Abstract
Analysis of viral genetic linkage can reveal generalized transmission patterns within a population. The HIV Prevention Trials Network 061 study evaluated HIV incidence among black men who have sex with men. HIV genotypes from 169 men who were HIV infected at enrollment and 23 men who seroconverted during the study were analyzed for genetic linkage. This analysis showed some associations of viral linkage with income, study site, and timing of infection.
Keywords: HIV transmission, HIV in men who have sex with men, HIV genetics, disparities in HIV
HIV Prevention Trials Network 061 (HPTN 061) study was a multisite randomized clinical trial that evaluated strategies for reducing HIV transmission in black men who have sex with men (MSM) and studied risk factors for HIV infection.1,2 Black MSM account for >20% of incident HIV infections in the United States in recent years, although only an estimated 12.7% of the American population described themselves as black in 2016.3,4 The concentrated HIV epidemic among black MSM highlights the importance of identifying reasons for high HIV incidence in this population, including sociobehavioral, structural, and biological factors associated with HIV infection.
This report analyzed viral genetic linkage among participants in HPTN 061 to explore factors associated with linked HIV infections in black MSM. We investigated the relationship of viral linkage (based on genetic similarity of viral sequences) with income, study site, and timing of infection. Although transmission networks cannot be inferred from linkage analysis, such assessments can provide insights about epidemic dynamics, because individuals whose HIV sequences are closely related are also likely to be more densely connected in a transmission network.
To evaluate strategies for recruitment and engagement of high-risk black MSM in HIV prevention research, HPTN 061 enrolled 1,553 self-identified black MSM in six U.S. cities: Atlanta (19%), Boston (15%), New York (20%), Los Angeles (19%), San Francisco (13%), and Washington, DC (14%).1,2 Participants were offered HIV testing, access to peer health navigation, education about safer sex practices, condoms, lubricants, and sexually transmitted infection testing. The number of participants who reported that they had HIV infection and were in care was capped at 10 per study site, because of the study's focus on HIV incidence. Participants were classified as having recent HIV infection at enrollment if either they had acute infection or “recent” infection according to a multiassay algorithm.5 Twenty-eight participants seroconverted after enrollment and during 1 year of follow-up. Participants infected at enrollment included both acute and recent infection at the time of enrollment. Income was classified as high (≥$30,000) or low (<$30,000).
Of 1,553 participants, 348 (22.4%) were HIV infected at enrollment; 90 of the latter were classified as newly diagnosed or previously diagnosed, based on self-reported data and retrospective testing for antiretroviral medication.6 HIV genotypes were obtained for 169 of the 171 men who were HIV infected at enrollment and had a viral load >400 copies/mL, and for 23 of the 28 men who acquired HIV infection during the HPTN 061 study.7
A matrix of genetic distances between sequences was generated using the APE package in R. Genetic distance was defined as the proportion of nucleotide positions where the nucleotides in the two sequences were different. Two sequences were considered to be linked if the genetic distance between the two sequences was <0.04.
Permutation tests were used to investigate the association of income, study site, and timing of infection (participants who were infected at enrollment vs. seroconverters) on probability of linkage. The test statistic represented the proportion of pairs of individuals in a given group who were linked to other individuals belonging either to the same or to different groups. The sampling distribution of the test statistic was obtained by repeatedly sampling from the observed linkage matrices without replacement. All p values are two-sided.
Participants in the low-income (LI) group were more likely to be linked to others in this group than to participants in the high-income (HI) group when information was combined across all study sites (p = .045). Three sites had a sufficient number of participants who provided HIV genotype data to be analyzed separately; the site-specific unadjusted analyses showed greater linkage within the LI group (p = .03 for New York; .11 for Atlanta; and .79 for Los Angeles), but no p values achieved significance at the .05 level after adjustment for multiple comparisons.
We next investigated the degree of linkage within and across study sites. Linkage was more likely within than across sites (p = .007). The unadjusted analyses for individual sites yielded the following results: for New York, Atlanta, San Francisco, Los Angeles, Boston, and Washington, DC, p values were .20, .75, .17, .07, .23, and .02, respectively. None of the tests reached statistical significance after adjusting for multiple comparisons, but the two sites with nominal p values (<.1; Los Angeles and Washington, DC) had more linkage within than across sites. In addition, linkage analysis demonstrated that participants who seroconverted during the study had sequences that were more likely to be linked to sequences from participants who were infected at enrollment than to other seroconverters (p = .04).
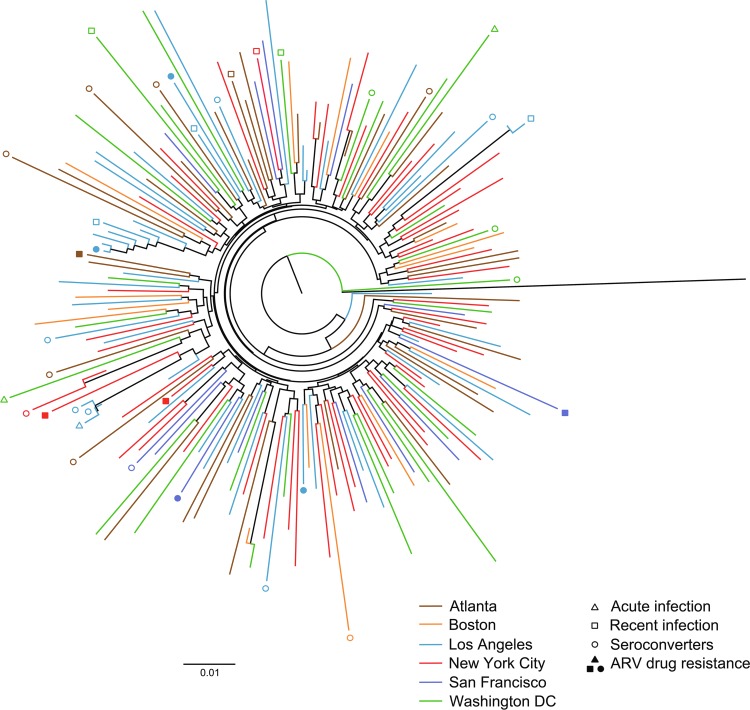
A phylogenetic tree (Fig. 1) reveals the complex relationship of HIV sequences from different study sites. No clustering pattern by site was revealed through visual inspection, although the greater probability of linkage within compared with across sites was noted in the statistical analysis. The phylogenetic tree also shows the relationship between HIV sequences from seroconverters and participants infected at enrollment. Participants who had recent infection at enrollment and participants who had HIV drug-resistant genotypes are also indicated.
FIG. 1.
Phylogenetic tree of HIV sequences from HIV Prevention Trials Network 061. The tree shows the relationship among HIV sequences from participants enrolled at different study sites. Data from participants who acquired HIV infection during the study (seroconverters) and from those with HIV drug resistance are noted in the figure.
Linkage analyses can reveal general patterns of transmission, even though they cannot be used to identify specific transmission chains. We found that the participants in the LI group tended to cluster more with each other than with those in the HI group. This pattern was observed for the entire cohort and among participants at sites with a large number of virally sequenced participants. Clustering reflects sampling of the study population and factors that affect probability of transmission, as well as transmission chains themselves; therefore, no direct inferences can be made about such chains. Our findings are consistent with those from other studies that describe the nature of income segregation, which can limit comingling of different groups and can lead to epidemics that are concentrated among subpopulations.8 Although phylogenetic analyses did not reveal any clustering within sites that was statistically significant, formal linkage analysis did demonstrate such clustering across participants in the entire study. We did not find any tendency for seroconverter infections to be linked to other seroconverters; instead, seroconverter infections were more likely to be linked to participants who were infected at study enrollment.
Acknowledgments
The authors thank Dr. Vlad Novitsky for his assistance in creating the genetic linkage matrix. Grant support for HPTN 061 was provided by the National Institute of Allergy and Infectious Disease (NIAID) and by the Office of AIDS Research of the U.S. National Institutes of Health (NIH; Cooperative Agreements U01-AI068613/UM1-AI068613, U01-AI068617/UM1-AI068617, and U01-AI068619/UM1-AI068619). Funding for these analyses was also provided by NIH grants NiAID R37 AI51164 and NIAID UM1 AI06941.
Author Disclosure Statement
I.C. contributed to this article in her personal capacity. The views expressed are her own and do not represent the views of the Health Resources and Services Administration or the United States Government. All other authors declare no competing financial interests exist.
References
- 1. Koblin BA, Mayer KH, Eshleman SH, et al. : Correlates of HIV acquisition in a cohort of Black men who have sex with men in the United States: HIV prevention trials network (HPTN) 061. PLoS One 2013;8:e70413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mayer KH, Wang L, Koblin B, et al. : Concomitant socioeconomic, behavioral, and biological factors associated with the disproportionate HIV infection burden among Black men who have sex with men in 6 U.S. cities. PLoS One 2014;9:e87298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prevention CfDCa. HIV/AIDS. Available at www.cdc.gov/AIDS, accessed May11, 2018
- 4. US Census Fact Finder. Available at https://factfinder.census.gov/faces/tableservices/jsf/pages/productview.xhtml?pid=ACS_16_1YR_CP05&prodType=table (2016) accessed August1, 2018
- 5. Brookmeyer R, Konikoff J, Laeyendecker O, Eshleman SH: Estimation of HIV incidence using multiple biomarkers. Am J Epidemiol 2013;177:264–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen I, Cummings V, Fogel JM, et al. : Low-level Viremia early in HIV infection. J Acquir Immune Defic Syndr 2014;67:405–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen I, Connor MB, Clarke W, et al. : Antiretroviral drug use and HIV drug resistance among HIV-infected black men who have sex with men: HIV Prevention Trials Network 061. J Acquir Immune Defic Syndr 2015;69:446–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pietila A: Not in My Neighborhood: How Bigotry Shaped a Great American City. Ivan R. Dee, Chicago, 2010 [Google Scholar]