Abstract
Background
Recent studies have demonstrated that Linc00152 is highly expressed in multiple cancer types and its genes show tumor-promoting characteristics. However, the efficacy and biological mechanism of Linc00152 in bladder cancer remains unclear.
Material/Methods
We study investigated the relative expression and promoter methylation of Linc00152 in 126 cases of bladder cancer tissues by qRT-PCR and Bisulfite sequencing PCR. qRT-PCR was used to assess the relative expression of Linc00152 in 4 human bladder cancer cell lines. To explore the biological properties of Linc00152, we performed cell growth and soft-agar colony-formation assays, flow cytometry analyses, wound-healing assay, and Transwell assay. Western blot analysis was used to detect the underlying mechanisms of Linc00152 in bladder cancer.
Results
We found that Linc00152 was highly expressed in 126 cases of bladder carcinoma tissues (p<0.001) and 4 cell lines (p<0.01), and Linc00152 is more commonly expressed in patients with advanced-stage cancer (p=0.021). Knockdown of Linc00152 by using siRNAs in bladder cancer cell lines (T24 and HT-1197) suppressed cell viability and growth by causing cell cycle arrest and apoptosis (p<0.001), as well as inhibiting cell migration and invasion (p<0.001). In addition, the quantitative RT-PCR and Western blot results suggest that knockdown of Linc00152 reduced Wnt/β-Catenin signaling (p<0.001).
Conclusions
This research shows that Linc00152 is highly expressed in patients with bladder cancer and the possible carcinogenic effect of Linc00152 in bladder cancer occurs through activating the Wnt/β-Catenin signaling pathway, and could be a new biomarker for diagnosis and prevention of this cancer.
MeSH Keywords: Cell Growth Processes; Gallbladder Neoplasms; Genes, Tumor Suppressor; RNA, Long Noncoding; Wnt Signaling Pathway
Background
Bladder cancer (BC) is a major cause of genitourinary cancer-related death among males worldwide. As reported by the American Cancer Society cancer statistics, bladder cancer leaded to 7% (62 380) of total male cancers and 4% (12 520) of all male cancer deaths in 2018 [1]. The incidence and mortality rates of BC are also increasing gradually in Asian countries [2]. Recent reports suggested that environmental factors and genetic and epigenetic alterations were responsible for BC tumorigenesis [3]. Although huge improvements have been made in the clinical diagnosis and multidisciplinary treatment of BC, reliable and specific prognosis of actionable biomarkers and targets in BC is still unavailable, implying the necessity for discovering a new and effective biomarker for the early prediction, progression, and prognosis of BC.
Long noncoding RNAs are protein-coding molecules that are over 200 nucleotides (>200 nt) in length and lack a detectable open reading frame [4,5]. It is reported that lncRNAs play a significant role in numerous of human cancers [6]. For instance, LncRNA-MALAT-1, which is highly expressed in bladder transitional cell carcinoma (BTCC), was found to play an important role in tumorigenesis and metastasis of BTCC, and the upregulation of MALAT1 were positively associated with advanced clinical and pathological stage and shorter survival of BTCC patients [7]. lncRNA-ZFAS1 (zinc finger antisense 1) was overexpressed in bladder cancer, and knockdown of ZFAS1 represses bladder cancer cell proliferation by upregulating the expression of KLF2 and NKD2, and inhibiting cell migration and invasion by downregulating ZEB1 and ZEB2 expression [8]. Thus, various lncRNAs were reported to be involved in carcinogenesis.
lncRNA- Linc00152 is an intergenic lncRNA mapped to 2p11.2, and previous studies have revealed its function as an oncogene in gastrointestinal cancer, clear cell renal cell carcinoma, hepatocellular carcinoma, and gallbladder carcinoma [9–12]. Linc00152 can also cause cancer cell proliferation and growth via the epidermal growth factor receptor (EGFR)/mammalian target of rapamycin (mTOR) signaling pathway, or promote cancer cell migration and invasion by regulating epithelial-to-mesenchymal transition (EMT) [11–14]. However, the Linc00152 expression and its related mechanism in BC has not yet been reported. Here, we explored the relative expression of Linc00152 in BC and assessed the clinicopathological features and biological regulation mechanisms of Linc00152 in BC.
Material and Methods
Cell culture and clinical samples
Four bladder cancer cell lines (T24, SW780, HT-1197, and HTB-9) and the normal urothelium cell line SV-HUC-1 were purchased from the American Type Culture Collection (Manassas, VA). All cell lines were incubated in RPMI 1640 with 10% fetal bovine serum (Life Sciences, USA) and antibiotics, and cultured in a humidified atmosphere at 37°C and 5% CO2.
Bladder cancer and adjacent non-cancerous tissue specimens were collected from 126 patients who underwent surgical resection of tumor lesions at the Chongqing University Cancer Hospital (Chongqing, China) between January 2012 and December 2016. All patients signed consent forms before surgery. The clinical characteristics of these patients are shown in Table 1. None of the patients recruited in the research received any chemotherapy, molecule-targeted therapy, or radiotherapy prior to surgical intervention. All patients provided signed informed consent. Among these patients, 58 (48.3%) were 50 years or older, 68 (51.6%) were younger than 50 years; 67 (53.2%) were men and 59 (46.8%) were women; and there were no significant differences between groups in age or sex. All samples were kept in liquid nitrogen and subsequently stored at −80°C until use. Pathological and clinical data of all the participants were obtained and are summarized in Table 1.
Table 1.
Association between lncRNA-Linc00152 expression and clinicopathological characteristics in bladder cancer patients.
| Parameters | Characteristics | Linc00152 expression | Linc00152 | P value |
|---|---|---|---|---|
| High | Low | |||
| Gender | Male | 32 | 35 | 0.213 |
| Female | 29 | 30 | ||
| Age | <50 | 27 | 41 | 0.312 |
| ≥50 | 22 | 36 | ||
| Tumor size | <3 cm | 36 | 41 | 0.293 |
| ≥3 cm | 22 | 27 | ||
| Lymph node metastasis | Negative | 44 | 58 | 0.014 |
| Positive | 22 | 2 | ||
| Histological grade | High grade | 31 | 30 | 0.021 |
| Low grade | 42 | 33 | ||
| Multiplicity | Single | 21 | 53 | 0.412 |
| Multiple | 18 | 34 |
The clinical staging was done using the TNM classification in 2010 AJCC. This research was approved by the Institutional Ethics Committees of Chongqing University Cancer Hospital (20120103).
RNA extraction and qRT-PCR
Total RNA extraction was carried out as described before [15]. RNA was extracted with TRIzol reagent (Life Technologies) and isolated RNA was quantified and then reverse-transcribed into cDNA by use of random primers for PCR. To assess relative Linc00152 expression, quantitative PCRs were applied using SYBR® Green PCR Master Mix (Thermo Fisher Scientific) [15]. The amplification reaction conditions were 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s for 32 cycles with GAPDP as an internal control. The experiments were repeated 3 times.
All primer sequences are listed in Table 2.
Table 2.
List of qRT-PCR primers used in this study.
| Primer | Sequence (5′-3′) | Product size (bp) | PCR cycles | Annealing temperature (°C) |
|---|---|---|---|---|
| c-MYC-F | CGCAAATGGGCGGTAGGCGTG | 165 | 40 | 60 |
| c-MYC -R | TAGAAGGCACAGTCGAGG3 | |||
| Ecad-F | TACACTGCCCAGGAGCCAGA | 103 | 40 | 60 |
| Ecad-R | TGGCACCAGTGTCCGGATTA | |||
| Vimentin-F | GACCAGCTAACCAACGACAA | 150 | 40 | 60 |
| Vimentin-R | GTCAACATCCTGTCTGAAAGAT | |||
| Ncad-F | CGAATGGATGAAAGACCCATCC | 174 | 40 | 60 |
| Ncad-R | GGAGCCACTGCCTTCATAGTCAA | |||
| Linc00152-F | TGAGAATGAAGGCTGAGGTGT | 116 | 40 | 60 |
| Linc00152-R | GCAGCGACCATCCAGTCATT | |||
| GAPDH-F | GGAGTCAACGGATTTGGT | 206 | 40 | 60 |
| GAPDH-R | GTGATGGGATTTCCATTGAT |
F – forward; R – reverse.
RNAi and transfection
The LINC00152 siRNAs and its negative vector (NC) siRNAs were purchased from RealGene (Nanjing, China) [16]. BC cell lines T24 and HT-1197 were transfected with 40 nM siRNA oligos introduced into cells, and controls were treated using Lipofectamine 3000 reagents (GENECHEM, Shanghai, China) following methods described in previous reports [17]. The cells were harvested and collected for RNA extraction or functional assays 48 h after the treatment of cells with siRNA, and qRT-PCR was first applied to determine the transfection efficiency. The siRNA oligos sequences targeting Linc00152 were:
Linc00152-si-1: 5′ GGGAAAUAAAUGACUGGAUdTdT 3′;
Linc00152-si-2: 5′ GGAGAUGAAACAGGAAGCUdTdT 3′;
Negative control (NC): 5′ UUCUCCGAACGUGUCACGUdTdT 3′.
Flow cytometry assay
Flow cytometry analysis was performed as described previously [18]. For cell cycle assays, cells were fixed in 75% ethanol at 4°C for 24 h, and then reacted with RNase and propidium iodide (PI) for about 30 min at 4°C in the dark. For cell cycle assays, Annexin V-FITC/PI staining was performed for apoptosis analysis according to the manufacturer’s instructions. The data were analyzed with CELL Quest software. Each experiment was repeated 3 times independently.
Cell viability assay
The cell proliferation was evaluated by CCK-8 assay. Linc00152-siRNAs and negative control (NC) cells were digested and seeded into 96-well tubes (2000 cells per well). After cells were cultured for 0, 24, 48, or 72 h, CCK-8 reagent (Beyotime, Beijing, China) was added into each well and then incubated for 3 h at 37°C. Absorbance readings at 490 nm were measured by a microplate reader (Thermo Scientific, USA). CCK-8 assay was performed 3 times.
Soft-agar colony-formation assay was performed as previously reported [18]. Cells were collected after transfecting for 48 h, cultured in RPMI-1640 containing 10% FBS and 0.4% agar, and then suspended in 10% FBS/RPMI 1640 containing 0.65% agar in a 6-well plate. Colonies were counted and photographed at 14 days after transfection. The experiment was repeated 3 times.
Wound-healing and Transwell assays
Cell migration was evaluated using a wound-healing assay. In brief, Linc00152-siRNAs and negative control (NC) cells (T24 and HT-1197) were seeded in 6-well plates until they were 90% confluent. A wound was created using a P-20 pipette tip, washed with PBS, and resuspended in RPMI 1640. Cultures were then photographed and assessed at low power using a phase-contrast microscope every 12 h (Leica DMI4000B, Bucks, UK). These experiments were performed 3 times.
We used 8-μm pore size Transwell chambers (Corning, USA) to measure cell migration and invasion. Linc00152-siRNAs and negative control (NC) cells (T24 and HT-1197) were digested, washed, and resuspended in serum-free medium, and added to the upper chamber, whereas the lower chambers contained 700 μl medium containing 10% FBS as a chemoattractant. After incubation at 37°C and 5% CO2 for 48 h, cells on the lower side of the insert membrane surface were fixed with 4% paraformaldehyde and stained with crystal violet. Non-migratory BC cells on the upper surface were wiped off with a cotton swab. Invaded cells were counted and photographed under a microscope in 5 random fields.
Protein extraction and Western blot analysis
Protein was harvested from the cells after transfection and lysed with RIPA buffer containing protease inhibitor (Beyotime, China). The concentration of proteins was measured using a BCA Protein Assay Kit (Pierce, USA). Equal amounts of proteins (40 μg) were separated with 10% SDS-PAGE (Pierce, Rockford, IL) and electrotransferred onto a PVDF membrane (Millipore). After being blocked with 5% non-fat milk at room temperature for 1 h, the membranes was treated with the following primary antibodies: Total β-catenin (1: 500, CST, USA), active-β-catenin (1: 500, Abcam, Cambridge, UK), cyclinD1 (1: 2000, Abcam), c-Myc (1: 1000, Abcam), p21 (1: 1000, Abcam), E-cadherin (1: 1000, Abcam), Vimentin (1: 1000, Abcam), and N-Cadherin (1: 1000, Abcam) overnight at 4°C. After being washed with PBST, the membranes were incubated with secondary antibodies at 37°C for 1 h. Protein blots were quantified using chemiluminescence method (Pierce, Rockford, IL) in accordance with the user’s manual.
Statistical analysis
SPSS version 17.0 software was used for statistical analyses and data were processed with GraphPad prism software. Data are expressed as mean ±SD, and differences between 2 independent groups were assessed using the t test. p<0.05 was considered to be statistically significant.
Results
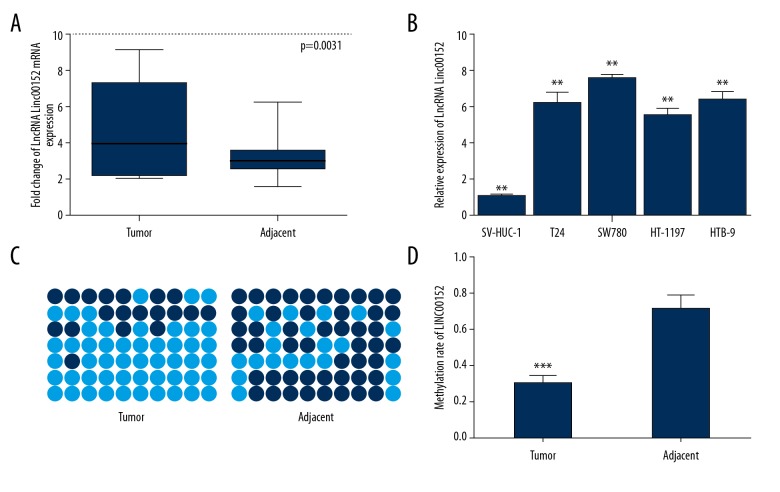
Linc00152 expression was upregulated and hypomethylated in BC tissues. To assess the relative Linc00152 expression in BC tumor tissues, we used qRT- PCR analysis of total RNA extracted from 126 pairs of BC specimens. The results revealed that the expression of Linc00152 was higher in cancerous bladder tissues than in normal adjacent paired tissues (Figure 1A, p=0.0031). We also assessed the expression levels of Linc00152 in BC cell lines, and results showed that Linc00152 expression was higher in BC cell lines than in SV-HUC-1 (Figure 1B, F=32.34, p<0.01). Methylation analysis was performed by Bisulfite sequencing PCR (BSP). We found that hypermethylation of Linc00152 promoter was significantly higher in paired tumor-adjacent tissues than in BC tumors (Figure 1C 1D, p<0.001), as reported in HCC [19].
Figure 1.
Expression and methylation status of Linc00152 in BC. (A) Expression of Linc00152 in a panel of bladder cancer tissues matched with normal adjacent bladder tissues measured by qRT-PCR with GAPDH as control (n=126, p=0.0031). (B) The relative expression of Linc00152 mRNA in BC and normal cell lines, ** p<0.01. (C, D) Bisulfite sequencing (BSP) analysis was used to examine methylation of CpG islands in Linc00152. The methylation level of the Linc00152 promoter in BC was downregulated compared with the normal adjacent bladder tissues (*** p<0.001).
Correlation between Linc00152 expression and its clinicopathological characteristics in BC
The BC clinical samples were divided into 2 groups according to median Linc00152 expression. As summarized in Table 1, we found significant correlations between higher expression of Linc00152 and histological grade and lymph node metastasis (P<0.05). We found no significant associations between Linc00152 expression and sex, age, tumor size, or tumor stage.
Knockdown of Linc00152 inhibited cell clonogenicity and proliferation
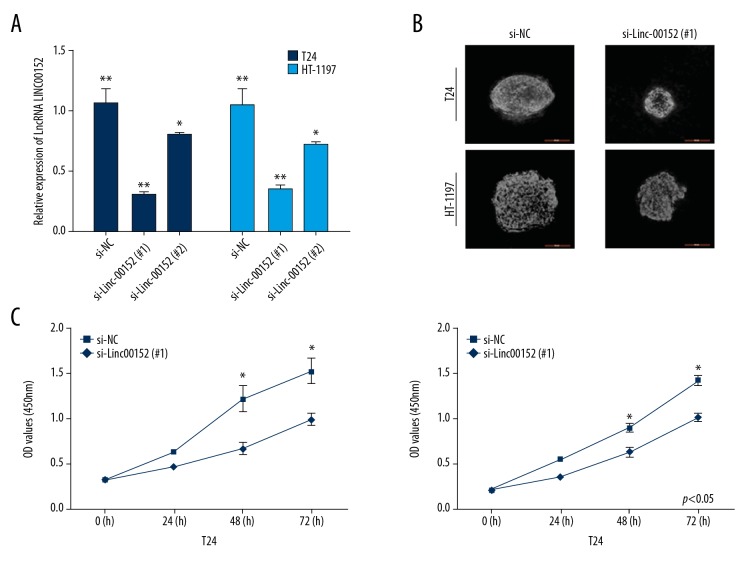
Soft-agar colony-formation and CCK-8 assays were used to determine the knockdown effects of Linc00152 on cell viability in T24 and HT-1197 cells. Two specific Linc00152-siRNAs were assessed for the transfection efficiency, and the results showed that si-Linc00152 (#1) and si-Linc00152 (#2) had weaker knockdown effects than in si-NC (Figure 2A), and si-Linc00152 (#1) had higher transfection efficiency than si-Linc00152 (#2) (Figure 2A, F=41.32, ** p<0.01). Thus, si-Linc00152 (#1) was chosen for further functional assays. Results clearly showed there were fewer colonies formed in si-Linc00152 (#1) groups than in vector groups (Figure 2B). We found decreased cellular proliferation after 48 h and 72 h in both BC cell lines with si- Linc00152 according to CCK-8 assay (p<0.05, Figure 2C).
Figure 2.
Knockdown of Linc00152 inhibited cell proliferation in T24 and HT-1197 cells. (A) BC cells transfected with si-NC and Linc00152 siRNAs. qRT-PCR was used to detect Linc00152 expression after 48-h transfection (** p<0.01, * p<0.05). (B) Representative soft-agar colony-formation. (C) CCK-8 assay for cellular proliferation of si-NC and si-Linc00152 (#1) -infected cell lines (* p<0.05).
Knockdown of Linc00152 suppress cell viability through causing cell apoptosis
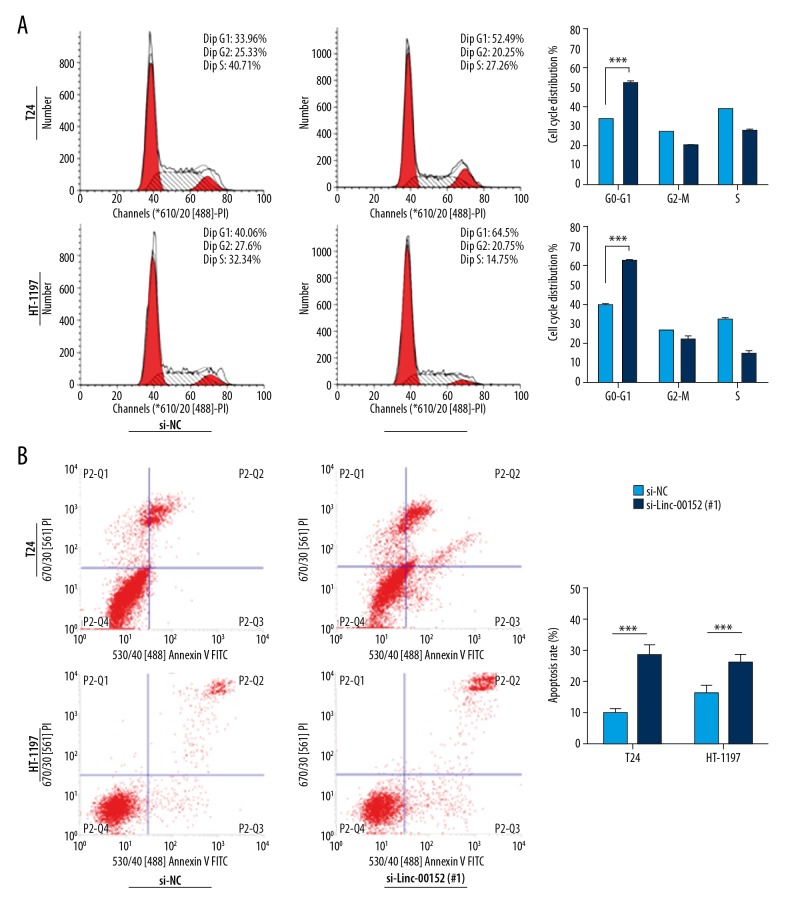
To evaluate the possible mechanism of Linc00152 in inhibiting cell proliferation, flow cytometric analyses of cell cycle and cell apoptosis were performed. We found that si-Linc00152 in T24 and HT-1197 significantly increased the percentage of G0–G1 cells by nearly 20% (Figure 3A, *** p<0.001). Meanwhile, changes between si-NC ad si-Linc in ‘G2-M’ and ‘S’-phases had no statistically significant difference. Significant differences in apoptosis were also observed between the control group and si-Linc00152 groups after transfection for 48 h (Figure 3B, *** p<0.001).
Figure 3.
Knockdown of Linc00152 induces cell cycle arrest and causes apoptosis. (A) Representative images of cell cycle distribution in T24 and HT-1197 cell lines. Right: statistical flow cytometry data. *** P<0.001 vs. control. (B) Detection of apoptosis with si-Linc00152 in T24 and HT-1197 cell lines. Right: statistical flow cytometry data. *** P<0.001 vs. control.
Knockdown of Linc00152 inhibited cellular migration and invasion
To ascertain whether knockdown of Linc00152 influences cell migration and invasion, we examined the influences of Linc00152 knockdown on cell migration by wound-healing assays. Results suggested that T24 and HT-1197 cells transfected with si-Linc00152 have less migrating ability than cells in the vector group (Figure 4A, ** p<0.01). Furthermore, we used the Transwell chamber system to assess cellular invasion ability and the results showed that the number cellular invasions significantly decreased with knockdown of Linc00152 by siRNA (Figure 4B, *** p<0.01).
Figure 4.
Linc00152 knockdown inhibited cell motility in T24 and HT-1197 cell lines. (A) The migration of T24 and HT-1197 cells was decreased by silncRNA-ATB-1, ** p<0.01. (B) The inhibitory effect of T24 and HT-1197 cells was decreased by si-Linc00152, *** p<0.001.
Knockdown of Linc00152 regulates the canonical Wnt in BC cells
We investigated whether the possible tumor-promotive mechanisms of Linc00152 are associated with Wnt/β-catenin signaling, as Wnt plays a vital role in regulating cancer cell proliferation and EMT [20]. Western blot assays and qRT-PCR were used to determine the expression of Wnt-related markers. Si-Linc00152 in T24 and HT-1197 cells significantly inhibited β-catenin expression levels instead of changing the expression of total β-catenin (Figure 5A), since β-catenin in cytoplasm plays an important part in regulating the downstream transcription factor T-cell factor/lymphocyte enhancer factor (TCF/LEF) [21]. We also found that the depletion of Linc00152 decreased numbers of β-catenin permanent target genes such as c-Myc, CCND1, and p21, and also upregulates epithelial mesenchymal transition-related E-cadherin and the downregulation of N-cadherin and Vimentin. qRT-PCR showed similar results (Figure 5B, *** p<0.001), indicating Linc00152 regulates BC progression through activation of the Wnt/β-Catenin signaling pathway.
Figure 5.
Knockdown of Linc00152 disrupted Wnt signaling in T24 and HT-1197 cell lines. (A) Western blot analysis was used to assess antibodies against total β-catenin, active β-catenin, and its downstream targets; β-actin was used as a control. (B) qRT-PCR was performed using c-Myc and its downstream targets.
Discussion
Increasing evidence proves that lncRNAs play vital roles in various malignant tumors [12,13]. Linc00152 is a recently discovered lncRNA located in 2p11.220. Linc00152 has been reported to be involved in multiple physiological and pathological processers such as colorectal cancer, gallbladder cancer, and breast cancer [11,12,22]. Recent studies have revealed that high expression of Linc00152 promotes cell proliferation, migration, and invasion [11,12,22]. However, the detailed role of Linc00152 and its related mechanisms involved in BC remain unclear. In this research, we focused on Linc00152 expression in BC cell lines and BC tissue samples and also sought to further reveal the functional role of Linc00152 in BC.
We found that Linc00152 expression was higher in bladder cancer tissues and cell lines compared to normal adjacent bladder tissues and the normal human bladder epithelial immortalized cell line SV-HUC-1. Additionally, hypermethylation of Linc00152 is common in BC and the expression of Linc00152 is inversely correlated with its promoter methylation. The increased expression of Linc00152 was associated with WHO grade (p=0.0027). These results suggest that Linc00152 is a potential prognostic biomarker and acts as a BC oncogene.
To investigate the function of Linc00152 in 2 BC cells, we used Linc00152 knockdown. It is reported that Linc00152 is related to cancer cell proliferation, cell cycle, cell apoptosis, epithelial-to-mesenchymal transition (EMT), cell migration/invasion, and other biological functions [11,12,22]. Thus, we mainly focused on these tumor-promotion activities. We found that siRNA-mediated knockdown of Linc00152 significantly inhibited cell viability, and the percentage of cells in G0/G1 phase and the apoptotic cell numbers were significantly increased in vitro. It has been reported that Linc00152 promotes cell migration/invasion in multiple cancers [11,12,22]. We also found Linc00152 downregulation significantly suppressed the migration and invasion of T24 and HT-1197 cells. These results are consistent with previous studies.
Multiple studies have shown that the canonical Wnt participates in a variety of cellular process, including cell growth, cell cycle, cell differentiation, and tumorigenesis [23,24]. Recent evidence reveals that the Wnt/β-catenin pathway is a good biomolecular target for BC diagnosis and treatment [25–29]. In the presence of Wnt signals, cytoplasmic β-catenin protein is activated and then accumulates in the cytoplasm and translocates to the nucleus, where it activates the transcription of Wnt downstream target genes, including cyclin D1, c-Myc, and MMPs [30–32]. Shan reported that Linc00152 regulates gastric cancer cell proliferation and metastasis by promoting β-catenin methylation, and thus activates the canonical Wnt signaling [33]. In the present study, we observed that decreased expression of Linc00152 causes decreased Wnt/β-catenin signaling in these 2 cancer cell types by affecting the distribution of β-catenin activity in the cell nucleus. In assessing the mechanisms of Linc00152, we found that Wnt/β-catenin downstream-targeted genes were also downregulated. EMT enables stem-like features in cancer cells, including metastasis, with loss of epithelial properties and gain of mesenchymal phenotype [34,35]. Remarkably, we found that with Linc00152 knockdown, some mesenchymal cellular hallmarks, including Vimentin and N-cadherin, were reduced, whereas E-cadherin was upregulated. Cumulatively, the above evidence shows the role of Linc00152 in regulation of Wnt/β-catenin signaling, an underlying mechanism that is responsible for bladder cancer progression.
Conclusions
Our findings suggest that Linc00152 is highly expressed in bladder cancer cells and that Si-Linc00152 suppresses cell viability, migration/invasion, and EMT in bladder cancer cells. We also found that Linc00152 stabilizes β-catenin and leads to abnormal activation of the Wnt/β-Catenin signaling pathway. Whether it can be used as a target gene for diagnosis and clinical treatment of bladder cancer requires more in vivo studies.
Footnotes
Source of support: Departmental sources
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China. Cancer Lett. 2013;401:63–71. doi: 10.1016/j.canlet.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 3.Mahdavifar N, Ghoncheh M, Pakzad R, et al. Epidemiology, incidence and mortality of bladder cancer and their relationship with the development index in the world. Asian Pac J Cancer. 2016;17:381–86. doi: 10.7314/apjcp.2016.17.1.381. [DOI] [PubMed] [Google Scholar]
- 4.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Greiman AK, Rosoff JS, Prasad SM. Association of Human Development Index with global bladder, kidney, prostate and testis cancer incidence and mortality. BJU Int. 2017;120:799–807. doi: 10.1111/bju.13875. [DOI] [PubMed] [Google Scholar]
- 7.Jiao D, Li Z, Zhu M, et al. LncRNA MALAT1 promotes tumor growth and metastasis by targeting miR-124/foxq1 in bladder transitional cell carcinoma (BTCC) Am J Cancer Res. 2018;8:748–60. [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H, Li G, Cheng B, et al. ZFAS1 functions as an oncogenic long non-coding RNA in bladder cancer. Biosci Rep. 2018;38(3) doi: 10.1042/BSR20180475. pii: BSR20180475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y, Tan C, Weng WW, et al. Long non-coding RNA Linc00152 is a positive prognostic factor for and demonstrates malignant biological behavior in clear cell renal cell carcinoma. Am J Cancer Res. 2016;6:285–99. [PMC free article] [PubMed] [Google Scholar]
- 10.Chen WM, Huang MD, Sun DP, et al. Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer. Oncotarget. 2016;7:9773–87. doi: 10.18632/oncotarget.6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Cai X, Chang L, et al. LINC00152 is a potential biomarker involved in the modulation of biological characteristics of residual colorectal cancer cells following chemoradiotherapy. Oncol Lett. 2018;15:4177–84. doi: 10.3892/ol.2018.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai Q, Wang Z, Wang S, et al. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial-mesenchymal transition by regulating HIF-1α via miR-138. Open Biol. 2107;7 doi: 10.1098/rsob.160247. pii: 160247. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Ji J, Tang J, Deng L, et al. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6:42813–24. doi: 10.18632/oncotarget.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiang T, Fan Y, Li C, et al. DACT2 silencing by promoter CpG methylation disrupts its regulation of epithelial-to-mesenchymal transition and cytoskeleton reorganization in breast cancer cells. Oncotarget. 2016;7:70924–35. doi: 10.18632/oncotarget.12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin X, Xiang T, Li L, et al. DACT1, an antagonist to Wnt/beta-catenin signaling, suppresses tumor cell growth and is frequently silenced in breast cancer. Breast Cancer Res. 2103;15:R23. doi: 10.1186/bcr3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen QN, Chen X, Chen ZY, et al. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with ezh2 and repressing il24 expression. Mol Cancer. 2017;16:17. doi: 10.1186/s12943-017-0581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong J, Liu Y, Jiang L, et al. High expression of long non-coding RNA lncRNA-ATB is correlated with metastases and promotes cell migration and invasion in renal cell carcinoma. Jpn J Clin Oncol. 2016;46:378–84. doi: 10.1093/jjco/hyv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao W, Peng T, Zhou Y. Long noncoding RNA activated by transforming growth factor-β promotes cancer development and is a prognostic marker in cervical cancer. J Cancer Res Ther. 2017;13:801–6. doi: 10.4103/jcrt.JCRT_256_17. [DOI] [PubMed] [Google Scholar]
- 19.Ji J, Tang J, Deng L, et al. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6:42813–24. doi: 10.18632/oncotarget.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shan Y, Ying R, Jia Z, et al. LINC00052 promotes gastric cancer cell proliferation and metastasis via activating the Wnt/β-catenin signaling pathway. Oncol Res. 2017;25:1589–99. doi: 10.3727/096504017X14897896412027. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Terasaki H, Saitoh T, Shiokawa K, et al. Frizzled-10, up-regulated in primary colorectal cancer, is a positive regulator of the WNT-β-catenin-TCF signaling pathway. Int J Mol Med. 2002;9:107–12. [PubMed] [Google Scholar]
- 22.Hu XL, Wang J, He W, et al. Down-regulation of lncRNA Linc00152 suppressed cell viability, invasion, migration, and epithelial to mesenchymal transition, and reversed chemo-resistance in breast cancer cells. Eur Rev Med Pharmacol Sci. 2018;22:3074–84. doi: 10.26355/eurrev_201805_15067. [DOI] [PubMed] [Google Scholar]
- 23.Dong ZC, Zhang D, Wang SB, et al. Target inhibition on GSK-3β by miR-9 to modulate proliferation and apoptosis of bladder cancer cells. Eur Rev Med Pharmacol Sci. 2018;22:3018–26. doi: 10.26355/eurrev_201805_15059. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida T, Sopko NA, Kates M, et al. Three-dimensional organoid culture reveals involvement of Wnt/β-catenin pathway in proliferation of bladder cancer cells. Oncotarget. 2018;9:11060–70. doi: 10.18632/oncotarget.24308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu X, Li Y, Zhao S, et al. LSINCT5 activates Wnt/β-catenin signaling by interacting with NCYM to promote bladder cancer progression. Biochem Biophys Res Commun. 2018;502:299–306. doi: 10.1016/j.bbrc.2018.05.076. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh HY, Shen CH, Lin RI, et al. Cyproheptadine exhibits antitumor activity in urothelial carcinoma cells by targeting GSK3β to suppress mTOR and β-catenin signaling pathways. Cancer Lett. 2016;370:56–65. doi: 10.1016/j.canlet.2015.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Du HF, Ou LP, Lv CK, et al. Expression of hepaCAM inhibits bladder cancer cell proliferation via a Wnt/β-catenin-dependent pathway in vitro and in vivo. Cancer Biol Ther. 2015;16:1502–13. doi: 10.1080/15384047.2015.1071732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao XW, Xiao JQ, Xu G, et al. CUL4B promotes bladder cancer metastasis and induces epithelial-to-mesenchymal transition by activating the Wnt/β-catenin signaling pathway. Oncotarget. 2017;8:77241–53. doi: 10.18632/oncotarget.20455. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Dang Y, Wei X, Xue L, et al. Long non-coding RNA in glioma: Target miRNA and signaling pathways. Clin Lab. 2018;64:887–94. doi: 10.7754/Clin.Lab.2018.180107. [DOI] [PubMed] [Google Scholar]
- 30.Ying Y, Tao Q. Epigenetic disruption of the WNT/beta-catenin signaling pathway in human cancers. Epigenetics. 2009;4:307–12. doi: 10.4161/epi.4.5.9371. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Ying J, Fan Y, et al. WNT5A antagonizes WNT/β-catenin signaling and is frequently silenced by promoter CpG methylation in esophageal squamous cell carcinoma. Cancer Biol Ther. 2010;10:617–24. doi: 10.4161/cbt.10.6.12609. [DOI] [PubMed] [Google Scholar]
- 32.Sebio A, Kahn M, Lenz HJ. The potential of targeting Wnt/β-catenin in colon cancer. Expert Opin Ther Targets. 2014;18:611–15. doi: 10.1517/14728222.2014.906580. [DOI] [PubMed] [Google Scholar]
- 33.Shan Y, Ying R, Jia Z, et al. LINC00052 promotes gastric cancer cell proliferation and metastasis via activating the Wnt/β-catenin signaling pathway. Oncol Res. 2017;25:1589–99. doi: 10.3727/096504017X14897896412027. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Huang Y, Li G, Wang K, et al. Collagen type VI alpha 3 chain promotes epithelial-mesenchymal transition in bladder cancer cells via transforming growth factor β (TGF-β)/Smad pathway. Med Sci Monit. 2018;24:5346–54. doi: 10.12659/MSM.909811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang M, Ren B, Li Z, et al. Expression of N-Myc downstream-regulated gene 2 in bladder cancer and its potential utility as a urinary diagnostic biomarker. Med Sci Monit. 2017;23:4644–49. doi: 10.12659/MSM.901610. [DOI] [PMC free article] [PubMed] [Google Scholar]