SUMMARY
Though sexual dimorphism is ubiquitous in animals, the means by which sex determination mechanisms trigger specific modifications to shared structures is not well understood. In C. elegans, tail tip morphology is highly dimorphic: While hermaphrodites have a whip-like, tapered tail tip, the male tail is blunt-ended and round. Here we show that the male-specific cell fusion and retraction that generate the adult tail are controlled by the previously undescribed doublesex-related DM gene dmd-3, with a secondary contribution from the paralogous gene mab-3. In dmd-3 mutants, cell fusion and retraction in the male tail tip are severely defective, while in mab-3; dmd-3 double mutants, these processes are completely absent. Conversely, expression of dmd-3 in the hermaphrodite tail tip is sufficient to trigger fusion and retraction. The master sexual regulator tra-1 normally represses dmd-3 expression in the hermaphrodite tail tip, accounting for the sexual specificity of tail tip morphogenesis. Temporal cues control the timing of tail remodeling in males by regulating dmd-3 expression, and Wnt signaling promotes this process by maintaining and enhancing dmd-3 expression in the tail tip. Downstream, dmd-3 and mab-3 regulate effectors of morphogenesis including the cell fusion gene eff-1. Together, our results reveal a regulatory network for male tail morphogenesis in which dmd-3 and mab-3 together occupy the central node. These findings indicate that an important conserved function of DM genes is to link the general sex determination hierarchy to specific effectors of differentiation and morphogenesis.
Keywords: sexual dimorphism, sex determination, sexual differentiation, sex differences, DM domain, DMRT, cell fusion, developmental timing, selector gene
INTRODUCTION
The presence of two morphologically distinct sexes is a nearly universal characteristic of animal species. To bring about sex differences in form, the sex-determination mechanism must regulate specific changes in differentiation and morphogenesis. However, largely as a result of the enormous variation in animal sex-determination pathways, the mechanisms that integrate sexual information with other developmental pathways remain poorly described.
In the nematode C. elegans, one of the most prominent sexual dimorphisms is in tail tip morphology. While the hermaphrodite tail tip is whip-like, the male tail is blunt-ended and harbors several copulatory structures (Sulston et al., 1980; Emmons, 2005). In larvae of both sexes, the tail tip comprises four nested larval cells, hyp8 through hyp11 (referred to here as hyp8–11). In hermaphrodites, this architecture remains static throughout development. In contrast, the male tail tip undergoes dramatic remodeling in the final (L4) larval stage (Sulston et al., 1980; Nguyen et al., 1999). During this morphogenesis, hyp8–11 fuse together and retract anteriorly to form a rounded tail tip. Subsequently, the distinct process of anterior tail retraction occurs, in which the entire tail is pulled anteriorly, leaving the elongated rays in their wake (Sulston et al., 1980; Nguyen et al., 1999).
Defects in male tail tip retraction result in a pointed or “leptoderan” (Lep) tail tip phenotype (Nguyen et al., 1999). Previous work has shown that mutations in both Wnt signaling and the heterochronic (developmental timing) pathways result in Lep phenotypes. Mutations in the Wnt gene lin-44 cause weak tail tip retraction defects (Zhao et al., 2002); additionally, loss of TLP-1, an Sp1-family Zn-finger factor that may act downstream of Wnt signaling, causes pronounced failure of tail tip morphogenesis (Zhao et al., 2002). In males carrying a gain-of-function allele of the heterochronic gene lin-41 (Slack et al., 2000), the retraction program is delayed, resulting in a partially unretracted tail tip. In contrast, lin-41(lf) males have an over-retracted tail, with premature retraction initiating in L3 (Del Rio-Albrechtsen et al., 2006). However, the means by which Wnt signaling and developmental timing converge on tail morphogenesis are not clear. Moreover, the mechanism that brings sex-specificity to tail morphogenesis is unknown.
All sex differences in C. elegans ultimately arise from sex chromosome content: XX in hermaphrodites, X0 in males (Brenner, 1974; Madl and Herman, 1979). Downstream of this primary cue, a regulatory hierarchy controls the activity of the master sexual regulator TRA-1A, a Gli-family transcriptional repressor (Hodgkin, 1987; Zarkower and Hodgkin, 1992). tra-1 activity is necessary and sufficient to generate essentially all somatic sexual dimorphism. Though TRA-1A is expressed in both sexes, it is fully active only in hermaphrodites, where it represses male-specific genes (Zarkower, 2006). Only three direct targets of TRA-1A in the soma are known: mab-3 in the intestine (Yi et al., 2000), egl-1 in the HSN neurons (Conradt and Horvitz, 1999), and ceh-30 in the CEM neurons (Peden et al., 2007; Schwartz and Horvitz, 2007). However, these targets account for only a small subset of sex-specific development and control single-cell-level processes (yolk production and cell death). In contrast, it is not known how tra-1 specifies sex-specific organogenesis, where sexual information must regulate cell fate, differentiation and morphogenesis.
Despite the great variety in sex-determination pathways of animal species, the conservation of DM family genes indicates that these mechanisms may derive from a common ancestor. The DM domain is an unusual DNA-binding Zn-finger initially identified in the Drosophila sex-determination gene doublesex and the C. elegans sexual differentiation gene mab-3 (Erdman and Burtis, 1993; Raymond et al., 1998). Genes of this family have since been implicated in sex-specific development across the animal kingdom. Interestingly, DM genes act at a variety of points in these pathways, from very early steps (e.g., DMY is the primary sex determining cue in Medaka (Matsuda et al., 2002; Matsuda et al., 2007)) to later sex-specific differentiation (e.g., DMRT1 is necessary for differentiation of testes and the germline in mice (Kim et al., 2007a; Kim et al., 2007b)). As a result, the nature of the ancestral, conserved function of DM genes in sex determination and differentiation remains unclear.
In C. elegans, only two of the eleven DM genes predicted from genome sequence, mab-3 and mab-23, have been characterized. As a direct target of tra-1, mab-3 represses yolk production in the male intestine (Shen and Hodgkin, 1988; Yi et al., 2000). mab-3 is also necessary for the male-specific expression of lin-32, a gene that triggers development of the male-specific sensory rays (Zhao and Emmons, 1995; Portman and Emmons, 2000), though this function seems to be indirectly regulated by tra-1 (Ross et al., 2005). mab-23 is also necessary for a variety of male-specific events, including ray sensory neuron patterning and male-specific muscle differentiation (Lints and Emmons, 2002). These sex-specific functions of mab-23 also seem to be indirectly regulated by tra-1. Whether additional DM genes control other sex-specific characteristics in C. elegans is unknown, as is the extent to which DM genes act as the primary effectors of tra-1 function.
Here, we find that a previously uncharacterized DM gene, dmd-3, is necessary for male-specific morphogenesis of the tail tip. Moreover, supplying dmd-3 to the hermaphrodite tail is sufficient to bring about male-like morphogenesis. By coordinating sexual, temporal and spatial information, dmd-3 occupies a critical node in the regulatory network that coordinates tail remodeling. In addition, mab-3 plays a secondary, partially redundant role in tail tip morphogenesis. dmd-3 and mab-3 trigger at least two independent processes necessary for morphogenesis, including the male-specific expression of the cell fusogen EFF-1. Together, our studies identify a critical role for two DM genes in a genetic mechanism that couples sex determination to the sex-specific modification of a set of shared cells.
MATERIALS AND METHODS
C. elegans genetics
Nematode culture was carried out as described (Brenner, 1974). The following mutations were used: lin-44(n1792), lin-41(bx42), lin-41(bx37), lin-41(ma104), mab-3(e1240), eff-1(ok1021), tra-1(e1099), pha-1(e2123), tlp-1(bx85), dmd-3(ok1327), dmd-3(tm2863), him-5(e1490), mab-23(bx118), and ozDf2. lin-41(bx37) and lin-41(bx42) were provided by D. Fitch (New York University) and dmd-3(tm2863) was provided by the National Bioresource Project (S. Mitani, Tokyo Women’s Medical University). All other mutants were obtained from the Caenorhabditis Genetics Center. All strains except those carrying tra-1(e1099) contained him-5(e1490) to increase the frequency of spontaneous males.
Transgenes
The following integrated transgenic strains were used in this study: syIs78 [AJM-1::GFP, unc-119(+)], fsIs2 [dmd-3::YFP, cc::GFP], fsIs3 [dmd-3::YFP, cc::GFP], fsIs7 [pUR13 (E(ht)ΔTRA-1::DMD-3::GFP), cc::GFP], fsIs9 [pUR12 (E(ht)::DMD-3::GFP), cc::GFP], fsIs10 [pUR15 (MAB-3::GFP), cc::GFP] and fsIs12 [pUR14 (E(ht)TRA-1-G→A::DMD-3::GFP), cc::GFP]. The following extrachromosomal transgenic strains were used in the study: fsEx110 [pUR18(E(ht)::nlsGFP, pBX1(pha-1+)], fsEx114 [pUR4 (E(ht)::GFP), pBX1 (pha-1(+))], fsEx118 [pUR5 (E(ht)ΔTRA-1::GFP), pBX1(pha-1(+))], fsEx135 [pJDC41(EFF-1::GFP [translational]), pBX1(pha-1(+))], fsEx136 [pJE3(eff-1::GFP [transcriptional]), fsEx154 [DMD-3::YFP, cc::GFP], fsEx182 [pUR25(E(ht)Δdmd-3Δmab-3::nlsGFP, pBX1(pha-1+)], fsEx183 [pUR17 (EFF-1::GFP::mCherry), pBX1(pha-1+)], fsEx184 [pUR27 (EFF-1Δdmd-3Δmab-3aΔmab-3b::GFP::mCherry), pBX1(pha-1+)]. pJDC41 and pJE3 were generously provided by W. Mohler (Mohler et al., 2002; del Campo et al., 2005). All conclusions drawn in the text were supported by additional independently-derived transgenes.
dmd-3 alleles
The dmd-3(ok1327) deletion removes 926 bp, including the 3´ end of the final dmd-3 exon, leaving a predicted mutant protein in which the 50 C-terminal residues are replaced with 5 novel amino acids. This deletion also removes much of the 3´ UTR of dmd-3. In dmd-3(tm2863), a 407-bp region that comprises much of exons 2 and 3 is replaced with a 10-bp insertion (Fig. 1A). This results in a predicted open reading frame encoding 55 N-terminal amino acids (including all but 6 amino acids of the first DM domain) followed by 12 novel amino acids and a stop codon. Thus, dmd-3(tm2863) is likely to be a molecular null allele. Both dmd-3(ok1327) and dmd-3(tm2863) are recessive and have essentially identical male tail defects.
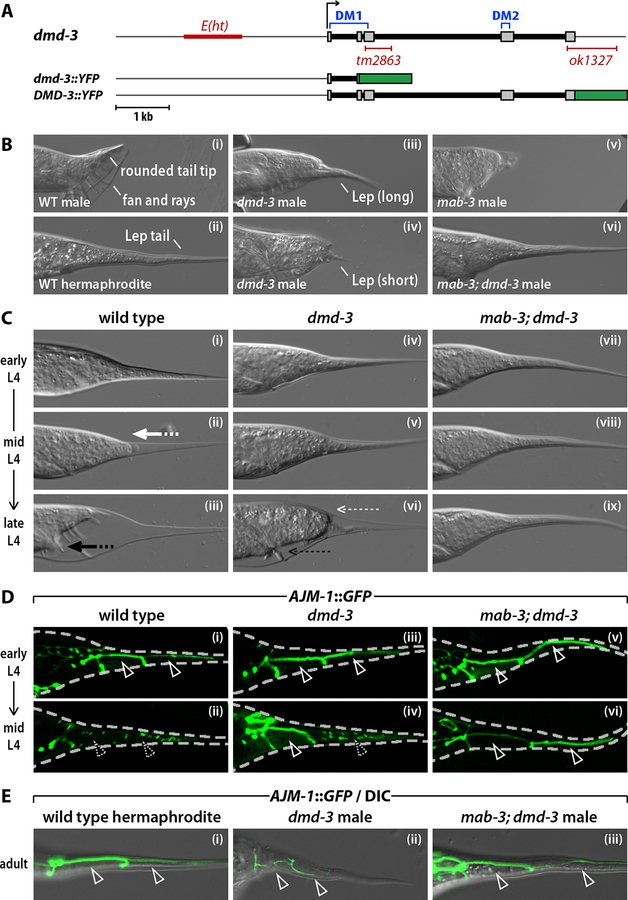
Figure 1. dmd-3 acts with mab-3 to direct cell fusion and retraction of the male tail.
(A) The dmd-3 locus and dmd-3 reporter genes. Thin grey lines indicate the dmd-3 promoter and 3´ UTR; grey boxes mark exons. The two DM domains, the E(ht) enhancer element and the deletions in the dmd-3(tm2863) and dmd-3(ok1327) alleles are indicated. (B) DIC images of the tails of a wild-type adult male (i), wild-type adult hermaphrodite (ii), dmd-3(ok1327) adult male (iii and iv), mab-3(e1240) adult male (v), and mab-3(e1240); dmd-3(ok1327) adult male (vi). Representative examples of the two classes (short and long) of Lep tails observed in dmd-3 males are shown (iii and iv). (C) Individual wild type (i-iii), dmd-3 mutant (iv-vi) and mab-3; dmd-3 double mutant (vii-ix) L4 males were observed periodically by DIC microscopy. Tail tip retraction is indicated with the bold white arrow; anterior tail retraction is indicated with the bold black arrow. Thin white and black arrows indicate the partial tail tip retraction and partial anterior tail retraction, respectively, seen in dmd-3 larvae. (D) Confocal images of wild type (i, ii), dmd-3 mutant (iii, iv) and mab-3; dmd-3 mutant L4 males (v, vi) expressing the apical junction marker AJM-1::GFP. The dashed line indicates the larval cuticle. Solid arrowheads mark intact cell boundaries; dashed arrowheads mark cell fusions. (E) AJM-1::GFP/DIC images of the unretracted tail tips of an adult hermaphrodite (i), an adult dmd-3 male (ii) and an adult mab-3; dmd-3 male (iii).
dmd-3 RNAi
Double stranded dmd-3 RNA was prepared as previously described (Fire et al., 1998) and injected into young adult hermaphrodites. F1 adult male and hermaphrodite offspring were examined for phenotypes.
DNA constructs
For the dmd-3::YFP transcriptional reporter, a genomic fragment from −4264 to +549 bp with respect to the dmd-3 start codon was amplified. For the DMD-3::YFP translational reporter, a genomic fragment from −4264 to +4674 bp was amplified. These fragments were fused to YFP coding sequence by overlap-extension PCR (Boulin et al., 2006). A transcriptional mab-3::GFP reporter (pUR11) was generated by cloning a mab-3 genomic fragment (−8786 to +12 bp) into pPD107.94. A translational MAB-3::GFP reporter (pUR15) was generated by inserting the mab-3 coding region (+13 to +3868 bp) into pUR11.
To generate E(ht)::GFP, the −2740 to −1595 region of the dmd-3 promoter was first cloned into pPD107.94 to generate pUR18. The NLS was then removed by digesting with KpnI and religating to generate E(ht)::GFP (pUR4).
To mutate the putative TRA-1A binding site within E(ht), the 3´ end of the TRA-1A site was replaced with a SalI site by cloning two PCR products into the SphI and XbaI sites in pPD107.94 (WT TRA-1A site: TTTCTGTGTGGGTGTTC, mutant site: TTTCTGTGTGTCGACTC). The NLS was removed to generate E(ht)ΔTRA-1::GFP (pUR5). A point mutation in the TRA-1A site was generated with the QuickChangeII-XL Site-Directed Mutagenesis Kit (Stratagene) using complementary primers that changed the pUR5 TRA-1A site to TTTCTGTGTGAGTGTTC to generate E(ht)TRA-1-G→A::GFP (pUR10).
To express dmd-3(+) from E(ht)::GFP, the dmd-3 coding sequence (with GAAAAA added upstream of the start codon to aid translation) was cloned into pUR4 to generate E(ht)::DMD-3::GFP (pUR12). To put DMD-3::GFP downstream of the mutant TRA-1 sites, the WT E(ht) fragment was removed from pUR12 and replaced with an SphI–XbaI fragment from pUR5 or pUR10 to generate E(ht)ΔTRA-1::DMD-3::GFP (pUR13) and E(ht)TRA-1-G→A::DMD-3::GFP (pUR14) respectively.
The putative DMD-3 and MAB-3 binding sites in E(ht), at −2516 and −2465, respectively, were mutated using pUR18 as the starting template. The putative DMD-3 site was changed from TGTAACA to TGGATCC and the putative MAB-3 site was changed from CCCAACA to CTCGAGA to generate pUR25.
To generate an operon containing the EFF-1::GFP translational reporter followed by an mCherry transcriptional reporter, an outron and mCherry sequence were inserted into the EFF-1::GFP translational reporter 24 bp downstream of the GFP stop codon and 105 bp upstream of the unc-54 3´ UTR. To generate this construct, a NotI site was inserted at this position by mutating pJDC41 from CCGGTCGC to GCGGCCGC to generate pUR16. The outron and mCherry coding sequence were amplified from pENTRY-SrfI-mCherry (a gift from J. White and E. Jorgensen) and cloned into the NotI site to generate pUR17.
The putative DMD-3 binding site (at position −4121 in the eff-1 promoter) and the two putative MAB-3 binding sites (−5501 and −3201) were mutated using pUR17 as the starting template. The putative DMD-3 site was changed from TGCAACA to TGCATGC and the putative MAB-3 sites were changed from CGCAACA to CGGATCC to generate pUR27. Unexpectedly, pUR27 also contained a 11-bp deletion (−3892 to −3882) that does not appear to affect EFF-1::GFP expression.
RT-PCR and RACE
The sequence of the dmd-3 cDNA was determined from RT-PCR products generated using Superscript III One-step RT-PCR w/Platinum Taq (Invitrogen). The 5´ end of the dmd-3 cDNA was determined by sequencing PCR products generated using the 5´RLM-RACE protocol from the First Choice RLM-Race Kit (Ambion).
Microscopy
Images were obtained using a Zeiss Axioplan 2 with epifluorescence illumination and ApoTome structured illumination (Carl Zeiss Microimaging) or by confocal microscopy using a Leica TCS NT. Digital images were processed using Adobe Photoshop. L4 larvae were staged according to linker cell migration, tail tip retraction and anterior tail retraction. In early L4, the linker cell has just completed migrating to the ventral side and the tail tip cells hyp8–11 are unfused and unretracted. In early mid-L4, the linker cell has progressed roughly halfway from its ventral turn to the hindgut and the tail tip is unfused and unretracted. By mid-L4, the linker cell has migrated completely to the hindgut and the tail tip is undergoing cell fusion but not retraction. In late mid-L4, the tail tip is fully fused and retraction is underway. In early late L4, the tail tip is fully retracted but anterior retraction has not yet begun. In late L4, anterior retraction is underway, generating elongated rays and the fan.
MH27 Antibody Staining
Mid-L4 fsIs7; him-5 and fsIs9; him-5 larvae were permeabilized by freeze-cracking (Hurd and Kemphues, 2003) and fixed with methanol (Miller and Shakes, 1995). Larvae were stained with the anti-AJM-1 antibody MH27 (Developmental Studies Hybridoma Bank, University of Iowa) (Francis and Waterston, 1991; Koppen et al., 2001) followed by Texas Red-labelled goat anti-mouse IgG (Jackson Immunoresearch).
RESULTS
dmd-3 and mab-3 are necessary for male-specific tail tip morphogenesis
We identified the previously uncharacterized C. elegans gene Y43F8C.10 in microarray studies of gene expression in the male tail sensory rays (Portman and Emmons, 2004). Y43F8C.10 is predicted to encode a 251-amino acid protein with two DM domains (Fig. 1A). We renamed this gene dmd-3, as the third characterized C. elegans DM-domain gene (Raymond et al., 1998; Lints and Emmons, 2002). Because DM domains have been implicated in sex-specific development, we examined hermaphrodites and males carrying the dmd-3 deletion alleles dmd-3(ok1327) and dmd-3(tm2863) (Fig. 1A). dmd-3 mutant hermaphrodites appeared morphologically wild type. In contrast, dmd-3(ok1327), dmd-3(tm2863) and dmd-3(RNAi) males displayed marked abnormalities in tail morphology. Most notably, they possessed partially unretracted Lep tail tips reminiscent of those of adult hermaphrodites (Fig. 1B, Table 1 and data not shown). This phenotype resulted from a developmental defect in tail tip retraction: the hypodermal cells hyp8–11 failed to pull back completely from the larval cuticle (Fig. 1C). In addition, the anterior region of the tail failed to retract completely in dmd-3 mutants (Figs 1B, C). We observed similar partial retraction defects in dmd-3(ok1327), dmd-3(tm2863), dmd-3(RNAi) and hemizygous dmd-3(ok1327)/ozDf2 males (Table 1 and data not shown), indicating that this is likely the null phenotype.
Table 1.
Tail tip retraction defects in adult males.a
| Genotypeb | n | Wild-type | Mab non-Lep | Lep (short) | Lep (long) | Un-retracted |
|---|---|---|---|---|---|---|
| Wild type | 51 | 100 | 0 | 0 | 0 | 0 |
| dmd-3(ok1327) | 136 | 0 | 0 | 38 | 62 | 0 |
| dmd-3(tm2863) | 59 | 0 | 0 | 44 | 56 | 0 |
| dmd-3(ok1327); fsIs9c | 35 | 86 | 14 | 0 | 0 | 0 |
| tra-1(e1099)d | 31 | 100 | 0 | 0 | 0 | 0 |
| tra-1(e1099); dmd-3(ok1327)d | 102 | 0 | 0 | 7 | 93 | 0 |
| mab-3(e1240) | 26 | 0 | 92 | 8 | 0 | 0 |
| mab-3(e1240); dmd-3(ok1327) | 60 | 0 | 0 | 0 | 0 | 100 |
| mab-3(e1240); dmd-3(ok1327); fsIs10e | 52 | 0 | 0 | 0 | 88 | 12 |
Numbers indicate the percentage of animals that fell into each classification. n, number of animals scored. “Mab non-Lep” indicates animals that did not display tail tip retraction defects but that did have other “male abnormal” (Mab) tail phenotypes. “Lep (short)” indicates animals with a short, partially retracted tail tip (e.g., Fig. 1Biv). “Lep (long)” indicates animals with a long, partially retracted tail tip (e.g., Fig. 1Biii). “Unretracted” indicates animals with a completely unretracted tail tip (e.g., Fig. 1Bvi).
All strains except those carrying tra-1(e1099) also included the mutation him-5(e1490).
fsIs9 is E(ht)::DMD-3::GFP.
Animals carrying tra-1(e1099) are XX pseudomales.
fsIs10 is MAB-3::GFP.
Because null mutations in the DM gene mab-3 have been reported to cause minor, low-penetrance tail tip retraction defects (Shen and Hodgkin, 1988), we constructed mab-3; dmd-3 double mutants (Fig. 1B,C and Table 1). Strikingly, the mab-3; dmd-3 adult male tail was almost identical to that of a hermaphrodite, with a long, smoothly tapered tail tip and no fan or rays. No tail tip or anterior retraction movements were seen in mab-3; dmd-3 L4 males. Thus, dmd-3 and mab-3 act in a partially redundant fashion to bring about male tail tip morphogenesis, with dmd-3 playing the primary role. We also examined mab-23; dmd-3 double mutants, but found no indication that mab-23 also acts redundantly in tail tip morphogenesis (data not shown). Interestingly, the consensus DNA binding site for DMD-3 as determined by in vitro site-selection experiments (M. Murphy and D. Zarkower, pers. comm.) closely matches that of MAB-3 (Yi and Zarkower, 1999), suggesting that the partial functional redundancy between dmd-3 and mab-3 arises from an ability to regulate common downstream targets.
Tail tip retraction is preceded by the male-specific fusion of hyp8–11 (Nguyen et al., 1999). In wild-type, 100% of mid-L4 males (n=28) showed hyp8–11 fusion, compared to 0% of hermaphrodites (n=41). However, in dmd-3 single mutant males, we observed cell fusion in only 44% of mid-L4 males (n=50), and these fusions usually occurred only between hyp9 and hyp10 (Fig. 1D). Again, the loss of mab-3 enhanced this phenotype, such that no hyp8–11 fusions were detectable in mid-L4 mab-3; dmd-3 males (0% of animals showed cell fusion; n=35) (Fig. 1D). In contrast, cell fusion occurred normally in mab-3 single mutants (100%; n=40). In both dmd-3 and mab-3; dmd-3 males, cell boundaries often persisted into adulthood (Fig. 1E). Thus dmd-3 acts with mab-3 to coordinate both tail tip cell fusion and retraction.
dmd-3 and mab-3 are expressed male-specifically in the tail tip coincident with retraction
To better understand how dmd-3 mediates tail tip morphogenesis, we constructed transcriptional (dmd-3::YFP) and translational (DMD-3::YFP) reporter genes (Fig. 1A). These reporters exhibited essentially identical cellular expression patterns, and DMD-3::YFP was able to rescue the dmd-3 tail phenotype (data not shown). In males, we found that dmd-3::YFP was expressed in a number of sexually dimorphic or sex-specific cells, including the tail tip, hindgut, B lineage, ray RnA neurons, and somatic gonad (Figs 2A, C). In contrast, hermaphrodites exhibited strong dmd-3::YFP expression only in the anchor cell (not shown), a hermaphrodite-specific somatic gonad cell that induces development of the vulva (Kimble, 1981). Non-sex-specific expression of these reporters was weak, occurring primarily in the body hypodermis. In addition, expression in phasmid neurons of both sexes was sometimes seen during L3 and L4 (not shown).
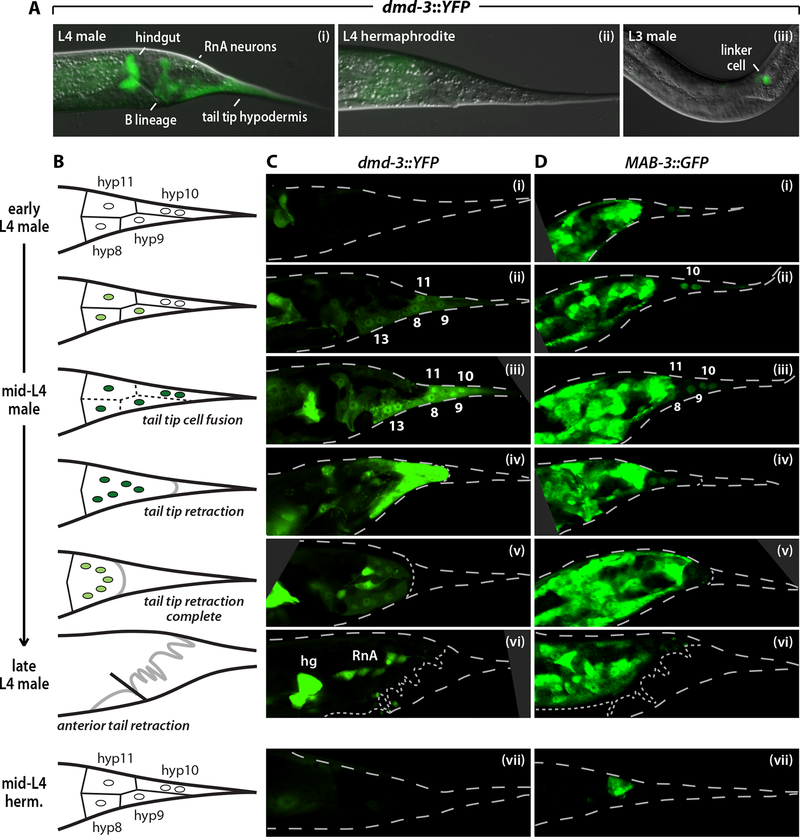
Figure 2. dmd-3 and mab-3 reporters are sex-specifically expressed in the tail tip hypodermis.
(A) dmd-3::YFP (fsIs2) expression in an L4 male (i) demonstrating expression in the hindgut, B lineage, tail tip (hyp8–11) and hyp13. The future position of the dmd-3-expressing RnA neurons is marked with a dashed white line. An L4 hermaphrodite carrying dmd-3::YFP (ii) demonstrates the male-specificity of expression. In late-L3 (iii) and L4 males, dmd-3::YFP is expressed in the linker cell of the developing male gonad. (B) Schematic diagrams of the progression of tail tip morphogenesis. Circles indicate tail tip hypodermal cells; green shading reflects dmd-3 expression. Solid lines between hypodermal cells indicate intact boundaries and dashed lines indicate cell fusion events. The thick black line represents the L4 cuticle, while the thick gray line indicates the newly formed adult cuticle. See Materials and Methods for a description of L4 staging criteria. (C, D) Confocal microscopy images of L4 males (i-vi) and hermaphrodites (vii) carrying dmd-3::YFP (fsIs2) (C) or MAB-3::GFP (fsIs10) (D). Numbers indicate tail tip hypodermal cells. “hg” and “RnA” indicate the hindgut and ray RnA neurons respectively. The larval cuticle is shown with dashed lines; dotted lines indicate the developing adult cuticle.
In hyp8–11, dmd-3::YFP expression was male-specific and coincided with morphogenesis (Figs 2B, C; see Materials and Methods for a description of L4 sub-stages). Tail tip expression initiated in early-mid L4 males, first in hyp8, hyp9 and hyp11, and shortly thereafter in hyp10 (Fig. 2C). We occasionally observed weak expression in hyp9 in late L3 males (see Fig. 4B). Expression levels peaked during tail tip retraction and decreased rapidly upon its completion. dmd-3 was also expressed in hyp13, a male-specific, bi-nucleated hypodermal cell thought not to have a role in tail tip morphogenesis (D.H.A. Fitch, pers. comm.). Importantly, dmd-3::YFP was not expressed in hermaphrodite hyp8–11 at any stage (Fig. 2Cvii). Thus dmd-3 expression parallels tail tip remodeling, consistent with a cell-autonomous, instructive role for dmd-3 in the control of morphogenesis.
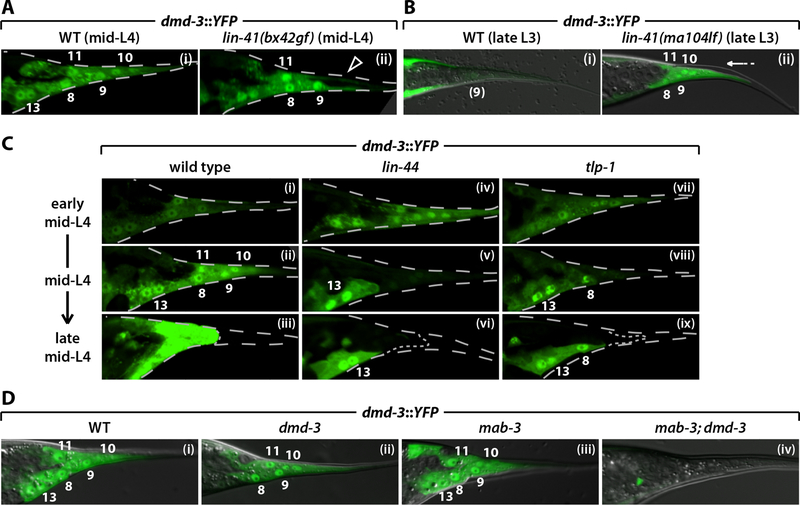
Figure 4. dmd-3 expression in the tail tip is regulated by lin-41, Wnt signaling and a positive feedback loop.
(A) dmd-3::YFP (fsIs3) expression in wild type (i) and lin-41(bx42gf) (ii) mid-L4 males. The arrowhead in (ii) indicates the absence of dmd-3::YFP expression in hyp10 in lin-41(bx42) males. (B) dmd-3::YFP (fsIs3) expression in wild type (i) and lin-41(ma104lf) (ii) late L3 males. The arrow in (ii) highlights the precocious tail tip retraction in lin-41(ma104) males. (C) dmd-3::YFP expression in wild type (i-iii), lin-44(n1792) (iv-vi) and tlp-1(bx85) (vii-ix) L4 males of the indicated stages. (D) dmd-3::YFP (fsIs2) expression in wild type (i), dmd-3(ok1327) (ii), mab-3(e1240) (iii), and mab-3(e1240); dmd-3(ok1327) (iv) mid-L4 males. In (A) and (C), the larval cuticle is shown with dashed lines. Dotted lines indicate the developing adult cuticle.
In contrast, an existing MAB-3::GFP translational reporter (Yi et al., 2000) was not reported to be expressed in the tail tip, nor could it rescue the tail tip retraction defects of mab-3; dmd-3 males (data not shown). We therefore generated a MAB-3::GFP translational reporter carrying 7.3 kb of additional upstream regulatory sequence. This transgene was able to rescue the tail tip retraction defects of dmd-3; mab-3 males to a dmd-3-like phenotype (Table 1). MAB-3::GFP was expressed in numerous cells of the male tail, including weak expression in hyp8–11 (Fig. 2D). In early L4 males, MAB-3::GFP was predominantly found only in hyp10, although hyp9 expression was occasionally observed. By mid-L4, we observed expression in all tail tip hypodermal cells. As with dmd-3, no MAB-3::GFP expression was detected in hyp8–11 in hermaphrodites at any stage (Fig. 2Dvii). Together, these results suggest that dmd-3 and mab-3 act in hyp8–11 to bring about male-specific morphogenesis.
dmd-3 is likely to be a direct tra-1 target
The male-specific expression of dmd-3 in two sets of cells present in both sexes—the tail tip hypodermis and the hindgut—indicated that dmd-3 could be a direct target of repression by TRA-1A in hermaphrodites. Consistent with this, we found that dmd-3 is both expressed during and is necessary for the tail tip retraction that occurs in tra-1(e1099) XX pseudomales (Fig. 3A and Table 1) (Hodgkin, 1987). Interestingly, the severity of the dmd-3 phenotype is slightly exaggerated in tra-1 pseudomales, though the reasons for this are unclear. Nevertheless, these data show that dmd-3 lies genetically and molecularly downstream of tra-1.
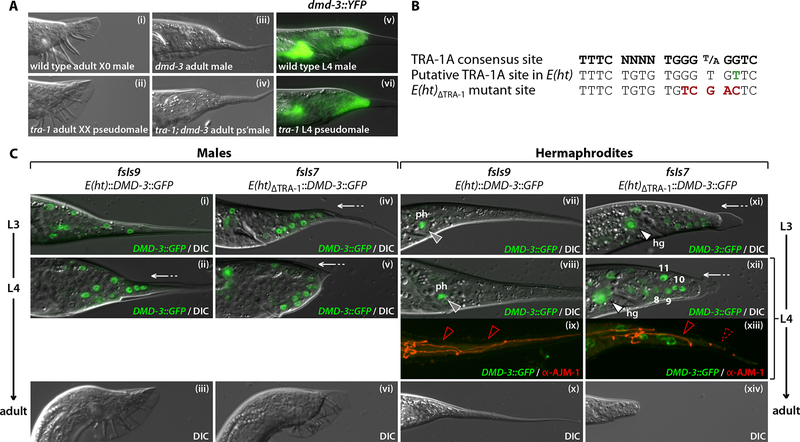
Figure 3. dmd-3 is likely a direct target of TRA-1A and is sufficient to trigger hermaphrodite tail tip retraction.
(A) DIC images of an adult wild type X0 male (i), an adult tra-1(e1099) XX pseudomale (ii), an adult dmd-3(ok1327) X0 male (iii) and an adult tra-1(e1099); dmd-3(ok1327) XX pseudomale (iv). Overlaid dmd-3::YFP/DIC images of a wildtype late mid-L4 X0 male (v) and a tra-1(e1099) late mid-L4 XX pseudomale (vi) expressing dmd-3::YFP (fsIs2). (B) Sequences of the TRA-1A consensus binding site (Zarkower and Hodgkin, 1993; Yi et al., 2000), the putative TRA-1A binding site in the dmd-3 promoter, and the ΔTRA-1 mutation. The single nucleotide difference between the consensus site and the dmd-3 site is shown in green; nucleotides mutated in ΔTRA-1 are shown in red. (C) DMD-3::GFP/DIC (i, ii, iv, v, vii, viii, xi, xii), DMD-3::GFP/MH27 antibody staining (red) (ix, xiii) and DIC (iii, vi, x, xiv) images of late L3 (i, iv, vii and xi), mid-L4 (ii, v, viii, ix, xii, and xiii) and adult (iii, vi, x, xiv) males (i-vi) and hermaphrodites (vii-xiv) carrying E(ht)::DMD-3::GFP (fsIs9) (i-iii and vii-x) or E(ht)ΔTRA-1::DMD-3::GFP (fsIs7) (iv-vi and xi-xiv). Expression of E(ht)ΔTRA-1::DMD-3::GFP in hermaphrodite tail tip hypodermal cells (numbers in xii) and hindgut (hg) (white arrowhead in xi and xii) is indicated. Grey arrowheads in (vii and viii) indicate expression in phasmid neurons (ph). Solid red arrowheads in (ix) and (xiii) mark intact hyp8–11 boundaries, while the dashed red arrowhead in (xiii) indicates cell fusion. White arrows in (ii, iv, v, xi and xiii) mark hyp8–11 retraction.
To ask how tra-1 regulates dmd-3, we first identified a ~1.1 kb region ~1.6 kb upstream of the dmd-3 start codon, E(ht), that was both necessary and sufficient to direct male-specific expression in the hindgut and tail tip (Figs 1A, 3C and data not shown) when placed upstream of the basal promoter Δpes-10 (Seydoux and Fire, 1994). Expression of DMD-3 under the control of E(ht)::Δpes-10 was sufficient to rescue the dmd-3 tail morphology defect (Table 1). Thus E(ht) contains cis-acting elements that direct the sexual and temporal regulation of dmd-3 in the tail tip hypodermis and hindgut.
E(ht) contains a nearly exact match to the TRA-1A consensus binding site (Zarkower and Hodgkin, 1993; Yi et al., 2000), varying at only one nucleotide (Fig. 3B). To disrupt this site, we replaced the central GGTGT with TCGAC to create E(ht)ΔTRA-1::DMD-3::GFP. Strikingly, this change led to clear DMD-3::GFP expression in the L4 hermaphrodite tail tip (93%; n=67 compared to 0%; n=51 for E(ht)::DMD-3::GFP) and hindgut (Fig. 3Cxii). In addition, a single point mutation (TGGG→TGAG) in the putative TRA-1A site led to similar expression in the L4 hermaphrodite tail tip (100%; n=40) and hindgut (data not shown). This specific G→A change has also been demonstrated to disrupt TRA-1A binding in the context of the egl-1 and ceh-30 promoters (Conradt and Horvitz, 1999; Schwartz and Horvitz, 2007). We cannot rule out the possibility that this site indirectly mediates sexual regulation of dmd-3 by TRA-1A. However, together with the finding that TRA-1A can bind to nearly identical sites in vitro (Zarkower and Hodgkin, 1993; Conradt and Horvitz, 1999; Yi et al., 2000), our results indicate that dmd-3 is very likely to be a direct target of repression by TRA-1A in the hermaphrodite tail tip and hindgut.
dmd-3 can bring about male-like tail tip morphogenesis in hermaphrodites
We next took advantage of the expression of E(ht)ΔTRA-1 in both sexes to ask whether providing DMD-3 to the hermaphrodite would be sufficient to masculinize the tail tip. While all adult hermaphrodites carrying the wild-type E(ht)::DMD-3::GFP transgene fsIs9 displayed normal whip-like tail tips (n=88), the mutant E(ht)ΔTRA-1::DMD-3::GFP transgene fsIs7 produced a male-like rounded tail tip in 94% of adult hermaphrodites (n=101) (Fig. 3Cxiv). Consistent with this, the tail tip hypodermal cells of L4 hermaphrodites carrying fsIs7 exhibited clear retraction-like movements and some cell fusion (Fig. 3Cxii, xiii). Thus, sexually dimorphic dmd-3 expression determines the sexual specificity of tail tip morphogenesis.
Unexpectedly, these mutations in E(ht) also disrupted the timing of dmd-3 expression. In both sexes, the expression of DMD-3::GFP from E(ht)ΔTRA-1 and E(ht)TRA-1-G→A initiated prematurely in L2 and L3 larvae (Fig. 3C and data not shown), suggesting that the regions mediating sexual and temporal input overlap in the dmd-3 promoter. In contrast, only minor premature expression was seen in L3 and early L4 males carrying the wild-type transgene; this effect likely results from increased positive autoregulation caused by DMD-3 overexpression (see below).
The premature expression of these transgenes also demonstrated that dmd-3 activity was sufficient to trigger retraction at an inappropriate time. fsIs7 induced precocious tail tip retraction in both male and hermaphrodite L3 larvae, such that essentially all fsIs7 L4 males and many L4 hermaphrodites had clearly pre-retracted tail tips (Fig. 3C), a phenotype not seen in fsIs9 L4 males. Furthermore, adult fsIs7 males exhibited a clear “over-retraction” phenotype (Del Rio-Albrechtsen et al., 2006) (Fig. 3Cvi). Thus, dmd-3 expression in the tail tip can provide an instructive cue for morphogenesis regardless of sex or developmental stage.
Wnt signaling and heterochronic genes regulate tail tip morphogenesis through dmd-3
Both Wnt and heterochronic genes are necessary for normal male tail tip morphogenesis (Zhao et al., 2002; Del Rio-Albrechtsen et al., 2006). However, the mechanisms underlying these functions are unknown. We therefore tested the possibility that these phenotypes result from misregulation of dmd-3. To ask whether the heterochronic gene lin-41 regulates dmd-3, we examined dmd-3::YFP in lin-41 mutants. Temporally delayed lin-41(bx42gf) males have a Lep phenotype (Del Rio-Albrechtsen et al., 2006). We found that dmd-3::YFP expression in these mutants initiated at the correct time in hyp8, hyp9 and hyp11, but was frequently absent from hyp10 even into late L4 (Fig. 4A). We observed this hyp10-specific expression defect in 89% of lin-41(bx42) mid-L4 males (n=27) and 33% of lin-41(bx37) mid-L4 males (n=48), but never in wild-type mid-L4 males (n=35). Interestingly, hyp10 is generally the only cell that fails to fuse in lin-41(gf) mutants (Nguyen et al., 1999; Del Rio-Albrechtsen et al., 2006). As the lack of dmd-3 expression specifically in hyp10 is characteristic of early-mid L4 wild-type males (Fig. 2Cii), we interpret the lin-41(gf) phenotype to be a defect in the maturation of dmd-3 expression. Conversely, we observed strong expression of dmd-3::YFP in the pre-retracting tail tips of lin-41(ma104lf) (Slack et al., 2000) L3 males (87%; n=15, compared to 0%; n=22 for wild-type) (Fig. 4B). This indicates that the early retraction in these animals (Del Rio-Albrechtsen et al., 2006) likely results from premature dmd-3 expression. Consistent with this, we found that the premature-retraction phenotype of lin-41(lf) males (95% L4 pre-retraction tail; n=80) was suppressed in lin-41; dmd-3 (13%; n=82) and lin-41; mab-3; dmd-3 (2%; n=54) mutants. Thus lin-41 controls the stage-specificity of tail tip morphogenesis by regulating dmd-3 expression.
We also examined the effects of a mutation in the Wnt ligand lin-44 (Herman et al., 1995) on dmd-3::YFP expression. In these animals, tail tip dmd-3::YFP expression initiated at the correct time, but was not sustained (Fig. 4C), such that we observed defects in dmd-3::YFP expression in some or all tail tip cells in 98% of mid-L4 lin-44 males (n=40), compared to 0% in wild-type (n=36). However, the Lep defect of lin-44 mutants is subtle, and we found that its severity was enhanced by a mutation in mab-3 (data not shown), suggesting that mab-3 can compensate for the reduction of dmd-3 expression in lin-44 males. Males carrying a mutation in the putative Wnt effector tlp-1 (Zhao et al., 2002) displayed a dmd-3::YFP expression defect similar to that of lin-44 males: initial expression was normal but it was not properly maintained in most mid-L4 males (75%, n=72) (Fig. 4C). These results indicate that tail tip dmd-3 expression is regulated in two distinct phases: an initial, Wnt-independent induction of dmd-3 expression, followed by Wnt-dependent maintenance and amplification.
An autoregulatory loop is important for tail tip morphogenesis
To determine whether dmd-3 autoregulation contributes to the maintenance phase of dmd-3 expression, we examined dmd-3::YFP in dmd-3, mab-3 and dmd-3; mab-3 mutants (Fig. 4D). We observed a subtle decrease in dmd-3::YFP expression in hyp8–11 in dmd-3 males. In contrast, mab-3 mutant males exhibited wild-type levels of dmd-3::YFP tail tip expression. More clearly, dmd-3::YFP expression was essentially abolished in hyp8–11 of mab-3; dmd-3 mid-L4 males. Therefore, dmd-3 and mab-3 are necessary for strong dmd-3 expression, and, at least in mab-3 mutants, dmd-3 has a positive autoregulatory function. To ask whether this autoregulation might occur through direct activation by DMD-3 itself, we identified and disrupted two candidate DMD-3/MAB-3 binding sites in the E(ht) region. However, mutating these sites did not result in a loss of hyp8–11 expression (not shown). Thus, dmd-3-dependent expression of dmd-3 may be mediated through intermediate regulators. As maintenance-phase expression of dmd-3 requires lin-44 and tlp-1, it is possible that dmd-3 activates a Wnt signal that then directly promotes dmd-3 expression.
dmd-3 and mab-3 activate sex-specific expression of the cell fusogen EFF-1
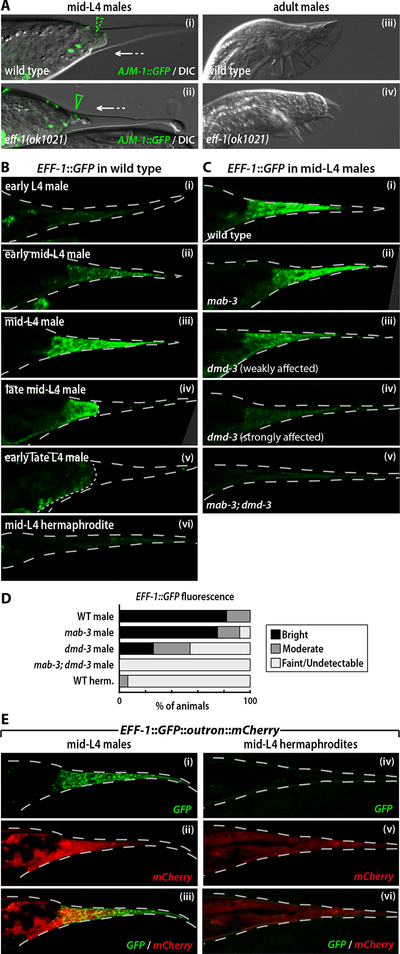
Though tail tip morphogenesis involves both cell fusion and retraction, it is unclear whether these two steps occur independently or, alternatively, if retraction is simply a consequence of cell fusion. To investigate this, we asked whether eff-1, the primary regulator of cell fusion in C. elegans (Mohler et al., 2002; Shemer et al., 2004; Podbilewicz et al., 2006), was necessary for hyp8–11 fusion and retraction. Consistent with previous findings (Mohler et al., 2002; Shemer and Podbilewicz, 2003), we detected no hyp8–11 fusion in males carrying the putative null allele eff-1(ok1021) (Fig. 5A). However, retraction proceeded with only subtle abnormalities in these animals, resulting in a non-Lep, blunt-ended tail (Fig. 5A). Thus the fusion of hyp8–11 is not a prerequisite for retraction, indicating that these two events are regulated in parallel.
Figure 5. EFF-1 is regulated by dmd-3 and mab-3.
(A) AJM-1::GFP (syIs78)/DIC and DIC images of wild type and eff-1(ok1021) L4 (i, ii) and adult (iii, iv) males. The dashed green arrowhead indicates cell fusion, while the solid green arrowhead marks intact cell boundaries. The white arrows indicate tail tip retraction. (B) Tail tip expression of an EFF-1::GFP translational reporter (fsEx135) in male (i-v) and hermaphrodite (vi) larvae. (C) Expression of EFF-1::GFP in wild type (i), mab-3 (ii), dmd-3 (iii and iv) and dmd-3; mab-3 (v) mid-L4 males. The examples in (iii) and (iv) represent the range of EFF-1::GFP expression levels seen in dmd-3 mutants. (D) Categorization of EFF-1::GFP fluorescence intensity in wild type (n=39), mab-3 (n=24), dmd-3 (n=61) and mab-3; dmd-3 (n=33) mid-L4 males based on confocal images. (E) Expression of EFF-1::GFP::outron::mCherry in a wild type mid-L4 male (i-iii) and hermaphrodite (iv-vi). GFP (i and iv), mCherry (ii and v), and GFP/mCherry overlays (iii and vi) are shown.
Consistent with the requirement of eff-1 for hyp8–11 fusion, we found that an EFF-1::GFP translational reporter (del Campo et al., 2005) was expressed in the male tail tip in a pattern that correlated closely with dmd-3::YFP expression. EFF-1::GFP was transiently expressed in hyp8–11 in mid-L4 males, peaking around the time of cell fusion, but was expressed only very weakly in the tail tip of L4 hermaphrodites (Fig. 5B,D). Interestingly, a transcriptional eff-1::GFP reporter lacking eff-1 coding sequence (Mohler et al., 2002) displayed only limited sex differences in expression (not shown). To further explore the mechanisms of eff-1 regulation, we generated a construct in which the eff-1 promoter drove expression of an artificial operon (Blumenthal, 2005; White et al., 2007) containing EFF-1::GFP coding sequence followed by an artificial “outron” and mCherry coding sequence (EFF-1::GFP::outron::mCherry). In this construct, mCherry fluorescence should reflect transcriptional regulation by the eff-1 promoter, while GFP fluorescence reveals the net influence of transcriptional and post-transcriptional controls on eff-1 expression. We observed mCherry expression in hyp8–11 of both sexes, but at lower levels in hermaphrodites than in males. In contrast, EFF-1::GFP expression was barely detectable in the hermaphrodite (Fig. 5E). Together, these results indicate that the male-specificity of tail tip syncytium formation arises from the regulation of EFF-1, and that this regulation likely occurs through both transcriptional and post-transcriptional mechanisms.
We next asked whether dmd-3 and mab-3 were necessary for the sexual dimorphism in EFF-1::GFP expression (Fig. 5C,D). We found that EFF-1::GFP levels were often reduced in the tail tips of mid-L4 dmd-3 males, consistent with their partial cell fusion defects. In mab-3; dmd-3 double mutants, EFF-1::GFP was always faint or absent, similar to the pattern observed in wild-type L4 hermaphrodites. However, in mab-3 single mutants, EFF-1::GFP was expressed at essentially wild type levels, in agreement with the observation that hyp8–11 fusion is unaffected in these mutants.
To ask whether the transcriptional regulation of eff-1 by dmd-3 and mab-3 might be direct, we searched the eff-1 promoter for candidate DMD-3 and MAB-3 binding sites. We identified three putative DMD-3/MAB-3 elements and disrupted them in the context of the EFF-1::GFP::outron::mCherry reporter. However, this resulted in no detectable change in GFP or mCherry fluorescence in mid-L4 larvae of either sex (data not shown), indicating that the transcriptional activation of eff-1 by DMD-3 and MAB-3 may not be direct.
DISCUSSION
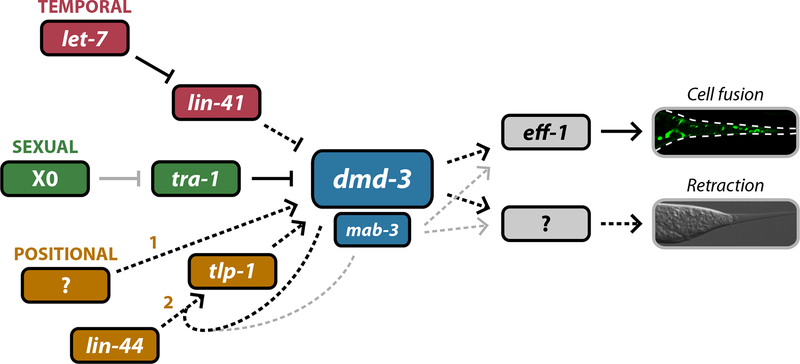
We have found that dmd-3, a previously undescribed member of the DM family, is both necessary and sufficient for male tail tip morphogenesis, a process that generates one of the most prominent sexual dimorphisms in the C. elegans soma. We also find a secondary, partially redundant role of the related gene mab-3 in this process. Together, our findings lead to a model in which dmd-3 instructively specifies tail tip morphogenesis by integrating multiple developmental signals and regulating at least two downstream events (Fig. 6). The temporal control of dmd-3 is specified by the heterochronic pathway through the regulator lin-41. (Mutations in the lin-41 regulator let-7 also cause Lep phenotypes (Del Rio-Albrechtsen et al., 2006), indicating that let-7 also acts in this pathway, though we have not tested this possibility directly.) Positional cues regulate dmd-3 through a Wnt pathway that includes the ligand LIN-44 and its downstream target tlp-1. Interestingly, this cue seems to be most important for the maintenance and amplification of dmd-3 expression; the initial positional or cell-type activator of dmd-3 remains unknown. Finally, the male-specificity of dmd-3 expression arises through regulation by the master sexual regulator TRA-1A.
Figure 6. dmd-3 and mab-3 occupy the central node of the tail tip retraction network.
Our results support a model in which multiple upstream regulatory pathways converge on dmd-3 and mab-3 to regulate the temporal (red), sexual (green) and cell-type (yellow) specificity of tail tip morphogenesis. Temporal specificity is imparted by the heterochronic pathway via let-7 and lin-41. Sexual specificity arises by the regulation of dmd-3 by tra-1, likely to be direct. At least two pathways can be thought of as cell-type determinants: the yellow 1 depicts the induction phase of dmd-3 expression in hyp8–11, while the yellow 2 indicates the maintenance and amplification phase. Downstream of dmd-3 and mab-3 lie multiple effectors of morphogenesis including eff-1. The targets that mediate hyp8–11 retraction are unknown, as is the upstream regulatory pathway that initiates dmd-3 expression (1) in the tail tip. Black arrows indicate regulatory events that are likely to be direct. The solid grey arrow indicates indirect regulation. Dashed arrows indicate steps for which the molecular mechanism is unknown. Thin grey arrows indicate that the function of mab-3 in tail tip morphogenesis is secondary to that of dmd-3.
The phenotype of mab-3; dmd-3 double mutants indicates that the functions of these two genes partially overlap. Though only a small percentage of mab-3 males have Lep defects, mab-3 enhances the phenotype of every animal in a dmd-3 background (Table 1). In addition, overexpression of mab-3(+) in a mab-3; dmd-3 double mutant can sometimes rescue animals to a nearly wild-type phenotype (not shown). As the in-vitro selected binding site for MAB-3 closely resembles that of MAB-3 (M. Murphy and D. Zarkower, pers. comm.) (Yi and Zarkower, 1999), we interpret this redundancy to reflect a partial overlap in the set of target genes that DMD-3 and MAB-3 can regulate.
The clear loss of sex-specificity in the hindgut and tail tip upon disruption of the putative TRA-1A site in dmd-3 provides strong evidence that dmd-3 is a direct target of TRA-1A. This site is a close match to the consensus site for binding in vitro, differing at only the +15 position (Zarkower and Hodgkin, 1993; Yi et al., 2000). We note that this +15 position is likely to be important for TRA-1A binding in vitro and in the TRA-1A target egl-1 (Zarkower and Hodgkin, 1993; Conradt and Horvitz, 1999). However, in contrast to egl-1, the TRA-1A site in dmd-3 matches the TRA-1A consensus at every other position, making it possible that the +15 site is less important in the context of the dmd-3 promoter. The C. briggsae dmd-3 ortholog, Cbr-dmd-3, also contains a strong match to the TRA-1A consensus site within a 50-bp conserved region of upstream sequence (not shown). Though we cannot rule out the possibility that sexual regulation of dmd-3 occurs indirectly through a site closely resembling that of TRA-1A, we consider this to be unlikely.
We believe that the sex-determination and heterochronic pathways likely converge on a common cis-element in dmd-3, since disruption of its TRA-1A site altered both sexual and temporal specificity of dmd-3 expression. These results support the short-range repression model proposed for TRA-1A function (Conradt and Horvitz, 1999; Yi et al., 2000; Zarkower, 2001), in which this factor acts locally to impart local sex-specificity to a single enhancer rather than to the entire locus. An alternative possibility, that sexual and temporal regulation are both mediated by TRA-1A, is unlikely, as precocious tail tip retraction is not observed in tra-1 XX pseudomales (Fig. 3A). Interestingly, a similar phenomenon has been observed in the Hox cluster gene egl-5: disruption of a putative upstream TRA-1A binding site was found to alter the sexual, temporal and spatial specificity of egl-5 expression in seam cells (Teng et al., 2004). Thus the overlap of TRA-1A sites with other regulatory elements may be a common property of sexually regulated genes.
In contrast to sexual regulation, our results indicate that the regulation of dmd-3 by lin-41 may be indirect. Previous work has indicated that lin-41 controls its targets post-transcriptionally (Slack et al., 2000). However, since mutating the E(ht) promoter fragment altered the timing of its expression, dmd-3 temporal control is likely to be mediated transcriptionally. Though other known effects of lin-41 on developmental timing are mediated through the transcription factor LIN-29 (Slack et al., 2000), lin-29 mutant males do not exhibit an unretracted tail tip (Euling et al., 1999). Thus it seems likely that an unidentified target of lin-41 (Del Rio-Albrechtsen et al., 2006) regulates dmd-3.
The phenotypes of lin-44 and tlp-1 mutants indicate that Wnt signaling is important for dmd-3 maintenance and amplification, but not for its initial expression. Mutation of tlp-1 leads to a defect in maintenance of dmd-3 expression and a pronounced Lep phenotype. In contrast, though loss of lin-44 leads to a similar dmd-3 expression defect (Fig. 4C), the tail tip retraction phenotypes of these animals are relatively subtle. This could indicate that residual dmd-3 expression in lin-44 mutants is still able to exert a significant level of function. Our finding that mab-3 can enhance the lin-44 phenotype indicates that while dmd-3 is regulated primarily through lin-44, a different Wnt ligand might act preferentially on mab-3. Both of these Wnt signals would likely act primarily through tlp-1.
Because of its relatively simplicity, tail tip retraction serves as an excellent model to explore the links between developmental signals and morphogenesis. We have found that dmd-3 and mab-3 trigger tail tip cell fusion by promoting expression of the fusogen EFF-1, likely indirectly, through both transcriptional and post-transcriptional mechanisms. Since cell fusion and retraction can vary independently in related nematode species (Fitch, 1997), and eff-1 mutant males clearly undergo retraction, dmd-3 and mab-3 must activate additional unknown effectors of morphogenesis. Furthermore, yet other genes are likely to mediate the effects of dmd-3 and mab-3 on the genetically separable process of anterior tail retraction.
How do these findings inform our understanding of the role of DM genes in sexual development? As discussed above, the surprising diversity in the nature of the sex-specific functions of DM factors has made it difficult to understand the basis for their conservation in these processes. Interestingly, dmd-3, mab-3 and dsx all function at the interface between the general sex determination hierarchy and the regulation of specific developmental events. Thus we suggest that the ancestral role of DM genes was to act as cell-autonomous determinants of sexual information, directly linking sex to the modulation of differentiation and morphogenesis. In a primitive system, the differential expression of these genes could have allowed them to act as “selector” genes of sexual information (Mann and Carroll, 2002), much as Hox cluster genes specify positional information. Once this critical function became fixed, upstream regulatory hierarchies could have evolved to allow more complex mechanisms of interpreting the primary sex-determining cue (Wilkins, 1995), giving rise to the present day roles of dmd-3, mab-3 and dsx. The selective expression of DM genes in one sex may also have allowed their functions to be captured in further downstream steps. Further exploration of this unique gene family will undoubtedly shed light onto the intersection of sex determination and developmental patterning.
ACKNOWLEDGMENTS
We are grateful to D. Fitch, S. Emmons, J. Wolff and D. Bohmann for their thoughtful feedback on the manuscript and to K.Y. Lee and S. McGregor for expert technical assistance. The C. elegans Gene Knockout Consortium, the National Bioresource Project of Japan, and the Caenorhabditis Genetics Center (funded by the NIH NCRR) generously provided C. elegans strains. We thank D. Zarkower and J. Wolff for their MAB-3::GFP reporters and for helpful discussions, D. Zarkower and M. Murphy for communicating unpublished data, A. Fire for vectors, W. Mohler and J. del Campo for eff-1 reporters, E. Jorgensen and J. White for their artificial-operon vector, D. Fitch for lin-41 mutants and helpful discussions, D. Hurd for assistance with immunofluorescence and members of the Portman lab for critical reading of the manuscript. This work was supported by University of Rochester startup funding to D.S.P. and by NIH T32CA009636, which supported the postdoctoral training of D.A.M.
REFERENCES
- Blumenthal T (2005). Trans-splicing and operons In WormBook, www.wormbook.org. [DOI] [PubMed] [Google Scholar]
- Boulin T, Etchberger JF and Hobert O (2006). Reporter gene fusions In WormBook, www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B and Horvitz HR (1999). The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell 98, 317–327. [DOI] [PubMed] [Google Scholar]
- del Campo JJ, Opoku-Serebuoh E, Isaacson AB, Scranton VL, Tucker M, Han M and Mohler WA (2005). Fusogenic activity of EFF-1 is regulated via dynamic localization in fusing somatic cells of C. elegans. Curr. Biol 15, 413–423. [DOI] [PubMed] [Google Scholar]
- Del Rio-Albrechtsen T, Kiontke K, Chiou S-Y and Fitch DH (2006). Novel gain-of-function alleles demonstrate a role for the heterochronic gene lin-41 in C. elegans male tail tip morphogenesis. Dev Biol 297, 74–86. [DOI] [PubMed] [Google Scholar]
- Emmons SW (2005). Male development In WormBook, www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman SE and Burtis KC (1993). The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 12, 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euling S, Bettinger JC and Rougvie AE (1999). The LIN-29 transcription factor is required for proper morphogenesis of the Caenorhabditis elegans male tail. Dev. Biol 206, 142–156. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE and Mello CC (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Fitch DH (1997). Evolution of male tail development in rhabditid nematodes related to Caenorhabditis elegans. Syst. Biol 46, 145–179. [DOI] [PubMed] [Google Scholar]
- Francis R and Waterston RH (1991). Muscle cell attachment in Caenorhabditis elegans. J. Cell Biol 114, 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman MA, Vassilieva LL, Horvitz HR, Shaw JE and Herman RK (1995). The C. elegans gene lin-44, which controls the polarity of certain asymmetric cell divisions, encodes a Wnt protein and acts cell nonautonomously. Cell 83, 101–110. [DOI] [PubMed] [Google Scholar]
- Hodgkin J (1987). A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes Dev. 1, 731–745. [DOI] [PubMed] [Google Scholar]
- Hurd DD and Kemphues KJ (2003). PAR-1 is required for morphogenesis of the Caenorhabditis elegans vulva. Dev. Biol 253, 54–65. [DOI] [PubMed] [Google Scholar]
- Kim S, Bardwell VJ and Zarkower D (2007a). Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev. Biol 307, 314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Namekawa SH, Niswander LM, Ward JO, Lee JT, Bardwell VJ and Zarkower D (2007b). A mammal-specific Doublesex homolog associates with male sex chromatin and is required for male meiosis. PLoS Genet. 3, e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J (1981). Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev. Biol 87, 286–300. [DOI] [PubMed] [Google Scholar]
- Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, Rongo C and Hardin JD (2001). Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat. Cell Biol 3, 983–991. [DOI] [PubMed] [Google Scholar]
- Lints R and Emmons SW (2002). Regulation of sex-specific differentiation and mating behavior in C. elegans by a new member of the DM domain transcription factor family. Genes Dev. 16, 2390–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl JE and Herman RK (1979). Polyploids and sex determination in Caenorhabditis elegans. Genetics 93, 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS and Carroll SB (2002). Molecular mechanisms of selector gene function and evolution. Curr. Opin. Genet. Dev 12, 592–600. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Shinomiya A, Kinoshita M, Suzuki A, Kobayashi T, Paul-Prasanth B, Lau EL, Hamaguchi S, Sakaizumi M and Nagahama Y (2007). DMY gene induces male development in genetically female (XX) medaka fish. Proc. Natl. Acad. Sci. USA 104, 3865–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey CE, Shibata N, Asakawa S, Shimizu N, Hori H, Hamaguchi S and Sakaizumi M (2002). DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417, 559–563. [DOI] [PubMed] [Google Scholar]
- Miller DM and Shakes DC. (1995). Immunofluorescence Microscopy. In: Caenorhabditis elegans: Modern biological analysis of an organism. [Google Scholar]
- Mohler WA, Shemer G, del Campo JJ, Valansi C, Opoku-Serebuoh E, Scranton V, Assaf N, White JG and Podbilewicz B (2002). The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev. Cell 2, 355–362. [DOI] [PubMed] [Google Scholar]
- Nguyen CQ, Hall DH, Yang Y and Fitch DH (1999). Morphogenesis of the Caenorhabditis elegans male tail tip. Dev. Biol 207, 86–106. [DOI] [PubMed] [Google Scholar]
- Peden E, Kimberly E, Gengyo-Ando K, Mitani S and Xue D (2007). Control of sex-specific apoptosis in C. elegans by the BarH homeodomain protein CEH-30 and the transcriptional repressor UNC-37/Groucho. Genes Dev. 21, 3195–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbilewicz B, Leikina E, Sapir A, Valansi C, Suissa M, Shemer G and Chernomordik LV (2006). The C. elegans developmental fusogen EFF-1 mediates homotypic fusion in heterologous cells and in vivo. Dev. Cell 11, 471–481. [DOI] [PubMed] [Google Scholar]
- Portman DS and Emmons SW (2000). The basic helix-loop-helix transcription factors LIN-32 and HLH-2 function together in multiple steps of a C. elegans neuronal sublineage. Development 127, 5415–5426. [DOI] [PubMed] [Google Scholar]
- Portman DS and Emmons SW (2004). Identification of C. elegans sensory ray genes using whole-genome expression profiling. Dev. Biol 270, 499–512. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J and Zarkower D (1998). Evidence for evolutionary conservation of sex-determining genes. Nature 391, 691–695. [DOI] [PubMed] [Google Scholar]
- Ross JM, Kalis AK, Murphy MW and Zarkower D (2005). The DM domain protein MAB-3 promotes sex-specific neurogenesis in C. elegans by regulating bHLH proteins. Dev Cell 8, 881–892. [DOI] [PubMed] [Google Scholar]
- Schwartz HT and Horvitz HR (2007). The C. elegans protein CEH-30 protects male-specific neurons from apoptosis independently of the Bcl-2 homolog CED-9. Genes Dev. 21, 3181–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G and Fire A (1994). Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development 120, 2823–2834. [DOI] [PubMed] [Google Scholar]
- Shemer G and Podbilewicz B (2003). The story of cell fusion: big lessons from little worms. Bioessays 25, 672–682. [DOI] [PubMed] [Google Scholar]
- Shemer G, Suissa M, Kolotuev I, Nguyen KC, Hall DH and Podbilewicz B (2004). EFF-1 is sufficient to initiate and execute tissue-specific cell fusion in C. elegans. Curr. Biol 14, 1587–1591. [DOI] [PubMed] [Google Scholar]
- Shen MM and Hodgkin J (1988). mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell 54, 1019–1031. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR and Ruvkun G (2000). The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5, 659–669. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Albertson DG and Thomson JN (1980). The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev. Biol 78, 542–576. [DOI] [PubMed] [Google Scholar]
- Teng Y, Girard L, Ferreira HB, Sternberg PW and Emmons SW (2004). Dissection of cis-regulatory elements in the C. elegans Hox gene egl-5 promoter. Dev. Biol 276, 476–492. [DOI] [PubMed] [Google Scholar]
- White JQ, Nicholas TJ, Gritton J, Truong L, Davidson ER and Jorgensen EM (2007). The sensory circuitry for sexual attraction in C. elegans males. Curr Biol 17, 1847–1857. [DOI] [PubMed] [Google Scholar]
- Wilkins AS (1995). Moving up the hierarchy: a hypothesis on the evolution of a genetic sex determination pathway. Bioessays 17, 71–77. [DOI] [PubMed] [Google Scholar]
- Yi W, Ross JM and Zarkower D (2000). mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development 127, 4469–4480. [DOI] [PubMed] [Google Scholar]
- Yi W and Zarkower D (1999). Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms. Development 126, 873–881. [DOI] [PubMed] [Google Scholar]
- Zarkower D (2006). Somatic sex determination In WormBook, www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D (2001). Establishing sexual dimorphism: conservation amidst diversity? Nat. Rev. Genet 2, 175–185. [DOI] [PubMed] [Google Scholar]
- Zarkower D and Hodgkin J (1992). Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell 70, 237–249. [DOI] [PubMed] [Google Scholar]
- Zarkower D and Hodgkin J (1993). Zinc fingers in sex determination: only one of the two C. elegans Tra-1 proteins binds DNA in vitro. Nucleic Acids Res. 21, 3691–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C and Emmons SW (1995). A transcription factor controlling development of peripheral sense organs in C. elegans. Nature 373, 74–78. [DOI] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Fitch DH and Herman MA (2002). TLP-1 is an asymmetric cell fate determinant that responds to Wnt signals and controls male tail tip morphogenesis in C. elegans. Development 129, 1497–1508. [DOI] [PubMed] [Google Scholar]