Abstract
Recently, L-4-cyanotryptophan has been shown to be an efficient blue fluorescence emitter, with the potential to enable novel applications in biological spectroscopy and microscopy. However, lack of facile synthetic routes to this unnatural amino acid limits its wide use. Herein, we describe an expedient approach to synthesize Fmoc protected L-4-cyanotryptophan with high optical purity (>99%). Additionally, we test the utility of this blue fluorophore in imaging cell-membrane-bound peptides and in determining peptide-membrane binding constants.
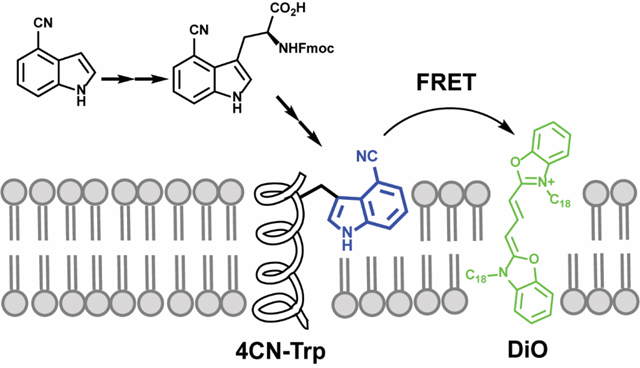
Graphical Abstract
Facile chemical synthesis of L-4CN-Trp and incorporation into a pHLIP peptide enabled FRET study on peptide-membrane interactions.
Fluorescence spectroscopy and microscopy are two of the most prevalent tools used in biochemistry, biophysics and biology, ranging from studying the conformational dynamics of proteins in vitro to imaging live cells.1–3 Since most biological molecules do not afford useful fluorescence properties in this regard, the majority of fluorescence measurements rely on the use of one or multiple extrinsic fluorophores. Therefore, there has been a continuous effort to develop new biological fluorescence reporters that can meet different requirements and goals. For example, over the past few decades many analogues of tryptophan (Trp), which is the most fluorescent native amino acid in proteins, have been examined and explored,4–16 for the purpose of identifying Trp-based, unnatural amino acid (UAA) fluorophores that have improved photophyiscal properties over Trp. While Trp is a useful and convenient intrinsic fluorescence probe of protein structure and dynamics, its application is limited to in vitro spectroscopic studies. This is because (1) its fluorescence quantum yield (QY) is relatively low (<0.2), (2) its fluorescence decay kinetics are complex, (3) its absorption spectrum is in the ultraviolet (UV) region (i.e., <300 nm), and (4) it has low photostability.
Recently, Hilaire et al.13 have shown that a simple derivative of Trp, L-4-cyanotryptophans (L-4CN-Trp), exhibits rather unique absorption and emission properties, making it useful in biological spectroscopic and imaging applications. In comparison to Trp, the much expanded utility of L-4CN-Trp is due to the following factors: (1) it has a larger fluorescence QY (>0.8 in aqueous solution) and, a longer fluorescence lifetime (ca. 13 ns); (2) its absorption and emission spectra are red-shifted, resulting in fluorescence excitation and detection conditions accessible by commercial fluorescence microscopes; and (3) it is more photostable. While the study of Hilaire et al. has validated the potential biological utility of this blue fluorescence amino acid, its wide use is limited by lack of a facile method to reliably synthesize L-4CN-Trp and the corresponding Fmoc- or Boc-protected L-4CN-Trp for solid-phase peptide synthesis.
Herein, we aim to describe a simple, high-yielding, and cost-effective synthetic route for L-4CN-Trp and Fmoc-L-4CN-Trp and demonstrate the utility of L-4CN-Trp fluorescence in the study of peptide-membrane interactions, under both in vitro and in vivo conditions. Indeed, several methods for syntheses of 4CN-Trp or its derivatives have been reported in the literature (Figure 1, Table S1, ESI†).13,17–21 For example, Bartoccini et al.20 described the synthesis of N-acetyl-4CN-Trp methyl ester from 4-pinacol boronated ester 1. However, the product was a racemic mixture and the yield was low. Recently, another racemic synthesis of N-acetyl-4CN-Trp from 4-cyanoindole 2 was reported.21 Since the experimental details were not described in that study, we attempted to reproduce its result by implementation of similar reaction conditions22–24 known for tryptophan analogues but the desired product was obtained in very low yield in our hands. In the study of Hilaire et al., a short synthetic route to produce 4CN-Trp methyl ester from 4-bromo-L-tryptophan 3 in optically pure form was described.13 However, this approach is not ideal in practice due to the low yield in the Pd-catalyzed cyanation step, the use of toxic reagent, and the high cost of the starting material. Alternatively, L-4CN-Trp can be enzymatically synthesized from 4-cyanoindole 2 and serine by tryptophan synthase (TrpS) or a variant of tryptophan synthase subunit B (TrpB) which was recently generated through directed evolution by Arnold and others.17–19 While very useful, this genetic approach is limited to labs with expertise in protein expression as well as the availability of the corresponding enzyme. Thus, devising an efficient and expedient chemical synthesis method of enantiomerically pure L-4CN-Trp is highly desirable.
Figure 1.
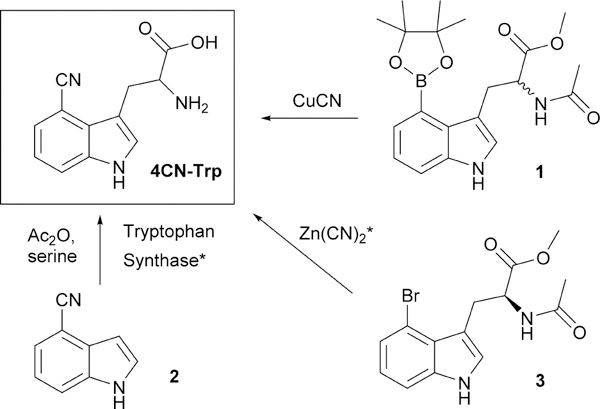
Previous synthetic approaches for 4CN-Trp and its derivatives. * denotes a route to enantiomerically enriched 4CN-Trp.
Among the available methods for preparation of enantiomerically enriched α-amino acids,25 enzymatic hydrolysis of racemic N-acyl amino acid via an acylase has the advantage of being able to mass-produce optically pure, non-proteogenic L-amino acid products.26,27 In addition, production of D-amino acid may be achievable by using of a D-acylase.28 Thus, we set to synthesize the required resolution substrate N-acyl amino acid 6 starting from the previously known amine 4 (Scheme 1) by conventional chemistry. The racemic amine 4 was easily prepared from 4-cyanoindole 2 following a well-established 3-step procedure (i.e., Mannich reaction, alkylation by ethyl nitroacetate, and Zn/AcOH reduction).29 This approach has the advantage of eliminating the use of toxic reagents and enables the preparation of the desired product on multi-gram scale with high efficiency. Next, acetylation provided the N-acetyl ester 5 in 92% yield and the mild hydrolysis by LiOH furnished the racemic N-acetyl carboxylic acid 6 in 89% yield. The pivotal step, enzymatic resolution of the racemic N-acetyl amino acid 6 proceeded smoothly by Amano acylase to provide the desired L-4CN-Trp 7. It is worth noting that the solubility of L-4CN-Trp 7 in aqueous media is negligible and the crude amino acid product could be easily separated from the unreacted material by centrifugation. Production of L-4CN-Trp 7 with high enantiomeric excess (over 99%) was confirmed by HPLC analysis of Nα-(2,4-Dinitro-5-fluorophenyl)-L-alaninamide (FDAA) derivatives (Figure S1, ESI†). Finally, Fmoc protection of L-4CN-Trp 7 under conventional reaction condition provided Fmoc-L-4CN-Trp 8 in good yield (62 % over the two steps).
Scheme 1.
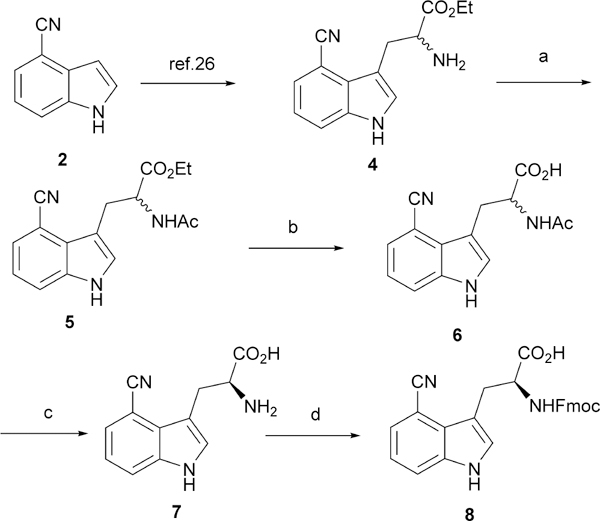
Synthetic route to L-4CN-Trp 7 and Fmoc-L-4CN-Trp 8. Reagents and conditions: a. AcCl, Et3N, 92%; b. LiOH, EtOH, 89%; c. Amano acylase, pH = 8.0; d. Fmoc-Osu, NaHCO3, 62% over c and d.
To further explore the biological utility of L-4CN-Trp, we incorporated it into the pH-(Low) Insertion Peptide (pHLIP) and studied the interaction of the mutant peptide with model and cell membranes using both fluorescence spectroscopic and imaging techniques. The pHLIP peptide (sequence: GGEQNPIYWARYADWLFTTPLLLLDLALLVDADEGT) is a pH-dependent, membrane-interacting peptide designed and well-characterized by Engelman and coworkers.30–34 The pHLIP peptide weakly binds to membranes as unstructured monomers at neutral pH, whereas at pH < 6.5 it forms a transmembrane α-helix. Specifically, we replaced the N-terminal Trp (i.e., Trp9) of pHLIP with L-4CN-Trp. Synthesis of this 4CN-Trp-containing pHLIP peptide (hereafter referred to as 4CN-Trp-pHLIP) via microwave assisted solid phase peptide synthesis using Fmoc-L-4CN-Trp 8 produced the desired pHLIP mutant with a yield that is comparable to that of synthesizing the wide-type pHLIP, hence providing additional support of the success of the synthesis protocol for 7 and 8.
The interaction between 4CN-Trp-pHLIP and membranes was first investigated via fluorescence microscopy (Figure 2). HeLa cells, which were incubated with 4CN-Trp-pHLIP (10 μM) in either pH 7.4 or pH 6.0 PBS buffer for 30 minutes and then thoroughly washed 3 times with the corresponding buffer clearly show that the fluorescence intensity of L-4CN-Trp increases with decreasing pH. These results, which are similar to those obtained with pHLIP peptides labeled with a fluorescent dye,31 not only show that the Trp to L-4CN-Trp mutation does not change the pH-responsive membrane-binding property of pHLIP, but also indicates that the fluorescence of L-4CN-Trp, which was collected through a Leica DM6000 widefield fluorescence microscope using a standard DAPI filter set (i.e., excitation and emission optical filters designed for the fluorescent dye DAPI, a commonly used stain for cell nucleus), is bright enough for biological imaging applications.
Figure 2.
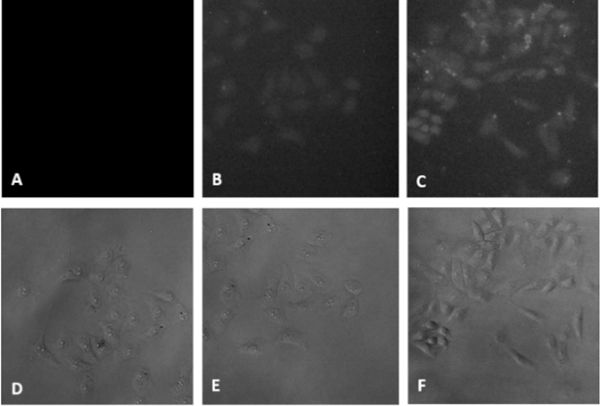
Widefield fluorescence images of HeLa cells treated with PBS buffer (A), L-4CN-Trp-pHLIP at pH 7.4 (B), and 4CN-Trp-pHLIP at pH 6.0 (C). D-F are their corresponding brightfield images.
Next, we sought to demonstrate that the fluorescence of L-4CN-Trp can be used to quantify the peptide-membrane interaction of interest, using 4CN-Trp-pHLIP as a model. The absorption spectrum of 3,3’-dioctadecyloxacarbocyanine perchlorate (DiO), a green lipophilic dye, overlaps significantly with the emission spectra of L-4CN-Trp (Figure S2, ESI†). This, along with the calculated ca. 62 Å Förster distance (R0) in MeOH (see details in ESI†), indicates that L-4CN-Trp and DiO can be used an efficient fluorescence resonance energy transfer (FRET) pair. Because DiO is a universal membrane stain,35 we propose that this FRET pair can be used to assess peptide-membrane interactions. Our working hypothesis is that binding of a L-4CN-Trp-containing peptide to a membrane that is stained with DiO will lead to an observable FRET signal that can be used to assess the underlying binding thermodynamics and kinetics. To test this hypothesis, we first compared the fluorescence spectra of 4CN-Trp-pHLIP (1.0 μM) collected in water (no DiO was added due to its negligible solubility) and in aqueous solutions containing DiO-stained (1%), 100-nm, large unilamellar vesicles (LUVs) made of POPC (1.0 mM) (Figure 3).
Figure 3.
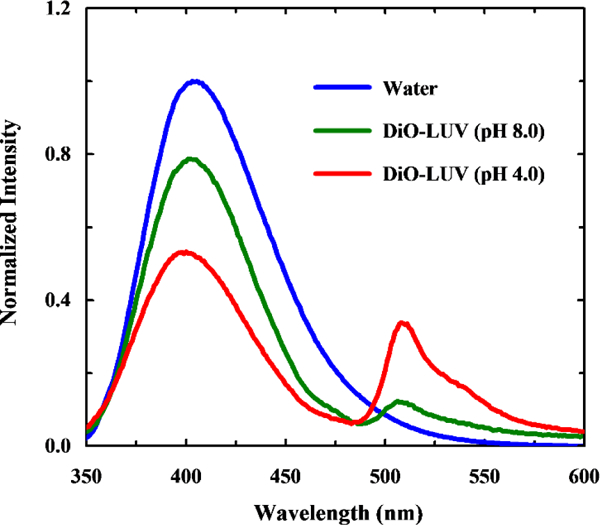
Normalized fluorescence spectra of 4CN-Trp-pHLIP collected in water and DiO stained POPC LUVs of different pH values, as indicated. The excitation wavelength was 320 nm.
When excited at 320 nm, where DiO has a negligible absorbance (Figure S2, ESI†), only the peptide-containing samples show observable FRET signals, indicating peptide binding. In addition, the FRET signal is significantly increased at acidic pH. Since the fluorescence intensity of the donor (i.e., L-4CN-Trp) is essentially independent of pH in the range of the experiments (Figure S3, ESI†), this result is therefore consistent with the designed pH-responsive behavior of pHLIP. To further determine the binding constant of 4CN-Trp-pHLIP to POPC membranes at pH 4.0, we collected the fluorescence spectra of a series of solutions containing 1.0 mM POPC (in the form of 100-nm LUVs stained with 1% DiO) and 4CN-Trp-pHLIP of varying concentrations ([4CN-Trp-pHLIP]). As indicated (Figure S4, ESI†), the FRET signal increases with increasing [4CN-Trp-pHLIP] and starts to level off at ca.30 μM, suggesting that the dissociation constant (Kd) in the range of 20–50 μM. Indeed, fitting the corresponding fluorescence signals (i.e., intensities at 512 nm) to a binding mode previously used by Engelman and co-workers,34 which assumed that 1 pHLIP peptide interacts with 50 lipid molecules, yielded a Kd of ca. 14 μM (Figure 4). This value is in agreement with that previously reported for pHLIP (i.e., Kd = 8 μM at low pH),36 hence validating the usefulness the 4CN-Trp-Dio FRET pair for quantitatively assessing peptide-membrane binding interactions.
Figure 4.
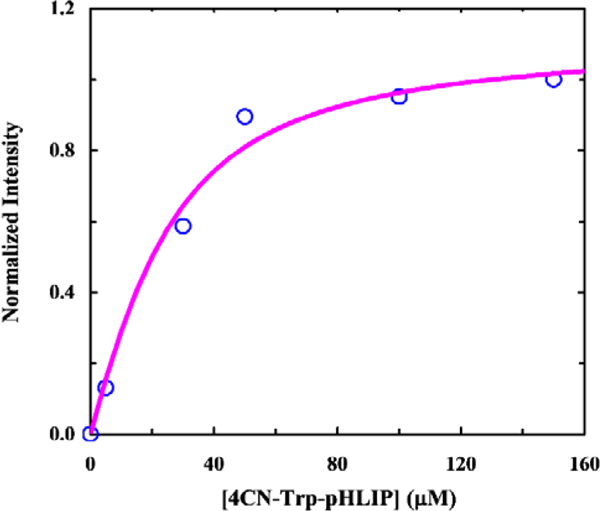
FRET intensity of DiO at 512 nm as a function of 4CN-Trp-pHLIP concentration. Smooth line is the fit of these data to a binding model described in text
While L-4CN-Trp can be incorporated into peptides via solid-phase peptide synthesis, its utility would be significantly expanded if it can also be genetically incorporated into proteins through the protein synthesis machinery of the cell. While the development of such a method is beyond the scope of the current study, we tested the toxicity of L-4CN-Trp to cells. This is because tryptophan analogues could intervene in the tryptophan metabolic pathway, leading to cell death. As indicated (Figure S5, ESI†), at 250 μM L-4CN-Trp does not show any significant growth inhibition of E. coli cells, suggesting that this UAA is amenable for cellular applications.
In conclusion, we developed an improved method for the selective synthesis of L-4CN-Trp and its Fmoc-protected version that can be used for incorporation of this blue fluorescence amino acid into polypeptides via solid phase peptide synthesis. In addition, we demonstrated that this UAA fluorophore is a viable fluorescence reporter to monitor binding of peptides to cell membranes. Moreover, we devised and validated a new fluorescence method to assess peptide-membrane interactions, which is based on the application of a FRET pair (R0 = 62 Å) consisting of L-4CN-Trp (donor) and a universal membrane stain, DiO (acceptor).
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Health grant R35GM122603 for K.K., H.T.K., W.F.D., and H.J., R01GM117593 for W.F.D., D-SPAN fellowship grant F99 NS108544–01 for I.A.A. and by a grant from The University Research Foundation of University of Pennsylvania to F.G. H.T.K. was supported by a Ruth L. Kirschstein NRSA Postdoctoral Fellowship (F32GM125217).
Footnotes
Conflicts of interest
There are no conflicts to declare.
Electronic Supplementary Information (ESI) available: experimental details on chemical synthesis, peptide synthesis, fluorescence measurement, HPLC analysis of optical purity and peptide, copy of 1H NMR and 13C NMR of 5,6, and 8 are available. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Zhang J, Chen S, Ma Y, Wang D, Zhang J, Wang Y, Li W, Yu Z, Zhang H, Yin F, Li Z, J. Mater. Chem. B, 2018, 6, 4065. [DOI] [PubMed] [Google Scholar]
- 2.Xia Q, Chen Z, Yu Z, Wang L, Qu J, Liu R, ACS Appl. Mater. Interfaces, 2018, 10, 17081. [DOI] [PubMed] [Google Scholar]
- 3.Li B, Wen H, Cui Y, Zhou W, Qian G, Chen B, Adv. Mater, 2016, 28, 8819. [DOI] [PubMed] [Google Scholar]
- 4.Smirnov AV, English DS, Rich RL, Lane J, Teyton L, Schwabacher AW, Luo S, Thornburg RW and Petrich JW, J. Phys. Chem. B, 1997, 101, 2758. [Google Scholar]
- 5.Lepthien S, Hoesl MG, Merkel L and Budisa N, Proc. Natl. Acad. Sci. U.S.A, 2008, 105, 16095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross JBA, Senear DF, Waxman E, Kombo BB, Rusinova E, Huang YT, Laws WR and Hasselbacher CA, Proc. Natl. Acad. Sci. U.S.A, 1992, 89, 12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toptygin D, Gronenborn AM and Brand L, Biophys J, 2005, 88, 161a. [Google Scholar]
- 8.Moroz YS, Binder W, Nygren P, Caputo GA and Korendovych IV, Chem. Commun, 2013, 49, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross JBA, Szabo AG and Hogue CWV, Method Enzymol, 1997, 278, 151. [DOI] [PubMed] [Google Scholar]
- 10.Lotte K, Plessow R and Brockhinke A, Photoch. Photobio. Sci, 2004, 3, 348. [DOI] [PubMed] [Google Scholar]
- 11.Talukder P, Chen SX, Roy B, Yakovchuk P, Spiering MM, Alam MP, Madathil MM, Bhattacharya C, Benkovic SJ and Hecht SM, Biochemistry, 2015, 54, 7457. [DOI] [PubMed] [Google Scholar]
- 12.Markiewicz BN, Mukherjee D, Troxler T and Gai F, J. Phys. Chem. B, 2016, 120, 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilaire MR, Ahmed IA, Lin CW, Jo H, DeGrado WF and Gai F, Proc. Natl. Acad. Sci. U.S.A, 2017, 114, 6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbas C, Gregoire BJ, Vanderesse R and Didierjean C, Acta Cryst. E, 2009, 65, O3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markiewicz BN, Mukherjee D, Troxler T and Gai F, J. Phys. Chem. B, 2016, 120, 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chalyavi F, Gilmartin PH, Schmitz AJ, Fennie MW and Tucker MJ, Angew. Chem., Int. Ed, 2018, 57, 7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boville CE, Romney DK, Almhjell PJ, Sieben M and Arnold FH, J. Org. Chem, 2018, 83, 7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romney DK, Murciano-Calles J, Wehrmuller JE and Arnold FH, J. Am. Chem. Soc, 2017, 139, 10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winn M, Francis D and Micklefield J, Angew. Chem., Int. Ed, 2018, 57, 6830–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartoccini F, Bartolucci S, Mari M and Piersanti G, Org. Biomol. Chem, 2016, 14, 10095–10100. [DOI] [PubMed] [Google Scholar]
- 21.van Wilderen LJGW, Brunst H, Gustmann H, Wachtveitl J, Broos J and Bredenbeck J, Phys. Chem. Chem. Phys, 2018, 20, 19906–19915. [DOI] [PubMed] [Google Scholar]
- 22.Yokoyama Y, Hikawa H, Mitsuhashi M, Uyama A, Hiroki Y and Murakami Y, Eur. J. Org. Chem, 2004, 2004, 1244. [Google Scholar]
- 23.Blaser G, Sanderson JM, Batsanov AS and Howard JAK, Tetrahedron Lett, 2008, 49, 2795. [Google Scholar]
- 24.Yokoyama Y, Hikawa H, Mitsuhashi M, Uyama A and Murakami Y, Tetrahedron Lett, 1999, 40, 7803. [Google Scholar]
- 25.Najera C and Sansano JM, Chem Rev, 2007, 107, 4584–4671. [DOI] [PubMed] [Google Scholar]
- 26.Duthaler RO, Tetrahedron, 1994, 50, 1539. [Google Scholar]
- 27.Chenault HK, Dahmer J and Whitesides GM, J. Am. Chem. Soc, 1989, 111, 6354. [Google Scholar]
- 28.Tsai YC, Lin CS, Tseng TH, Lee H and Wang YJ, Enzyme Microb. Technol, 1992, 14, 384. [DOI] [PubMed] [Google Scholar]
- 29.Jian L. L. Guo; Nargund Ravi P.; Pasternak Alexander; Yang Lihu; Ye Zhixiong, PCT Int. Appl, 2010051177, 06 May 2010. [Google Scholar]
- 30.Hunt JF, Rath P, Rothschild KJ and Engelman DM, Biochemistry, 1997, 36, 15177. [DOI] [PubMed] [Google Scholar]
- 31.Reshetnyak YK, Andreev OA, Lehnert U and Engelman DM, Proc. Natl. Acad. Sci. U.S.A, 2006, 103, 6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andreev OA, Dupuy AD, Segala M, Sandugu S, Serra DA, Chichester CO, Engelman DM and Reshetnyak YK, Proc. Natl. Acad. Sci. U.S.A, 2007, 104, 7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reshetnyak YK, Segala M, Andreev OA and Engelman DM, Biophys J, 2007, 93, 2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoonens M, Reshetnyak YK and Engelman DM, Biophys J, 2008, 95, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honig MG and Hume RI, J. Cell Biol, 1986, 103, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider CA, Rasband WS and Eliceiri KW, Nat. Methods, 2012, 9, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


