Abstract
In this work, we report selection of covalent linker on anomeric position of N-azidoacetylmannosamine (ManNAz) for caging its metabolic process. We synthesized DT-diaphorase responsive metabolic precursor HQ-NN-AAM using the optimized linker. The caged metabolite showed responsiveness to DT-diaphorase in vitro, resulting in metabolic incorporation of azido sugar on cell surface in multiple cell lines.
TOC image
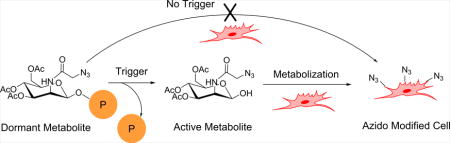
The research of metabolic glycoengineering has evolved rapidly over the last decades as it utilizes endogenous enzymes to process unnatural glycan substrates and introduces chemical functional groups onto proteoglycans and glycoproteins.1–3 Being combined with bioorthogonal Click chemistry,4, 5 these chemical anchors serve as powerful tools to understand and study glycans both on cell surfaces and in the cytoplasm, which result in improved understanding about proteomics,6–8 cell-cell interaction,9 and cell metastasis.10 Metabolic incorporation of unnatural sugar have opened up new directions in applied science, especially drug delivery and tumor imaging.11–15 One of the most studied unnatural sugar N-azidoacetylmannosamine (ManNAz) shares the same metabolic process with N-acetylmannosamine.6 The native hexose undergoes several steps of enzymatic processing and yields cytidine monophosphate-sialic acid (CMP-Neu5Ac) which is conjugated to cell surface glycans through post-translational modification in Golgi apparatus.3 This metabolic process is shared by most mammalian cell types, so the use of ManNAz to selective label specific types of cells has been proven to be difficult without interfering with their cellular metabolic machinery.3
Currently, there are a few reports on controlling the metabolic labelling of ManNAz through different strategies. Cellular uptake levels determine the amount of potential unnatural sugars that can be metabolized in specific cells. Thus, researchers have developed delivery methods to enhance cellular uptake16, 17 or spatial availability of ManNAz towards cells of interest.13 Since ManNAz needs to go through metabolic processes inside cells, modification with caging groups on different positions of the unnatural hexose, namely anomeric14 or C-6 hydroxy groups,18, 19 would provide extra control on its metabolic process. The metabolic precursor usually consists of three components: a metabolic labeling agent, a self-immolative linker and a trigger-cleavable cage group.20 These metabolic precursors have been demonstrated to have great potential in cancer imaging19 and targeted delivery of drugs.14 However, in some cases, the self-immolative linker has compromised stability in physiological condition, which results in cleavage of the caged group even when their respective trigger was absent.18, 19 For example, it is reported that a caged precursor with peptide conjugated to Ac3ManNAz through carbonate bond on C-6 position showed nonenzymatic carbonate hydrolysis in the absence of trigger enzyme in degradation studies.18 Clearly, it is challenging to achieve trigger specific metabolic labeling with these existing linkers which can be cleaved nonspecifically in cellular environment. Our group previously developed a selective labeling technique which demonstrated that using ether linkages on the anomeric position of ManNAz successfully controlled the labeling process of ManNAz,14 achieving selective metabolic labeling of cancer cells based on their different enzyme activity. However, the anomeric ether bond formation requires strong acidic condition, which hinders incorporation of acid labile caging groups such as Boc-protected oligopeptides. Therefore, it is important to design metabolic precursors with controlled decaging processes and feasibility to incorporate wide range of trigger responsive functionalities.
Herein, we report a new metabolic precursor (Scheme 1), HQ-NN-AAM, which contains a DT-diaphorase (DTD) responsive trimethyl quinone moiety (HQ)21, 22 and a self-immolative dimethylethylenediamine (NN) linker attached to the anomeric position of ManNAz through N-methyl carbamate linkage.23, 24 The linkage was selected from our study of unspecific cleavage of various common linkers in typical cellular environments. The self-immolative linker also enables mild synthetic condition and avoids harsh acidic glycosylation reactions to couple trigger groups onto the anomeric position. HQ-NN-AAM was utilized to label multiple cell lines and incorporation of azido groups was achieved on cell surfaces. The labelling activity was suppressed when DTD activity was inhibited, demonstrating the trigger-specific metabolic activity of HQ-NN-AAM.
Scheme 1.
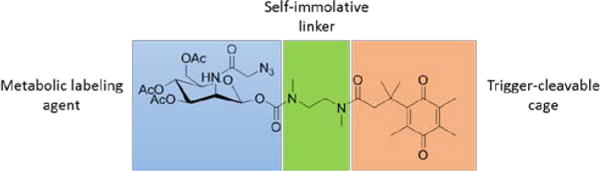
Scheme of three components of caged metabolite HQ-NN-AAM
To achieve controlled labeling with trigger responsiveness, two main requirements of the caging groups have to be met. First, the caged precursor should be efficiently de-caged under the designated trigger condition. Second, the cage group must resist non-specific degradation in a complex cellular environment when the trigger is absent (Figure 1a). We synthesized four model compounds (Figure 1b) to study suitable types of bond linkage on anomeric position of ManNAz that can resist cleavage from the complex subcellular environment. The anomeric hydroxyl position of ManNAz were caged with acetyl (Ac-AAM, ester linkage), ethoxycarbonyl (Et-O-AAM, carbonate linkage), N-butylcarbamate (Bu-N-AAM, carbamate linkage), and N-methyl-N-((Bocmethylamido)ethyl)carbamate (Boc-NN-AAM, N-methyl carbamate linkage), respectively. We incubated these four caged sugars with a panel of six different cell lines at concentration of 50 μM for three days. The cells lines include Bxpc-3 (pancreatic cancer cells), Miapaca-2 (pancreatic cancer cells), A549 (lung cancer cells), MDA-MB-231 (breast cancer cells), HEK293 (embryonic kidney cell), and IMR90 (fibroblast cells). The azide labeling in all cells were analyzed by flow cytometry using DBCO-Cy5 as a probe. The cell surface azido glycan density was quantified through average Cy5 fluorescence intensity per cell. The labeling results (Figure 1c) showed that the ester linkage (Ac-AAM) and the carbonate linkage (Et-O-AAM) could not block the metabolic process, resulting in high metabolic labeling level in all six cell lines presumably due to the non-specifically hydrolysis of the ester and the carbonate bonds by intracellular esterses.2, 25 Carbamate linkage (Bu-N-AAM), on the other hand, showed partial block of the labeling, with reduced Cy5 fluorescence as compared to Ac-AAM and Et-O-AAM at the same sugar concentration. The N-methyl carbamate linkage (Boc-NN-AAM) showed negligible labeling in all six cell lines, indicating the promise of the linker for blocking the metabolic process of ManNAz. As shown by typical flow cytometry trace (Figure 1d), the cells treated with Boc-NN-AAM showed similar fluorescence as compared to cells without any treatment, indicating the signal was predominantly contributed by passive uptake of DBCO-Cy5. On the other hand, cells treated with Ac-AAM or Et-O-AAM displayed higher fluorescence signal distribution, indicating predominant role of cell surface azido group attaching DBCO-Cy5 through the Click reaction.
Figure 1.
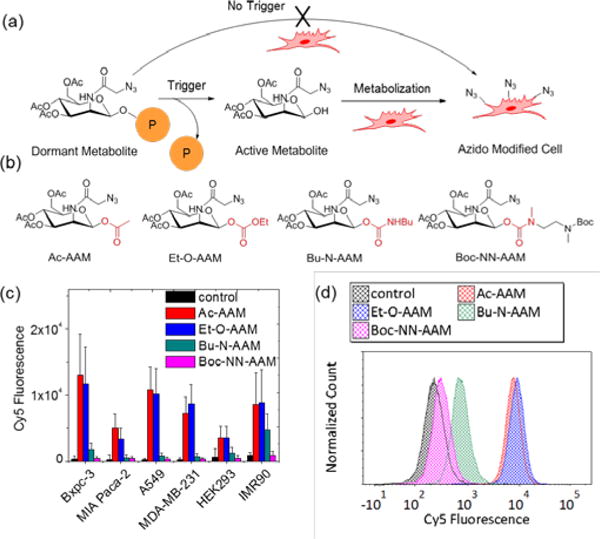
Metabolic labeling of N-azidoacetylmannosamine is blocked by modification of N-methyl carbamate linker on 1-hydroxyl group. a) Schematic illustration of controlled metabolic labeling by caging strategy. The dormant sugar activation depended solely on cellular trigger signal. b) Chemical structures of four control metabolic precursors with different linkers. c) Metabolic labeling of control metabolic precursors (50 μM) in multiple cell lines. d) Representative flow cytometry histogram of metabolic labeling of control precursors in MDA-MB-231 cells.
DTD has been reported to be overexpressed in multiple cancer cell lines and its high activity has been associated with hypoxia and cancer cell aggressiveness.26 Incorporating previous selected best performing N-methyl carbamate linker, we designed a DTD responsive metabolic precursor, HQ-NN-AAM (Scheme 1). The conjugate consists of three major components, the HQ moiety which can be specifically reduced by DTD,21 NN linker with N-methyl carbamate linkage which can self-cyclize when the end amine is exposed,23 and the metabolic active precursor of triacetylated ManNAz (Figure 2a). The synthetic route (Scheme 2) started with HQ-COOH which was prepared according to literature by N-bromosuccinimide (NBS) oxidizing the lactone precursor.21 HQ-COOH was coupled with mono-Boc protected N,N′-dimethylethylenediamine with N,N′-dicyclohexylcarbodiimide (DCC), N-hydroxysuccinimide (NHS) and 4-dimethylaminopyridine (DMAP). The Boc protecting group was then removed in TFA/DCM. Ac3ManNAz-OH was synthesized according to literature report.14 It was then activated using 4-nitrophenol chloroformate to NB-O-AAM and coupled with HQ-NN under mild basic conditions.
Figure 2.
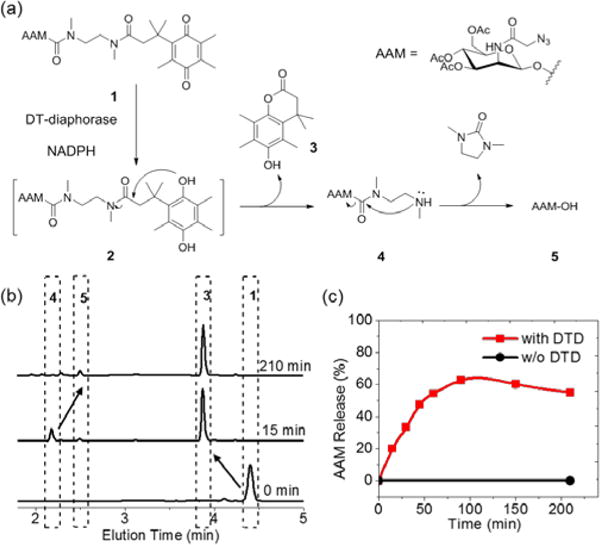
DTD-responsive release of Ac3ManNAz-OH from HQ-NN-AAM. a) Schematic illustration of cascade degradation of HQ-NN-AAM under DTD trigger. b) HPLC trace of HQ-NN-AAM (1), compound 3 (15 min), compound 4 (15 min) and Ac3ManNAz-OH (AAM-OH, 210 min). c) Accumulative release of Ac3ManNAz-OH from HQ-NN-AAM with (red) or without (black) DTD. The slightly lower release of AAM was due to further hydrolysis of AAM-OH.
Scheme 2.
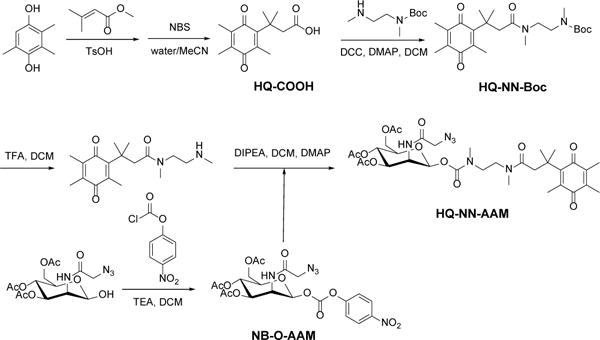
Synthesis of HQ-NN-AAM
After obtaining the caged sugar, we examined the trigger-responsive decaging process in physiological conditions of HQ-NN-AAM by HPLC. The designed decaging mechanism is shown in Figure 2a. After the reduction to hydroquinone by DTD from the trimethylquinone structure, the cage group undergoes cyclization into lactone and exposed the amine group of the NN linker, which also cyclizes into a five membered cyclic urea and expose the anomeric hydroxy group on ManNAz, finishing the decaging cascade process. We characterized this degradation process in mimicked physiological condition (37°C, pH = 7.4). HQ-NN-AAM was first reduced in the presence of DTD and NADPH,27 resulting in hydroquinone 2. The hydroquinone structure cyclized into compounds 3 and 4 in less than 15 min. The products were characterized by LC-MS and HPLC (Figure 2b). Compound 4 then undergoes pH dependent cyclization (Figure S1) and yielded the active metabolic reagent Ac3ManNAz-OH (AAM-OH) (Figure 2b). The accumulative release of AAM into the reaction mixture reached plateau of 60 % at around 2 h and slightly decreased afterwards, presumably because of the further hydrolysis of the pendant acetyl groups with the increased hydrophilicity. In comparison, NADPH was not able to trigger the decaging process of HQ-NN-AAM in the absence of DTD and there was only negligible degradation from hydrolysis of the pendant acetyl groups (Figure 2c, black curve). These results demonstrated that HQ-NN-AAM was able to respond specifically to enzymatic degradation cues and release active metabolite efficiently within a few hours.
We examined the metabolic labeling of HQ-NN-AAM in cell lines with reported high DTD activity. We used MDA-MB-23126 and Bxpc-328 as model cell lines. HQ-NN-AAM showed efficient labeling in cells and the cell surface azide incorporation was confirmed by flow cytometry analysis (Figure 3a). Boc-NN-AAM, which shares the same NN linker with inert Boc end group, showed minimal azide labeling while HQ-NN-AAM was able to efficiently label cells evidenced by the strong DBCO-Cy5 fluorescence (Figure 3b). The metabolic labeling of azido sugar was also confirmed by laser scanning confocal microscopy (Figure 3d). The Cy5 fluorescent intensity and distribution of cells incubated with same concentration of Boc-NN-AAM and HQ-NN-AAM showed distinct differences. The continuous red fluorescence on cell surface (co-localization with membrane stain, Figure S2) resulted from HQ-NN-AAM incubation indicated successful metabolic incorporation of azido sugar because of the decaging of HQ moiety in cells. On the other hand, cells treated with Boc-NN-AAM only showed weak punctuated red fluorescence signal inside cells, which was attributed to background cellular uptake of DBCO-Cy5. The metabolic labeling of HQ-NN-AAM was further validated using western blot (Figure S3). Cell surface azido concentration showed a positive correlation to HQ-NN-AAM concentration ranging from 100 nM to 50 μM (Figure S4). We also performed labeling kinetic studies in MDA-MB-231 cells (Figure S5). HQ-NN-AAM showed similar labeling kinetics as compared to literature reported Ac4ManNAz kinetics,14 reaching an azido concentration plateau at around 48 hours of incubation. This result indicated that the triggered releasing process of HQ-NN-AAM did not delay the metabolic process of ManNAz to cell surface glycan, which was presumably due to the relatively fast release of AAM-OH from HQ-NN-AAM in physiological condition.
Figure 3.
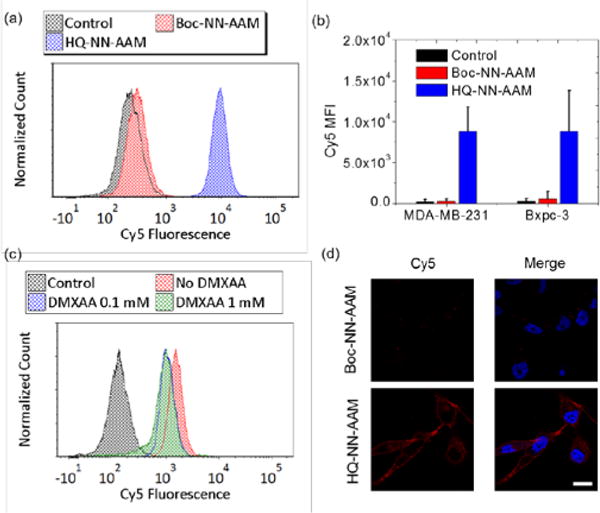
HQ-NN-AAM metabolic labelled cells. a) Flow cytometry histogram of metabolic labeling of HQ-NN-AAM (50 μM) and Boc-NN-AAM (50 μM) in MDA-MB-231 cells. b) Quantification of azido labeling in MDA-MB-231 cells and Bxpc-3 cells c) Flow cytometry histogram of metabolic labeling of HQ-NN-AAM (50 μM) with co-incubation of DTD inhibitor in MDA-MB-231 cells. d) Confocal image of HQ-NN-AAM and Boc-NN-AAM treated MDA-MB-231 cells. Azido groups were labeled with DBCO-Cy5 (red) for visualization. Nucleus were stained with Hoechst dye (blue). Scale bar = 20 μM
We next demonstrated the specificity of HQ-NN-AAM in response to endogenous DTD using inhibition assay. 5,6-Dimethylxanthenone-4-acetic acid (DMXAA) was used as an inhibitor of enzymatic activity of DTD.27 The surface azide concentration of MDA-MB-231 cells from incubation with HQ-NN-AAM reduced after the pre-treatment with DMXAA (Figure 3c), while the labeling of Ac4ManNAz was not interfered by the inhibitor (Figure S6). Western blot also confirmed the reduction of total azido sugar glycosylation level in cells with DMXAA treatment (Figure S7). The residue metabolic labeling at high concentration of inhibitor was presumably attributed to the non-specific reduction of HQ moiety by other reductase in cellular environments.
Conclusions
In this work, we demonstrate that the N-methyl carbamate linker incorporated on the anomeric position of ManNAz successfully controlled the metabolic property of an unnatural sugar. The NN linker completely blocked its metabolic processes when caged and fully resumed its metabolic activity after being decaged. We synthesized HQ-NN-AAM which was reduced by DTD/NADPH and underwent a subsequent self-immolative process to yield the active metabolic species. The incorporation of unnatural sugars in cell surface glycans was confirmed using HQ-NN-AAM as a labeling reagent. The in vitro labeling experiments demonstrated the specificity of HQ-NN-AAM that responded to DTD. This work not only demonstrated DTD responsive metabolic labeling of the caged precursor, but also provides deep insight into developing future metabolically active species that respond to endogenous cues in cells.
Supplementary Material
Acknowledgments
J.C. acknowledges support from the United States National Institute of Health Grant NIH-R21 1R21CA198684 and 1R01CA207584. Research reported in this publication was supported by the National Institute of Biomedical Imaging And Bioengineering of the National Institutes of Health under Award Number T32EB019944. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here].
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Saxon E, Bertozzi CR. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 2.Prescher JA, Dube DH, Bertozzi CR. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 3.Cheng B, Xie R, Dong L, Chen X. Chembiochem. 2016;17:11–27. doi: 10.1002/cbic.201500344. [DOI] [PubMed] [Google Scholar]
- 4.Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR. Proc Natl Acad Sci U S A. 2010;107:1821–1826. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jewett JC, Bertozzi CR. Chem Soc Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laughlin ST, Bertozzi CR. Nat Protoc. 2007;2:2930–2944. doi: 10.1038/nprot.2007.422. [DOI] [PubMed] [Google Scholar]
- 7.Xie R, Dong L, Huang R, Hong S, Lei R, Chen X. Angew Chem Int Ed. 2014;53:14082–14086. doi: 10.1002/anie.201408442. [DOI] [PubMed] [Google Scholar]
- 8.Huang ML, Purcell SC, Verespy S, Wang YN, Godula K. Biomater Sci. 2017;5:1537–1540. doi: 10.1039/c7bm00289k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hudak JE, Canham SM, Bertozzi CR. Nat Chem Biol. 2014;10:69–75. doi: 10.1038/nchembio.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woods EC, Yee NA, Shen J, Bertozzi CR. Angew Chem Int Ed. 2015;54:15782–15788. doi: 10.1002/anie.201508783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koo H, Lee S, Na JH, Kim SH, Hahn SK, Choi K, Kwon IC, Jeong SY, Kim K. Angew Chem Int Ed. 2012;51:11836–11840. doi: 10.1002/anie.201206703. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, Koo H, Na JH, Han SJ, Min HS, Lee SJ, Kim SH, Yun SH, Jeong SY, Kwon IC, Choi K, Kim K. ACS Nano. 2014;8:2048–2063. doi: 10.1021/nn406584y. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Gauthier M, Kelly JR, Miller RJ, Xu M, O’Brien WD, Jr, Cheng J. Angew Chem Int Ed. 2016;55:5452–5456. doi: 10.1002/anie.201509601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Wang R, Cai K, He H, Liu Y, Yen J, Wang Z, Xu M, Sun Y, Zhou X, Yin Q, Tang L, Dobrucki IT, Dobrucki LW, Chaney EJ, Boppart SA, Fan TM, Lezmi S, Chen X, Yin L, Cheng J. Nat Chem Biol. 2017;13:415–424. doi: 10.1038/nchembio.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu XJ, Tian YP, Yu MZ, Lin BJ, Han JH, Han SF. Biomater Sci. 2014;2:1120–1127. doi: 10.1039/c4bm00028e. [DOI] [PubMed] [Google Scholar]
- 16.Xie R, Hong S, Feng L, Rong J, Chen X. J Am Chem Soc. 2012;134:9914–9917. doi: 10.1021/ja303853y. [DOI] [PubMed] [Google Scholar]
- 17.Xie R, Dong L, Du Y, Zhu Y, Hua R, Zhang C, Chen X. Proc Natl Acad Sci U S A. 2016;113:5173–5178. doi: 10.1073/pnas.1516524113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang PV, Dube DH, Sletten EM, Bertozzi CR. J Am Chem Soc. 2010;132:9516–9518. doi: 10.1021/ja101080y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shim MK, Yoon HY, Ryu JH, Koo H, Lee S, Park JH, Kim JH, Lee S, Pomper MG, Kwon IC, Kim K. Angew Chem Int Ed. 2016;55:14698–14703. doi: 10.1002/anie.201608504. [DOI] [PubMed] [Google Scholar]
- 20.Zelzer M, Todd SJ, Hirst AR, McDonald TO, Ulijn RV. Biomater Sci. 2013;1:11–39. doi: 10.1039/c2bm00041e. [DOI] [PubMed] [Google Scholar]
- 21.Liu P, Xu J, Yan D, Zhang P, Zeng F, Li B, Wu S. Chem Commun. 2015;51:9567–9570. doi: 10.1039/c5cc02149a. [DOI] [PubMed] [Google Scholar]
- 22.Kwon N, Cho MK, Park SJ, Kim D, Nam SJ, Cui L, Kim HM, Yoon J. Chem Commun. 2017;53:525–528. doi: 10.1039/c6cc08971b. [DOI] [PubMed] [Google Scholar]
- 23.Saari WS, Schwering JE, Lyle PA, Smith SJ, Engelhardt EL. J Med Chem. 1990;33:97–101. doi: 10.1021/jm00163a016. [DOI] [PubMed] [Google Scholar]
- 24.Alouane A, Labruere R, Le Saux T, Schmidt F, Jullien L. Angew Chem Int Ed. 54:7492–7509. doi: 10.1002/anie.201500088. [DOI] [PubMed] [Google Scholar]
- 25.Quiroga AD, Lehner R. Trends Endocrinol Metab. 2011;22:218–225. doi: 10.1016/j.tem.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y, Zhang Y, Wu Q, Cui X, Lin Z, Liu S, Chen L. J Exp Clin Cancer Res. 2014;33:14–22. doi: 10.1186/1756-9966-33-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips RM. Biochem Pharmacol. 1999;58:303–310. doi: 10.1016/s0006-2952(99)00092-1. [DOI] [PubMed] [Google Scholar]
- 28.Ji M, Jin A, Sun J, Cui X, Yang Y, Chen L, Lin Z. Oncol Lett. 2017;13:2996–3002. doi: 10.3892/ol.2017.5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


