Abstract
Due to the invasive procedure associated with Pap smears for diagnosing cervical cancer and the conservative culture of developing countries, identifying less invasive biomarkers is of great interest. Quantitative label-free mass spectrometry was performed to identify potential biomarkers in the urine samples of patients with cervical cancer. This technique was used to study the differential expression of urinary proteomes between normal individuals and cancer patients. The alterations in the levels of urinary proteomes in normal and cancer patients were analyzed by Progenesis label-free software and the results revealed that 60 proteins were upregulated while 73 proteins were downregulated in patients with cervical cancer. This method could enrich high molecular weight proteins from 100 kDa. The protein-protein interactions were obtained by Search Tool for the Retrieval of Interacting Genes/Proteins analysis and predicted the biological pathways involving various functions including cell-cell adhesion, blood coagulation, metabolic processes, stress response and the regulation of morphogenesis. Two notable upregulated urinary proteins were leucine-rich α-2-glycoprotein (LRG1) and isoform-1 of multimerin-1 (MMRN1), while the 3 notable downregulated proteins were S100 calcium-binding protein A8 (S100A8), serpin B3 (SERPINB3) and cluster of differentiation-44 antigen (CD44). The validation of these 5 proteins was performed by western blot analysis and the biomarker sensitivity of these proteins was analyzed individually and in combination with receiver operator characteristic curve (ROC) analysis. Quantitative mass spectrometry analysis may allow for the identification of urinary proteins of high molecular weight. The proteins MMRN1 and LRG1 were presented, for the first time, to be highly expressed urinary proteins in cervical cancer. ROC analysis revealed that LRG1 and SERPINB3 could be individually used, and these 5 proteins could also be combined, to detect the occurrence of cervical cancer.
Keywords: urinary biomarkers, cervical cancer, label-free, leucine-rich α-2-glycoprotein, multimerin-1, S100A8, serpin B3, cluster of differentiation 44 antigen
Introduction
Cervical cancer is the fourth most frequent cause of mortality in women worldwide (1). It has a prevalence of 20% in developed countries, whereas there is a high incidence of invasive cases in developing countries, including Thailand, due to the lack of cancer prevention strategies, leading to ~90% cervical cancer-associated mortalities (2–4). This type of cancer can take from one to numerous years to progress from a precancerous state to cancer since it is a slow progressive disease. The precancerous state is known as cervical intraepithelial neoplasia (CIN) and is categorized according to the cell abnormality levels (CIN I, II and III) (5). Human papillomavirus (HPV) is the main etiological factor of cervical cancer (6). It can be classified into many types; the high-risk HPV types (including HPV 16, 18 and 52) are commonly associated with cervical cancer.
In the clinic, there are several methods of treatment for cervical cancer, including surgery (7,8), chemotherapy (hormone), radiotherapy or various combinations of these methods (9–12). However, the mortality rate of cervical cancer is still high (13). Currently, several screening tests for cervical cancer are available, including conventional Pap smears, liquid-based cytology and HPV DNA testing. From previous studies, a combination of liquid-based cytology and DNA testing has demonstrated a high sensitivity in cervical cancer screening. However, this technique requires cervical swabs, which are complicated and highly invasive when compared with the collection of urine. Therefore, a less invasive protocol is required for diagnosis. The detection of urinary proteins is a good alternative that may lead to the discovery of cancer biomarkers (14).
Proteomic techniques, including two-dimensional gel electrophoresis, mass spectrometry (MS) and validation by immunodetection, have been used to identify protein biomarkers of diseases, especially in different types of cancers (15). Recently, MS-based proteomics has become an increasingly powerful technique, not only for the high-throughput identification of large numbers of proteins, but also for protein quantification (16). Protein quantification can increase the amount of important information obtained from identified proteomes, and can compare protein expressions across various types of samples or treatments. Using MS for comparative protein quantification typically employs stable isotope incorporation, but recently comparative quantification by label-free liquid chromatography (LCn)-MS proteomics data has provided an alternative approach (17).
The aim of the present study was to employ the quantitative label-free MS technique to compare the profiles of protein expression between the urine of normal HPV negative patients and cervical cancer patients. We did not include preinvasive diseases as cell abnormality levels (CIN I, II and III). The results may elucidate the protein-protein interactions and the potential urinary detection biomarkers of cervical cancer.
Materials and methods
Collection and preparation of samples
Cytobrush and urine samples were obtained from cervical cancer patients and healthy volunteers at the Chulabhorn Hospital from July 2014 to April 2015. All participants received detailed information regarding the study objectives and gave written informed consent. The present study was approved by the Ethical Review Board of the Chulabhorn Hospital (Bangkok, Thailand; no. 31/2554). All participants were subsequently performed HPV genotyping analysis and normal routine cervical cancer screening at Chulabhorn Hospital. The samples for HPV genotyping were obtained using a cytobrush by gynecologists or well-trained general practitioners during pelvic examinations at Chulabhorn Hospital. The first morning urine samples were collected and fractionated within 24 h. Each specimen was collected in 10 ml conical tubes and the protease inhibitor cocktails (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) were added. The detail of the 13 urine samples from HPV-negative females who were diagnosed as non-cervical cancer was shown in Table IA. While, the other 24 urine samples were collected from females with cervical cancer at different stages (S1B1, SIB2, SIIA, SIIA1, SIIB, SIIIB, SIVA and SIVB) as shown in Table IB.
Table I.
Clinicopathological data of normal individuals and patients with cervical cancer.
| A, Normal | ||||
|---|---|---|---|---|
| Sample no. | Age, years | HPV genotype | CA state | Pathology information |
| N1 | 47 | Negative | – | – |
| N2 | 35 | Negative | – | – |
| N3 | 49 | Negative | – | – |
| N4 | 49 | Negative | – | – |
| N5 | 52 | Negative | – | – |
| N6 | 52 | Negative | – | – |
| N7 | 42 | Negative | – | – |
| N8 | 44 | Negative | – | – |
| N9 | 40 | Negative | – | – |
| N10 | 44 | Negative | – | – |
| N11 | 53 | Negative | – | – |
| N12 | 55 | Negative | – | – |
| N13 | 64 | Negative | – | – |
| B, Cancer | ||||
| Sample no. | Age, years | HPV genotype | CA state | Pathology information |
| C1 | 38 | Negative | IB1 | Adenocarcinoma in situ |
| C2 | 44 | 33 | IB1 | Squamous cell carcinoma, non-keratinizing moderately differentiated |
| C3 | 51 | 16 | IIA | Squamous cell carcinoma, non-keratinizing moderately differentiated |
| C4 | 50 | 68 | IIA | Adenocarcinoma, moderately-poorly differentiated |
| C5 | 56 | Negative | IIA1 | Squamous cell carcinoma, papillary type |
| C6 | 51 | 59 | IIB | Squamous cell keratinizing carcinoma |
| C7 | 54 | 16 | IIB | Adenocarcinoma, endocervical |
| C8 | 64 | 18 | IIB | Squamous cell carcinoma, non-keratinizing, moderately-poorly |
| differentiated | ||||
| C9 | 50 | 33, LR11 | IIB | Squamous cell carcinoma, keratinizing type, moderately differentiated |
| C10 | 49 | 16 | IIB | Squamous cell carcinoma |
| C11 | 64 | Negative | IIB | Adenocarcinoma, endometrial moderately differentiated, metastasis |
| C12 | 42 | 16 | IIIB | Squamous cell carcinoma, non-keratinizing moderately differentiated |
| C13 | 61 | 16 | IIIB | Squamous cell carcinoma, non-keratinizing well-moderately |
| differentiated | ||||
| C14 | 50 | Negative | IIIB | Squamous cell carcinoma, keratinizing type |
| C15 | 58 | 31 | IIIB | Squamous cell carcinoma, non-keratinizing moderately differentiated, |
| metastasis | ||||
| C16 | 49 | 52,33 | IVA | Squamous cell carcinoma, non-keratinizing moderately differentiated |
| C17 | 49 | 18 | IVB | Adenocarcinoma, Poorly differentiated |
| C18 | 59 | 52 | IVB | Squamous cell carcinoma, non-keratinizing, moderate differentiated |
| C19 | 59 | 18 | IIB | Adenocarcinoma, endocervical like, metastasis |
| C20 | 57 | 53 | IB2 | Squamous cell carcinoma, keratinizing poorly differentiated |
| C21 | 33 | 16 | IIB | Squamous cell carcinoma, non-keratinizing moderately differentiated |
| C22 | 55 | Negative | IIB | Endometrium biopsy, poorly differentiated |
| C23 | 60 | 16 | IIB | Squamous cell carcinoma |
| C24 | 61 | LR 72 | IIB | Adenocarcinoma, poorly differentiated, metastasis |
Cytology examination
The Bethesda 2001 report system was used as the standard protocol for classification of all cervical cytology slides (18). The different grades, such as squamous cell carcinoma and adenocarcinoma, were determined using the cervical cytology slides by qualified pathologists via a normal routine at Chulabhorn Hospital.
HPV genotyping
The Linear array HPV testing (Roche Diagnostics Indianapolis, IN, USA) was employed to identify HPV genotypes. This kit was for identification of 37 HPV genotypes including 12 high-risk, 8 probable high-risk and 17 low-risk types. Briefly, 450 bp fragments of the L1 region of HPV were first amplified by polymerase chain reaction, followed by hybridization using a reverse line blot system for the simultaneous detection of up to 37 HPV genotypes (i.e., genotypes 6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 55, 56, 58, 59, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72, 73, 81, 82, 83, 84, IS39, and CP6108) (19).
Sample preparation
Urine samples were stored at 4°C and then centrifuged at 1,120 × g for 5 min at 4°C to remove cell debris. The supernatants were precipitated with 50% v/v acetone for 16 h followed by centrifugation at 12,000 × g for 10 min at 4°C. The pellets were kept at −80°C until further use.
In solution digestion
The pellets of urine samples from 5 normal (N9-N13) and from 6 cervical cancer patients at different stages (C19-C24) were resuspended in 50 mM ammonium bicarbonate (NH4HCO3) separately and the protein concentration was determined by spectrophotometry using the Bradford method (20). The samples were pooled by obtaining 5 µg from each of the 5 normal urine samples and 5 µg from each of the 6 cancer urine samples. A total of 5 µg of the pooled samples were separately reduced with 100 mM DTT (10 mM final concentration) for 5 min at 95°C and alkylated with 1/10 volume of 200 mM iodoacetamide prior to incubation for 30 min in the dark at room temperature. Digestion was performed overnight at 37°C by adding 1:50 (w/w) of sequencing grade trypsin (Promega Corporation, Madison, WI, USA) to the protein in each sample. The digestion reaction was stopped by adding formic acid to reach a 1% final concentration and the samples were dried completely by SpeedVac.
LC-MS/MS analysis
To prepare samples for label-free LC-MS/MS quantification, the digested samples were dissolved in 0.1% formic acid in H2O and separated on a nanoACQUITY system (Waters Corporation, Milford, MA, USA). All pooled urine samples were run in triplicate. The samples were injected into a nanoACQUITY UPLC column (1.7 µm BEH, 75 µm × 200 mm C18) at a flow rate of 300 nl/min. The column temperature was maintained at 40°C. The LC gradient (1–50% B in 70 min, 50–90% B in 5 min, followed by 15 min on 90% B) was performed using 0.1% formic acid in H2O as solvent A and 0.1% formic acid in ACN as solvent B. The eluting peptides were analyzed directly via MS/MS on an amaZon speed ion trap mass spectrometer (Bruker Corporation, Billerica, MA, USA) equipped with a captive-electrospray ion source. The positive mode was used with a spray voltage of 1,300 V and the capillary temperature was set at 150°C. Mass spectra were acquired from 400–1,400 m/z using parameters optimized at 922 m/z with a target of 500,000 set for ion charge control and a maximum acquisition time of 100 msec. The scan range was 50–3,000 m/z. MS/MS data were processed by Bruker Compass 1.4 software (Bruker Corporation).
Data analysis and label-free LC-MS quantitative profiling
Progenesis label-free LC-MS software version 3.1 (Nonlinear Dynamics, Ltd., Newcastle upon Tyne, UK) was used to process the raw data obtained from LC-MS/MS and to calculate the significant changes. The retention time of each sample was aligned and the reference sample was set up. For all of the replicates, their retention times were aligned to the established reference and the intensities of the peak were then normalized. Three criteria were used to filter all data prior to exporting the MS/MS output files to identify proteins using Mascot software (www.matrixscience.com), including: i) An analysis of variance (ANOVA) with Tukey's post hoc test score shown between experimental groups of P≤0.05; ii) featured mass peaks with +2, +3 and +4 as the charge states; and iii) only data with >1 isotopes or peptides. From the aforementioned Progenesis software, all of the exported MS/MS spectra were generated using a Mascot generic file. Mascot software version 2.4.0 was used for the identification of peptides. The SwissProt human protein database (Matrix Science, Ltd., London, UK; www.matrixscience.com) program was set up as follows: The MS/MS mass tolerance was set at 0.6 Da, peptide mass tolerance was set to 1.2 Da, carbamidomethylation was set as a fixed modification and ≤1 missed cleavages were allowed. A false discovery rate threshold of 1% was applied and identification of two or more unique peptides and two or more peptides were required for positive identification, respectively.
Characterization, classification, functional and protein- protein interactions analysis
Venn Diagrams web tool (http://www.interactivenn.net) was used to compare the expression of proteins from cervical cancer patients between identified proteins by LC-MS/MS and the database of Human Proteome Atlas (www.proteinatlas.org/humanproteome/pathology/cervical+cancer). The classification and functional analysis of differential expressed proteins was performed using the PANTHER database (www.pantherdb.org/). The Gene Ontology (GO) standard was employed to categorize proteins according to ‘Molecular function’ and ‘Biological process’. The Search Tool for Retrieval of Interacting Genes/Proteins (STRING) database (string-db.org) version 9.0 was used to obtain protein-protein interactions. Protein mode from STRING was analyzed, and the interactions from upregulated and downregulated urinary expressed proteins were identified.
Western blot analysis
The expression levels of proteins were analyzed by western blot analysis. Leucine-rich α-2-glycoprotein (LRG1), multimerin-1 (MMRN1), S100 calcium-binding protein A8 (S100A8), serpin B3 (SERPINB3) and cluster of differentiation-44 antigen (CD44) were selected to be validated for expression. The pellets of 8 normal (N1-N8) and 18 different stages (C1-C18) of cancer urine samples were resuspended in 50 mM ammonium bicarbonate (NH4HCO3). A total of 20 µg of urine samples were diluted in sample buffer (50 mM Tris, pH 6.8, 10% glycerol, 2% SDS and 0.01% bromophenol blue) and resolved by SDS-PAGE. Following SDS-PAGE, the proteins were transferred onto 0.20 µm polyvinylidene difluoride membranes (Pall Life Sciences, Port Washington, NY, USA). The membranes were blocked with 5% (w/v) non-fat dry milk in TBST [50 mM Tris, pH 8.0, 150 mM NaCl, and 0.1% Tween-20 (v/v)] for 1 h. Membranes were washed 10 min for 3 times in TBST and incubated at 4°C overnight with the following primary antibodies: Mouse anti-LRG1 (1:2,000; ab57992; Abcam, Cambridge, UK), rabbit anti-MMRN1 (1:2,000; ab130585; Abcam), rabbit anti-S100A8 (1:1,000; ab92331; Abcam), rabbit anti-CD44 (1:2,000; ab51037; Abcam) and rabbit anti-SerpinB3 (1:5,000; ab126752; Abcam). Then the membranes were washed 3 times in TBST, and incubated with horseradish peroxidase-conjugated anti-mouse (P0260) or anti-rabbit (P0217) antibodies secondary antibody (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) at room temperature for 1 h. Following washing 3 times with TBST, the reaction was detected by chemiluminescence with the WesternBright ECL detection kit (Advansta Inc., Menlo Park, CA, USA). Darkroom development techniques were employed to expose and acquire images of the membranes.
Statistical analysis
The receiver operating characteristic (ROC) curve and area under the curve (AUC) were used to determine the performance of the five individual potential biomarkers by employing GraphPad Prism software version 7 (GraphPad Software, Inc., La Jolla, CA, USA). The combination of the five potential biomarkers was also analyzed using logistic regression with SPSS software version 11 (IBM Corp., Armonk, NY, USA).
Results
Analysis of the altered urinary proteins expression
The body filtrates from urine such as water, electrolytes, salts, nitrogenous waste and proteins are derived from the plasma, kidneys and urogenital tract. The amount of urinary protein excretion normally is <150 mg/day and needs to be concentrated for use in experiments (21). Label-free quantitative proteomics is the best choice for analyzing samples with a low level of proteins. The pooled urinary samples from 5 normal individuals (N9-N13) and 6 cervical cancer patients (C19-C24) were analyzed by LC-MS/MS, as shown in Table IIA and B. Significant changes were calculated using Progenesis label-free LC-MS software version 3.1. A list of proteins with 2 or more matched peptides was compiled. A total 133 proteins with 1% FDR were identified. A total of 60 upregulated and 73 downregulated urinary proteins were expressed when comparing normal and cervical cancer samples. Serotransferrin (TF) was observed to be the most significantly increased protein (2.91-fold, P=0.000004) while Kininogen-1 (KNG1) was the most significantly decreased protein (1.73-fold, P=0.000003). The results also revealed that LRG1, low-density lipoprotein receptor-related protein 2 (LRP2), Myosin light chain kinase (MYLK), SPARC-like 1, Zinc finger CCHC domain-containing protein 8, MMRN1 and nectin-2 were notable proteins that had >2 fold expression while aminopeptidase N had downregulated protein expression (5.31 fold) and another 10 proteins had between 3.0–4.33 fold altered expression including protein S100A8, trefoil factor 2, apolipoprotein E (APOE), apolipoprotein D (APOD), DNA (cytosine-5)-methyltransferase 3B (DNMT3B), CD44, SERPINB3, macrophage colony-stimulating factor 1 (CSF1), epidymal secretory protein E1 and ribonuclease pancreatic. The large molecular weight proteins of >100 kDa were obtained using this method, including LRP2 (521.6 kDa), MYLK (210.7 kDa) and MMRN1 (138 kDa). These proteins of interest (LRG1, MMRN1, S100A8, SERPINB3 and CD44) were selected to confirm their expression by western blot analysis.
Table II.
List of differentially expressed urinary proteins in normal individuals and patients with cervical cancer, identified by relative quantitative label-free mass spectrometry analysis.
| A, Upregulated urinary proteins in patients with cervical cancer with ANOVA P≤0.05 and Max fold change ≥1.5 | ||||||||
|---|---|---|---|---|---|---|---|---|
| No. | Gene name | Accession | Description | Peptide count | Score | ANOVA P-value | Fold-change | MW(kDa)/pI |
| 1 | TF | TRFE_HUMAN | Serotransferrin | 15 | 2,566.45 | 0.000004 | 2.91 | 77.06/6.81 |
| 2 | ORM1 | A1AG1_HUMAN | α-1-acid glycoprotein 1 | 4 | 656.38 | 0.000005 | 2.39 | 23.72/4.93 |
| 3 | LRG1 | A2GL_HUMAN | Leucine-rich α-2-glycoprotein | 3 | 456.41 | 0.000007 | 2.72 | 38.38/6.45 |
| 4 | LRP2 | LRP2_HUMAN | Low-density lipoprotein receptor-related protein 2 | 17 | 1,119.49 | 0.000009 | 4.34 | 521.59/4.89 |
| 5 | MYLK | MYLK_HUMAN | Myosin light chain kinase, smooth muscle | 7 | 288.88 | 0.000013 | 3.28 | 210.71/5.85 |
| 6 | ORM2 | A1AG2_HUMAN | α-1-acid glycoprotein | 4 | 584.86 | 0.00003 | 2.64 | 23.87/5.03 |
| 7 | PDCD11 | RRP5_HUMAN | Protein RRP5 homolog | 8 | 272.23 | 0.000031 | 2.48 | 208.70/8.99 |
| 8 | APOA1 | APOA1_HUMAN | Apolipoprotein A-I | 2 | 436.83 | 0.000036 | 2.99 | 30.77/5.56 |
| 9 | FAM109B | SESQ2_HUMAN | Sesquipedalian-2 | 4 | 35.46 | 0.000048 | 1.8 | 28.33/6.66 |
| 10 | ALB | ALBU_HUMAN | Serum albumin | 30 | 4,048.64 | 0.000063 | 1.98 | 71.31/5.92 |
| 11 | CPM | CBPM_HUMAN | Carboxypeptidase M | 2 | 111.23 | 0.000337 | 1.95 | 50.93/6.94 |
| 12 | A1BG | A1BG_HUMAN | α-1B-glycoprotein | 4 | 797.85 | 0.000407 | 2.04 | 54.79/5.56 |
| 13 | SPARCL1 | SPRL1_HUMAN | SPARC-like protein 1 | 9 | 136.75 | 0.000421 | 3.28 | 75.20/4.71 |
| 14 | LYST | LYST_HUMAN | Lysosomal-trafficking regulator | 12 | 440.97 | 0.000459 | 1.78 | 429.13/6.15 |
| 15 | ZCCHC8 | ZCHC8_HUMAN | Zinc finger CCHC domain-containing protein 8 | 2 | 50.3 | 0.000489 | 4.78 | 78.57/4.79 |
| 16 | DNAH10 | DYH10_HUMAN | Dynein heavy chain 10, axonemal | 21 | 610.61 | 0.00054 | 1.73 | 514.84/5.64 |
| 17 | CHD6 | CHD6_HUMAN | Chromodomain-helicase-DNA-binding protein 6 | 14 | 341.78 | 0.000555 | 1.87 | 305.41/5.90 |
| 18 | SYNE1 | SYNE1_HUMAN | Nesprin-1 | 42 | 1,118.05 | 0.000586 | 1.87 | 1011.08/5.37 |
| 19 | HPX | HEMO_HUMAN | Hemopexin | 6 | 727.65 | 0.00061 | 2.29 | 51.67/6.55 |
| 20 | WDFY2 | WDFY2_HUMAN | WD repeat and FYVE domain-containing protein 2 | 6 | 97.64 | 0.000612 | 2.94 | 45.15/6.46 |
| 21 | SOX6 | SOX6_HUMAN | Transcription factor SOX-6 | 6 | 137.57 | 0.000684 | 2.14 | 91.92/7.65 |
| 22 | CHGB | SCG1_HUMAN | Secretogranin-1 | 2 | 73.98 | 0.000973 | 2.99 | 78.27/5.02 |
| 23 | CPE | CBPE_HUMAN | Carboxypeptidase E | 4 | 37.93 | 0.00107 | 2.19 | 53.15/5.03 |
| 24 | MAP3K7 | M3K7_HUMAN | Mitogen-activated protein kinase kinase kinase 7 | 8 | 168.58 | 0.00111 | 2.19 | 67.19/6.69 |
| 25 | DCLK1 | DCLK1_HUMAN | Serine/threonine-protein kinase DCLK1 | 6 | 137.52 | 0.00125 | 1.68 | 82.22/8.84 |
| 26 | GC | VTDB_HUMAN | Vitamin D-binding protein | 11 | 703.57 | 0.0013 | 1.59 | 54.52/5.40 |
| 27 | KRT17 | K1C17_HUMAN | Keratin, type I cytoskeletal 17 | 5 | 123.3 | 0.00131 | 2.13 | 48.10/4.97 |
| 28 | – | KV105_HUMAN | Immunoglobulin κ variable 1–5 | 2 | 82.6 | 0.00149 | 1.74 | 12.78/8.49 |
| 29 | MYO18B | MY18B_HUMAN | Unconventional myosin-XVIIIb | 21 | 644.74 | 0.00168 | 1.66 | 285.18/6.49 |
| 30 | MACF1 | MACF1_HUMAN | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 | 32 | 996.53 | 0.00173 | 1.82 | 838.30/5.28 |
| 31 | FGG | FIBG_HUMAN | Fibrinogen γ chain | 3 | 373.78 | 0.00215 | 1.83 | 51.51/5.37 |
| 32 | C3 | CO3_HUMAN | Complement C3 | 10 | 740.32 | 0.00228 | 1.73 | 187.14/6.02 |
| 33 | CP | CERU_HUMAN | Ceruloplasmin | 8 | 1,240.04 | 0.00273 | 1.54 | 122.98/5.44 |
| 34 | AZGP1 | ZA2G_HUMAN | Zinc-α-2-glycoprotein | 5 | 1,217.5 | 0.00316 | 1.55 | 34.46/5.71 |
| 35 | FGB | FIBB_HUMAN | Fibrinogen β-chain | 2 | 172.98 | 0.00317 | 1.75 | 55.92/8.54 |
| 36 | PVRL2 | NECT2_HUMAN | Nectin-2 | 3 | 95.27 | 0.00352 | 3.37 | 55.74/4.74 |
| 37 | IL16 | IL16_HUMAN | Pro-interleukin-16 | 11 | 208.63 | 0.00371 | 1.82 | 141.75/8.34 |
| 38 | UCMA | UCMA_HUMAN | Unique cartilage matrix-associated protein | 2 | 66.89 | 0.00441 | 1.78 | 16.56/5.47 |
| 39 | PGK2 | PGK2_HUMAN | Phosphoglycerate kinase 2 | 3 | 92.54 | 0.00489 | 1.95 | 44.79/8.74 |
| 40 | H2BFWT | H2BWT_HUMAN | Histone H2B type W-T | 3 | 44.94 | 0.00489 | 1.72 | 19.61/10.69 |
| 41 | HP | HPT_HUMAN | Haptoglobin | 3 | 1,055.7 | 0.0061 | 1.72 | 45.20/6.13 |
| 42 | APOA4 | APOA4_HUMAN | Apolipoprotein A-IV | 6 | 70.31 | 0.00638 | 1.64 | 45.39/5.28 |
| 43 | – | IGHG3_HUMAN | Ig γ-3 chain C region | 2 | 584.82 | 0.00751 | 1.56 | 41.28/8.23 |
| 44 | R3HCC1L | R3HCL_HUMAN | Coiled-coil domain-containing protein R3HCC1L | 3 | 156.15 | 0.00789 | 1.53 | 87.88/5.01 |
| 45 | FN1 | FINC_HUMAN | Fibronectin | 12 | 647.44 | 0.00855 | 2 | 262.62/5.46 |
| 46 | HEXA | HEXA_HUMAN | β-hexosaminidase subunit alpha | 2 | 72.64 | 0.00857 | 1.59 | 61.12/5.04 |
| 47 | AKAP4 | AKAP4_HUMAN | A-kinase anchor protein 4 | 4 | 151.22 | 0.00885 | 1.76 | 94.47/6.56 |
| 48 | PREX1 | PREX1_HUMAN | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | 8 | 231.71 | 0.02 | 2.19 | 186.20/6.03 |
| 49 | RPS27A | RS27A_HUMAN | Ubiquitin-40S ribosomal protein S27a | 3 | 63.39 | 0.02 | 1.93 | 17.96/9.68 |
| 50 | AMY2A | AMYP_HUMAN | Pancreatic α-amylase | 3 | 1,106.07 | 0.02 | 1.89 | 57.70/6.60 |
| 51 | – | KV320_HUMAN | Immunoglobulin κ variable 3–20 | 2 | 200.67 | 0.02 | 1.77 | 12.55/4.85 |
| 52 | SERPINA7 | THBG_HUMAN | Thyroxine-binding globulin | 3 | 243.84 | 0.02 | 1.65 | 46.32/5.87 |
| 53 | DNAH3 | DYH3_HUMAN | Dynein heavy chain 3, axonemal | 23 | 639.86 | 0.02 | 1.61 | 470.77/6.04 |
| 54 | MMRN2 | MMRN2_HUMAN | Multimerin-2 | 3 | 79.01 | 0.02 | 1.54 | 104.40/5.56 |
| 55 | AFM | AFAM_HUMAN | Afamin | 6 | 335.44 | 0.03 | 1.65 | 69.06/5.64 |
| 56 | FAM208B | F208B_HUMAN | Protein FAM208B | 18 | 427.39 | 0.03 | 1.65 | 268.84/5.61 |
| 57 | PTPRS | PTPRS_HUMAN | Receptor-type tyrosine-protein phosphatase S | 14 | 335.72 | 0.03 | 1.53 | 217.04/6.06 |
| 58 | MMRN1 | MMRN1_HUMAN | Multimerin-1 | 3 | 225.55 | 0.04 | 2.01 | 138.11/8.15 |
| 59 | COL4A2 | CO4A2_HUMAN | Collagen α-2(IV) chain | 8 | 171.58 | 0.05 | 1.71 | 167.55/8.89 |
| 60 | PLAU | UROK_HUMAN | Urokinase-type plasminogen activator | 2 | 116.86 | 0.05 | 1.5 | 48.50/8.78 |
| B, Downregulated urinary proteins in patients with cervical cancer with ANOVA P≤0.05 and Max fold change ≥1.5 | ||||||||
| No. | Gene name | Accession | Description | Peptide count | Score | ANOVA P-value | Fold-change | MW(kDa)/pI |
| 1 | KNG1 | KNG1_HUMAN | Kininogen-1 | 13 | 1,338.67 | 0.000003 | 1.73 | n/a |
| 2 | S100A8 | S10A8_HUMAN | Protein S100-A8 | 2 | 378.86 | 0.000003 | 3.42 | n/a |
| 3 | ANPEP | AMPN_HUMAN | Aminopeptidase N | 6 | 403.62 | 0.000005 | 5.31 | n/a |
| 4 | APOE | APOE_HUMAN | Apolipoprotein E | 4 | 61.1 | 0.000027 | 3.04 | n/a |
| 5 | SPP1 | OSTP_HUMAN | Osteopontin | 4 | 801.94 | 0.000047 | 2.81 | n/a |
| 6 | LAMP1 | LAMP1_HUMAN | Lysosome-associated membrane glycoprotein 1 | 4 | 209.2 | 0.00005 | 2.1 | n/a |
| 7 | APOD | APOD_HUMAN | Apolipoprotein D | 2 | 373.48 | 0.000052 | 3.62 | n/a |
| 8 | LPHN1 | AGRL1_HUMAN | Adhesion G protein-coupled receptor L1 | 11 | 164.47 | 0.000056 | 1.69 | n/a |
| 9 | UMOD | UROM_HUMAN | Uromodulin | 11 | 1,580 | 0.000057 | 2.1 | n/a |
| 10 | DNMT3B | DNM3B_HUMAN | DNA (cytosine-5)-methyltransferase 3B | 2 | 67.24 | 0.000069 | 3.41 | n/a |
| 11 | TFF2 | TFF2_HUMAN | Trefoil factor 2 | 2 | 86.69 | 0.000074 | 4.33 | n/a |
| 12 | ACTBL2 | ACTBL_HUMAN | β-actin-like protein 2 | 5 | 188.69 | 0.000093 | 1.77 | n/a |
| 13 | CTSD | CATD_HUMAN | Cathepsin D | 7 | 403.41 | 0.000193 | 2.09 | n/a |
| 14 | GAA | LYAG_HUMAN | Lysosomal α-glucosidase | 7 | 1,018.11 | 0.00022 | 2.33 | n/a |
| 15 | TNC | TENA_HUMAN | Tenascin | 6 | 271.43 | 0.000327 | 1.85 | n/a |
| 16 | CD44 | CD44_HUMAN | CD44 antigen | 4 | 483.38 | 0.000369 | 3.12 | n/a |
| 17 | CSF1 | CSF1_HUMAN | Macrophage colony-stimulating factor 1 | 2 | 198.48 | 0.000435 | 3.55 | n/a |
| 18 | CD248 | CD248_HUMAN | Endosialin | 5 | 234.21 | 0.000596 | 2.6 | n/a |
| 19 | ARSA | ARSA_HUMAN | Arylsulfatase A | 4 | 511.64 | 0.000624 | 2.91 | n/a |
| 20 | CTSB | CATB_HUMAN | Cathepsin B | 2 | 264.96 | 0.000639 | 2.14 | n/a |
| 21 | ANK3 | ANK3_HUMAN | Ankyrin-3 | 49 | 1,009.24 | 0.000851 | 1.7 | n/a |
| 22 | PSAP | SAP_HUMAN | Prosaposin | 3 | 361.86 | 0.000852 | 2.17 | n/a |
| 23 | – | LV321_HUMAN | Immunoglobulin lambda variable 3–21 | 2 | 154.16 | 0.000969 | 2.13 | n/a |
| 24 | BHMT | BHMT1_HUMAN | Betaine--homocysteine S-methyltransferase 1 | 7 | 289.37 | 0.001 | 1.94 | n/a |
| 25 | COL15A1 | COFA1_HUMAN | Collagen α-1(XV) chain | 2 | 179.35 | 0.00104 | 2.04 | n/a |
| 26 | IGLL5 | IGLL5_HUMAN | Immunoglobulin lambda-like polypeptide 5 | 2 | 385.74 | 0.00107 | 1.59 | n/a |
| 27 | CDH13 | CAD13_HUMAN | Cadherin-13 | 4 | 369.61 | 0.00117 | 1.67 | n/a |
| 28 | PTGDS | PTGDS_HUMAN | Prostaglandin-H2 D-isomerase | 5 | 333.35 | 0.00121 | 1.91 | n/a |
| 29 | COL6A1 | CO6A1_HUMAN | Collagen α-1(VI) chain | 7 | 715.12 | 0.00138 | 2.01 | n/a |
| 30 | FOLR1 | FOLR1_HUMAN | Folate receptor α | 5 | 190.49 | 0.00142 | 1.81 | n/a |
| 31 | CD55 | DAF_HUMAN | Complement decay-accelerating factor | 3 | 500.96 | 0.00148 | 1.82 | n/a |
| 32 | NPC2 | NPC2_HUMAN | Epididymal secretory protein E1 | 2 | 209.65 | 0.00151 | 3.11 | n/a |
| 33 | PROCR | EPCR_HUMAN | Endothelial protein C receptor | 3 | 291 | 0.00156 | 1.74 | n/a |
| 34 | AHSG | FETUA_HUMAN | Alpha-2-HS-glycoprotein | 5 | 229.87 | 0.00162 | 1.51 | n/a |
| 35 | ABCB5 | ABCB5_HUMAN | ATP-binding cassette sub-family B member 5 | 7 | 217.25 | 0.00162 | 2.16 | n/a |
| 36 | CADM4 | CADM4_HUMAN | Cell adhesion molecule 4 | 5 | 454.21 | 0.00174 | 1.63 | n/a |
| 37 | RNASE1 | RNAS1_HUMAN | Ribonuclease pancreatic | 4 | 267.25 | 0.00177 | 3.15 | n/a |
| 38 | WFDC2 | WFDC2_HUMAN | WAP four-disulfide core domain protein 2 | 2 | 237.59 | 0.00215 | 2.21 | n/a |
| 39 | F5 | FA5_HUMAN | Coagulation factor V | 10 | 340.57 | 0.0023 | 1.65 | n/a |
| 40 | PRNP | PRIO_HUMAN | Major prion protein | 4 | 159.7 | 0.00244 | 2.18 | n/a |
| 41 | RETN | RETN_HUMAN | Resistin | 2 | 175.19 | 0.00254 | 1.68 | n/a |
| 42 | KLHL4 | KLHL4_HUMAN | Kelch-like protein 4 | 3 | 97.09 | 0.00335 | 2.16 | n/a |
| 43 | SERPING1 | IC1_HUMAN | Plasma protease C1 inhibitor | 6 | 567.51 | 0.00389 | 1.6 | n/a |
| 44 | PARP14 | PAR14_HUMAN | Poly [ADP-ribose] polymerase 14 | 4 | 210.84 | 0.00415 | 1.68 | n/a |
| 45 | CRYL1 | CRYL1_HUMAN | Lambda-crystallin homolog | 4 | 84.49 | 0.00432 | 1.6 | n/a |
| 46 | C16orf62 | CP062_HUMAN | UPF0505 protein C16orf62 | 4 | 94.21 | 0.00456 | 1.89 | n/a |
| 47 | GALNS | GALNS_HUMAN | N-acetylgalactosamine-6-sulfatase | 4 | 121.68 | 0.00543 | 1.67 | n/a |
| 48 | SERPINA4 | KAIN_HUMAN | Kallistatin | 8 | 133.52 | 0.00559 | 1.71 | n/a |
| 49 | OSCAR | OSCAR_HUMAN | Osteoclast-associated immunoglobulin-like receptor | 3 | 189.22 | 0.00559 | 2.14 | n/a |
| 50 | AMBP | AMBP_HUMAN | Protein AMBP | 8 | 1,211.69 | 0.00628 | 1.76 | n/a |
| 51 | CPQ | CBPQ_HUMAN | Carboxypeptidase Q | 6 | 355.87 | 0.00656 | 1.64 | n/a |
| 52 | ANGPTL2 | ANGL2_HUMAN | Angiopoietin-related protein 2 | 5 | 195.93 | 0.00678 | 1.55 | n/a |
| 53 | LAIR1 | LAIR1_HUMAN | Leukocyte-associated immunoglobulin-like receptor 1 | 4 | 302.08 | 0.00754 | 1.66 | n/a |
| 54 | PIP | PIP_HUMAN | Prolactin-inducible protein | 2 | 286.9 | 0.00785 | 1.52 | n/a |
| 55 | SPRR3 | SPRR3_HUMAN | Small proline-rich protein 3 | 5 | 263.95 | 0.00984 | 3.37 | n/a |
| 56 | MXRA8 | MXRA8_HUMAN | Matrix-remodeling-associated protein 8 | 2 | 188.15 | 0.00991 | 1.6 | n/a |
| 57 | WDFY3 | WDFY3_HUMAN | WD repeat and FYVE domain-containing protein 3 | 15 | 358.34 | 0.01 | 1.51 | n/a |
| 58 | CDH2 | CADH2_HUMAN | Cadherin-2 | 2 | 329.32 | 0.01 | 1.6 | n/a |
| 59 | ACP2 | PPAL_HUMAN | Lysosomal acid phosphatase | 6 | 380.73 | 0.01 | 1.62 | n/a |
| 60 | ATG2A | ATG2A_HUMAN | Autophagy-related protein 2 homolog A | 5 | 91.97 | 0.01 | 1.85 | n/a |
| 61 | CRNN | CRNN_HUMAN | Cornulin | 2 | 277.69 | 0.01 | 2.04 | n/a |
| 62 | MT1G | MT1G_HUMAN | Metallothionein-1G | 2 | 111.25 | 0.01 | 2.06 | n/a |
| 63 | ESAM | ESAM_HUMAN | Endothelial cell-selective adhesion molecule | 5 | 79.85 | 0.01 | 2.14 | n/a |
| 64 | TNRC18 | TNC18_HUMAN | Trinucleotide repeat-containing gene 18 protein | 9 | 266.88 | 0.02 | 1.51 | n/a |
| 65 | APP | A4_HUMAN | Amyloid β A4 protein | 4 | 227.47 | 0.02 | 1.57 | n/a |
| 66 | SLC3A2 | 4F2_HUMAN | 4F2 cell-surface antigen heavy chain | 4 | 151.92 | 0.02 | 1.58 | n/a |
| 67 | TTR | TTHY_HUMAN | Transthyretin | 4 | 212.41 | 0.02 | 1.64 | n/a |
| 68 | VPS18 | VPS18_HUMAN | Vacuolar protein sorting-associated protein 18 homolog | 3 | 87.62 | 0.03 | 1.54 | n/a |
| 69 | S100P | S100P_HUMAN | Protein S100-P | 3 | 103.16 | 0.03 | 1.63 | n/a |
| 70 | ZNF805 | ZN805_HUMAN | Zinc finger protein 805 | 3 | 82.25 | 0.03 | 2.83 | n/a |
| 71 | LYVE1 | LYVE1_HUMAN | Lymphatic vessel endothelial hyaluronic acid receptor 1 | 4 | 396.4 | 0.04 | 1.52 | n/a |
| 72 | RNF113A | R113A_HUMAN | RING finger protein 113A | 4 | 126.64 | 0.04 | 1.82 | n/a |
| 73 | SERPINB3 | SPB3_HUMAN | Serpin B3 | 3 | 108 | 0.04 | 1.9 | n/a |
Characterization, classification, functional and protein- protein interactions analysis
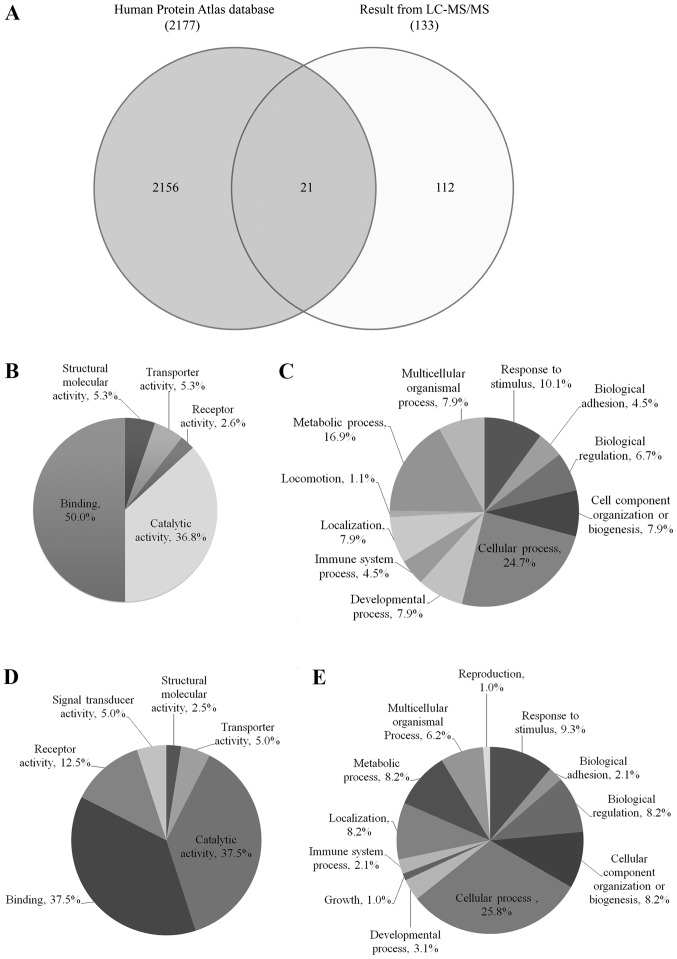
Venn diagram comparing protein expression from cervical cancer between identified proteins by LC-MS/MS and the database of Human Protein Atlas was shown in Fig. 1A. Twenty-one identified proteins were overlapped with The Human Protein Atlas Database, including ABCB5, AMY2A, ANGPTL2, C3, CD44, CD55, COL6A1, CRNN, CRYL1, DNAH3 ESAM, GALNS, HEXA, KRT17, MACF1, MXRA8, PGK2, PIP, PTGDS, SERPINB3 and WFDC2. Interestingly, 112 identified proteins have never been reported in the database of Human Protein Atlas for cervical cancer.
Figure 1.
Protein expression. (A) Venn diagram comparing the total differential protein expression from cervical cancer between identified proteins by LC-MS/MS and the Human Protein Atlas Database. Differentially expressed urinary proteins from normal individuals and patients with cervical cancer were analyzed by PANTHER classification based on (B and D) molecular function and (C and E) biological processes: (B and C) 60 upregulated proteins and (D and E) 73 downregulated proteins. LC, liquid chromatography; MS, mass spectrometry.
The up- and downregulated urinary proteins from Table IIA and B were analyzed by GO with the PANTHER database system to obtain ‘Molecular function’ and ‘Biological process’ data, as presented in pie charts in Fig. 1B-E. The molecular functions of 60 upregulated proteins from Fig. 1B were classified into 5 groups: Binding (50%), catalytic activity (36.8%), structural molecular activity (5.3%), transporter activity (5.3%) and receptor activity (2.6%), while the biological processes of these proteins (Fig. 1C) were classified into 11 groups with the highest percentage observed for cellular process (24.7%), followed by metabolic process (16.9%) and response to stimulus (10.1%). The molecular functions of the 73 downregulated proteins, as presented in Fig. 1D, were classified into 6 groups and were mainly involved in binding (37.5%) and catalytic activity (37.5%), while the biological processes of these proteins (Fig. 1E) were classified into 12 groups and were mainly involved in cellular process (25.8%) and response to stimulus (9.3%).
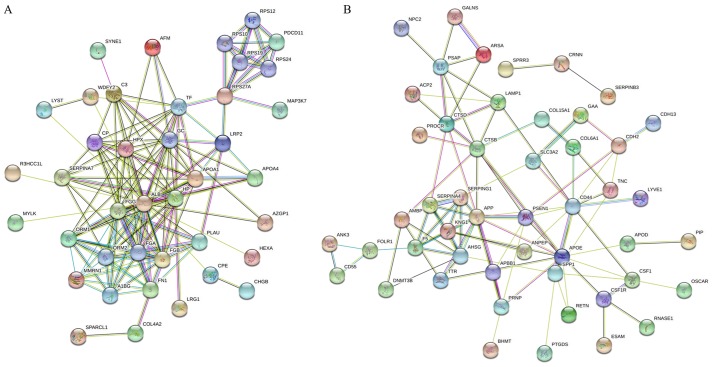
The STRING database was used for analysis of the protein-protein interactions and was able to predict the similar functions of proteins by accessing various free resources. The ‘molecular function’, ‘cellular component’ and ‘biological process’ from GO and KEGG pathways enrichment analyses were obtained. The present study analyzed the 60 and 73 identified proteins that were up- and downregulated, respectively (Table II), when the pooled urinary samples of normal individuals and patients with cervical cancer were compared. The results for the interaction map of the upregulated proteins was presented in Fig. 2A. The results for the associations between these proteins revealed that 40 of the 60 proteins had good interactions. The big cluster was involved in protein binding functions from molecular function (GO), metabolic process, blood coagulation, cell-cell adhesion, regulation of cell morphogenesis and cell motility from biological process (GO). A total of 19 proteins were involved in binding including, albumin (ALB), fibrinogen β-chain (FGB), fibrinogen γ-chain (FGG), fibronectin (FN1), haptoglobin, nesprin-1, zinc-α-2-glycoprotein, vitamin D-binding protein, TF, LRP2, protein RRP5 homolog and mitogen-activated protein kinase kinase kinase 7 (MAP3K7) while 12 proteins were involved in blood coagulation, such as fibrinolysis, and fibrin clot formation including, ALB, FGG, FGB, FN1, TF, MMRN1, apolipoprotein A, urokinase-type plasminogen activator, complement C3, ceruloplasmin and lysosomal-trafficking regulator. There were some proteins of interest that were involved in multiple functions: For example, MAP3K7 was not only involved in binding functions but was also involved in cell-cell adhesion and metabolic process; and microtubule-actin cross-linking factor 1 was involved in metabolic processes, the regulation of cell morphogenesis for differentiation and the regulation of anatomical structure morphogenesis. MMRN1 was also involved in the stress response and cell adhesion function.
Figure 2.
Search Tool for Retrieval of Interacting Genes/Proteins analysis. A network of interacting proteins: (A) 60 upregulated urinary proteins and (B) 73 downregulated urinary proteins from patients with cervical cancer.
Fig. 2B presents the STRING analysis of 73 downregulated proteins from the same comparison. The results revealed that 49 of the 73 proteins had good interactions. The big cluster was involved in different protein functions such as stress-response, cell adhesion, leukocyte migration, wounding response and extracellular matrix organization from biological process (GO). A total of 20 proteins were involved in the stress response with fold change range of 2 to 3.62 including DNMT3B (3.41-fold), APOD (3.62-fold), CSF1 (3.55-fold) and APOE (3.04-fold). Some proteins were involved in multiple functions: For example, CD44 and S100A8 were involved in all functions associated with biological process while MMRN1 were involved in only the stress response and cell adhesion function.
Validation of the identified potential urine markers
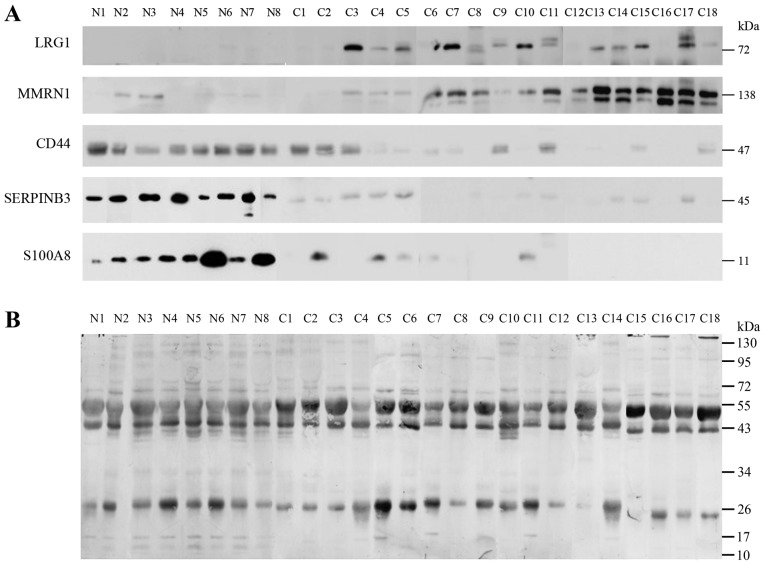
Notable proteins were selected to confirm the association with the urine of normal individuals and patients with different stages of cervical cancer including 8 normal (N1-N8), 2 samples of stage IB1 (C1-C2), 9 samples of stage II (C3-C11), 4 samples of stage IB1 (C12-C15) and 3 samples of stage IVB (C16-C18), as determined by western blot analysis. The expression of LRG1, MMRN1, S100A8, SERPINB3 and CD44 were confirmed as shown in Fig. 3. LRG1 and MMRN1 were upregulated in stages IIB, IIIB and IVB of cervical cancer while S100A8, SERPINB3 and CD44 had reduced expression in stage I and in some stage II cancer samples. Notably, CD44 was highly expressed in normal and stage IB1, and had low expression in stages II, III and IV.
Figure 3.
Western blot analysis. (A) Levels of LRG1, MMRN1, CD44, SERPINB3 and S100A8 in the urine of normal (n=8) individuals and patients with different stages (C1-C2=stage I; C3-C11=stage II; C12-C15=stage III; C16-C18=stage IV) of cervical cancer were studied by western blot analysis. (B) The loading controls were subsequently prepared by staining the membrane with Coomassie blue following western blotting. LRG1, leucine-rich α-2-glycoprotein; MMRN1, multimerin-1; SERPINB3, serpin B3; CD44, cluster of differentiation 44 antigen.
Sensitivity of potential biomarkers
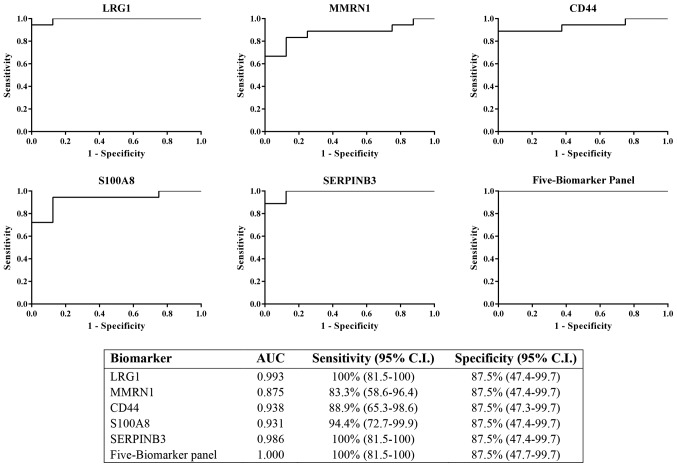
The results from immunodetection by western blot analysis revealed that LRG1, MMRN1, CD44, S100A8 and SERPINB3 could be potential biomarkers for cervical cancer. All five proteins were tested individually and in combination for sensitivity and specificity using logistic regression on SPSS software and ROC curves, respectively. The ROC curves were plotted for these proteins to discriminate between cervical cancer (all stages) and control urine samples (Fig. 4). The results showed the AUC of LRG1, MMRN1, CD44, S100A8 and SERPINB3 were 0.993, 0.785, 0.938, 0.931 and 0.986, respectively. LRG1 and SERPINB3 had the highest AUC among all 5 proteins. The combination of the 5 biomarker panel was calculated and the value of the AUC was increased to 1 (100% sensitivity, 87.5% specificity), which was better than LRG1 or SERPINB3 individually.
Figure 4.
ROC curve of the 5 individual potential biomarkers (LRG1, MMRN1, CD44, S100A8 and SERPINB3) was calculated to determine the area under the curve, sensitivity (95% CI) and specificity (95% CI). The ROC for the 5-biomarker panel was analyzed by combining the 5 potential biomarkers. ROC, receiver operating characteristic; LRG1, leucine-rich α-2-glycoprotein; MMRN1, multimerin-1; SERPINB3, serpin B3; CD44, cluster of differentiation 44 antigen; CI, confidence interval; AUC, area under the curve.
Discussion
Proteomics is a useful technique to identify for cancer biomarkers. Urinary proteomic analysis for biomarker discovery of cervical cancer has been reported previously (15). There have also been reports in cervical cancer cell lines of the deregulation of cytoskeletal-associated proteins (22). Two-dimensional gel electrophoresis followed by MS has been used for a decade and non-gel based-MS has now become popular so label-free shotgun proteomics has been a good choice to produce very effective results for the global profiles of the samples. In the present study, quantitative label-free MS analysis was employed to identify the differences in the expression of urinary proteins in pooled cancer and normal samples. Progenesis QI software was used to analyze the differential expression between these pooled samples. The following criteria was used in this experiment: i) An ANOVA score ≤0.05; ii) proteins with ≥2 matched peptides; and iii) a Mascot score of ≥40. The present study identified a total of 133 proteins (1% FDR).
Blood coagulation and fibrinolysis is one of the main functions in the urinary proteins of cervical cancer. The proteins of interest were coagulation factor V, FGB, FGG, KNG1, MMRN1, LRP2 and MYLK. Following validation via western blot analysis with 8 normal urine samples, (2 of stage I, 9 of stage II, 4 of stage III and 3 of stage IV separately), the proteins LRG1, MMRN1, S100A8, SERPINB3 and CD44 were revealed to be potential multiple biomarkers for cervical cancer. The upregulation of LRG1 and MMRN1 in stages II, III and IV and the downregulation of S100-A8 and SERPINB3 in stages I, II, III and IV were of interest. In addition, CD44 was highly expressed in normal samples and stage IB1, and had reduced expression in stages II, III and IV. Following the immunodetection results, ROC curve and logistic regression analysis were employed to analyze all 5 potential biomarkers. The results of the individual test for discriminatory ability using ROC analysis revealed the highest AUC of 0.993 and 0.986 for LRG1 and SERPINB3, respectively. When the panel of 5 biomarkers was combined, the value of the AUC was increased from ~0.99 in 2 potentially good biomarkers (LRG1 and SERPINB3) to 1 with 100% sensitivity and 87.5% specificity. On the other hand, combination of the panel of 5 biomarkers improved the distinguished power between two diagnostic groups (normal/cancer). Since an individual biomarker is potentially a good diagnostic marker for cervical cancer, the panel of 5 biomarkers is a better option for the identification of cervical cancer patients.
To the best of our knowledge, MMRN1, for the first time, was revealed to be highly expressed in urinary samples of cervical cancer. This protein was also shown to be an isoform-1 of MMRN1 due to the spectrum of peptides belonging to this isoform; for example, KIENLTSAVNSLNFIIK (residue 677–693) and NTDNIIYPEEYSSCSR (residue 1,032–1,047) belong to isoform-1. The MMRN-1 precursor has a molecular weight of 135.75–136 Da. This protein was reported to be a potential serum biomarker in multiple myeloma as it decreased in patients with cancer (23), and was downregulated in non-small cell lung cancer (24). MMRN1 was also revealed to serve an important role in the storage and stabilization of factor V in platelets and was reported to have a function in the extracellular matrix or as an adhesive protein.
LRG1, which serves a role in cell survival, human malignancies, proliferation and inhibits apoptosis, was upregulated in various types of cancers including hepatocellular carcinoma, pancreatic cancer and ovarian cancer (25–27). The expression of LRG1 was 2.72-fold upregulated in the urine of patients with cervical cancer when compared with normal samples; to the best of our knowledge, this is the first time this protein has been associated with this type of cancer. The immunodetection of the expression of LRG1 was present only in the cervical cancer stages II to IV.
The expressions of S100A8, CD44 and SERPINB3 were downregulated when compared with the urine of normal and different stages of cancer samples as determined by label free MS and confirmation via western blotting.
Protein S100A8/A9 or calprotactin, a member of the S100 family, is a heterogenous multimer of the subunit S100A8 and S100A9. It is an EF-hand calcium-binding protein and has been shown to be involved in cell cycle regulation, differentiation, invasion and extracellular matrix cell adhesion (28,29). This protein expression is upregulated in various types of cancers by differential regulation in malignancies and is dependent on the origin of cells and tissues. The adenocarcinomas were diagnosed via the occurrence of tumorigenesis. Examples of these tissues include prostate (30), gastric (31), thyroid (32), colorectal (33), ovarian (34), bladder (35) and pancreatic ductal adenocarcinoma (36). By contrast, downregulated expression of S100A8/A9 was observed in tissues of squamous cell carcinomas of head and neck (HNSCC) (28), cervical (37), oral, oropharyngeal, esophageal and nasopharyngeal (38,39). Loss of S100A8/A9 in tumors was reported to be strongly associated with higher grades, poor squamous differentiation and poorer survival of patients with HNSCC (40). The present study demonstrated no S100A8 expression in cancer patients, which was in agreement with an earlier report (28,40,41).
CD44 was first observed on the surface of granulocytes, T-lymphocytes and cortical thymocytes by Dalchau et al (42). It is a protein of the multifunctional family of the type I transmembrane and is involved in the adhesion, migration and metastasis of cells. The changes in CD44 expression serve a role in malignant tumors. A previous study reported that the loss of CD44 expression predicted higher advanced stages of oral squamous cell carcinoma (43). The reduction of CD44 expression was reported to be involved in high invasiveness of tongue SCC and lead to the metastasis of cervical lymph node (44). The expression of CD44 in the urine of bladder cancer and urothelial carcinoma patients was also reported previously and further studies were performed in the combination with other proteins (45,46). The present results revealed that there was a lower expression of CD44 in the urine samples of stages II, III and IV cervical cancer patients but high expression was observed in normal and stage I patients. Some studies have reported CD44 as a biomarker for cervical cancer from tissue samples (47) and they revealed that a lower expression of CD44 was an indicator of high invasiveness of tumors by increasing the rate of cervical cancer lymph node metastasis. The mediation of CD44 on constitutive type I receptor signaling was also observed in cervical cancer cells (48).
SERPINB3, a serine protease inhibitor member, has been detected in many types of cells including leukocytes, normal epithelium and tumors of epithelial origins for example epithelium of cervix uterine, tonsils, esophagus and tongue. SERPINB3 was also expressed in the amniotic fluid, saliva and respiratory secretions of healthy people (49). SERPINB3 has been found in the serum of patients with many types of SCC including HNSCC (50) and in patients with psoriasis (51). It is involved in the regulation of apoptosis via many mechanisms (52). In Matrigel assays, when cancer cell invasion was decreased, SERPINB3 was overexpressed (53). The present results revealed that SERPINB3 was overexpressed in all 9 normal urine samples but had lower expression in stage I samples, and in 3 out of 9 stage II samples and some of stage III, 2 out of 5 samples of stage III and 1 out of 3 samples of stage IV.
Our study aimed to demonstrate different urinary protein profile between normal HPV negative and cervical cancer patients, so we did not include preinvasive diseases as CIN1, CIN2 or CIN3. We evaluated the potential biomarkers (MMRN1, LRG1, S100A8, SERPINB3 and CD44 using immunodetection with different stages (stage I to stage IV) of cervical cancer. For further study, we will collect all types of samples including precancerous state and confirm whether these proteins could detect in early stage of cervical cancer or not.
In conclusion, quantitative MS analysis could identify the up- and downregulated proteins when comparing the urine of normal individuals and cervical cancer patients. To the best of our knowledge, MMRN1 and LRG1 were detected for the first time in cervical cancer. MMRN1, LRG1, S100A8, SERPINB3 and CD44 were selected for validation by western blot analysis. The results revealed that MMRN1 and LRG1 were upregulated, while S100A8, SERPINB3 and CD44 were downregulated in cancer patients. ROC analysis demonstrated that LRG1 and SERPINB3 could be used alone, while these 5 proteins could also be combined to detect the occurrence of cervical cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Chulabhorn Research Institute (grant no. BC 2008-02) and Chulabhorn Royal Academy (grant no. 31/2554).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DC and KWa performed the experiments including mass spectrometry, progenesis analysis, western blot analysis and data analysis, and helped with manuscript preparation. CS and CW designed the study, developed the methodology and interpreted the results. PS, SK and PDNA made substantial contributions to the analysis and interpretation of data, wrote part of manuscript and performed the literature search. KS, JC and CV performed the statistical and bioinformatics analyses. DC, KWa, PS, CW, KS, JC and CV prepared the figures. CS wrote the manuscript and prepared the original draft. JS supervised the quality of the study design and gave final approval of the manuscript prior to submission. NS performed the clinical data collection and provided the HPV genotyping data. NK and NP performed the patient recruitment procedures, as well as the surgical operations and treatments on the enrolled subjects. KWi performed the urine sample analyses and data collection. WU was involved in sample and data collection. TS performed the histological examination and provided the pathology information. JS, CA and NMP provided technical assistance and contributed to the study design. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethical Review Board of the Chulabhorn Hospital (no. 31/2554), and written informed consent was obtained from the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers CD, Rebelo M, Parkin DM, Forman D, Bray F. 11 [Internet] Lyon, France: International Agency for Research on Cancer; 2013. [Dec 10;2016 ]. GLOBOCAN 2012 v1.0, cancer incidence and mortality worldwide: IARC cancer base No. [Google Scholar]
- 3.von Knebel Doeberitz M, Reuschenbach M, Schmidt D, Bergeron C. Biomarkers for cervical cancer screening: The role of p16(INK4a) to highlight transforming HPV infections. Expert Rev Proteomics. 2012;9:149–163. doi: 10.1586/epr.12.13. [DOI] [PubMed] [Google Scholar]
- 4.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 5.Ostör AG. Natural history of cervical intraepithelial neoplasia: A critical review. Int J Gynecol Pathol. 1993;12:186–192. doi: 10.1097/00004347-199304000-00018. [DOI] [PubMed] [Google Scholar]
- 6.Van Keer S, Pattyn J, Tjalma WAA, Van Ostade X, Ieven M, Van Damme P, Vorsters A. First-void urine: A potential biomarker source for triage of high-risk human papillomavirus infected women. Eur J Obstet Gynecol Reprod Biol. 2017;216:1–11. doi: 10.1016/j.ejogrb.2017.06.036. [DOI] [PubMed] [Google Scholar]
- 7.Ng HT, Yen MS, Chao KC, Chen CY, Yuan CC. Radical hysterectomy: Past, present, and future. Eur J Gynaecol Oncol. 2005;26:585–588. [PubMed] [Google Scholar]
- 8.Hoffman MS. Extent of radical hysterectomy: Evolving emphasis. Gynecol Oncol. 2004;94:1–9. doi: 10.1016/j.ygyno.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 9.Umanzor J, Aguiluz M, Pineda C, Andrade S, Erazo M, Flores C, Santillana S. Concurrent cisplatin/gemcitabine chemotherapy along with radiotherapy in locally advanced cervical carcinoma: A phase II trial. Gynecol Oncol. 2006;100:70–75. doi: 10.1016/j.ygyno.2005.07.132. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka T, Kokawa K, Umesaki N. Preoperative chemotherapy with irinotecan and mitomycin for FIGO stage IIIb cervical squamous cell carcinoma: A pilot study. Eur J Gynaecol Oncol. 2005;26:605–607. [PubMed] [Google Scholar]
- 11.Linghu H, Xu XR, Mei YY, Tang JY, Tang LD, Sun T. Response of early stage bulky cervical squamous carcinoma to preoperative adjuvant chemotherapy. Chin Med Sci J. 2004;19:116–119. [PubMed] [Google Scholar]
- 12.Candelaria M, Garcia-Arias A, Cetina L, Dueñas-Gonzalez A. Radiosensitizers in cervical cancer. Cisplatin and beyond. Radiat Oncol. 2006;1:15. doi: 10.1186/1748-717X-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goto T, Kino N, Shirai T, Fujimura M, Takahashi M, Shiromizu K. Late recurrence of invasive cervical cancer: Twenty years' experience in a single cancer institute. J Obstet Gynaecol Res. 2005;31:514–519. doi: 10.1111/j.1447-0756.2005.00343.x. [DOI] [PubMed] [Google Scholar]
- 14.Anderson NG, Anderson NL, Tollaksen SL. Proteins of human urine. I. Concentration and analysis by two-dimensional electrophoresis. Clin Chem. 1979;25:1199–1210. [PubMed] [Google Scholar]
- 15.Aobchey T, Niamsup H, Siriaree S, Sookkheo B, Boonyapranai K, Chen ST. Proteomic analysis of candidate prognostic urinary marker for cervical cancer. J Proteomics Bioinform. 2013;6:245–251. doi: 10.4172/jpb.1000287. [DOI] [Google Scholar]
- 16.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong JW, Cagney G. An overview of label-free quantitation methods in proteomics by mass spectrometry. Methods Mol Biol. 2010;604:273–283. doi: 10.1007/978-1-60761-444-9_18. [DOI] [PubMed] [Google Scholar]
- 18.Apgar BS, Zoschnick L, Wright Jr TC. The 2001 Bethesda system terminology. Am Fam Physician. 2003;68:1992–1998. [PubMed] [Google Scholar]
- 19.Kantathavorn N, Mahidol C, Sritana N, Sricharunrat T, Phoolcharoen N, Auewarakul C, Teerayathanakul N, Taepisitpong C, Saeloo S, Sornsamdang G, et al. Genotypic distribution of human papillomavirus (HPV) and cervical cytology findings in 5906 Thai women undergoing cervical cancer screening programs. Infect Agent Cancer. 2015;10:7. doi: 10.1186/s13027-015-0001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Beetham R, Cattell WR. Proteinuria: Pathophysiology, significance and recommendations for measurement in clinical practice. Ann Clin Biochem. 1993;30:425–434. doi: 10.1177/000456329303000502. [DOI] [PubMed] [Google Scholar]
- 22.Pappa KI, Lygirou V, Kontostathi G, Zoidakis J, Makridakis M, Vougas K, Daskalakis G, Polyzos A, Anagnou NP. Proteomic analysis of normal and cancer cervical cell lines reveals deregulation of cytoskeleton-associated proteins. Cancer Genomics Proteomics. 2017;14:253–266. doi: 10.21873/cgp.20036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang HT, Tian EB, Chen YL, Deng HT, Wang QT. Proteomic analysis for finding serum pathogenic factors and potential biomarkers in multiple myeloma. Chin Med J (Engl) 2015;128:1108–1113. doi: 10.4103/0366-6999.155112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Välk K, Vooder T, Kolde R, Reintam MA, Petzold C, Vilo J, Metspalu A. Gene expression profiles of non-small cell lung cancer: Survival prediction and new biomarkers. Oncology. 2010;79:283–292. doi: 10.1159/000322116. [DOI] [PubMed] [Google Scholar]
- 25.He X, Wang Y, Zhang W, Li H, Luo R, Zhou Y, Liao CL, Huang H, Lv X, Xie Z, He M. Screening differential expression of serum proteins in AFP-negative HBV-related hepatocellular carcinoma using iTRAQ-MALDI-MS/MS. Neoplasma. 2014;61:17–26. doi: 10.4149/neo_2014_001. [DOI] [PubMed] [Google Scholar]
- 26.Furukawa K, Kawamoto K, Eguchi H, Tanemura M, Tanida T, Tomimaru Y, Akita H, Hama N, Wada H, Kobayashi S, et al. Clinicopathological significance of leucine-rich α2-glycoprotein-1 in sera of patients with pancreatic cancer. Pancreas. 2015;44:93–98. doi: 10.1097/MPA.0000000000000205. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Xie X, Nie S, Buckanovich RJ, Lubman DM. Altered expression of sialylated glycoproteins in ovarian cancer sera using lectin-based ELISA assay and quantitative glycoproteomics analysis. J Proteome Res. 2013;12:3342–3352. doi: 10.1021/pr400169n. [DOI] [PubMed] [Google Scholar]
- 28.Khammanivong A, Wang C, Sorenson BS, Ross KF, Herzberg MC. S100A8/A9 (calprotectin) negatively regulates G2/M cell cycle progression and growth of squamous cell carcinoma. PLoS One. 2013;8:e69395. doi: 10.1371/journal.pone.0069395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva EJ, Argyris PP, Zou X, Ross KF, Herzberg MC. S100A8/A9 regulates MMP-2 expression and invasion and migration by carcinoma cells. Int J Biochem Cell Biol. 2014;55:279–287. doi: 10.1016/j.biocel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermani A, Hess J, De Servi B, Medunjanin S, Grobholz R, Trojan L, Angel P, Mayer D. Calcium-binding proteins S100A8 and S100A9 as novel diagnostic markers in human prostate cancer. Clin Cancer Res. 2005;11:5146–5152. doi: 10.1158/1078-0432.CCR-05-0352. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Chang EW, Wong SC, Ong SM, Chong DQ, Ling KL. Increased myeloid-derived suppressor cells in gastric cancer correlate with cancer stage and plasma S100A8/A9 proinflammatory proteins. J Immunol. 2013;190:794–804. doi: 10.4049/jimmunol.1202088. [DOI] [PubMed] [Google Scholar]
- 32.Ito Y, Arai K, Nozawa R, Yoshida H, Hirokawa M, Fukushima M, Inoue H, Tomoda C, Kihara M, Higashiyama T, et al. S100A8 and S100A9 expression is a crucial factor for dedifferentiation in thyroid carcinoma. Anticancer Res. 2009;29:4157–4161. [PubMed] [Google Scholar]
- 33.Duan L, Wu R, Ye L, Wang H, Yang X, Zhang Y, Chen X, Zuo G, Zhang Y, Weng Y, et al. S100A8 and S100A9 are associated with colorectal carcinoma progression and contribute to colorectal carcinoma cell survival and migration via Wnt/β-catenin pathway. PLoS One. 2013;8:e62092. doi: 10.1371/journal.pone.0062092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott HW, Lindner H, Sarg B, Mueller-Holzner E, Abendstein B, Bergant A, Fessler S, Schwaerzler P, Zeimet A, Marth C, Illmensee K. Calgranulins in cystic fluid and serum from patients with ovarian carcinomas. Cancer Res. 2003;63:7507–7514. [PubMed] [Google Scholar]
- 35.Minami S, Sato Y, Matsumoto T, Kageyama T, Kawashima Y, Yoshio K, Ishii J, Matsumoto K, Nagashio R, Okayasu I. Proteomic study of sera from patients with bladder cancer: Usefulness of S100A8 and S100A9 proteins. Cancer Genomics Proteomics. 2010;7:181–189. [PubMed] [Google Scholar]
- 36.El Gammal AT, Sturm JH, Pinnschmidt HO, Hofmann BT, Bellon E, Ghadban T, Melling NT, Bachmann KA, Izbicki J, Bockhorn M, et al. Protein S100A8/A9: A potential new biomarker for pancreatic diseases. Int J Clin Endocrinol Metab. 2017;3:23–28. doi: 10.17352/ijcem.000025. [DOI] [Google Scholar]
- 37.Tugizov S, Berline J, Herrera R, Penaranda ME, Nakagawa M, Palefsky J. Inhibition of human papillomavirus type 16 E7 phosphorylation by the S100 MRP-8/14 protein complex. J Virol. 2005;79:1099–1112. doi: 10.1128/JVI.79.2.1099-1112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong JP, Ding F, Zhou CN, Wang XQ, Miao XP, Wu M, Liu ZH. Loss of myeloid-related proteins 8 and myeloid-related proteins 14 expression in human esophageal squamous cell carcinoma correlates with poor differentiation. World J Gastroenterol. 2004;10:1093–1097. doi: 10.3748/wjg.v10.i8.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Cai Y, Xu H, Zhao J, Xu X, Han YL, Xu ZX, Chen BS, Hu H, Wu M, Wang MR. Expression of MRP14 gene is frequently down-regulated in Chinese human esophageal cancer. Cell Res. 2004;14:46–53. doi: 10.1038/sj.cr.7290201. [DOI] [PubMed] [Google Scholar]
- 40.Argyris PP, Slama ZM, Ross KF, Khammanivong A, Herzberg MC. Calprotectin and the initiation and progression of head and neck cancer. J Dent Res. 2018;97:674–682. doi: 10.1177/0022034518756330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khammanivong A, Sorenson BS, Ross KF, Dickerson EB, Hasina R, Lingen MW, Herzberg MC. Involvement of calprotectin (S100A8/A9) in molecular pathways associated with HNSCC. Oncotarget. 2016;7:14029–14047. doi: 10.18632/oncotarget.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalchau R, Kirkley J, Fabre JW. Monoclonal antibody to a human leukocyte-specific membrane glycoprotein probably homologous to the leukocyte-common (L-C) antigen of the rat. Eur J Immunol. 1980;10:737–744. doi: 10.1002/eji.1830101003. [DOI] [PubMed] [Google Scholar]
- 43.Kosunen A, Pirinen R, Ropponen K, Pukkila M, Kellokoski J, Virtaniemi J, Sironen R, Juhola M, Kumpulainen E, Johansson R, et al. CD44 expression and its relationship with MMP-9, clinicopathological factors and survival in oral squamous cell carcinoma. Oral Oncol. 2007;43:51–59. doi: 10.1016/j.oraloncology.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 44.Mostaan LV, Khorsandi MT, Sharifian SM, Shandiz FH, Mirashrafi F, Sabzari H, Badiee R, Borghei H, Yazdani N. Correlation between E-cadherin and CD44 adhesion molecules expression and cervical lymph node metastasis in oral tongue SCC: Predictive significance or not. Pathol Res Pract. 2011;207:448–451. doi: 10.1016/j.prp.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 45.Urquidi V, Kim J, Chang M, Dai Y, Rosser CJ, Goodison S. CCL18 in a multiplex urine-based assay for the detection of bladder cancer. PLoS One. 2012;7:e37797. doi: 10.1371/journal.pone.0037797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arville B, O'Rourke E, Chung F, Amin M, Bose S. Evaluation of a triple combination of cytokeratin 20, p53 and CD44 for improving detection of urothelial carcinoma in urine cytology specimens. Cytojournal. 2013;10:25. doi: 10.4103/1742-6413.123784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao S, Zhou Y, Jiang J, Yuan L, Xue M. CD44 affects the expression level of FOS-like antigen 1 in cervical cancer tissues. Mol Med Rep. 2014;9:1667–1674. doi: 10.3892/mmr.2014.2010. [DOI] [PubMed] [Google Scholar]
- 48.Wobus M, Kuns R, Wolf C, Horn LC, Köhler U, Sheyn I, Werness BA, Sherman LS. CD44 mediates constitutive type I receptor signaling in cervical carcinoma cells. Gynecol Oncol. 2001;83:227–234. doi: 10.1006/gyno.2001.6369. [DOI] [PubMed] [Google Scholar]
- 49.Kato H. Expression and function of squamous cell carcinoma antigen. Anticancer Res. 1996;16:2149–2153. [PubMed] [Google Scholar]
- 50.Sun Y, Sheshadri N, Zong WX. SERPINB3 and B4: From biochemistry to biology. Semin Cell Dev Biol. 2017;62:170–177. doi: 10.1016/j.semcdb.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamanaka S, Ujihara M, Numa F, Kato H. Serum level of squamous cell carcinoma antigen as a new indicator of disease activity in patients with psoriasis. Arch Dermatol. 1997;133:393–395. doi: 10.1001/archderm.133.3.393. [DOI] [PubMed] [Google Scholar]
- 52.Vidalino L, Doria A, Quarta S, Zen M, Gatta A, Pontisso P. SERPINB3, apoptosis and autoimmunity. Autoimmun Rev. 2009;9:108–112. doi: 10.1016/j.autrev.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 53.Nakashima T, Yasumatsu R, Kuratomi Y, Masuda M, Kuwano T, Toh S, Umezaki T, Cataltepe S, Silverman GA, Komune S. Role of squamous cell carcinoma antigen 1 expression in the invasive potential of head and neck squamous cell carcinoma. Head Neck. 2006;28:24–30. doi: 10.1002/hed.20293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.