Abstract
GROWTH-REGULATING FACTORs (GRFs) are sequence-specific DNA-binding transcription factors that regulate various aspects of plant growth and development. GRF proteins interact with a transcription cofactor, GRF-INTERACTING FACTOR (GIF), to form a functional transcriptional complex. For its activities, the GRF-GIF duo requires the SWITCH2/SUCROSE NONFERMENTING2 chromatin remodeling complex. One of the most conspicuous roles of the duo is conferring the meristematic potential on the proliferative and formative cells during organogenesis. GRF expression is post-transcriptionally down-regulated by microRNA396 (miR396), thus constructing the GRF-GIF-miR396 module and fine-tuning the duo’s action. Since the last comprehensive review articles were published over three years ago, many studies have added further insight into its action and elucidated new biological roles. The current review highlights recent advances in our understanding of how the GRF-GIF-miR396 module regulates plant growth and development. In addition, I revise the previous view on the evolutionary origin of the GRF gene family.
Keywords: Evolution, GIF, GRF, miR396, Organ growth
INTRODUCTION
Transcription factors control gene expression and thus regulate the patterns of plant growth and development. The number of transcription factors in Arabidopsis thaliana (Arabidopsis hereafter) has been estimated to be more than 2,000, which is comparable to that in humans (1–4). Besides the large numbers, a significant portion of them are present only in plants (4, 5). One class of plant-specific transcription factors, GROWTH-REGULATING FACTOR (GRF), was first identified in rice and Arabidopsis (the notion of ‘plant-specific’ needs revision, as described in the last section below), and found to exist in multiple homologous copies: Arabidopsis and rice have nine and twelve members, respectively (6–8). Later, GRF proteins were found to interact with GRF-INTERACTING FACTORs (GIFs) in Arabidopsis, which form a small family of three members: AtGIF1 (aka ANGUSTIFOLIA3, AN3), AtGIF2, and AtGIF3 (9, 10). Since then, many studies have demonstrated that GRFs and GIFs are bona fide interacting partner proteins that form a functional unit, and that the GRF-GIF complex plays essential roles in various aspects of plant growth and development (for review, see 11–13). It has also been well documented that microRNA396 (miR396) post-transcriptionally regulates the majority of GRF members and fine-tunes their expression, thus controlling GRF-GIF-dependent processes (14, 15).
It has been more than three years since the last comprehensive review articles were published on the GRF-GIF-miR396 module (11–13). During that time, many reports have been published, elucidating its new biological roles and identifying its downstream and upstream genes as well as target cis-elements. The current review highlights recent studies that have increased the understanding of the regulatory module. It also revises the previous view on the evolutionary origin of the GRF gene family.
WHAT ARE GRF AND GIF?
GRF proteins contain two highly conserved QLQ and WRC domains in the N-terminal half (6–8). The QLQ domain consists of highly conserved Gln-Leu-Gln (QX3LX2Q) and neighboring residues. The QLQ domain provides an interface for interacting with GIFs (9, 10). The WRC domain consists of the conserved spacing of three Cys and one His residues (CX9CX10CX2H, simply the C3H motif), which acts as a DNA-binding domain (DBD) (6, 7, 16–18). The C-terminal regions of GRF proteins are highly variable in length and composition of amino acid residues, and they function as a transactivation domain (7–10). AtGRFs with truncated C-termini have been shown to lose their transactivation activities, while OsGRF10 (rice) and ZmGRF10 (maize) with short C-termini have also exhibited no activities (9, 19, 20).
GIFs were identified by their capability interacting with GRFs and characterized as transcription cofactors with no DBD (9, 10). The interacting partnership between almost all members of the two protein families has been demonstrated in all plants tested (19–25). GIF proteins have the highly conserved SNH domain in the N-terminus that directly interacts with the GRF QLQ domain. The C-terminal regions of GIFs exert transactivation activities and are rich in Gln (Q) and Gly (G), and are thus called the QG domain. GIF genes are more ancient in terms of evolutionary origin than GRFs, and they exist in major lineages of eukaryotes, including humans, in which they are called SYT (synovial translocation protein), aka SS18 (synovial sarcoma associated protein) (26, 27).
POTENTIAL CIS-ELEMENTS BOUND BY GRF AND GIF
As transcription factors with the WRC DBD, GRFs are expected to regulate the expression of downstream target genes and bind to specific regulatory cis-elements in them. A GRF-targeting cis-element (GTE), TGTCAGG, was first identified in the promoter of DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) in Arabidopsis: AtGRF7 bound to the GTE and led to the repression of DREB2A expression (Table 1) (17). AtGRF9 bound to the promoter region of a bZIP transcription factor gene, OBP3-RESPONSIVE GENE3 (ORG3), whose promoter contains a potential GTE, CTGACA (28); rice GRFs (OsGRF6 and/or OsGRF10), to a GTE (TGTGTTG) of OsJMJ706 (a JMJD2 family jmjC gene) and OsCR4 (a gene for crinkly4 receptor-like kinase), upregulating their expression (19); OsGRF6, to CGSMR in the promoters of AUXIN RESPONSE FACTOR2 (ARF2), ARF7, and an YUCCA-like gene, whose expression is up-regulated by OsGRF6 overexpression (29).
Table 1.
Potential cis-elements bound by GRFs and AtGIF1/AN3
| Proteins | cis-elements | Target genes | Transcriptional regulation |
|---|---|---|---|
| AtGRF7 | TGTCAGGa | DREB2A | −b |
| AtGRF9 | CTGACA | ORG3 | + |
| OsGRF6 | TGTGTTG | OsJMJ706 | + |
| OsGRF9 | OsCR4 | + | |
| OsGRF6 | CGSMRc | ARF2 | + |
| ARF7 | + | ||
| YUCCA-like | + | ||
| AtGIF1/AN3 | CACGTG | COL5 | + |
| GAGAGAGA | COL5 | + | |
| HEC1 | + | ||
| TGTCAGA | PLT1 | − |
Nucleotide sequences read from the 5′ to 3′ direction.
Minus and plus symbolize up- and down-regulation of target gene expression, respectively.
S indicates G and C; M, A and C; R, A and G.
Aside from GTEs, chromatin immunoprecipitation assays (ChIP) revealed that AtGIF1/AN3 proteins were strongly associated with the G-box and GAGA elements in the Arabidopsis genome, and that these elements were found to reside in the promoters of some target genes, including CONSTANS-LIKE5 (COL5) and HECATE1 (HEC1; Table 1) (23). Another ChIP assay using AtGIF1/AN3 as bait revealed the strong enrichment of a promoter region of PLETHORA1 (PLT1), which contains a cis-element, TGTCAGA (30). Since AtGIF1/AN3 lacks a DBD, its association with cis-elements is made possible when it works in concert with transcription factors with DBDs, such as GRFs. Therefore, the high similarity between the elements found in DREB2A and PLT1, which were associated with AtGRF7 and AtGIF1/AN3, respectively, may not be the result of a coincidence. Nevertheless, the systematic inference and experimental verification of a canonical or consensus GTE still seems to be premature, or GTEs may be variable depending on the classes of GRFs and/or plant species. Indeed, the first GTE found in DREB2A was associated only with AtGRF7, and not with any other AtGRF members (17).
MOLECULAR FUNCTION OF THE GRF-GIF DUO IN THE TRANSCRIPTIONAL REGULATION
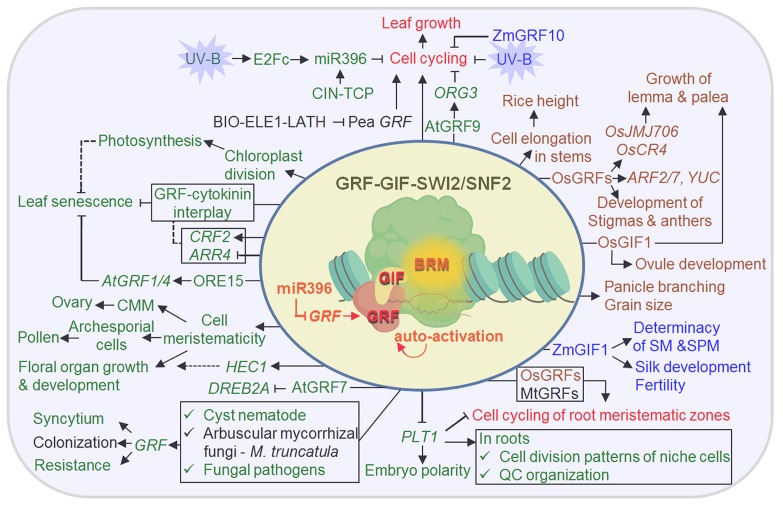
As mentioned above, ChIP assays revealed that AtGIF1/AN3 and maize GIF1/AN3 (ZmGIF1/ZmAN3) proteins were associated with the promoter regions of certain target genes (23, 25). The assays also showed that these associations were not limited to those known target genes, but detected widely over the whole genome of Arabidopsis, suggesting that AtGIF1/AN3 may be a key transcription cofactor acting together with GRFs and/or other transcription factors. Consistently with the notion, a series of tandem affinity purification (TAP) and co-immunoprecipitation (co-IP) approaches revealed that GIF1/AN3 proteins of Arabidopsis and maize were co-purified with the components of SWI2/SNF2 chromatin-remodeling complexes, including the core SWI2/SNF2 chromatin-remodeling ATPases, such as BRAHMA (BRM) and SPLAYED (SYD) (21, 23, 24). Upregulation of AtGIF1/AN3 target genes also required intact activities of BRM. These results give rise to the notion that GIF1/AN3 transcription cofactors may recruit both SWI2/SNF2 complexes and GRFs to GTEs, thus activating or repressing target genes (Fig. 1).
Fig. 1.
Schematic summary of molecular and biological functions of the GRF-GIF duo. The core and common molecular features of the duo are depicted in the circle, in which the GRF-GIF duo associated with the SWI2/SNF2 complex performs transcriptional regulation of target genes, including its own (auto-activation), and miR396 post-transcriptionally represses GRF expression. The biological functions common to eudicots and monocots are shown in red, i.e., the promotion of leaf growth via the regulation of cell cycling and promotion of cell cycling in root meristematic zones; the biological functions confirmed in Arabidopsis and other eudicots are shown in green and black, respectively; the biological functions validated in rice and maize are shown in brown and blue, respectively. The solid arrows and block bars indicate experimentally verified promotive and inhibitory actions, respectively, while the dotted ones indicate speculated possibilities.
The results and notion are consistent with the fact that the human GIF homolog, SYT, directly interacts with the human BRM and its homolog (31, 32), as well as the fact that TAP experiments using the human SYT as bait also retrieved the components of human SWI2/SNF2 chromatin-remodeling complexes (33). The result suggests that the interaction between GIF1/AN3 and the SWI2/SNF2 complex may be mediated via direct interaction between GIF1/AN3 and plant BRM homologs, and that the interaction between GIF1/AN3 and the SWI2/SNF2 complex is not only evolutionarily conserved in metazoans and plants, but also essential for transcriptional regulation, despite the fact that metazoans lack GRFs.
Some additional interesting features of the GRF-GIF action are that they activate their own transcription through a positive feedback loop in Arabidopsis, rice, and maize, likely by forming the GRF-GIF-SWI2/SNF2 complex at the promoter sites of target GRFs and GIFs (Fig. 1; for detailed information, see 11, 25); and that Arabidopsis and rice GIF1 proteins translocate between different cell layers through plasmodesmata, thus coordinating the cell proliferation activities of different cell layers (34, 35).
REGULATORY ROLES OF THE GRF-GIF-MIR396 MODULE IN LEAF GROWTH
Roles in cell proliferation of dicot leaves
GRF and GIF genes are highly expressed in almost all meristematic tissues, including leaf and floral organ primordia. Loss-of-function mutants of AtGRFs and AtGIFs had small and narrow leaves and petals, whereas their overexpressors developed larger ones (7, 9, 10, 36, 37). Determination of cellular parameters elucidated that AtGRFs and AtGIFs are positive regulators of cell proliferation in leaf primordia, providing cells with a meristematic potential or meristematicity.
In Arabidopsis, miR396 species, products of AtMIR396a and AtMIR396b genes, target and induce the cleavage of AtGRF mRNA species with the exceptions of AtGRF5 and AtGRF6 mRNAs, which lack the miR396 target site (14, 15). Therefore, the overexpression of AtMIR396 (35S:AtMIR396) resulted in post-transcriptional down-regulation of target AtGRFs, resulting in small and narrow leaves along with a reduced number of cells. By contrast, the expression of At-rGRFs, which were manipulated to be resistant to miR396 by altering their target sites, induced an enhancement of cell proliferation, consequently resulting in large leaves (15, 21, 22, 38).
The function of the GRF-miR396 module holds up for other eudicot plants as well. The overexpression of Arabidopsis and Populus trichocarpa MIR396s in tobacco plants has been shown to result in small and narrow leaves (38–40). Arabidopsis or Brassica napus plants overexpressing B. napus and B. rapa GRFs have also shown the development of enlarged leaves with more cells, along with increased expression of a set of cell cycle genes (41–43).
It has been shown that the AtGIF family controls both the rate and duration of cell division (37, 44). An increase in the AtGIF1/AN3 activity not only enhanced, but also prolonged the expression of a marker gene for the G-to-M transition in cell cycle, CYCB1;1, in the leaf primordium (23), whereas decreases in GIF activities were accompanied by reductions in the expressions of cell cycle-related genes (37). Similarly, the overexpression of AtMIR396 (35S:AtMIR396) and At-rGRF3 resulted in reductions and increases in CYCB1;1 expression (15, 21), respectively, indicating that the GRF-GIF duo plays a critical role in regulating cell cycle activities, consequently maintaining and supplying meristematic cells for cell proliferation in leaf primordia. Further study is needed as to what signaling pathway downstream of the GRF-GIF duo leads to the regulation of cell proliferation-mediating genes, including cell cycle-related genes.
The movement of the cell cycle arrest front (AF) from the distal to proximal regions of Arabidopsis leaf primordia during the early stages of leaf growth has been well documented (45). 35S:AtMIR396 induced the precocious AF movement toward the leaf proximal region, reducing cell proliferation activities and accelerating cell expansion (an indication of differentiation in plants) in the distal region behind AF, whereas an enhancement of the AtGRF5 activity exerted the opposite effects on those cellular processes, stimulating leaf growth (15, 46). Similarly, enhanced activities of AtGIF1/AN3 delayed the AF movement (23), and the distribution patterns of AtGIF expression were consistent with the AF movement (10, 47, 48), suggesting that the GRF-GIF-miR396 module is a crucial regulator of the AF movement. Gupta and Nath analyzed various types of leaf growth polarity present in 75 eudicot species, including the distal-proximal type of Arabidopsis, and found that the patterns of leaf growth polarity are tightly coupled with the abundance patterns of miR396 species and GRF mRNAs: regions of active cell proliferation are positively correlated with abundance of GRF mRNAs, whereas regions of cessation of cell proliferation and commencement of cell differentiation are positively correlated with miR396 abundance (49). The results suggest that, in eudicots, the patterns of cell proliferation and differentiation are controlled by the GRF-GIF-miR396 module.
A pea transcriptional complex consisting of BIGGER ORGANS (BIO) and ELEPHANT EAR LIKE LEAF1 (ELE1) negatively regulates leaf growth and interacts with a WUSCHEL-related transcription factor, LATHYDROIDES (LATH) (50). LATH has been shown to directly bind to a promoter region of a pea GRF, indicating that the negative regulator complex of leaf growth exerts it function through the repression of GRF expression. Arabidopsis PEAPOD (PPD) genes, orthologs of pea ELE1, are negative regulators of cell proliferation in leaves (51), giving rise to the possibility that PPDs may directly repress AtGRF expression in order to exert their negative role in the regulation of cell meristematicity.
CINCINNATA-like TCP (CIN-TCPs) transcription factors control the transition from cell proliferation to expansion during leaf morphogenesis and act as growth repressors (for review, see 52). The overexpression of Arabidopsis CIN-TCPs directly enhances AtMIR396 expression, leading to precocious declines in GRF-GIF expression and leaf growth (15, 53). On the other hand, multiple loss-of-function mutations, tcp2 tcp4 (tcp2/4) and tcp2/4/10, promoted leaf growth by increasing cell proliferation (54). These results suggest that certain negative regulators of leaf cell proliferation, including PPDs and TCPs, may exert their function, directly or indirectly and at least partially, through the repression of GRF-GIF expression.
It should be noted that not all Arabidopsis GRFs seem to act as positive regulators of leaf growth. The loss-of-function Atgrf9 mutation enhanced cell cycle activities, resulting in large leaves, whereas AtGRF9 overexpression reduced those activities, resulting in small leaves (28, 55). This indicates that AtGRF9 negatively regulates leaf growth via the suppression of cell proliferation in leaf primordia. The authors also showed that the negative effect of AtGRF9 on cell proliferation was, at least partially, mediated by the regulation of a target gene, which encodes ORG3 (aka bHLH039). AtGRF9 directly activated ORG3 expression, and loss-of-function org3 mutants developed large leaves with more cells, whereas ORG3 overexpressors had small leaves with fewer cells. No additive effect on leaf growth was found in the org3 Atgrf9 double mutant, and the enhancement of leaf growth by org3 was not negated by AtGRF9 overexpression. It remains unclear how the specific GRF member exerts the opposite function. In contrast, Horiguchi et al. observed a very slight increase in the leaf size of the Atgrf9 mutant, but this increase was not statistically significant (10). Furthermore, it has been reported that AtGRF9 overexpressors produced only slightly enlarged leaves and that the same Atgrf9 mutant allele did not contribute to changes in leaf size (22). This incongruity should be reconciled in the future.
Roles in cell proliferation and expansion of monocot leaves
It appears that monocot GRFs and GIFs primarily act as positive regulators of cell proliferation in leaves. makiba3 (mkb3), a loss-of-function mutant allele of rice GIF1 (OsGIF1), caused a reduction in the number of leaf epidermal cells, producing small leaves, whereas MKB3 overexpression resulted in the opposite phenotypes, indicating that OsGIF1/MKB3 acts as a positive regulator of cell proliferation in the leaf organ as well (35). Similarly, CRISPR/CAS9-induced loss-of-function mutations of OsGIF1 (C/C-Osgif1) reduced leaf size, whereas 35S:OsGIF1 increased leaf size (56). mkb3 and C/C-Osgif1 mutants also shared other phenotypes, such as leaf rolling and reductions in the length of stem internodes. Based on the analysis of subepidermal cells in the internodes and main veins of leaf blades, the latter attributed the change in leaf size to the change in cell size, rather not in cell numbers. However, the latter also suggested a role of OsGIF1 in regulating cell proliferation of some tissues, such as specialized epidermal cells of the leaf blade, proposing that both the cell proliferation and expansion processes are under the control of OsGIF1, likely depending on the cell types. 35S:OsMIR396d and Osgrf6 rice plants showed reductions in cell length of the stem internode, leaf collar, and leaf sheath, indicating that lack of GRFs leads to defects in the cell elongation process in rice (57).
The overexpression of ZmGRF10, which lacks the C-terminal transactivation domain, led to a reduction in the size of maize leaves by decreasing cell proliferation, as it could execute a dominant negative effect by competing with other ZmGRFs with transactivation activities, suggesting that the other ZmGRFs may act as positive regulators of cell proliferation (20, 24). Consistently with the notion, the overexpression of Zm-rGRF1, a miR396-resistant version, increased the number of dividing cells in the leaf division zone, producing longer leaves (24). Intriguingly, however, Zm-rGRF1 overexpression prolonged the duration of cell cycling of those dividing cells, and thus the increase in leaf length was not as large as expected based on the increase in the number of dividing cells. Inversely, loss-of-function Zmgif1 mutants developed shorter and narrower vegetative and ear leaves than the wild type, and the size of their epidermal cells were larger, which is indicative of a reduction in cell numbers and thus a defect in the cell proliferation process. All things considered, therefore, the timing of the transition from the meristematic state to differentiating state in the leaf organ is governed by the GRF-GIF-miR396 module, in consequence determining the leaf size and shape in both eudicot and monocot plants.
It should be noted that, like loss-of-function mutant leaves of Arabidopsis GRFs and GIFs as well as 35S:AtMIR396, the rice mkb3 and Zmgif1 leaves developed larger cells, partially compensating for a reduced leaf size (25, 35). The compensation syndrome, which here I do not elaborate on, has been well documented (58).
Leaf senescence in Arabidopsis
It has been reported that increases in the GRF activities of Arabidopsis, B. napus, and B. rapa stimulate photosynthetic activities, resulting in an abundance of photosynthetic assimilates or seed oil (21, 41, 42, 59). The increases were concomitant with increases in total chlorophyll content and the rate of chloroplast division (41, 46). Activation of the GRF-GIF duo delayed leaf senescence, whereas their down-regulation accelerated it (21, 46). During dark-induced leaf senescence, the expression of specific marker genes for leaf senescence was markedly suppressed by 35S:At-rGRF3 but enhanced by 35S:AtMIR396 (21). The suppressive role of GRFs in leaf senescence may be explicable in light of the GRF-cytokinin interplay, as 35S:AtGRF5 increases the sensitivity of leaves to cytokinins (46). Cytokinins are well known to act not only as potent stimulators of cell proliferation but also as specific suppressors of leaf senescence (for review, see 60). The nature of the GRF-cytokinin interplay requires further investigation, although AtGIF1/AN3 directly activated CYTOKININ RESPONSE FACTOR2 (CRF2) and repressed ARABIDOPSIS RESPONSE REGULATOR4 (ARR4) (23).
Recently, an Arabidopsis gain-of-function mutant, oresara15-1D (ore15-1D), was shown to delay leaf senescence and promote organ growth (61). ORE15 encodes a transcription factor belonging to the PLATS family (PLANT A/T-RICH SEQUENCE- AND ZINC-BINDING PROTEIN). The suppression of leaf senescence by ore15-1D was accompanied by reductions in the expression of senescence marker genes, while the promotion of leaf growth by ore15-1D was coupled with the upregulation of AtGRF5 and AtGIF1/AN3 expression as well as cell cycle genes. The ORE15 transcription factor directly bound to the promoters of AtGRF1 and AtGRF4. In contrast to ore15-1D, ore15 null mutations functioned inversely in most of those physiological and molecular phenotypes. The findings demonstrate that ORE15 is not only a negative regulator of leaf senescence, but also a positive regulator of leaf cell proliferation. Therefore, ORE15 provides a genetic link mediating both of the processes, and the dual function of ORE15 is likely manifested, in part, through control of the GRF-GIF-miR396 module. Indeed, the an3 mutation promoted leaf senescence in the presence of ore15, but nullified the effect of ore15-1D. Both the leaf cell proliferation and senescence events occur temporally separated in Arabidopsis, i.e., at the primordial and mature stages, respectively. Therefore, it remains to be addressed in the future how the ORE15-GRF-GIF pathway regulates both of the cellular processes.
REGULATORY ROLES IN ROOT GROWTH AND DEVELOPMENT
It was recently demonstrated that AtGRFs are required for the transition of stem cells into transit-amplifying cells in the root meristem (62). Briefly, the abolishment of AtGRF activities by 35S:MIR396 suppressed the activities of cell cycle markers in root tips, reducing the root elongation rate and root length, whereas the overexpression of At-rGRF3 exhibited opposite effects on the marker activities. Unexpectedly, however, the final length of the 35S:At-rGRF3 root was short. The contradiction may be comprehensible in light of an additional function of AtGRFs in the root: ectopic AtGRF expression interferes with the normal patterning of cell divisions in the stem cell niche and organization of the quiescent center (QC). Nevertheless, it is intriguing that the heterologous overexpression of At-rGRF3 in B. oleracea resulted in longer roots than the wild type (63).
AtGIF1/AN3 also plays crucial roles in QC organization, which are, interestingly, independent of GRF activities (30). The AtGIF1/AN3 action was shown to be mediated, at least in part, by regulating the expression patterns of PLT1, as PLT1 was ectopically expressed in the an3 mutant. As mentioned above, PLT1 is one of the direct targets of AtGIF1/AN3. The report suggests that, as AtGIF1/AN3 lacks a DBD and its role in QC organization is independent of GRF, its targeting to PLT1 should be associated with another transcription factor, likely in concert with the SWI2/SNF2 chromatin-remodeling complex. The result and its implication are consistent with the roles of AtGIF1/AN3 in suppressing ectopic PLT1 expression during the development of embryonic polarity: if PLT1 not suppressed, the apical regions of embryos, which are presumed to develop into cotyledons, are converted to ectopic roots, as observed in the an3 han double mutant (64). HAN (HANABA TARANU) encodes a GATA transcription factor (65). In the legume plant Medicago truncatula, deactivation of MtGRFs by MtMIR396a and RNA interference (RNAi) inhibited root growth, due to reductions in both the cell cycling activity and the numbers of transit-amplifying cells in the meristematic zone of the root, though it did not affect the organization of the root apical meristem (66).
Regarding monocot root growth, the roots of 35S:OsMIR396d rice plants possessed fewer cells in the G2/M phase, suggesting that a lack of OsGRF activities may lead to reductions in cell cycling activities in rice roots (57). It should be noted, however, that the reduced cell cycling activity did not affect the root length, although 35S:OsMIR396d boosted the brassinosteroid (BR)-induced inhibition of root growth. By contrast, a loss-of-function rice mutant, Osgrf6, had shorter roots than the wild type (57). Taken together, the GRF-GIF duo generally seems to be required for root growth in both eudicot and monocot plants. However, this notion may not hold up straightforwardly, since one needs to separately analyze the duo’s effects on cell cycling activities in the transition zone of the root meristem as well as the organization of the stem cell niche.
REGULATORY ROLES IN THE DEVELOPMENT OF FLORAL ORGANS IN ARABIDOPSIS AND MONOCOTS
The Arabidopsis gif1/2/3 triple mutant displayed severe defects in the growth and development of all floral organs (67). Most conspicuously, the mutant gynoecium was split into two carpels along the medial regions, because the primordial replum cells of carpel margin meristems (CMMs) failed to maintain their meristematicity, precociously differentiating into papillar cells and thus not completely accomplishing the carpel fusion process. The mutant gynoecium either completely lacked or showed poor development in all internal tissues of the ovary (ovules, the septum, and the transmitting tract), which are all derived from CMMs. The gif triple mutant also had malformed anthers with no development of microsporangia bearing pollen grains, because the archesporial cells and their progeny lost meristematicity. Taken together, these results indicate that AtGIFs are essential factors for the establishment of the reproductive competency.
Since GRFs and GIFs form a functional unit for transcriptional regulation, the deactivation of AtGRFs is expected to cause similar floral aberrancies. Indeed, some strong 35S:MIR396 lines frequently developed single-carpel gynoecia, instead of two, and, on rare occasions, split gynoecia (22, 68). Recently, the strong deactivation of both AtGIF and AtGRF by gif 35S:MIR396 and grf multiple mutations allowed for further insight into the roles of AtGRFs in floral organ development. Those mutants completely aborted the pluripotent CMMs and archesporial cells of the anther (69). Strikingly, the mutant gynoecium developed no ovary at all, forming a rod-shaped gynoecium only with the stigma, style, and replum: the interior and exterior tissues of the gynoecial body were entirely filled in and covered with replum tissues. It is therefore obvious that AtGRFs are essential factors for the meristematic competency of formative cells in floral tissues, as are AtGIFs. Furthermore, the results showed that the lack of CMM development allows for the replum cells to infiltrate the whole gynoecial body, suggesting a developmental antagonism between the ovary and replum. GRF and GIF proteins are abundantly localized in the formative tissues of gynoecium and anther primordia, and the localization patterns of both proteins match exactly (67, 69). It has been shown that HEC1 is a direct target gene of AtGIF1/AN3 (23), thus giving rise to the possibility that it may mediate the duo’s action in floral organ growth and development.
The rod-shaped gynoecium phenotypes of those mutants were exacerbated by the pinoid-3 mutation and N-1-naphthylphthalamic acid, which is indicative of interplay between the GRF-GIF duo and polar auxin transport (69). It is noteworthy that, although the floral organ phenotypes of gif1/2/3 and gif 35S:MIR396 overlap on a broad scale, some of the details differ: the former predominantly displayed split gynoecia and yet developed the ovary, though poor, whereas the latter completely failed to form the ovary. In addition, the grf1/2/3/5 quadruple mutant, the strongest among the grf mutants obtained, mostly lost their ovary, but hardly developed split gynoecia. The results suggest that, in addition to their common pathway, AtGRFs and AtGIFs may have their own specific roles in the regulation of cell proliferation and differentiation during gynoecium development. The down-regulation of tobacco GRFs by AtMIR396 and PtMIR396 caused aberrant floral organs, which were reminiscent of the grf phenotypes, suggesting that the functionality of the GRF-GIF duo is conserved in eudicot floral organs (39, 40).
35S:OsMIR396 and Osgrf6/10 double mutants frequently produced aberrant floral organs: open husks, long sterile lemmas, and/or anomalous numbers of the stigma and anther (19). OsCR4 and Osjmj706 were shown to be directly activated by OsGRF10, thus mediating, at least in part, the roles of OsGRFs in floral organ development: the open-husk phenotype was also induced by the deactivation of OsCR4 and Osjmj706 (70, 71); 35S:OsJMJ706 partly rescued the floral defects of 35S:OsMIR396 (19).
Rice GIF1/MKB3 has been shown to be involved in floral organ development, as spikelets of the mkb3 mutant exhibited morphological abnormalities: the shapes of the lemma and palea were distorted, and the width of the palea was significantly reduced (35). The mkb3 mutant was not able to complete the ovule formation and integument elongation processes, and also produced no pollen or abnormal pollen, similar to the Arabidopsis gif mutants.
The maize gif1 mutant also showed many defects in floral organs: it is male and female sterile; it produced multiple silks, or pistils, per floret; and its nucellus protruded out of the carpel, as seen in Arabidopsis gif mutants (25). Interestingly, in the Zmgif1 mutants, extra numbers of spikelet meristems (SMs) were initiated from spikelet pair meristems (SPMs) in both ears and tassels, indicating that the axillary meristems lose their determinate nature, and thus ZmGIF1 is involved in promoting determinacy of the inflorescence. The situation seems to be contrary to that of leaf growth, in which Zmgif1 leaf cells are less meristematic, producing small leaves with fewer cells, as mentioned previously. Rice and maize GIF1 mRNAs are highly expressed in floral organ primordia, SMs, and SPMs (25, 35). In conclusion, the GRF-GIF duo of both eudicot and monocot plants plays essential roles in the growth and development of floral organs, thus warranting successful reproduction. Additionally, depending on different evolutionary pathways, it may have co-opted a switch function balancing the determinacy and indeterminacy of spikelet meristems in monocots.
REGULATORY ROLES IN SCULPTING PLANT ARCHITECTURE OF MONOCOT PLANTS
Roles in regulation of stem elongation and plant height
The deactivation of OsGRFs by loss-of-function mutations, 35S:OsMIR396, and RNAi resulted in semi-dwarf rice plants (18, 19, 29, 57). On the other hand, a rice dominant quantitative trait locus (QTL), GRAIN SIZE ON CHROMOSOME2 in the Baodali line (GS2-BDL) caused a slight increase in height with significantly longer leaves (72). The GS2-BDL locus corresponds to OsGRF4, whose transcripts lost its miR396 target site by a mutation in it, thus increasing its transcripts level but not affecting the amino acid sequence of the OsGRF4 protein and thus its function (Osgrf4-1DGS2-BDL hereafter for simplicity). Of note, however, no significant changes in those phenotypes were detected in Osgrf4-1DGS2-JDL, which contained the same kind of a gain-of-function mutation from the GRAIN SIZE AND WEIGHT2 QTL (GS2) in Judali as Osgrf4-1DGS2-BDL (73). The reduced height of 35S:OsMIR396d rice was due to the short internodes with compromised cell elongation. 35S:OsMIR396d also increased the degree of the leaf angles, because the cell elongation of the adaxial side of the leaf collar was less affected than that of the abaxial side. Taken together, this indicates that the rice height is controlled by the GRF-miR396 module. The compromised and differential elongation of stem intermodal and leaf collar cells of 35S:OsMIR396d was shown to be intimately linked with the signaling and biosynthetic pathways of the phytohormones BR and gibberellin (GA): OsBZR1, a key transcription activator of BR signaling, directly activated OsMIR396d expression, while OsGRF6 promoted GA biosynthesis and signaling. Inversely, Osgrf4-1DGL2 stimulated seedling growth and reduced leaf angles, and the central negative regulator in BR signaling, OsGSK2, physically interacted with OsGRF4, inhibiting OsGRF4 expression (74).
Both the rice mkb3 and Zmgif1 mutants exhibited dwarf phenotypes due to shortened internodes, indicating that, like GRFs, GIFs are also involved in the regulation of stem elongation, and thus plant height (25, 35). Unexpectedly, however, the overexpression of Zm-rGRF1 resulted in dwarfism (24), likely due to a perturbation in the stem elongation process due to its nature of a strong ectopic expression.
Based on results derived from the deactivation of monocot GRF and GIF genes, it is clear that the compromised cell elongation process is a primary cause of short internodes, suggesting that the GRF-GIF-miR396 module is involved in the regulation of cell elongation in stem growth, rather than cell proliferation. It is noteworthy, however, that those studies have focused on cell elongation of internode regions. Given that the first GRF member, OsGRF1, was identified in the intercalary meristem of rice plants, which supplies internode tissues with new cells (6), examining the cell cycling activities in the intercalary meristem of the rice and maize mutants could provide further insight into the role of the GRF-GIF-miR396 module in the regulation of monocot stem elongation.
The Arabidopsis inflorescence stem showed a bi-phasic growth pattern in response to different dosages of gif mutations: the gif1 single mutant developed longer stems than the wild type, whereas the gif triple mutant had much shorter ones (37), although its nature was not investigated in detail. Interestingly, Arabidopsis roots also showed a bi-phasic pattern: gif1 roots, longer; gif1/2/3, shorter (30).
Roles in regulation of grain size and panicle development of monocots
In monocot plants, the activities of the GRF-GIF-miR396 module affected the grain size and architecture of panicles, such as the length and number of branches as well as spikelet numbers. The up-regulation of OsGRFs by 35S:OsGRF6, Os-rGRF6, and a target mimicry of miR396 (35S:MIM396) increased the numbers of panicle branches and spikelets, resulting in high yield, whereas the down-regulation of OsGRFs by 35S:OsMIR396d and RNAi caused the opposite phenotypes (29). The report suggested that regulation of the axillary branches and spikelet numbers by OsGRFs appeared to be mediated by stimulated auxin biosynthesis and signaling. The dominant Osgrf4-1D mutations and OsGRF overexpression also markedly increased grain size and panicle length, consequently producing more grains with increased weight (72, 73, 75, 76). The deactivation of OsGRFs by loss-of-function approaches impaired those yield traits (19, 76). As expected, the OsGIF1 function in the panicle traits parallels that of OsGRFs. The mkb3 and C/C-Osgif1 mutants had shorter branches and/or reduced size and weight of grains, whereas the overexpression of OsGIF1 increased both grain size and weight (35, 56, 73, 75). Therefore, it is clear that the GRF-GIF duo acts as a positive regulator of grain size and panicle development in rice. Maize gif1 mutants also displayed severe defects in the inflorescence architecture: reduced lengths of tassels and ears as well as reduced numbers of tassel branches, but increased numbers of short branches in the ear (25).
Evidence indicates a role of eudicot GRFs in determining seed size. 35S:AtGRF1 and 35S:AtGRF5 have been shown to increase seed size, albeit not always accompanied by increases in seed weight (77). Arabidopsis plants overexpressing BnGRF2a and BrGRFs as well as B. napus plants overexpressing BrGRFs all developed large seeds with increased weight (41–43). The promotive effect on seed growth may be closely associated with the increases in photosynthetic activities and senescence retardation by GRFs.
REGULATORY ROLES IN PLANT-PATHOGEN INTERACTION AND IN RESPONSES TO UV-B LIGHT
Syncytium formation occurring in Arabidopsis roots by an infective cyst nematode (Heterodera schachtii) was deterred by 35S:AtMIR396 and the grf1/2/3 triple mutation, indicating that AtGRFs are required for the reprogramming processes of root cells, such as changes in cell fate, re-differentiation, and cell proliferation (78). This leads to an interesting, evolutionary question of how the parasite wired up the GRF-miR396 module in order to induce the nourishment source tissue. Similarly, in M. truncatula, 35S:MtMIR396 reduced the frequency of colonization by arbuscular mycorrhizal fungi, whereas 35S:MIM396 frequently reversed it (66). This indicates that the GRF-miR396 module promotes (sym)biotic associations with microbes in the rhizosphere.
Genes involved in the regulation of defense responses and disease resistance were found to be enriched in the potential target candidates of AtGRF1 and AtGRF3 (79). In support of that, Arabidopsis plants expressing 35S:MIR396 enhanced the susceptibility to infection, thus increasing fungal biomass, whereas 35S:MIM396 plants displayed broad resistance to fungal pathogens with concomitant activation of defense responses, indicating that GRFs help deter pathogenic organisms (80).
In Arabidopsis leaves, UV-B light induced the accumulation of miR396 and thus reduced the abundance of AtGRF mRNA, resulting in repressed cell proliferation (81). Therefore, the GRF-miR396 module mediates, at least in part, the UV-B-repression of leaf growth, and also likely provides a protective mechanism against UV-B light, as plant cells with UV-B-damaged DNA are not to proliferate. Arabidopsis E2Fc transcription factor acts upstream of AtMIR396, probably activating, directly or not, the expression of AtMIR396 or of genes that encode proteins involved in the processing of miR396 precursors (82). UV-B light induced the accumulation of miR396 in maize leaves as well, and caused a reduction in cell proliferation and a shortened growth zone (83). These results suggest that both dicot and monocot plants may have adopted the parallel molecular apparatus in order to cope with the detrimental effect of UV-B light.
THE EVOLUTIONARY GENESIS OF GRFs
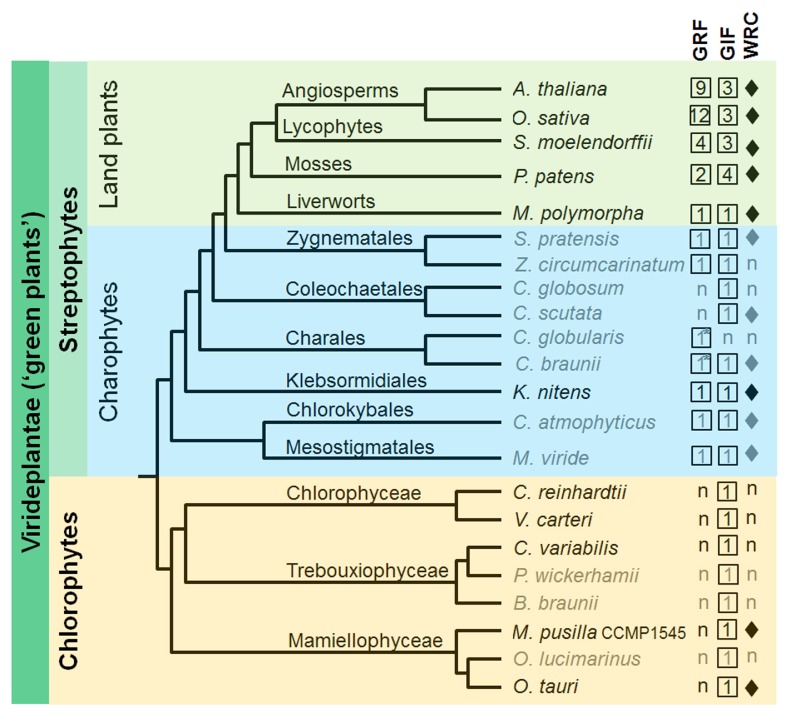
It has been described in the last review articles that GRFs are land plant-specific genes, since genomic and transcriptomic resources available then revealed their presence only in plants (land plants or embryophytes) but not in metazoans, fungi, and protists, including ‘green algae’ (11, 12). The ‘green algae’ are members of chlorophytes and charophytes that are paraphyletic to land plants (Fig. 2). However, a recent paper reported the presence of a single GRF gene in the genome sequence of a charophyte, Klebsormidium nitens (previously known as K. flaccidum) (84). This prompted me to scrutinize other charophycean sequences that have been recently deposited in public databases and I found single GRF genes in the genomes or transcriptomes of almost all orders of charophycean algae, but not in coleochaetales (Chaetosphaeridium globosum and Coleochaete scutata), probably because of insufficient coverages of their transcriptomic sequences (Fig. 2). However, I have still not been able to detect the presence of GRF in any chlorophytes. This sequence profile calls for a revision of the previous notion regarding the evolutionary origin of GRFs: GRFs are not land plant-specific transcription factors but streptophyte-specific. Streptophytes comprise both charophytes and land plants, and are paraphyletic to chlorophytes (Fig. 2) (85, 86). Therefore, it is likely that a GRF gene may have evolutionarily emerged in an ancestral charophyte after its divergence from chlorophytes, and that an ancestral land plant inherited and duplicated it, thus diversifying its function to meet the biological complexity of or to give rise to the complexity in ensuing lineages of land plants.
Fig. 2.
Phylogenetic relationships among ‘green plants’ and the presence of GRF-GIF genes. Depicted are the relationships among the three lineages of ‘green plants’: chlorophytes, charophytes, and land plants. The numbers of GRF and GIF are indicated in black boxes, while diamond bullets indicate the presence of the WRC domain. Species depicted in gray tone only have whole transcriptomic resources, but no whole genome sequenced. n designates ‘not present’; asterisks indicate that the GRF genes are predicted to encode three consecutive WRC domains after the QLQ domain.
How could the GRF gene have been invented in an ancestral charophyte? This question may remain unanswered for years. One may speculate that an ancient QLQ domain derived from the N-terminus of the SWI2/SNF2 chromatin-emodeling ATPases (BRM and its homologs) have acquired the WRC domain, resulting in an ancestral GRF gene (9, 11). SWI2/SNF2 ATPases are universally present in eukaryotes, including viridiplantae, and they play essential roles in the chromatin remodeling process (11, 87, 88). According to the Pfam profile, QLQ domains exist in 66 different architectures with 2303 entries (http://pfam.xfam.org, PF08880). Half of the entry proteins have the QLQ domain together with WRC as GRF proteins; roughly the other half together with the SNF2_N domain of the SWI2/SNF2 ATPases; and only a few entries are together with other kinds of domains. These combinatorial structures with QLQ are compatible with the notion that the SWI2/SNF2 ATPase QLQ domain might be co-opted into an ancient GRF gene.
The Pfam profile also reveals that the WRC domain, which contains the DNA-binding C3H motif, is present in streptophytes and Mamiellophyceae, but is not present in Chlorophyceae and Trebouxiophyceae or any other organisms (Fig. 2; http://pfam.xfam.org, PF08879). WRC domains exist in 26 different architectures with 1984 entries: more than half of the entry proteins have the WRC domain together with the QLQ domain as GRFs, a quarter are mostly uncharacterized proteins with a single WRC domain but with no associated known domains, and the rest have single or multiple WRC domains associated with other kinds of known domains. Interestingly, the GRFs of Chara globularis and Chara braunii belonging to Charales have a QLQ domain followed by three consecutive WRC domains (Fig. 2). This profiling suggests that the evolutionary swapping of the WRC domain might have frequently occurred in virideplantae (“green plants”). Therefore, it is tempting to speculate that QLQ and WRC domains might have been co-opted into a GRF protein in an ancestral charophyte.
The origin of GIF genes is much more ancient than that of GRFs, as they exist in most eukaryotes, such as virideplantae, and metazoans, and not in fungi and protists other than ‘green algae’ (11, 12). GIFs are present in the genomes of a charophyte (K. nitens) and chlorophytes (Chlamydomonas reinhardtii, Ostreococcus lucimarinus, and Ostreococcus tauri) (84). Additionally, this profiling identified GIFs in the genomes or transcriptomic sequences of chlorophytes (Botryococcus braunii, Chlorella variabilis NC64, and Prototheca wickerhamii) and charophytes (Chaetosphaeridium globosum and Chlorokybus atmophyticus; Fig. 2).
Both genomic and cDNA sequences were available for some of those algal GIFs (C. reinhardtii, V. cateri, and K. nitens), allowing for the construction of their exon-intron structures. I found that the SNH domains of those three algae and land plants are encoded in the first three exons with conserved intron positions and phases (data not shown). The analysis suggests that the structure of GIF genes has been highly conserved during the evolutionary path of chlorophytes, charophytes, and land plants. Therefore, it is a plausible hypothesis that the GRF-GIF partnership was established in an ancestral charophyte. It would be interesting to explore whether charophycean GRFs and GIFs interact together; then if so, what the biological role and molecular function of the duo are, especially in terms of evolution, and what chlorophytic GIFs do in the absence of the canonical partner protein GRF.
In summary, the GRF-GIF-miR396 module plays essential roles in the growth and development of angiosperms. It regulates the meristematic potential of primordial cells during leaf growth, determining the final size and shape of the leaf organ. The GRF-GIF duo is a prerequisite for floral organ development, and thus enables the production of the formative cells, such as CMMs and egg cells as well as microsporangia and sperm cells. It is also involved in the regulation of leaf longevity and photosynthetic efficiency in mature leaves. Importantly, the monocot GRF-GIF duo also promoted the yield traits, such as grain size and panicle architecture, warranting crop productivity. Finally, the GRF gene has a charophycean origin, so studies on GRFs of basalmost land plants and charophytes could shed light on their significance in the evolution-developmental history of a main lineage of life, the streptophyte.
ACKNOWLEDGEMENTS
This work was supported by the Korea Research Foundation Grants, 2015R1D1A1A01059934 and 2018R1D1A1B07050016. I apologize to all colleagues whose work could not be cited due to space constraints.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 2.Guo A, He K, Liu D, et al. DATF: A Database of Arabidopsis transcription factors. Bioinformatics. 2005;21:2568–2569. doi: 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- 3.Riaño-Pachón DM, Ruzicic S, Dreyer I, Mueller-Roeber B. PlnTFDB: an integrative plant transcription factor database. BMC Bioinformatics. 2007;8:42. doi: 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitsuda N, Ohme-Takagi M. Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 2009;50:1232–1248. doi: 10.1093/pcp/pcp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez D. Plant transcription factors: evolutionary, structural and functional aspects. 1st ed. Academic Press; 2015. [Google Scholar]
- 6.van der Knaap E, Kim JH, Kende H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000;122:695–704. doi: 10.1104/pp.122.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim JH, Choi D, Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003;36:94–104. doi: 10.1046/j.1365-313X.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- 8.Choi D, Kim JH, Kende H. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L) Plant Cell Physiol. 2004;45:897–904. doi: 10.1093/pcp/pch098. [DOI] [PubMed] [Google Scholar]
- 9.Kim JH, Kende H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci U S A. 2004;101:13374–13379. doi: 10.1073/pnas.0405450101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horiguchi G, Kim G, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005;43:68–78. doi: 10.1111/j.1365-313X.2005.02429.x. [DOI] [PubMed] [Google Scholar]
- 11.Kim JH, Tsukaya H. Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J Exp Bot. 2015;66:6093–6107. doi: 10.1093/jxb/erv349. [DOI] [PubMed] [Google Scholar]
- 12.Omidbakhshfard MA, Proost S, Fujikura U, Mueller-Roeber B. Growth-Regulating Factors (GRFs): A small transcription factor family with important functions in plant biology. Mol Plant. 2015;8:998–1010. doi: 10.1016/j.molp.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez RE, Ercoli MF, Debernadi JM, Palatnik J. Growth-regulating factors, a transcription factor family regulating more than just plant growth. In: Gonzalez DH, editor. Plant Transcription Factors. Chapter 17. Elsevier; The Netherlands: 2016. pp. 269–280. [DOI] [Google Scholar]
- 14.Liu D, Song Y, Chen Z, Yu D. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol Plant. 2009;136:223–236. doi: 10.1111/j.1399-3054.2009.01229.x. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development. 2010;137:103–112. doi: 10.1242/dev.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osnato M, Stile MR, Wang Y, et al. Cross talk between the KNOX and ethylene pathways is mediated by intron-binding transcription factors in barley. Plant Physiol. 2010;154:1616–1632. doi: 10.1104/pp.110.161984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JS, Mizoi J, Kidokoro S, et al. Arabidopsis GROWTH-REGULATING FACTOR7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell. 2012;24:3393–3405. doi: 10.1105/tpc.112.100933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuijt SJ, Greco R, Agalou A, et al. Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 2014;164:1952–1966. doi: 10.1104/pp.113.222836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, Guo S, Xu Y, et al. OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 2014;165:160–174. doi: 10.1104/pp.114.235564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu L, Zhang D, Xue M, Qian J, He Y, Wang S. Overexpression of the maize GRF10, an endogenous truncated GRF protein, leads to reduction in leaf size and plant height. J Integr Plant Biol. 2014;56:1053–1063. doi: 10.1111/jipb.12220. [DOI] [PubMed] [Google Scholar]
- 21.Debernardi JM, Mecchia MA, Vercruyssen L, et al. Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 2014;79:413–426. doi: 10.1111/tpj.12567. [DOI] [PubMed] [Google Scholar]
- 22.Liang G, He H, Li Y, Wang F, Yu D. Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 2014;164:249–258. doi: 10.1104/pp.113.225144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vercruyssen L, Verkest A, Gonzalez N, et al. ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell. 2014;26:210–229. doi: 10.1105/tpc.113.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelissen H, Eeckhout D, Demuynck K, et al. Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell. 2015;27:1605–1619. doi: 10.1105/tpc.15.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D, Sun W, Singh R, et al. GRF-interacting factor1 (gif1) regulates shoot architecture and meristem determinacy in maize. Plant Cell. 2018;30:360–374. doi: 10.1105/tpc.17.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thaete C, Brett D, Monaghan P, et al. Functional domains of the SYT and SYT-SSX synovial sarcoma translocation proteins and co-localization with the SNF protein BRM in the nucleus. Human Mol Gen. 1999;8:585–591. doi: 10.1093/hmg/8.4.585. [DOI] [PubMed] [Google Scholar]
- 27.de Bruijn DR, van Kessel GA. Common origin of the human synovial sarcoma associated SS18 and SS18L1 gene loci. Cytogenet Genom Res. 2006;112:222–226. doi: 10.1159/000089874. [DOI] [PubMed] [Google Scholar]
- 28.Omidbakhshfard MA, Fujikura U, Olas JJ, Xue GP, Balazadeh S, Mueller-Roeber B. GROWTH-REGULATING FACTOR 9 negatively regulates Arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. PLoS Genet. 2018;14:e1007484. doi: 10.1371/journal.pgen.1007484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao F, Wang K, Liu Y, et al. Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat Plants. 2016;2:15196. doi: 10.1038/nplants.2015.196. [DOI] [PubMed] [Google Scholar]
- 30.Ercoli MF, Ferela A, Debernardi JM, Perrone AP, Rodriguez RE, Palatnik JF. GIF transcriptional coregulators control root meristem homeostasis. Plant Cell. 2018;30:347–359. doi: 10.1105/tpc.17.00856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagai M, Tanaka S, Tsuda M, et al. Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha. Proc Natl Acad Sci U S A. 2001;98:3843–3848. doi: 10.1073/pnas.061036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perani M, Ingram CJ, Cooper CS, Garrett MD, Goodwin GH. Conserved SNH domain of the proto-oncoprotein SYT interacts with components of the human chromatin remodelling complexes, while the QPGY repeat domain forms homo-oligomers. Oncogene. 2003;22:8156–8167. doi: 10.1038/sj.onc.1207031. [DOI] [PubMed] [Google Scholar]
- 33.Middeljans E, Wan X, Jansen PW, Sharma V. SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes. PLoS One. 2012;7:e33834. doi: 10.1371/journal.pone.0033834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawade K, Horiguchi G, Usami T, Hirai MY, Tsukaya H. ANGUSTIFOLIA3 signaling coordinates proliferation between clonally distinct cells in leaves. Curr Biol. 2013;23:788–792. doi: 10.1016/j.cub.2013.03.044. [DOI] [PubMed] [Google Scholar]
- 35.Shimano S, Hibara K-I, Furuya T, Arimura S-I, Tsukaya H, Itoh J-I. Conserved functional control, but distinct regulation of cell proliferation in rice and Arabidopsis leaves revealed by comparative analysis of GRF-INTERACTING FACTOR1 orthologs. Development. 2018;145 doi: 10.1242/dev.159624. dev159624. [DOI] [PubMed] [Google Scholar]
- 36.Kim JH, Lee BH. GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J Plant Biol. 2006;49:463–468. doi: 10.1007/BF03031127. [DOI] [Google Scholar]
- 37.Lee BH, Ko JH, Lee S, Lee Y, Pak JH, Kim JH. The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 2009;151:655–668. doi: 10.1104/pp.109.141838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Gu X, Xu D, et al. miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J Exp Bot. 2011;62:761–773. doi: 10.1093/jxb/erq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang F, Liang G, Liu D, Yu D. Arabidopsis miR396 mediates the development of leaves and flowers in transgenic tobacco. J Plant Biol. 2009;52:475–481. doi: 10.1007/s12374-009-9061-7. [DOI] [Google Scholar]
- 40.Baucher M, Moussawi J, Vandeputte OM, et al. A role for the miR396/GRF network in specification of organ type during flower development, as supported by ectopic expression of Populus trichocarpa miR396c in transgenic tobacco. Plant Biol. 2013;15:892–898. doi: 10.1111/j.1438-8677.2012.00696.x. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Hua W, Yang HL, et al. The BnGRF2 gene (GRF2-like gene from Brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. J Exp Bot. 2012;63:3727–3740. doi: 10.1093/jxb/ers066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong JK, Oh S-W, Kim JH, Lee SB, Suh EJ, Lee Y-H. Overexpression of Brassica rapa GROWTH-REGULATING FACTOR genes in Arabidosis thaliana increases organ growth by enhancing cell proliferation. J Plant Biotechnol. 2017;44:271–286. doi: 10.5010/JPB.2017.44.3.271. [DOI] [Google Scholar]
- 43.Hong JK, Suh EJ, Lee SB, Yoon H-J, Lee Y-H. Effects of overexpression of Brasscia rapa GROWTH-REGUALTING FACTOR genes on B. napus organ size. Kor J Breed Sci. 2018;50:378–386. doi: 10.9787/KJBS.2018.50.4.378. (in Korean) [DOI] [Google Scholar]
- 44.Ferjani A, Yano S, Horiguchi G, Tsukaya H. Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol. 2007;144:988–999. doi: 10.1104/pp.107.099325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol. 1999;215:407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
- 46.Vercruyssen L, Tognetti VB, Gonzalez N, et al. GROWTH REGULATING FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiol. 2015;167:817–832. doi: 10.1104/pp.114.256180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ichihashi Y, Kawade K, Usami T, Horiguchi G, Takahashi T, Tsukaya H. Key proliferative activity in the junction between the leaf blade and leaf petiole of Arabidopsis. Plant Physiol. 2011;157:1151–1162. doi: 10.1104/pp.111.185066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee BH, Kim JH. Spatio-temporal distribution patterns of GRF-INTERACTING FACTOR expression and leaf size control. Plant Signal Behav. 2014;9:e29697. doi: 10.4161/psb.29697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta MD, Nath U. Divergence in patterns of leaf growth polarity is associated with the expression divergence of miR396. Plant Cell. 2015;27:2785–2799. doi: 10.1105/tpc.15.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Liu W, Zhu Y, et al. BIGGER ORGANS and ELEPHANT EAR-LIKE LEAF1 control organ size and floral organ internal asymmetry in pea. J Exp Bot. 2019;70:179–191. doi: 10.1093/jxb/ery352. [DOI] [PubMed] [Google Scholar]
- 51.White DWR. PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:13238–13243. doi: 10.1073/pnas.0604349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarvepalli K, Gupta MD, Challa KR, Nath U. Molecular cartography of leaf development — role of transcription factors. Curr Opin Plant Biol. 2019;47:22–31. doi: 10.1016/j.pbi.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 53.Schommer C, Debernardi JM, Bresso EG, Rodriguez RE, Palatnik JF. Repression of cell proliferation by miR319-regulated TCP4. Mol Plant. 2014;7:1533–1544. doi: 10.1093/mp/ssu084. [DOI] [PubMed] [Google Scholar]
- 54.Bresso EG, Chorostecki U, Rodriguez RE, Palatnik JF, Schommer C. Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiol. 2017;176:1694–1708. doi: 10.1104/pp.17.00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arvidsson S, Pérez-Rodríguez P, Mueller-Roeber B. A growth phenotyping pipeline for Arabidopsis thaliana integrating image analysis and rosette area modeling for robust quantification of genotype effects. New Phytol. 2011;191:895–907. doi: 10.1111/j.1469-8137.2011.03756.x. [DOI] [PubMed] [Google Scholar]
- 56.He Z, Zeng J, Ren Y, et al. OsGIF1 positively regulates the sizes of stems, leaves and grains in rice. Front Plant Sci. 2017;8:1730. doi: 10.3389/fpls.2017.01730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang Y, Liu H, Guo S, et al. OsmiR396d miRNA affects gibberellin and brassinosteroid signaling to regulate plant architecture. Plant Physiol. 2018;176:946–959. doi: 10.1104/pp.17.00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsukaya H. Mechanism of leaf-shape determination. Ann Rev of Plant Biol. 2006;57:477–496. doi: 10.1146/annurev.arplant.57.032905.105320. [DOI] [PubMed] [Google Scholar]
- 59.Gonzalez N, De Bodt S, Sulpice R, et al. Increased leaf size, different means to an end. Plant Physiol. 2010;153:1261–1279. doi: 10.1104/pp.110.156018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zürcher E, Müller B. Cytokinin synthesis, signaling and function—advances and new insights. Int Rev Cell Mol Bio. 2016;324:1–38. doi: 10.1016/bs.ircmb.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Kim JH, Kim J, Jun SE, et al. ORESARA15, a PLATZ transcription factor, mediates leaf growth and senescence in Arabidopsis. New Phytol. 2018;220:609–623. doi: 10.1111/nph.15291. [DOI] [PubMed] [Google Scholar]
- 62.Rodriguez RE, Ercoli MF, Debernardi J, et al. MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. Plant Cell. 2015;27:3354–3366. doi: 10.1105/tpc.15.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beltramino M, Ercoli MF, Debernardi JM, et al. Robust increase of leaf size by Arabidopsis thaliana GRF3-like transcription factors under different growth conditions. Sci Rep. 2018;8:13447. doi: 10.1038/s41598-018-29859-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanei M, Horiguchi G, Tsukaya H. Stable establishment of cotyledon identity during embryogenesis in Arabidopsis by ANGUSTIFOLIA3 and HANABA TARANU. Development. 2012;139:2436–2446. doi: 10.1242/dev.081547. [DOI] [PubMed] [Google Scholar]
- 65.Zhao Y, Medrano L, Ohashi K, et al. HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell. 2004;16:2586–2600. doi: 10.1105/tpc.104.024869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bazin J, Khan GA, Combier J, et al. miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 2013;74:920–934. doi: 10.1111/tpj.12178. [DOI] [PubMed] [Google Scholar]
- 67.Lee BH, Wynn AN, Franks RG, Hwang Y, Lim J, Kim JH. The Arabidopsis thaliana GRF- INTERACTING FACTOR gene family plays an essential role in control of male and female reproductive development. Dev Biol. 2014;386:12–24. doi: 10.1016/j.ydbio.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 68.Pajoro A, Madrigal P, Muino JM, et al. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014;15:R41. doi: 10.1186/gb-2014-15-3-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee S-J, Lee BH, Jung J-H, Park SK, Song JT, Kim JH. GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR specify meristematic cells of gynoecia and anthers. Plant Physiol. 2018;176:717–729. doi: 10.1104/pp.17.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun Q, Zhou DX. Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci U S A. 2008;105:13679–13684. doi: 10.1073/pnas.0805901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pu C, Ma Y, Wang J, et al. Crinkly4 receptor-like kinase is required to maintain the interlocking of the palea and lemma and fertility in rice, by promoting epidermal cell differentiation. Plant J. 2012;70:940–953. doi: 10.1111/j.1365-313X.2012.04925.x. [DOI] [PubMed] [Google Scholar]
- 72.Hu J, Wang Y, Fang Y, et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant. 2015;8:1455–1465. doi: 10.1016/j.molp.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 73.Duan P, Ni S, Wang B, et al. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat Plants. 2016;2:1. doi: 10.1038/nplants.2015.203. [DOI] [PubMed] [Google Scholar]
- 74.Che R, Tong H, Shi B, et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants. 2016;2:15195. doi: 10.1038/nplants.2015.195. [DOI] [PubMed] [Google Scholar]
- 75.Li S, Gao F, Xie K, et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotech J. 2016;14:2134–2146. doi: 10.1111/pbi.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun P, Zhang W, Wang Y, et al. OsGRF4 controls grain shape, panicle length and seed shattering in rice. J Integr Plant Biol. 2016;58:836–847. doi: 10.1111/jipb.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Daele I, Gonzalez N, Vercauteren I, et al. A comparative study of seed yield parameters in Arabidopsis thaliana mutants and transgenics. Plant Biotech J. 2012;10:488–500. doi: 10.1111/j.1467-7652.2012.00687.x. [DOI] [PubMed] [Google Scholar]
- 78.Hewezi T, Maier TR, Nettleton D, Baum TJ. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol. 2012;159:321–335. doi: 10.1104/pp.112.193649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu J, Rice JH, Chen N, Baum TJ, Hewezi T. Synchronization of developmental processes and defense signaling by growth regulating transcription factors. PLoS One. 2014;9:e98477. doi: 10.1371/journal.pone.0098477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soto-Suárez M, Baldrich P, Weigel D, Rubio-Somoza I, Segundo BS. The Arabidopsis miR396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens. Sci Rep. 2017;7:44898. doi: 10.1038/srep44898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Casadevall R, Rodriguez RE, Debernardi JM, Palatnik JF, Casati P. Repression of growth regulating factors by the microRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. Plant Cell. 2013;25:3570–3583. doi: 10.1105/tpc.113.117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gómez MS, Ferreyra MLF, Sheridan M, Casati P. Arabidopsis E2Fc is required for the DNA damage response under UV-B radiation epistatically over the microRNA396 and independently of E2Fe. Plant J. 2018;97:749–764. doi: 10.1111/tpj.14158. [DOI] [PubMed] [Google Scholar]
- 83.Fina JP, Casadevall R, AbdElgawad H, et al. UV-B inhibits leaf growth through changes in Growth Regulating Factors and gibberellin levels. Plant Physiol. 2017;174:1110–1126. doi: 10.1104/pp.17.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bowman JL, Kohchi T, Yamato K, et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 2017;171:287–304. doi: 10.1016/j.cell.2017.09.030. [DOI] [PubMed] [Google Scholar]
- 85.Delwiche CF, Cooper ED. The evolutionary origin of a terrestrial flora. Curr Biol. 2015;25:R899–R910. doi: 10.1016/j.cub.2015.08.029. [DOI] [PubMed] [Google Scholar]
- 86.Timme RE, Delwiche CF. Uncovering the evolutionary origin of plant molecular processes: comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol. 2010;10:96. doi: 10.1186/1471-2229-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Han S-K, Wu M-F, Cui S, Wagner D. Roles and activities of chromatin remodeling ATPases in plants. Plant J. 2015;83:62–77. doi: 10.1111/tpj.12877. [DOI] [PubMed] [Google Scholar]
- 88.Koster MJ, Snel B, Timmers HT. Genesis of chromatin and transcription dynamics in the origin of species. Cell. 2015;161:724–736. doi: 10.1016/j.cell.2015.04.033. [DOI] [PubMed] [Google Scholar]