Abstract
Hepatocellular carcinoma (HCC) ranks the third major cause of cancer-associated mortality globally. Numerous studies have attempted to elucidate the underlying mechanisms of HCC using various biomarkers. In the present study, two expression profiles datasets from Gene Expression Omnibus (GSE76427 and GSE84402) and data associated with liver cancer samples from The Cancer Genome Atlas (TCGA) were downloaded for integrated analysis. Five differentially expressed genes (DEGs) exhibiting high expression, including ubiquitin-conjugating enzyme 2C (UBE2C), topoisomerase II α, pituitary tumor transforming gene 1, glypican-3 and polycomb-repressive complex 1, were selected and considered as candidate genes. Enrichment analysis demonstrated that these genes were associated with Gene Ontology terms of cellular components and molecular functions, including regulation of apoptosis, stabilization of p53 and Anaphase Promoting Complex/Cyclosome (APC/C) (APC/C:Cdc20)-mediated degradation of Securin. The expression profiles of these genes in HCC, other human malignancies and different human HCC cell lines were validated using GSE14520, GSE3500 and TCGA data. The results confirmed the upregulation of UBE2C in tissues from patients with HCC or other human malignancies and human liver cancer cell lines, compared with the expression levels in the corresponding adjacent non-tumor tissues and cell lines, respectively. Patients with HCC who exhibited an increased messenger RNA level of UBE2C exhibited a significantly shorter survival time. The results of the present study suggest that the overexpression of UBE2C may be used as a novel prognostic biomarker of HCC.
Keywords: hepatocellular carcinoma, ubiquitin-conjugating enzyme 2C, bioinformatics, tumor target
Introduction
Hepatocellular carcinoma (HCC) is the main pathological type of liver cancer. The incidence of HCC in developed countries has significantly increased in the past few decades (1). Due to its complexity, heterogeneity and high recurrence following surgical resection, HCC ranks the second or third major cause of cancer-associated mortalities in the world (2,3). The fact that the diagnosis of HCC primarily depends on the serologic alterations that occur in advanced HCC restricts the therapeutic options for HCC (4). Thus, the identification of specific molecular targets for early diagnosis and timely treatment for HCC is imperative (5).
With the extensive application of gene chip technologies, abundant expression profile information and screening of differentially expressed genes (DEGs), biomarkers in tumor tissues could be detected efficiently by integrating publicly available datasets (3,6,7). Several studies based on the integrated analysis of microarray data have provided valuable insights to the underlying molecular mechanisms of diseases (7–10). For instance, Gan et al (10) reported that microRNA (miR)-145-5p was associated with lymph node metastasis in non-small cell lung cancer through revealing that the expression levels of miR-145-5p were significantly lower in these tissues compared with those in healthy tissues following a meta-analysis and integrated analysis of microarray data. Guo et al (7) performed an integrated bioinformatics analysis of four colorectal cancer (CRC) expression profiles in Gene Expression Omnibus (GEO) and identified 31 key candidate genes in CRC. Similarly, Liang et al (3) and Zhang et al (11) analyzed a publicly available data of HCC in GEO and The Cancer Genome Atlas (TCGA) database, and explored the diagnostic role of miR-133a-3p and miR-224-5p in HCC, respectively. These findings provide novel and valuable directions for further research into various cancer types, including HCC. It was proposed that a thorough re-analysis using integrated bioinformatics methods combined with the newest datasets would be innovative, and may provide novel additional insights into the underlying molecular mechanisms of HCC.
The GEO database is a public database of microarray profile founded by the National Center for Biotechnology Information and provides access to high-throughput screening of abnormally expressed genes in cancerous tissues (7). Numerous studies have explored microarray data profiling (8,9). However, although reliable molecular biomarkers have been clinically observed in patients with HCC, the underlying molecular mechanisms of HCC remain to be clarified.
The present study identified a number of DEGs in HCC tissues by integrated analysis of the two newest datasets (GSE76427 and GSE84402) from the GEO database and HCC samples in TCGA (12,13). Functional enrichment was conducted on key candidate genes with the DEGs threshold of | log2 (fold change) |≥1.5 in the GEO data or fold change ≥10 in the TCGA data and P≤0.001. GSE14520 and GSE3500 (14,15), and the TCGA dataset with expression profiles of cancer and adjacent non-cancerous tissues were used for validation of candidate genes. In addition, overall survival (OS) analysis of candidate genes was performed to analyze the potential of using such genes as prognostic biomarkers of HCC. The present study may provide novel insights into the molecular mechanism of HCC and may serve as a reference for clinical studies in HCC.
Materials and methods
Microarray data information
The gene expression profiles of GSE76427, GSE84402, GSE14520 and GSE3500 were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/), and gene expression data for HCC and adjacent non-cancerous tissue were obtained. The newest datasets available (GSE76427 and GSE84402) were based on the platforms GPL10558 (Illumina HumanHT-12 v4.0 Expression BeadChip; Illumina, Inc., San Diego, CA, USA) and GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array; Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA), respectively. GSE76427 data included 115 HCC primary tumor tissues and 52 adjacent non-tumor tissues (submission date, 30th December 2015) (12). GSE84402 included 14 HCC tissues and 14 adjacent non-tumor tissues (submission date, 14th July 2016) (13). GSE14520 was based on the GPL571 (Affymetrix Human Genome U133A 2.0 Array; Affymetrix; Thermo Fisher Scientific, Inc.) and GPL3921 platforms (Affymetrix HT Human Genome U133A Array; Affymetrix; Thermo Fisher Scientific, Inc.). GSE14520 included 66 tumor and paired non-tumor samples (submission date, 22nd January 2009) (16). GSE3500 data was based on 13 platforms (GPL2648, GPL2649, GPL2831, GPL2868, GPL2906, GPL2935, GPL2938, GPL2948, GPL3007, GPL3008, GPL3009, GPL3010 and GPL3011), and included 102 primary HCC tissues (from 82 patients), 74 non-tumor tissues (from 72 patients), seven benign tumor tissues (three adenoma and four focal nodular hyperplasia), 10 metastatic cancer tissues, and 10 HCC cell lines (14).
Another expression profile of liver cancer was obtained from the TCGA database by Cancer RNA-seq Nexus (CRN) online (http://syslab4.nchu.edu.tw/) using University of California, Santa Cruz Refseq Gene Array, which consisted of 84 HCC tissues and 42 adjacent non-tumor tissues. GSE76427, GSE84402 and TCGA data were used for the identification of DEGs. The GSE14520 and GSE3500 datasets and TCGA data were used for the validation of expression profiles. The details and patient information of the datasets in GEO and TCGA are listed in Table I.
Table I.
Detailed information of the datasets in GEO and TCGA patients.
| GEO ID | GSE14520 | GSE3500 | GSE76427 | GSE84402 | TCGA |
|---|---|---|---|---|---|
| Total no. of patients | 22 | 82 | Not mentioned | 14 | 371 |
| Total no. of samples | 132 | 193 | 167 | 14 | 373 |
| No. of non-tumor samples | 66 (paired non-tumor samples) | 74 (non-tumor samples) | 52 (adjacent non-tumor tissues) | 14 (>5 cm laterally from the edge of the cancerous) region | 42 (healthy tissues) |
| No. of primary HCC samples | 66 (tumor samples) | 102 (tumor samples from HBV+ patients) | 115 (primary tumor tissues) | 14 (percentage of tumor cells >70%) | 84 (tumor samples) |
| Tumor types | Hepatocellular carcinoma | Hepatocellular carcinoma | Hepatocellular carcinoma | Hepatocellular carcinoma | Hepatocellular carcinoma |
| Grading of tumors (TNM stage) | No subdivision | No subdivision | No subdivision | No subdivision | Stage II |
| Pathological grade | No subdivision | No subdivision | No subdivision | No subdivision | No subdivision |
| Comment | N.A. | N.A. | Percentage of HCC patients with HBV infection and cirrhosis were 46% and 54%. | N.A. | Using CRN (Cancer RNA-Seq Nexus tool) for analysis. |
GEO, Gene Expression Omnibus; TCGA, The Cancer Genome Atlas; HCC, hepatocellular carcinoma; TNM, tumor node metastasis; HBV, hepatitis B virus.
Identification of overexpressed DEGs in HCC tissues
The raw data from the downloaded datasets GSE76427 and GSE84402 as well as TCGA data were analyzed following integrated transformation and correlation analysis using Funrich (version 3.3; http://www.funrich.org/) and Morpheus software (https://software.broadinstitute.org/morpheus). Data processing was conducted by two professional analysts. DEGs between human HCC tissues and paired non-tumor or healthy tissues were defined using the Student's t-test with a cut-off criterion of P<0.001 and log2 (fold change) |≥1.5 in GEO data or fold change |≥10 in TCGA data. Upon overlapping with Funrich software, candidate genes with highly overexpressed levels in HCC tissues were identified in the three datasets.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of candidates
Functional enrichment analysis of GO and KEGG pathways was parsed using the online tool Database for Annotation Visualization and Integrated Discovery (https://david.ncifcrf.gov/), which contains integrated gene visualization. Funrich software was also utilized to identify functional enrichment under a threshold of P<0.05.
Construction of protein-protein interaction (PPI) network and heatmap analysis
DEGs-encoded proteins and PPI network information data were obtained using the Search Tool for the Retrieval of Interacting Genes (STRING) database (http://string-db.org). Cytoscape software (version 3.7.0; http://cytoscape.org/) was used for visualization of the interactions among the candidate DEGs. Heatmap hot spots with gradient from red to blue color, which were generated from the TCGA dataset, were used to show the relative expression level of key candidate genes in HCC.
Validation of the aberrant expression of UBE2C in HCC based on GEO and TCGA datasets
The expression profiles of candidate genes in DEGs were validated using another two GEO data (GSE14520 and GSE3500) with the online tool Oncomine (https://www.oncomine.org/resource/login.html), which functions as a profiler for intuitive expression of the GEO database. An independent sample Student's t-test was used to compare the DEG levels between HCC and non-cancerous tissues. The online tool Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle) and Firebrowse (http://firebrowse.org/) were employed to evaluate the relative transcription level of candidate genes among human malignancies and different human HCC cell lines according to the data obtained from the GEO and TCGA databases.
Survival association of candidate genes with clinicopathological parameters of patients with HCC
Survival analysis of patients with high expression of candidate DEGs in TCGA HCC data was performed using Kaplan-Meier estimator survival curves with several clinicopathological characteristics taken into consideration, including sex and pathological stage. Data was obtained from cBioPortal (http://www.cbioportal.org) and Kaplan-Meier Plotter (http://kmplot.com/analysis/) was used for analysis. A threshold of P<0.05 was used to set the cut-off criterion.
Results
Identification of DEGs and overexpressed candidate genes in human HCC tissues
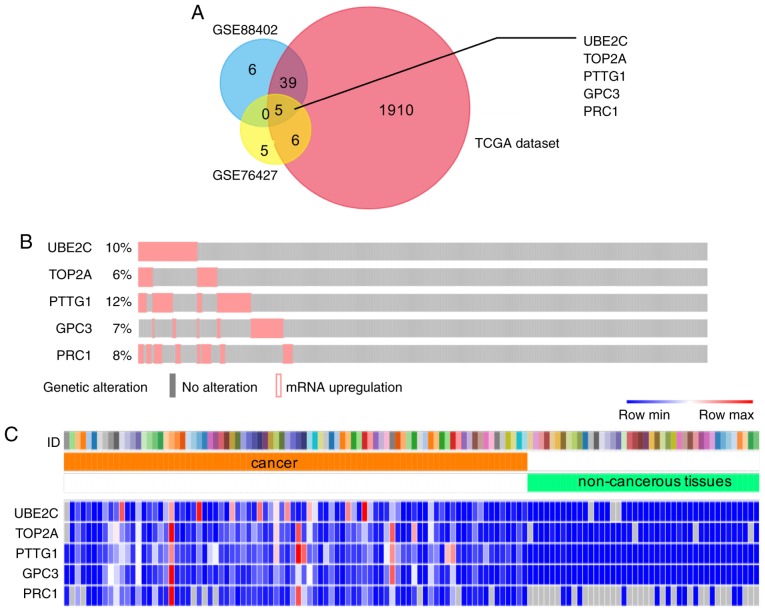
To date, the GEO database contains the most comprehensive public microarray/gene data resources, while TCGA processes the largest quantity of cancer genes. The gene expression profile of HCC and adjacent non-cancerous tissues was downloaded from GEO (GSE76427 and GSE84402) and TCGA. Under the inclusion criteria, a total of 1,650 and 1,960 DEGs were identified from GSE76427, GSE84402 and TCGA data, respectively. Using the FunRich software, five upregulated genes were identified [ubiquitin-conjugating enzyme 2C (UBE2C), topoisomerase II α (TOP2A), pituitary tumor transforming gene 1 (PTTG1), glypican-3 (GPC3) and polycomb-repressive complex 1 (PRC1)], which were overlapped in three groups and were considered as candidate genes for selection of HCC biomarkers (Fig. 1A). The OncoPrint created from 373 HCC tissues in the TCGA database using available data from the cBioPortal website indicated that 27% (100/373) of clinical cases exhibited gene upregulation in patients with HCC (Fig. 1B). A heatmap of these five DEGs in the TCGA data was visualized using STRING and Morpheus software (Fig. 1C), which partly revealed the significant discrepancy between human HCC and healthy tissues.
Figure 1.
DEGs expression profile in the GEO and TCGA datasets. (A) Venn chart of significantly overexpressed genes in the GSE76427, GSE88402 and TCGA datasets. Five significantly overexpressed genes were overlaped in three datasets, including ubiquitin-conjugating enzyme 2C, topoisomerase II α, pituitary tumor transforming gene 1, glypican-3 and polycomb-repressive complex 1. (B) Genetic alteration of these five genes in patients with in TCGA datasets. Pink samples represent upregulation. CBioPortal was used to obtain the transcription level by messenger RNA sequencing analysis of 373 samples in 371 patients with hepatocellular carcinoma. TCGA OncoPrint portal was used. Z-score±2.0 was used as a threshold for aberrant expression in TCGA RNA-Seq V2 data. (C) Heatmap of DEGs in TCGA datasets. Hot spots ranged from red (high expression) to blue (low expression). Search Tool for the Retrieval of Interacting Genes was used to obtain TCGA data, while Morpheus was used to process and visulize data. DEGs were defined with P<0.001 and |log2 (fold change) |≥1.5 in GEO data or fold Change >10 in TCGA data. GEO, Gene Expression Omnibus; TCGA, The Cancer Genome Atlas; DEGs, differentially expressed genes.
GO and KEGG pathway enrichment analysis for DEGs
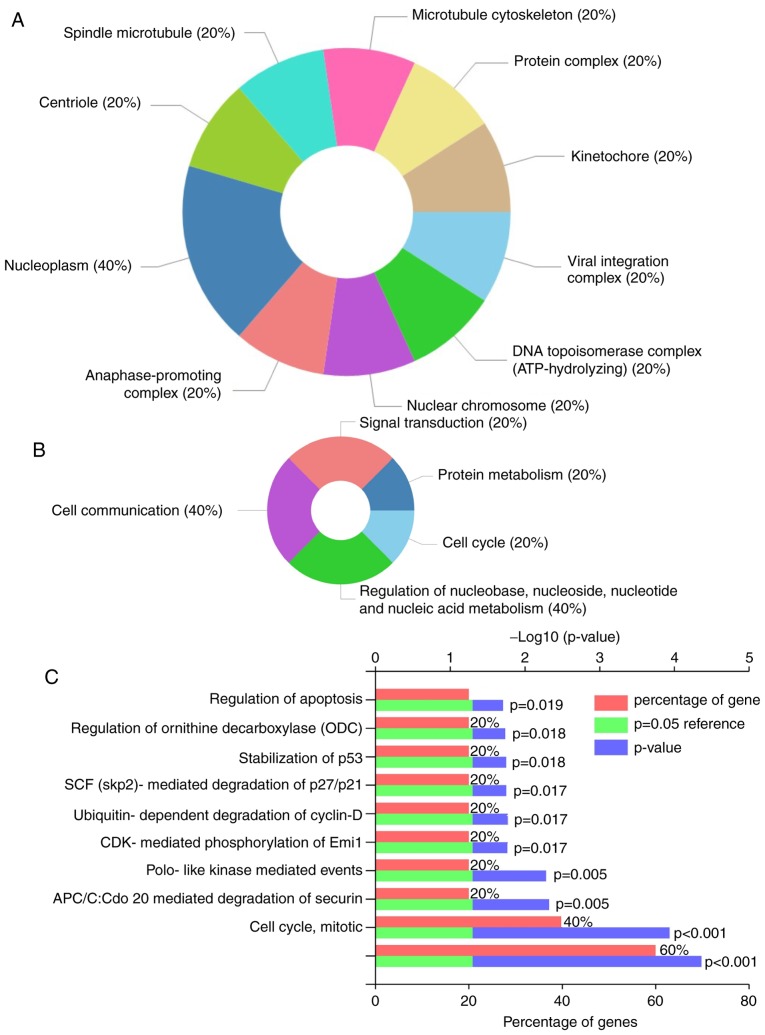
The five overlapped upregulated candidated genes were subjected to functional enrichment analysis via GO. It was observed that they were primarily enriched in: i) Cellular components, including nucleoplasm, nuclear chromosome and kinetochore and DNA topoisomerase complex (ATP-hydrolyzing) (Fig. 2A); and ii) biological processes, including cell communication, signal transduction and cell cycle (Fig. 2B). Candidate genes were associated with various functions, including apoptosis regulation, stabilization of p53 and CDK-mediated phosphorylation, and the removal of cell division cycle 6 (Fig. 2C). These results suggested that these five candidate genes serve important biological roles during HCC development.
Figure 2.
Functional enrichment analysis of DEGs. Charts of DEGs enriched for (A) cellular components, (B) biological processes and (C) molecular functions in Gene Ontology analysis using the online tool Database for Annotation Visualization and Integrated Discovery and Funrich software. P<0.05 was considered to indicate a statistically significant difference. DEGs, differentially expressed genes.
KEGG pathway enrichment analysis revealed that PTTG1 was enriched in two pathways, including ‘Oocyte meiosis’ and ‘Cell cycle’ (P<0.05) (data not shown).
Construction of PPI network
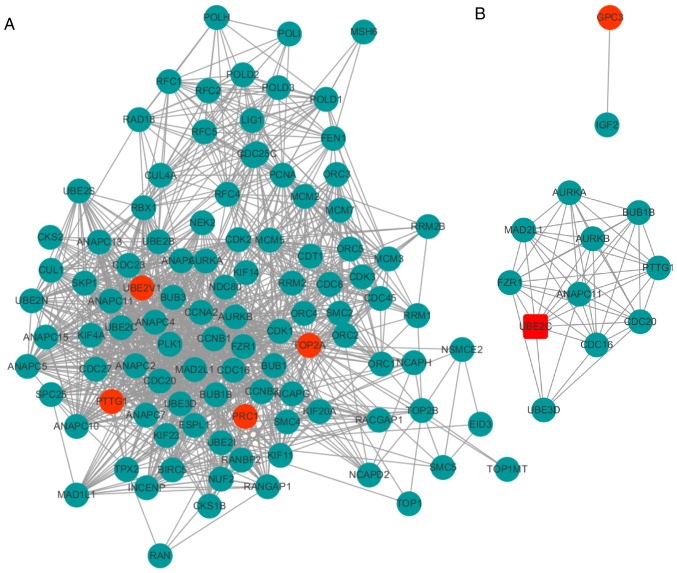
Five DEGs and their interactions were included in a PPI network, in which there were 105 nodes (proteins) and 1,036 edges (interactions) (Fig. 3A). In the integrated PPI network, a module consisting of UBE2C and PTTG1, was identified, which included 11 nodes and 50 edges (Fig. 3B). Based on the expression levels of these five genes, it was observed that UBE2C exhibited relatively higher expression levels compared with other genes in HCC tissues from TCGA. Thus, UBE2C was considered a candidate biomarker for HCC.
Figure 3.
Protein-protein interaction network for selected DEGs. (A) Integrated network for the five candidated genes (UBE2C, topoisomerase II α, pituitary tumor transforming gene 1, glypican-3 and polycomb-repressive complex 1) and their interactions using Search Tool for the Retrieval of Interacting Genes. (B) The module including UBE2C. Nodes indicate proteins and edges represented as lines between DEGs indicate interactions. Red nodes indicate the DEGs discussed in the present manuscript and green nodes indicate other genes. DEGs, differentially expressed genes; UBE2C, ubiquitin-conjugating enzyme 2C.
Validation of UBE2C overexpression in HCC tissues and cell lines
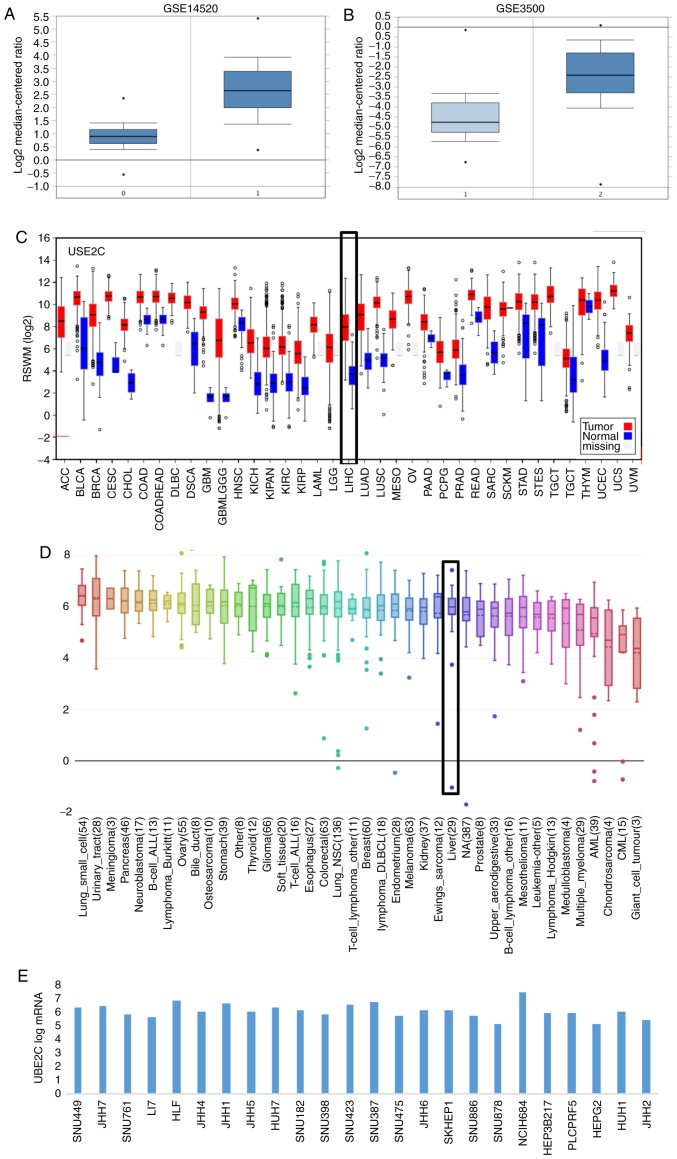
To verify the overexpression of UBE2C in HCC tissues, the GSE14520 and GSE3500 data were downloaded, and the expression profile of UBE2C was analyzed. As expected, the significant overexpression of UBE2C in HCC tissues compared with adjacent non-cancerous tissues was validated in the datasets GSE14520 and GSE3500 (Fig. 4A and B), as well as in other various human malignant tumors, compared with healthy or adjacent non-cancerous tissues in data from the GEO and TCGA databases (Fig. 4C and D). High expression levels of UBE2C in different human liver cell lines was further demonstrated (Fig. 4E). These results revealed that UBE2C serves important roles in the development of HCC and other cancer types, and may function as an oncogene in tumorigenesis.
Figure 4.
Validation of UBE2C overexpression in HCC tissues, cell lines and other cancer tissues in data from GEO and TCGA databases. Validation of UBE2C overexpression in (A) GSE14520 and (B) GSE3500 data. Comparisons between HCC tissues (T) and adjacent non-cancerous tissues (N) were acquired using Oncomine. UBE2C exhibited singnificantly higher expression according to the Student's t-test (P=1.17×10−65 and P=1.06×10−25). (C) UBE2C is overexpressed in numerous human malignant tumors in the TCGA database according to FireBrowse. (D) UBE2C is overexpressed in multiple human malignant tumors in the GEO database uisng CCLE. (E) A relatively high transcriptional level of UBE2C was obseved in liver cancer cell lines. Data are obtained through analysis of the GEO database using the CCLE website. GEO, Gene Expression Omnibus; TCGA, The Cancer Genome Atlas; HCC, hepatocellular carcinoma; UBE2C, ubiquitin-conjugating enzyme 2C; CCLE, Cancer Cell Line Encyclopedia.
Kaplan-Meier analysis of DEGs and UBE2C in HCC tissues
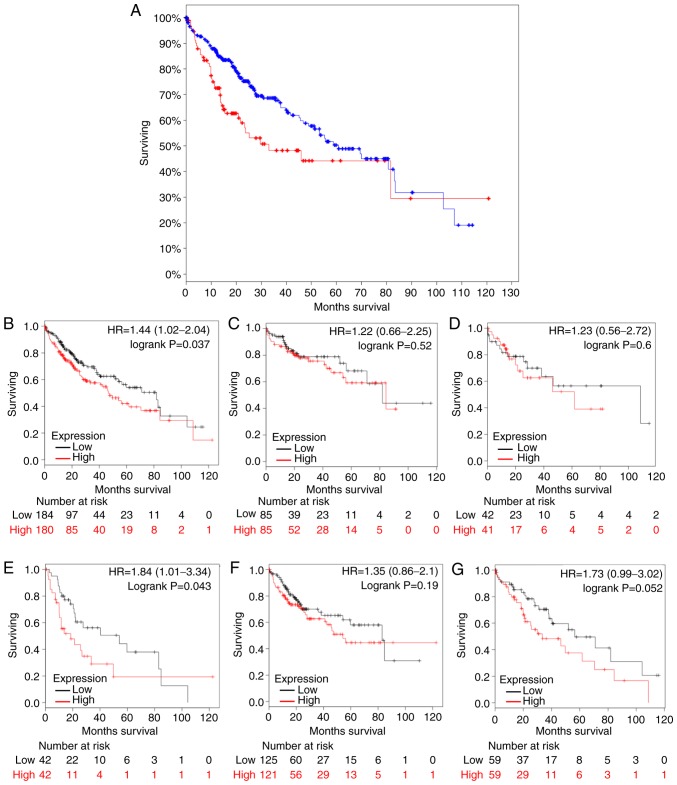
Kaplan-Meier analysis was conducted for five DEGs (UBE2C, TOP2A, PTTG1, GPC3 and PRC1). Survival analysis indicted that patients with overexpression of these five DEGs exhibited significantly significantly shorter survival times (P=0.0127) compared with patients with relatively low expression levels (Fig. 5A). Analysis of UBE2C in cohorts with different clinical characteristics revealed that patients with higher UBE2C expression exhibited short survival times [hazard ratio (HR)=1.44; confidence interval (CI)=1.02–2.04; log-rank P=0.037] (Fig. 5B), and patients in stage III HCC with high UBE2C expression had relatively short survival times compared with patients in stage III HCC with low UBE2C expression (HR=1.84; CI=1.01–3.34; log-rank P=0.043) (Fig. 5C-F). Stage IV was ignored due to low sample size (n<10). Overexpression of UBE2C may serve as a novel indicator of reduced survival time in patients with diagnosed HCC. Thus, UBE2C may be used as a prognostic biomarker for HCC.
Figure 5.
Kaplan-Meier estimator analysis of candidate genes in patients with HCC. (A) Kaplan-Meier plots patients with HCC with five differentially expressed genes (UBE2C, topoisomerase II α, pituitary tumor transforming gene 1, glypican-3 and polycomb-repressive complex 1) overexpression with data recruited from cBioPortal (P=0.0127). The blue line represents HCC patients with relatively low UBE2C, TOP2A, PTTG1, GPC3 and PRC1 mRNA expression, and the red line represents HCC patients with high expression of the aforementioned genes. Log-rank analysis revealed that patients with HCC and high mRNA expression exhibited significant short survival times. (B-G) Kaplan-Meier plots of different clinicopathological features associated with UBE2C overexpression. The black line represents UBE2C low expression and the red line represents UBE2C overexpression. The X and Y axes indicate survival rate and overall survival time (days), respectively. (B) All patients. (C) Patients in clinical stage I. (D) Patients in clinical stage II. (E) Patients in clinical stage III. (F) Male and (G) female patients with HCC. Log-rank analysis revealed that patients with HCC in clinical stage III with high UBE2C mRNA expression exhibited significantly lower survival times compared with patients with low UBE2C mRNA expression. Log rank P-value and 95% confidence intervals of the hazard ratio were used for statistical analysis. HCC, hepatocellular carcinoma; mRNA, messenger RNA; UBE2C, ubiquitin-conjugating enzyme 2C.
Discussion
The overall survival of patients with HCC is markedly short, and morbidity is increasing in developed and developing contries. In USA, 22,000 newly diagnosed cases occurred annually, and 18,000 patients succumbed to HCC (4). Approximately 1,000,000 patients in the world are diagnosed with HCC each year (17). Numerous studies and clinical trails had attempted to uncover the molecular mechanisms of HCC (18–20). The present study integrated and thoroughly re-analyzed three datasets of HCC. UBE2C and four other overexpressed DEGs (TOP2A, PTTG1, GPC3 and PRC1) were identified to be associated with HCC. Enrichment analysis revealed that these genes were important for HCC development via different signaling pahtways. Expression validation analysis using GEO and TCGA data demonstrated that these genes were associated with HCC pathogenesis, and survival analysis revaled that the overexpression of these five genes, particularly UBE2C, was important for survival of patients with HCC.
Formerly known as UBCH10, UBE2C is essential for ubiquitination and inactivation of protein activity (21). Normal expression of UBE2C guarantees normal physiology functions, such as cell cycle progression and programmed cell death (22,23). However, high expression levels of UBE2C frequently leads to the destruction of essential proteins, thus disrupting the function of mitotic cyclins, spindle checkpoint control and euploidy status of cells (24,25). Numerous studies have identified the overexpression of UBE2C in several types of cancer, such as human cervical carcinoma, bladder cancer and breast cancer (21,26,27). It has been previously reported that UBE2C demonstrated potential in becoming a promising cancer biomarker (28). Inhibition of UBE2C, by contrast, suppressed tumor cell proliferation, inhibited tumorigenesis and sensitized cancer cells to radiation (21,27,28). Zhang et al (29) reported that the inhibtion of oncogenic miR-17/20a suppressed gastic cancer cell proflieration by downregulating UBE2C. These studies, as well as the finding that UBE2C was upregualted in HCC tiusses compared with controls in the present study, suggest that the overexpression of UBE2C may be associated with the tumorigenesis of HCC.
A previous study revealed the molecular structure of anaphase-promoting complex/cyclosome (APC/C)-coactivator complexes under cryo-electron microscopy (30). The union of APC/C complexes with UBE2C promotes ubiquitination, whereas early mitotic inhibitor-1 (Emi1) constrains this effect. Deubiquitination enzymes are activated as soon as UBE2C is separated from APC/C, but the process is slow, with unclear molecular patterns (31). In the present study, GO analysis revealed that the upregulated UBE2C, TOP2A, PTTG1, GPC3 and PRC1 genes were primarily involved in cell cycle, cell communication and protein metabolism biological processes. Paclitaxel (PTX) is a widely used microtubule-poisoning drug in anti-neoplastic strategies, which triggers cell death by activating the spindle assembly checkpoint (SAC) (32). Cancer cells senstive to PTX do not achieve SAC, and undergo mitosis by degradation of APC/C and cyclin B, which is called mitotic catastrophe, thus leading to mitotic slippage or cell apoptosis (33). High expression of UBE2C has been demonstrated to override SAC (24). Since opposite characteristics are observed for UBE2C and Emi1 when interacting with APC/C, the exact role of Emi, as well as its expression levels and significance in PTX-treated HCC, require further study.
In summary, the present study confirmed that the high-expression level of UBE2C as well as other four genes (TOP2A, PTTG1, GPC3 and PRC1) was associated with poor overall survival of patients with HCC. Overexpression of UBE2C may serve as a novel indicator of short survival in patients with HCC. Furher studies should foucus on the potential use of UBE2C as a poor prognostic factor for HCC. Whether UBE2C overexpression serves a specific role in the growth of HCC, its inhibtion may block the growth of HCC in vivo or in vitro.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Postgraduate Research & Practice Innovation Program of Jiangsu Province (grant no. KYCX18_1462), National Natural Science Foundation of China (grant no. 81700392), Nanjing Health Youth Talent Training Project in 13th Five-Year (grant no. QRX17113) and Natural Science Foundation of Jiangsu Province Youth Fund Project (grant no. BK20150106).
Availability of data and materials
The datasets analyzed in the present study are all available on NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).
Authors' contributions
ZW, YL, XS and BX designed the study. ZW and SQ wrote the manuscript. XL, QL, JH, JZ, ZHW, SQ and AS performed the bioinformatics analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Neumann H, Longo D, Fauci A, Kasper D, Hauser S, Jameson J, Loscalzo J. Harrison's Principles of Internal Medicine. 2011 [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Liang HW, Yang X, Wen DY, Gao L, Zhang XY, Ye ZH, Luo J, Li ZY, He Y, Pang YY, Chen G. Utility of miR-133a-3p as a diagnostic indicator for hepatocellular carcinoma: An investigation combined with GEO, TCGA, meta-analysis and bioinformatics. Mol Med Rep. 2018;17:1469. doi: 10.3892/mmr.2017.8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang N, Ekanem NR, Sakyi CA, Ray SD. Hepatocellular carcinoma and microRNA: New perspectives on therapeutics and diagnostics. Adv Drug Deliv Rev. 2015;81:62–74. doi: 10.1016/j.addr.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 5.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Y, Bao Y, Ma M, Yang W. Identification of key candidate genes and pathways in colorectal cancer by integrated bioinformatical analysis. Int J Mol Sci. 2017;18:E722. doi: 10.3390/ijms18040722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang L, Gao L, Zou XP, Huang ML, Chen G, Li JJ, Cai XY. Diagnostic significance and potential function of miR-338-5p in hepatocellular carcinoma: A bioinformatics study with microarray and RNA sequencing data. Mol Med Rep. 2018;17:2297–2312. doi: 10.3892/mmr.2017.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng T, Wang D, Chen J, Chen K, Yu G, Chen Q, Liu Y, Yan S, Zhu L, Zhou H, et al. AF119895 regulates NXF3 expression to promote migration and invasion of hepatocellular carcinoma through an interaction with miR-6508-3p. Exp Cell Res. 2018;363:129–139. doi: 10.1016/j.yexcr.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Gan TQ, Xie ZC, Tang RX, Zhang TT, Li DY, Li ZY, Chen G. Clinical value of miR-145-5p in NSCLC and potential molecular mechanism exploration: A retrospective study based on GEO, qRT-PCR, and TCGA data. Tumour Biol. 2017;39:1010428317691683. doi: 10.1177/1010428317691683. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Huang L, Liang H, Zhang R, Chen G, Pang Y, Feng Z. Clinical value and potential targets of miR-224-5p in hepatocellular carcinoma validated by a TCGA- and GEO- based study. Int J Clin Exp Pathol. 2017;10:9970–9989. [PMC free article] [PubMed] [Google Scholar]
- 12.Grinchuk OV, Yenamandra SP, Iyer R, Singh M, Lee HK, Lim KH, Chow PK, Kuznetsov VA. Tumor-adjacent tissue co-expression profile analysis reveals pro-oncogenic ribosomal gene signature for prognosis of resectable hepatocellular carcinoma. Mol Oncol. 2018;12:89–113. doi: 10.1002/1878-0261.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, Deng X, Jin H, Wang N, Wang C, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, Walker R, Jia HL, Ye QH, Qin LX, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–966. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, Wang XW. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhanasekaran R, Limaye A, Cabrera R. Hepatocellular carcinoma: Current trends in worldwide epidemiology, risk factors, diagnosis, and therapeutics. Hepat Med. 2012;4:19–37. doi: 10.2147/HMER.S16316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo X, Wang Z, Zhang J, Xu Q, Hou G, Yang Y, Dong C, Liu G, Liang C, Liu L, et al. Upregulated KPNA2 promotes hepatocellular carcinoma progression and indicates prognostic significance across human cancer types. Acta Biochim Biophys Sin (Shanghai) 2019;51:285–292. doi: 10.1093/abbs/gmz003. [DOI] [PubMed] [Google Scholar]
- 19.Tong H, Liu X, Li T, Qiu W, Peng C, Shen B, Zhu Z. INTS8 accelerates the epithelial-to-mesenchymal transition in hepatocellular carcinoma by upregulating the TGF-beta signaling pathway. Cancer Manag Res. 2019;11:1869–1879. doi: 10.2147/CMAR.S184392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aziz K, Limzerwala JF, Sturmlechner I, Hurley E, Zhang C, Jeganathan KB, Nelson G, Bronk S, Velasco RF, van Deursen EJ, et al. Ccne1 overexpression causes chromosome instability in liver cells and liver tumor development in Mice. Gastroenterology. 2019 doi: 10.1053/j.gastro.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bose MV, Gopisetty G, Selvaluxmy G, Rajkumar T. Dominant negative Ubiquitin-conjugating enzyme E2C sensitizes cervical cancer cells to radiation. Int J Radiat Biol. 2012;88:629–634. doi: 10.3109/09553002.2012.702299. [DOI] [PubMed] [Google Scholar]
- 22.Liu YC. Ubiquitin ligases and the immune response. Annu Rev Immunol. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- 23.Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a Physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad USA. 2009;106:18213–11821. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–926. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 25.van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188:83–83. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morikawa T, Kawai T, Abe H, Kume H, Homma Y, Fukayama M. UBE2C is a marker of unfavorable prognosis in bladder cancer after radical cystectomy. Int J Clin Exp Pathol. 2013;6:1367–1374. [PMC free article] [PubMed] [Google Scholar]
- 27.Rawat A, Gopal G, Selvaluxmy G, Rajkumar T. Inhibition of ubiquitin conjugating enzyme UBE2C reduces proliferation and sensitizes breast cancer cells to radiation, doxorubicin, tamoxifen and letrozole. Cell Oncol (Dordr) 2013;36:459–467. doi: 10.1007/s13402-013-0150-8. [DOI] [PubMed] [Google Scholar]
- 28.Hao Z, Zhang H, Cowell J. Ubiquitin-conjugating enzyme UBE2C: Molecular biology, role in tumorigenesis, and potential as a biomarker. Tumor Biol. 2012;33:723–730. doi: 10.1007/s13277-011-0291-1. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Han T, Wei G, Wang Y. Inhibition of microRNA-17/20a suppresses cell proliferation in gastric cancer by modulating UBE2C expression. Oncol Rep. 2015;33:2529–2536. doi: 10.3892/or.2015.3835. [DOI] [PubMed] [Google Scholar]
- 30.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie C, Powell C, Yao M, Wu J, Dong Q. Molecules in focus: Ubiquitin-conjugating enzyme E2C: A potential cancer biomarker. Int J Biochem Cell Biol. 2014;47:113–118. doi: 10.1016/j.biocel.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 32.Rowinsky EK, Donehower RC. Paclitaxel (taxol) N Engl J Med. 1995;332:1004–1014. doi: 10.1056/NEJM199504133321507. [DOI] [PubMed] [Google Scholar]
- 33.Gascoigne KE, Taylor SS. Article: Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–123. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed in the present study are all available on NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).