Abstract
MicroRNA (miR)-196a is upregulated in various types of malignancy, including esophageal squamous cell carcinoma (ESCC); however, its role in ESCC is currently unclear. The present study aimed to investigate the biological role and molecular mechanism of miR-196a in ESCC. The expression levels of miR-196a in 25 tumor tissues and adjacent non-tumor tissues from patients with ESCC were measured by reverse transcription-quantitative polymerase chain reaction. In addition, miR-196a expression levels were assessed in the human normal esophageal epithelial cell line Het-1A and the ESCC cell line EC109. The effects of miR-196a on the proliferation, apoptosis, invasion and migration of EC109 cells were determined by MTT, flow cytometry and Transwell assays, respectively. A luciferase reporter assay and western blotting were performed to confirm the target gene of miR-196a, and to explore the molecular mechanism underlying the effects of miR-196a on regulation of ESCC cell phenotypes. The results demonstrated that miR-196a was markedly upregulated in ESCC tissues and EC109 cells. In addition, miR-196a downregulation suppressed EC109 cell proliferation, invasion and migration, but did not affect apoptosis. Annexin A1 (ANXA1) was demonstrated to be a direct target gene of miR-196a. ANXA1 protein knockdown reversed the effects of miR-196a inhibition on EC109 cell proliferation, invasion and migration. Furthermore, alongside the downregulation of miR-196a and the increase in ANXA1 expression, cyclooxygenase 2 (COX2), matrix metalloproteinase (MMP)-2 and Snail were downregulated, and E-cadherin was upregulated in EC109 cells. The results of the present study suggested that miR-196a may act as an oncogene in ESCC, and that miR-196a may regulate the proliferation, invasion and migration of ESCC cells by targeting ANXA1. Subsequently, ANXA1 may further modulate the expression levels of COX2, MMP-2, Snail and E-cadherin. In conclusion, the miR-196a/ANXA1 axis may represent a potential therapeutic target in ESCC.
Keywords: esophageal squamous cell carcinoma, microRNA-196a, Annexin A1, cell proliferation, cell invasion, cell migration
Introduction
Esophageal cancer, including esophageal adenocarcinoma and esophageal squamous cell carcinoma (ESCC), is one of the most common types of cancer worldwide (1). In Asia, ESCC is the main type of esophageal cancer (2). Due to the lack of an early diagnostic method for ESCC, the mortality rate for ESCC is very high; the 5-year overall survival rate is <25% (3). Therefore, a detailed study on the mechanisms underlying the development and progression of ESCC is required, in order to improve the prevention, diagnosis and treatment of this disease.
MicroRNAs (miRNAs/miRs) are a large class of small non-coding RNAs that regulate gene expression by targeting mRNAs, and induce mRNA degradation or translational suppression (4). In the human genome, >1,000 miRNAs have been identified, which are thought to regulate ~30% of all genes (5). It has previously been reported that miRNA dysregulation is involved in the development and progression of cancer (6). Previous studies have revealed that miR-196a is upregulated in various types of cancer, including ESCC (7–9). Further studies have demonstrated that miR-196a promotes tumor progression and acts as an oncogene in some types of cancer (10,11). For example, it has been reported that miR-196a promotes cell proliferation and invasion by targeting homeobox A5 in non-small cell lung cancer (12). However, the role of miR-196a in ESCC remains unclear.
The present study demonstrated that miR-196a was significantly upregulated in human esophageal cancer tissue samples and in the ESCC cell line EC109. In addition, downregulation of miR-196a suppressed proliferation, invasion and migration of EC109 cells. The mechanism by which miR-196a affected the EC109 cell phenotype was further investigated, and the results revealed that miR-196a acted by targeting Annexin A1 (ANXA1). These results suggested that miR-196a may be a potential therapeutic target in ESCC.
Materials and methods
Clinical sample collection
ESCC and adjacent non-cancerous tissues were harvested from 25 patients (age, 58.3±6.2 years; male, n=18; female, n=7) from the Second Xiangya Hospital, Central South University (Changsha, China) between June 2016 and December 2016. All patients underwent esophageal cancer resection prior to chemotherapy. After resection, tumor tissues and adjacent non-tumor tissues were collected and stored at −80°C, and malignancy was confirmed by pathologists. All experiments were approved by the Ethics Committees of the Second Xiangya Hospital of Central South University. All patients provided written informed consent for their participation.
Cell culture
The human normal esophageal epithelial cell line Het-1A and the ESCC cell line EC109 were purchased from Shanghai Institutes for Biological Sciences (Shanghai, China). Het-1A cells were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and EC109 cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.). All media were supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were maintained at 37°C in a humidified incubator containing 5% CO2.
Detection of miR-196a by reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from tissues, Het-1A cells and EC109 cells was isolated with TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and were then converted to cDNA with a PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocol. SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) was used for the RT-qPCR assays to detect the expression levels of miR-196a in tissues and cells. The qPCR reaction was run at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec for melting, and 60°C for 1 min for annealing/extension. RNU6 was used as the endogenous control gene to normalize the expression levels of miR-196a. The qPCR primers for miR-196a and U6 were as follows: miR-196a, forward 5′-GCGCCCTAGGTAGTTTCATGTT-3′, reverse, 5′-GTGCAGGGTCCGAGGT-3′; and U6, forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′. Primers were purchased from YRgene Co., Ltd. (Changsha, China). The RT-qPCR assays were performed in triplicate and alterations in miR-196a expression were calculated using the 2−ΔΔCq method (ABI 7500 Software v2.0.1; Applied Biosystems; Thermo Fisher Scientific, Inc.) (13).
Transfection of cells with the miR-196a inhibitor
The miR-196a inhibitor (sense: 5′-CCCAACAACAUGAAACUACCUA-3′) and a negative control (NC) inhibitor (sense: 5′-CAGUACUUUUGUGUAGUACAA-3′) were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). EC109 cells were plated in 6-well plates at 3×105 cells/well and were cultured for 24 h. The miR-196a inhibitor or inhibitor NC were transfected into EC109 cells at a final concentration of 150 nM using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. A total of 48 h post-transfection, cells were harvested for further analysis.
Cell proliferation assay
EC109 cells were seeded in 96-well plates at a density of 5,000 cells/well. The miR-196a inhibitor or inhibitor NC were then transfected into the cells at a final concentration of 150 nM. After 24-, 48- and 72-h transfection, cell proliferation was measured using the MTT Assay kit (Beyotime Institute of Biotechnology, Haimen, China), according to the manufacturer's protocol. Each experiment was performed three times.
Flow cytometric analysis of apoptosis
EC109 cell apoptosis was measured using an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) Apoptosis Detection kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). Briefly, 5×105 cells were harvested with 0.25% EDTA-free trypsin, washed twice with PBS and resuspended in 500 µl binding buffer. Cells were then incubated with 5 µl Annexin V-FITC and 5 µl PI for 15 min at room temperature in the dark, and apoptosis was detected with a NovoCyte Flow Cytometer (ACEA Biosciences, Inc., San Diego, CA, USA). Each sample was assessed in triplicate.
Transwell cell invasion and migration assays
The invasive and migratory abilities of EC109 cells were measured using Transwell assay. Briefly, cells were harvested and resuspended in serum-free medium. For the migration assay, 2.5×104 cells were seeded into the upper chamber of a Transwell insert placed in a 24-well plate (8 µm pore; Corning Inc., Corning, NY, USA). For the invasion assay, 5×104 cells were seeded into the upper chamber of a Transwell insert coated with Matrigel (BD Biosciences, San Jose, CA, USA). Medium containing 10% FBS was added into the bottom chamber. After 24-h incubation at 37°C, cells on the upper surface of the membrane were removed, and the invaded or migrated cells on the lower surface of the membrane were fixed with 100% methanol for 15 min at 4°C and were then stained with 0.1% crystal violet for 15 min at room temperature. The number of invaded or migrated cells was counted under a light microscope.
Luciferase reporter assay
The possible target genes of miR-196a were predicted with TargetScan 7.1 (http://www.targetscan.org) and ANXA1 was selected for further analysis. A dual-luciferase miRNA target expression plasmid containing the wild-type 3′-untranslated region fragment of ANXA1, named pYr-MirTarget-ANXA1-3U, and a corresponding mutant reporter plasmid containing the mutant miR-196a target site, named pYr-MirTarget-ANXA1-3U-Mut, were purchased from YRgene Co., Ltd. The miR-196a mimics (sense, 5′-UAGGUAGUUUCAUGUUGUUGGG-3′; and antisense, 5′-CAACAACAUGAAACUACCUAUU-3′) and NC mimics (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′; and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′) were purchased from Shanghai GenePharma Co., Ltd. Briefly, 293 cells (Shanghai Institutes for Biological Sciences) were seeded in 96-well plates at a density of 5×103 cells/well and were co-transfected with pYr-MirTarget-ANXA1-3U or pYr-MirTarget-ANXA1-3U-Mut and miR-196a mimics or mimics NC (50 nM) using Lipofectamine® 2000. After 48-h transfection, cells were harvested and a luciferase reporter assay was performed using the Dual-Luciferase Reporter Assay kit (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol. Each experiment was performed three times.
Western blotting
Total protein was extracted from EC109 cells using lysis buffer (Beyotime Institute of Biotechnology), and protein concentrations were measured with a Bicinchoninic Acid Protein Assay kit (Beyotime Institute of Biotechnology). The proteins (50 µg) were separated by 10% SDS-PAGE and were then transferred to a 0.22-µm nitrocellulose membrane (EMD Millipore, Billerica, MA, USA). After blocking with 5% skimmed milk for 2 h at room temperature, membranes were incubated with primary antibodies against ANXA1 (cat. no. 32934S; Cell Signaling Technology, Inc., Danvers, MA, USA), cyclooxygenase 2 (COX2; cat. no. ab15191; Abcam, Cambridge, MA, USA), matrix metalloproteinase (MMP)-2 (cat. no. ab97779; Abcam), Snail (cat. no. 3879S; Cell Signaling Technology, Inc.), E-cadherin (cat. no. 3195S; Cell Signaling Technology, Inc.) and β-actin (cat. no AT0001; CMCTAG, Inc., Milwaukee, WI, USA) (all dilutions, 1:1,000) overnight at 4°C with gentle agitation. Membranes were further incubated for 1 h at room temperature with the corresponding horseradish peroxidase-conjugated secondary antibodies: Goat anti-rabbit secondary antibody for ANXA1, COX2, MMP-2, Snail and E-cadherin (cat. no. sc-2004; 1:5,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or goat anti-mouse secondary antibody for β-actin (cat. no. sc-2005; 1:5,000; Santa Cruz Biotechnology, Inc.). The protein bands were visualized using an enhanced chemiluminescence system (Beyotime Institute of Biotechnology) and were semi-quantified using ImageJ2× software (National Institutes of Health, Bethesda, MD, USA). β-actin was used as a loading control for normalization.
Transfection of cells with ANXA1 small interfering (si)RNA
ANXA1 siRNA (sense, 5′-GAGAGAUCUGGCCAAAGACTT-3′; and antisense, 5′-GUCUUUGGCCAGAUCUCUCTT-3′) or siRNA NC (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′; and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′) was purchased from Shanghai GenePharma Co., Ltd. and was co-transfected into EC109 cells with miR-196a inhibitor or inhibitor NC using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The final concentrations of ANXA1 siRNA, miR-196a inhibitor or inhibitor NC were 150 nM. After 48-h transfection, cells were harvested, and western blotting, MTT and Transwell assays were performed.
Effects of miR-196a mimics and ANXA1 siRNA
To confirm the effectiveness of miR-196a mimics, 293 cells were transfected with miR-196a mimics or mimics NC and were harvested a total of 48 h post-transfection. Subsequently, the expression levels of miR-196a were detected in 293 cells using RT-qPCR. RNU6 was used as an endogenous control to normalize the expression levels of miR-196a. Furthermore, to confirm the knockdown efficiency of ANXA1 siRNA, EC109 cells were transfected with ANXA1 siRNA or siRNA NC, and were harvested after 48 h of transfection. Subsequently, the mRNA expression levels of ANXA1 in EC109 cells were measured by RT-qPCR. GAPDH was used as an endogenous control to normalize the mRNA expression levels of ANXA1. The qPCR primers for ANXA1 and GAPDH were as follows: ANXA1 forward, 5′-CTAAGCGAAACAATGCACAGC-3′; and reverse, 5′-CCTCCTCAAGGTGACCTGTAA-3′; and GAPDH forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′; and reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Statistical analysis
Data are presented as the means ± standard deviation of at least three independent experiments. The Student's t-test was used for the comparison of two groups. Comparisons of means among multiple groups were determined using one-way analysis of variance followed by Tukey's post hoc test. All statistical analyses were performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-196a is upregulated in human esophageal cancer clinical tissues and ESCC cells
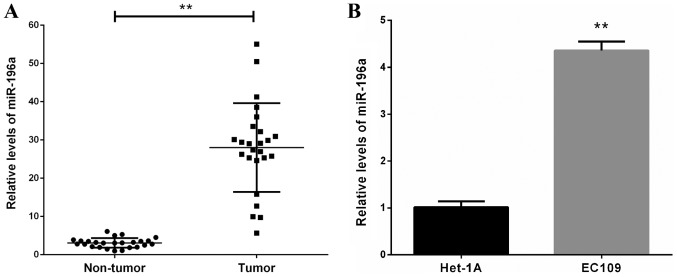
The expression levels of miR-196a in 25 human esophageal cancer clinical tissues and corresponding non-cancerous tissues were measured by RT-qPCR. The results revealed that the expression levels of miR-196a were markedly upregulated in tumor tissues compared with in adjacent non-tumor tissues (Fig. 1A). Furthermore, miR-196a expression levels were significantly increased in the ESCC EC109 cell line compared with in the Het-1A line cell (Fig. 1B). These data indicated that upregulation of miR-196a may be involved in the development and progression of ESCC.
Figure 1.
miR-196a is upregulated in human esophageal cancer clinical tissues and the EC109 cell line. (A) RT-qPCR analysis of miR-196a expression in 25 human esophageal cancer clinical tissues and corresponding non-cancerous tissues. (B) RT-qPCR analysis of miR-196a expression in EC109 and Het-1A cell lines. Experiments were performed in triplicate and data are expressed as the means ± standard deviation. **P<0.01. miR-196a, microRNA-196a; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
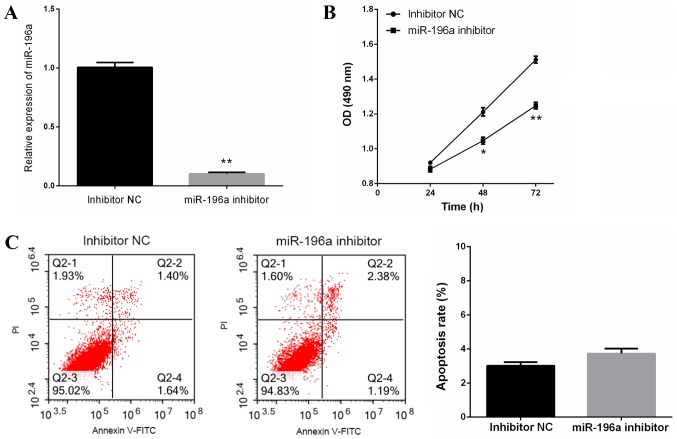
miR-196a knockdown suppresses cell proliferation without affecting apoptosis of EC109 cells
To explore the role of miR-196a in ESCC progression, miR-196a inhibitor or inhibitor NC were transfected into EC109 cells, and cell proliferation and apoptosis were measured. miR-196a inhibitor-induced miR-196a knockdown in EC109 cells was confirmed by RT-qPCR (Fig. 2A). The results of the MTT assay revealed that miR-196a downregulation significantly inhibited EC109 cell proliferation (Fig. 2B); however, flow cytometric analysis revealed that miR-196a downregulation did not affect EC109 cell apoptosis (Fig. 2C). These data suggested that miR-196a downregulation inhibited EC109 cell proliferation, but was not associated with EC109 cell apoptosis.
Figure 2.
Downregulation of miR-196a suppresses cell proliferation without affecting apoptosis of EC109 cells. (A) Reverse transcription-quantitative polymerase chain reaction analysis of miR-196a expression in miR-196a inhibitor- and inhibitor NC-transfected EC109 cells. The results revealed that miR-196a was downregulated following miR-196a inhibition in EC109 cells. (B) Cell proliferation of miR-196a inhibitor- or inhibitor NC-transfected EC109 cells was measured using the MTT assay. After 48-h transfection, EC109 cell proliferation was significantly inhibited following miR-196a inhibition. (C) Cell apoptosis of miR-196a inhibitor- or inhibitor NC-transfected EC109 cells was measured by flow cytometric analysis. There was no significant difference in the apoptosis rate between the two groups of cells. Experiments were performed in triplicate and data are expressed as the means ± standard deviation. *P<0.05, **P<0.01 vs. inhibitor NC. FITC, fluorescein isothiocyanate; miR-196a, microRNA-196a; NC, negative control; OD, optical density; PI, propidium iodide.
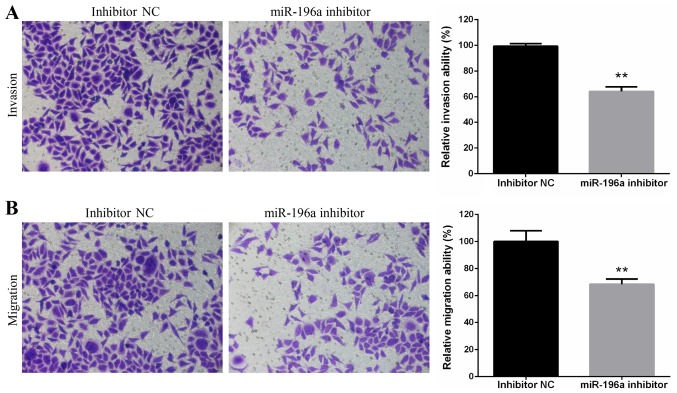
Downregulation of miR-196a suppresses the invasion and migration of EC109 cells
The invasive and migratory ability of EC109 cells transfected with miR-196a inhibitor or inhibitor NC were assessed using Transwell invasion and migration assays. The results demonstrated that miR-196a silencing significantly suppressed EC109 cell invasion (Fig. 3A) and migration (Fig. 3B).
Figure 3.
Downregulation of miR-196a suppresses the invasion and migration of EC109 cells. (A) Invasive ability of miR-196a inhibitor- or inhibitor NC-transfected EC109 cells was determined by Transwell cell invasion assay. Magnification, ×200. (B) Migratory ability of EC109 cells transfected with miR-196a inhibitor or inhibitor NC was determined by Transwell cell migration assay. Magnification, ×200. Experiments were performed in triplicate and data are expressed as the means ± standard deviation. **P<0.01 vs. inhibitor NC. miR-196a, microRNA-196a; NC, negative control.
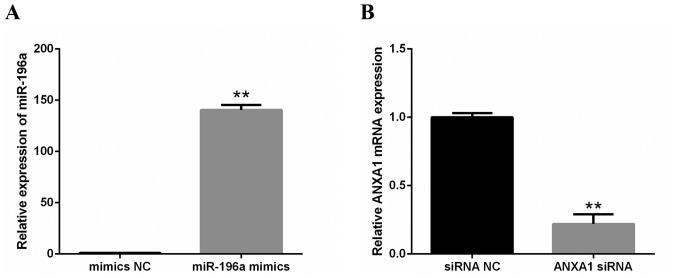
Effects of miR-196a mimics and ANXA1 siRNA
The effectiveness of miR-196a mimics and ANXA1 siRNA were confirmed by RT-qPCR. The results revealed that miR-196a was markedly upregulated by miR-196a mimics in 293 cells (Fig. 4A) and the mRNA expression levels of ANXA1 were significantly silenced by ANXA1 siRNA in EC109 cells (Fig. 4B).
Figure 4.
Effects of miR-196a mimics and ANXA1 siRNA. (A) miR-196a expression was significantly upregulated by miR-196a mimics in 293 cells. (B) The mRNA expression level of ANXA1 was significantly reduced by ANXA1 siRNA in EC109 cells. Experiments were performed in triplicate and data are presentedd as the mean ± standard deviation. **P<0.01. miR-196a, microRNA-196a; ANXA1, Annexin A1; NC, negative control.
miR-196a regulates the proliferation, invasion and migration of EC109 cells by targeting ANXA1
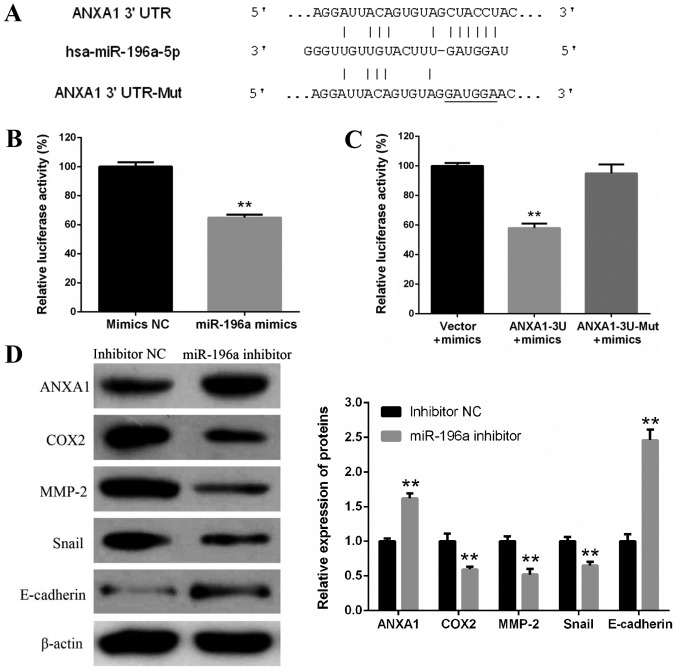
To explore the molecular mechanism underlying the regulatory effects of miR-196a on EC109 cell proliferation, invasion and migration, possible target genes of miR-196a were predicted with TargetScan 7.1. ANXA1 was selected for further analysis, due to its complementary structure to miR-196a (Fig. 5A). A luciferase reporter assay was performed to further confirm whether ANXA1 was a direct target gene of miR-196a. pYr-MirTarget-ANXA1-3U or pYr-MirTarget-ANXA1-3U-Mut and miR-196a mimics or mimics NC were co-transfected into 293 cells. The results of the luciferase activity assay demonstrated that miR-196a mimics significantly reduced the luciferase activity of pYr-MirTarget-ANXA1-3U, but did not affect the luciferase activity of pYr-MirTarget-ANXA1-3U-Mut (Fig. 5B and C). In addition, western blotting revealed that miR-196a knockdown via the miR-196a inhibitor increased the protein expression levels of ANXA1 in EC109 cells (Fig. 5D). These results indicated that ANXA1 may be a direct target gene of miR-196a. In addition, following the increase in ANXA1 protein expression, COX2, MMP-2 and Snail proteins were significantly downregulated, whereas E-cadherin protein was markedly upregulated (Fig. 5D).
Figure 5.
ANXA1 is a direct target gene of miR-196a. (A) Human ANXA1 3′UTR fragments containing a wild-type or Mut miR-196a target site were cloned downstream of the luciferase reporter gene. (B) 293 cells were co-transfected with pYr-MirTarget-ANXA1-3U and miR-196a mimics or mimics NC. After 48-h transfection, dual-luciferase activity was detected. **P<0.01 vs. mimic NC. (C) Luciferase activity assay revealed that miR-196a mimics reduced the luciferase activity of pYr-MirTarget-ANXA1-3U, but not pYr-MirTarget-ANXA1-3U-Mut. **P<0.01 vs. vector control. (D) Western blotting of ANXA1, COX2, MMP-2, Snail and E-cadherin protein expression in EC109 cells transfected with miR-196a inhibitor or inhibitor NC. β-actin was used as a control. **P<0.01 vs. NC. Experiments were performed in triplicate and data are expressed as the means ± standard deviation. 3′UTR, 3′-untranslated region; ANXA1, Annexin A1; COX2, cyclooxygenase 2; miR-196a, microRNA-196a; MMP2, matrix metalloproteinase 2; Mut, mutant; NC, negative control.
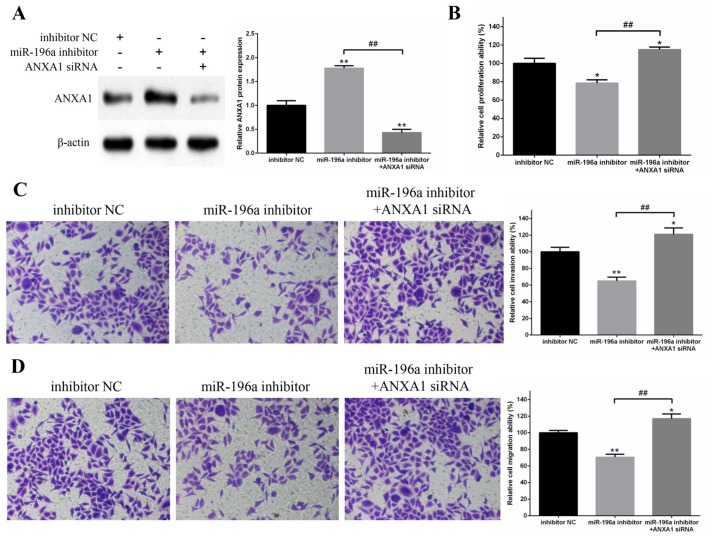
To confirm whether miR-196a regulated EC109 cell proliferation, invasion and migration via ANXA1, ANXA1 protein expression was knocked down via siRNA, whereas miR-196a was inhibited via the miR-196a inhibitor (Fig. 6A). The results revealed that miR-196a downregulation significantly inhibited EC109 cell proliferation, invasion and migration; however, ANXA1 protein knockdown reversed these inhibitory effects (Fig. 6B-D). These results suggested that miR-196a may regulate EC109 cell proliferation, invasion and migration by targeting ANXA1.
Figure 6.
Knockdown of ANXA1 can reverse the inhibitory effects of miR-196a downregulation on the proliferation, invasion and migration of EC109 cells. (A) Protein expression levels of ANXA1 in EC109 cells were measured by western blotting. β-actin was used as a control. The results demonstrated that ANXA1 protein expression was significantly upregulated by miR-196a inhibitor and markedly downregulated by ANXA1 siRNA. (B) Proliferation of EC109 cells was detected by MTT assay. The miR-196a inhibitor suppressed the proliferation of EC109 cells, whereas co-transfection with ANXA1 siRNA reversed this inhibitory effect. (C) Invasive and (D) migratory abilities of EC109 cells were determined by Transwell cell invasion and migration assays. Magnification, ×200. Downregulation of miR-196a significantly suppressed the invasion and migration of EC109 cells, whereas knockdown of ANXA1 reversed these inhibitory effects. Experiments were performed in triplicate and data are expressed as the means ± standard deviation. *P<0.05, **P<0.01 vs. inhibitor NC; ##P<0.01, as indicated. ANXA1, Annexin A1; miR-196a, microRNA-196a; NC, negative control; siRNA, small interfering RNA.
Discussion
Previous studies have reported that miR-196a is upregulated in various types of cancer, including gastric cancer, gastrointestinal stromal tumor, colon cancer, pancreatic cancer, non-small-cell lung carcinoma, laryngeal cancer and cervical carcinoma (14–16). Further studies have revealed that miR-196a is likely to act as an oncogene in some types of cancer, since it can promote cancer cell proliferation, invasion and migration, and suppress tumor cell apoptosis (17,18). For example, miR-196a promotes cell proliferation in gastric cancer by targeting cyclin-dependent kinase inhibitor 1B (19). In colorectal cancer, miR-196a stimulates the oncogenic phenotype of tumor cells (20). It has also been reported that miR-196a inhibits cell apoptosis, and promotes cell proliferation and invasion by targeting the inhibitor of growth family member 5 in pancreatic cancer (21). Furthermore, Maru et al (18) reported that miR-196a is upregulated in esophageal adenocarcinoma, and Fendereski et al (9) described an overexpression of miR-196a in ESCC. However, to the best of our knowledge, few studies have examined the role of miR-196a in regulation of the ESCC phenotype, and the role of miR-196a in ESCC remains unclear.
The present study demonstrated that miR-196a was markedly upregulated in ESCC tissues and EC109 cells compared with in non-cancerous tissues and normal cells, respectively. These data suggested that miR-196a may function as an oncogene in ESCC. Furthermore, the regulatory role of a miR-196a inhibitor on EC109 cell phenotype was examined, and the results revealed that miR-196a downregulation inhibited proliferation, invasion and migration of EC109 cells. These results further suggested that miR-196a may be involved in the development and progression of ESCC.
The molecular mechanisms underlying the regulatory effects of miR-196a on the EC109 cell phenotype were investigated. The results of the luciferase reporter assay and western blotting demonstrated that ANXA1 was a direct target gene of miR-196a. ANXA1, a member of the Annexin family, is a calcium-dependent phospholipid binding protein (22). It is involved in inflammation, cell proliferation, apoptosis, tumorigenesis and tumor progression (23–25), and is aberrantly expressed in various types of malignancy. It is overexpressed in breast, liver and pancreatic cancer, whereas it is downregulated in nasopharyngeal carcinoma, thyroid carcinoma, prostatic cancer and ESCC (26–28). Previous studies have demonstrated that ANXA1 is likely to function as an anti-oncogene in some types of cancer, including ESCC. For example, upregulation of ANXA1 expression inhibits cell growth and induces cell apoptosis in DU145 human prostate cancer cells (29). Xia et al (30) and Hu et al (31) reported that ANXA1 is significantly downregulated in ESCC. Paweletz et al (32) discovered that a loss of ANXA1 is involved in the tumorigenesis of esophageal and prostate carcinoma. Álvarez-Teijeiro et al (33) reported that ANXA1 is downregulated in head and neck squamous cell carcinoma (HNSCC) and is a direct target of miR-196a/b in HNSCC-derived cell lines; however, the authors did not investigate the effects of ANXA1 or miR-196a/b on HNSCC cell phenotypes and the corresponding molecular mechanism. In the present study, miR-196a directly regulated the protein expression levels of ANXA1, and miR-196a downregulation increased ANXA1 expression and inhibited proliferation, invasion and migration of EC109 cells. Furthermore, ANXA1 protein knockdown reversed the inhibitory effects of miR-196a on EC109 cell proliferation, invasion and migration. These results, combined with previous findings, suggested that miR-196a may regulate the proliferation, invasion and migration of ESCC cells by targeting ANXA1.
COX2, MMP-2 and Snail were downregulated and E-cadherin was upregulated in EC109 cells transfected with miR-196a inhibitor. These results further suggested that miR-196a may indeed affect ESCC cell phenotypes.
COX2 is overexpressed in various types of cancer, including ESCC, and is involved in the development and progression of tumors (34). Numerous studies reported that COX2 inhibition suppresses proliferation, invasion and migration of tumor cells (35,36). Gao et al (37) revealed that the loss of COX2 following ANXA1 overexpression inhibits cell proliferation and invasion in gastric cancer cells. Hannon et al (38) and Croxtall et al (39) also reported that ANXA1 negatively regulates COX2 expression. In the present study, COX2 expression was decreased alongside the upregulation of ANXA1 in EC109 cells. This finding suggested that miR-196a may regulate cell proliferation in ESCC by targeting ANXA1, which may further modulate COX2 expression.
E-cadherin is a cell adhesion molecule and a key marker of epithelial-mesenchymal transition, which can suppress tumor invasion and migration (40). In addition, Snail can decrease the expression of E-cadherin by binding to the E-box of the E-cadherin promoter (41). Furthermore, MMP-2 can promote the invasion and migration of tumor cells by degrading the extracellular matrix (42). MMPs can also weaken the effect of E-cadherin by mediating E-cadherin ectodomain shedding (43). Previous studies have reported that MMP-2 and Snail are overexpressed in various types of cancer, including ESCC, whereas E-cadherin is downregulated (40–42). It has also been reported that COX2 downregulation decreases MMP-2 and Snail expression, and increases E-cadherin expression, in tumor cells, and that these alterations result in suppression of tumor cell invasion and migration (44,45). These findings, combined with the results of the present study, provided evidence supporting the hypothesis that miR-196a may regulate the invasion and migration of ESCC cells by targeting ANXA1, which may further modulate MMP-2, Snail and E-cadherin expression by regulating COX2 expression.
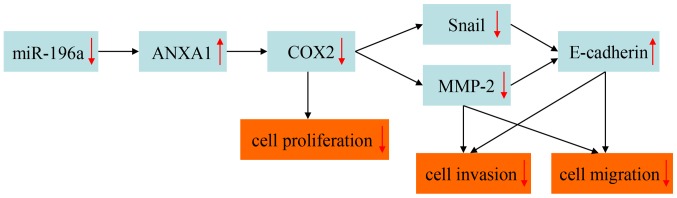
In conclusion, the present study demonstrated that miR-196a was upregulated in ESCC tumor tissues and cells, and may function as an oncogene in ESCC. In addition, miR-196a downregulation inhibited cell proliferation, invasion and migration, but did not affect apoptosis of EC109 cells. The molecular mechanism by which miR-196a may regulate EC109 cell phenotypes was also investigated. The hypothetical mechanism of regulation is presented in Fig. 7. To the best of our knowledge, this mechanism has not yet been reported in the literature. Further studies investigating the associated cell signaling pathways are required. In particular, whether miR-196a induces the degradation of ANXA1 mRNA and the molecular mechanism underlying miR-196a-induced ANXA1 mRNA degradation remain to be explored. Furthermore, the role of ANXA1 in regulating COX2 expression levels remains to be investigated.
Figure 7.
Hypothetical molecular mechanism by which miR-196a may regulate EC109 cell phenotypes. ANXA1, Annexin A1; COX2, cyclooxygenase 2; miR-196a, microRNA-196a; MMP2, matrix metalloproteinase 2.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CH and JP designed the study. CH, LL, XW and YZ performed the experiments. JH and DL analyzed the data. CH wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The Ethics Committees of the Second Xiangya Hospital, Central South University approved the study, and all patients provided written informed consent prior to their inclusion.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kamangar F, Qiao YL, Schiller JT, Dawsey SM, Fears T, Sun XD, Abnet CC, Zhao P, Taylor PR, Mark SD. Human papillomavirus serology and the risk of esophageal and gastric cancers: Results from a cohort in a high-risk region in China. Int J Cancer. 2006;119:579–584. doi: 10.1002/ijc.21871. [DOI] [PubMed] [Google Scholar]
- 2.Lagergren J. Oesophageal cancer in 2014: Advances in curatively intended treatment. Nat Rev Gastroenterol Hepatol. 2015;12:74–75. doi: 10.1038/nrgastro.2014.213. [DOI] [PubMed] [Google Scholar]
- 3.Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 6.Fendler A, Jung M, Stephan C, Honey RJ, Stewart RJ, Pace KT, Erbersdobler A, Samaan S, Jung K, Yousef GM. miRNAs can predict prostate cancer biochemical relapse and are involved in tumor progression. Int J Oncol. 2011;39:1183–1192. doi: 10.3892/ijo.2011.1128. [DOI] [PubMed] [Google Scholar]
- 7.Darda L, Hakami F, Morgan R, Murdoch C, Lambert DW, Hunter KD. The role of HOXB9 and miR-196a in head and neck squamous cell carcinoma. PLoS One. 2015;10:e0122285. doi: 10.1371/journal.pone.0122285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li HL, Xie SP, Yang YL, Cheng YX, Zhang Y, Wang J, Wang Y, Liu DL, Chen ZF, Zhou YN, Wu HY. Clinical significance of upregulation of mir-196a-5p in gastric cancer and enriched KEGG pathway analysis of target genes. Asian Pac J Cancer Prev. 2015;16:1781–1787. doi: 10.7314/APJCP.2015.16.5.1781. [DOI] [PubMed] [Google Scholar]
- 9.Fendereski M, Zia MF, Shafiee M, Safari F, Saneie MH, Tavassoli M. MicroRNA-196a as a potential diagnostic biomarker for esophageal squamous cell carcinoma. Cancer Invest. 2017;35:78–84. doi: 10.1080/07357907.2016.1254228. [DOI] [PubMed] [Google Scholar]
- 10.Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki Y, et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126–1136. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- 11.Lu YC, Chang JT, Liao CT, Kang CJ, Huang SF, Chen IH, Huang CC, Huang YC, Chen WH, Tsai CY, et al. OncomiR-196 promotes an invasive phenotype in oral cancer through the NME4-JNK-TIMP1-MMP signaling pathway. Mol Cancer. 2014;13:218. doi: 10.1186/1476-4598-13-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu XH, Lu KH, Wang KM, Sun M, Zhang EB, Yang JS, Yin DD, Liu ZL, Zhou J, Liu ZJ, et al. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer. 2012;12:348. doi: 10.1186/1471-2407-12-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Hui AB, Shi W, Boutros PC, Miller N, Pintilie M, Fyles T, McCready D, Wong D, Gerster K, Waldron L, et al. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest. 2009;89:597–606. doi: 10.1038/labinvest.2009.12. [DOI] [PubMed] [Google Scholar]
- 15.Kong X, Du Y, Wang G, Gao J, Gong Y, Li L, Zhang Z, Zhu J, Jing Q, Qin Y, Li Z. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci. 2011;56:602–609. doi: 10.1007/s10620-010-1285-3. [DOI] [PubMed] [Google Scholar]
- 16.Liu P, Xin F, Ma CF. Clinical significance of serum miR-196a in cervical intraepithelial neoplasia and cervical cancer. Genet Mol Res. 2015;14:17995–18002. doi: 10.4238/2015.December.22.25. [DOI] [PubMed] [Google Scholar]
- 17.Jin C, Zhang Y, Li J. Upregulation of miR-196a promotes cell proliferation by downregulating p27kip1 in laryngeal cancer. Biol Res. 2016;49:40. doi: 10.1186/s40659-016-0100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maru DM, Singh RR, Hannah C, Albarracin CT, Li YX, Abraham R, Romans AM, Yao H, Luthra MG, Anandasabapathy S, et al. MicroRNA-196a is a potential marker of progression during Barrett's metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940–1948. doi: 10.2353/ajpath.2009.080718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun M, Liu XH, Li JH, Yang JS, Zhang EB, Yin DD, Liu ZL, Zhou J, Ding Y, Li SQ, et al. miR-196a is upregulated in gastric cancer and promotes cell proliferation by downregulating p27(kip1) Mol Cancer Ther. 2012;11:842–852. doi: 10.1158/1535-7163.MCT-11-1015. [DOI] [PubMed] [Google Scholar]
- 20.Schimanski CC, Frerichs K, Rahman F, Berger M, Lang H, Galle PR, Moehler M, Gockel I. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;15:2089–2096. doi: 10.3748/wjg.15.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu M, Du Y, Gao J, Liu J, Kong X, Gong Y, Li Z, Wu H, Chen H. Aberrant expression miR-196a is associated with abnormal apoptosis, invasion, and proliferation of pancreatic cancer cells. Pancreas. 2013;42:1169–1181. doi: 10.1097/MPA.0b013e3182962acb. [DOI] [PubMed] [Google Scholar]
- 22.Xiao Y, Ouyang C, Huang W, Tang Y, Fu W, Cheng A. Annexin A1 can inhibit the in vitro invasive ability of nasopharyngeal carcinoma cells possibly through Annexin A1/S100A9/Vimentin interaction. PLoS One. 2017;12:e0174383. doi: 10.1371/journal.pone.0174383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugimoto MA, Vago JP, Teixeira MM, Sousa LP. Annexin A1 and the resolution of inflammation: Modulation of neutrophil recruitment, apoptosis and clearance. J Immunol Res. 2016;2016:8239258. doi: 10.1155/2016/8239258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boudhraa Z, Bouchon B, Viallard C, D'Incan M, Degoul F. Annexin A1 localization and its relevance to cancer. Clin Sci (Lond) 2016;130:205–220. doi: 10.1042/CS20150415. [DOI] [PubMed] [Google Scholar]
- 25.Suh YE, Raulf N, Gäken J, Lawler K, Urbano TG, Bullenkamp J, Gobeil S, Huot J, Odell E, Tavassoli M. MicroRNA-196a promotes an oncogenic effect in head and neck cancer cells by suppressing Annexin A1 and enhancing radioresistance. Int J Cancer. 2015;137:1021–1034. doi: 10.1002/ijc.29397. [DOI] [PubMed] [Google Scholar]
- 26.Huang X, Wang Q, Li T, Li F, Lang J. The negative expression of Annexin A1 in esophageal squamous cell carcinoma is associated with poor outcome. Int J Radiat Oncol. 2013;87(Suppl):S288–S289. doi: 10.1016/j.ijrobp.2013.06.755. [DOI] [Google Scholar]
- 27.Shen D, Nooraie F, Elshimali Y, Lonsberry V, He J, Bose S, Chia D, Seligson D, Chang HR, Goodglick L. Decreased expression of Annexin A1 is correlated with breast cancer development and progression as determined by a tissue microarray analysis. Hum Pathol. 2006;37:1583–1591. doi: 10.1016/j.humpath.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Hsiang CH, Tunoda T, Whang YE, Tyson DR, Ornstein DK. The impact of altered Annexin I protein levels on apoptosis and signal transduction pathways in prostate cancer cells. Prostate. 2006;66:1413–1424. doi: 10.1002/pros.20457. [DOI] [PubMed] [Google Scholar]
- 29.Mu D, Gao Z, Guo H, Zhou G, Sun B. Sodium butyrate induces growth inhibition and apoptosis in human prostate cancer DU145 cells by up-regulation of the expression of Annexin A1. PLoS One. 2013;8:e74922. doi: 10.1371/journal.pone.0074922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia SH, Hu LP, Hu H, Ying WT, Xu X, Cai Y, Han YL, Chen BS, Wei F, Qian XH, et al. Three isoforms of Annexin I are preferentially expressed in normal esophageal epithelia but down-regulated in esophageal squamous cell carcinomas. Oncogene. 2002;21:6641–6648. doi: 10.1038/sj.onc.1205818. [DOI] [PubMed] [Google Scholar]
- 31.Hu N, Flaig MJ, Su H, Shou JZ, Roth MJ, Li WJ, Wang C, Goldstein AM, Li G, Emmert-Buck MR, Taylor PR. Comprehensive characterization of Annexin I alterations in esophageal squamous cell carcinoma. Clin Cancer Res. 2004;10:6013–6022. doi: 10.1158/1078-0432.CCR-04-0317. [DOI] [PubMed] [Google Scholar]
- 32.Paweletz CP, Ornstein DK, Roth MJ, Bichsel VE, Gillespie JW, Calvert VS, Vocke CD, Hewitt SM, Duray PH, Herring J, et al. Loss of Annexin 1 correlates with early onset of tumorigenesis in esophageal and prostate carcinoma. Cancer Res. 2000;60:6293–6297. [PubMed] [Google Scholar]
- 33.Álvarez-Teijeiro S, Menéndez ST, Villaronga MÁ, Pena-Alonso E, Rodrigo JP, Morgan RO, Granda-Díaz R, Salom C, Fernandez MP, García-Pedrero JM. Annexin A1 down-regulation in head and neck squamous cell carcinoma is mediated via transcriptional control with direct involvement of miR-196a/b. Sci Rep. 2017;7:6790. doi: 10.1038/s41598-017-07169-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Li P, Zhang ST, You H, Jia JD, Yu ZL. COX-2 mRNA expression in esophageal squamous cell carcinoma (ESCC) and effect by NSAID. Dis Esophagus. 2008;21:9–14. doi: 10.1111/j.1442-2050.2007.00697.x. [DOI] [PubMed] [Google Scholar]
- 35.Shao Y, Li P, Zhu ST, Yue JP, Ji XJ, He Z, Ma D, Wang L, Wang YJ, Zong Y, et al. Cyclooxygenase-2, a potential therapeutic target, is regulated by miR-101 in esophageal squamous cell carcinoma. PLoS One. 2015;10:e0140642. doi: 10.1371/journal.pone.0140642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao Y, Li P, Zhu ST, Yue JP, Ji XJ, Ma D, Wang L, Wang YJ, Zong Y, Wu YD, Zhang ST. miR-26a and miR-144 inhibit proliferation and metastasis of esophageal squamous cell cancer by inhibiting cyclooxygenase-2. Oncotarget. 2016;7:15173–15186. doi: 10.18632/oncotarget.7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Y, Chen Y, Xu D, Wang J, Yu G. Differential expression of ANXA1 in benign human gastrointestinal tissues and cancers. BMC Cancer. 2014;14:520. doi: 10.1186/1471-2407-14-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hannon R, Croxtall JD, Getting SJ, Roviezzo F, Yona S, Paul-Clark MJ, Gavins FN, Perretti M, Morris JF, Buckingham JC, Flower RJ. Aberrant inflammation and resistance to glucocorticoids in Annexin 1−/− mouse. FASEB J. 2003;17:253–255. doi: 10.1096/fj.02-0239fje. [DOI] [PubMed] [Google Scholar]
- 39.Croxtall JD, Newman SP, Choudhury Q, Flower RJ. The concerted regulation of cPLA2, COX2, and lipocortin 1 expression by IL-1beta in A549 cells. Biochem Biophys Res Commun. 1996;220:491–495. doi: 10.1006/bbrc.1996.0432. [DOI] [PubMed] [Google Scholar]
- 40.Dohadwala M, Yang SC, Luo J, Sharma S, Batra RK, Huang M, Lin Y, Goodglick L, Krysan K, Fishbein MC, et al. Cyclooxygenase-2-dependent regulation of E-cadherin: Prostaglandin E(2) induces transcriptional repressors ZEB1 and snail in non-small cell lung cancer. Cancer Res. 2006;66:5338–5345. doi: 10.1158/0008-5472.CAN-05-3635. [DOI] [PubMed] [Google Scholar]
- 41.Chen Z, Liu M, Liu X, Huang S, Li L, Song B, Li H, Ren Q, Hu Z, Zhou Y, Qiao L. COX-2 regulates E-cadherin expression through the NF-κB/Snail signaling pathway in gastric cancer. Int J Mol Med. 2013;32:93–100. doi: 10.3892/ijmm.2013.1376. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Ma J, Guo Q, Duan F, Tang F, Zheng P, Zhao Z, Lu G. Overexpression of MMP-2 and MMP-9 in esophageal squamous cell carcinoma. Dis Esophagus. 2009;22:664–667. doi: 10.1111/j.1442-2050.2008.00928.x. [DOI] [PubMed] [Google Scholar]
- 43.McGuire JK, Li Q, Parks WC. Matrilysin (matrix metalloproteinase-7) mediates E-cadherin ectodomain shedding in injured lung epithelium. Am J Pathol. 2003;162:1831–1843. doi: 10.1016/S0002-9440(10)64318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujii R, Imanishi Y, Shibata K, Sakai N, Sakamoto K, Shigetomi S, Habu N, Otsuka K, Sato Y, Watanabe Y, et al. Restoration of E-cadherin expression by selective Cox-2 inhibition and the clinical relevance of the epithelial-to-mesenchymal transition in head and neck squamous cell carcinoma. J Exp Clin Cancer Res. 2014;33:40. doi: 10.1186/1756-9966-33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurihara Y, Hatori M, Ando Y, Ito D, Toyoshima T, Tanaka M, Shintani S. Inhibition of cyclooxygenase-2 suppresses the invasiveness of oral squamous cell carcinoma cell lines via down-regulation of matrix metalloproteinase-2 production and activation. Clin Exp Metastasis. 2009;26:425–432. doi: 10.1007/s10585-009-9241-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.