Abstract
Water stress, in a climate change scenario is one of the major threats for sustainable rice productivity. Combining drought resistance with yield and desirable economic traits is the most promising solution for the researchers. Although several studies resulted in the identification of QTLs for drought resistance in rice, but none of them serve as a milestone. Therefore, there is always a quest to find the new QTLs. The present investigation was carried out to map QTLs involved in drought resistance and yield related parameter in a cross of IR55419-04 and Super Basmati. An F2 population of 418 individuals was used as the mapping population. The raised nursery was transplanted in lyzimeters. Two extreme sets of tolerant (23 Nos.) and sensitive (23 Nos.) individuals were selected based on total water uptake under water stress conditions. Two hundred thirty microsatellite markers staggered on the whole genome were used for identifying polymorphic markers between the two parents. The selected 73 polymorphic microsatellites were used to genotype individuals and were scattered on a distance of 1735 cM on all 12 linkage groups. QTL analysis was performed by using the WinQTL Cartographer 2.5 V. A total of 21 QTLs were detected using composite interval mapping. The QTLs relating to drought tolerance at the vegetative stage were found on chromosome 1. Novel genomic regions were detected in the marker interval RM520-RM143 and RM168-RM520. The region has a significant QTL qTWU3.1 for total water uptake. Root morphological trait QTLs were found on chromosome 3. QTLs responsible for additive effects were due to the alleles of the IR55419-04. These novel QTLs can be used for marker assisted breeding to develop new drought-tolerant rice varieties and fine mapping can be used to explore the functional relationship between the QTLs and phenotypic traits.
Keywords: rice, water uptake, mapping population, SSR markers, composite interval mapping
Introduction
Rice (Oryza sativa L.) is one of the most important food crops in the world. It covers about 9% of the earth’s arable land. It is a source of 21% of global human per capita energy and 15% of per capita protein and makes up a quality food for almost half of the world population (Xu et al. 2011). Aromatic rice is famous in Asia and has gained broader acceptance in Europe and the United States (Ahmad et al. 2005). In an aromatic group of rice, Basmati is very popular and is cultivated solely in the foothills of the Himalayas, the Uttar Pradesh and Haryana regions of India and the Punjab province of Pakistan and India (Nagaraju et al. 2002). Basmati rice is a very well-known type of rice in the world due to the unique characteristics like long grain, aroma, and intermediate amylose content.
Rice is cultivated in deep water, irrigated, rainfed upland and rainfed lowland (Poehlman and Sleper 1995) whatever the case of its cultivation is, it requires plenty of water to give the optimum yield. The recent climate shift has urged the scientific community to develop strategies to address this issue with the help of scientific advancements. QTL mapping for the different traits and their introgression in cultivated varieties in the past few decades resolved several problems. The main issue with these achievements was their regional acceptability.
Currently number of scientists developed population for QTL mapping for almost all traits, including drought, submergence, grain quality traits, vascular bundle related traits, and bacterial leaf blight (Bernier et al. 2009, Dixit et al. 2014, Sandhu et al. 2014, Shamsudin et al. 2016, Singh et al. 2016, Zhai et al. 2018). To the best of our knowledge, no study is conducted in which Super Basmati used as one of the parents. Super Basmati is a popular Basmati rice variety developed by Rice Research Institute, Kala Shah Kaku, Punjab (Pakistan) and released in 1996 for general cultivation. The variety is liked by the rice growers of the Punjab province and is being cultivated in the famous Basmati rice belt known as “Kallar Tract”. Super Basmati variety covers major (>80%) area of the total Basmati rice area in the Punjab province. The variety is also very popular among millers, rice traders and consumers for its extra-long grain, excellent taste and typical Basmati aroma. Moreover, it is also preferred in the international market and fetches a considerable amount of foreign exchange for the country since its approval.
Pakistan, in the past few years is facing a challenging situation regarding climatic shift. These shifts resulted in uncertainty and unpredictability for grower of rice. In Pakistan, rice occupies second position after wheat and is grown on an area of 2.3 million hectares. The share of Pakistan in total world rice trade is about 11%, while by quantity it is 8.9%. Rice contributes 5.7% of the total agricultural value and 1.1% to GDP. Therefore, it is of prime importance to increase or at least maintain the yield as the climatic shift is a big challenge for its yield sustainability.
These environmental stresses pave the way for biotic and abiotic stresses. It has been observed that among all stresses drought so far is the main abiotic stress for rice, and many efforts have been done to improve rice yield under drought stress (Guan et al. 2010, Xiao et al. 2007). Thus, the best approach to increase the rice production is to develop new drought tolerant rice varieties with high yielding potential.
Phenotypic characters such as root dry weight, shoot dry weight, culm length, leaf area etc. have a strong correlation with drought. Many QTLs were identified for drought tolerance related traits such as leaf water potential, root dry weight, plant osmotic potential, root morphology and leaf rolling, shoot dry weight, root hair length, root thickness, deep root mass and total root number (Henry et al. 2015, Jongdee et al. 2002, Li et al. 2015, 2017, Liu et al. 2005, Qu et al. 2008a, 2008b, Terra et al. 2016).
The present study was designed for identification of novel QTLs for enhancement of drought tolerance in a cross between Super Basmati and IR55419-04. In turn, the drought-tolerant introgression lines developed through this research effort will be utilized in the future rice breeding programs in the country. Ultimately, sustainable rice production will bring more opportunities for the prosperity of the country through food security and poverty alleviation.
Materials and Methods
Parental materials and population development
The two parents were selected based on the study reported by Sabar and Arif (2014) in order to develop the mapping population. Super Basmati is an aromatic rice variety sensitive to water stress at the vegetative stage with a long grain. Whereas IR55419-04 is a coarse grain rice variety of the indica group. It has a stiff stem, is drought tolerant, but has short grain.
Three dates of parental lines, Super Basmati and IR55419-04, were planted in a crossing block at the research area of the National Institute for Biotechnology and Genetic Engineering (NIBGE), Faisalabad, Pakistan. Super Basmati was used as a female parent while making a cross with donor IR55419-04. Emasculation of the female plant was done in the afternoon using vacuum emasculator at 500 mmHg and pollination was performed in the next morning with proper tagging. At maturity, successful F1 seeds were harvested and stored properly. To raise F1 seedlings, the seeds were placed on a filter paper in petri plates at 30°C in the incubator. One week old seedlings were transferred from the petri plates to the raised wet beds in the field. Genotyping for heterogeneity conformity of the F1 using two to four polymorphic microsatellites was also done before transplantation. Thirty days after sowing, the seedlings were transplanted in the field. Subsequently, these individuals were allowed to be selfed and the F2 seeds were harvested at maturity. An F2 population was used as a mapping population.
Experimental site and F2 phenotyping
The experiment was conducted in the greenhouse at the International Rice Research Institute (IRRI), Los Banos, Philippines (14°11′ N 121°15′ E, 21 m above sea level) during the dry season. The bulk seeds harvested from F1 plants were sown in the seedling trays with covets (cup-shaped) filled 80% with soil. Each covets carried a small hole at the bottom allowing water to enter inside. In each covet, only one seed was dribbled manually and was covered with a thin soil layer. The trays were placed on the flat leveled surface of a tank where the water could remain stagnant so that it could only enter from the covet bottom hole through capillary action. The seedlings were irrigated by maintaining the water level of the tank at 50% covets sink. The raised seedlings were transferred into the pots 20 days after seeding (DAS). The pots were made of 105 cm long 19 cm diameter PVC pipes lined with plastic bags, filled with homogenized IRRI upland farm soil and sealed at the bottom. Each pot was drilled for making three holes near the bottom of the PVC pipe sides to allow drainage. These holes were plugged with rubber corks not allowing the drainage. The lower portion (80%) of the pots was filled with a bulk density equivalent to 1.15 g/cm3 of dry soil by compacting the soil even after every 5 cm during the filling process (Bernier et al. 2009). To settle down the soil, the pots were saturated with water for a few days before transplanting the seedlings. The upper (20%) portion was filled with lowland irrigated soil. The soil level was set aside 5 cm below the edge of the pots. The pots were painted white to reduce the soil temperature from the surroundings. The total number of pots for the F2 population was 418. For each of the two parental genotypes and a drought-sensitive check variety IR64, there were 30 pots (15 for well-watered and 15 for water stress). The pots were divided into six blocks. Each block was protected with 2.45 cm thick polyester film/polyester fiber non-woven fabric flexible composite material sheet to minimize the heat effect on the surrounding pots. Phosphorus and potassium were applied at 40 kg ha−1 as a basal dose. The equivalent of 40 kg ha−1 of nitrogen was applied at 10 days after transplanting (DAT) of seedlings. The pots of two parents (tolerant and sensitive) were watered thrice a week to maintain the soil near field capacity for the whole duration of the experiment in the well-watered (control) treatment and for 29 days at 40 DAS in the water stress treatment to F2 population and their two parents.
Phenotypic data collection
The water consumption of water-stressed plants was measured by weighing pots the day following the last irrigation i.e., 41 DAS. During the stress treatment, other readings were recorded at 48, 55, 62 and 69 DAS (once in a week). Evapo-transpiration (water consumption) was calculated as the difference of the initial pot weight and the pot weight at different times (Bernier et al. 2009). Soil evaporation was minimized by covering the pots opening with polythene plastic sheets having a hole in the middle for the plant shoot to come out. Aboveground biomass was sampled at 69 DAS by cutting the plants at the soil surface level. The plant harvested shoots were dried by keeping them in the oven at 70°C for 72 hours and their weights were measured with the help of a top loading balance. The data for a number of tillers per plant and plant height were also recorded as described in the revised Standard Evaluation System for rice (SES 2002). For the measurement of the root parameters, the plastic bags were pulled out from the pots and the soil was then cut into three sections i.e., 0–30 cm depth, 30–60 cm depth and 60–100 cm depth. Within every soil section, roots were carefully washed to remove the soil. The washed roots were preserved in plastic bags containing 70% ethanol solution at 4°C until the completion of the scanning process. Root volume, root surface area, root diameter and root length of 60–100 cm depth section were measured by a scanner (EPSON, V700) and the recorded data were analyzed by using WinRHIZO Software, Canada. Roots and shoots were dried by keeping them in oven at 70°C for 72 hours. The root and shoot dry weight was then obtained by weighing on the scale. The leaf area (LA) of the parental genotypes, under well-watered treatment, was measured by the leaf area meter. For leaf area estimation of the F2 population, a non-destructive method was used. The digital picture of each plant was captured by using a digital camera (Power-Shot G7, Canon) set at one meter distance from the pot. A paper comprising of 100 cm2 area was used as a calibration reference and a three-digit number on white paper was placed just by the side of the top of the pot. The JPG images of the plants were taken at each weighing and were subjected to analysis using ImageJ software.
Genotyping
CTAB method with little modification was used for the genomic DNA extraction from 21 days old leaves of the plants. For measurements of DNA concentration and quality, NanoDrop spectrophotometer (DE 19810 U.S.A.) was used and as per quantification, the working dilutions (25 ng/μl) of the template DNA were prepared.
Polymerase chain reaction (PCR) was performed in 15 μl reaction mix containing 2 μl of 20–25 ng DNA template, 1.5 μl 10x TB buffer [containing 200 mM Tris-HCl pH 8.3, 500 mM KCl, 15 mM MgCl2], 1.2 μl of 1 mM dNTP, 0.80 μl of Taq DNA polymerase (5 Unit/μl) 0.50 μl each of 5 μM forward and reverse primers using a G-Storm GS1 thermal cycler (AlphaMetrix Biotech GmbH, Rodermark, Germany).
Mini vertical polyacrylamide gels for high throughput manual genotyping (CBS Scientific Co. Inc., CA, USA) were used. The samples allowed to run for about 1.5–3.5 hours at 100 volts depended on the size of the PCR fragment. The polyacrylamide gel was stained with SYBR Safe 10% solution in an opaque staining tray with lid for 30 minutes. For visualizing the gel, gel documentation system Alpha Imager 1220 (Alpha Innotech, CA, USA) was used.
Molecular data analysis
Evenly distributed SSR markers were used to identify polymorphic markers to differentiate the two parents. A molecular map was constructed with microsatellites to position the polymorphic markers relative to each other on the map using Map Manager QTX. QTL analysis was performed with microsatellite mapping data using WinQTL Cartographer 2.5 V (Wang et al. 2001) to generate the genetic linkage map. Composite interval mapping (CIM) was employed using the selected F2 plants to locate the more accurate association and precise position of putative QTLs. The QTLs were designated based on the nomenclature procedure suggested by McCouch et al. (1997) and McCouch and CGSNL (2008). In the suggested nomenclature, “q” stands for QTL and “Ht, Till, SDW”, refer different traits like plant height, tillers per plant, stem dry weight, the number at the end of the locus name defines the chromosome number on which the QTL has been mapped.
Results
Frequency distribution of physiological and agronomic traits
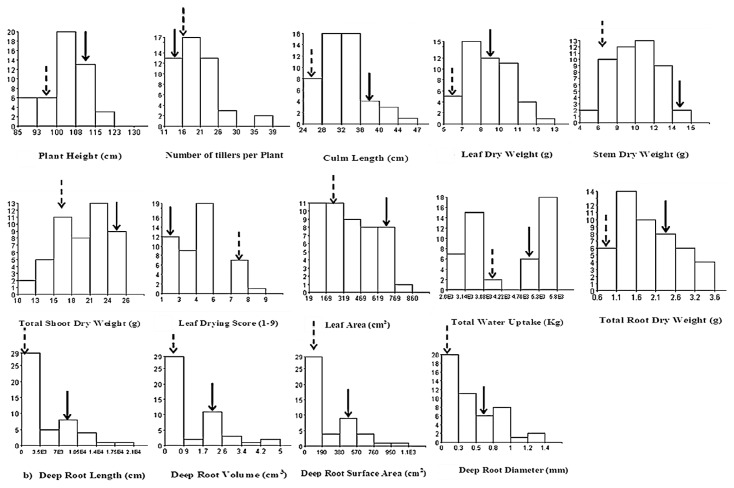
The two parents and their F2 population comprising of 418 plants were subjected to severe water stress for four weeks at the vegetative stage (Fig. 1). Whole F2 population gave normal frequency distribution for total water uptake. The mean total water uptake for IR55419-04 was 4956 g, while that of Super Basmati was 3711 g. The variation for total water uptake in the F2 plants ranged from 2675 to 5743 g. The two extremes i.e., high and low (23 each) were selected on the basis of this trait.
Fig. 1.
F2 population screening at severe water stress.
The parental lines, IR55419-04 and Super Basmati achieved the plant height of 111.2 and 100.0 cm, respectively. Selected F2 plants showed the plant height in the range from 85.4 to 120.2 cm. More number of taller plants (14 out of selected 46 plants) as compared the donor parent IR55419-04 was observed. A high frequency (71.7%) of the tillers per plant in a skewed manner amongst the F2 individuals showed greater value as compared to sensitive parent Super Basmati. Similarly, the variation for culm length of the selected plants was in the range from 24.9 to 47.0 cm. The frequency of transgressive segregates, for leaf dry weight and stem dry weight was nearly in a normal distribution. The variation in total shoot dry weight of F2 plants was in the range of 9.4 to 25.7 g showing the transgressive segregation for the parameter. The frequency distribution of this trait seems to follow a bimodal distribution that is negatively skewed (skewness = −0.300). The two parents revealed extreme responses to water stress regarding the leaf drying score, as Super Basmati is drought sensitive (final score = 7) and IR55419-04 is tolerant (final score = 1). Slightly skewed data towards tolerant class was observed for leaf drying score than would be expected in a normal distribution. About 22 individuals of the F2 population had a range 19.3–319 cm2, 24 plants ranged from 319.1–852 cm2, and parents IR55419-04 (703.9 cm2) and Super Basmati (312.4 cm2) for leaf area measured. Frequency distribution of each trait among plants of the mapping population and their parents has shown in Fig. 2.
Fig. 2.
Frequency distribution of selected two extremes of the F2 population for water stress-related traits at vegetative stage. Filled and dotted arrows indicate the phenotypic value of the parents IR55409-4 and Super Basmati, respectively.
Correlation studies of drought-related parameters
Correlation analysis was performed on 14 different plant traits including roots, shoot, and leaves (Table 1). All the root related traits like total root dry weight, deep root length, deep root volume, deep root surface area and deep root diameter were found to have a strong correlation (at P < 0.001) having correlation coefficient in the range of 0.725–0.820 with total water uptake. Leaf dry weight, total shoot dry weight, and stem dry weight were the other traits having a significant correlation with total water uptake towards positive side. The relationship among leaf drying (an indicator of water stress level) and root traits was observed to be significant but negative. A strong correlation was observed among all the root traits with each other with positive impact. Although plant height was not significantly associated with root traits but has a strong association with stem length that was positively correlated with all the root traits except deep root length. A negative but significant association between tillers per plant and leaf area was also recorded.
Table 1.
Matrix for Pearson Correlation coefficient (r) showing the simple linear relationship among various drought-related rice plant parameters
| PLHT | TILL | CL | LDW | SDW | TSDW | LFDG | TWU | LFA | TRDW | DRL | DRVOL | DRSA | DRD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PLHT | 1 | |||||||||||||
| TILL | −0.264ns | 1 | ||||||||||||
| CL | 0.567*** | −0.355* | 1 | |||||||||||
| LDW | 0.204ns | 0.511*** | −0.053ns | 1 | ||||||||||
| SDW | 0.286ns | 0.200ns | 0.441** | 0.590*** | 1 | |||||||||
| TSDW | 0.322* | 0.298* | 0.303* | 0.835*** | 0.929*** | 1 | ||||||||
| LFDG | −0.100ns | 0.206ns | −0.173ns | −0.265ns | −0.544*** | −0.503*** | 1 | |||||||
| TWU | 0.172ns | −0.195ns | 0.270ns | 0.441** | 0.638*** | 0.667*** | −0.708*** | 1 | ||||||
| LFA | 0.236ns | −0.367* | 0.258ns | 0.327* | 0.487*** | 0.521*** | −0.749*** | 0.803*** | 1 | |||||
| TRDW | 0.053ns | 0.041ns | 0.368* | 0.315* | 0.704*** | 0.626*** | −0.454** | 0.725*** | 0.424** | 1 | ||||
| DRL | 0.128ns | −0.096ns | 0.275ns | 0.413** | 0.471*** | 0.547*** | −0.488*** | 0.807*** | 0.587*** | 0.681*** | 1 | |||
| DRVOL | 0.202ns | −0.189ns | 0.435** | 0.284ns | 0.500*** | 0.516*** | −0.479*** | 0.810*** | 0.563*** | 0.73*** | 0.942*** | 1 | ||
| DRSA | 0.168ns | −0.140ns | 0.353* | 0.361** | 0.494*** | 0.543*** | −0.491*** | 0.820*** | 0.587*** | 0.717*** | 0.989*** | 0.981*** | 1 | |
| DRD | 0.163ns | −0.114ns | 0.429** | 0.298* | 0.482*** | 0.508*** | −0.410** | 0.769*** | 0.470*** | 0.696*** | 0.885*** | 0.952*** | 0.925*** | 1 |
Abbreviations: PLHT = Plant height; TILL = Tillers per plant; CL = Culm length; LDW = Leaf dry weight; SDW = Stem dry weight; TSDW = Total shoot dry weight; LFDG = Leaf drying; TWU = Total water uptake; LFA = Leaf area; TRDW = Total root dry weight; DRL = Deep root length (Below 60 cm); DRVOL = Deep root volume (Below 60 cm); DRSA = Deep root surface area (Below 60 cm); DRAD = Deep root average diameter (Below 60 cm).
Significance at P < 0.05;
Significance at P < 0.01;
Significance at P < 0.001.
Genetic linkage mapping and QTL analysis
Two hundred thirty SSR markers were used to identify the polymorphism between the two parents. Seventy-three microsatellites (31.7%) scattered on 1735 cM distance on all 12 linkage groups (chromosomes) were found polymorphic (co-dominant).
The identified polymorphic SSR markers were then applied on 46 F2 individuals of the two extreme classes (23 plants from drought-tolerant extreme and 23 plants from drought-sensitive extreme) in the population. All the 73 polymorphic markers displayed clear banding pattern. A molecular map was constructed with these microsatellite mapping data to position the markers relative to each other on the map using Map Manager QTX.
The total size of the linkage map was 1735 cM and the average interval size was 23.7 cM. Few gaps with large distances were persisted mainly on chromosomes 5, 6, 8 and 9. The analysis was performed with microsatellite mapping data using WinQTL Cartographer 2.5 V (Wang et al. 2001) to generate the genetic linkage map.
QTLs identified for different plant traits
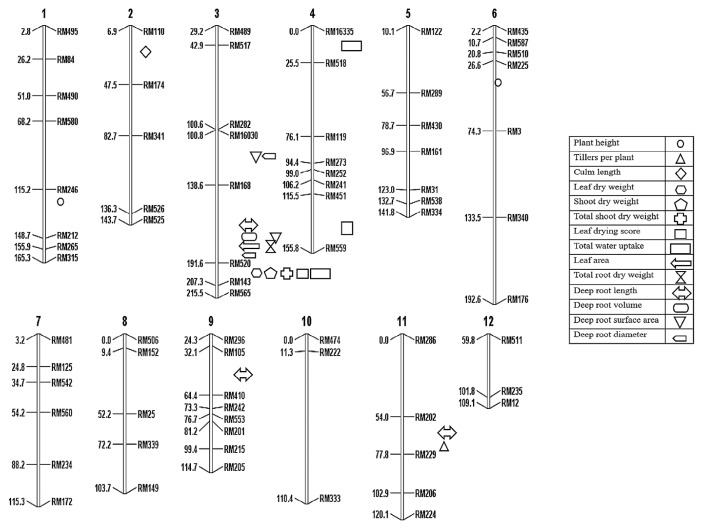
Composite interval mapping (CIM) was employed using the F2 population to locate the more accurate association and precise position of putative QTLs. A total of 21 QTLs were detected using CIM. The QTLs relating to tolerance to water stress conditions at vegetative stage were found on, chromosomes ch1 (qHt1), ch2 (qCmL2), ch3 (qLDW3, qSDW3, qTSDW3, qLDW3, qTWU3, qLA3, qTRDW3, qDRL3, qDRV3, qDRSA3, qDRD3, qDRSA3, qDRD3), ch4 (qTWU4), ch6 (qHt6), ch9 (qDRL9) and ch11 (qTill11, qDRL11). A summary of the statistics of all significant QTLs (LOD ≥3.0) are presented in Table 2 and are located on chromosomes in Fig. 3.
Table 2.
QTLs detected for water stress tolerance related traits based on composite interval mapping in selected extreme plants of the F2 population derived from the cross Super Basmati/IR55419-04
| Trait name | QTL name | Position | Ch. number | Marker interval | LOD value | R2 (%) | Additive effect |
|---|---|---|---|---|---|---|---|
| Plant height | qHt1 | 164.2 | 1 | RM212-RM265 | 3.6 | 19.5 | −5.6 |
| qHt6 | 66.01 | 6 | RM587-RM510 | 5.3 | 25.0 | −7.6 | |
| Tillers per plant | qTill11 | 69.2 | 11 | RM202-RM229 | 4.0 | 19.5 | 3.1 |
| Culm length | qCmL2 | 34.0 | 2 | RM110-RM174 | 4.7 | 29.5 | −0.3 |
| Leaf dry weight | qLDW3 | 246.3 | 3 | RM520-RM143 | 3.8 | 34.0 | 0.4 |
| Stem dry weight | qSDW3 | 244.3 | 3 | RM520-RM143 | 10.7 | 7.5 | −2.4 |
| Total shoot dry weight | qTSDW3 | 238.3 | 3 | RM520-RM143 | 11.7 | 6.9 | −4.7 |
| Leaf drying score | qLDS3 | 234.3 | 3 | RM520-RM143 | 5.3 | 4.5 | 0.7 |
| qLDS4 | 109.5 | 4 | RM451-RM559 | 4.8 | 12.5 | 2.2 | |
| Total water uptake | qTWU3 | 238.3 | 3 | RM520-RM143 | 27.0 | 9.96 | −299.150 |
| qTWU4 | 4.4 | 4 | RM119-RM518 | 5.6 | 2.45 | −236.494 | |
| Leaf area | qLA3 | 234.3 | 3 | RM168-RM520 | 9.7 | 17.12 | 207.195 |
| Total root dry weight | qTRDW3 | 236.3 | 3 | RM168-RM520 | 8.3 | 20.901 | −0.778 |
| Deep root length | qDRL3 | 226.3 | 3 | RM168-RM520 | 7.2 | 29.51 | −0.010 |
| qDRL9 | 50.0 | 9 | RM105-RM410 | 3.5 | 9.114 | −0.080 | |
| qDRL11 | 56.0 | 11 | RM202-RM229 | 3.3 | 3.857 | −0.100 | |
| Deep root volume | qDRV3 | 232.3 | 3 | RM168-RM520 | 6.7 | 25.784 | −1.829 |
| Deep root surface area | qDRSA3.1 | 136.7 | 3 | RM16030-RM168 | 4.1 | 13.624 | 48.625 |
| qDRSA3.2 | 228.3 | 3 | RM168-RM520 | 7.7 | 32.096 | −220.250 | |
| Deep root diameter | qDRD3.1 | 136.8 | 3 | RM16030-RM168 | 4.7 | 18.346 | 0.082 |
| qDRD3.2 | 242.3 | 3 | RM168-RM520 | 5.2 | 25.430 | −0.292 |
Fig. 3.
A genetic linkage map of rice showing detected QTLs on different chromosomes. Chromosome number is at the top, the map distances in centi Morgan (cM) at the left side and SSR markers names at the right side of each linkage group.
Plant height
Two QTLs qHt1 and qHt6 were identified for plant height that explains 19.5%, and 25% of the phenotypic variance (R2), respectively. QTL qHt1 on chromosome 1 (LOD value 3.6) was localized at interval RM212-RM265 (peak marker RM212) and IR55419-04 allele contributed in the phenotypic performance. Another QTL qHt6.2 was present on chromosome 6 at marker interval RM587-RM510 (66.01 cM) showed the phenotypic effect on plant height is coming from IR55419-04 alleles. The LOD value of this QTL was 5.3.
Number of tillers per plant
Only one region was identified with a significant LOD score of 4.0 for the number of tillers per plant on chromosome 11. For this QTL, Super Basmati alleles had a positive effect on tillering ability and explained 19.5% of the phenotypic variation. The QTL qTill11 for the number of tillers per plant was located between RM202-RM229 marker interval sited at 69.2 cM on chromosome 11.
Culm length
A QTL was spotted on chromosome 2 that was significantly associated with culm length with LOD values 4.7. IR55419-04 alleles contributed (29.5%) for culm length at chromosome 2. The QTL qCmL2 was placed between RM110-RM174.
Leaf dry weight
A region was identified on chromosome 3 with LOD scores 3.8 for leaf dry weight. QTL qLDW3 (Super Basmati allele) had a positive effect and explained 34% of the phenotypic variance for leaf dry weight. The QTL was co-localized in the marker interval RM520-RM143, on the chromosome 3 (246.3 cM).
Stem dry weight
On chromosome 3, one QTL qSDW3 was associated with stem dry weight with a significant LOD score of 10.7. The marker locus RM520-RM143 was found responsible for this QTL that explained about 7.5% of the phenotypic variation.
Total shoot dry weight
One QTL was located on chromosome 3 that was significantly associated with the total shoot dry weight having LOD value 11.7. IR55419-04 allele contributed an additive effect of 4.7 for this QTL with the phenotypic variance of 6.9% and was located between RM520-RM143.
Leaf drying score
Two genomic regions were identified on chromosomes 3 and 4 with LOD scores 5.3 and 4.8 at marker intervals RM520-RM143 and RM451-RM559 for leaf drying score respectively. For the QTL qLDS3, Super Basmati alleles had a positive effect with phenotypic variances 4.5% and 12.5% for leaf drying scores on chromosomes 3 and 4, respectively.
Total water uptake
For total water uptake, one genomic region in the marker interval RM520-RM143 was detected with a significant QTL qTWU3 on the long arm of the chromosome 3 with LOD score 27.0 that explained about 9.96% of the phenotypic variation. Another locus between RM119-RM518 was identified on chromosome 4 that shared about 2.45% in phenotypic variance. The additive effect of both the QTLs was due to the alleles of the IR55419-04 with the values 299.15 and 236.49, respectively.
Leaf area
One QTL qLA3 was located on chromosome 3 that was significantly associated with leaf area with LOD value 9.7. Super Basmati alleles contributed (207.2) for the QTLs with the phenotypic variance of 17.12% and were located between RM168-RM520.
Total root dry weight
One significant QTL was detected through composite interval mapping on the long arm of chromosome 3 between RM168-RM520 for total root dry weight, with the positive alleles contributed by IR55419-04. This QTL accounted for about 20.1% of the total variation in total root dry weight.
Deep root length
Three genomic regions co-localized in marker intervals RM168-RM520, RM105-RM410 and RM202-RM229 with LOD values of 7.2, 3.5 and 3.3 respectively were identified on chromosomes 3, 9 and 11, respectively. All the alleles of IR55419-04 contributed positively explaining 29.5%, 9.1% and 3.86% of the total phenotypic variation for deep root length.
Deep root volume
On chromosome 3, one QTL qDRV3 was associated with deep root volume with significant LOD score of 6.7. The marker locus RM168-RM520 was found responsible for this QTL that explained about 25.78% of the phenotypic variation.
Deep root surface area
The two regions were identified on chromosome 3 with LOD scores 4.1 and 7.7 at marker intervals RM16030-RM168 and RM168-RM520 for deep root surface area, respectively. For the QTL qDRSA3.1, Super Basmati allele had a positive effect with phenotypic variance 13.62%, whereas IR55419-04 was the contributor of the alleles for the QTL qDRSA3.2 with the phenotypic variance of 32.096% on the same chromosome 3.
Deep root diameter
Two significant QTLs were detected on the long arm of chromosome 3 between RM16030-RM168 and RM168-RM520 for deep root diameter with the positive alleles contributed by Super Basmati and IR55419-04, respectively. These QTLs accounted for about 18.35% and 25.43% of the total variation in deep root diameter, respectively.
Discussion
Climate shift is becoming one of the major concern for sustainable agriculture production including rice. To cope with climatic shift, rice breeders have increased their efforts to breed new rice varieties by utilizing currently applied technologies such as marker assisted breeding along with conventional methods. Marker assisted breeding is dependent upon DNA markers tightly linked with identified QTLs for traits of interest such as drought in the climate change scenario. Bi-parental QTL mapping resulted in identification of many QTLs in rice, but to explore potential drought tolerant QTLs is a dire need due to diverse characteristics of Basmati rice.
Physiological and agronomic traits analysis
Screening for drought tolerance targeting the root traits is laborious, complicated and not easy under field conditions due to logistical and methodological issues of root study. Although, hydroponic studies of root phenotyping (Price and Tomos 1997, Steudle 2000, Topp et al. 2013) are easy but the results could not be translated successfully in breeding varieties because most of these identified QTLs were detected in quite different environment as experienced in field. That’s why we used PVC pipes of bigger diameter with bulk density closer to the field by compacting the soil while filling to make the soil of root zone in accordance with field conditions so that the findings can be translated in improving the drought tolerance.
Drought stress screening of parents and F2 for physiological and agronomic traits showed that these traits are highly correlated to each other at significant levels of 0.001 and 0.05 (Table 1). Marri et al. (2005) reported that plant height has a significant negative correlation with number of tillers, which are in harmony with the finding of current study. The possible reason could be the utilization of maximum energy produce by existing dividing tissue rather than to initiate new division in the form of new tillers. Our results depicting negative but strong association of leaf drying with total water uptake are in agreement with the findings of Courtois et al. (2000) and Yue et al. (2005). The three traits; deep root length, deep root volume and deep root surface area responded with each other in significantly positive manner as reported by Subashri et al. (2009). A positive association of total water uptake with roots related traits i.e., deeper root length, deep root volume and deep root surface area clearly showed that root system of the plant is certainly essential for extraction of water from deeper soil layers under drought stress conditions. Further, the presence of deep roots under water deficit environment were observed and was the response in search of moisture for maintaining turgor of the tissues (Gowda et al. 2011). Many other researchers also reported that under drought stress, genotypes modified root distribution and penetrate in the deeper soil to extract water for their survival and growth (Allah et al. 2010, Lobet et al. 2014, Puckridge and O’Toole 1980, Uga et al. 2008). In addition, a positive and highly significant relationship of shoot dry weight with all deep roots related traits strengthened the finding of Zhang et al. (2008). Shoot biomass is an indicator of ultimate grain yield as it translates into the emergence of panicles that bear grains. High shoot biomass under drought indicates the better growth due to higher plant water status maintained through increased water uptake by roots (Levitt 1980). The presence of positive inter relationships of deep root average diameter, deep root length and root dry weight was proved to be obvious under water limited environment as these results are in accordance with the earlier reports (Kano et al. 2011, Qu et al. 2008a, 2008b). Root architecture adaptations under water deficit conditions plays an important role in developing deeper roots in soil (Bernier et al. 2008). Li et al. (2005) reported that root diameter and deep root thickness can be used to improve drought tolerance in rice.
Genetic linkage mapping
Genetic map covering 1735 cM genomic region was constructed by 73 SSR markers with an average marker interval of 23.7 cM which is slightly more than the minimum required interval (20 cM) for QTL mapping (Lander and Botstein 1989). However, for selective genotyping more specific markers were used for mapping. Linkage maps of variable lengths of 1781.5, 1645.1 and 1650 cM by using 213 markers, 130 SSR markers and 9303 SNP markers were constructed by Solis et al. (2017), Gu et al. (2011) and De Leon et al. (2016) respectively with different marker spacings. These difference in linkage map length and spacing among studies was results of variation in genetic background of selected parents, marker types used, number of polymorphic markers found and recombination rate in the developed mapping population.
QTL analysis by selective genotyping
Selective genotyping is a highly cost-effective and more powerful way than simple interval mapping for identification of novel QTLs controlling the specific trait. There is one disadvantage of this technique is false positive QTLs due to insufficient marker density and small size of the population (Sun et al. 2010). Phansak et al. (2016) adapted selective genotyping approach in soybean F2 population for seed protein and oil contents QTLs detection and demonstrated that selective genotyping strategy was highly successful. Keeping in view the power and advantages of the technique, we adopted this approach and 21 QTLs were identified controlling morphological and physiological traits under study at vegetative stage by choosing 6% of population extremes based on total water uptake during stress period. According to Ayoub and Mather (2002) 5 to 10% population extremes were sufficient for selective genotyping.
In our study, two QTLs for plant height were identified under water stress condition, one on chromosome 1 and one on chromosome 6, however, the LOD value and phenotypic variance of IR55419-04 alleles for QTL qHt6 were higher than qHt1. The QTL qHt1 identified at the interval of RM212 and RM265 was also detected by Septiningsih et al. (2003) at RM315 locus near to identified RM265. Ye et al. (2012) has also reported QTLs controlling plant height located on chromosome 1. Two QTLs associated with plant height were detected by using SNP markers on chromosome 6 by McNally et al. (2009). The reduction in plant height has been observed as the impact of drought stress at vegetative stage resulting in relative yield penalty (Ahmadikhah and Marufinia 2016). One QTL qTill11 on chromosome 11 was identified for tiller number per plant at a marker interval of RM202-RM229 which was previously reported by Septiningsih et al. (2003) at the same position. The allele of Super Basmati contributed for this locus indicating that tillering ability of this line is either high or tillering was completed before the commencement of earlier than the stress period.
Only one QTL was detected under the severe stress condition (29 days stress) for total shoot dry weight at the lower tip of chromosome 3 at the marker position of RM520. Kamoshita et al. (2002) also found the shoot biomass QTL on the lower tip of chromosome 3 by using RFLP and AFLP markers.
Overall, two QTLs were detected controlling leaf drying under a severe drought condition on chromosomes 3 and 4 with LOD scores of 5.3 and 4.8, respectively. Leaf drying QTL on the chromosome 3 was found between the interval of marker RM520-RM143 with a peak at RM520. Similar QTL was reported by Yue et al. (2005) at the locus RM520. They also identified other QTLs for this trait on different chromosomes.
Three QTLs (qDRL3, qDRL9 and qDRL11) were identified for deep root length (below 60 cm) on chromosomes 3, 9 and 11 under stress condition. Qu et al. (2008a) reported deep root length QTL on the tip of chromosome 6 at the marker position of RM454 very near to RM520. The QTL affects deep root length was identified 10 cM distance at RM278 marker position apart from the detected QTL qDRL9 at marker interval RM105-RM410. Kamoshita et al. (2002) reported deep root length on chromosome 11 at RM167 marker position near to identified QTL qDRL11 at an interval of RM202-RM229. Yue et al. (2005) also reported similar QTL on chromosome 11 at the same marker position of RM 229.
One QTL for deep root volume (below 60 cm) was identified on chromosome 3 of the selected extremes lines based on the total water uptake. In contrast, root volume QTL was identified by Qu et al. (2008a) using SSR markers at chromosome 4. Yue et al. (2005) also reported deep root volume under drought QTL on chromosome 4 at RM470 marker position at the similar position of RM451.
Maximum number of QTLs were found at RM520 marker locus under the severe drought stress condition for leaf dry weight, stem dry weight, total shoot dry weight, leaf drying, total water uptake, leaf area, total root dry weight, deep root length, deep root volume, deep root surface area and root diameter. All these QTLs were not previously reported at this locus, however, this locus was identified for yield under drought in different previous studies (Yue et al. 2005).
In the net shell, composite interval mapping resulted in the identification of 21 QTLs. A major genomic region at RM520 was detected with a significant QTL (qTWU3.1) for total water uptake (LOD = 27.0) and some associated root morphological traits on chromosome 3. This major QTL can be used for MAS to develop new drought-tolerant rice varieties. The major water uptake QTL linked with RM520 needs this research push up for the identification of underlying genes and to explore the functional relationship between the QTLs and phenotypic trait.
Rice genotypes having deep roots and higher root diameter produce more aboveground biomass that helps in survival under water stress conditions. In earlier studies, root related traits have been suggested to be of high consideration due to the fact that plant extracts water and nutrition from the soil layers through roots. High root numbers, deep root length and deep root volume enhance physical strength, improved root system architecture, increased nutrients/soil moisture absorption from deeper layers, penetration ability, hydraulic conductivity, root system size and branching density thus minimizing the risk of deficient water status of the plant tissues and resulting in increased tolerance under drought through avoidance mechanism (Kanbar et al. 2009, Steele et al. 2007, Venuprasad et al. 2002). This study had shown that total water uptake and root-related traits are of high importance under water stress environment and their identified QTLs must be given a real focus for the improvement of drought tolerance in future rice varieties. Recycling of drought and floods in a shorter period of time is the outcome of prevailing climate change scenario. Thus, developing water stress tolerant plant varieties is a big challenge for the breeders to meet the higher food demand and ensure food security to human beings. Our findings will be helpful to solve this situation.
Supplementary Information
Acknowledgments
We thank Higher Education Commission, Pakistan for providing financial support. We also acknowledge the technical support, assistance and guidance during this research work by Dr. Rachid Serraj, Senior Scientist, Crop and Environmental Sciences Division, IRRI, Philippines. Valuable suggestions of Dr. Henry Amelia, the leader of Drought Physiology Group at IRRI, Philippines are also acknowledged.
Literature Cited
- Ahmadikhah, A. and Marufinia, A. (2016) Effect of reduced plant height on drought tolerance in rice. 3 Biotech 6: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allah, A., Badawy, S.A., Zayed, B. and El-Gohary, A. (2010) The role of root system traits in the drought tolerance of rice (Oryza sativa L.). World Acad. Sci. Eng. Technol. 68: 1378–1382. [Google Scholar]
- Ayoub, M. and Mather, D. (2002) Effectiveness of selective genotyping for detection of quantitative trait loci: an analysis of grain and malt quality traits in three barley populations. Genome 45: 1116–1124. [DOI] [PubMed] [Google Scholar]
- Bernier, J., Atlin, G.N., Serraj, R., Kumar, A. and Spaner, D. (2008) Breeding upland rice for drought resistance. J. Sci. Food Agric. 88: 927–939. [Google Scholar]
- Bernier, J., Serraj, R., Kumar, A., Venuprasad, R., Impa, S., Veeresh Gowda, R.P., Oane, R., Spaner, D. and Atlin, G. (2009) The large-effect drought-resistance QTL qtl12.1 increases water uptake in upland rice. Field Crops Res. 110: 139–146. [Google Scholar]
- Courtois, B., McLaren, G., Sinha, P., Prasad, K., Yadav, R. and Shen, L. (2000) Mapping QTLs associated with drought avoidance in upland rice. Mol. Breed. 6: 55–66. [Google Scholar]
- De Leon, T.B., Linscombe, S. and Subudhi, P.K. (2016) Molecular dissection of seedling salinity tolerance in rice (Oryza sativa L.) using a high-density GBS-based SNP linkage map. Rice (N Y) 9: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit, S., Singh, A., Cruz, M.T.S., Maturan, P.T., Amante, M. and Kumar, A. (2014) Multiple major QTL lead to stable yield performance of rice cultivars across varying drought intensities. BMC Genet. 15: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda, V.R., Henry, A., Yamauchi, A., Shashidhar, H. and Serraj, R. (2011) Root biology and genetic improvement for drought avoidance in rice. Field Crops Res. 122: 1–13. [Google Scholar]
- Gu, J., Yin, X., Struik, P.C., Stomph, T.J. and Wang, H. (2011) Using chromosome introgression lines to map quantitative trait loci for photosynthesis parameters in rice (Oryza sativa L.) leaves under drought and well-watered field conditions. J. Exp. Bot. 63: 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Y., Serraj, R., Liu, S., Xu, J., Ali, J., Wang, W., Venus, E., Zhu, L. and Li, Z. (2010) Simultaneously improving yield under drought stress and non-stress conditions: a case study of rice (Oryza sativa L.). J. Exp. Bot. 61: 4145–4156. [DOI] [PubMed] [Google Scholar]
- Henry, A., Mallikarjuna Swamy, B.P., Dixit, S., Torres, R.D., Batoto, T.C., Manalili, M., Anantha, M., Mandal, N. and Kumar, A. (2015) Physiological mechanisms contributing to the QTL-combination effects on improved performance of IR64 rice NILs under drought. J. Exp. Bot. 66: 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongdee, B., Fukai, S. and Cooper, M. (2002) Leaf water potential and osmotic adjustment as physiological traits to improve drought tolerance in rice. Field Crops Res. 76: 153–163. [Google Scholar]
- Kamoshita, A., Wade, L., Ali, M., Pathan, M., Zhang, J., Sarkarung, S. and Nguyen, H. (2002) Mapping QTLs for root morphology of a rice population adapted to rainfed lowland conditions. Theor. Appl. Genet. 104: 880–893. [DOI] [PubMed] [Google Scholar]
- Kanbar, A., Toorchi, M. and Shashidhar, H. (2009) Relationship between root and yield morphological characters in rainfed low land rice (Oryza sativa L.). Cereal Res. Commun. 37: 261–268. [Google Scholar]
- Kano, M., Inukai, Y., Kitano, H. and Yamauchi, A. (2011) Root plasticity as the key root trait for adaptation to various intensities of drought stress in rice. Plant Soil 342: 117–128. [Google Scholar]
- Lander, E.S. and Botstein, D. (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt, J. (1980) Responses of plants to environmental stresses, Vol. 2 Academic Press, New York. [Google Scholar]
- Li, J., Han, Y., Liu, L., Chen, Y., Du, Y., Zhang, J., Sun, H. and Zhao, Q. (2015) qRT9, a quantitative trait locus controlling root thickness and root length in upland rice. J. Exp. Bot. 66: 2723–2732. [DOI] [PubMed] [Google Scholar]
- Li, X., Guo, Z., Lv, Y., Cen, X., Ding, X., Wu, H., Li, X., Huang, J. and Xiong, L. (2017) Genetic control of the root system in rice under normal and drought stress conditions by genome-wide association study. PLoS Genet. 13: e1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Mu, P., Li, C., Zhang, H., Li, Z., Gao, Y. and Wang, X. (2005) QTL mapping of root traits in a doubled haploid population from a cross between upland and lowland japonica rice in three environments. Theor. Appl. Genet. 110: 1244–1252. [DOI] [PubMed] [Google Scholar]
- Liu, H., Zou, G., Liu, G., Hu, S., Li, M., Yu, X., Mei, H. and Luo, L. (2005) Correlation analysis and QTL identification for canopy temperature, leaf water potential and spikelet fertility in rice under contrasting moisture regimes. Chin. Sci. Bull. 50: 317–326. [Google Scholar]
- Lobet, G., Couvreur, V., Meunier, F., Javaux, M. and Draye, X. (2014) Plant water uptake in drying soils. Plant Physiol. 164: 1619–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri, P.R., Sarla, N., Reddy, L.V. and Siddiq, E. (2005) Identification and mapping of yield and yield related QTLs from an Indian accession of Oryza rufipogon. BMC Genet. 6: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, K.L., Childs, K.L., Bohnert, R., Davidson, R.M., Zhao, K., Ulat, V.J., Zeller, G., Clark, R.M., Hoen, D.R., Bureau, T.E.et al. (2009) Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc. Natl. Acad. Sci. USA 106: 12273–12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraju, J., Kathirvel, M., Kumar, R.R., Siddiq, E. and Hasnain, S.E. (2002) Genetic analysis of traditional and evolved Basmati and non-Basmati rice varieties by using fluorescence-based ISSR-PCR and SSR markers. Proc. Natl. Acad. Sci. USA 99: 5836–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phansak, P., Soonsuwon, W., Hyten, D.L., Song, Q., Cregan, P.B., Graef, G.L. and Specht, J.E. (2016) Multi-population selective genotyping to identify soybean (Glycine max (L.) Merr.) seed protein and oil QTLs. G3 (Bethesda) 6: 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlman, J. and Sleper, D. (1995) Methods in plant breeding. Breeding Field Crops: 172–174. [Google Scholar]
- Price, A.H. and Tomos, A. (1997) Genetic dissection of root growth in rice (Oryza sativa L.). II: mapping quantitative trait loci using molecular markers. Theor. Appl. Genet. 95: 143–152. [Google Scholar]
- Puckridge, D. and O’Toole, J. (1980) Dry matter and grain production of rice, using a line source sprinkler in drought studies. Field Crops Res. 3: 303–319. [Google Scholar]
- Qu, Y., Mu, P., Zhang, H., Chen, C.Y., Gao, Y., Tian, Y., Wen, F. and Li, Z. (2008a) Mapping QTLs of root morphological traits at different growth stages in rice. Genetica 133: 187–200. [DOI] [PubMed] [Google Scholar]
- Qu, Y.Y., Mu, P., Li, X.Q., Tian, Y.X., Wen, F., Zhang, H.L. and Li, Z.C. (2008b) QTL mapping and correlations between leaf water potential and drought resistance in rice under upland and lowland environments. Acta Agron. Sci. 34: 198–206. [Google Scholar]
- Sabar, M. and Arif, M. (2014) Phenotypic response of rice (Oryza sativa) genotypes to variable moisture stress regimes. Int. J. Agric. Biol. 16: 32–40. [Google Scholar]
- Sandhu, N., Singh, A., Dixit, S., Cruz, M.T.S., Maturan, P.C., Jain, R.K. and Kumar, A. (2014) Identification and mapping of stable QTL with main and epistasis effect on rice grain yield under upland drought stress. BMC Genet. 15: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Septiningsih, E., Prasetiyono, J., Lubis, E., Tai, T., Tjubaryat, T., Moeljopawiro, S. and McCouch, S. (2003) Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor. Appl. Genet. 107: 1419–1432. [DOI] [PubMed] [Google Scholar]
- Shamsudin, N.A.A., Swamy, B.M., Ratnam, W., Cruz, M.T.S., Sandhu, N., Raman, A.K. and Kumar, A. (2016) Pyramiding of drought yield QTLs into a high quality Malaysian rice cultivar MRQ74 improves yield under reproductive stage drought. Rice (N Y) 9: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R., Singh, Y., Xalaxo, S., Verulkar, S., Yadav, N., Singh, S., Singh, N., Prasad, K., Kondayya, K., Rao, P.R.et al. (2016) From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci. 242: 278–287. [DOI] [PubMed] [Google Scholar]
- Solis, J., Gutierrez, A., Mangu, V., Sanchez, E., Bedre, R., Linscombe, S. and Baisakh, N. (2017) Genetic mapping of quantitative trait loci for grain yield under drought in rice under controlled greenhouse conditions. Front. Chem. 5: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele, K., Virk, D.S., Kumar, R., Prasad, S.C. and Witcombe, J.R. (2007) Field evaluation of upland rice lines selected for QTLs controlling root traits. Field Crop Res. 101: 180–186. [Google Scholar]
- Steudle, E. (2000) Water uptake by roots: effects of water deficit. J. Exp. Bot. 51: 1531–1542. [DOI] [PubMed] [Google Scholar]
- Subashri, M., Robin, S., Vinod, K., Rajeswari, S., Mohanasundaram, K. and Raveendran, T. (2009) Trait identification and QTL validation for reproductive stage drought resistance in rice using selective genotyping of near flowering RILs. Euphytica 166: 291–305. [Google Scholar]
- Sun, Y., Wang, J., Crouch, J.H. and Xu, Y. (2010) Efficiency of selective genotyping for genetic analysis of complex traits and potential applications in crop improvement. Mol. Breed. 26: 493–511. [Google Scholar]
- Terra, T.G.R., Rodrigues, H.S., Rangel, P.H.N., Tomaz, R.S., Cruz, C.D. and Borém, A. (2016) QTLs identification for characteristics of the root system in upland rice through DNA microarray. Acta Sci. Agron. 38: 457–466. [Google Scholar]
- Topp, C.N., Iyer-Pascuzzi, A.S., Anderson, J.T., Lee, C.-R., Zurek, P.R., Symonova, O., Zheng, Y., Bucksch, A., Mileyko, Y., Galkovskyi, T.et al. (2013) 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc. Natl. Acad. Sci. USA 110: E1695–E1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga, Y., Okuno, K. and Yano, M. (2008) QTLs underlying natural variation in stele and xylem structures of rice root. Breed. Sci. 58: 7–14. [Google Scholar]
- Venuprasad, R., Shashidhar, H.E., Hittalmani, S. and Hemamalini, G.S. (2002) Tagging quantitative trait loci associated with grain yield and root morphological traits in rice (Oryza sativa L.) under contrasting moisture regimes. Euphytica 128: 293–300. [Google Scholar]
- Wang, S., Basten, C., Gaffney, P. and Zeng, Z. (2001) Windows QTL Cartographer version 2.5. North Carolina State University. Bioinformatics Research Center, Raleigh. [Google Scholar]
- Xiao, B., Huang, Y., Tang, N. and Xiong, L. (2007) Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 115: 35–46. [DOI] [PubMed] [Google Scholar]
- Xu, Q., Yuan, X.P., Yu, H.Y., Wang, Y.P., Tang, S.X. and Wei, X.H. (2011) Mapping QTLs for drought tolerance at seedling stage in rice using doubled haploid population. Rice Sci. 18: 23–28. [Google Scholar]
- Ye, C., Argayoso, M.A., Redoña, E.D., Sierra, S.N., Laza, M.A., Dilla, C.J., Mo, Y., Thomson, M.J., Chin, J., Delaviña, C.B.et al. (2012) Mapping QTL for heat tolerance at flowering stage in rice using SNP markers. Plant Breed. 131: 33–41. [Google Scholar]
- Yue, B., Xiong, L., Xue, W., Xing, Y., Luo, L. and Xu, C. (2005) Genetic analysis for drought resistance of rice at reproductive stage in field with different types of soil. Theor. Appl. Genet. 111: 1127–1136. [DOI] [PubMed] [Google Scholar]
- Zhai, L., Zheng, T., Wang, X., Wang, Y., Chen, K., Wang, S., Wang, Y., Xu, J. and Li, Z. (2018) QTL mapping and candidate gene analysis of peduncle vascular bundle related traits in rice by genome-wide association study. Rice (N Y) 11: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Li, H., Li, Z. and Wang, J. (2008) Interactions between markers can be caused by the dominance effect of quantitative trait loci. Genetics 180: 1177–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.