystbg-J/CrlCrlj) mice were purchased from Charles River Laboratories, Japan (Sizuoka, Japan). A CCS of soft tissue cell line, HS-MM, was previously established, characterized, and maintained as a stock in our laboratory (15,16). HS-MM cells (1.2×106) were subcutaneously injected into the soft tissue of the thigh of 12-week-old SCID-Beige mice. In another independent experiment, 1.0×106 HS-MM cells were similarly injected into 10-week-old SCID-Beige mice. Tumor volume was measured by calipers using the following equation: tumor volumes (mm3) = 4/3π × [a/2] × [b/2]2, where ‘a’ and ‘b’ correspond to the longest and shortest diameter measured twice a week. After five weeks later, when tumor volumes reached near 1.0 cm3, mice were randomly divided into two groups (n=4 and n=3 for each group), and these mice were intraperitoneally inoculated with or without 3 mg of anti-CD151 antibody (clone 50-6). Two weeks later, mice were sacrificed to examine the extent of metastasis. The animals were euthanized after anesthesia, and every effort was made to minimize suffering. The xenografts and metastatic tissues were excised, formalin fixed, paraffin embedded, and sectioned for histopathological analysis.
Immunofluorescence staining
Immunofluorescence staining was performed as previously described (17). Briefly, cells were incubated with a murine anti-CD151 antibody (clone 11G5a) for 1 h at 4°C, washed with PBS twice, and then incubated with Alexa Fluor 488-conjugated anti-mouse antibody (1:200 dilution) for 30 min at 4°C. After re-washing with PBS, the cells were analyzed with a Guava EasyCyte cell analyzer (Guava Technologies, Inc., Hayward, CA, USA). Guava easyCyte™ flow cytometry system software was used to obtain the one parameter log histogram.
Immunoblotting
Immunoblotting was performed according to a previously described method (18), with the modification proposed by Towbin et al (19). Samples were analyzed by electrophoresis on SDS-containing polyacrylamide gels under reducing conditions. The separated proteins were then transferred to polyvinylidene difluoride membranes (EMD Millipore Co., Billerica, MA, USA) and probed with various antibodies. Immunoreactivity was assessed using the Western Blotting Detection Kit (Promega, Madison, WI, USA). The immunoblot band was quantified by densitometry using LI-COR C-DiGit Blot Scanner imaging software version 3.1 (LI-COR Biosciences, Lincoln, NE, USA) and was normalized to the GAPDH band as previously reported (20).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
cDNA synthesis from total RNA, and subsequent PCR experiments were performed using the Reverse Transcription Polymerase Chain Reaction Kit (Takara Bio, Inc., Otsu, Japan) according to manufacturer instructions and as previously described (18). Briefly, the first strand cDNA was synthesized using a random 6-mer primer. The specific reaction conditions were 42°C for 30 min, followed by 95°C for 5 min, and then finally 5°C for 5 min.
qPCR reactions were performed using the FastStart Essential DNA Green Master Mix according to the manufacturer's instructions (Roche Diagnostics, GmbH, Mannheim, Germany) using a LightCycler (Roche Diagnostics) as previously described (21). cDNA (2 µl each) was diluted with PCR mix containing a 0.2 pmol of primer to a final volume of 20 µl. The following qPCR primers were used for the real-time RT-PCR: CD151 forward 5′-CATCGCTGGTATCCTCG-3′ and reverse 5′-CTCGCTGCCCACAAAG-3′, and GAPDH forward 5′-GAAATCCCATCACCATCTTCCAGG-3′ and reverse 5′-GAGCCCCAGCCTTCTCCATG-3′. The run protocols used included a denaturation program (95°C for 10 min), an amplification and quantification program repeated 45 times (95°C for 10 sec, 60°C for 10 sec, 72°C for 15 sec), and a melting curve program (60-95°C with a heating rate of 0.1°C per second and a continuous fluorescence measurement).
To ensure that SYBR Green was not incorporated, resulting in primer dimers or non-specific amplification in the qPCR runs, the CD151 and GAPDH PCR products were analyzed by polyacrylamide gel electrophoresis in preliminary experiments. Single intense bands at their respective expected sizes were observed. The samples were cultured in triplicates, and the expression of each target gene was analyzed with a LightCycler system using the 2−ΔΔCT method described by Livak and Schmittgen (22). For each triplicate set, the ΔCT values were normalized to GAPDH expression in both control and target cells. The values of the target group were then calculated as the fold change relative to the mean values of the control group, i.e., original HS-MM cells or HS-MMhigh cells treated by a green fluorescent protein (GFP)-siRNA duplex (control; set to 1.0). Following this, the standard deviations were calculated for the triplicate sets, and the fold changes for the target genes were plotted/calculated.
siRNA-mediated gene silencing
The detailed procedure followed for gene silencing has been described previously (23). In this study, we used the FlexiTube GeneSolution GS977, which is composed of 4 siRNAs: SI02777257, SI02777250, SI04434570, and SI03070018 (Qiagen, Inc., Valencia, CA, USA), to silence the CD151 gene. A green fluorescent protein (GFP)-siRNA duplex with the target sequence 5′-CGGCAAGCUGACCCUGAAGUUCAU-3′ was used as a non-silencing control. The siRNAs were transfected into cells with Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.). At 72 h post transfection, the cells were used for subsequent experiments.
Cell proliferation and Matrigel invasion assays
Cell proliferation was evaluated by counting the number of viable cells as previously described (24). Briefly, 1×104 cells were cultured in triplicates in standard 35-mm tissue culture dishes (BD Falcon; BD Biosciences, San Jose, CA, USA). After 24, 48, and 72 h, the live cells were counted. The assay was performed in triplicates and repeated twice.
The invasiveness of the cultured cells was determined using 24-well Corning BioCoat Matrigel Invasion Chamber Plates (Discovery Labware Inc., Bedford, MA, USA) according to the manufacturer's protocol and as described previously (23). Briefly, 5×104 cells were placed in the upper compartment of the invasion chamber. After 48 h of incubation with DMEM containing 10% (lower chamber) or 2% (upper chamber) FBS, the non-invading cells were gently removed from the filter by swiping with a cotton-tipped swab. The cells on the lower surface of the filter were counted under a microscope.
Statistical analysis
We repeated experiments twice to evaluate CD151 expression in HS-MM and HS-MMhigh cells. Xenotransplant assay of anti-CD151 inoculation was repeated twice. To assess the involvement of CD151 in invasion activity, we performed two independent experiments. The zymography assay was repeated three times and quantification of enzymatic activity assay was repeated twice. Statistical analysis was performed by Student's t-test for unpaired observations or ANOVA using Tukey's test. Findings with P<0.05 were considered significant.
Zymography analysis
Functional activity of matrix metalloproteinase-9 (MMP-9) was evaluated by gelatin zymography, as previously described by Heussen and Dowdle (25). Briefly, cells were incubated with serum-free DMEM for 24 h. Supernatant of culture medium was collected and mixed with non-reducing SDS sample buffer. After SDS-PAGE on 10% polyacrylamide gels containing 1 mg/ml gelatin, SDS was removed from the gels by incubating in 2.5% Triton X-100 for 1 h at room temperature. Subsequently, the gels were incubated in a buffer containing 50 mM Tris-HCl, 5 mM CaCl2, pH 7.6, for 24 h at 37°C, and stained with Coomassie blue R 250 (0.25%). Proteolytic activities of MMP-9 were detected as clear bands against the blue background of stained gelatin.
MMP-9 activity quantification assay
The MMP-9 total activity (already active plus latent MMP-9) was assayed using QuickZyme Human MMP-9 Activity Assay kit (QuickZyme BioSciences, Leiden, The Netherlands) according to the manufacturer's instructions. Briefly, culture supernatant MMP-9 was captured by specific antibody to MMP-9. After treatment with p-aminophenyl mercuric acetate, active MMP-9 altered pro-detection enzyme to active detection enzyme. Subsequently, this active detection enzyme recognized peptide substrate to form colored product, which was measured at 405 nm. Assay was performed in triplicate.
Cytostaining
Cells were fixed with 4% (m/v) paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 10% goat serum. Subsequently, cells were incubated with 1 µg/ml rabbit anti-phospho-SMAD3 (Ser423/425) and murine anti-CD151 (clone 11G5a) antibodies at room temperature for 1 h. After washing with PBS, cells were incubated with 1:200 diluted Alexa Fluor 555-conjugated anti-rabbit antibody and Alexa Fluor 488-conjugated anti-mouse antibody (Invitrogen; Thermo Fisher Scientific, Inc.). After staining, images were acquired with the help of a confocal laser scanning microscope (Leica TCS SP8; Leica Microsystems GmbH, Wetzlar, Germany).
Results
Increased CD151 expression in HS-MM clear cell sarcoma cells with high metastatic activity
As previously reported, metastasis of HS-MM cells to lung, liver, and lymph nodes was observed in xenoplanted SCID-Beige mice (13). Notably, peritoneal dissemination was also found in xenoplanted mice. Serial in vivo passaging of peritoneally disseminated tumor cells accelerated metastasis following implantation. After four passages, we obtained HS-MM cells with high-metastatic activity, and these cells were designated HS-MMhigh.
As recent advances indicate that CD151 enhances metastasis of various malignant tumors, we examined the expression status of original HS-MM and HS-MMhigh cells.
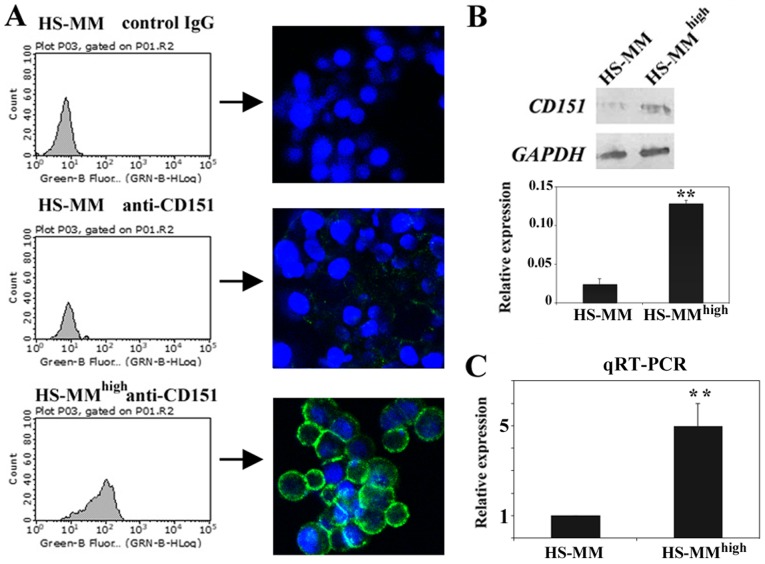
Immunofluorescent staining followed by cell analyzer analysis revealed that HS-MMhigh cells expressed significantly higher levels of cell surface CD151 compared with those of original HS-MM cells (Fig. 1A). Subsequent immunoblotting also demonstrated that CD151 protein levels were more abundant in the cell lysates of HS-MMhigh cells than they were from those derived from HS-MM cells (Fig. 1B). RT-qPCR showed that transcription of the CD151 gene is significantly upregulated in HS-MMhigh cells compared to HS-MM cells (Fig. 1C). We concluded that CD151 was quite abundantly expressed in HS-MMhigh cells relative to expression in the original HS-MM cells, and this was increase in expression was detectable at the cell membrane surface, in total proteins, and at the transcript level. This experiment was performed in duplicate, and both times we obtained similar results.
Figure 1.
Expression of CD151 in HS-MMhigh cells correlates with metastatic activity compared to HS-MM cells. (A) Original HS-MM cells exhibited weak immunoreactivity at the cell surface with specific antibody to CD151. By contrast, the surface of HS-MMhigh cells was strongly stained with anti-CD151 antibody. Staining was analyzed using a Guava EasyCyte cell analyzer and accompanying software to obtain a one-parameter log histogram. Immunocytostaining results are also shown. CD151 immunoreactivity visualized with green-fluorescent Invitrogen Alexa Fluor 488 and blue 4′,6-diamidino-2-phenylindole nuclear counterstain. (B) Representative immunoblotting using HS-MM and HS-MMhigh cells. Immunoblotting showed that HS-MMhigh cells abundantly expressed CD151 compared to original HS-MM cells. The equal intensity of the GAPDH band indicated equal protein loading. Semiquantitation was done as described in the Methods. The intensity ratio of each band to GAPDH is shown. Data are expressed as mean ± SD (n=3; Student's t-test, **P<0.01 vs. counterpart). (C) Reverse transcription-quantitative polymerase chain reaction also demonstrated that HS-MMhigh cells expressed higher levels of CD151 mRNA than the original HS-MM cells. The value for the HS-MMhigh groups (n=3) was calculated as the fold-change relative to the mean value for the HS-MM group (control set to 1.0). Standard deviations were then computed for the triplicate sets. Student's t-test was performed to confirm the significant differences, **P<0.01 vs. counterpart. CD151, cluster of differentiation 151; SD, standard deviation.
Anti-CD151 antibody treatment suppressed metastasis of HS-MM cells in xenoplanted SCID-Beige mice
As demonstrated in Fig. 2A, HS-MMhigh cells showed high levels of invasion in the absence of anti-CD151 antibody inoculation (Fig. 2A, Mock). High numbers of HS-MMhigh cells within lymphatic vessels are indicated by arrows, and a single inoculation of anti-CD151 antibody markedly impaired the infiltration of HS-MMhigh cells into lymphatic vessels (Fig. 2A, anti-CD151). We did observe a small number of HS-MMhigh cells in lymphatic vessels following treatment as indicated by arrows; however, anti-CD151 antibody did not alter the tumor volume of HS-MMhigh cells at the injection site as demonstrated in Fig. 2B (Student's t-test. P=0.37). In contrast, administration of anti-CD151 antibody significantly decreased total weights of collected, disseminated mesenteric tumors (Student's t-test, P=0.0036).
Figure 2.
Effect of anti-CD151 antibody on lymphatic invasion of HS-MMhigh cells in the xenoplantation mouse model. Anti-CD151 antibody reduced lymphatic invasion at the xenoplant site in severe combined immunodeficient-beige mice and the rate of intraperitoneal dissemination of HS-MMhigh cells without affecting local tumor growth. (A) Robust lymphatic infiltration of HS-MMhigh cells was evident beneath the local growth site (indicated by arrow). Inoculation of anti-CD151 antibody almost entirely abolished the lymphatic infiltration of HS-MMhigh cells. Note a few cells in the lymphatic vessels (indicated by arrow). Original magnification, ×100 (Mock and anti-CD151) and ×400 (anti-CD151, inserted figure). (B) No significant alteration of local growth of HS-MMhigh cells was found in the xenoplant assay. (C) Administration of anti-CD151 antibody significantly decreased the total weights of the disseminated mesenteric tumors. Data are expressed as mean ± SD (n=4 for anti-CD151 group, n=3 for Mock group; Student's t-test, **P<0.01 vs. counterpart). CD151, cluster of differentiation 151; SD, standard deviation.
Representative data using an individual experiment is shown. We also obtained similar results from another independent experiment, in which 1.0×106 HS-MMhigh cells were injected into 10-week-old SCID-Beige mice (n=3 for each group). Given this, it is clear that anti-CD151 antibody treatment suppressed metastasis of HS-MMhigh cells in xenoplanted SCID-Beige mice.
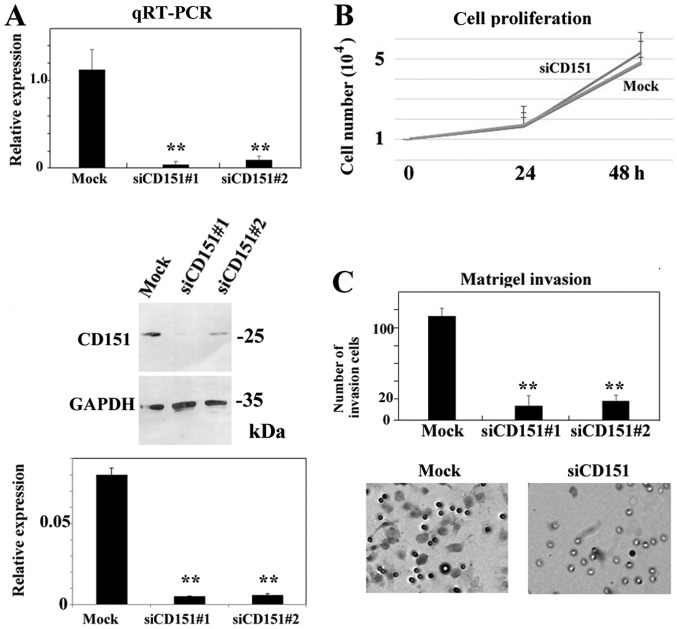
siRNA-mediated gene silencing of CD151 decreased the Matrigel-invasion activity of HS-MMhigh cells
Further, we investigated if CD151 plays a role in the invasion of HS-MMhigh cells by using a Matrigel invasion assay. Successful downregulation of CD151 mRNA levels (>90% as determined by RT-qPCR) was achieved using any of 4 siRNAs (SI02777257, SI02777250, SI04434570, and SI03070018; Qiagen). Both SI02777257 (indicated by siCD151#1) and SI02777250 (indicated by siCD151#2) siRNA treatments decreased CD151 protein expression. Representative data are shown in Fig. 3A. Downregulation of CD151 using SI02777257 did not affect the cell growth of HS-MMhigh cells in vitro (Student's t-test, P=0.54, Fig. 3B). We obtained similar results using SI02777250 siRNA. In contrast, downregulation of CD151 using SI02777257 (indicated by siCD151#1) or SI02777250 (indicated by siCD151#2) significantly reduced Matrigel invasion activity (ANOVA, Tukey's test. P<0.05, Fig. 3C).
Figure 3.
siRNA-mediated gene-silencing of CD151 does not affect cell proliferation, but significantly decreases Matrigel invasion activity of HS-MMhigh in vitro. (A) Both SI02777257 (indicated by siCD151#1) and SI02777250 (indicated by siCD151#2) siRNA treatments reduced CD151 expression at both mRNA and protein level. Reverse transcription-quantitative-polymerase chain reaction indicated that both SI02777257 and SI02777250 significantly reduced CD151 expression. Immunoblotting also demonstrated little CD151 protein in SI02777257 and SI02777250 siRNA treated HS-MMhigh cells. The intensity ratio of each band to GAPDH is shown. The data represent the mean ± SD of triplicate assays. ANOVA, Tukey's t-test. was performed to confirm the significant differences, **P<0.01 vs. Mock group. (B) Representative cell proliferation assay using siRNA targeting CD151. At 24 h, the cell number was 1.67±0.47 (Mock), 1.67±0.67 (SI02777257), 1.73±0.35 (SI02777250). Respective numbers at 48 h were 5.33±0.75 (Mock), 4.77±0.30 (SI02777257), and 4.83±1.07 (SI02777250). The data represent mean ± SD in triplicate assays. ANOVA, Tukey's t-test. P>0.5). Data using Mock and SI02777257 siRNAs are shown in this cell proliferation line graph. (C) SI02777257 and SI02777250 siRNAs specific to CD151 significantly reduced Matrigel invasion activity of HS-MMhigh cells. The number of invading cells was 113.7±6.42 (Mock), 13.7±8.33 (SI02777257, indicated by #1), and 19.3±11.0 (SI02777250, indicated by #2). Data from triplicate assay are expressed as the mean ± SD (n=3; ANOVA, Tukey's t-test. **P<0.01 vs. Mock group). A representative image of Matrigel invasion assay using Mock and SI02777257 siRNAs are shown. Cells that migrated to the undersurface of the membrane are shown. Original magnification, ×100. CD151, cluster of differentiation 151; SD, standard deviation; siRNA, small interfering RNA.
Our results indicate that silencing of CD151 decreased the Matrigel invasion activity of metastatic HS-MMhigh cells without affecting cell proliferation. Representative data are shown in Fig. 3. We also obtained similar results from another independent experiment.
siRNA-mediated gene silencing of CD151 decreased MMP-9 activity and phosphorylation status of in SMAD3 of HS-MMhigh cells
CD151 correlates with MMP-9 activity in melanoma, osteosarcoma, hepatocellular carcinoma, and a number of other malignant tumors (26–29). Additionally, recent evidence indicates that CD151 appears to increase phosphorylation status of SMAD3, an event that can lead to epithelial-mesenchymal transition in renal cell carcinoma (3). Given this, we investigated if CD151 participates in MMP-9 activation and active phosphorylation status of SMAD3.
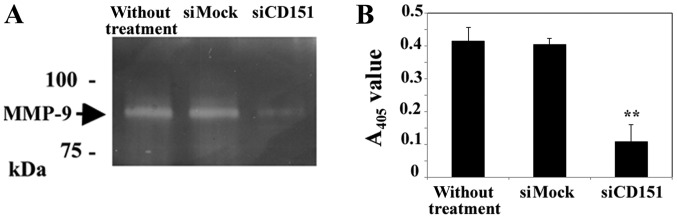
Results of a gelatin zymography and quantification assay indicated that siRNA-mediated downregulation of CD151 decreased MMP-9 activation in HS-MMhigh cells. The MMP-9 active band in CD151 downregulated HS-MMhigh cells is weak when compared to that of control cells (Fig. 4A). Furthermore, the present quantification assay demonstrated that the amount of already active plus latent MMP-9 was decreased by downregulation of CD151 (Fig. 4B).
Figure 4.
Association of CD151 with MMP-9 expression in representative gelatin zymograms of culture supernatants of siRNA-mediated CD151 silencing and control siRNA-treated HS-MMhigh cells. (A) MMP-9 active band was weakly evident in CD151-downregulated HS-MMhigh cells (indicated by siCD151) compared with control cells without siRNA-treatment or with control siRNA (indicated by siMock). (B) MMP-9 was reduced by the downregulation of CD151. The results shown were obtained using SI02777257 siRNA (Qiagen) to silence the CD151 gene. Similar results were obtained using SI02777250 siRNA. The data represent mean ± SD of triplicate assays. ANOVA and Tukey's test was performed to confirm the significant differences, **P<0.01 vs. si-Mock group. CD151, cluster of differentiation 151; MMP-9, matrix metalloproteinase-9; SD, standard deviation; siRNA, small interfering RNA; MMP, matrix metallopeptidase.
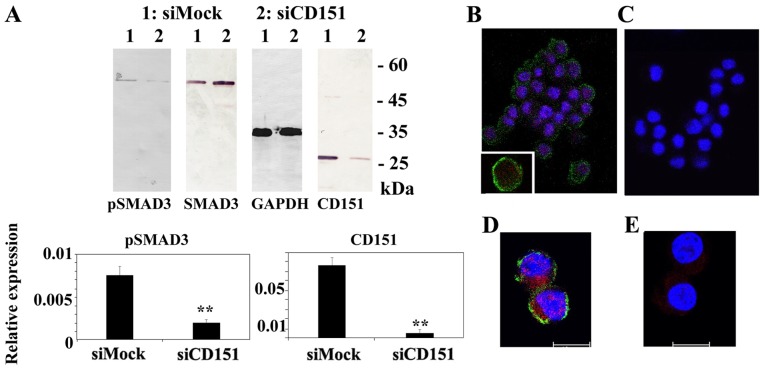
Additionally, downregulation of CD151 impaired the phosphorylation status and nuclear transport of SMAD3 (Fig. 5). Immunoblot analysis demonstrated little or no phospho-SMAD family member 3 (pSMAD3), band in CD151 knockout HS-MMhigh cells. Immunocytostaining indicated that HS-MMhigh cells exhibited CD151 immunoreactivity at the cell membrane surface (green staining) and pSMAD3 reactivity in both cytoplasm and nucleus (red staining). In contrast, downregulation of CD151 reduced the immunoreactivity of both CD151 and pSMAD3.
Figure 5.
(A-E) Silencing of CD151 protein decreased phosphorylation status and nuclear transport of SMAD3. (A) Immunoblotting demonstrated little or no pSMAD3 in CD151 knockout HS-MMhigh cells. The intensity of the GAPDH reference did not change. Semi quantitation was performed as described in the Materials and methods. The intensity ratio of each band to GAPDH is shown. The data represent mean ± SD in triplicate assays. Student's t-test was performed to confirm the significant differences, **P<0.01 vs. counterpart. The pSMAD3/SMAD3 ratio was 0.17±0.07 (Mock) and 0.016±0.007 (CD151 downregualtion group) in triplicate assay (Student's t-test, P<0.01). (B and D) HS-MMhigh cells exhibited CD151 immunoreactivity at the cell membrane surface (green staining; Alexa Fluor 488) and pSMAD3 was detected in both the cytoplasm and nucleus (red staining; Alexa Fluor 555). (C and E) Downregulation of CD151 diminished immunoreactivity of CD151 or pSMAD immunoreactivity. All nuclei were stained with 4′,6-diamidino-2-phenylindole (blue). Scale bar, 10 µm. Original magnification, ×100 (B and C) and ×400 (D and E). The results shown were obtained using SI02777257 siRNA (Qiagen) to silence the CD151 gene. Similar results were obtained using SI02777250 siRNA. pSMAD3, phospho-SMAD family member 3; CD151, cluster of differentiation 151; SD, standard deviation.
Representative data using SI02777257 siRNA is presented in Figs. 4 and 5. Similar results were obtained using another siRNA, SI02777250, to silence the CD151 gene.
Discussion
CCS is a rare, high-grade malignant soft tissue tumor (8,12). This tumor was previously known as ‘melanoma of soft part’ (9), but it is now thought to be derived from the neural crest (30,31). Most CCS tumors are characterized by a t(12;22)(q13;q12) translocation harboring an EWS-ATF1 fusion gene (32). The resulting fusion protein directly activates expression of the melanocyte master transcription factor that drives the same down-stream pathways in both CCS and melanoma (33). As a result, this chimeric molecular alteration leads to significant clinicopathological parallels between CCS and melanoma.
Surgical resection, if necessary, in combination with adjuvant radiotherapy is the clinical mainstay for treatment of patients diagnosed with CCS. Robust lymphatic metastatic activity, however, often impedes effective treatment, and currently the overall survival rate for patients diagnosed with this malignancy is approximately 60% for 5 years. In this context, it is important to understand the molecular mechanisms, underlying the elevated lymphatic metastatic activity of CCS.
The present study strongly suggests an important role for CD151 in lymphatic invasion, and we identify potent suppressor activity of anti-CD151 antibody in the context of lymphatic metastases using an orthotropic metastatic model of CCS. Specifically, we demonstrate that CD151 is highly expressed in HS-MMhigh cells, and that these cells possessed an elevated potential for lymphatic invasion and metastasis compared to that of the original HS-MM cells. Lymphatic infiltration of HS-MMhigh cells was also found at transplant sites in the control mice (Fig. 2A, Mock), but was absent at these sites in anti-CD151 treated mice (Fig. 2A, anti-CD151). Additionally, treatment with anti-CD151 significantly abrogated intraperitoneal lymph node metastasis (Fig. 2C). We expect that the combination of local surgical resection with anti-CD151 treatment may provide significant improvement in clinical outcomes for patients diagnosed with CCS.
We also studied the molecular pathway by which CD151 influenced lymphatic invasion and lymph node metastasis in our Xenoplant model. Metastasis is a complex process composed of consecutive steps, including initiation of local invasion, lymphovascular invasion, migration to a distant site, extravasation, formation of a micrometastasis, and, finally, the development of overt metastatic tumors (19). Certain steps, especially initial steps or those that occur during local invasion and at the end of the cascade, appear to be more challenging for tumor cells (20). The tetraspanin molecule CD151 is believed to be a ‘facilitator’ of tumor metastasis. The role of this molecule in the promotion of early stage metastasis, particularly invasion and intravasation at the primary tumor site, has been demonstrated using in vivo models (21). Consistent with these experimental findings, clinicopathological studies also demonstrated that overexpression of CD151 is associated with poor prognosis, and there is significant correlation between overexpression of CD151 and lymph node metastasis of oesophageal squamous cell carcinoma (22), breast cancer (23), and pancreatic ductal adenocarcinoma (24).
In this study, silencing of CD151 impaired the Matrigel invasion activity of HS-MMhigh cells through downregulation of activated MMP-9. MMP-9 is a well-recognized key enzyme that functions in the proteolytic degradation of the extracellular matrix during tumor invasion (25). Homophilic interactions of CD151 upregulate MMP-9 expression in human melanoma cells. Anti-CD151 antibody may impair the homophilic interactions of CD151, resulting in decreased expression of MMP-9 and abrogation of the initiation of lymphatic vessel invasion.
Additionally, the present study demonstrated that downregulation of CD151 affected the phosphorylation status of SMAD3. Phosphorylation, followed by the nuclear import of SMAD3, is a key signal for the induction of EMT (26,27). This process not only facilitates tumor invasion, but also contributes to tumor progression via pleiotropic pathobiological mechanisms such as the acquisition of cancer stem cell features (28).
Our results suggest that anti-CD151 antibody treatment may impair activation of MMP-9, thus decreasing the lymphatic invasion activity of HS-MMhigh cells. It is also likely that reduced levels of phosphorylated SMAD3 may contribute to reduced lymphatic invasion by abrogating EMT activity.
As CD151 possesses a tetraspanin with a long extracellular protein domain, CD151 may be an ideal candidate for use in antibody-based target therapies to regulate CCS metastasis.
Numerous monoclonal antibodies have been successfully generated to regulate various malignant tumors; however, applications for monoclonal antibodies in the context of soft tissue sarcoma regulation remain limited. This is surprising, given that breast cancer, lymphoma, and gastric cancer outcomes can all be significantly improved by the use of newly developed antibody-based therapies. Recently, a monoclonal antibody called Olaratumab that targets platelet-derived growth factor receptor (PDGFR)-α (34), when used in combination with doxorubicin received accelerated FDA approval based on improvement of overall survival compared with doxorubicin monotherapy in a phase 1b/2 trial.
The monoclonal antibody 50-6, which is used in the present study, was initially generated by subtractive immunization, and this antibody inhibited the metastasis of a human epidermoid carcinoma cell, HEp-3, in a chicken embryo assay (14). Another monoclonal antibody, SFA1.2B4, was also found to suppress the in vivo pulmonary metastasis of the CD151-overexpressing human colon cancer cell line RPMI4788 induced by direct intravenous challenge to BALB/c nu/nu mice (35). These findings may indicate that anti-CD151 antibodies may be of clinical benefit to patients not only suffering from CSS, a rare soft part tumor, but also those diagnosed with other malignant tumors.
We believe that anti-CD151 antibody treatment may impair activation of MMP-9 to decrease the lymphatic invasion activity of HS-MMhigh cells. Additionally, reduced amounts of phosphorylated SMAD3 may also inhibit lymphatic invasion by abrogating EMT activity. Currently, experiments to delineate the mechanisms by which anti-CD151 antibody treatment can abrogate lymphatic invasion and metastasis are underway.
In conclusion, the present findings indicate that CD151 may provide a therapeutic target to regulate CCS metastasis. Antibody-based therapy combined with surgical resection could provide an effective treatment approach for patients diagnosed with CCS.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the Ministry of Education of Japan (grant nos. KAKEN 15K08361 and 15K19051).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
CS and TT participated in the design of the study, the data interpretation and manuscript drafting. KK, YK, YH and YE performed the experiments. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The experimental protocol was approved by the Animal Care Committee of Gifu Graduate School of Gifu, Japan (approval no. H30-32).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Romanska HM, Berditchevski F. Tetraspanins in human epithelial malignancies. J Pathol. 2011;223:4–14. doi: 10.1002/path.2779. [DOI] [PubMed] [Google Scholar]
- 2.Zöller M. Tetraspanins: Push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009;9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 3.Yu Y, Liang C, Wang S, Zhu J, Miao C, Hua Y, Bao M, Cao Q, Qin C, Shao P, Wang Z. CD151 promotes cell metastasis via activating TGF-β1/Smad signaling in renal cell carcinoma. Oncotarget. 2018;9:13313–13323. doi: 10.18632/oncotarget.24028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Wang C, Zhou Z, Sun M, Zhou C, Chen J, Yin F, Wang H, Lin B, Zuo D, et al. CD151-mediated adhesion is crucial to osteosarcoma pulmonary metastasis. Oncotarget. 2016;7:60623–60638. doi: 10.18632/oncotarget.11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ke AW, Zhang PF, Shen YH, Gao PT, Dong ZR, Zhang C, Cai JB, Huang XY, Wu C, Zhang L, et al. Generation and characterization of a tetraspanin CD151/integrin α6β1-binding domain competitively binding monoclonal antibody for inhibition of tumor progression in HCC. Oncotarget. 2016;7:6314–6322. doi: 10.18632/oncotarget.6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer TD, Martínez CH, Vasquez C, Hebron KE, Jones-Paris C, Arnold SA, Chan SM, Chalasani V, Gomez-Lemus JA, Williams AK, et al. Integrin-free tetraspanin CD151 can inhibit tumor cell motility upon clustering and is a clinical indicator of prostate cancer progression. Cancer Res. 2014;74:173–187. doi: 10.1158/0008-5472.CAN-13-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng P, Wang YH, Si M, Gu JH, Li P, Lu PH, Chen MB. Tetraspanin CD151 as an emerging potential poor prognostic factor across solid tumors: A systematic review and meta-analysis. Oncotarget. 2017;8:5592–5602. doi: 10.18632/oncotarget.13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enzinger FM. Clear-cell sarcoma of tendons and aponeuroses. An analysis of 21 cases. Cancer. 1965;18:1163–1174. doi: 10.1002/1097-0142(196509)18:9<1163::AID-CNCR2820180916>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 9.Chung EB, Enzinger FM. Malignant melanoma of soft parts. A reassessment of clear cell sarcoma. Am J Surg Pathol. 1983;7:405–413. doi: 10.1097/00000478-198307000-00003. [DOI] [PubMed] [Google Scholar]
- 10.Andreou D, Tunn PU. Sentinel node biopsy in soft tissue sarcoma. Recent Results Cancer Res. 2009;179:25–36. doi: 10.1007/978-3-540-77960-5_3. [DOI] [PubMed] [Google Scholar]
- 11.Andreou D, Boldt H, Werner M, Hamann C, Pink D, Tunn PU. Sentinel node biopsy in soft tissue sarcoma subtypes with a high propensity for regional lymphatic spread-results of a large prospective trial. Ann Oncol. 2013;24:1400–1405. doi: 10.1093/annonc/mds650. [DOI] [PubMed] [Google Scholar]
- 12.Mavrogenis A, Bianchi G, Stavropoulos N, Papagelopoulos P, Ruggieri P. Clinicopathological features, diagnosis and treatment of clear cell sarcoma/melanoma of soft parts. Hippokratia. 2013;17:298–302. [PMC free article] [PubMed] [Google Scholar]
- 13.Egawa Y, Saigo C, Kito Y, Moriki T, Takeuchi T. Therapeutic potential of CPI-613 for targeting tumorous mitochondrial energy metabolism and inhibiting autophagy in clear cell sarcoma. PLoS One. 2018;13:e0198940. doi: 10.1371/journal.pone.0198940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Testa JE, Brooks PC, Lin JM, Quigley JP. Eukaryotic expression cloning with an antimetastatic monoclonal antibody identifies a tetraspanin (PETA-3/CD151) as an effector of human tumor cell migration and metastasis. Cancer Res. 1999;59:3812–3820. [PubMed] [Google Scholar]
- 15.Sonobe H, Furihata M, Iwata J, Ohtsuki Y, Mizobuchi H, Yamamoto H, Kumano O. Establishment and characterization of a new human clear-cell sarcoma cell-line, HS-MM. J Pathol. 1993;169:317–322. doi: 10.1002/path.1711690307. [DOI] [PubMed] [Google Scholar]
- 16.Sonobe H, Takeuchi T, Taguchi T, Shimizu K, Iwata J, Furihata M, Ohtsuki Y. Further characterization of the human clear cell sarcoma cell line HS-MM demonstrating a specific t(12;22)(q13;q12) translocation and hybrid EWSR1/ATF-1 transcript. J Pahol. 1999;187:594–597. doi: 10.1002/(SICI)1096-9896(199904)187:5<594::AID-PATH277>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi T, Kuro-o M, Miyazawa H, Ohtsuki Y, Yamamoto H. Transgenic expression of a novel thymic epithelial cell antigen stimulates abberant development of thymocytes. J Immunol. 1997;159:726–733. [PubMed] [Google Scholar]
- 18.Takeuchi T, Adachi Y, Sonobe H, Furihata M, Ohtsuki Y. A ubiquitin ligase, skeletrophin, is a negative regulator of melanoma invasion. Oncogene. 2006;25:7059–7069. doi: 10.1038/sj.onc.1209688. [DOI] [PubMed] [Google Scholar]
- 19.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kito Y, Saigo C, Takeuchi T. Novel transgenic mouse model of polycystic kidney disease. Am J Pathol. 2017;187:1916–1922. doi: 10.1016/j.ajpath.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Morikawa A, Takeuchi T, Kito Y, Saigo C, Sakuratani T, Futamura M, Yoshida K. Expression of beclin-1 in the microenvironment of invasive ductal carcinoma of the breast: Correlation with prognosis and the cancer-stromal interaction. PLoS One. 2015;10:e0125762. doi: 10.1371/journal.pone.0125762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi T, Adachi Y, Nagayama T. A WWOX-binding molecule, transmembrane protein 207, is related to the invasiveness of gastric signet-ring cell carcinoma. Carcinogenesis. 2012;33:548–554. doi: 10.1093/carcin/bgs001. [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi T, Misaki A, Liang SB, Tachibana A, Hayashi N, Sonobe H. Expression of T-cadherin (CDH13, H-cadherin) in human brain and its characteristics as a negative growth regulator of epidermal growth factor in neuroblastoma cells. J Neurochem. 2000;74:1489–1497. doi: 10.1046/j.1471-4159.2000.0741489.x. [DOI] [PubMed] [Google Scholar]
- 25.Heussen C, Dowdle EB. Electrophoretic analysis of plasminogen activators in polyacrylamide gels containing sodium dodecyl sulfate and copolymerized substrates. Anal Biochem. 1980;102:196–202. doi: 10.1016/0003-2697(80)90338-3. [DOI] [PubMed] [Google Scholar]
- 26.Hong IK, Jin YJ, Byun HJ, Jeoung DI, Kim YM, Lee H. Homophilic interactions of Tetraspanin CD151 up-regulate motility and matrix metalloproteinase-9 expression of human melanoma cells through adhesion-dependent c-Jun activation signaling pathways. J Biol Chem. 2006;281:24279–24292. doi: 10.1074/jbc.M601209200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Wang F, Li Q, Zhang H, Cui Y, Ma C, Zhu J, Gu X, Sun Z. CD151 knockdown inhibits osteosarcoma metastasis through the GSK-3β/β-catenin/MMP9 pathway. Oncol Rep. 2016;35:1764–1770. doi: 10.3892/or.2015.4517. [DOI] [PubMed] [Google Scholar]
- 28.Shi GM, Ke AW, Zhou J, Wang XY, Xu Y, Ding ZB, Devbhandari RP, Huang XY, Qiu SJ, Shi YH, et al. CD151 modulates expression of matrix metalloproteinase 9 and promotes neoangiogenesis and progression of hepatocellular carcinoma. Hepatology. 2010;52:183–196. doi: 10.1002/hep.23661. [DOI] [PubMed] [Google Scholar]
- 29.Li P, Zeng H, Qin J, Zou Y, Peng D, Zuo H, Liu Z. Effects of tetraspanin CD151 inhibition on A549 human lung adenocarcinoma cells. Mol Med Rep. 2015;11:1258–1265. doi: 10.3892/mmr.2014.2774. [DOI] [PubMed] [Google Scholar]
- 30.Mii Y, Miyauchi Y, Hohnoki K, Maruyama H, Tsutsumi M, Dohmae K, Tamai S, Konishi Y, Yamanouchi T. Neural crest origin of clear cell sarcoma of tendons and aponeuroses. Ultrastructural and enzyme cytochemical study of human and nude mouse-transplanted tumours. Virchows Arch A Pathol Anat Histopathol. 1989;415:51–60. doi: 10.1007/BF00718604. [DOI] [PubMed] [Google Scholar]
- 31.Yamada K, Ohno T, Aoki H, Semi K, Watanabe A, Moritake H, Shiozawa S, Kunisada T, Kobayashi Y, Toguchida J, et al. EWS/ATF1 expression induces sarcomas from neural crest-derived cells in mice. J Clin Invest. 2013;123:600–610. doi: 10.1172/JCI63572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zucman J, Delattre O, Desmaze C, Epstein AL, Stenman G, Speleman F, Fletchers CD, Aurias A, Thomas G. EWS and ATF-1 gene fusion induced by t(12;22) translocation in malignant melanoma of soft parts. Nat Genet. 1993;4:341–345. doi: 10.1038/ng0893-341. [DOI] [PubMed] [Google Scholar]
- 33.Davis IJ, Kim JJ, Ozsolak F, Widlund HR, Rozenblatt-Rosen O, Granter SR, Du J, Fletcher JA, Denny CT, Lessnick SL, et al. Oncogenic MITF dysregulation in clear cell sarcoma: Defining the MiT family of human cancers. Cancer Cell. 2006;9:473–484. doi: 10.1016/j.ccr.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 34.Antoniou G, Lee ATJ, Huang PH, Jones RL. Olaratumab in soft tissue sarcoma-Current status and future perspectives. Eur J Cancer. 2018;92:33–39. doi: 10.1016/j.ejca.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 35.Kohno M, Hasegawa H, Miyake M, Yamamoto T, Fujita S. CD151 enhances cell motility and metastasis of cancer cells in the presence of focal adhesion kinase. Int J Cancer. 2002;97:336–343. doi: 10.1002/ijc.1605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.