Abstract
Actin-depolymerizing factor (ADF)/cofilins are essential regulators of actin filament turnover. Several ADF/cofilin isoforms are found in multicellular organisms, but their biological differences have remained unclear. Herein, we show that three ADF/cofilins exist in mouse and most likely in all other mammalian species. Northern blot and in situ hybridization analyses demonstrate that cofilin-1 is expressed in most cell types of embryos and adult mice. Cofilin-2 is expressed in muscle cells and ADF is restricted to epithelia and endothelia. Although the three mouse ADF/cofilins do not show actin isoform specificity, they all depolymerize platelet actin filaments more efficiently than muscle actin. Furthermore, these ADF/cofilins are biochemically different. The epithelial-specific ADF is the most efficient in turning over actin filaments and promotes a stronger pH-dependent actin filament disassembly than the two other isoforms. The muscle-specific cofilin-2 has a weaker actin filament depolymerization activity and displays a 5–10-fold higher affinity for ATP-actin monomers than cofilin-1 and ADF. In steady-state assays, cofilin-2 also promotes filament assembly rather than disassembly. Taken together, these data suggest that the three biochemically distinct mammalian ADF/cofilin isoforms evolved to fulfill specific requirements for actin filament dynamics in different cell types.
INTRODUCTION
Actin is a highly conserved and ubiquitous protein found in probably all eukaryotic cells. In muscle cells actin filaments assemble into highly ordered, relatively stable structures that together with myosin form muscle cells' basic contractile apparatus. In nonmuscle cells actin filaments are highly dynamic and participate in a range of processes such as cell polarization and movement, cytokinesis, and endocytosis. The dynamics of actin filaments are tightly regulated, both spatially and temporally, by a large number of actin-binding proteins (Ayscough, 1998; Sheterline, 1998).
ADF/cofilins form a family of actin monomer- and filament-binding proteins (reviewed in Bamburg, 1999), whose activities are fundamental to cells because ADF/cofilin-inactivating mutations are lethal (Moon et al., 1993; McKim et al., 1994; Gunsalus et al., 1995). ADF/cofilins localize to regions of rapid actin dynamics, such as yeast cortical actin patches, neuronal growth cones, and the leading edge and ruffling membranes of motile cells (Bamburg and Bray, 1987; Yonezawa et al., 1987; Moon et al., 1993; Nagaoka et al., 1995). They are central in the dynamics of yeast's cortical actin cytoskeleton (Lappalainen and Drubin, 1997) and in Listeria actin tails (Carlier et al., 1997; Rosenblatt et al., 1997), where ADF/cofilin is among the minimal set of proteins required for motility of this intracellular pathogen (Loisel et al., 1999).
ADF/cofilins' most important physiological function is to depolymerize filaments from their pointed ends, thereby increasing actin dynamics (Carlier et al., 1997). Under physiological conditions ADF/cofilins bind ADP-actin monomers and filaments with higher affinity than ATP-actin (Maciver and Weeds, 1994; Carlier et al., 1997; Blanchoin and Pollard, 1998, 1999). ADF/cofilins bind to actin filaments in a cooperative manner (Hawkins et al., 1993; Hayden et al., 1993) and the binding induces actin filaments to twist by ∼5o/subunit (McGough et al., 1997), changing the filaments' thermodynamic stability (McGough and Chiu, 1999). ADF/cofilins also have a weak filament-severing activity, which increases the amount of filament ends, and thus their turnover (Maciver et al., 1991; Moriyama and Yahara, 1999; Chan et al. 2000).
Unicellular eukaryotes, such as yeast, have only one ADF/cofilin protein, whereas multicellular organisms can have several ADF/cofilin isoforms (Lappalainen et al., 1998). Caenorhabditis elegans alternatively splices their unc-60 gene to express two different ADF/cofilins: Unc-60A and Unc-60B (Ono and Benian, 1998; Ono et al., 1999). There are three ADF/cofilin genes in maize; two are expressed solely in pollen, and the third is expressed in vegetative tissues (Lopez et al., 1996).
Mammals and birds have several ADF/cofilins. So far, two different ADF/cofilins have been reported to exist in mice, pigs, and chickens (reviewed in Lappalainen et al., 1998), whereas three ADF/cofilins are found in humans (Ogawa et al., 1990; Hawkins et al., 1993; Thirion et al., 2001). The two porcine ADF/cofilins have a wide tissue distribution and display some biochemical differences in actin cosedimentation assays (Moriyama et al., 1990). The two mouse ADF/cofilins, on the other hand, have different expression patterns. One isoform is expressed in several tissues and the other is found only in muscles and testes (Ono et al., 1994). However, the possible biochemical differences between these two mouse ADF/cofilin isoforms have not been characterized so far.
Although individual ADF/cofilins have been extensively studied for more than two decades, no comprehensive studies have been carried out to elucidate why multicellular organisms, such as mammals have multiple ADF/cofilin isoforms. This lack of knowledge about differences between ADF/cofilin isoforms hampers the interpretation and comparison of various results concerning these actin-binding proteins. In this study, we compared the expression patterns as well as cell biological and biochemical properties of the three mouse ADF/cofilin isoforms. These ADF/cofilin isoforms have distinct expression patterns and display a number of quantitative biochemical differences.
MATERIALS AND METHODS
Plasmid Construction
DNA fragments corresponding to the open reading frames of the three mouse ADF/cofilins were amplified from mouse cDNA by using oligonucleotides that created NcoΙ and XhoΙ sites at the 5′ and 3′ ends of the polymerase chain reaction (PCR) fragment, respectively. These fragments were digested and ligated to pGAT2 (Peränen et al., 1996) to create plasmids pPL92 (cofilin-1), pPL93 (cofilin-2), and pPL94 (ADF). DNA fragments comprising the complete open reading frame of cofilin-1 and cofilin-2 and a region encoding amino acids 1–124 of ADF were amplified by PCR with oligonucleotides, creating EcoRΙ (5′) and XhoΙ (3′) sites, digested, and ligated into pBSΙΙKS to create plasmids pPL65 (ADF, encoding for amino acids 1–124), pPL66 (cofilin-1) and pPL67 (cofilin-2). To express tagged versions of ADF/cofilins in mammalian cells, pPL92, pPL93, and pPL94 plasmids were used as templates for PCR with oligonucleotides, creating NotΙ (5′) and HindΙΙΙ (3′) sites. These fragments were then ligated into pCMV-Tag1 (Stratagene, La Jolla, CA), resulting in a plasmid where the open reading frame of ADF/cofilin is followed by C-terminal c-myc tag (EQKLISEEDL). The fragments containing ADF/cofilin and the c-myc tag were digested from pCMV-Tag1 with SacΙ and KpnΙ and ligated to pEGFP-N1 (CLONTECH, Palo Alto, CA) to create plasmids pPL108 (cofilin-1-myc), pPL110 (cofilin-2-myc) and pPL112 (ADF-myc). In cells, these plasmids drive the expression of myc-tagged full-length ADF/cofilin under the control of the cytomegaloviral promoter.
Northern Blotting
Mouse ADF/cofilin cDNA probes were prepared from plasmids pPL65, pPL66, and pPL67 as described for mouse twinfilin (Vartiainen et al., 2000). ADF/cofilin probes were hybridized to commercial mouse multiple tissue and mouse embryo multiple tissue Northern blots (CLONTECH) according to manufacturer's instructions, and the Northern blot filters were exposed on a PhosphorImager screen for 2 h. β-Actin controls were used to ensure equal amounts of RNA.
Whole Mount In Situ Hybridization
To prepare the probes, plasmids pPL65 (ADF, amino acids 1–124), pPL66 (cofilin-1), and pPL67 (cofilin-2) were linearized with EcoRΙ for antisense probes or with XhoΙ for sense probes. We labeled 1 μg of linearized DNA with the DIG-labeling kit (Amersham Biosciences AB, Uppsala, Sweden). The whole mount in situ hybridization of 9.5-d-old mouse embryos was performed as described in Henrique et al. (1995). We used the sense probes as controls to show specificity of our antisense probes.
Radioactive In Situ Hybridization
The antisense probes of ∼600 base pairs were obtained by linearizing the plasmids pPL65 (ADF), pPL66 (cofilin-1), and pPL67 (cofilin-2) with EcoRI and transcribing with T3 RNA polymerase. For sense probes, XhoI and T7 RNA polymerase were used. In situ hybridization on tissue sections was performed using 35S-UTP-labeled riboprobes as described previously (Vainio et al., 1991; Rice et al., 2000).
Cell Culture and Immunofluorescence
HeLa cells were maintained in modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were transfected with 1 μg of desired plasmid, by using the Roche Molecular Biochemicals (Mannheim, Germany) FuGENE 6 transfection reagent according to manufacturer's instructions. Indirect immunofluorescence was carried out as described in Vartiainen et al. (2000). The monoclonal anti-myc antibody (9E10) was used at 1:500 dilution and fluorescein isothiocyanate- and tetramethylrhodamine B isothiocyanate-conjugated phalloidin (Molecular Probes, Eugene, OR) at 1:300. Fluorescent secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA) were used at 1:250 dilutions.
Protein Expression and Purification
Mouse ADF/cofilins were expressed as glutathione S-transferase (GST) fusion proteins as described for mouse twinfilin (Vartiainen et al., 2000). GST fusion proteins were enriched from the lysis supernatant with glutathione agarose beads (Sigma, St. Louis, MO). ADF/cofilins were cleaved off the GST by 0.05 mg/ml thrombin and further purified by a Superdex-75 HiLoad gel filtration column (Amersham Biosciences AB). The peak fractions that eluted from the column at 75 ml were pooled and concentrated in a Centricon 10 to ∼500 μM. Rabbit muscle actin was prepared from acetone powder as described in Pardee and Spudich (1982) and human platelet actin was purchased from Cytoskeleton.
Cosedimentation Assays
Aliquots (40 μl) of actin were diluted to desired concentrations in G-buffer (20 mM Tris pH 7.0–8.5, 0.2 mM ATP, 0.2 mM dithiothreitol [DTT], 0.2 mM CaCl2) and polymerized for 30 min by the addition of 5 μl of 10× initiation mix (20 mM MgCl2, 10 mM ATP, 1 M KCl). Five microliters of ADF/cofilins in G-buffer were added to actin filaments and incubated for 30 min. We sedimented actin filaments by centrifuging the samples for 30 min in a Beckman Optima MAX Ultracentrifuge at 217,000 × g by using a TLA100 rotor. All steps were carried out at room temperature. Equal proportions of supernatants and pellets were loaded onto SDS-13.5% polyacrylamide gels, the gels were stained with Coomassie Blue, and scanned with FluorS-Imager (Bio-Rad, Hercules, CA). The intensities of actin and ADF/cofilin bands were quantified with QuantityOne program (Bio-Rad).
Depolymerization/Fragmentation of Alexa 488-Actin Filaments
Alexa 488-actin (50% labeled; Molecular Probes) was polymerized in 20 mM Tris pH 8.0, 0.2 mM ATP, 0.2 mM DTT, 0.2 mM CaCl2, 2 mM MgCl2, 0.1 M KCl for 45 min. Three microliters of 3.3 μM ADF/cofilins was added onto 2 μl of polymerized 5 μM Alexa 488-actin and the mixture was incubated at room temperature for 20 s. Then 15 μl of mounting medium (Mowiol; Calbiochem, Meudon, France) was added on samples. Aliquots (5.5 μl) were placed on a glass slide, coverslipped, and immediately observed under a light microscope.
Measurements of Treadmilling Rate of Actin Filaments
The steady-state rate of actin filament turnover was monitored by the decrease in the fluorescence of ε-ADP bound to F-actin after addition of ATP (Carlier et al., 1997). ε-ATP-G-actin (20 μM) was polymerized at pH 8.0 in the presence of 100 μM ε-ATP. Samples (60 μl) of ε-ATP-F-actin were preincubated in the presence of 5 μM ADF/cofilins for 5 min and the decrease in fluorescence of ε-ATP (excitation at 350 nm, emission at 410 nm) was monitored with a BioLogic MOS-250 fluorometer after addition of 10 μl of 5 mM ATP.
Nucleotide Exchange Assay
The fluorescence signal provided by ε-ATP bound to G-actin was applied to measure the rate of nucleotide exchange of actin. G-buffer (60 μl; 10 mM Tris, pH 8.0, 2 mM CaCl2, 2 mM DTT, 100 μM ε-ATP) was mixed with ε-ATP-G-actin (2.5 μM) and ADF/cofilins (2.5/5 μM). This was mixed with 15 μl of 10 mM ATP and the reaction was followed in a BioLogic MOS-250 fluorescence spectrophotometer at an excitation of 350 nm and emission of 410 nm.
Binding of ADF/Cofilins to Actin Monomers
The change in the fluorescence of NBD-labeled G-actin was used to monitor the binding of ADF/cofilins to actin monomers (Carlier et al., 1997). Actin was labeled by NBD-Cl as described in Detmers et al. (1981) and Weeds et al. (1986). ADP-actin was prepared by incubating NBD-actin with hexokinase-agarose beads (Sigma) and glucose for 2 h at 4oC (Pollard, 1986). The final concentration of actin in these assays was 0.2 μM and the ADF/cofilin concentrations were varied from 0.05 to 20 μM. Experiments were carried out at room temperature in F-buffer containing 2 mM Tris-HCl pH 8.0, 0.1 mM CaCl2, 0.1 mM DTT, 0.2 mM ADP or ATP, 1 mg/ml bovine serum albumin, 2 mM MgCl2, and 0.1 M KCl. The normalized enhancement of fluorescence, as determined by the equation
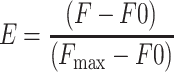 |
was measured at each concentration of ADF/cofilin. The data were analyzed using SigmaPlot software and fitted using the following equation:
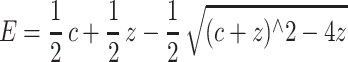 |
where
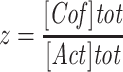 |
and
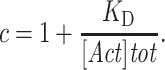 |
Miscellaneous
SDS-PAGE was carried out by using the buffer system of Laemmli (1970). Protein concentrations were determined with a Hewlett Packard 8452A diode array spectrophotometer by using calculated extinction coefficients for mouse ADF/cofilins at 280 nm (cofilin-1, ε = 13,490 M−1 cm−1; cofilin-2, ε = 17,840 M−1 cm−1, and ADF, ε = 12,270 M−1 cm−1) and for actin at 290 nm (ε = 26 600 M−1 cm−1). Concentrations of ADF/cofilins were also estimated from SDS-PAGE by quantifying the intensities of Coomassie-stained protein bands with FluorS-Imager and QuantityOne software (Bio-Rad).
RESULTS
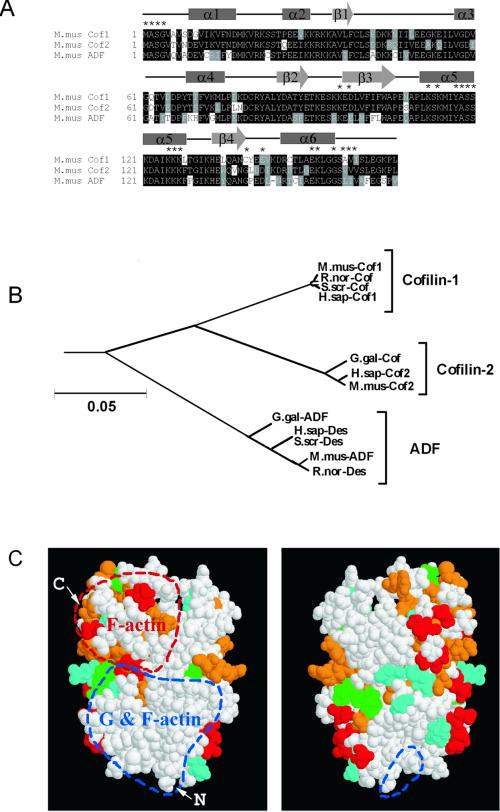
We identified three ADF/cofilins in mice from >30 ADF/cofilins present in the public sequence databases (Figure 1A). Two were the known muscle and nonmuscle cofilins (Swiss-Prot P45591 and P18760) and the third (EMBL AB025406) was highly homologous to human destrin, chicken ADF, and porcine destrin. Phylogenetic analysis of all known mammalian and avian ADF/cofilins divided these proteins into three distinct subclasses, which we have named cofilin-1, cofilin-2, and ADF (Figure 1B). Mouse cofilin-1 and cofilin-2 are ∼80% identical and both are ∼70% identical to mouse ADF. We mapped the unique residues for these isoforms on a space-filling model of human destrin (Hatanaka et al., 1996) and found that most of the variable residues are outside ADF/cofilins' known actin-binding surfaces (Figure 1C).
Figure 1.
Comparison of the three mouse ADF/cofilin sequences. (A) Sequence alignment of the three mouse ADF/cofilins. The positions of the secondary structure elements based on the nuclear magnetic resonance structure of human destrin (Hatanaka et al., 1996) are indicated with boxes (α-helixes) and arrows (β-sheets). Asterisks above the sequences indicate residues that have been implicated in actin binding (Lappalainen et al. 1997; Ono et al. 1999, 2001; Van Troys et al. 2000). (B) Phylogenetic tree of all known mammalian and avian ADF/cofilins. The neighbor joining tree was produced by the Clustal-X software. A bar showing 5% divergence is included. Protein names, database, and accession numbers for the sequences, respectively, are listed below. Mus musculus cofilin-1: Swiss-Prot, P18760, Rattus norvegicus cofilin, Swiss-Prot, P45592, S. scrofa cofilin: GenBank, M20866, Homo sapiens cofilin-1: EMBL, X95404, Gallus gallus cofilin: GenBank, M55659, H. sapiens cofilin-2: EMBL, AF134802, M. musculus cofilin-2: Swiss-Prot, P45591, G. gallus ADF: GenBank, J02912, H. sapiens destrin: PIR, A54184, S. scrofa destrin: DDBJ, D90053, M. musculus ADF: EMBL, AB025406, R. norvegicus destrin: PIR, JE0223. (C) Space-filling model of human destrin (Hatanaka et al., 1996) in two different orientations (rotated 180o horizontally). Residues that correspond to variable amino acids between all three ADF/cofilins are indicated in red. The variable residues specific for cofilin-1, cofilin-2, and ADF are indicated in green, cyan, and orange, respectively. Regions that participate in actin monomer- and filament binding, and those involved only in actin filament binding are circled by red and blue dashed lines, respectively. Arrows mark the amino and carboxy terminus of the protein.
Expression Patterns of the Three ADF/Cofilins
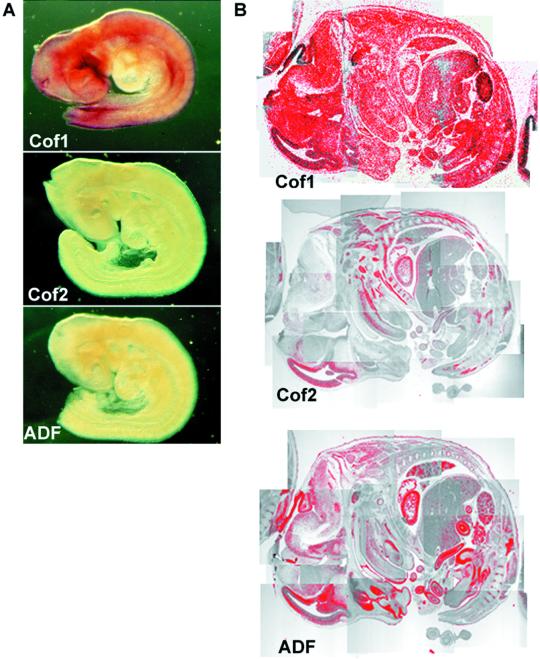
To elucidate the expression patterns of the three mouse ADF/cofilins we carried out Northern blot and in situ hybridization analyses on embryonic and adult mice. Cofilin-1 mRNA was uniformly expressed in embryonic day (E) 9.5 mice by whole mount in situ hybridization (Figure 2A), but neither cofilin-2 nor ADF was detected under the same conditions. We did not detect anything with respective sense probes, indicating that these hybridizations were specific (our unpublished data). In E14 embryos, cofilin-1 mRNA was intensely expressed in all cell types by radioactive in situ hybridizations of tissue sections (Figure 2B). At this stage cofilin-2 mRNA appeared in developing muscles such as the tongue and tail. On the other hand, ADF mRNA was detected in the brain and epithelial tissues such as in intestines and skin.
Figure 2.
Expression of ADF/cofilins during development. (A) Whole mount in situ hybridization of E9.5 mouse embryos. Cofilin-1 mRNA (top) is expressed throughout the embryo, whereas only very weak cofilin-2 (middle) or ADF (bottom) expression can be detected under the same conditions. (B) In situ hybridization of E14 mouse embryo section. Cofilin-1 mRNA (top) shows ubiquitous expression. Expression of cofilin-2 (middle) is detected in developing muscles, such as in the tongue and tail. ADF (bottom) is expressed in developing epithelial cells, e.g., in intestine and skin.
A Northern blot analysis of 7–17-d-old mouse embryos confirmed that cofilin-1 mRNA is expressed at a constant level during development and that cofilin-2 and ADF are expressed at lower levels than cofilin-1 (our unpublished data). A Northern blot analysis of adult tissues with ADF/cofilin cDNA probes showed that the ADF/cofilins are variably expressed in many different organs (Figure 3). Cofilin-1's 1.4-kb mRNA is highly expressed in brain and liver; moderately expressed in heart, spleen, lungs, kidneys, and testes; and entirely absent in skeletal muscle, even although cofilin-1 is expressed in all embryonic cells (Figure 2, A and B). The only isoform expressed in skeletal muscle is cofilin-2, which is also in heart, liver, and testes, and at lower amounts in other organs. The two transcripts of 1.8 and 3 kb seen in cofilin-2 blot are a consequence of selective use of two polyadenylation signals, resulting in size difference of the 3′-noncoding sequence (Ono et al., 1994). ADF's 1.8-kb mRNA is also expressed in most organs but not in skeletal muscle (Figure 3). The blots were also hybridized with a β-actin control probe to ensure that each lane contained equal amounts of RNA (our unpublished data).
Figure 3.
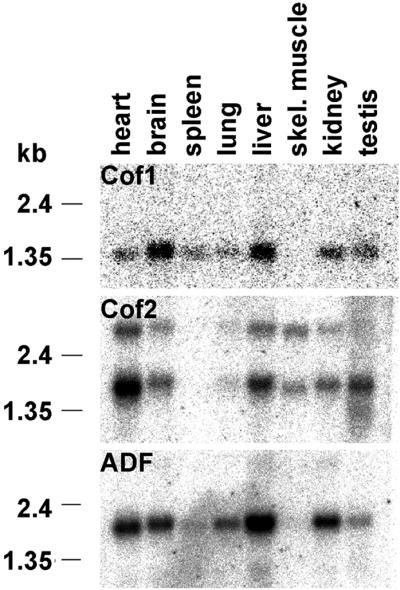
Northern blot analyses of ADF/cofilin expression in adult mice. Cofilin-1 mRNA (top) is expressed at high levels in brain and liver; at moderate amounts in heart, spleen, lung, kidney, and testis; but is not expressed in skeletal muscle. The only isoform expressed in skeletal muscle is cofilin-2 (middle), which is also expressed at moderate levels in heart, liver, and testis, and at lower amounts in other organs. ADF (bottom) is expressed strongly in liver and at moderate levels in other organs, but is not found in skeletal muscle.
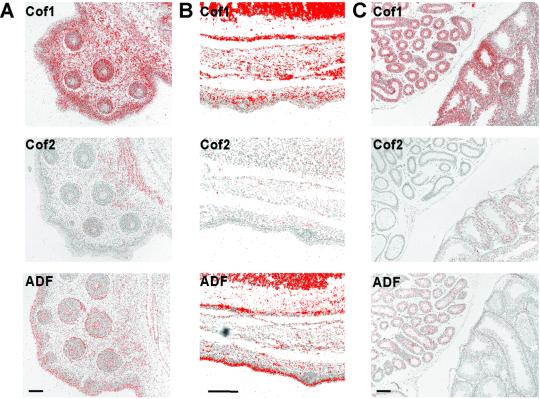
Although Northern blot analysis shows that the three ADF/cofilins are coexpressed in many organs, they segregate to different tissues and cell types. In situ hybridization of E15 embryos' whisker pads (Figure 4A) showed that cofilin-2 is expressed in the developing muscles behind the whisker pad, whereas ADF is in the epithelial cells of the whisker follicles. Furthermore, ADF is expressed in the skin epidermis (Figure 4, A and B). Cofilin-1 shows ubiquitous expression in these sections. We also studied adult testes by in situ hybridization because it is among the organs where all three ADF/cofilins are expressed (Figure 4C). Cofilin-1 is found throughout both testes and epididymides, although its expression levels in seminiferous tubules varied depending on the stage of spermatogenesis. Cofilin-2 expression is restricted to the seminiferous tubules, whereas ADF is found only in the epididymal epithelia. Taken together, these data show that cofilin-1 is expressed in many tissues and in most of their cell types. Interestingly, cofilin-2 and ADF are mainly restricted to muscle and epithelial tissues, respectively. Cofilin-1 and ADF mRNAs were coexpressed in several tissues as were cofilin-1 and cofilin-2. However, expression of ADF and cofilin-2 mRNAs is not generally found in the same tissues.
Figure 4.
Expression of ADF/cofilins in different tissues. (A) In situ hybridization of an E15 mouse embryo's whisker pad. Cofilin-1 mRNA (top) is expressed all cell types of the whisker pad, whereas cofilin-2's (middle) expression is confined to the muscles behind the pad. ADF (bottom) is expressed in the epidermis and in the epithelia of whisker hair follicles. (B) In situ hybridization of E16 mouse embryo's back skin. Cofilin-1 (top) is found throughout the skin, whereas ADF (bottom) is concentrated especially to the outermost layers of the epidermis. Cofilin-2 (middle) is not expressed in the skin. (C) In situ hybridization of adult mouse testes. Cofilin-1 (top) is expressed in the testes and the epididymides. Cofilin-2 (middle) is expressed in the testes, but not in the epididymides. On the contrary, ADF (bottom) is expressed in the epididymal epithelia but not in the testis. Bar, 100 μm.
We also studied the intracellular localization of the three ADF/cofilins. Myc-tagged ADF/cofilins were diffusely cytosolic in transfected cultured cells and also accumulated in actin-rich filopodia and lamellipodia; similar to endogenous cofilin-1 detected by polyclonal antibodies (our unpublished data).
Biochemical Properties of Mouse ADF/Cofilins
Because the three mouse ADF/cofilins do not appear to have any detectable cell biological differences, we next tested whether they would show any biochemical differences. We expressed all three ADF/cofilins as GST-fusion proteins in Escherichia coli. GST was subsequently cleaved off by thrombin and recombinant proteins were further purified by gel filtration chromatography. The purified proteins were monomeric and fully soluble. All ADF/cofilins contained their own N-terminal methionine and an N-terminal extension of serine and glycine (see MATERIALS AND METHODS). Because the three mouse ADF/cofilins had clearly distinct expression patterns, we also tested whether the tissue distribution would reflect their specificity for a certain actin isoform. In these assays we used muscle (α) and platelet (β + γ) actin.
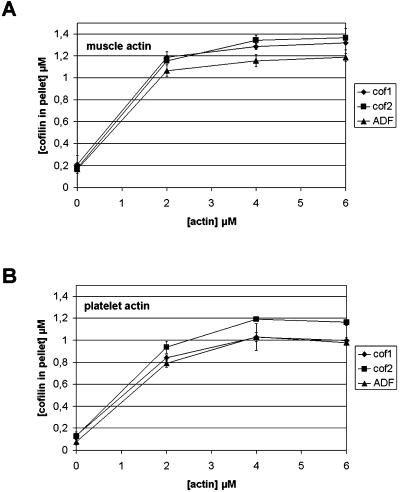
We used cosedimentation assays to measure the interactions of ADF/cofilins with muscle (Figure 5A) and platelet (Figure 5B) actin filaments. All ADF/cofilins bound equally well to both actins and the binding was nearly saturated at ∼2 μM actin. More ADF/cofilins cosedimented with muscle than platelet actin, perhaps reflecting muscle actin's greater resistance to depolymerization (see below).
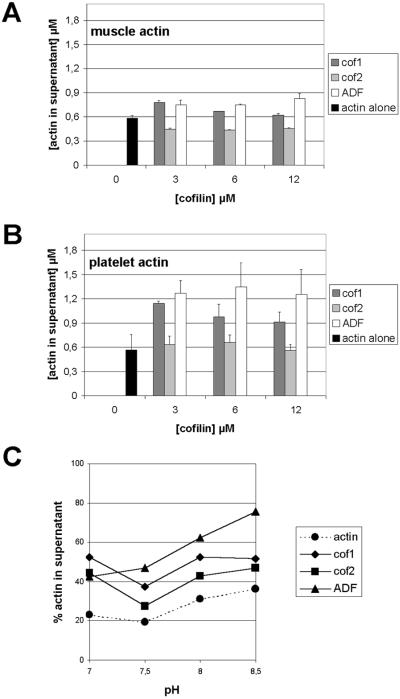
Figure 5.
Binding of mouse ADF/cofilins to actin filaments. ADF/cofilins (1.5 μM) were mixed with 0, 2, 4, or 6 μM muscle (A) or platelet (B) actin at pH 7.5 and actin filaments were sedimented by centrifugation. The amount of ADF/cofilins in the pellet fraction was quantified from three independent experiments. All ADF/cofilins bound equally well to muscle and platelet actin filaments. The small differences in the levels of cosedimented ADF/cofilins result from the differences in their ability to promote actin filament disassembly (Figure 6).
We determined ADF/cofilins' abilities to disassemble actin filaments by cosedimentation assays with actin at 3 μM and varying ADF/cofilins from 0 to 12 μM. Cofilin-1 and ADF both disassembled actin filaments: cofilin-1 most efficiently at 3 μM, whereas ADF was more efficient at higher concentrations (Figure 6). Cofilin-2 was very different compared with the other two ADF/cofilins. In this assay, cofilin-2 was unable to increase the amount of monomeric actin, and when the assay was carried out with muscle actin, it even slightly promoted the formation of filaments (Figure 6A). Although mouse ADF/cofilins did not show any actin isoform specificity in these assays, our data indicate that the muscle actin and platelet actin themselves have quantitative differences in their dynamics. In the absence of ADF/cofilins the two actins sedimented to a similar extent, but platelet actin appeared to be more susceptible for ADF/cofilin-induced filament disassembly than muscle actin (Figure 6, A and B).
Figure 6.
Ability of ADF/cofilins to shift actin to the monomeric fraction in actin filament sedimentation assay. Muscle (3 μM) (A) or platelet actin (B) were mixed with 0, 3, 6, or 12 μM ADF/cofilins, actin filaments were sedimented by centrifugation, and the amount of actin in the supernatant and pellet fractions was quantified from three independent experiments. This assay was carried out at pH 7.5. Both cofilin-1 and ADF promote significant increases in the amount of monomeric actin and the ability of ADF to disassemble actin filaments is more pronounced with higher protein concentrations. In contrast to cofilin-1 and ADF, cofilin-2 is not able to promote actin filament disassembly. ADF/cofilins shifted more platelet actin (B) to the monomeric pool than muscle actin (A). (C) pH dependency of actin filament disassembly by mouse ADF/cofilins was studied with 3 μM platelet actin and 3 μM ADF/cofilins at pH 7.0–8.5. The ability of cofilin-1 and cofilin-2 to increase the amount of actin monomers is not greatly affected by pH. In contrast, the ability of ADF to disassemble actin filaments is significantly increased at higher pH.
Many ADF/cofilin family members have been shown to bind and depolymerize actin filaments in a pH-dependent manner (Yonezawa et al., 1985; Hawkins et al., 1993; Hayden et al., 1993; Maciver et al., 1998). Therefore, we carried out a sedimentation assay with 3 μM platelet actin and 3 μM ADF/cofilins at pH 7.0–8.5 (Figure 6C). Increasing the pH from 7.5 to 8.5 slightly depolymerizes actin even in the absence of ADF/cofilins, and adding cofilin-1 or cofilin-2 did not affect actin disassembly above this background; however, adding ADF significantly increased the amount of monomeric actin at pH 8.5, suggesting that ADF's actin disassembly activity is affected by an increase in pH.
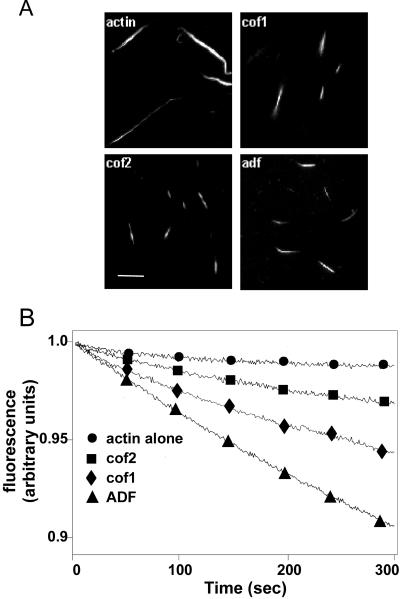
Because cofilin-2 was not able to increase the amount of monomeric actin, we next examined whether this was due to its incapability to depolymerize or fragment actin filaments. We first carried out a visual assay, where we observed the depolymerization of Alexa 488-actin filaments under light microscope (Figure 7A). All ADF/cofilins efficiently depolymerized or fragmented actin, shortening filaments from ∼12 to ∼4 μm. The effect of ADF/cofilins on the actin filament turnover was also examined more quantitatively by following the release of actin-bound ε-ATP from filaments. All three ADF/cofilins increased the actin filament turnover in this assay. However, ADF was the most efficient isoform, whereas cofilin-2 promoted the smallest increase in filament dynamics (Figure 7B).
Figure 7.
Depolymerization/fragmentation of actin filaments by ADF/cofilins. (A) Labeled Alexa 488-actin (50%) was polymerized at pH 8.0 and mixed with ADF/cofilins to yield a final concentration of 2 μM for both proteins. Samples were visualized ∼40 s after mixing the two proteins. All three mouse ADF/cofilins shortened filaments, suggesting that they depolymerized/fragmented actin filaments in this assay. Bar 5, μM. (B) Turnover of actin filaments in the presence and absence of ADF/cofilins was measured by following the release of ε-ATP from the filaments at pH 8.0 after an ATP chase. The final concentrations of actin and ADF/cofilins in this assay were 17 and 4.3 μM, respectively. The decrease in the fluorescence of F-actin–bound ε-ATP represents the turnover of actin filaments.
Previous studies have shown that ADF/cofilins from different species decrease the rate of nucleotide exchange upon binding to actin monomers (Hayden et al. 1993; Hawkins et al. 1993). The ability of the three mouse ADF/cofilins to inhibit the exchange of ε-ATP for ATP was measured by using 2 μM actin monomers and 2 or 4 μM ADF/cofilins. Although the three mouse ADF/cofilins differ in their abilities to promote filament turnover and actin filament disassembly (Figures 6 and 7), they all promoted the inhibition of the nucleotide exchange on actin monomers to a similar extent (our unpublished data).
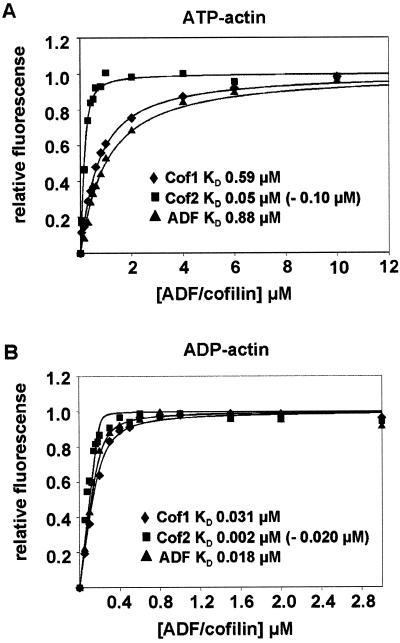
We next determined the affinity of each ADF/cofilin to actin monomers. The binding of mouse ADF/cofilins to actin monomers results in an enhancement of NBD-actin monomer fluorescence. The extent of the increase in NBD-actin fluorescence versus ADF/cofilin concentration displayed a saturating behavior and this enabled us to calculate the KD values for ADF/cofilin–actin monomer complexes. All three mouse ADF/cofilins bound ADP-actin monomers (Figure 8B) with higher affinity than ATP-actin monomers (Figure 8A). The KD values for ADP-actin ranged from 2 to 20 nM for cofilin-2 to 31 nM for cofilin-1. Taking into account the experimental error due to the substoichiometric ADF/cofilin concentrations used in these assays, all three mouse ADF/cofilins appear to have similar affinities for ADP-actin monomers. The two alternative KD values given for cofilin-2 result from two independent methods applied for determination of its concentration. The lower values, KD (ATP-actin) = 0.050 μM and KD (ADP-actin) = 0.002 μM, were obtained when the concentration of cofilin-2 was measured on the basis of its calculated extinction coefficient at 280 nm (ε = 17 840 M−1 cm−1). This method was also used for determination of concentrations of cofilin-1 and ADF. However, because cofilin-2 typically stained with higher intensity with Coomassie Brilliant Blue on SDS-polyacrylamide gels than equal concentrations (as measured using calculated extinction coefficients) of cofilin-1 and ADF, we also quantified the concentration of cofilin-2 from Coomassie-stained gels by comparison with cofilin-1. This method resulted in somewhat higher values for the dissociation constants of the cofilin-2-actin monomer complex (KD for ADP-actin = 0.02 μM and for ATP-actin = 0.10 μM) compared with those values obtained by using the calculated extinction coefficient. Cofilin-2's affinity for actin monomers is most likely between these two values.
Figure 8.
Interaction of ADF/cofilins with actin monomers. The increase in fluorescence of 0.2 μM NBD-labeled MgATP-G-actin (A) or Mg-ADP-G-actin (B) was measured at different concentrations of ADF/cofilins under physiological ionic conditions at pH 8.0. Symbols are data and lines are calculated binding curves. Each data point on the graph is a mean value of three independent experiments. Dissociation constants (KD) derived from the binding curves are indicated in the figure. The two values given for cofilin-2 result from two independent methods that were applied for determination of the concentration of this protein.
These assays also showed that cofilin-1 and ADF bind ATP-actin monomers with 20–90 times lower affinity (KD = 0.60–0.88 μM) than ADP-actin monomers (KD = 0.02–0.03 μM). This difference is of the same order of magnitude than with plant and Acanthamoeba ADF/cofilins, but mouse ADF's and cofilin-1's affinities are ∼5–10-fold higher than previously reported for plant, human, and Acanthamoeba ADFs (Carlier et al., 1997; Blanchoin and Pollard, 1998; Ressad et al., 1998). Interestingly, cofilin-2 binds ATP-actin monomers with 5–10-fold higher affinity than the two other mouse ADF/cofilin isoforms, and the difference between its affinities for ADP- and ATP-actin monomers is also smaller (Figure 8).
DISCUSSION
Our phylogenetic analysis (Figure 1B) groups all known mammalian and avian ADF/cofilins into three subgroups. To clarify the nomenclature of these proteins, we named these subgroups cofilin-1, cofilin-2, and ADF. These mammalian isoforms arose from two duplications of the ancestral ADF/cofilin gene. The first duplication yielded ADF and “cofilin,” and “cofilin” then duplicated to give cofilin-1 (the nonmuscle isoform) and cofilin-2 (the muscle isoform) (Figure 1B). These three classes of ADF/cofilins are in all mammals and birds, although chicken currently has only cofilin-2 and ADF (Abe et al., 1990; Adams et al., 1990).
Mouse ADF/Cofilins Show Cell-Type–specific Expression Patterns
We found that the three ADF/cofilins are expressed in distinct tissues in mice in agreement with previous findings that mouse cofilin-1 is expressed in many adult tissues, whereas cofilin-2 is only in skeletal muscles, heart, and testes (Ono et al., 1994). Cofilin-1 is present in embryonic skeletal muscle but postnatally disappears and is replaced by cofilin-2 in terminally differentiated myogenic cells (Ono et al., 1994; Obinata et al., 1997; Mohri et al., 2000). A C. elegans ADF/cofilin, Unc-60B, also participates in muscle development (Ono et al., 1999). In contrast to our observations, previous studies by Moriyama et al. (1990) suggested that neither cofilin-1 nor ADF would be expressed in liver. However, these differences most likely result from differences between the experimental setups in these two studies.
We found cofilin-2 in organs other than skeletal muscle by Northern blots, but in situ hybridizations showed that cofilin-2 is confined mainly to the muscle cells in these organs (Figures 2 and 4). One exception was the testes, where cofilin-2 was detected in developing sperm cells. The other exception was the neuroepithelium of developing brain. It is also important to note that although the Northern blot analysis indicates that ADF and cofilin-1 are expressed in same organs, their expression patterns are different by in situ hybridizations. ADF is specific for epithelial and endothelial tissues, whereas cofilin-1 is ubiquitous. The distinct expression patterns of ADF/cofilins raises many questions: Because more than one ADF/cofilin can be found in a single cell, are they in distinct intracellular locations and are their activities regulated by different mechanisms? Does the tissue distribution correlate with specificity toward a specific actin isoform? Are the mammalian ADF/cofilins biochemically different?
The localizations of myc-tagged ADF/cofilins in cultured cells were similar to each other and to endogenous ADF/cofilins (Bamburg and Bray, 1987; Yonezawa et al., 1987). Although these ADF/cofilins lack differences in their basic cell biology, they may behave differently in specific cells and situations. Actin disassembles and accumulates in the furrow of dividing myoblasts when cofilin is microinjected, but ADF has no effects (Obinata et al., 1997); and only ADF's distribution in Swiss 3T3 cells is changed by pH (Berstein et al., 2000). The second example agrees with our findings that only ADF's actin disassembly activity is affected by pH.
The Three ADF/Cofilin Isoforms Have Different Effects on Actin Filament Dynamics
The three ADF/cofilins are not specific for muscle or platelet actin in actin filament binding and disassembly assays (Figures 5 and 6). However, all ADF/cofilins disassemble platelet actin more efficiently than muscle actin (Figure 6). This difference has not been reported previously, but it agrees with the finding that muscle actin filaments are less dynamic than actin in other cells (Sanger et al., 1984).
The three ADF/cofilins in mice are biochemically distinct: the muscle-specific cofilin-2 is less efficient at actin disassembly than cofilin-1 and ADF (Figure 6). Analogously, the muscle-specific ADF/cofilin, Unc-60B, in C. elegans is also less efficient at actin disassembly than the ubiquitous Unc-60A (Ono and Benian, 1998). It is, however, important to note that the actin filament disassembly activity measured by these sedimentation assays is a combination of several different parameters, including actin filament binding, depolymerization and severing, monomer sequestering, and the ability to promote filament assembly. Therefore, direct actin filament depolymerization and actin monomer binding assays were carried out.
Visual depolymerization/severing assays showed that all three mouse ADF/cofilins, including cofilin-2, are able to depolymerize actin filaments (Figure 7A). Results from this assay also indicate that all three ADF/cofilins have some actin filament-severing activity, because within the 40-s incubation with ADF/cofilins, the average length of filaments decreased ∼8 μm. This would correspond to dissociation rate (koff) of 70 s−1 at the pointed end of the filament. This is significantly higher value than previously reported for ADF/cofilin-assisted filament depolymerization (koff = 5 s−1) (Carlier et al., 1997). It remains to be shown whether the three mouse ADF/cofilins induce actin filament severing with different efficiencies from each other. Although all three ADF/cofilins increased filament treadmilling in our assays, they did this to different extents (Figure 7B). Cofilin-2 was the least efficient isoform, which can at least partly explain why cofilin-2 could not bring actin into monomeric form in actin filament sedimentation assays.
Interaction of Mouse ADF/Cofilins with Actin Monomers
Our results (Figure 8) agree with findings that the ADF/cofilins of Acanthamoeba, plants, and humans bind ADP-actin monomers with higher affinities (KD 100–200 nM) than ATP-actin monomers (4–8 μM) (Carlier et al., 1997; Blanchoin and Pollard, 1998; Ressad et al., 1998). However, our affinities are ∼5–10 times greater: mouse cofilin-1 has a KD for ATP-actin of 590 nM and KD for ADP-actin of 31 nM; ADF has a KD for ATP-actin of 880 nM and KD for ADP-actin of 18 nM. These differences may be due to how we made our ADF/cofilin constructs. Cleaving our GST-ADF/cofilins with thrombin added an N-terminal Ser-Gly to the native ADF/cofilins. In other studies ADF/cofilins were expressed from pET or equivalent vectors, where translation starts at the protein's methionine codon. In E. coli this methionine is usually removed (Hawkins et al., 1993), and its absence may affect the actin binding of these recombinant ADF/cofilins. The first four amino acids are important in actin–ADF/cofilin interactions (Lappalainen et al., 1997; Wriggers et al., 1998). An alternative is that the N-terminal Ser-Gly increases the recombinant ADF/cofilins' affinities for actin monomers.
A key difference that we found is that cofilin-2 has a 5–10 times higher affinity for ATP-actin monomers than ADF and cofilin-1, which results in a smaller difference between its affinities for ADP- and ATP-actin monomers. This may affect its ability to promote actin filament assembly, because computer simulations and experimental data suggest that the association rate constant of ADF/cofilin-ATP-actin monomer complex at the barbed ends of filaments is higher than the one of ATP-actin monomer alone (Carlier et al., 1997; Sept et al., 1999).
Biological Roles of Mouse ADF/Cofilins
The three ADF/cofilins are adapted to promote actin filament dynamics in specific mouse cell types (Table 1). Cofilin-2 is the only ADF/cofilin in mature mouse muscles (Figure 3; Obinata et al., 1997), has a high affinity for ATP-actin monomers, and promotes filament assembly rather than disassembly, unlike ADF and cofilin-1. Although recent studies suggest that actin dynamics at thin filament ends in cardiac myocytes is comparable to actin dynamics in relatively stable actin filament structures in other cells (Littlefield et al., 2001), the turnover of the entire actin filaments in muscle cells is likely to be very slow. Therefore, muscle may not require an ADF/cofilin that would increase actin dynamics efficiently. Also the muscle actin is more resistant to ADF/cofilin-induced actin filament disassembly than platelet actin (Figure 6). In contrast, ADF is the most active ADF/cofilin and is found in polarized epithelial and nervous tissues. Polarization depends on a dynamic actin cytoskeleton and benefits from an efficient promoter of actin dynamics. Epithelia and endothelia are also subjected to physical damage and wounding, and repair requires rapid changes in the actin cytoskeleton so that cells can move. Finally, epithelia have cell surface modifications and specialized junctions that are linked to the actin cytoskeleton and are constantly maintained and modified. Cofilin-1 is a biochemical intermediate between cofilin-2 and ADF and may promote actin dynamics in less specialized cell types.
Table 1.
Expression patterns and biochemical properties of mouse ADF/cofilins
| Expression | Filament binding | Increase actin monomers | Actin isoform specificity | Depolymerization | Affinity for actin
monomers (μM)
|
||
|---|---|---|---|---|---|---|---|
| ATP | ADP | ||||||
| Cofilin-1 | Most cell types | +++ | ++ | no | ++ | 0.59 | 0.031 |
| Cofilin-2 | Muscle tissues | +++ | +/− | no | + | 0.05–0.10 | 0.002–0.020 |
| ADF | Epithelial tissues | +++ | +++ | no | +++ | 0.88 | 0.018 |
In conclusion, specialized cells are a hallmark of multicellular organisms and have different requirements for the regulation of their actin filament dynamics. We suggest that ADF/cofilin genes have duplicated and the resulting three ADF/cofilins fulfill actin-regulatory demands in multicellular organisms.
ACKNOWLEDGMENTS
We thank Drs. Kirsi Sainio and Hannu Sariola for help with testis in situ hybridizations. Dr. Alan Weeds is acknowledged for advice on actin monomer-binding assays. This study was supported by grants from Academy of Finland and Biocentrum Helsinki (to P.L.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01-07-0331. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–07-0331.
REFERENCES
- Abe H, Endo T, Yamamoto K, Obinata T. Sequence of cDNAs encoding actin depolymerizing factor and cofilin of embryonic chicken skeletal muscle: two functionally distinct actin-regulatory proteins exhibit high structural homology. Biochemistry. 1990;29:7420–7425. doi: 10.1021/bi00484a010. [DOI] [PubMed] [Google Scholar]
- Adams ME, Minamide LS, Duester G, Bamburg JR. Nucleotide sequence and expression of a cDNA encoding chick brain actin depolymerizing factor. Biochemistry. 1990;29:7414–7420. doi: 10.1021/bi00484a009. [DOI] [PubMed] [Google Scholar]
- Ayscough KR. In vivo functions of actin-binding proteins. Curr Opin Cell Biol. 1998;10:102–111. doi: 10.1016/s0955-0674(98)80092-6. [DOI] [PubMed] [Google Scholar]
- Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, Bray D. Distribution and cellular localization of actin depolymerizing factor. J Cell Biol. 1987;105:2817–2825. doi: 10.1083/jcb.105.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berstein BW, Painter WB, Chen H, Minamide LS, Abe H, Bamburg JR. Intracellular pH modulation of ADF/cofilin proteins. Cell Motil. 2000;47:319–336. doi: 10.1002/1097-0169(200012)47:4<319::AID-CM6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Interaction of actin monomers with Acanthamoebaactophorin (ADF/cofilin) and profilin. J Biol Chem. 1998;273:25106–25111. doi: 10.1074/jbc.273.39.25106. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoebaactophorin (ADF/Cofilin) with actin filaments. J Biol Chem. 1999;274:15538–15546. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AY, Bailly M, Zebda N, Segall JE, Condeelis JS. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J Cell Biol. 2000;148:531–542. doi: 10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmers P, Weber A, Elzinga M, Stephens RE. 7-Chloro-4-nitrobenzeno-2-oxa-1,3-diazole actin as a probe for actin polymerization. J Biol Chem. 1981;256:99–105. [PubMed] [Google Scholar]
- Gunsalus KC, Bonaccorsi S, Williams E, Verni F, Gatti M, Goldberg ML. Mutations in twinstar, a Drosophilagene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka H, Ogura K, Moriyama K, Ichikawa S, Yahara I, Inagaki F. Tertiary structure of destrin and structural similarity between two actin-regulating protein families. Cell. 1996;85:1047–1055. doi: 10.1016/s0092-8674(00)81305-7. [DOI] [PubMed] [Google Scholar]
- Hawkins M, Pope B, Maciver SK, Weeds AG. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry. 1993;32:9985–9993. doi: 10.1021/bi00089a014. [DOI] [PubMed] [Google Scholar]
- Hayden SM, Miller PS, Brauweiler A, Bamburg JR. Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochemistry. 1993;32:9994–10004. doi: 10.1021/bi00089a015. [DOI] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lappalainen P, Drubin DG. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P, Kessels MM, Cope MJ, Drubin DG. The ADF homology (ADF-H) domain: a highly exploited actin-binding module. Mol Biol Cell. 1998;9:1951–1959. doi: 10.1091/mbc.9.8.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol. 2001;3:544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigellausing pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Lopez I, Anthony RG, Maciver SK, Jiang CJ, Khan S, Weeds AG, Hussey PJ. Pollen specific expression of maize genes encoding actin depolymerizing factor-like proteins. Proc Natl Acad Sci USA. 1996;93:7415–7420. doi: 10.1073/pnas.93.14.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK, Pope BJ, Whytock S, Weeds AG. The effect of two actin depolymerizing factors (A.D.F/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human A.D.F., but not of Acanthamoebaactophorin. Eur J Biochem. 1998;256:388–397. doi: 10.1046/j.1432-1327.1998.2560388.x. [DOI] [PubMed] [Google Scholar]
- Maciver SK, Weeds AG. Actophorin preferentially binds monomeric ADP-actin over ATP-bound actin: consequences for cell locomotion. FEBS Lett. 1994;347:251–256. doi: 10.1016/0014-5793(94)00552-4. [DOI] [PubMed] [Google Scholar]
- Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A, Chiu W. ADF/cofilin weakens lateral contacts in the actin filament. J Mol Biol. 1999;291:513–519. doi: 10.1006/jmbi.1999.2968. [DOI] [PubMed] [Google Scholar]
- McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Matheson C, Marra MA, Wakarchuk MF, Baillie DL. The Caenorhabditis elegansunc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol Gen Genet. 1994;242:346–357. doi: 10.1007/BF00280425. [DOI] [PubMed] [Google Scholar]
- Mohri K, Takano-Ohmuro H, Nakashima H, Hayakawa K, Endo T, Hanaoka K, Obinata T. Expression of cofilin isoforms during development of mouse striated muscles. J Musc Res Cell Motil. 2000;21:49–57. doi: 10.1023/a:1005682322132. [DOI] [PubMed] [Google Scholar]
- Moon AL, Janmey PA, Louie KA, Drubin DG. Cofilin is an essential component of the yeast cortical cytoskeleton. J Cell Biol. 1993;120:421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K, Nishida E, Yonezawa N, Sakai H, Matsumoto S, Iida K, Yahara I. Destrin, a mammalian actin-depolymerizing protein, is closely related to cofilin. Cloning and expression of porcine brain destrin cDNA. J Biol Chem. 1990;265:5768–5773. [PubMed] [Google Scholar]
- Moriyama K, Yahara I. Two activities of cofilin, severing and accelerating directional depolymerization of actin filaments, are affected differentially by mutations around the actin-binding helix. EMBO J. 1999;18:6752–6761. doi: 10.1093/emboj/18.23.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka R, Kusano K, Abe H, Obinata T. Effects of cofilin on actin filamentous structures in cultured muscle cells. Intracellular regulation of cofilin action. J Cell Sci. 1995;108:581–593. doi: 10.1242/jcs.108.2.581. [DOI] [PubMed] [Google Scholar]
- Obinata T, Nagaoka-Yasuda R, Ono S, Kusano K, Mohri K, Ohtaka Y, Yamashiro S, Okada K, Abe H. Low molecular-weight G-actin binding proteins involved in the regulation of actin assembly during myofibrillogenesis. Cell Struct Funct. 1997;22:181–189. doi: 10.1247/csf.22.181. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Tashima M, Yumoto Y, Okuda T, Sawada H, Okuma M, Maruyama Y. Coding sequence of human placenta cofilin cDNA. Nucleic Acids Res. 1990;18:7169. doi: 10.1093/nar/18.23.7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Baillie DL, Benian GM. UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegansbody wall muscle. J Cell Biol. 1999;145:491–502. doi: 10.1083/jcb.145.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Benian GM. Two Caenorhabditis elegansactin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. J Biol Chem. 1998;273:3778–3783. doi: 10.1074/jbc.273.6.3778. [DOI] [PubMed] [Google Scholar]
- Ono S, McGough A, Pope BJ, Tolbert VT, Bui A, Pohl J, Benian GM, Gernert KM, Weeds AG. The C-terminal tail of UNC-60B (actin depolymerization factor/cofilin) is critical for maintaining its stable association with F-actin and is implicated in the second actin-binding site. J Biol Chem. 2001;276:5952–5958. doi: 10.1074/jbc.M007563200. [DOI] [PubMed] [Google Scholar]
- Ono S, Minami N, Abe H, Obinata T. Characterization of a novel cofilin isoform that is predominantly expressed in mammalian skeletal muscle. J Biol Chem. 1994;269:15280–15286. [PubMed] [Google Scholar]
- Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- Peränen J, Rikkonen M, Hyvönen M, Kääriäinen L. T7 vectors with a modified T7lac promoter for expression of proteins in Escherichia coli. Anal Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Rate constants for the reaction of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressad F, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D, Carlier MF. Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins. Comparison of plant and human ADFs and effect of phosphorylation. J Biol Chem. 1998;273:20894–20902. doi: 10.1074/jbc.273.33.20894. [DOI] [PubMed] [Google Scholar]
- Rice DB, Aberg T, Chan Y, Tang Z, Kettunen PJ, Pakarinen L, Maxon RE, Thesleff I. Integration of FGF and TWIST in calvarial bone and suture development. Development. 2000;127:1845–1855. doi: 10.1242/dev.127.9.1845. [DOI] [PubMed] [Google Scholar]
- Rosenblatt J, Agnew BJ, Abe H, Bamburg JR, Mitchison TJ. Xenopus actin depolymerizing factor/cofilin (XAC) is responsible for the turnover of actin filaments in Listeriamonocytogenes tails. J Cell Biol. 1997;136:1323–1332. doi: 10.1083/jcb.136.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger JW, Mittal B, Sanger JM. Analysis of myofibrillar structure and assembly using fluorescently labeled contractile proteins. J Cell Biol. 1984;98:825–833. doi: 10.1083/jcb.98.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sept D, Elcock AH, McCammon JA. Computer simulations of actin polymerization can explain the barbed-pointed end asymmetry. J Mol Biol. 1999;294:1181–1189. doi: 10.1006/jmbi.1999.3332. [DOI] [PubMed] [Google Scholar]
- Sheterline P. Actin. Protein Profile. 4th ed. Oxford, UK: Oxford University Press; 1998. [Google Scholar]
- Thirion C, Stucka R, Mendel B, Gruhler A, Jaksch M, Nowak KJ, Binz N, Laing NG, Lochmuller H. Characterization of human muscle type cofilin (CFL2) in normal and regenerating muscle. Eur J Biochem. 2001;268:3473–3482. doi: 10.1046/j.1432-1327.2001.02247.x. [DOI] [PubMed] [Google Scholar]
- Vainio S, Jalkanen M, Vaahtokari A, Sahlberg C, Mali M, Bernfield M, Thesleff I. Expression of syndecan gene is induced early, is transient, and correlates with mesenchymal cell proliferation during tooth organogenesis. Dev Biol. 1991;147:322–333. doi: 10.1016/0012-1606(91)90290-j. [DOI] [PubMed] [Google Scholar]
- Van Troys M, Dewitte D, Verschelde J-L, Goethals M, Vandekerckhove J, Ampe C. The competitive interaction of actin and PIP2with actophorin is based on overlapping target sites: design of gain-of-function mutant. Biochemistry. 2000;39:12181–12189. doi: 10.1021/bi000816c. [DOI] [PubMed] [Google Scholar]
- Vartiainen M, Ojala PJ, Auvinen P, Peranen J, Lappalainen P. Mouse A6/twinfilin is an actin monomer-binding protein that localizes to the regions of rapid actin dynamics. Mol Cell Biol. 2000;20:1772–1783. doi: 10.1128/mcb.20.5.1772-1783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeds AG, Harris H, Gratser W, Gooch J. Interactions of pig plasma gelsolin with G-actin. Eur J Biochem. 1986;161:77–84. doi: 10.1111/j.1432-1033.1986.tb10126.x. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Tang JX, Azuma T, Marks PW, Jamney P. Cofilin and gelsolin segment-1: molecular dynamics simulation and biochemical analysis predict a similar actin binding mode. J Mol Biol. 1998;282:921–932. doi: 10.1006/jmbi.1998.2048. [DOI] [PubMed] [Google Scholar]
- Yonezawa N, Nishida E, Koyasu S, Maekawa S, Ohta Y, Yahara I, Sakai H. Distribution among tissues and intracellular localization of cofilin, a 21 kDa actin-binding protein. Cell Struct Funct. 1987;12:443–452. doi: 10.1247/csf.12.443. [DOI] [PubMed] [Google Scholar]
- Yonezawa N, Nishida E, Sakai H. pH control of actin polymerization by cofilin. J Biol Chem. 1985;260:14410–14412. [PubMed] [Google Scholar]