Abstract
Environmental stress-induced phosphorylation of eIF2α inhibits protein translation by reducing the availability of eIF2-GTP-tRNAiMet, the ternary complex that joins initiator tRNAMet to the 43S preinitiation complex. The resulting untranslated mRNA is dynamically routed to discrete cytoplasmic foci known as stress granules (SGs), a process requiring the related RNA-binding proteins TIA-1 and TIAR. SGs appear to be in equilibrium with polysomes, but the nature of this relationship is obscure. We now show that most components of the 48S preinitiation complex (i.e., small, but not large, ribosomal subunits, eIF3, eIF4E, eIF4G) are coordinately recruited to SGs in arsenite-stressed cells. In contrast, eIF2 is not a component of newly assembled SGs. Cells expressing a phosphomimetic mutant (S51D) of eIF2α assemble SGs of similar composition, confirming that the recruitment of these factors is a direct consequence of blocked translational initiation and not due to other effects of arsenite. Surprisingly, phospho-eIF2α is recruited to SGs that are disassembling in cells recovering from arsenite-induced stress. We discuss these results in the context of a translational checkpoint model wherein TIA and eIF2 are functional antagonists of translational initiation, and in which lack of ternary complex drives SG assembly.
INTRODUCTION
Stress granules (SGs) are discrete cytoplasmic foci at which untranslated mRNAs accumulate in cells subjected to environmental stress (Nover et al., 1983, 1989; Scharf et al., 1998; Kedersha et al., 1999, 2000). In mammalian cells, the assembly of SGs is initiated by the phosphorylation of eIF2α (Kedersha et al., 1999), a translation initiation factor that loads the initiator tRNA onto the small ribosomal subunit (reviewed by Berlanga et al., 1998; Gray and Wickens, 1998). Phosphorylation of eIF2α inhibits protein translation and promotes the accumulation of untranslated mRNA (Farrell et al., 1977; Srivastava et al., 1998). The related RNA-binding proteins TIA-1 and TIAR act downstream of the phosphorylation of eIF2α to recruit these untranslated mRNAs to SGs (Kedersha et al., 1999). By sequestering untranslated mRNA from the translational machinery, SGs have been proposed to determine the duration of stress-induced translational arrest (Kedersha et al., 1999). Because heat-shock transcripts are excluded from the SGs found in heat-shocked plant cells (Nover et al., 1983), this mechanism might explain, in part, the preferential translation of heat-shock transcripts in stressed cells.
Although the sequestration of mRNA at SGs is likely to contribute to stress-induced translational arrest, SGs are not passive repositories of untranslated mRNA. The different effects of drugs that stabilize or destabilize polysomes (i.e., emetine and puromycin, respectively) on the assembly of SGs indicate that mRNA can move between polysomes and SGs (Kedersha et al., 2000). Although the rate at which mRNA moves in and out of SGs has not been directly measured, poly(A)+ binding protein I (PABP-I), a marker of poly(A)+ RNA, rapidly shuttles in and out of SGs (∼50% of SG-associated PABP-I is replaced every 20 s). The shuttling of TIA-1 is ∼10 times faster than that of PABP-I (∼50% of SG-associated TIA-1 is replaced every 2 s). Given these kinetics, and considering that a dominant negative mutant of TIA-1 prevents SG assembly altogether (Kedersha et al., 1999), it appears that TIA-1 escorts untranslated mRNA to SGs. The dynamic shuttling of SG-associated mRNA and protein led us to postulate that SGs may be sites of mRNA triage at which untranslated mRNAs are sorted and processed for either reinitiation, degradation, or packaging into stable nonpolysomal messenger ribonucleoprotein (mRNP) complexes (Kedersha et al., 2000).
The ability of phospho-eIF2α to drive mRNA from polysomes to SGs suggests that impaired translational initiation might contribute to the assembly of SGs. The highly dynamic nature of SG proteins revealed by measuring fluorescent recovery after photobleaching of green fluorescent protein (GFP)-TIA-1 and GFP-PABP suggested that biochemical purification of intact SGs might be impossible. We therefore used immunofluorescence microscopy to determine whether known translation initiation factors are components of SGs. With the exception of eIF2, components of the 48S preinitiation complex (i.e., eIF3, eIF4E, eIF4G, and small, but not large, ribosomal subunits) are selectively recruited to arsenite- or eIF2α(S51D)-induced SGs. SGs can also be induced by energy starvation under conditions that do not induce eIF2α phosphorylation, suggesting that the proximal trigger for SG assembly may be the reduced availability of eIF2-GTP-tRNAiMet rather than eIF2α phosphorylation per se. We propose a model wherein eIF2-deficient 48S complexes assembled in stressed cells are recruited by TIA-1/TIAR into SGs.
MATERIALS AND METHODS
Cell Lines
COS7 and DU145 cell lines were obtained from American Type Culture Collection (Rockville, MD) and cultured in DMEM with 10% fetal bovine serum, in a CO2 incubator using 7% CO2.
Antibodies and Reagents
Anti-TIA-1/R hybridoma (3E6) was raised in our laboratory as described previously (Taupin et al., 1995). Antibodies against hemagglutinin (HA) were obtained from Berkeley Antibody (Berkeley, CA). Antibodies against phospho-eIF2α and HSP27 were obtained from StressGen Biotechnologies (British Columbia, Victoria, Canada). A different polyclonal antibody against phospho-eIF2α was obtained from Research Genetics (Huntsville, AL) and gave similar results to those obtained with the StressGen antibody. Monoclonal antibodies against eIF4E, eIF4G, and eIF5 were obtained from Transduction Laboratories (Lexington, KY). Polyclonal affinity-purified goat antibodies against TIA-1, eIF-2α, and eIF4G, and rabbit antibodies against eIF5 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit antibody against PHAS-I (eIF4E-BP) was obtained from Zymed Laboratories (South San Francisco, CA). Human auto-antisera against ribosomal P antigen was obtained from ImmunoVision (Springdale, AR). Affinity-purified goat polyclonal antibody against eIF3 was a kind gift from John Hershey (University of California, Davis, Davis, CA). Anti-HuR was a kind gift from Imed Gallouzi (Yale University, New Haven, CT). Anti-heterogeneous nuclear ribonucleoprotein (hnRNP) C and anti-PABP (monoclonal 10E10) were a kind gift from Gideon Dreyfuss (University of Pennsylvania, School of Medicine, Philadelphia, PA). Anti-eIF-2α monoclonal antibody was a kind gift from Dr. Richard Panniers (National Institutes of Health, Bethesda, MD). Antibodies against eIF2Bε were generously provided by Scot Kimball (Pennsylvania State University College of Medicine, Hershey, PA). Affinity-purified antibodies against small ribosomal subunit proteins S3a and S19 were described previously (Lutsch et al., 1990). Affinity-purified antibodies against ribosomal proteins L5 and L37 were prepared from antisera obtained from a rabbit (L5) or a sheep (L37) immunized with purified proteins from rat liver ribosomes. Antisera were fractionated by 35% ammonium sulfate precipitation, dialyzed against phosphate-buffered saline (PBS), and purified against ribosomal proteins bound to Sepharose. The bound antibodies were eluted using 3.0 M sodium thiocyanate in PBS, dialyzed against PBS, and tested for subunit specificity by immunoblotting against purified ribosomal subunits. Secondary antibody conjugates (donkey anti-mouse, rabbit, goat, or human; all ML grade) conjugated to Cy2 or Cy3 were obtained from Jackson Immunoresearch Laboratories (West Grove, PA). Sodium arsenite, oligomycin, 2-deoxyglucose, carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), emetine, diphenyleneiodonium chloride (DPI), and puromycin were purchased from Sigma (St. Louis, MO).
Immunofluorescence and In Situ Fluorescence Staining
Cells were grown on coverslips, treated with arsenite or other drugs as indicated in the figure legends, fixed, and stained for either protein as described previously (Kedersha et al., 1999), or processed for fluorescent in situ poly(A)+ RNA detection in combination with immunofluorescence protein detection. For in situ staining, cells were fixed in 4% paraformaldehyde, postfixed and permeabilized in absolute methanol, and equilibrated with 50 mM Tris buffer pH 7.5. Cells were hybridized to an end-labeled biotinylated oligo-dT (50 nucleotides; Invitrogen, Carlsbad, CA) by using a commercially available kit (mRNA Locator-Hyb kit; Ambion, Austin, TX), at 45° for 4 h. After washing with buffers supplied in the kit, the cells were equilibrated with 2× SSC containing 0.1% Triton X-100 (Sigma), supplemented with goat antibodies against anti-TIA-1 (Santa Cruz Biotechnology) and containing Alexa-594–labeled strepavidin (Molecular Probes, Eugene, OR) diluted 1/2000. After a 1-h incubation, cells were washed in 2× SSC and then incubated for 1 h in donkey anti-goat Cy antibody (1/200 dilution) and 0.5 μg/ml Hoechst dye. Cells were washed and mounted, and specimens were viewed using a Nikon Eclipse 800 microscope. Images were digitally captured using a charge-coupled device-SPOT RT digital camera and compiled using Adobe Photoshop software (version 5.5; Adobe Systems, Mountain View, CA).
Sucrose Gradient Analysis
DU145 cells were plated and used within 48 h of plating, at ∼90% confluence. Monolayers were washed with Hanks' balanced salt solution, incubated in Hanks' balanced salt solution containing 10 μg/ml cycloheximide for 5 min, and then scrape harvested and centrifuged. Pellets were lysed in 0.5 ml of ice-cold lysis buffer (140 mM KCl, 1 mM dithiothreitol, 20 mM Tris pH 8, 5 mM MgCl2, 0.5% NP-40, 0.5 U/ml RNAsin [Promega, Madison, WI], 10 mM cycloheximide) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 5 μM leupeptin, 5 mM benzamidine, 5 μg/ml aprotinin), phosphotase inhibitors (1.0 mM sodium vanadate and 50 mM sodium fluoride), and 0.5 mM GMP-PNP (Roche Molecular Biochemicals, Indianapolis, IN). The cells suspension was mechanically disrupted using 12 strokes of a Teflon-glass homogenizer at low speed on ice. Nuclei were pelleted at low speed, and the postnuclear supernatant was subjected to microfuge centrifugation for 20 min at 14,000 × g. The resulting supernatant was then layered onto preformed 10–47% linear sucrose gradients (made up in 140 mM KCl, 1 mM dithiothreitol, 20 mM Tris-HCl pH 8, 5 mM MgCl2) over a 60%–0.5-ml sucrose cushion in 11-ml tubes. Centrifugation was performed at 40,000 rpm for 2 h 45 min by using a Beckman SW40Ti rotor. Gradients were eluted from the top by using a Brandel elution system (Brandel, Gaithersburg, MD). The eluate was continuously monitored at 254 nm using an ISCO UA5 UV monitor (ISCO, Lincoln, NE). Fractions were collected from the top of the gradient. Aliquots of individual fractions were acetone precipitated to remove sucrose and to concentrate the samples, resuspended in SDS-sample buffer, and processed for Western blot analysis.
Plasmid Construction
The plasmids encoding wild-type and the mutant eIF2α (S51D) were a gift from Dr. Randal Kaufman (University of Michigan, Ann Arbor, MI). The coding region of each plasmid was cloned in frame into the pMT2-HA vector by using a polymerase chain reaction (PCR) strategy. The coding region was amplified from PETF-VA-eIF2αwt or PETF-VA-eIF2S51D mutant for 30 cycles (94°C for 1 min, 50°C for 1 min, and 74°C for 1 min) using Tli polymerase (Promega) and primers with EcoRI and XbaI cloning sites (GGGAATTCATGCCGGGTCTAAGTTGTAGATTTTA and GCTCTAGATTAATCTTCAGCTTTGGCT), respectively. The inserts were cut with EcoRI and XbaI and cloned in-frame with an HA tag in pMT2 that was similarly cut. The plasmid encoding pCDNA3-HA-eIF2β was obtained by using PCR to amplify the coding region from pGEX-2T-eIF2β, a generous gift from Drs. Supratik Das and Umadas Maitra (Albert Einstein College of Medicine, Yeshiva University, Bronx, NY) with the primers CGCGGATCCATGTCTGGGGACGAGATG and CCGGAATTCTTAGTTAGCTTTGGCACG. The PCR product was cloned into pCDNA3 in-frame with the N-terminal HA tag at the BamHI and EcoRI sites. The plasmid encoding pCDNA3 HA-eIF3p48/int6 was constructed by in-frame ligation into pcDNA3 N-terminal HA of the ClaI modified, high-fidelity pfu polymerase-amplified open reading frame (ORF), starting from the second codon, by using human PBL cDNA and the primers AGAGAGATCGATCCTACTGGAGACTTTGATTCG and AGAGA GATCGATAC-CGAGTACACAGTGGCAGCT. The final clones were verified by sequencing.
COS Cell Transfections
COS7 cells were transfected using SuperFect (QIAGEN, Valencia, CA) according to the manufacturer's instructions. Cells plated in six-well plates (2 × 105 cells/well plated 20 h before transfection) were transfected for 2–4 h then trypsinized and replated into parallel plates for immunofluorescence (24-well plates containing 11-mm coverslips).
RESULTS
Small, but not Large, Ribosomal Subunits Are Components of SGs
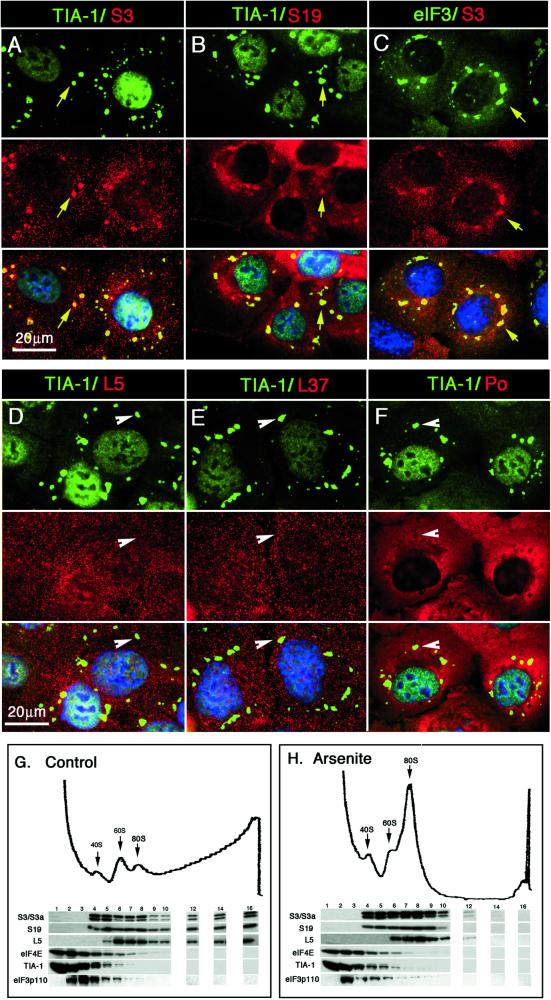
Stress-induced phosphorylation of the translation initiation factor eIF2α has profound effects on protein translation. Although the translation of most mRNAs is reduced, the translation of mRNAs encoding heat-shock proteins and stress-induced transcription factors (e.g., GCN4 in yeast and ATF4 in mammals; Hinnebusch, 1996, 1997; Harding et al., 2000) is enhanced in response to phospho-eIF2α. In plant cells, the untranslated mRNAs that accumulate in heat-stressed cells are concentrated at discrete cytoplasmic foci known as heat-shock granules (Nover et al., 1989). In mammalian cells, poly(A)+ RNA accumulates at similar cytoplasmic foci in response to environmental stress (Kedersha et al., 1999). It remains to be determined whether mammalian SGs are sites at which untranslated mRNAs are sequestered from the translational machinery or sites at which selected mRNAs are actively translated during stress. To distinguish between these possibilities, we used two-color immunofluorescence microscopy to determine whether ribosomes are components of mammalian SGs. Arsenite-stressed DU145 cells were probed using antibodies reactive with TIA-1/R and either ribosomal protein S3/3a (Figure 1A), ribosomal protein S19 (Figure 1B), ribosomal protein L5 (Figure 1D), ribosomal protein L37 (Figure 1E), or ribosomal protein Po (Figure 1F). As found previously, arsenite treatment causes most of the cytoplasmic TIA-1 and TIAR to accumulate at SGs in these cells (Figure 1, A, B, D–F; top, green). In paired views of the same fields, antibodies specific for small ribosomal subunit proteins S3/3a and S19 (Figure 1, A and B; middle, red) reveal that a significant fraction of the small ribosomal subunits accumulates at the TIA-1/R+ SGs (yellow arrows; merged views are shown in the bottom panels). The subset of small ribosomal subunits present in SGs is also coincident with eIF3 (Figure 1C, green), suggesting that the small ribosomal subunits found at SGs are components of preinitiation complexes. Consistent with this notion, components of the large ribosomal subunit (L5, L37, and Po) do not accumulate at TIA-1/R+ SGs (Figure 1, D–F; middle, red). The merged views (Figure 1, bottom) confirm the absence of large ribosomal proteins at TIA-1/R+ SGs (arrowheads reveal green SGs). These results suggest that small, but not large, ribosomal subunits are components of mammalian SGs.
Figure 1.
Small, but not large ribosomal subunits, are recruited to SGs. DU145 cells were exposed to 1 mM sodium arsenite for 30 min and allowed to recover for 2 h to generate SGs of maximal size. Cells were fixed and stained for TIA-1 to reveal SGs (green, panels A, B, D, E, and F), and counterstained with affinity-purified antibodies against different ribosomal proteins: ribosomal protein S3/3a (A and C, red), ribosomal protein S19 (B, red), ribosomal protein L5 (D, red), and ribosomal protein L37 (E, red). (F) Localization of ribosome Po proteins (red) detected using a human auto-antisera. Affinity-purified antisera against eIF3 was used to reveal the colocalization of eIF3 (C, green) with SG-associated ribosomal S3/3a (C, red). Yellow arrows indicate recruitment of ribosomal proteins to SGs; white arrowheads indicate SGs to which large ribosomal subunit proteins are not recruited. Bar, 20 μm. Sedimentation of S20 extracts of control (G) versus arsenite-treated (H) DU145 cells were resolved on 10–47% sucrose gradients, eluted from the top, and the elution profile at OD254 was recorded. Fractions were collected and subjected to SDS-PAGE and Western blotting by using antibodies to components of the small (e.g., S3/S3a and S19) or large (e.g., L5, L37, and Po) ribosomal subunit, as well as the eIF4 component eIF4E, TIA-1, and eIF3.
To address the issue of whether TIA-1 associates with small ribosomal subunits using biochemical means, we fractionated lysates from DU145 cells cultured in the absence or presence of arsenite (0.5 mM, 30 min) on 10–47% sucrose gradients, and analyzed individual fractions for the presence of small and large ribosomal subunits, eIF3, eIF4E, and TIA-1 by immunoblotting. Control cell lysates yielded UV254 absorbance profiles containing the expected peaks corresponding to 40S and 60S ribosomal subunits as well as intact 80S ribosomes (Figure 1G). Fractions at higher densities contain polysomes that include both small and large ribosomal subunits (fractions 12–16). In lysates prepared from arsenite-treated cells, polysomes have completely dispersed, and the 80S peak has correspondingly increased. The major fraction of both eIF4E and TIA-1 is found in the soluble material which does not enter the gradient, but substantial amounts of each protein cosediment with the fractions containing 40–60S ribosomal subunits (Figure 1, G and H, fractions 4–6). As expected of a protein component of a particle with a mass of ∼600,000 (Hershey et al., 1996), eIF3p110 is not found among soluble proteins at the top of the gradient (i.e., fraction 1). Roughly half of eIF3 appears in fractions 2 and 3, apparently not associated with preinitiation complexes. Most of the remaining eIF3 is present in fractions 4 and 5, coincident with small ribosomal subunits. Surprisingly, the distribution of all of these proteins within the sucrose gradient is not significantly altered by arsenite-induced oxidative stress, despite their relocalization into SGs (Figure 1, A–C). Although we do not know whether SGs maintain their structural integrity during cell fractionation and sucrose gradient centrifugation, the only fractions that contain all SG constituents are fractions 4 and 5, which sediment at ∼50S. If a subset of SG components survives sucrose gradient centrifugation, these components are likely to migrate at the position expected of the 48S preinitiation complex.
Components of 48S Preinitiation Complex Are Selectively Recruited to SGs
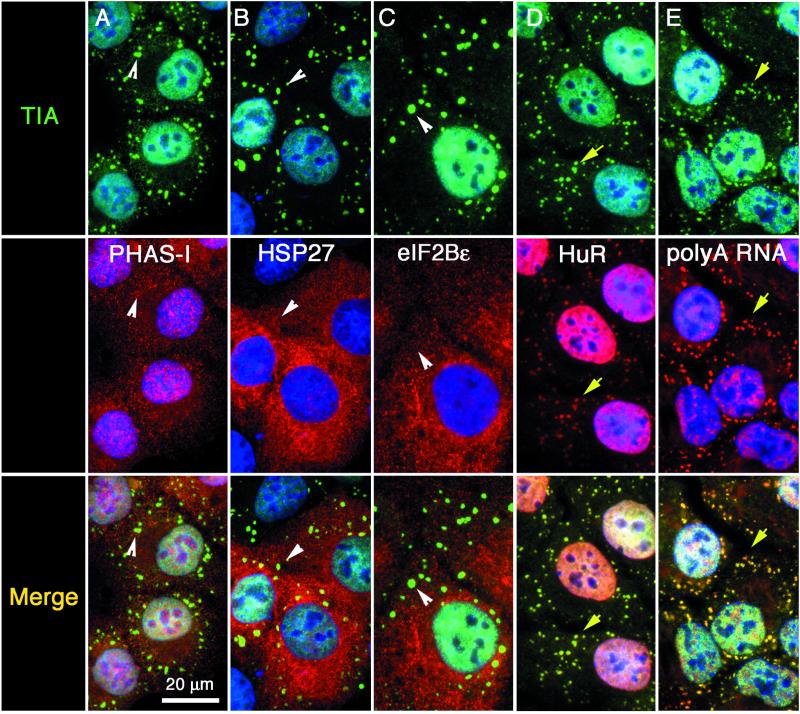
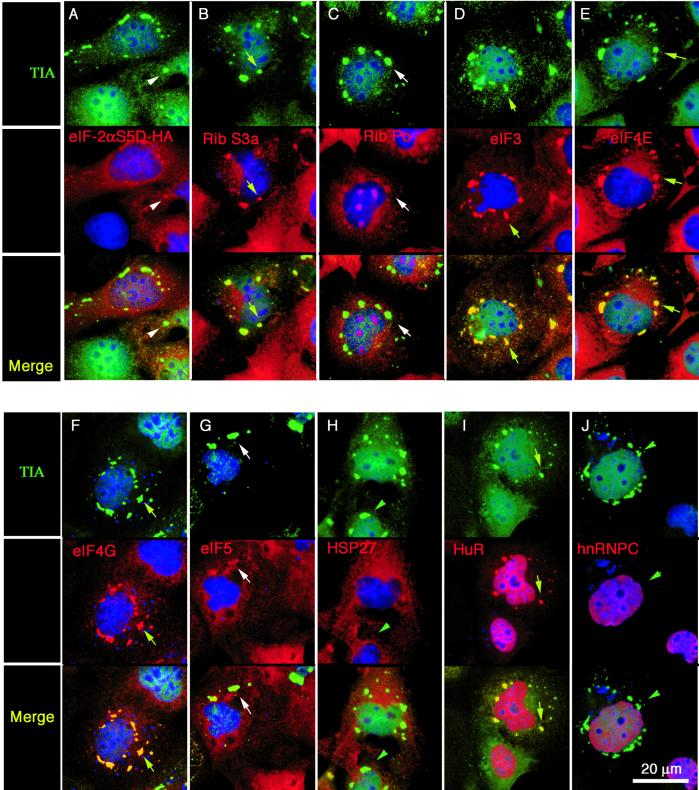
The absence of large ribosomal subunits eliminates the possibility that SGs are sites of privileged translation during stress. Because only the eIF3-colocalized fraction of the total detectable small ribosomal subunits was associated with SGs, we reasoned that the SG-associated small ribosomal subunits might represent 48S preinitiation complexes (which include mRNA and the small, but not the large, ribosomal subunit). We therefore used immunofluorescence microscopy to compare the subcellular localization of TIA-1 or TIAR+ SGs and individual components of the translation initiation apparatus in arsenite-stressed (0.5 mM, 1 h) DU145 cells. In the survey shown in Figures 2 and 3, the top panels show the localization of TIA-1/R+ SGs (green). In paired views of the same fields, the middle panels show the localization of the indicated translation initiation factors (red). The bottom panels show the merged images. Arrows indicate TIA-1+ SGs to which initiation factors are recruited in the paired views, whereas arrowheads indicate TIA-1+ SGs to which the indicated translation initiation factors are not recruited. This analysis reveals that eIF3, eIF4E, eIF4G, the RNA-binding protein HuR, and poly(A)+ RNA are highly concentrated at SGs, whereas eIF-2α, eIF5, PHAS-I/eIF4E-BP1, HSP27, and eIF2Bε do not accumulate at SGs. HSP27, which was previously shown to be present in heat-shock–induced SGs, is not a component of arsenite-induced SGs (Figure 3B, red). To ensure antibody specificity, two different commercial antibodies were used against eIF-4E and eIF-4G and gave identical results (our unpublished data). To confirm that eIF3, but not eIF2, is a component of SGs, we constructed expression vectors encoding HA-eIF2α, HA-eIF2β, and HA-p48/int-6 (a subunit of eIF3), so as to detect these different subunits by using the same anti-HA antibody. In COS transfectants, immunofluorescence microscopy with anti-HA revealed that each of these recombinant proteins is distributed between nuclear and cytoplasmic compartments (Figure 4, A–C; top, green). The subcellular localization of TIA-1 is shown in paired views of the same fields (Figure 4, A–C; middle, red). The merged views (Figure 4, A–C) are shown in the bottom panels. In untreated cells, there is little or no colocalization of either HA-eIF2α or HA-eIF2β with TIA-1 (Figure 4, A and B). In contrast, HA-p48 eIF3 accumulates at cytoplasmic foci that include TIA-1 in ∼50% of the transfected cells (Figure 4C, arrows). In response to arsenite-induced oxidative stress, HA-p48eIF3 (Figure 4F, arrows), but not HA-eIF2α (Figure 4D, arrowheads) or HA-eIF2β (Figure 4E, arrowheads), is recruited to TIA-1+ SGs (Figure 4, D–F; middle, red) in 100% of the transfected cells. These results confirm that eIF3, but not eIF2, is a component of SGs and supports the concept that SGs are sites at which eIF2/ternary complex-deficient preinitiation complexes accumulate during stress.
Figure 2.
Components of the 48S preinitiation complex are recruited to stress granules. DU145 cells were treated with arsenite (0.5 mM, 1 h) and processed for multicolor immunofluorescence microscopy by using antibodies reactive with TIA-1/R (green), the indicated initiation factor or other protein (red), and DNA (blue). Each vertical column shows views of the same field. Top row: TIA-1 or TIAR (green). Middle row (red): eIF2α (A), eIF3 (B), eIF4E (C), eIF4G (D), and eIF5 (E). Bottom row, merged views. Yellow arrows point out individual stress granules in which both the red and the green signals overlap; white arrowheads indicate the position of SGs that only contain TIA. Nuclei are stained blue. Bar, 20 μm.
Figure 3.
Other factors present or absent from stress granules. DU145 cells were stimulated with arsenite as described in Figure 2 and processed for immunofluorescence. Each vertical column shows views of the same field. Top row: TIA-1 or TIAR (green). Middle row (red): PHAS-I (eIF4E-binding protein) (A), HSP27 (B), eIF2Bε (C), and HuR (D); poly(A)+ RNA (E). Bottom row, merged views. Yellow arrows point out individual stress granules in which both the red and the green signals overlap; white arrowheads indicate the position of SGs that only contain TIA. Nuclei are stained blue. Bar, 20 μm.
Figure 4.
Recombinant HA-eIF2α, and HA-eIF2β are not recruited to SGs; recombinant HA-eIF3p48 is recruited to SGs. COS cells were transiently transfected with HA-eIF2α (A and D; green), HA-eIF2β (B and E; green); or HA-eIF3p48/int6 (C and F; green). Untreated cells (A–C) or arsenite-treated cells (D–F) were fixed and stained for HA (green, all panels) and TIA-1 (red, all panels). Arrows indicate SGs in which the HA-tagged protein is recruited to the SG; arrowheads indicate SGs where no recruitment of the HA-tagged protein is detected. Bar, 20 μm.
Coordinate Recruitment and Dispersal of eIFs during Assembly and Disassembly of SGs
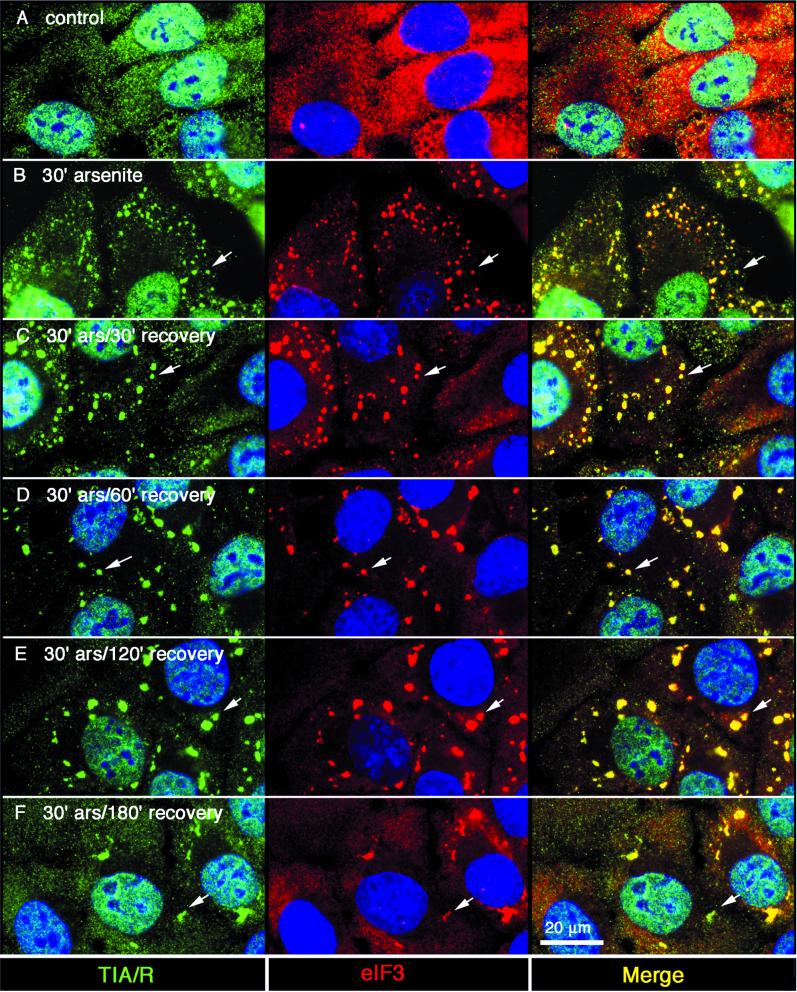
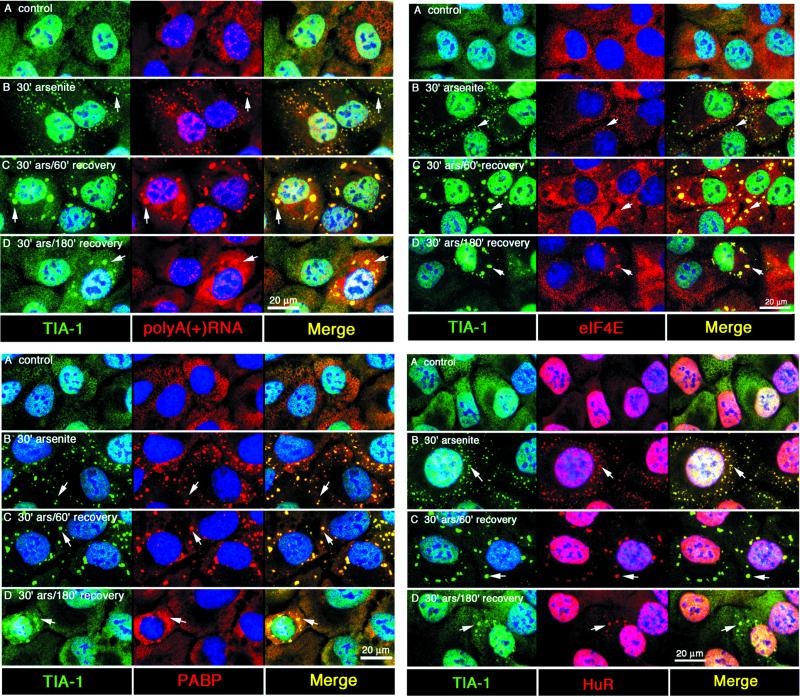
The above-mentioned compositional analysis suggested that poly(A)+ mRNA is recruited to SGs as a component of a modified 48S preinitiation complex. If this is true, individual components of the mRNP complex should be coordinately recruited to, and dispersed from, assembling and disassembling SGs. In cells pulsed with a sublethal dose of arsenite, SGs that assemble in response to the initial stress spontaneously disperse as cells recover (Kedersha et al., 1999). We used this system to determine whether SG components are coordinately recruited to, and dispersed from, SGs. DU145 cells were pulsed with a sublethal dose of arsenite (1.0 mM, 30 min) and then allowed to recover in media without arsenite. Cells were fixed at different times and SG composition was assessed by using immunofluorescence. Nascent SGs are small and dispersed throughout the cell; they later coalesce into larger, perinuclear structures as previously shown using GFP-TIA-1 in live cells (Kedersha et al., 2000). In unstressed cells, eIF3 is diffusely present in the cytoplasm (Figure 5A; red), whereas TIA-1/R is distributed between the nucleus and the cytoplasm in a micropunctate pattern (Figure 5, row A; detected using an antibody that sees both TIA-1 and TIAR, shown in green). In cells subjected to arsenite treatment, eIF3 (red) is recruited to SGs with similar kinetics as TIA-1/R (green) during both stress and recovery (Figure 5, B–F). Even during the earliest stages of SG assembly (i.e., after 30 min of exposure to arsenite; Figure 5B), eIF3 is present in the smallest SGs, coincident with SGs containing TIA-1/R (Figure 5B, arrows); this colocalization appears yellow in the merged view. After the removal of arsenite, eIF3 and TIA-1/R continue to accumulate at SGs, which become less numerous but progressively enlarge over the first 2 h of the recovery phase. During the 3rd h of recovery, SGs are disassembled, allowing TIA-1/R and eIF3 to return to their original subcellular locales (Figure 5F). Thus, eIF3 is recruited to SGs in parallel with TIA-1/R during SG assembly and disassembly, and is recruited to SGs very early during their assembly. Similar time course experiments were performed to determine whether poly(A)+ RNA, eIF4E, PABP, and the ELAV protein HuR are coordinately recruited to SGs with TIA-1 and TIAR. As shown in the abbreviated analysis in Figure 6, each of these mRNP components is coordinately recruited to, and dispersed from, SGs. Similar results were seen with eIF4G and small ribosomal subunit S3 (our unpublished data). These results suggest that poly(A)+ RNA moves in and out of SGs as a component of a modified 48S preinitiation complex.
Figure 5.
Kinetics of the colocalization of TIAR and eIF3 at stress granules. DU145 cells were treated with 1.0 mM sodium arsenite for 30 min, washed, and then further incubated in the absence of arsenite (recovery) for the indicated times. Each row depicts parallel views of the same field. TIA-1/R appears green, eIF3 appears red, and coincidence appears yellow (right column: merge). Control (A), 30-min arsenite treatment without recovery (B), 30-min arsenite followed by 30-min recovery (C); 30-min arsenite followed by 1-h recovery (D); 30-min arsenite followed by 2-h recovery (E); and 30-min arsenite followed by 3-h recovery (F). Nuclei are stained blue. Bar, 20 μm.
Figure 6.
Coordinate recruitment of poly(A)+ RNA, PABP-I, eIF4E, and HuR to SGs during SG assembly and disassembly. DU145 cells were arsenite-stressed and allowed to recover as in Figure 5. Top left, TIA-1 (green) versus poly(A) (red); bottom left, TIA-1 (green), PABP (red); Top right, TIA-1 (green), eIF4E (red); bottom right, TIA-1(green), HuR (red). In all panels, blue represents DNA (Hoechst staining). Bar, 20 μm. Merged views are shown in the right panels of each series. Arrows indicate SGs in each view.
Recruitment of Translation Initiation Factors to eIF2α (S51D)-induced SGs
Arsenite has complex effects on cells that are not limited to the activation of eIF2α kinases (Bernstam and Nriagu, 2000). We therefore determined the composition of SGs that are assembled in COS cells transfected with a phosphomimetic mutant of eIF2α (i.e., S51D), which was previously shown to induce the assembly of TIA-1+ SGs (Kedersha et al., 1999). Like arsenite-induced SGs, HA-tagged eIF2α (S51D)-induced SGs are dispersed by cycloheximide or emetine (our unpublished data), suggesting that they are in dynamic equilibrium with polysomes as are arsenite-induced SGs. As shown using antibodies reactive with the HA tag (Figure 7A; red), recombinant HA-eIF2α (S51D) is distributed diffusely throughout the cell, but a minor fraction of it is clearly localized to the TIA-1+ SGs that it induces (Figure 7A; green; white arrow points out SG). HA-eIF2α (S51D)-induced SGs strongly recruit ribosomal protein S3a (Figure 7B; red), eIF3 (Figure 7D; red), eIF4E (Figure 7E; red), and eIF4G (Figure 7F; red), because these proteins are clearly concentrated at SGs, which appear yellow in the merged views shown at the bottom of each column (yellow arrows). Weaker, partial recruitment of ribosome Po antigen (Figure 7C; red) and eIF5 (Figure 7G; red) is observed (white arrows), in contrast to results obtained with arsenite-induced SGs (Figures 1F and 2E, respectively), suggesting that these factors may be minor or transient SG components that are only detectable when eIF2α is continuously phosphorylated. HSP27 (Figure 7H; red) and eIF2B (our unpublished data) are not recruited to HA-eIF2α (S51D)-induced SGs. These results confirm earlier studies (Kedersha et al., 1999) reporting that HSP27 is a component of SGs induced by heat shock, but not by other stimuli. Although HSP27 has been reported to coaggregate with eIF4G and form insoluble cytosolic heat-shock granules, these were reported to exclude eIF4E, eIF3, and PABP (Cuesta et al., 2000) based on sedimentation, suggesting that heat-shock granules are distinct from SGs. The RNA-binding protein HuR was reported to form cytosolic aggregates in response to heat shock (Gallouzi et al., 2000), leading us to examine its recruitment to SGs. We find that HuR is recruited to HA-eIF2α (S51D)-induced TIA-1+ SGs (Figure 7I; red, yellow arrows) and is also recruited to arsenite-induced TIA-1+ SGs (Figure 3D; red). As expected, the nonshuttling hnRNP C is not recruited to HA-eIF2α (S51D)-induced SGs (Figure 7J; red) nor to arsenite-induced SGs (our unpublished data). Thus, the minimal “core” SG induced by expression of phosphomimetic eIF2α contains eIF3, eIF4E, eIF4G, and HuR in addition to the previously reported TIA-1, TIAR, PABP-I, and poly(A)+ RNA (Kedersha et al., 1999).
Figure 7.
Composition of eIF2α (S51D)-induced stress granules. COS cells were transiently transfected with a plasmid encoding recombinant phosphomimetic eIF2α (S51D) to induce the assembly of stress granules. After 28 h, cells were processed for two-color immunofluorescence microscopy by using antibodies reactive with TIA-1 or TIAR (green) and the indicated translation factors (red). Each column contains paired views of the same field, with the merged image at the bottom. Shown in red are HA-eIF2α (S51D) (A); small ribosomal protein S3a (B); large ribosomal subunit protein Po (C); eIF3 (D); eIF4E (E); eIF4G (F); eIF5 (G); HSP27 (H); HuR (I); and hnRNP C (J). SGs appear yellow/orange when the indicated translation factor colocalizes with TIA-1 (eIF3, eIF4E, eIF4G, ribosomal subunit S3, HuR; yellow arrows) or green when the indicated translation factor does not colocalize with TIA-1 (e.g., HSP27, hnRNP C; green arrowheads). Minimal colocalization is seen with HA-eIF2α (S51D), ribosome P antigen, and eIF-5 (white arrows). Nuclei are stained blue. Bar, 20 μm.
The association of small, but detectable, amounts of phosphomimetic HA-eIF2α (S51D) with the SGs that it induces (Figure 7A) suggested that phospho-eIF2α, which constitutes a subset of total eIF2α, might be selectively recruited to SGs (as opposed to total eIF2α, which did not appear to be a component of arsenite-induced nascent SGs as shown in Figures 2A and 4D). To examine this possibility, we used antibodies specific for phospho-eIF2α to monitor to its localization during SG assembly/disassembly (Figure 8). In the absence of stress, the phospho-eIF2α antibodies reveal nonspecific nuclear staining (Figure 8A; red). This weak nuclear staining was detected using antibodies from two different commercial sources and probably represents weak cross-reactivity against the very abundant nuclear phosphoproteins (e.g., histones). Although 30 min of arsenite treatment (Figure 8B) clearly elevates the cytoplasmic phospho-eIF2α signal, there is no colocalization with TIA-1+ SGs (Figure 8B, white arrows; SGs appear green in the merged field). After 30 min of recovery, TIA-1+ SGs increase in size (Figure 8C; green, arrows), but very little phospho-eIF-2α is present at the SGs (Figure 8C; red). As recovery progresses, and the total amount of phospho-eIF-2α decreases, the residual phospho-eIF2α is quantitatively found at SGs (Figure 8, D and E). The levels of cytoplasmic phospho-eIF2α detected by immunofluorescence parallel those shown by Western blot on identically treated samples (Figure 8G), and similar results were obtained using antibodies from two different commercial sources. It therefore appears that although eIF2α phosphorylation initiates SG assembly, little if any phospho-eIF2α is physically present within nascent SGs. It is only during recovery, when the amount of total phospho-eIF2α is reduced relative to the amount produced during the initial 30-min exposure (Figure 8G, compare lane E with lane B) and only a minor percentage of the cells have SGs, that phospho-eIF2α is localized to the SG. That this result is real and not due to some nonspecific effects of the antibody is supported by the detection of a small but consistent amount of phophomimetic HA-eIF2αS51D at SGs (Figure 8H), as detected with anti-HA antibodies. The limited colocalization of the phosphomimetic HA-eIF2αS51D at SGs suggests that these are compositionally similar to “late” SGs rather than “early” ones.
Figure 8.
Phospho-eIF2α is only present in stress granules during recovery. DU145 cells were treated with arsenite and allowed to recover as described in Figure 5 and then stained for TIA-1 and phospho-eIF2α. Left column, TIA-1 (green) and DNA (blue); middle column, phospho-eIF2α(red) and DNA (blue); right column, merge. Control (A), 30-min arsenite treatment without recovery (B), 30-min arsenite followed by 30-min recovery (C), 30-min arsenite followed by 1-h recovery (D), 30-min arsenite followed by 2-h recovery (E), and 30-min arsenite followed by 3-h recovery (F). Bars, 20 μm. (G) Immunoblot of equal amounts of total protein lysates from cells treated identically as described above, probed with phospho-eIF2α antibody. (H) Phospho-mimetic HA-eIF2αS51D was expressed in COS and detected with anti-HA (green). Cells were counterstained for TIA-1 (red) and DNA (blue). Arrows indicate position of SG, to which a minor but detectable fraction of the HA-tagged protein is localized. Bar, 20 μm.
Energy Depletion Induces SGs without Inducing Phosphorylation of eIF2α
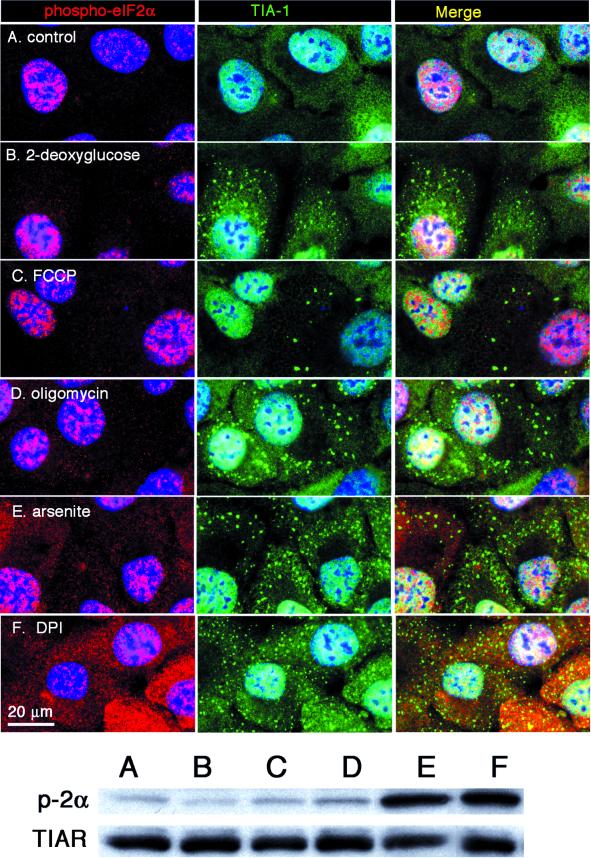
Phosphorylation of eIF2α arrests translation by limiting the amount of eIF2-GTP-tRNAiMet ternary complex; this occurs because eIF2(αP)GDP acts as competitive inhibitor of eIF2B, the guanine nucleotide exchange factor that allows eIF2 to exchange GDP for GTP. Only eIF2-GTP can bind met-tRNAi and yield the eIF2-GTP-tRNAiMet ternary complex that is required for initiation (reviewed by Trachsel, 1996). The exchange of eIF2-GDP for eIF2-GTP is also impaired by a low cellular GTP/GDP ratio in vitro, leading to ternary complex deficiency and impaired translation (Walton and Gill, 1976; Hucul et al., 1985). We therefore treated cells with a variety of drugs that deplete intracellular energy stores to determine whether energy starvation could induce SGs independently of eIF2α phosphorylation. Brief treatment with inhibitors of glycolysis (2-deoxyglucose; Figure 9B), mitochondrial membrane potential (ionophore FCCP; Figure 9C), and the mitochondrial ATPase (oligomycin; Figure 9D) induce the assembly of SGs. Double staining for phospho-eIF2α (Figure 9; red) reveals that these SGs are assembled in cells in which there is no detectable increase in eIF2α phosphorylation (Figure 9, B–D; red) relative to the control (Figure 9A; red). In contrast, both arsenite (Figure 9E) and DPI, a specific inhibitor of flavin-dependent oxidoreductase (Figure 9F), induce SGs concurrently with elevated phospho-eIF2α. Immunofluorescence was used to verify that SGs induced by energy starvation contain other SG markers including poly(A)+ RNA and eIF3 (our unpublished data). These treatments were insufficient to induce apoptosis, e.g., washing out the drugs allowed the cells to recover. Western blot analysis with phosphospecific anti-eIF2α antibodies confirms that 2-deoxyglucose, FCCP, and oligomycin do not induce the phosphorylation of eIF2α (Figure 9, bottom; antibodies reactive with TIAR were used to confirm equal loading between samples). We conclude that energy depletion can induce SGs without increasing eIF2α phosphorylation. Energy starvation and concurrent reduced ATP/GTP levels would affect on many facets of translation in addition to ternary complex formation, such as amino acyl-tRNA charging, eIF4A, eIF1A, and the termination events. However, the strong link between SG assembly and phosphorylation of phospho-eIF2α leads us to favor the possibility that reduced levels of GTP result in reduced levels of the eIF2-GTP-tRNAiMet ternary complex, and that the lack of ternary complex may drive SG formation, rather than increased phospho-eIF2α per se.
Figure 9.
Induction of SGs by energy depletion without inducing eIF2α phosphorylation. DU145 cells were treated with 2-deoxyglucose (B), FCCP (C), oligomycin (D), arsenite (E), or DPI (F) in either glucose-free media (B, C, D, and F) or regular media (E) for 30 min. Cells were then fixed and stained or lysed directly in SDS-buffer for PAGE. (Top) Immunofluorescence showing TIA-1 in green and phospho-eIF2α in red. Nuclei are stained blue. Bars, 20 μm. (Bottom) Immunoblot of total cell lysates treated as described above, probed with phospho-eIF2α and TIAR (as loading control).
DISCUSSION
Previous work from our lab and others (Nover et al., 1989; Kedersha et al., 1999, 2000) has established that SGs are highly dynamic cytoplasmic foci at which poly(A)+ RNA, TIA-1, TIAR, and PABP-I transiently accumulate in response to stress-induced phosphorylation of eIF2α. Both the RNA and protein components of SGs are in equilibrium with polysomes, as evinced by the dispersal of SGs upon treatment of cells with drugs that stabilize polysomes (e.g., emetine and cycloheximide), and by the enhanced assembly of SGs in cells treated with drugs that destabilize polysomes (e.g., puromycin). Herein, we demonstrate that SGs are not only in equilibrium with polysomes but also that they contain many components of the classical cap-dependent 48S preinitiation complex, including eIF3, eIF4E, eIF4G, and small (but not large) ribosomal subunits. Excluded from SGs are eIF2B, eIF5, HSP27, the eIF4E-binding protein PHAS-I, and surprisingly, eIF2. The SGs assembled in response to enforced expression of the S51D phospho-mimetic mutant of eIF2α have a similar composition as those induced by stress, suggesting that phosphorylation of eIF2α is sufficient to recruit these eIFs to SGs. The ability of metabolic poisons to induce the assembly of SGs without inducing the phosphorylation of eIF2α suggests that the SG-initiating effects of phospho-eIF2α are indirect. Both metabolic poisons and phospho-eIF2α reduce the levels of eIF2-GTP-tRNAiMet, however, suggesting that it is the lack of ternary complex that initiates SG formation rather than phosphorylation of eIF2α alone, although other explanations are possible. The major components of SGs [i.e., TIA-1/R, poly(A)+ mRNA, PABP-I (Kedersha et al., 1999), HuR, eIF3, eIF4E, and eIF4G (this study)], assemble into/disassemble from SGs with similar kinetics, suggesting that they are coordinately recruited to the SG as a complex. SG components that survive sucrose gradient sedimentation appear to migrate at 40–50S, consistent with our contention that SGs are composed of eIF2-deficient preinitiation complexes.
Eukaryotic translational initiation in vitro (reviewed by Dever, 1999; Pestova and Hellen, 1999; Hershey and Merrick, 2000) begins with the binding of eIF3 to the 40S ribosomal subunit. The subsequent recruitment of eIF1A and eIF2-GTP-tRNAiMet forms a 43S complex that binds to the mRNA/eIF4E/eIF4G/PABP mRNP to produce the canonical 48S preinitiation complex. In stressed cells, components of the 48S preinitiation complex (with the glaring exception of eIF2) are coordinately recruited to SGs. These results suggest that ternary complex-deficient “noncanonical” 48S complexes (hereafter referred to as 48S* complexes) are core constituents of SGs. The existence of some type of eIF2-deficient 48S preinitiation complex has been proposed to explain the preferential translation of stress-induced transcription factors such as ATF4 in mammals (Harding et al., 2000) and GCN4 in yeast (Hinnebusch, 1996, 1997; Harding et al., 2000), both of which are preferentially translated during conditions of eIF2α phosphorylation. Their transcripts possess long 5′ untranslated regions, which contain multiple small upstream ORFs, which prevent translation by causing scanning ribosomes to terminate before they reach the initiator codon of the stress-induced transcription factor. In stressed cells, the reduced availability of eIF2-GTP-tRNAiMet has been proposed to allow the assembly of eIF2-deficient 48S preinitiation complexes that scan past the upstream ORFs. When these eIF2-deficient 48S complexes acquire eIF2-GTP-tRNAiMet before they reach the initiation codon of the transcription factor, translation can begin. Both the induction of SG assembly by phospho-eIF2α and the composition of SGs suggest that they are comprised of 48S* complexes; whether these are identical to those proposed to regulate the translation of ATF4 and GCN4 remains to be determined.
We previously demonstrated that mRNA is in a dynamic equilibrium between polysomes and SGs (Kedersha et al., 2000), because treatment of SG-containing cells with emetine, a drug that “freezes” ribosomes on their mRNA, shifts the distribution of TIA-1 toward the polysome fractions in sucrose gradients prepared from arsenite-stressed cells (Kedersha et al., 2000). If eIF2-deficient 48S* preinitiation complexes comprise the basic unit of SGs, the mechanism whereby this occurs becomes apparent. The assembly of a TIA-1 containing scanning-incompetent/eIF2-deficient preinitiation complex at the 5′ end of a polysomal mRNA will allow elongating ribosomes to “run off” the transcript, effectively converting the mRNA from a polysome into a 48S* mRNP. Emetine treatment prevents this, allowing small amounts of TIA-1 to be shifted into the polysome region of the gradient (Kedersha et al., 2000). In the absence of emetine, the 48S* complexes are routed into SGs by the self-aggregating prion-like domain of TIA-1. Within the SG “triage” environment, the structure and composition of individual mRNPs could determine whether mRNAs are repacked into mRNPs, reinitiated, silenced, or degraded (Kedersha et al., 2000). The recruitment of the mRNA-stabilizing ELAV protein HuR to SGs supports this model, because both mRNA-stabilizing and -destabilizing proteins would be required at mRNA triage sites to determine the fates of individual transcripts.
Our data suggest that TIA-1/R and eIF2-GTP-tRNAiMet may act as functional antagonists to regulate the equilibrium between polysomes and SGs. This concept is supported by the following experimental observations: 1) the phosphomimetic eIF2α mutant (S51D) that reduces the concentration of active ternary complex promotes the assembly of SGs (Kedersha et al., 1999); 2) a nonphosphorylatable eIF2α mutant (S51A) that prevents the stress-induced depletion of active ternary complex inhibits arsenite-induced assembly of SGs (Kedersha et al., 1999); 3) overexpression of TIA-1ΔRRM, a truncation mutant that sequesters endogenous TIA-1 and TIAR, prevents SG assembly and promotes the expression of cotransfected reporter genes (Kedersha et al., 2000); and 4) overexpression of TIA-1 represses the expression of cotransfected reporter genes (Kedersha et al., 2000). The effects of recombinant TIA-1 and TIA-1ΔRRM are observed in the absence of stress, suggesting that these proteins might regulate the equilibrium between polysomal and nonpolysomal mRNPs in both stressed and unstressed cells.
The finding that phospho-eIF2α is selectively recruited only to disassembling SGs is remarkable, because it suggests that phospho-eIF2α may be required for SG disassembly. Because ternary complexes composed of phospho-eIF2α can initiate a single round of translation in vitro (reviewed by Trachsel, 1996), it is possible that such phospho-eIF2α ternary complexes are actually required to displace TIA-1/R from 48S* complexes and allow the restoration of normal initiation. Such a mechanism could explain the observed weak recruitment of ribosomal Po and eIF5 to eIF2α (S51D)-induced SGs. In this case, the SG-localized fraction of phosphomimetic eIF2α would be present as ternary complex and initiate translation. Although the extent to which the S51D mutant can be incorporated into ternary complex is not known, most of the SGs it induces are slowly dispersed upon cycloheximide treatment (our unpublished data), suggesting that their component mRNAs are in equilibrium with polysomes.
In addition to their role as organizers of SGs during environmental stress, TIA-1 and TIAR have been shown to selectively repress the translation of TNF-α transcripts. In macrophages lacking TIA-1, the polysome profile of tumor necrosis factor-α (TNF-α) transcripts is shifted such that the percentage of TNF-α transcripts associated with polysomes is increased compared with that of wild-type macrophages (Piecyk et al., 2000). This result suggests that TIA-1 represses the translation of TNF-α by promoting the assembly of a nonpolysomal mRNP complex. We suggest that this complex is structurally equivalent to the 48S* mRNP. Because TIA-1 binds to an AU-rich element in the 3′-untranslated region of TNF-α transcripts, the probability that it will interact with an assembling preinitiation complex will be markedly increased compared with other transcripts. The tethering of TIA-1 to TNF-α transcripts would increase the percentage of these transcripts found in nonpolysomal mRNP complexes, dampen the expression of TNF-α, and increase the sensitivity of these transcripts to regulatory control. In a similar manner, hnRNP K/E1, proteins that are tethered to the 3′-untranslated region of 15-lipoxygenase transcripts, induce the assembly of a translationally incompetent 48S preinitiation complex (Ostareck et al., 2001). The hnRNP K/E1 proteins act by preventing the joining of the 60S ribosomal subunit (Ostareck et al., 2001), while allowing the small ribosomal subunit to scan to the initiation codon, at which point the scanning is halted and the transcript is silenced. Because eIF2-GTP-tRNAiMet is required for the initiator codon recognition, it is likely that TIA-1-induced 48S* complexes promote translational silencing via a different mechanism. Future experiments are needed to resolve this issue.
Assessing translational control mechanisms in the context of the intact cell is complicated relative to the in vitro systems by the multifunctional nature of shuttling proteins such as TIA-1, TIAR, and HuR, which may perform different but related functions in the nucleus and the cytoplasm. Recent reports indicate that TIA-1 regulates specific mRNA splicing events (Del Gatto-Konczak et al., 2000; Forch et al., 2000; Le Guiner et al., 2001), suggesting that an indirect function of SG assembly in the cytoplasm may be the altered splicing of specific nuclear transcripts, owing to the relocalization of TIA-1 from the nucleus to SGs. Indeed, the stress-induced relocalization of the shuttling protein hnRNP A1 from the nucleus to the cytoplasm appears to regulate pre-mRNA splicing in vivo by such a mechanism (van der Houven van Oordt et al., 2000). The ability of TIA-1/R to recruit eIF-2-deficient 48S* complexes to SGs demonstrates the modular and versatile nature of mRNA binding proteins, and emphasizes that many aspects of mRNA metabolism occur concurrent with the movement of mRNA between organelles or subcellular domains.
ACKNOWLEDGMENTS
We thank members of the Anderson laboratory for helpful discussions. We thank Drs. Supratik Das, Gideon Dreyfuss, Imed Gallouzi, John Hershey, Randal Kaufman, Scot Kimball, Umadas Maitra, and Richard Panniers for generously providing many of the reagents used in this study. P.A. was supported by National Institutes of Health grant AI-33600 and a Biomedical Science Grant from the Arthritis Foundation.
Abbreviations used:
- eIF
eukaryotic initiation factor
- PABP-I
poly(A)+ binding protein I
- SG
stress granule
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–05-0221. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–05-0221.
REFERENCES
- Berlanga J, Herrero S, DeHaro C. Characterization of the hemin-sensitive eukaryotic initiation factor 2a kinase from mouse nonerythroid cells. J Cell Biol. 1998;273:32340–32346. doi: 10.1074/jbc.273.48.32340. [DOI] [PubMed] [Google Scholar]
- Bernstam L, Nriagu J. Molecular aspects of arsenic stress. J Toxicol Environ Health B Crit Rev. 2000;3:293–322. doi: 10.1080/109374000436355. [DOI] [PubMed] [Google Scholar]
- Cuesta R, Laroia G, Schneider RJ. Chaperone Hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 2000;14:1460–1470. [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F, Bourgeois CF, Le Guiner C, Kister L, Gesnel MC, Stevenin J, Breathnach R. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol Cell Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE. Translation initiation: adept at adapting. Trends Biochem Sci. 1999;24:398–403. doi: 10.1016/s0968-0004(99)01457-7. [DOI] [PubMed] [Google Scholar]
- Farrell PJ, Balkow K, Hunt T, Jackson RJ, Trachsel H. Phosphorylation of initiation factor elF-2 and the control of reticulocyte protein synthesis. Cell. 1977;11:187–200. doi: 10.1016/0092-8674(77)90330-0. [DOI] [PubMed] [Google Scholar]
- Forch P, Puig O, Kedersha N, Martinez C, Granneman S, Seraphin B, Anderson P, Valcarcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci USA. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–2108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Hershey JW, Asano K, Naranda T, Vornlocher HP, Hanachi P, Merrick WC. Conservation and diversity in the structure of translation initiation factor EIF3 from humans and yeast. Biochimie. 1996;78:903–907. doi: 10.1016/s0300-9084(97)86711-9. [DOI] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC. The pathway and mechanism of initiation of protein synthesis. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 33–88. [Google Scholar]
- Hinnebusch AG. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 199–244. [Google Scholar]
- Hinnebusch AG. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- Hucul JA, Henshaw EC, Young DA. Nucleoside diphosphate regulation of overall rates of protein synthesis acting at the level of initiation. J Biol Chem. 1985;260:15585–15591. [PubMed] [Google Scholar]
- Kedersha N, Cho M, Li W, Yacono P, Chen S, Gilks N, Golan D, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guiner, C., Lejeune, F., Galiana, D., Kister, L., Breathnach, R., Stevenin, J., and Del Gatto-Konczak, F. (2001). TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. (in press). [DOI] [PubMed]
- Lutsch G, Stahl J, Kärgel H-J, Noll F, Bielka H. Immunoelectron microscopic studies on the location of ribosomal proteins on the surface of the 40S ribosomal subunit from rat liver. Eur J Cell Biol. 1990;51:140–150. [PubMed] [Google Scholar]
- Nover L, Scharf K, Neumann D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol Cell Biol. 1983;3:1648–1655. doi: 10.1128/mcb.3.9.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck D, Ostareck-Lederer A, Shatsky I, Hentze M. Lipoxygenase mRNA silencing in erythroid differentiation: the 3′ UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. Ribosome recruitment and scanning: what's new? Trends Biochem Sci. 1999;24:85–87. doi: 10.1016/s0968-0004(99)01356-0. [DOI] [PubMed] [Google Scholar]
- Piecyk M, et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Heider H, Hohfeld I, Lyck R, Schmidt E, Nover L. The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol. 1998;18:2240–2251. doi: 10.1128/mcb.18.4.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Kumar K, Kaufman R. Phosphorylation of eIF2alpha mediates apoptosis in response to activation of the double stranded RNA-dependent protein kinase. J Biol Chem. 1998;273:2416–2423. doi: 10.1074/jbc.273.4.2416. [DOI] [PubMed] [Google Scholar]
- Taupin JL, Tian Q, Kedersha N, Robertson M, Anderson P. The RNA-binding protein TIAR is translocated from the nucleus to the cytoplasm during Fas-mediated apoptotic cell death. Proc Natl Acad Sci USA. 1995;92:1629–1633. doi: 10.1073/pnas.92.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel H. Binding of initiator methionyl-tRNA to ribosomes. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 113–138. [Google Scholar]
- van der Houven van Oordt W, Diaz-Meco MT, Lozano J, Krainer AR, Moscat J, Caceres JF. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J Cell Biol. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton GM, Gill GN. Preferential regulation of protein synthesis initiation complex formation by purine nucleotides. Biochim Biophys Acta. 1976;447:11–19. doi: 10.1016/0005-2787(76)90090-3. [DOI] [PubMed] [Google Scholar]