Summary
Restoration of barrier tissue integrity after injury is dependent on the function of immune cells and stem cells (SCs) residing in the tissue. In response to skin injury, hair follicle stem cells (HFSCs), normally poised for hair generation, are recruited to the site of injury and differentiate into cells that repair damaged epithelium. We used a stem cell fate mapping approach to examine the contribution of regulatory T (Treg) cells to epidermal barrier repair after injury. Depletion of Treg cells impaired skin barrier regeneration and was associated with a Th17 inflammatory response and failed HFSC differentiation. In this setting, damaged epithelial cells preferentially expressed the neutrophil chemoattractant CXCL5, and blockade of CXCL5 or neutrophil depletion restored barrier function and stem cell differentiation after epidermal injury. Thus, Treg cell regulation of localized inflammation enables HFSC differentiation and thereby skin barrier regeneration, with implications for the maintenance and repair of other barrier tissues.
Graphical Abstract
eTOC
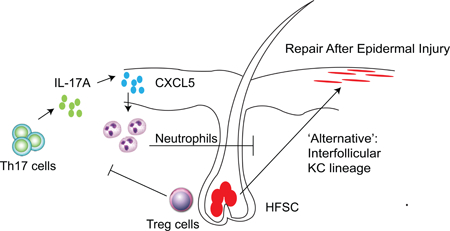
In response to skin injury, hair follicle stem cells (HFSCs) differentiate into epithelial cells that contribute to the repair of damaged epithelium. Mathur et al. show that regulatory T cells facilitate HFSC differentiation via the control of the local inflammatory environment and specifically, the prevention of an over-exuberant Th17 and neutrophil response mediated by CXCL5.
Introduction
Specific epidermal SC compartments contribute to maintaining skin barrier integrity over the lifetime of mammals by replacing cells that are lost during homeostatic turnover or after skin injury. Stem cells located within the basal layer of the epidermis contribute to the maintenance of the skin barrier, while hair follicle stem cells (HFSCs), located within the permanent portion of the hair follicle bulge region, contribute to cyclic rounds of hair generation (Blanpain and Fuchs, 2006). In the steady-state, these stem cell pools generate epithelium that have distinct biologic functions. However, after injury, HFSCs are recruited to support regeneration of the damaged epithelium (Ito et al., 2005; Levy et al., 2007). The rapid response of these cells ensures re-establishment of barrier function, thereby limiting infection and insensible water loss (Ito et al., 2005). Thus, HFSCs are normally poised for hair regeneration, but can differentiate into epithelial cells that facilitate barrier repair.
While mechanisms that control HFSC function during hair generation are fairly well established, the specific cell types and molecular pathways that govern HFSC lineage commitment to cells of the interfollicular stratified epithelium during epidermal repair are largely unknown. In contrast to hair follicle cycling, epithelial injury in skin is a highly inflammatory process (Gregorio et al., 2010). Thus, it is plausible that tissue-resident immune cells influence HFSC lineage fate decisions during epidermal regeneration after injury.
We have previously shown that Treg cells localize to the hair follicle niche in the steady-state. In the absence of skin injury, Treg cell expression of the Notch ligand, Jagged-1 promotes HFSC proliferation and differentiation during hair generation. These findings suggest that Jag1+ Treg cells influence HFSC niche signals that are required for efficient hair generation (Ali et al., 2017). Here, we examined the impact of Treg cells in the HFSC response to acute epithelial injury. We found that Treg cells control a specific IL-17-CXCL5-neutrophil axis of inflammation during barrier repair. Treg cell mediated control of this inflammatory axis facilitated the differentiation of HFSCs into epithelial cells necessary for repair of the epidermis after injury.
Results
Model of Epidermal Barrier Regeneration.
We set out to determine if Treg cells influence SC biology during skin barrier repair. To do so, we employed a well-established model of subacute skin injury (Supplementary Figure 1a) (Gregorio et al., 2010). In this model, the stratum corneum is physically disrupted through repeated applications of adhesive tape (i.e., tape stripping) while leaving the underlying dermis and subcutaneous tissues relatively unaffected. This insult incites a highly orchestrated and evolutionary conserved program of epidermal regeneration, characterized by keratinocyte proliferation and HFSC differentiation, culminating in a restoration of barrier function (Elias, 2005). Water loss through the skin (i.e., transepidermal water loss; TEWL) is a specific measure of barrier integrity, where excessive losses indicate poor stratum corneum function (Gregorio et al., 2010; Jin et al., 2009; Sano et al., 2005). Return to baseline levels of water loss following tape stripping signified a complete restoration of the skin barrier and occurred within 6 days after injury (Supplementary Figure 1b). Consistent with recovery, expression of key epidermal differentiation genes that are necessary for stratum corneum formation were diminished early after injury and restored to basal levels over time (Supplementary Figure 1c) (Elias, 2005). During the recovery phase, Treg cells in skin significantly accumulated, reaching peak numbers approximately 7 days after injury (Supplementary Figure 1d & 1e). While Treg cells maximally accumulated 7 days after injury, these cells were highly activated early during barrier repair indicated by peak expression of the proliferation marker Ki-67 on day 2 of recovery (Supplementary Figure 1f & 1g). We have previously shown that Treg cells preferentially localize to hair follicles in mouse and human skin (Ali et al., 2017; Gratz et al., 2013; Scharschmidt et al., 2015). While Treg cells accumulated in the tissue during barrier regeneration, 50–95% of cells remained localized within 20 µm of a HF throughout the repair process (Supplementary Figure 1h &1i). Collectively, these results indicate that Treg cells accumulate during epidermal repair and remain in close proximity to HFs.
Tregs Facilitate Skin Barrier Regeneration.
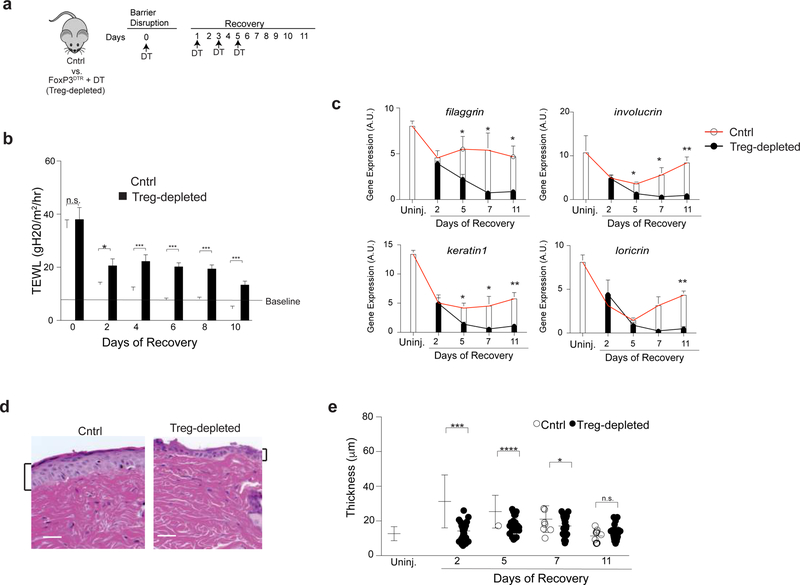
To determine if Treg cells influence barrier regeneration, we utilized mice transgenic for the Diphtheria toxin receptor under the control of the FoxP3 promoter (FoxP3DTR) (Kim et al., 2007). These mice allowed for selective and inducible depletion of Treg cells in skin following administration of diphtheria toxin (DT) (Supplementary Figure 2a). When compared to control mice (either DT-treated WT or untreated FoxP3DTR littermates), FoxP3DTR mice treated with DT (i.e., Treg cell-depleted mice) during the recovery phase had sustained TEWL throughout the entire period of epidermal regeneration (Figure 1b). In addition, Treg cell-depleted mice had reduced expression of epidermal differentiation genes required for stratum corneum formation, accompanied by a marked attenuation of epidermal thickening when compared to control mice (Figure 1c–e and Supplementary Figure 2b). In contrast, Treg cell depletion alone had no effect on skin barrier function in the absence of epidermal injury, as evidenced by no change in TEWL between Treg cell-sufficient and Treg cell-depleted animals (Supplementary Figure 2c). Taken together, these results suggest that Treg cells facilitate epidermal repair, but are dispensable for maintaining barrier homeostasis.
Figure 1. Regulatory T cells facilitate epidermal regeneration after injury.
(a) Schematic showing diphtheria toxin (DT) administration schedule after skin barrier disruption. Cntrl animals are DT-treated WT mice or FoxP3DTR littermates not given DT.
(b) Transepidermal water loss (TEWL) on the indicated days of recovery.
(c) qRT-PCR of epidermal differentiation genes normalized to β2m during barrier regeneration from the back affected back skin of Cntrl and Treg cell-depleted mice.
(d) Representative histology 2 days after barrier injury and
(e) quantification of epidermal thickness of affected back skin of Cntrl and Treg cell-depleted mice throughout the course of barrier repair. Results in b are representative of > 10 experiments (n=2–4 mice per group). Results in d & e are representative of 3 experiments (n = 3 mice per group) Scale bar in d is 50 μm. See also Supplementary Figures 1 and 2.
Tregs Preferentially Attenuate IL-17 Associated Inflammation Early During Epidermal Regeneration.
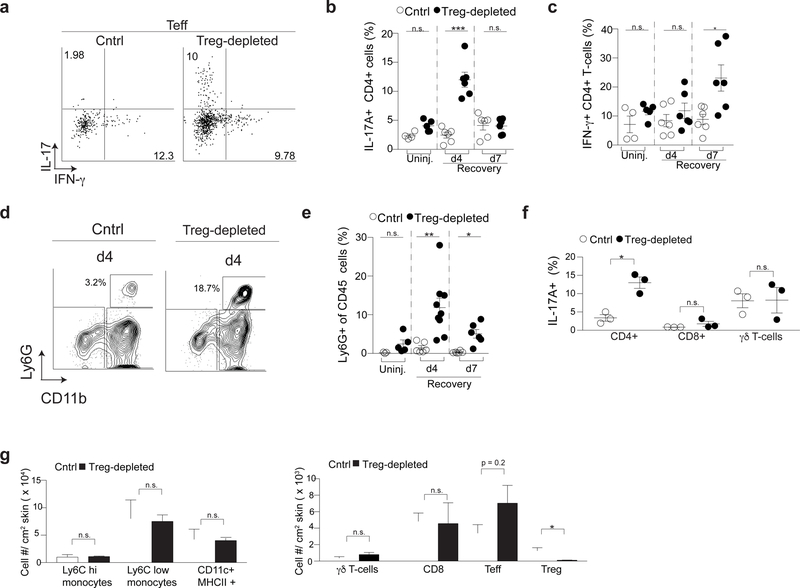
We hypothesized that Treg cells facilitate epidermal regeneration by preferentially regulating specific inflammatory pathways. To determine which immune pathways Treg cells control, we comprehensively quantified immune cell infiltrates in skin of Treg cell-depleted and Treg cell-sufficient control mice throughout the skin barrier recovery period. After epidermal injury in Treg cell-sufficient control mice, the percentage of IL-17A and IFN-γ producing CD4+ cells were maintained at similar levels to that observed in uninjured mice (Figure 2a–c). In contrast, Treg cell-depleted mice had a preferential accumulation of IL-17A-producing CD4+ T cells (i.e., Th17 cells) peaking early during barrier recovery, which was followed by an accumulation of IFNγ-producing CD4+ T cells later during recovery (i.e., Th1 cells; Figure 2a–c). Consistent with the early accumulation of Th17 cells, we observed an increased accumulation of neutrophils early in the recovery phase in Treg cell-depleted mice (Figure 2d & 2e). There were no differences in IL-17A or IFN-γ production from cutaneous CD8+ or γδ T cells between Treg cell-sufficient and Treg cell-depleted animals, and we did not observe any appreciable amounts of Th2 cytokines (IL-5 and IL-13) being produced by Teff cells (Figure 2f & Supplementary Figure 3). No differences in the number of myeloid mononuclear cells (CD11b+ Ly6Chigh and CD11b+ Ly6Clow), dendritic cells (CD11chigh MHC II+), CD8+ T cells or γδ T cells were observed between Treg cell-sufficient and Treg cell-depleted mice early during epidermal regeneration (Figure 2f–g & Supplementary Figure 3a). However, there was a reproducible, but non-statistically significant trend towards an increased percentage of CD4+FoxP3- effector T cells (Teffs) in the skin of Treg cell-depleted mice compared to controls (Figure 2g). Taken together, these results indicate that Treg cells preferentially limit Th17 and neutrophil accumulation early during barrier repair and Th1 cells later in this process.
Figure 2. Regulatory T cells preferentially regulate Th17 and neutrophil accumulation in skin early during barrier repair.
(a) Representative flow cytometry plots of IL-17A+ CD4+ (Th17) and IFN-γ+ CD4+ (Th1) T cells in skin following PMA/ionomycin stimulation of single cell suspensions 4 days after skin barrier disruption.
(b) Quantification of Th17 and (c) Th1 cells of Treg cell-depleted mice following skin injury compared to Treg cell-sufficient cntrls (Cntrl) at the indicated times of barrier recovery.
(d) Representative flow cytometry plots of CD11b+ Ly6G+ cells (neutrophils) in the skin 4 days after injury. Plots are pregated on live, CD45+ cells.
(e) Percent of CD45+CD11b+Ly6G+ neutrophils in skin at the indicated times of barrier recovery.
(f) Percent of IL-17+ CD4+, CD8+ and γδ T cells in the skin of Cntrl and Treg cell-depleted mice following PMA/ionomycin stimulation of single cell suspensions 4 days after skin barrier disruption.
(g) Absolute number of the indicated cell types in the skin of Cntrl and Treg cell-depleted mice 4 days after injury.
d4 – day 4; d7 – day 7; Uninj- uninjured skin; For all relevant panels, error bars are +/−SEM. n.s. – no significance; *p < 0.05; **p < 0.01; ***p < 0.001 according to a Student’s t-test. (n=3 mice per group) Data are pooled (b,c,e) or representative (f-h) of two independent experiments. (See also Supplementary Figure 3).
Hair Follicle Stem Cells are Recruited to Repair the Epidermis After Injury.
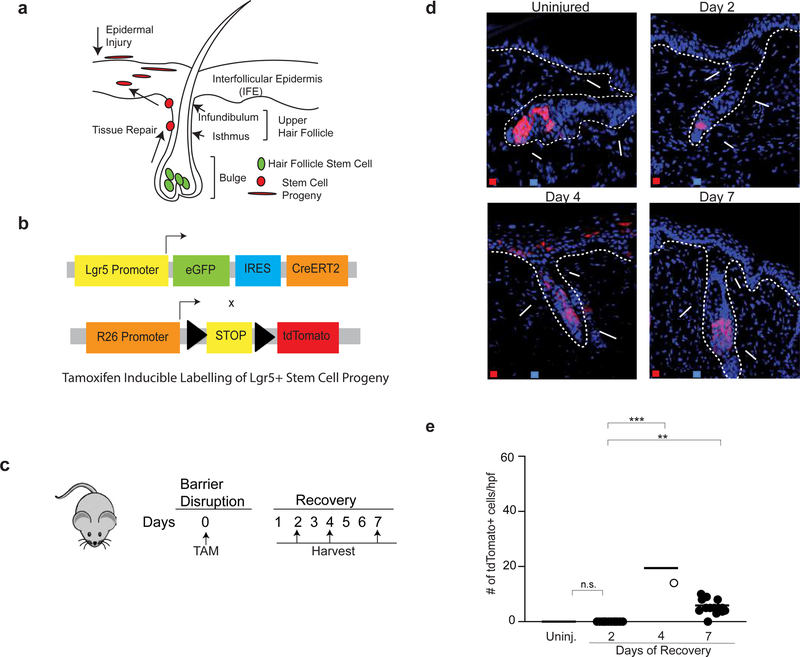
Hair follicle stem cells play a major role in repairing the epidermis after injury (Ito and Cotsarelis, 2008; Ito et al., 2005). Given that Treg cells facilitate epidermal regeneration (Figure 1), localize to the HFSC niche, (Gratz et al., 2013; Sanchez Rodriguez et al., 2014) and promote HFSC differentiation during hair follicle cycling, (Ali et al., 2017) we hypothesized that Treg cells facilitate epithelial repair by influencing the function of HFSCs. To test this, we utilized a Leucine-rich G-protein coupled receptor (Lgr5) lineage-tracing approach, where Lgr5 labels a subset of HFSCs (Jaks et al., 2008). Following skin injury, a subset of cells derived from HFSCs emigrate from the hair follicle bulge and participate in the repair of the upper hair follicle and interfollicular epidermis (IFE) (Summarized in Figure 3a) (Brownell et al., 2011; Ito et al., 2005; Tumbar et al., 2004). We used Lgr5-tdTomato (Lgr5-tdTom) reporter mice, which allows for the permanent labeling of Lgr5+ cells and their progeny with tdTomato (tdTom) after administration of tamoxifen (Jaks et al., 2008) (Figure 3b). This enabled us to track Lgr5-derived progeny by immunofluorescence (IF) microscopy and isolate labeled cells for flow cytometric and transcriptome analyses. To determine if Lgr5+ stem cells contribute to epidermal repair, Lgr5-tdTom mice were injected with tamoxifen on day 0 of epidermal injury and migration of tdTomato+ cells was quantified by IF microscopy 2, 4 and 7 days after barrier injury (Figure 3c). Lgr5-derived cells migrated into the upper hair follicle and IFE, peaking 4 days after injury (Figure 3d & 3e). In contrast, Lgr5-derived cells in uninjured mice remained restricted to the hair follicle bulge (Figure 3d & 3e). To ensure that tamoxifen used to label Lgr5 cells specifically labeled HFSCs and not an epidermal cell population that might transiently express Lgr5 in response to inflammation, Lgr5-tdTomato mice were treated with tamoxifen 7 days before epidermal injury (Supplementary Figure 4a). This allowed for labelling of Lgr5 cells well in advance of barrier disruption. Based on the known kinetics of tamoxifen metabolism in mice (Robinson et al., 1991; Wilson et al., 2014), this longer time frame between stem cell labelling and skin barrier injury ensured that any non-HFSCs that might transiently express Lgr5 upon skin injury were not ‘erroneously’ labelled and traced. Following tamoxifen labeling, and skin barrier injury 7 days later, animals were harvested 2,4 and 7 days after barrier injury. Similar to our results in Figure 3c, Lgr5-derived cells again migrated into the IFE peaking 4 days after injury (Supplementary Figure 4b & 4c). These results demonstrate that Lgr5 faithfully labels HFSCs despite inflammation caused by skin barrier disruption.
Figure 3. Lgr5-derived hair follicle stem cells contribute to epidermal repair.
(a) Diagram of hair follicle anatomy. Following epidermal injury, cells derived from HFSCs located in the bulge region migrate into the upper hair follicle (isthmus and infundibulum) and interfollicular epidermis (IFE) to participate in epithelial repair.
(b) Schematic of Lgr5-tdTom mice. Lgr5 drives expression of eGFP and a tamoxifen inducible Cre (CreERT2). These mice are crossed to R26-tdTomato mice. Injection with tamoxifen allows for inducible and permanent labeling of Lgr5+ stem cells and their progeny.
(c) The back skin of Lgr5-tdTom mice were injured (as in Supplementary Figure 1a) and injected with tamoxifen on day 0. Mice were harvested on days 2,4 and 7 of recovery and compared to uninjured Lgr5-traced mice.
(d) Representative high-power images of Lgr5-traced cells in the bulge, upper hair follicle and IFE during barrier regeneration compared to uninjured control.
(e) Quantification of Lgr5-derived cells in the IFE during epidermal repair. Error bars are +/− S.E.M. n.s.- no significance; **p < 0.01; *** p<0.001. n=2–4 mice per group. IFE – Interfollicular epidermis; Infund. – Infundibulum.
Treg Cells Promote HFSC Migration and Differentiation During Epidermal Regeneration
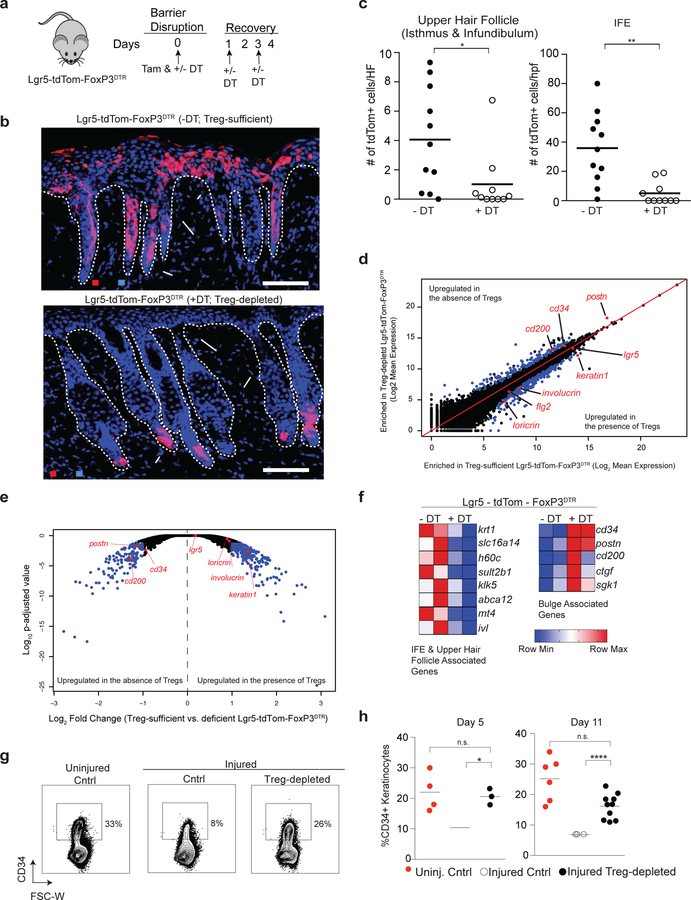
To determine if Treg cells promote the egress of keratinocytes derived from HFSCs during epidermal repair, we crossed Lgr5-tdTom mice to FoxP3DTR mice. The resultant Lgr5-tdTom-FoxP3DTR strain enabled comprehensive quantification of Lgr5-expressing SC progeny in the presence or absence of Treg cells. The epidermis of Lgr5-tdTom-FoxP3DTR mice was injured by repetitive tape stripping. On day 0 of the recovery phase, Lgr5-expressing HFSCs were labeled with tamoxifen, and Treg cells depleted by administration of DT. Experimental mice were compared to tamoxifen labeled, non-DT treated Lgr5-tdTom-FoxP3DTR littermate controls (i.e., Treg cell-sufficient mice) (Figure 4a). The migration of Lgr5 stem cell progeny in the presence and absence of Treg cells was quantified four days after injury, a time where Th17 and neutrophil influx is maximally attenuated by Treg cells (Figure 2a–e) and Lgr5-derived cells maximally contribute to epidermal repair (Figure 3d & 3e). In Treg cell-sufficient mice, we consistently observed Lgr5-derived cell emigration from the hair follicle bulge to regions of the upper hair follicle and IFE after injury (Figure 4b & 4c). In striking contrast, Lgr5-derived cells from Treg cell-depleted animals remained almost entirely confined to the hair follicle bulge region (Figure 4b & 4c), resembling the localization pattern seen in uninjured animals (Figure 3d). To ensure that DT itself did not affect the cellular pool or migration of Lgr5 traced cells, FoxP3DTR negative / Lgr5-tdTom mice were treated as in Figure 2a and harvested 4 days later. The contribution of Lgr5 traced cells into the IFE was comparable between DT treated and untreated mice (Supplementary Figure 4d & 4e). These results indicate that diphtheria toxin alone does not affect the migration of Lgr5-derived cells during barrier regeneration. Collectively, our results highlight a critical role for Treg cells in promoting the migration of HFSC progeny to repair injured epidermis.
Figure 4. Treg cells facilitate HFSC migration and differentiation during epidermal regeneration.
(a) Schematic showing DT and tamoxifen (Tam) administration in Lgr5-tdTom-FoxP3DTR mice during skin barrier recovery for experiments described in panels b-f. Mice are compared to tamoxifen-labeled, non-DT treated littermates and harvested 4 days after skin barrier disruption.
(b) Representative images demonstrating the patterns of stem cell progeny (tdTomato+ cell) localization in the skin of tamoxifen-labeled, Lgr5 stem cell lineage tracing mice after injury. Mice are either sufficient (-DT) or depleted (+DT) of Treg cells. (c) Quantification of Lgr5-derived cell (tdTomato+) localization in the upper hair follicle (isthmus and infundibulum; left panel) and IFE (right panel). tdTomato+/ HF and tdTomato+/hpf represent cell numbers normalized per hair follicle and per high power field respectively. See also Supplementary Figure 4.
(d) FACS purified tdTom+ cells from DT-treated (i.e. Treg cell-depleted) or untreated Lgr5-tdTom-FoxP3DTR (i.e. Treg cell-sufficient) mice were analyzed by RNA-sequencing. Comparison plots of normalized gene expression of tdTomato+ cells isolated from Treg cell-sufficient and Treg cell-depleted Lgr5-tdTom-FoxP3DTR mice. Blue dots represent genes with a p-adjusted value < 0.05 and > 2-fold difference in gene expression between groups.
(e) Volcano plot comparing the p-adjusted value versus fold change for tdTomato+ cells isolated from Treg cell-sufficient and Treg cell-deficient Lgr5-tdTom-FoxP3DTR mice. Blue dots represent genes with a p-adjusted value < 0.05 and > 2 fold differences in gene expression between groups.
(f) Heat map of gene transcripts of tdTomato+ cells isolated from Treg cell-sufficient and Treg cell-depleted Lgr5-tdTom-FoxP3DTR mice focusing on differentially expressed genes with a padj. value < 0.05, fold change differences > 2 and predominant expression in either the bulge or IFE/upper hair follicle in adult mice.(Joost et al., 2016)
(g) Cntrl and FoxP3DTR mice were treated as diagramed in Figure 1a and compared to uninjured WT mice. Representative plots of CD34+ cells in the epidermis 11 days after epidermal injury. Plots are pre-gated on live, CD45-negative epidermal cells.
(h) Percentage of CD34+ epidermal cells in uninjured cntrl mice compared to barrier-disrupted cntrl and Treg cell-depleted mice 5 and 11 days after injury. See also Supplementary Figure 5.
For all relevant panels, error bars are +/− SEM. *p < 0.05; **p < 0.01; **** p<0.0001 by Student’s t-test or by one-way ANOVA for panel h. Results in b-c are representative of > 8 independent experiments (n=2 to 4 mice per group); d-f are representative of 2 independent experiments (n=2 mice per group) Results of h (right panel) are pooled data from 3 independent experiments. (n=2–4 mice per group) and representative of 2 experiments (left panel). Scale bar in b is 100 µm; Red-tdTomato+ Lgr5 stem cell progeny; Blue – DAPI.
We hypothesized that Treg cells promote the migration of Lgr5-derived cells into the upper epidermis after injury by facilitating their differentiation towards keratinized epithelial cells. To test this, we FACS sorted Lgr5-derived tdTomato+ cells during the recovery phase from Treg cell-sufficient and Treg cell-depleted mice and performed whole transcriptome RNA-sequencing (Figure 4d–f). Recently, the heterogeneity of murine epidermal transcriptomes has been dissected into distinct follicular and interfollicular subsets at the single cell level. (Joost et al., 2016) Utilizing this resource, we assessed all differentially regulated genes (>2 fold change; padj < 0.05) in our dataset for transcripts preferentially expressed in the IFE, upper hair follicle and bulge region. Lgr5-derived cells from Treg cell-sufficient mice preferentially expressed IFE and upper hair follicle signatures (Figure 4d–f). Increased expression of genes such as keratin 1 and involucrin suggest these cells are skewed toward a terminally differentiated keratinized epithelium, a phenotype essential for barrier function and consistent with the localization pattern of Lgr5 derived cells by IF (Figure 4b). In contrast, Lgr5-derived cells from Treg cell-depleted mice expressed higher levels of bulge-associated genes, including cd34, cd200, and postn (Figure 4d–f).
Of note, Lgr5-derived cells from mice sufficient in Treg cells lost expression of the stem cell marker, CD34, as they migrated and differentiated into keratinized epithelial cells (Figure 4f). Loss of CD34 expression in HFSCs has previously been correlated with rapid keratinocyte differentiation (Castilho et al., 2009). Consistent with this finding, epidermal disruption in WT mice resulted in the progressive loss of CD34+ keratinocytes during repair compared to uninjured controls (Figure 4g & 4h; Supplementary Figure 5a–5b). In contrast, Treg cell depletion during barrier repair resulted in the retention of a higher proportion of CD34-expressing keratinocytes when compared to Treg cell-sufficient controls (Figure 4g & 4h; Supplementary Figure 5a–5b), suggesting a relative retention of bulge stem cell identity throughout barrier repair. Progressive loss of CD34 expression during epithelial repair was not only observed from the total pool of keratinocytes, but also specifically observed in Lgr5-traced cells as well (Supplementary Figure 5c–5d). In contrast, mice lacking in Treg cells retained a higher proportion of Lgr5-traced cells expressing CD34 (Supplementary Figure 5c–5d). Taken together, these results suggest that the loss of CD34+ keratinocytes throughout skin barrier recovery reflects a loss of HFSC identity as these cells differentiate and contribute to the repair of the epidermis and that Treg cells play a critical role in this process.
While Treg cells influenced HFSC migration and differentiation during epidermal repair, we observed that the lack of Treg cells minimally affected HFSC proliferation, as measured by Ki-67 staining of Lgr5-labelled cells during skin barrier repair (Supplementary Figure 5e–5f). There was a reproducible but non-statistically significant trend toward a reduced number of Ki-67+ Lgr5-derived cells in mice lacking Treg cells four days after epidermal injury. Furthermore, there were minimal differences in the expression of cell cycle associated genes of Lgr5-traced cells from mice sufficient or lacking in Treg cells (Supplementary Figure 5g). Taken together, these results indicate that Treg cells play a major role in facilitating the migration and differentiation of HFSC derived cells in attempts to repair the epithelium after injury and plays a minor role in the proliferative potential of HFSCs during barrier regeneration.
Treg Cells Preferentially Regulate CXCL5 Expression from Interfollicular Epidermal Cells During Repair.
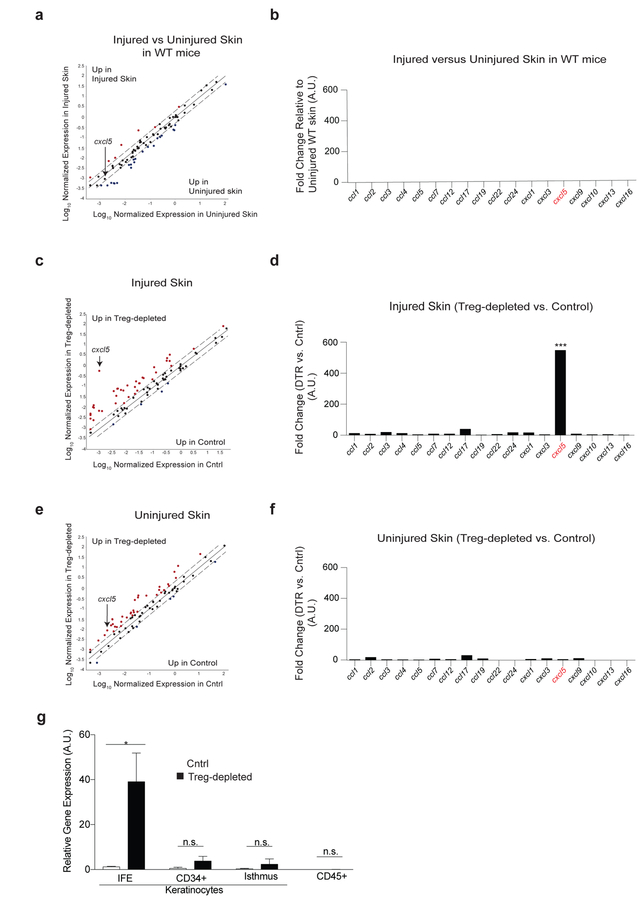
We next set out to elucidate the cellular and molecular mechanisms responsible for Treg-cell mediated control over HFSC function. We were intrigued by the preferential suppression of Th17 cell and neutrophil accumulation in skin mediated by Treg cells early during the barrier recovery period (Figure 2a–e). Inflammation has been shown to enhance the regenerative capacity of tissues by clearing apoptotic cells and debris, as well as activating mechanisms of tissue remodeling (Aurora and Olson, 2014; Karin and Clevers, 2016). However, excessive inflammation can lead to further tissue damage through the release of mediators that inhibit regeneration or tissue stem cell activation, such as IL-1, TNF-α, IL-6 and IL-17 family members (Aurora and Olson, 2014; Doles et al., 2012; Karin and Clevers, 2016). We hypothesized that Treg cells regulate specific immune cell recruitment in skin, thereby enabling the epidermis to effectively regenerate after injury. To test this hypothesis, we took a discovery-based approach to determine which chemokine ligands were preferentially regulated by Treg cells in skin during epidermal barrier recovery. Utilizing a qRT-PCR-based chemokine expression array, we first determined which C-C and C-X-C chemokine ligands were expressed in skin during epidermal repair. Four days after epidermal injury we observed minimal differences in expression of specific chemokines between the skin of injured and uninjured WT mice (Figure 5a & 5b). This result is consistent with the minimal inflammatory cell recruitment observed in both injured and uninjured WT mice (Figure 2a–e) and suggests that in the presence of Treg cells, there is minimal immune cell recruitment to skin at this time of barrier recovery. In contrast, there was a pronounced and highly preferential increase in expression of the chemokine cxcl5 in Treg cell-depleted mice (Figure 5c & 5d). Injured mice whose Treg cells were depleted during the recovery phase had >200-fold increase in cxcl5 in skin when compared to injured Treg cell-sufficient control mice, with minimal increases in expression of other chemokines (Figure 5c & 5d). Interestingly, selective regulation of cxcl5 expression by Treg cells was only observed during the period of barrier regeneration, as there was low-level expression of several chemokines (including cxcl5) in Treg cell-depleted mice compared to Treg cell-sufficient controls in the absence of injury (Figure 5e & 5f). To determine the predominant cellular sources of CXCL5, epidermal cells from control and Treg cell-depleted mice were FACS sorted into specific keratinocyte and immune cell populations as previously described (Nagao et al., 2012) (and shown in Supplementary Figure 5a & Figure 5g). CXCL5 was quantified from purified epidermal cell populations using qRT-PCR. Notably, IFE keratinocytes were the dominant source of CXCL5 in Treg cell-depleted mice compared to barrier injured controls. Taken together, these data suggest that Treg cells preferentially suppress CXCL5 expression from IFE keratinocytes during epidermal regeneration.
Figure 5. Treg cells preferentially regulate cxcl5 expression during epidermal barrier repair.
Cntrl and DT-administered FoxP3DTR mice (Treg cell-depleted) were treated as in Figure 1a, harvested 4 days after skin injury and compared to uninjured mice.
(a) Scatter plot and (b) quantification of chemokines by qRT-PCR array of uninjured WT vs. epidermal injured WT mice.
(c) Scatter plot and (d) quantification of chemokines from injured cntrl vs. injured Treg cell-depleted mice.
(e) Plot and (f) chemokine quantification of uninjured cntrl and uninjured Treg cell-depleted mice.
(g) cxcl5 expression from the indicated sort purified epidermal cells (See also Supplementary Figure 3a for gating strategy).
Representative of 2 independent experiments (n=3 mice per group) Data are +/− SEM *** p< 0.001 comparing cxcl5 expression in panel d versus f by Student’s t-test.
Neutralization of the IL-17-CXCL5-Neutrophil Axis Partially Restores HFSC Activation During Barrier Repair in the Absence of Treg cells.
CXCL5 is an epithelial-derived chemokine that can be induced by IL-17A (Guilloteau et al., 2010) and is a known regulator of neutrophil accumulation into tissues, including the lung and gut (Balamayooran et al., 2012; Rousselle et al., 2013). High expression of this chemokine in skin of Treg cell-depleted mice correlated with a large accumulation of Th17 cells and neutrophils observed early during barrier repair (Figure 2a–e).
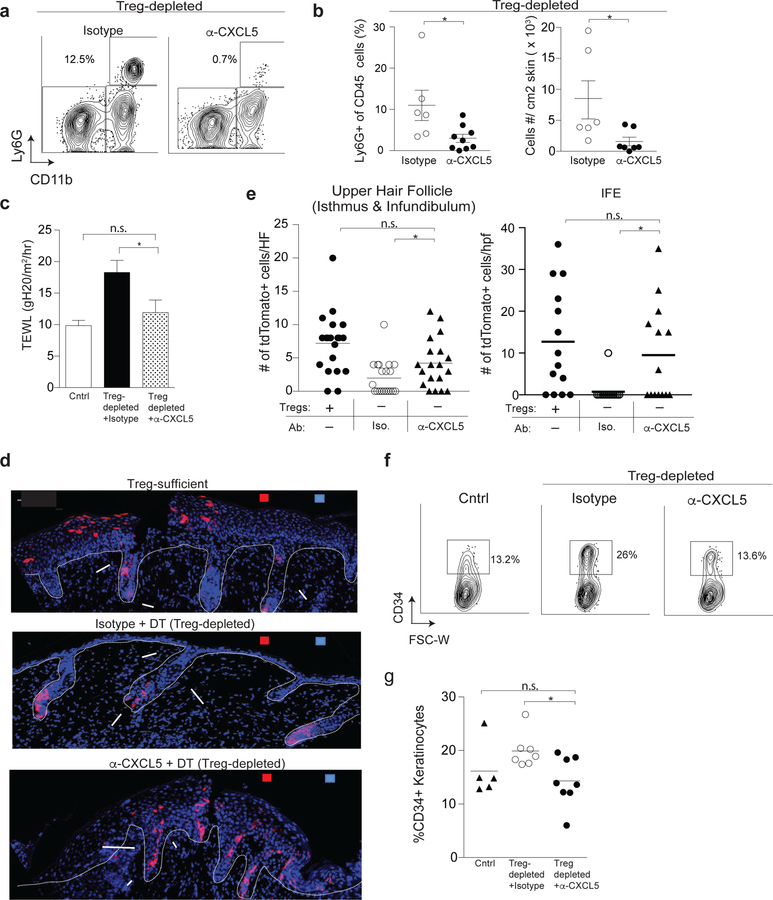
To determine if regulation of CXCL5 expression is a major mechanism by which Treg cells facilitate epidermal regeneration, we neutralized CXCL5 function in Treg cell-depleted mice using an anti-CXCL5 monoclonal antibody (α-CXCL5 mab) (Jia et al., 2016; Rousselle et al., 2013). Treg cell-depleted mice were administered either α-CXCL5 mab or isotype control antibody during the recovery phase after epidermal injury. Neutrophil accumulation, skin barrier function, and HFSC differentiation were quantified 4 days after injury. Neutralization of CXCL5 significantly reduced the percentage and absolute number of neutrophils infiltrating skin during epithelial regeneration (Figure 6a & 6b). In addition, neutralization of this single chemokine in Treg cell-depleted mice resulted in partial restoration of TEWL (Figure 6c). To determine if CXCL5 neutralization in Treg cell-depleted animals could restore Lgr5-derived cell migration during repair, we induced epidermal injury in Lgr5-tdTom-FoxP3DTR mice, depleted Treg cells, and treated with α-CXCL5 mab or isotype control. After labeling Lgr5 stem cells with tamoxifen, we tracked the localization of Lgr5 cell progeny in the epidermis. Neutralization of CXCL5 partially restored the egress of Lgr5 stem cell progeny into the upper hair follicle and IFE (Figure 6d & 6e). Consistent with these results, treatment with α-CXCL5 mab resulted in reduced keratinocyte expression of the stem cell marker CD34 (Figure 6f & 6g), indicating a restoration of HFSC differentiation (see above, Figure 4f–h and Supplementary Figure 5). Taken together, these results indicate that the predominant pathway regulated by Treg cells during skin barrier recovery involves CXCL5 and that neutralization of this chemokine is capable, at least in part, of restoring the defects in HFSC differentiation and skin barrier function observed in the absence of Treg cells. To determine if keratinocytes might directly respond to CXCL5 to limit stem cell activation in Treg cell-depleted mice, the expression of the CXCL5 receptor, cxcr2, was quantified in both Lgr5-traced cells (Supplementary Figure 6a) and in bulk keratinocytes (Supplementary Figure 6b) in the presence or absence of Treg cells. These results revealed minimal to no expression of cxcr2 in keratinocytes, indicating that this chemokine is most likely not directly influencing epidermal cell function in our model.
Figure 6. CXCL5 neutralization restores skin barrier function and HFSC differentiation in the absence of Treg cells.
FoxP3DTR mice were treated as in Figure 1a. Mice were co-administered α-CXCL5 mab or isotype control with DT on days 0,1 and 3 and harvested on day 4.
(a) Representative flow cytometry plots of Ly6G+ CD11b+ neutrophils in skin. Plots are pregated on live CD45+ cells.
(b) Percent and absolute number of Ly6G+ CD11b+ neutrophils in skin.
(c) Transepidermal water loss (TEWL) 4 days after epidermal injury in Treg cell-depleted mice treated with either α-CXCL5 or isotype control antibody compared to injured Treg cell-sufficient cntrl mice.
(d) Lgr5-tdTom-FoxP3DTR mice were treated as in Figure 2a and co-administered α-CXCL5 or isotype control mAb with DT on days 0,1 and 3 of recovery. Mice were harvested on day 4. Representative IF images of Lgr5-derived cell (tdTomato+) localization in the epidermis compared to Treg cell-sufficient Lgr5-tdTom-FoxP3DTR mice. (e) Quantification of Lgr5-derived cell localization in the epidermis of the indicated strains and conditions 4 days after skin injury. tdTomato+/ HF and tdTomato+/hpf are cell numbers normalized per hair follicle and per high power field respectively
(f) Representative plots and
(g) quantification of CD34+ HFSCs in the epidermis of Cntrl and FoxP3DTR mice treated with α-CXCL5 or isotype control. For all relevant panels, error bars are +/−SEM. *p < 0.05. Results of panels a-f are representative of 3–5 independent experiments (n=2–4 mice per group). Results of g are pooled data from 3 independent experiments. Scale bar in d is 100µm; Red- tdTomato+ Lgr5 stem cell progeny; Blue – DAPI.
IL-17A can promote CXCL5 expression in damaged epithelial tissues and promotes neutrophil recruitment during inflammation. (Balamayooran et al., 2012; Guilloteau et al., 2010; Mei et al., 2012) Thus, in addition to CXCL5, we hypothesized that regulation of Th17 cells and neutrophils play a role in Treg cell-mediated augmentation of HFSC function during barrier regeneration. To test this, the epidermis of Lgr5-tdTom-Foxp3DTR mice was disrupted, Lgr5 cells were labeled with tamoxifen, and Treg cells were depleted as in Figure 4a. On the days of DT administration, mice were either co-injected with an IL-17A neutralizing mAb (α-IL-17) or a neutrophil depleting mAb (α-Gr1). Mice were harvested on day 4 and compared to littermate controls either sufficient or depleted of Treg cells. IL-17A neutralization or co-depletion of neutrophils in Treg cell-depleted mice partially rescued the migration defect of Lgr5-derived cells into the IFE and TEWL during epidermal repair (Supplementary Figure 6 c–e). Taken together, our results suggest that a dominant function of Treg cells during epidermal barrier repair is to regulate the CXCL5-IL-17A-neutrophil axis.
Discussion:
Our results demonstrated that Treg cells play a vital role in restoring skin barrier integrity after epidermal injury. These data add to a growing body of work demonstrating a critical role for Treg cells in tissue regeneration following acute injury. Others have shown that Treg cells accumulate in damaged skeletal muscle (Burzyn et al., 2013; Castiglioni et al., 2015; Panduro et al., 2018; Villalta et al., 2014), lung (Arpaia et al., 2015), heart (Dobaczewski et al., 2010; Weirather et al., 2014) and central nervous system (Dombrowski et al., 2017) following injury. These cells act to both limit inflammation and promote tissue healing through the secretion of regenerative factors (Burzyn et al., 2013).
Here, we highlight a mechanism by which Treg cells promoted an alternative fate decision for HFSCs, normally poised for hair generation, to differentiate into stratified epithelium necessary for skin barrier repair. While the activating and inhibitory signals that influence HFSC function during hair generation are well characterized (Morrison and Spradling, 2008), the specific mechanisms that govern HFSC differentiation during epithelial repair are less well defined. Signals that balance HFSC quiescence and differentiation come from micro-environmental cues from a broad array of cell types within the stem cell environment. (Brownell et al., 2011; Festa et al., 2011; Hsu et al., 2011; Keyes et al., 2013) We demonstrate that Treg cells, like other cells within the HF stem cell niche, regulate key SC functions. This was in part achieved by fine-tuning immune cell composition in the skin after barrier injury. We observed that neutralization of CXCL5, IL-17A and neutrophil depletion partially restored barrier function and Lgr5-derived cell migration into the IFE in the absence of Treg cells. The fact that these processes were not completely restored suggests that multiple partially redundant mechanisms may be at play and/or antibody-mediated neutralization of CXCL5 or IL-17A may be incomplete. Nevertheless, our findings highlight a dominant role for Treg cells in suppressing the CXCL5/Th17/neutrophil axis to facilitate HFSC lineage commitment after epidermal injury.
Treg cells that reside in non-lymphoid organs promote tissue homeostasis by controlling inflammation, a ‘traditional’ function of Treg cells. However, these cells can function through mechanisms that are independent of their ability to regulate immune cells (Burzyn et al., 2013). Recently, we have shown that Treg cells in skin facilitate HFSC differentiation during hair regeneration (Ali et al., 2017). In this relatively non-inflammatory context, Treg cells promote ‘classical’ HFSC differentiation towards hair follicle keratinocyte lineages, at least in part, through direct interactions with HFSCs. In contrast to hair follicle cycling, epidermal injury is highly inflammatory. In this context, Treg cells regulated a specific inflammatory module mediated by CXCL5 and promoted HFSC differentiation toward IFE keratinocytes. Taken together, these results suggest that Treg cells in peripheral tissues can utilize multiple mechanisms to influence stem cell fate commitment depending upon the inflammatory context and specific demands of the tissue. Whether Treg cells directly interact with other lymphocytes, myeloid cells and/or keratinocytes to influence HFSC fate during epidermal regeneration is currently unknown and is the focus of future investigations.
While our results show that Treg cells diverted HFSC lineage commitment to repair the epidermis after injury, we cannot completely exclude a role for Treg cells in influencing the IFE or other stem / progenitor cell compartments during barrier regeneration. In addition to suppressing CXCL5 expression, it is possible that Treg cells promote the secretion of specific factors from IFE cells that act to mediate HFSC migration out of the bulge after epidermal injury. In addition, it is likely that Treg cells act on other stem cell populations in skin, such as IFE stem cells, that may act immediately after epidermal injury. To determine how Treg cells globally influence regenerative programs in the skin, future studies should be aimed at systematically dissecting the heterogeneity of skin Treg cells in epithelial repair programs. It may be that Treg cells in skin are relatively homogenous and the same cells utilize different mechanisms to mediate different functions in this tissue in different contexts (i.e., HF cycling vs. epidermal repair). Alternatively, it may be that Treg cells in skin are heterogeneous and different subsets of cells mediate different functions in specific settings. Currently, we do not have a comprehensive appreciation of the heterogeneity of Treg cells in murine skin and thus can only speculate on how these cells utilize different mechanisms to influence epithelial stem cell function. Nevertheless, our data clearly shows a role for these cells in controlling a specific pathway of immune activation that facilitated HFSC plasticity after skin injury, which was necessary to promptly re-establish barrier function in skin.
Pharmacologic and cell-based therapeutic approaches to augment Treg cells are promising new modalities to treat human autoimmune and chronic inflammatory diseases.(Bluestone et al., 2006; Spence et al., 2015) Several skin diseases, such atopic dermatitis, are characterized by both chronic tissue inflammation and compromised skin barrier function. Interestingly, patients with dysfunctional Treg cells develop skin disease that closely resemble atopic dermatitis.(Halabi-Tawil et al., 2009; Martín-Santiago et al., 2013; Nieves et al., 2004) Our results suggest that Treg cell augmentation may have benefit in both suppressing skin inflammation and restoring epidermal barrier function in patients with atopic dermatitis and similar skin disorders.
STAR METHODS:
CONTACT FOR REAGENT AND RESOURCE SHARING:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael Rosenblum (Michael.Rosenblum@ucsf.edu)
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Mice.
Wild type C57BL/6 and FoxP3DTR mice were purchased from The Jackson Laboratory. The generation of Lgr5-EGFP-Ires-CreERT2 mice and the Rosa26-CAG-LSL-tdTomato are described elsewhere.(Barker et al., 2007; Madisen et al., 2010) These two strains were crossed and resultant Lgr5-tdTom transgenic mice were provided by the laboratory of Ophir Klein (Plaks et al., 2013). Lgr5-tdTom mice were bred to FoxP3DTR mice to develop the Lgr5-tdTom-FoxP3DTR strain. Animal experiments were performed on 7–12 week old mice. Mice were maintained through routine breeding at the University of California San Francisco (UCSF) School of Medicine in a specific pathogen free facility. All animal experiments were performed in accordance with guidelines established by Laboratory Animal Resource Center (LARC) at UCSF and all experimental plans and protocols were approved by IACUC beforehand.
METHOD DETAILS
Mechanical injury of mouse skin and transepidermal water loss measurements.
Mice backs were shaved and rested for 18–24 hours. Baseline transepidermal water loss (TEWL; Tewameter TM 300, Khazaka Electronic) was measured on four quadrants of back skin. Mechanical injury was applied by tape stripping (Shurtape) 5–7 times per day for three consecutive days to achieve a TEWL measurement between 40–75 gmH20/m2/hr averaged over four quadrants of back skin by the last day of epidermal injury (day 0). During the skin recovery phase, TEWL measurements were taken every other day for up to eleven days. Mice were sedated with isofluorane during shaving, mechanical injury, and TEWL measurements.
Administration of diphtheria toxin, neutralizing mAbs, and tamoxifen.
Treg cells were depleted from FoxP3DTR or Lgr5-tdTom-FoxP3DTR mice by i.p. injection of DT (30–50 mg/kg body weight; Sigma-Aldrich) on two consecutive days (day 0 and day 1 of recovery) and every other day (Kim et al., 2007) for up to four injections. Tissues were harvested on the indicated days as described in Results. Mice were compared to age and gender matched DT-treated WT mice or non-DT treated littermates. In experiments using Lgr5-tdTom-FoxP3DTR mice, Cre recombinase was activated with tamoxifen (Sigma-Aldrich; 2.5 mg i.p. dissolved in corn oil) on day −1 and 0. Mice were treated with a rat anti-mouse CXCL5 monoclonal neutralizing antibody (R&D systems; mAb 433) (Rousselle et al., 2013) rat anti-mouse Gr1 monoclonal neutralizing antibody (Sigma; RB6–685), rat anti-mouse IL-17A monoclonal neutralizing antibody (R&D systems; mAb 421)(Choi et al., 2016) or rat IgG2B isotype control (R&D systems; mAb 0061). Each mouse was administered 40 µg of the indicated antibody by i.p. injection on days 0,1 and 3 and harvested on day 4.
Cell preparation from tissues and stimulation for intracellular cytokine staining.
Single-cell suspensions of skin draining lymph nodes were mechanically dissociated through a 100 µm filter and 2.5 cm2 dorsal skin was processed as previously described.(Gratz et al., 2014) Single cells were washed in tissue culture media and filtered. Cells were counted using an automated cell counter (NucleoCounter NC 200; Chemomtec) to determine the absolute number of specific cell populations per unit area of skin by flow cytometry. 2–3 × 106 single cells were stained for flow cytometry or cultured for intracellular cytokine staining using a PMA & ionomycin cell stimulation cocktail (Tonbo Biosciences). For experiments using epidermal cell suspensions, 2.5 cm2 of back skin was harvested. The skin was mechanically defatted using forceps. Skin was placed dermis side down in a well of a 6-well tissue culture plate with 1.2 mL of Trypsin (0.5%; Gibco) and placed in a 37° C CO2 incubator for 1 hour. The epidermis was gently disassociated from the underlying dermis using forceps. Single cells were filtered, counted, and stained for flow cytometry.
Flow cytometry.
Single-cell suspensions prepared above were pelleted and incubated with anti-CD16/anti-CD32 (UCSF Antibody Core Facility or BD Bioscences; 2.4G2) in PBS. Cells were washed and stained with Ghost Viability dye (Tonbo Biosciences) in PBS. Following a wash in PBS, cells were stained for surface markers in PBS containing 2% FCS. For intracellular staining, cells were fixed and permeabilized with a FoxP3 buffer set (eBioscences). Samples were run on a Fortessa analyzer (BD Biosciences) in the UCSF Flow Cytometry Core and collected using FACS Diva software (BD Biosciences). Flow cytometry data were analyzed using FlowJo software (Treestar). Fluorophore-conjugated antibodies specific for mouse surface and intracellular antigens were purchased from eBiosciences, BD Biosciences and Biolegend and as detailed in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Mouse/Rat FoxP3 eFluor 450 | eBioscience | Cat# 48–5773-82 |
| Anti-Human/Mouse Ki-67 PE-Cy7 | BD Biosciences | Cat # 561283 |
| Rabbit anti-GFP | Invitrogen | Cat# A11122 |
| Anti-mouse/human ITGA6 | BD Biosciences | Cat# 555734 |
| Anti-Mouse CD45 Alexa Fluor 700 | eBioscience | Cat# 56–0451-82 |
| Brilliant Violet 605 anti-mouse Ly-6A/E (Sca-1) Antibody | Biolegend | Cat# 108133 |
| Anti-Mouse CD34 Alexa Fluor 647 | BD Biosciences | Cat# 560233 |
| Anti-Mouse CD326 (EpCAM) APC-eFluor 780 | eBioscience | Cat# 47–5791-82 |
| Anti-Human/Mouse CD49f (Integrin a 6) FITC | eBioscience | Cat# 11–0495-82 |
| Anti-Mouse TCR gamma delta PerCP-Cy 5.5 | Biolegend | Cat# 118117 |
| Brilliant Violet 605 anti-mouse CD8a Antibody | Biolegend | Cat # 100743 |
| Brilliant Violet 650 anti-mouse CD4 Antibody | Biolegend | Cat# 100545 |
| Anti-Mouse CD11b APC eFluor 780 | eBioscience | Cat# 47–0118-42 |
| Brilliant Violet 650 anti-mouse CD11c Antibody | Biolegend | Cat# 117339 |
| Anti-Mouse Ly-6G PE-Cy7 | BD Biosciences | Cat# 560601 |
| Brilliant Violet 605 anti-mouse Ly-6C Antibody | Biolegend | Cat# 128035 |
| Purified Rat Anti-Mouse CD16/CD32 | BD Biosciences | Cat# 553141 |
| Anti-Mouse IFN-gamma FITC | BD Biosciences | Cat #554411 |
| Anti-Mouse MHC Class II (I-A/I-E) eFluor 450 | eBioscience | Cat # 48–5321-80 |
| Anti-Mouse IL-5 PE | BD Biosciences | Cat # 554395 |
| Anti-Mouse IL-13 eFluor 450 | eBiosciences | Cat # 48–7133-82 |
| Anti-Mouse IL-17A Pe-Cy7 | Biolegend | Cat # 506921 |
| Living Colors DsRed Polyclonal Antibody | Clontech | Cat # 632496 |
| Goat anti-Rabbit Alexa 555 F’ab Fragment IgG | Invitrogen | Cat # A-21430 |
| Anti-Mouse Ki-67 FITC | eBiosciences | Cat # 11–5698-82 |
| In vivo mAb anti-CXCL5 | R&D Systems | Cat # mAb 433 |
| In vivo mAb anti-Gr1 | Sigma-Aldrich | Cat # MABF474 |
| In vivo mAb anti-IL-17A | R&D Systems | Cat # mAb 421 |
| In vivo mAb rat IgG2B isotype control | R&D Systems | Cat # mAb 0061 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase from Clostridium histolyticum, Type XI | Sigma-Aldrich | Cat # C9407 |
| DNase | Sigma-Aldrich | Cat # DN25 |
| Hyaluronidase from bovine testes | Sigma-Aldrich | Cat # H3506 |
| Cell Stimulation Cocktail (500X) | Tonbo Biosciences | Cat # TNB-4975 |
| Diphtheria Toxin from Corynebacterium diphtheriae | Sigma-Aldrich | Cat # D0564 |
| Shurtape | Adhesive Tape | Amazon.com |
| Tamoxifen | Sigma-Aldrich | Cat # T5648 |
| Ghost Dye™ Violet 510 Live/Dead Stain | Tonbo Biosciences | Cat #13–0870-T100 |
| 0.5% Trypsin-EDTA (10x), no phenol red | ThermoFisher | Cat # 1540054 |
| Critical Commercial Assays | ||
| PureLink® RNA Mini Kit | ThermoFisher | Cat # 12183018A |
| RNeasy® Fibrous Tissue Kit | Qiagen | Cat # 74704 |
| iScript Kit Advanced cDNA Synthesis Kit for RT-qPCR | Bio-Rad | Cat #1725038 |
| SsoAdvanced™ Universal SYBR® Green Supermix | Bio-Rad | Cat # 1725270 |
| TaqMan Gene Expression Master Mix | ThermoFisher | Cat# 4369510 |
| Cxcr2 TaqMan Assay | ThermoFisher | Cat # Mm00438258_m1 |
| Cxcl5 TaqMan Assay | ThermoFisher | Cat # Mm00436451_g1 |
| Actb TaqMan Assay | ThermoFisher | Cat # Mm01205647_g1 |
| Beta 2 microglobulin TaqMan Assay | ThermoFisher | Cat # Mm00437762_m1 |
| Gapdh TaqMan Assay | ThermoFisher | Cat # Mm00484668_m1 |
| RT2 Profiler PCR Array | Qiagen | Cat # PAMM-150Z |
| Deposited Data | ||
| Raw Data Files for RNA-sequencing of Lgr5-derived cells | NCBI Gene Expression Omnibus | GEO: TBD |
| Experimental Models: Organisms/Strains | ||
| B6.129(Cg)-Foxp3tm3(DTR/GFP)Ayr/J (Foxp3DTR) | The Jackson Laboratory | Cat # 016958 |
| B6.Cg-Foxp3tm2Tch/J (Foxp3GFP) | The Jackson Laboratory | Cat # 006772 |
| C57BL/6J wild type (WT) | The Jackson Laboratory | Cat # 000664 |
| B6.Lgr5-EGFP-IRES-CreERT2/Rosa26-tdTomato | Klein Lab | (Plaks et al., 2013) |
| Oligonucleotides | ||
| involucrin For: 5’–ATGTCCCATCAACACACACTG - 3’; Rev: 5’-TGGAGTTGGTTGCTTTGCTTG-3’ | IDT | This paper |
| loricrin (For: 5’ - GCGGATCGTCCCAACAGTATC −3’ ; Rev: 5’ –TGAGAGGAGTAATAGCCCCCT - 3’) | IDT | This paper |
| filaggrin (For: 5’ – CTAGAGGGCATGAGTGTAGTCA – 3’ Rev: 5’ - CAAGACTGGACAGTTGGCTGG – 3’); | IDT | This paper |
| keratin 1 (For: 5’ – GAGCAGATCAAGTCACTCAATGA - 3’; Rev: 5’ - CCCATT TGGTTTGTAGCACCT - 3’) | IDT | This paper |
| Software and Algorithms | ||
| TopHat | (Trapnell et al., 2009) | https://ccb.jhu.edu/software/tophat/index.shtml |
| SAMtools | (Li et al., 2009) | http://www.htslib.org |
| DESeq2 | (Anders and Huber, 2010) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| HTSeq | (Anders et al., 2015) | http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html |
| R Statistical Computing Software | The R Foundation | https://www.r-project.org/ |
| GraphPad Prism | GraphPad Software, Inc. | http://www.graphpad.com/scientific-software/prism/ |
| FlowJo | FlowJo, LLC | https://www.flowjo.com/solutions/flowjo |
| RT2 Profiler Array Online Software | Qiagen | https://dataanalysis.sabiosciences.com/pcr/arrayanalysis |
| Spatial signature patterns of Lgr5-derived cells | (Joost et al., 2016) | http://linnarssonlab.org/epidermis |
| ImageJ | NIH | Version 1.50i |
RNA sequencing analysis and qRT-PCR.
For the analysis of barrier function genes and chemokines, 30 mg of back skin was homogenized in a tissue lyser (gentleMACS; Miltenyi Biotec). RNA was isolated with the RNeasy fibrous tissue kit (Qiagen) and used to synthesize cDNA with the iScript cDNA synthesis kit (Biorad). Message levels of barrier function genes were determined using a SYBER Green assay (SSo Advanced Universal SYBER kit; Biorad). Cycle number of duplicate or triplicate samples were normalized to the expression of the endogenous control β2m. Primer sequences for the expression of barrier function and control genes are as follows: involucrin (For: 5’–ATGTCCCATCAACACACACTG - 3’; Rev: 5’-TGGAGTTGGTTGCTTTGCTTG-3’); loricrin (For: 5’ - GCGGATCGTCCCAACAGTATC −3’ ; Rev: 5’ –TGAGAGGAGTAATAGCCCCCT - 3’); filaggrin (For: 5’ – CTAGAGGGCATGAGTGTAGTCA – 3’ Rev: 5’ - CAAGACTGGACAGTTGGCTGG – 3’); keratin 1 (For: 5’ – GAGCAGATCAAGTCACTCAATGA - 3’; Rev: 5’ - CCCATT TGGTTTGTAGCACCT - 3’); β2m (For: 5’ – TTCTGGTGCTTGTCTCACTGA – 3’; Rev 5’ – CAGTATGTTCGGCTTCCCATTC - 3’). A mouse chemokine expression array (Qiagen; RT2 Prolifer PCR Array PAMM150-Z) was used in experiments to detect chemokine expression patterns from skin. All other real time qPCR experiments utilized FAM labelled gene expression reagents from Thermo Fisher / Applied Biosystems. For RNA-sequencing of Lgr5-derived cells, tdTomato+ sorted cell populations from epidermal preparations were flash frozen in liquid nitrogen and sent overnight on dry ice to Expression Analysis, Quintiles (Morrisville, NC). RNA samples were converted into cDNA libraries using the Illumina TruSeq Stranded mRNA sample preparation kit. (Illumina). RNA was isolated by Expression Analysis using Qiagen RNeasy Spin Column and was quantified via Nanodrop ND-8000 spectrophotometer. RNA quality was checked by Agilent Bioanalyzer Pico Chip. cDNA was created from 220 pg of input RNA with the SMARTer Ultra Low input kit and sequenced to a 25M read depth with Illumina RNASeq. Reads were aligned to Ensembl mg GRCm38.p4 reference genome with TopHat software (v. 2.0.12). SAM files were generated with SAMtools from alignment results. Read counts were obtained with htseq-count (0.6.1p1) with the union option. Differential expression was determined using the R/Bioconducter package DESeq2. All differentially expressed genes (> 2 fold; p<0.05) were sequentially evaluated for specific localization patterns of expression within the bulge region, interfollicular epidermis or upper hair follicle(Joost et al., 2016) (http://linnarssonlab.org/epidermis). Using this resource, all genes were sequentially probed and aligned to spatial expression patterns described in this manuscript. Our gene set was grossly partitioned into IFE/UHF versus bulge-associated genes.
Histology and immunofluorescence microscopy.
For histopathology, skin tissue was fixed in 10% formalin and paraffin-embedded. Tissue was stained with hematoxylin and eosin (H&E) by the University of California San Francisco Mouse Pathology Core. H&E quantifications of epidermal hyperplasia were performed using ImageJ64 software (NIH, USA). For immunofluorescent tissue staining, dorsal skin from FoxP3GFP, Lgr5-tdTom, or Lgr5-tdTom-FoxP3DTR mice was first fixed in 2% PFA for 4 hours, washed with PBS and left in 30% sucrose overnight before embedding in OCT and freezing in a cooled isopentane solution. 10 or 12 µm sections were prepared on SuperFrost slides (VWR). For detection of tdTomato+ cells, slides were stained with rabbit dsRed Polyclonal antibody at 1:500 (Clontech, 632496). Primary signal was amplified with Goat anti-Rabbit Alexa 555 F’ab Fragment IgG (1:500; Invitrogen). For Ki-67 staining, slides were stained with an anti-Ki-67 monoclonal Ab at 1:50 (eBiosciences, SolA15). Slides were then washed in PBS and mounted with DAPI containing medium before imaging on a standard fluorescent microscope.
QUANTIFCATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using Prism software package 6.0 (GraphPad). p values were calculated using two-tailed unpaired or paired Student’s t-test or one-way ANOVA and as indicated in the Figure Legends. Mice cohort size was designed to be sufficient to enable accurate determination of statistical significance and no animals were excluded from the statistical analysis. Mice were assigned to treatment or control groups randomly. All in vivo experiments used at least two independent cohorts. RNA-seq experiments were conducted using 2–4 biological samples (as indicated in figure legends) from the indicated cohorts.
DATA AND SOFTWARE AVAILABILITY
NA sequencing data has been deposited in NCBI.
Supplementary Material
Highlights.
Treg cells promote epidermal regeneration after injury
Treg cells control a CXCL5-IL-17 axis of inflammation during epidermal repair
Treg cell control of CXCL5 and IL-17 diverts HFSC differentiation toward IFE cells
CXCL5 or IL-17 neutralization restores HFSC differentiation in Treg depleted mice
Acknowledgements:
The authors thank Jason Cyster and Mark Kaplan for their critical review of the manuscript, Ophir Klein for providing Lgr5-tdTom mice, and Carlos Benetiz for assistance with animal husbandry. Flow Cytometry data was generated in the UCSF Parnassus Flow Cytometry Core which is supported by the Diabetes Research Center (DRC) grant, NIH P30 DK063720. Histology was performed with assistance from the UCSF Mouse Pathology Core which is supported by NIH 5P30CA082103–15. A.N.M is supported by a Dermatology Foundation Career Development Award and NIH-K08 AR070910. This work was primarily funded by M.D.R.’s grants: NIH K08-AR062064, NIH DP2-AR068130, Burroughs Wellcome Fund CAMS-1010934, and NIH R21-AR066821.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
References:
- Ali N, Zirak B, Rodriguez RS, Pauli ML, Truong H-A, Lai K, Ahn R, Corbin K, Lowe MM, Scharschmidt TC, et al. (2017). Regulatory T Cells in Skin Facilitate Epithelial Stem Cell Differentiation. Cell 169, 1119–1129.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinforma. Oxf. Engl 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Green JA, Moltedo B, Arvey A, Hemmers S, Yuan S, Treuting PM, and Rudensky AY (2015). A Distinct Function of Regulatory T Cells in Tissue Protection. Cell 162, 1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurora AB, and Olson EN (2014). Immune modulation of stem cells and regeneration. Cell Stem Cell 15, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamayooran G, Batra S, Cai S, Mei J, Worthen GS, Penn AL, and Jeyaseelan S (2012). Role of CXCL5 in leukocyte recruitment to the lungs during secondhand smoke exposure. Am. J. Respir. Cell Mol. Biol 47, 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. [DOI] [PubMed] [Google Scholar]
- Blanpain C, and Fuchs E (2006). Epidermal stem cells of the skin. Annu. Rev. Cell Dev. Biol 22, 339–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluestone JA, St Clair EW, and Turka LA (2006). CTLA4Ig: bridging the basic immunology with clinical application. Immunity 24, 233–238. [DOI] [PubMed] [Google Scholar]
- Brownell I, Guevara E, Bai CB, Loomis CA, and Joyner AL (2011). Nerve-derived sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells. Cell Stem Cell 8, 552–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyn D, Benoist C, and Mathis D (2013). Regulatory T cells in nonlymphoid tissues. Nat. Immunol 14, 1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglioni A, Corna G, Rigamonti E, Basso V, Vezzoli M, Monno A, Almada AE, Mondino A, Wagers AJ, Manfredi AA, et al. (2015). FOXP3+ T Cells Recruited to Sites of Sterile Skeletal Muscle Injury Regulate the Fate of Satellite Cells and Guide Effective Tissue Regeneration. PloS One 10, e0128094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho RM, Squarize CH, Chodosh LA, Williams BO, and Gutkind JS (2009). mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell 5, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, Hoeffer CA, Littman DR, and Huh JR (2016). The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 351, 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobaczewski M, Xia Y, Bujak M, Gonzalez-Quesada C, and Frangogiannis NG (2010). CCR5 signaling suppresses inflammation and reduces adverse remodeling of the infarcted heart, mediating recruitment of regulatory T cells. Am. J. Pathol 176, 2177–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doles J, Storer M, Cozzuto L, Roma G, and Keyes WM (2012). Age-associated inflammation inhibits epidermal stem cell function. Genes Dev 26, 2144–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski Y, O’Hagan T, Dittmer M, Penalva R, Mayoral SR, Bankhead P, Fleville S, Eleftheriadis G, Zhao C, Naughton M, et al. (2017). Regulatory T cells promote myelin regeneration in the central nervous system. Nat. Neurosci 20, 674–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias PM (2005). Stratum corneum defensive functions: an integrated view. J. Invest. Dermatol 125, 183–200. [DOI] [PubMed] [Google Scholar]
- Festa E, Fretz J, Berry R, Schmidt B, Rodeheffer M, Horowitz M, and Horsley V (2011). Adipocyte lineage cells contribute to the skin stem cell niche to drive hair cycling. Cell 146, 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz IK, Truong H-A, Yang SH-Y, Maurano MM, Lee K, Abbas AK, and Rosenblum MD (2013). Memory regulatory T cells require IL-7 and not IL-2 for their maintenance in peripheral tissues. J. Immunol. Baltim. Md 1950 190, 4483–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz IK, Rosenblum MD, Maurano MM, Paw JS, Truong H-A, Marshak-Rothstein A, and Abbas AK (2014). Cutting edge: Self-antigen controls the balance between effector and regulatory T cells in peripheral tissues. J. Immunol. Baltim. Md 1950 192, 1351–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio J, Meller S, Conrad C, Di Nardo A, Homey B, Lauerma A, Arai N, Gallo RL, Digiovanni J, and Gilliet M (2010). Plasmacytoid dendritic cells sense skin injury and promote wound healing through type I interferons. J. Exp. Med 207, 2921–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilloteau K, Paris I, Pedretti N, Boniface K, Juchaux F, Huguier V, Guillet G, Bernard F-X, Lecron J-C, and Morel F (2010). Skin Inflammation Induced by the Synergistic Action of IL-17A, IL-22, Oncostatin M, IL-1{alpha}, and TNF-{alpha} Recapitulates Some Features of Psoriasis. J. Immunol. Baltim. Md 1950. [DOI] [PubMed] [Google Scholar]
- Halabi-Tawil M, Ruemmele FM, Fraitag S, Rieux-Laucat F, Neven B, Brousse N, De Prost Y, Fischer A, Goulet O, and Bodemer C (2009). Cutaneous manifestations of immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome. Br. J. Dermatol 160, 645–651. [DOI] [PubMed] [Google Scholar]
- Hsu Y-C, Pasolli HA, and Fuchs E (2011). Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, and Cotsarelis G (2008). Is the hair follicle necessary for normal wound healing? J. Invest. Dermatol 128, 1059–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, and Cotsarelis G (2005). Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med 11, 1351–1354. [DOI] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, and Toftgard R (2008). Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet 40, 1291–1299. [DOI] [PubMed] [Google Scholar]
- Jia H, Sodhi CP, Yamaguchi Y, Lu P, Martin LY, Good M, Zhou Q, Sung J, Fulton WB, Nino DF, et al. (2016). Pulmonary Epithelial TLR4 Activation Leads to Lung Injury in Neonatal Necrotizing Enterocolitis. J. Immunol. Baltim. Md 1950 197, 859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Oyoshi MK, Le Y, Bianchi T, Koduru S, Mathias CB, Kumar L, Le Bras S, Young D, Collins M, et al. (2009). IL-21R is essential for epicutaneous sensitization and allergic skin inflammation in humans and mice. J. Clin. Invest 119, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joost S, Zeisel A, Jacob T, Sun X, La Manno G, Lonnerberg P, Linnarsson S, and Kasper M (2016). Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity. Cell Syst 3, 221–237.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, and Clevers H (2016). Reparative inflammation takes charge of tissue regeneration. Nature 529, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes BE, Segal JP, Heller E, Lien W-H, Chang C-Y, Guo X, Oristian DS, Zheng D, and Fuchs E (2013). Nfatc1 orchestrates aging in hair follicle stem cells. Proc. Natl. Acad. Sci. U. S. A 110, E4950–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Rasmussen JP, and Rudensky AY (2007). Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol 8, 191–197. [DOI] [PubMed] [Google Scholar]
- Levy V, Lindon C, Zheng Y, Harfe BD, and Morgan BA (2007). Epidermal stem cells arise from the hair follicle after wounding. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 21, 1358–1366. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinforma. Oxf. Engl 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Santiago A, Hervás JA, Hervás D, Rosell A, Caimari M, de Carlos JC, and Matamoros N (2013). Diagnostic value of the skin lesions in immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome. Pediatr. Dermatol 30, e221–222. [DOI] [PubMed] [Google Scholar]
- Mei J, Liu Y, Dai N, Hoffmann C, Hudock KM, Zhang P, Guttentag SH, Kolls JK, Oliver PM, Bushman FD, et al. (2012). Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J. Clin. Invest 122, 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, and Spradling AC (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao K, Kobayashi T, Moro K, Ohyama M, Adachi T, Kitashima DY, Ueha S, Horiuchi K, Tanizaki H, Kabashima K, et al. (2012). Stress-induced production of chemokines by hair follicles regulates the trafficking of dendritic cells in skin. Nat. Immunol 13, 744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves DS, Phipps RP, Pollock SJ, Ochs HD, Zhu Q, Scott GA, Ryan CK, Kobayashi I, Rossi TM, and Goldsmith LA (2004). Dermatologic and immunologic findings in the immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome. Arch. Dermatol 140, 466–472. [DOI] [PubMed] [Google Scholar]
- Panduro M, Benoist C, and Mathis D (2018). Treg cells limit IFN-γ production to control macrophage accrual and phenotype during skeletal muscle regeneration. Proc. Natl. Acad. Sci. U. S. A 115, E2585–E2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaks V, Brenot A, Lawson DA, Linnemann JR, Van Kappel EC, Wong KC, de Sauvage F, Klein OD, and Werb Z (2013). Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep 3, 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SP, Langan-Fahey SM, Johnson DA, and Jordan VC (1991). Metabolites, pharmacodynamics, and pharmacokinetics of tamoxifen in rats and mice compared to the breast cancer patient. Drug Metab. Dispos. Biol. Fate Chem 19, 36–43. [PubMed] [Google Scholar]
- Rousselle A, Qadri F, Leukel L, Yilmaz R, Fontaine J-F, Sihn G, Bader M, Ahluwalia A, and Duchene J (2013). CXCL5 limits macrophage foam cell formation in atherosclerosis. J. Clin. Invest 123, 1343–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Rodriguez R, Pauli ML, Neuhaus IM, Yu SS, Arron ST, Harris HW, Yang SH-Y, Anthony BA, Sverdrup FM, Krow-Lucal E, et al. (2014). Memory regulatory T cells reside in human skin. J. Clin. Invest 124, 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano S, Chan KS, Carbajal S, Clifford J, Peavey M, Kiguchi K, Itami S, Nickoloff BJ, and DiGiovanni J (2005). Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat. Med 11, 43–49. [DOI] [PubMed] [Google Scholar]
- Scharschmidt TC, Vasquez KS, Truong H-A, Gearty SV, Pauli ML, Nosbaum A, Gratz IK, Otto M, Moon JJ, Liese J, et al. (2015). A Wave of Regulatory T Cells into Neonatal Skin Mediates Tolerance to Commensal Microbes. Immunity 43, 1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharschmidt TC, Vasquez KS, Pauli ML, Leitner EG, Chu K, Truong H-A, Lowe MM, Sanchez Rodriguez R, Ali N, Laszik ZG, et al. (2017). Commensal Microbes and Hair Follicle Morphogenesis Coordinately Drive Treg Migration into Neonatal Skin. Cell Host Microbe 21, 467–477.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence A, Klementowicz JE, Bluestone JA, and Tang Q (2015). Targeting Treg signaling for the treatment of autoimmune diseases. Curr. Opin. Immunol 37, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinforma. Oxf. Engl 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, and Fuchs E (2004). Defining the Epithelial Stem Cell Niche in Skin. Science 303, 359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalta SA, Rosenthal W, Martinez L, Kaur A, Sparwasser T, Tidball JG, Margeta M, Spencer MJ, and Bluestone JA (2014). Regulatory T cells suppress muscle inflammation and injury in muscular dystrophy. Sci. Transl. Med 6, 258ra142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirather J, Hofmann UDW, Beyersdorf N, Ramos GC, Vogel B, Frey A, Ertl G, Kerkau T, and Frantz S (2014). Foxp3+ CD4+ T cells improve healing after myocardial infarction by modulating monocyte/macrophage differentiation. Circ. Res 115, 55–67. [DOI] [PubMed] [Google Scholar]
- Wilson CH, Gamper I, Perfetto A, Auw J, Littlewood TD, and Evan GI (2014). The kinetics of ER fusion protein activation in vivo. Oncogene 33, 4877–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NA sequencing data has been deposited in NCBI.