Abstract
Pseudomonas syringae is a gram‐negative bacterial pathogen that causes disease on more than 100 different plant species, including the model plant Arabidopsis thaliana. Dissection of the Arabidopsis thaliana–Pseudomonas syringae pathosystem has identified many factors that contribute to successful infection or immunity, including the genetics of the host, the genetics of the pathogen, and the environment. Environmental factors that contribute to a successful interaction can include temperature, light, and the circadian clock, as well as the soil environment. As silicon‐amended Resilience soil is advertised to enhance plant health, we sought to examine the extent to which this soil might affect the behavior of the A. thaliana–P. syringae model pathosystem and to characterize the mechanisms through which these effects may occur. We found that plants grown in Si‐amended Resilience soil displayed enhanced resistance to bacteria compared to plants grown in non‐Si‐amended Sunshine soil, and salicylic acid biosynthesis and signaling were not required for resistance. Although silicon has been shown to contribute to broad‐spectrum resistance, our data indicate that silicon is not the direct cause of enhanced resistance and that the Si‐amended Resilience soil has additional properties that modulate plant resistance. Our work demonstrates the importance of environmental factors, such as soil in modulating interactions between the plant and foliar pathogens, and highlights the significance of careful annotation of the environmental conditions under which plant–pathogen interactions are studied.
Keywords: Arabidopsis thaliana, environment, host resistance, Pseudomonas syringae, salicylic acid, soil
1. INTRODUCTION
Pseudomonas syringae, a gram‐negative, rod‐shaped bacterium, infects a wide range of species, including the model plant, Arabidopsis thaliana. Under cool moist conditions, P. syringae causes substantial disease and is spread to adjacent plants or leaves by rain splash (Hirano & Upper, 2000). Disease depends on compatibility among the host, pathogen, and environment, in an interaction termed the disease triangle (Agrios, 2005). Host genetics determine whether the plant is resistant or susceptible to the pathogen, and pathogen genetics determine whether the pathogen is virulent or nonvirulent to a specific host (Dangl, Horvath, & Staskawicz, 2013; Dangl & Jones, 2001; Vleeshouwers & Oliver, 2014). Environmental factors play a substantial role in contributing to the outcome of a plant–pathogen interaction, and these conditions can vary depending on the plant and pathogen under study.
Plants protect themselves from pathogens with three layers of induced defenses. In the first line of defense, the plant utilizes transmembrane pattern recognition receptors (PRRs) to recognize highly conserved pathogen‐associated molecular patterns (PAMPs), such as bacterial flagellin (Zipfel et al., 2004). PRR‐triggered immunity (PTI) results in the activation of mitogen‐activated protein kinase cascades, production of reactive oxygen species, induction of pathogenesis‐related genes, and deposition of callose and stomatal closure (Macho & Zipfel, 2014). To suppress PTI, pathogens such as Pseudomonas syringae can use the type III secretion system (T3SS) to mobilize type III‐secreted effector proteins (T3SEs) across the cell wall and into the host cell (Gohre & Robatzek, 2008; Grant, Fisher, Chang, Mole, & Dangl, 2006; Lewis, Desveaux, & Guttman, 2009; Mudgett, 2005; Xin & He, 2013). Common targets of T3SEs are PRRs, mitogen‐activated protein kinase (MAPK) cascades associated with PTI, vesicular trafficking pathways involved in the transport of antimicrobial compounds, and hormone signaling pathways (Block & Alfano, 2011; Feng & Zhou, 2012; Grant et al., 2006; Lewis et al., 2009; Mudgett, 2005; Shigenaga & Argueso, 2016; Speth, Lee, & He, 2007; Zhou & Chai, 2008). In the second line of defense, the plant uses nucleotide‐binding leucine‐rich repeat receptors (NLRs, also called Resistance proteins) which can recognize specific effectors resulting in effector‐triggered immunity (ETI) (Jones, Vance, & Dangl, 2016; Schreiber, Baudin, Hassan, & Lewis, 2016). ETI often culminates in a hypersensitive response (HR), characterized by localized programmed cell death at the infection site (Heath, 2000). In the third line of defense, plants are primed to respond quickly to subsequent pathogen exposure, resulting in systemic acquired resistance (SAR) (Fu & Dong, 2013; Gao, Kachroo, & Kachroo, 2014; Gao, Zhu, Kachroo, & Kachroo, 2015). SAR allows plants to more effectively fend off a subsequent pathogen attack and induces the production of salicylic acid (SA), a hormone which plays a central role in immune responses. While SA is a major contributor to systemic broad‐spectrum resistance in plants, complex interactions with other plant hormones also contribute to immunity (Shigenaga & Argueso, 2016).
Environmental factors such as light, temperature, humidity, water availability, and soil both separately and in combination profoundly affect the molecular mechanisms underlying plant defense responses to pathogens (Bostock, Pye, & Roubtsova, 2014; Hua, 2013; Smirnova et al., 2001; Suzuki, Rivero, Shulaev, Blumwald, & Mittler, 2014). Light is necessary for immunity to viral, bacterial, and fungal pathogens, including the development of the HR and SA‐mediated defense responses (Chandra‐Shekara et al., 2006; Griebel & Zeier, 2008; Lozano & Sequeira, 1970; Roden & Ingle, 2009). Ambient temperature influences PTI and ETI responses. Plants favor PTI responses at higher temperatures (23–32°C) which inhibit bacterial effector secretion and promote bacterial proliferation (Cheng et al., 2013; van Dijk et al., 1999). Plants activate ETI signaling at lower temperatures (10–23°C) when bacteria secrete effectors to promote virulence (Cheng et al., 2013; van Dijk et al., 1999). Both light and temperature are important external cues that entrain the plant circadian clock, which also plays an integral role in coordinating immune responses to pathogens. For both bacterial and fungal pathogens, plants are more susceptible when infected at subjective morning as opposed to at night (Bhardwaj, Meier, Petersen, Ingle, & Roden, 2011; Lu, McClung, & Zhang, 2017; Wang et al., 2011). Mutations in core clock genes abolish temporal variation in resistance levels, suggesting that clock genes play an important role in plant immunity (Bhardwaj et al., 2011; Lu et al., 2017; Wang et al., 2011). Lastly, abiotic and biotic soil factors affect plant resistance to disease. Abiotic factors such as water availability or nutrient availability in the soil can impact plant immunity (Bostock et al., 2014; De Coninck, Timmermans, Vos, Cammue, & Kazan, 2015; Suzuki et al., 2014; Thalineau et al., 2016). The soil microbiome can provide the host with protection from disease (Bakker, Doornbos, Zamioudis, Berendsen, & Pieterse, 2013; Pieterse et al., 2014), by competing with the pathogen for host nutrients, interacting antagonistically with the pathogen (antibiosis) or eliciting induced systemic resistance (ISR).
We identified enhanced resistance to P. syringae in Arabidopsis grown on commercial soil enriched with silicon. Using the Arabidopsis–P. syringae pathosystem, we investigated the effects of silicon‐amended soil (Resilience) compared to non‐silicon‐amended soil (Sunshine) and characterized the mechanisms that underlie these effects. We show that Si‐amended Resilience soil enhances plant resistance to multiple strains of the foliar pathogen, P. syringae, when it is inoculated into the apoplast and to a moderately virulent P. syringae strain that is spray‐inoculated onto the leaf surface. Plants impaired in PTI still exhibited enhanced resistance, and ETI responses are normal in plants grown in amended Resilience soil. In addition, the observed resistance is mediated by SA‐independent pathways. Interestingly, this work demonstrates that the soil environment can influence plant resistance to a foliar pathogen and points to the importance of carefully documented conditions for plant–pathogen interactions.
2. MATERIALS AND METHODS
2.1. Bacterial strains and routine culture conditions
Pseudomonas syringae was grown in either King's broth (KB) or minimal medium for induction of the type III secretion system (Huynh, Dahlbeck, & Staskawicz, 1989). Antibiotics were used at the following concentrations: 50 μg/ml kanamycin, 50 μg/ml rifampicin (PtoDC3000, PmaM6C∆E, and PtoDC3000∆hrcC), 300 μg/ml streptomycin (PcalES4326), and 50 μg/ml cycloheximide. PtoDC3000 carrying HopZ1a under its native promoter with a C‐terminal in‐frame HA epitope tag was previously described (Lewis, Abada, Ma, Guttman, & Desveaux, 2008).
2.2. Plant growth conditions
Arabidopsis thaliana plants were grown under 9 hrs of light (~130 microeinsteins m−2 s−1) and 15 hrs of darkness at 22°C in Sunshine #1 (Sun) or Sunshine Resilience #1 (Res) soil (Sun Gro Horticulture) supplemented with 20:20:20 fertilizer. Sunshine #1 soil is the previous formulation of Resilience and is not amended with silicon. Sunshine #1 soil must be ordered as a custom formulation. Resilience soil is reported to be amended with 1.7 mM silicon (Sun Gro Horticulture); however, its contents are patented. Assays used ecotype Col‐0 as the wild‐type background, the following mutants: npr1‐1 (Cao, Bowling, Gordon, & Dong, 1994), ics1 (Dewdney et al., 2000; Nawrath & Metraux, 1999; Wildermuth, Dewdney, Wu, & Ausubel, 2001), or the transgenic line nahG (Delaney et al., 1994). Plants were grown for 5–6 weeks before evaluating them for silicon accumulation, HR, or in bacterial growth assays; 5–6 pools of three individuals were used for silicon accumulation experiments. A total of 8–10 individuals were used for bacterial growth assays; 17–30 individuals were used for biomass accumulation experiments.
2.3. P. syringae HR and in planta growth assays
Pseudomonas syringae was resuspended in 10 mM MgCl2 to an optical density at 600 nm of 0.1 (~5 × 107 cfu/ml) for HR assays, or an optical density at 600 nm of 0.8 (~4.0 × 108 cfu/ml) for spray growth assays, or diluted to a concentration of 1 × 105 cfu/ml for growth assays by pressure infiltration. For HR and growth assays, bacteria were hand‐infiltrated into the leaf using a needleless syringe as described previously (Katagiri, Thilmony, & He, 2002). For HR, the plants were infiltrated in the late afternoon and maintained under 24 hrs of light. The HR was scored at 16–20 hrs. For infiltrated growth assays, four disks (total of 1 cm2) were harvested, ground in 10 mM MgCl2, and plated on KB with rifampicin or streptomycin and cycloheximide on days 0, 3, or 7 (PmaM6C∆E) to count colonies. For spray assays, the bacterial inoculum included 0.02% Silwet L‐77, and the plants were sprayed until the leaf surface was completely wet. The plants were covered with a humidified dome for 3 or 7 (PmaM6C∆E) days. On day 3 or 7 (PmaM6C∆E), the leaves were sterilized in 70% ethanol for 10 s and rinsed in sterile H2O for 10 s before harvesting, grinding, and plating as above. For infiltrated growth assays or spray assays, the plants were inoculated in the morning and kept in the growth chamber under short‐day conditions during the infections.
2.4. Pseudomonas syringae protein expression
Pseudomonas syringae cultures were grown overnight in KB containing kanamycin and rifampicin, pelleted, and washed in minimal media. Bacteria were resuspended in minimal media supplemented with potassium silicate (adjusted to pH 5.7 with H3PO4) at a final concentration of 1 μM, 10 μM, 100 μM, 500 μM, or 1 mM. Cultures were incubated with shaking at 28°C overnight to induce the type III secretion system (Huynh et al., 1989). An aliquot of 1.3 ml of each culture was pelleted, resuspended in 50 μl of 1× Laemmli loading dye, and boiled for 5 min, and 5 μl was separated on 12% sodium dodecyl sulfate‐polyacrylamide (SDS‐PAGE) gels. Proteins were transferred to nitrocellulose membranes and detected with HA antibodies (Roche) by chemiluminescence (GE Healthcare).
2.5. Silicon analysis
Sequential digestion steps (acid and alkaline) were conducted in vessels for a CEM Mars 6 Microwave Digestor (Buckingham, UK). Oven‐dried plant samples (approximately 100 mg) were digested by microwave‐assisted digestion (set at 1,300 W) using 5 ml of 1 M HNO3 plus 5 ml of H2O2 (30% v/v) (Barros, de Souza, Schiavo, & Nobrega, 2016). The heating program was as follows: 5 min to reach 120°C with a hold for 5 min, 5 min to reach 160°C with a hold for 5 min, and 3 min to reach 210°C for 5 min. The vessels were then removed and cooled to ambient temperature. Each vessel was then amended with 5 ml of 1.5 M NaOH and heated as follows: 10 min to reach 150°C with a 5 min hold; 10 min to reach 200°C with a 10 min hold. After cooling, the vessels were amended with 750 μl of 14 M HNO3. Final volumes were adjusted to 50 ml prior to analysis by inductively coupled plasma mass spectrometry (ICP‐MS; Agilent 7500ce) for Si. The digest Si content was quantified against a four‐point calibration curve that had been previously evaluated for linearity and accuracy. Analytical blanks, matrix blanks, and calibration verification samples were included in each sequence.
2.6. Soil analyses
Analyses were performed by the Cornell Nutrient Analysis Laboratory using standard methods.
2.7. Statistical analyses
The data were analyzed using two‐tailed homoscedastic t tests in Minitab 17. A significance level of α = 0.05 was chosen for all statistical analyses.
3. RESULTS
3.1. Resistance to virulent P. syringae strains in the apoplast is enhanced in plants grown in silicon‐amended Resilience soil
To investigate whether Arabidopsis plants grown in silicon‐amended Resilience soil or non‐silicon‐amended Sunshine soil show differential growth of virulent P. syringae strains, we carried out bacterial growth assays with two different strains, P. syringae pv. tomato DC3000 (PtoDC3000) or P. cannabina pv. alisalensis ES4326 (PcalES4326, formerly P. syringae pv. maculicola ES4326 PmaES4326). Bacteria were pressure‐infiltrated into the leaves of 5‐week‐old Arabidopsis plants that were grown in Sunshine (non‐silicon‐amended) or Sunshine soil amended with silicon (hereafter Resilience), and the bacterial populations were monitored at 1 hr (day 0) and 3 days after infiltration (day 3). In plants grown on Sunshine soil, both strains grew to high levels (~log 6–7), indicating that the pathogens were able to cause disease (Figure 1a, b). In plants grown in Resilience soil, both strains exhibited statistically significant reductions in growth (0.5–1 log) (Figure 1a, b).
Figure 1.
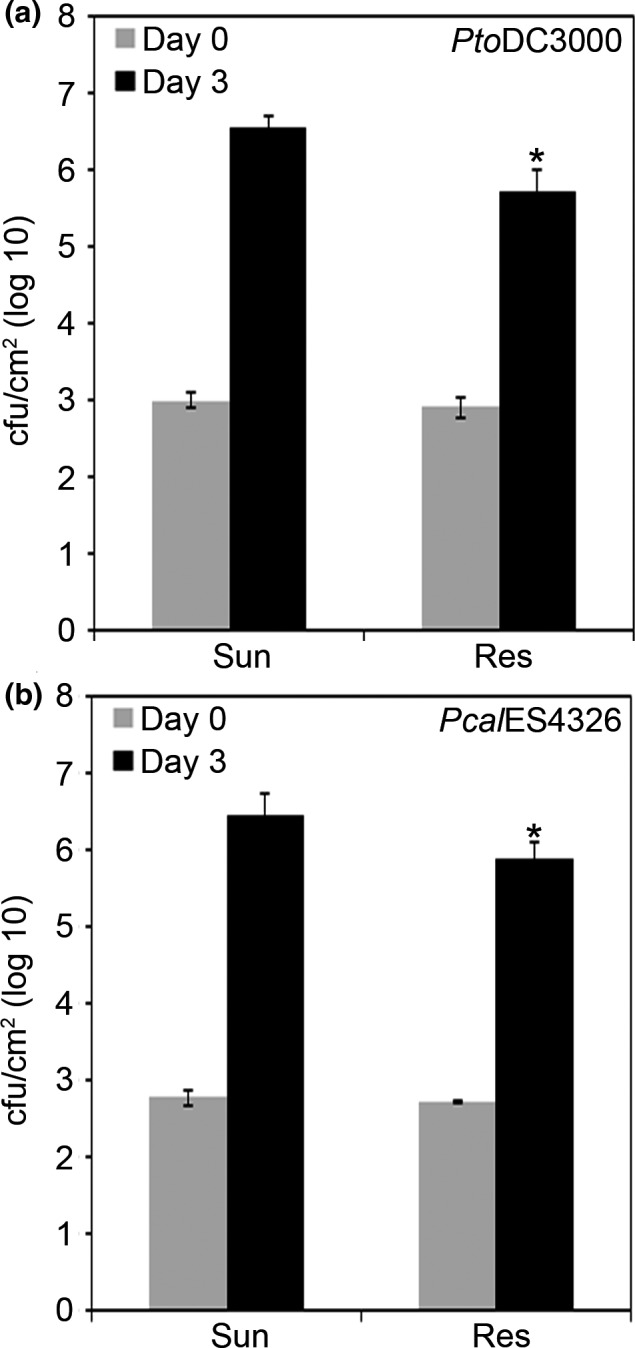
Pseudomonas syringae strains exhibit reduced virulence after pressure infiltration into plants grown on Resilience soil. (a) P. syringae pv. tomato DC3000 (Pto DC3000) was syringe infiltrated into Arabidopsis ecotype Col‐0 leaves, with a suspension containing 1 × 105 CFU/ml. Bacterial counts were determined 1 hr postinfection (day 0) and 3 days postinfection (day 3). (b) P. cannabina pv. alisalensis ES4326 (Pcal ES4326) was syringe infiltrated as described above. Error bars indicate the standard deviation. Two‐tailed homoscedastic t tests were performed to test for significant differences. Significant differences are shown with an asterisk (*p < .001). Experiments were repeated three times with similar results
To test whether plants grown in Resilience or Sunshine soil exhibited differential entry of bacteria into the leaf, we also infected Arabidopsis plants by spray inoculation, which requires the bacteria to swim into the leaf through the stomata. Bacteria were sprayed onto the leaves of 5‐week‐old Arabidopsis plants, and the endophytic bacterial populations were monitored 3 days after inoculation. We observed high levels of bacterial growth (~log 7–8) with PtoDC3000 (Figure 2a) or PcalES4326 (Figure 2b), in plants grown on Sunshine or Resilience soil. To determine whether the high bacterial titers associated with PtoDC3000 and PcalES4326 may have overcome the subtle resistance benefit conferred by growth of the plants in Resilience soil, we also carried out spray assays with the moderately virulent P. syringae pv. maculicola M6C∆E (hereafter PmaM6C∆E) strain (Rohmer, Kjemtrup, Marchesini, & Dangl, 2003). As PmaM6C∆E is not highly infectious when sprayed onto the leaves, we maintained the plants in high humidity conditions and monitored the endophytic bacterial populations 7 days after inoculation. We observed a significant reduction of 0.5–1 log bacterial growth in plants grown with on Resilience soil compared to plants grown on Sunshine soil (Figure 2c). Therefore, plants grown in Resilience soil showed resistance against bacteria inoculated into the apoplast or onto the surface of the leaf, indicating that bacterial entry was not affected by the plants' soil environment. The observed resistance was dependent on the bacterial titer and could be overcome by highly virulent strains.
Figure 2.

Highly virulent P. syringae strains show similar virulence in plants grown in Sunshine or Resilience soil after spray infiltration, while a moderately virulent P. syringae strain exhibits restricted bacterial growth on plants grown in Resilience soil. Strains were spray‐inoculated onto Arabidopsis ecotype Col‐0 leaves, with a suspension containing ~4.0 × 108 CFU/ml and 0.02% Silwet. Error bars indicate the standard deviation. Two‐tailed homoscedastic t tests were performed to test for significant differences. (a) Bacterial counts for P. syringae pv. tomato DC3000 (Pto DC3000) were determined 3 days postinfection. No significant differences were observed. Experiments were repeated three times with similar results. (b) Bacterial counts for P. cannabina pv. alisalensis ES4326 (Pcal ES4326) were determined 3 days postinfection. No significant differences were observed. Experiments were repeated three times with similar results. (c) Bacterial counts for P. syringae pv. maculicola M6C∆E (PmaM6C∆E) were determined 7 days postinfection. Significant differences are shown with an asterisk (*p < .05). Experiments were repeated two times with similar results
3.2. Pattern‐triggered immunity (PTI) does not contribute to enhanced resistance in plants grown in Resilience soil
To determine whether plants grown in Resilience soil are affected in PRR‐triggered immune responses, we carried out bacterial growth assays with a strain of P. syringae pv. tomato DC3000∆hrcC (hereafter PtoDC3000∆hrcC) that is unable to secrete type III effector proteins, due to a mutation in a structural component of the type III secretion system (Roine et al., 1997). We tested wild‐type Col‐0 and the fls2 mutant which lacks the PRR necessary for recognition of bacterial flagellin (Gomez‐Gomez & Boller, 2000; Gomez‐Gomez, Felix, & Boller, 1999). We found that both Col‐0 and fls2 plants grown in Resilience soil supported ~0.4–0.5 log less bacterial growth than plants grown in Sunshine soil (Figure 3), indicating that PTI does not contribute to the restriction of bacterial growth. As expected, PtoDC3000∆hrcC growth was slightly greater in fls2 compared to Col‐0 on both soil types (Figure 3), as the flagellin peptide is not recognized. These data indicate that the plants grown in Resilience soil have enhanced resistance that is not PTI‐dependent.
Figure 3.

PTI does not contribute to the enhanced resistance observed in plants grown in Resilience soil. P. syringae pv. tomato DC3000∆hrcC (Pto DC3000∆hrcC) was syringe infiltrated into Arabidopsis ecotype Col‐0 or fls2 leaves, with a suspension containing 1 × 105 CFU/ml. Bacterial counts were determined 1 hr postinfection (day 0) and 3 days postinfection (day 3). Error bars indicate the standard deviation. Two‐tailed homoscedastic t tests were performed to test for significant differences. Significant differences between genotypes on the same soil type are shown with an asterisk (*p < .05), and significant differences between soil types for the same genotype are shown with a theta (θ p < .05). The experiment was repeated two times with similar results
3.3. Plants grown in Resilience soil exhibit typical effector‐triggered immunity (ETI)
We also investigated whether plants grown in Resilience soil are impacted in ETI by conducting hypersensitive response (HR) assays with HopZ1a, a well‐characterized bacterial effector that causes a strong immune response in Arabidopsis ecotype Col‐0 (Lewis, Wu, Guttman, & Desveaux, 2010; Lewis et al., 2008). The HR is visualized as silvering and flattening of the infiltrated half‐leaf within 16–20 hrs postinfiltration (Lewis et al., 2008, 2010). Plants grown in Sunshine or Resilience soil showed similarly strong HRs in the same time frame (16–20 hrs postinfiltration) (Figure 4a), suggesting that the soil in which plants are grown does not affect ETI.
Figure 4.
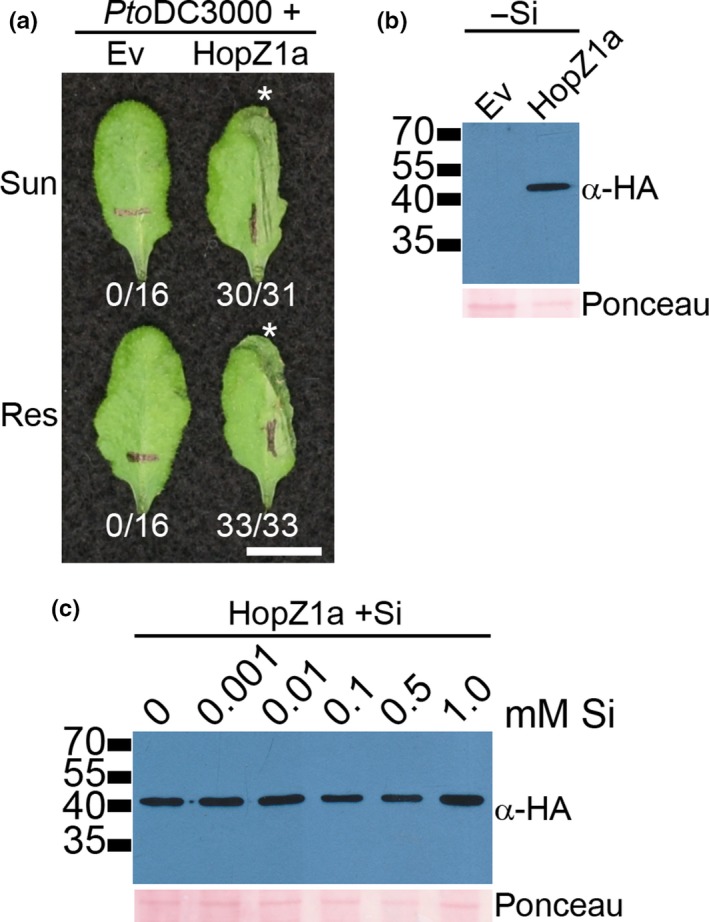
Plants grown in Resilience soil show normal ETI responses, and P. syringae effector production is not affected by silicon. (a) Half‐leaves of Arabidopsis ecotype Col‐0 were infiltrated with 5 × 107 CFU/ml of P. syringae pv. tomato DC3000 expressing empty vector (Pto DC3000 (Ev)) or HopZ1a with a C‐terminal HA tag under the control of its endogenous promoter (Pto DC3000 (hopZ1a)). Photographs were taken 22 hrs after infiltration. The HR is indicated with an asterisk. The number of leaves showing an HR is shown below each treatment. Scale bar = 1 cm. (b) Immunoblot analysis of HopZ1a‐HA protein expression in P. syringae pv. tomato DC3000 (Pto DC3000). Pto DC3000 carrying an empty vector (Ev) or HA‐tagged HopZ1a (42.1 kDa) was grown in minimal media to induce the type III secretion system. Equal amounts of proteins were separated on 12% SDS‐PAGE gels, blotted onto nitrocellulose, and probed with HA antibodies. The Ponceau red‐stained blot was used as the loading control. (c) Immunoblot analysis of HopZ1a‐HA protein expression in P. syringae pv. tomato DC3000 (Pto DC3000) as in part b. Pto DC3000 carrying HA‐tagged HopZ1a was grown in minimal media, with different concentrations of potassium silicate pH 5.7 as indicated
3.4. P. syringae effector HopZ1a is induced regardless of silicon treatment
Vivancos, Labbe, Menzies, and Belanger (2015) recently suggested that silicon might affect effector secretion or function in the apoplast. As Resilience soil is amended with silicon, we investigated whether silicon could interfere with effector expression. As the amount of effector proteins produced during P. syringae infection in planta is exceedingly low, we tested whether silicon could impact effector expression in vitro using minimal medium that induces the type III secretion system (Huynh et al., 1989; Lewis et al., 2008) and minimal medium containing a range of different concentrations of potassium silicate at pH 5.7. The pH of the minimal medium and the carbon source are particularly important for effector production (Huynh et al., 1989). We examined protein expression of T3SE HopZ1a tagged with a hemagglutinin (HA) tag, as we have previously demonstrated that it, like other T3SEs, is secreted and translocated into plant cells (Lewis et al., 2008). HopZ1a was expressed in P. syringae grown in standard minimal media lacking potassium silicate and visualized by Western blot analysis (Figure 4b; Lewis et al., 2008). We tested whether HopZ1a production was affected when P. syringae was grown in minimal medium containing 1 μM to 1 mM potassium silicate, as the maximum solubility of silicon is 1.7 mM. Similar levels of HopZ1a protein were detected across the range of potassium silicate concentrations (Figure 4c). This suggests that silicon does not impair effector production in vitro.
3.5. Plants grown in Resilience soil do not accumulate more silicon
Resilience soil is reported to be Sunshine soil that is amended with 1.7 mM silicon, but is patented so its contents are not readily ascertained. Silicon has been extensively characterized for its beneficial effects on disease resistance to biotrophic and necrotrophic pathogens, as well as on abiotic stress (Datnoff, Deren, & Snyder, 1997; Epstein, 1994, 1999; Guerriero, Hausman, & Legay, 2016; Liang, Nikolic, Belanger, Gong, & Song, 2015a, 2015b; Van Bockhaven et al., 2015). However, Arabidopsis has been reported to accumulate low levels of silicon and to lack silicon influx transporters (Ghanmi, McNally, Benhamou, Menzies, & Belanger, 2004). To determine whether there was differential uptake of silicon in plants grown in Sunshine or Resilience soil, we conducted inductively coupled plasma mass spectrometry (ICP‐MS), which allows for the quantification of trace elements (Kroukamp, Wondimu, & Forbes, 2016). Plants grown in Sunshine versus Resilience soil accumulated similar levels of silicon (Figure 5a), although the range of silicon concentrations was much broader in the plants grown in Resilience soil. These data suggest that silicon is not responsible for the enhanced resistance to P. syringae and that additional unknown factors in Resilience soil contribute to plant resistance to P. syringae infection.
Figure 5.
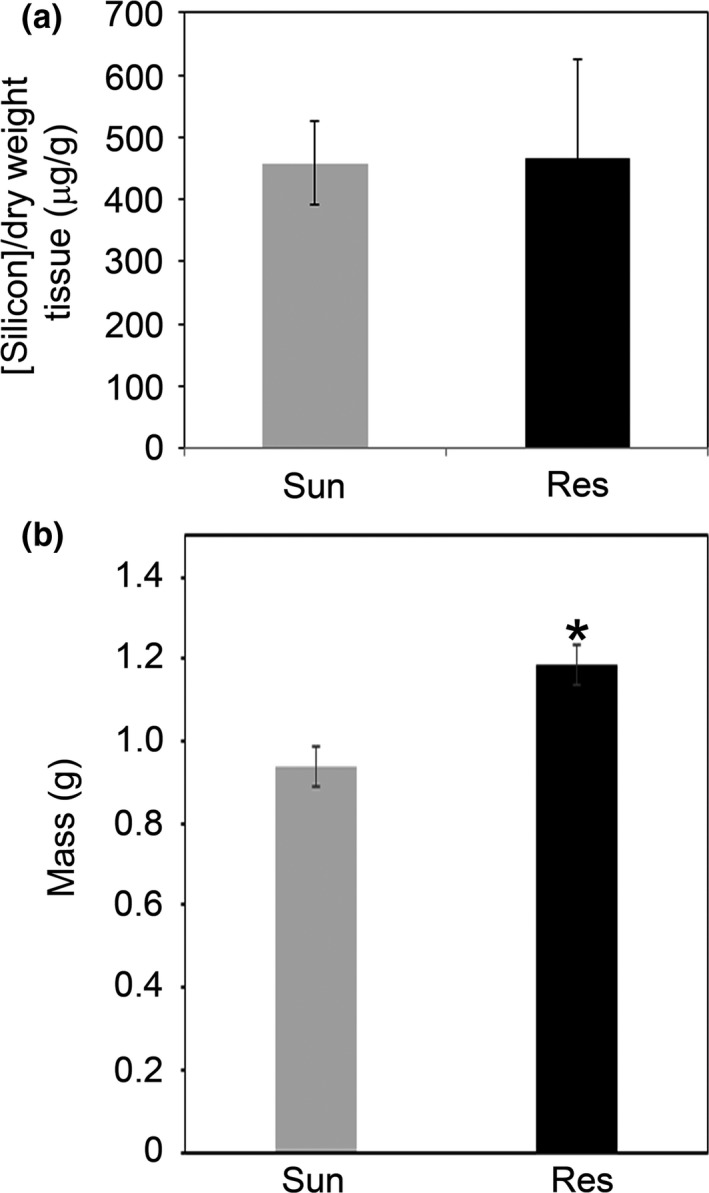
Plants grown in Resilience soil do not accumulate more silicon but do accumulate more biomass. Arabidopsis ecotype Col‐0 plants were initially grown on 0.5× Murashige and Skoog plates for 10 days, before being transplanted to Sunshine or Resilience soil. (a) The average level of silicon accumulation after 5–6 weeks is shown. Error bars indicate the standard error from the mean of six individuals. Two‐tailed homoscedastic t tests were performed to test for significant differences. No significant differences were observed. The experiment was repeated two times with similar results. (b) The average aerial biomass for each plant, 5–6 weeks after transplanting, is shown. Error bars indicate the standard error from the mean of 26 individuals. Two‐tailed homoscedastic t tests were performed to test for significant differences. Significant differences are shown with an asterisk (*p < .05). The experiment was repeated three times with similar results
We observed that Arabidopsis plants were frequently bigger when grown on Resilience soil compared to Sunshine soil. To determine whether Arabidopsis would exhibit enhanced biomass accumulation on Resilience soil, we grew Arabidopsis seedlings on Resilience or Sunshine soil for 5–6 weeks under short‐day conditions. To avoid any effects of soil type on germination, we sowed out Arabidopsis seed on Murashige and Skoog media, incubated the plates at 4°C to synchronize seed germination, and transplanted 10‐day‐old seedlings to Resilience or Sunshine soil. The average aerial biomass was significantly different between the two treatments. Plants grown on Sunshine soil had an average biomass of 0.94 g, while plants grown on Resilience soil had an average biomass of 1.18 g, representing a 25% increase in biomass (Figure 5b).
To identify other potential differences between Sunshine and Resilience soils, we carried out comprehensive soil analysis. Sunshine and Resilience soils contained similar amounts of total carbon, total nitrogen, and cation exchange capacity, which measures the soil's capacity to bind cations (Table 1). Both contained a high percentage of organic matter, and Resilience soil contained more organic matter (~74%) than Sunshine soil (~65%) (Table 1) using the loss on ignition method (Nelson & Sommers, 1996). Sunshine soil was slightly less acidic (pH 6.02) than Resilience soil (pH 5.25). Sunshine soil contained less moisture (7.99%) than Resilience soil (12.16%) (Table 1). We also measured macronutrients, micronutrients, and heavy metals in the two soil types, for bioavailable elements and for total element content (Table 2). Resilience soil contained more total calcium (Res 32,363 mg/kg vs. Sun 19,166 mg/kg), and more total sodium (Res 1349 mg/kg vs. Sun 794 mg/kg), compared to Sunshine soil; however, the bioavailable calcium and sodium contents were similar. Resilience soil contained more total potassium (Res 2,185 mg/kg vs. Sun 1,494 mg/kg), more total magnesium (Res 13,471 mg/kg vs. Sun 2,766 mg/kg), and more total sulfur (Res 5,273 mg/kg vs. Sun 4,547 mg/kg); however, bioavailable potassium, magnesium, and sulfur were higher in Sunshine soil (K 1,427 mg/kg, Mg 4,034 mg/kg, and S 2,643 mg/kg) versus Resilience soil (K 1,118 mg/kg, Mg 1,751 mg/kg, S 2,135 mg/kg). Bioavailable silicon was also higher in Resilience soil (45.01 mg/kg) compared to Sunshine soil (37.16 mg/kg). The observed differences were not particularly striking; however, our data suggest the differences in the soil itself contribute to the enhanced resistance.
Table 1.
Properties of Sunshine and Resilience soil
| Moisture (%) | pH | Cation exchange capacitya | Organic matter (LOI, %) | Organic matter (%) | Total N (%) | Total C (%) | |
|---|---|---|---|---|---|---|---|
| Sunshine | 7.99 | 6.02 | 27.24 | 64.91 | 45.20 | 0.88 | 34.45 |
| Resilience | 12.16 | 5.25 | 30.88 | 74.05 | 51.60 | 0.93 | 37.69 |
LOI, loss on ignition method.
Cation exchange capacity (CEC) measures the soil's capacity to hold cations, particularly potassium, calcium, magnesium, and sodium.
Table 2.
Elemental analysis of Sunshine and Resilience soil
| mg/kg | Al | As | B | Ba | Be | Ca | Cd | Co | Cr | Cu | Fe | K | Li | Mg |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioavailable elementsa | ||||||||||||||
| Sunshine | 10.09 | 0.74 | 1.88 | 5.80 | – | 11,337 | 0.05 | 0.10 | 0.25 | 0.23 | 6.17 | 1,427 | – | 4,034 |
| Resilience | 12.05 | 0.75 | 1.48 | 6.07 | – | 12,004 | 0.06 | 0.11 | 0.19 | 0.12 | 6.39 | 1,118 | – | 1,751 |
| Total elements in soilb | ||||||||||||||
| Sunshine | 1,123 | 1 | 12.1 | 38 | 0 | 19,166 | 0 | 0.7 | 1 | 12 | 1240 | 1,494 | 5.9 | 2,766 |
| Resilience | 575 | 0 | 15.2 | 27 | 0 | 32,263 | 0 | 0.4 | 1 | 13 | 926 | 2,185 | 5.6 | 13,471 |
| mg/kg | Mn | Mo | Na | Ni | P | Pb | S | Se | Si | Sr | Ti | V | Zn | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioavailable elementsa | ||||||||||||||
| Sunshine | 27.31 | 0.16 | 359 | 0.30 | 446 | 0.38 | 2,643 | – | 37.16 | 28.96 | – | – | 5.55 | |
| Resilience | 32.09 | 0.17 | 286 | 0.29 | 512 | 0.32 | 2,135 | – | 45.01 | 30.58 | – | – | 4.56 | |
| Total elements in soilb | ||||||||||||||
| Sunshine | 112.8 | 2.7 | 794 | 1 | 668 | 1 | 4,547 | 0 | – | 141.6 | 12.5 | 0.6 | 31 | |
| Resilience | 150.4 | 2.1 | 1349 | 0 | 660 | 1 | 5,273 | 0 | – | 142.0 | 9.2 | 0 | 29 | |
Elements are available for uptake by plants.
Total elements in soil include soluble and insoluble portions.
3.6. Plants grown on Resilience soil are not primed for salicylic acid‐dependent immune responses
To investigate whether SA might contribute to priming of defenses in plants grown in Sunshine or Resilience soil, we conducted bacterial growth assays on Arabidopsis lines (npr1, nahG, or ics1/sid2) that are affected in SA production or signaling. As all three lines support higher bacterial growth than Col‐0 (Cao et al., 1994; Delaney et al., 1994; Lewis et al., 2010; Nawrath & Metraux, 1999), we used the moderately virulent PmaM6C∆E strain so that we could quantify impaired or enhanced virulence (Rohmer et al., 2003) and pressure‐infiltrated the bacteria. NPR1 is a master regulator of SA‐dependent defenses (Cao et al., 1994; Fu & Dong, 2013). nahG is a transgenic Arabidopsis line carrying the bacterial salicylate hydroxylase enzyme and cannot accumulate SA (Delaney et al., 1994). ICS1/SID2 is required for synthesis of SA in the chloroplast (Dewdney et al., 2000; Nawrath & Metraux, 1999; Wildermuth et al., 2001). In Col‐0, we observed ~log 6 growth in plants grown in Sunshine soil but only ~log 4.5 growth in plants grown in Resilience soil (Figure 6). As previously reported, npr1, nahG, and ics1/sid2 lines were more susceptible to bacterial infections (Cao et al., 1994; Delaney et al., 1994; Lewis et al., 2010; Nawrath & Metraux, 1999). In npr1, nahG, and ics1/sid2 lines, we observed ~log 6.5–7.5 growth in plants grown in Sunshine soil compared to ~log 5–6 growth in plants grown in Resilience soil (Figure 6). As the npr1, nahG, and ics1/sid2 lines grown in Resilience soil still exhibited ~0.5–1.5 log lower bacterial growth compared to the same lines grown in Sunshine soil, this indicates that these SA‐related genes are not required for the restriction in bacterial growth. These results suggest that SA signaling does not contribute to enhanced resistance in plants grown in Resilience soil.
Figure 6.

Plants grown in Resilience soil are not primed for salicylic acid‐dependent immune responses. P. syringae pv. maculicola M6C∆E (PmaM6C∆E) was syringe infiltrated into Arabidopsis ecotype Col‐0 leaves, nahG, npr1, or ics1 mutants, with a suspension containing 1 × 105 CFU/ml. Bacterial counts were determined 1 hr postinfection (day 0) and 3 days postinfection (day 3). Error bars indicate the standard deviation. Two‐tailed homoscedastic t tests were performed to test for significant differences in bacterial growth within a genotype on the two soil types. Significant differences are shown with an asterisk (*p < .001). The experiment was repeated two times with similar results
4. DISCUSSION
Although environmental effects on host resistance to pathogens have been well‐characterized, there has been little systematic evaluation of the contribution of soil to resistance and defense signaling in a well‐characterized pathosystem. We found that Arabidopsis plants grown in a silicon‐amended soil (Resilience) exhibit enhanced resistance toward the foliar pathogen P. syringae (Figure 1) and investigated how resistance was influenced by the type of soil in which the plants were grown. We observed enhanced resistance when PtoDC3000 or PcalES4326, two highly virulent strains of P. syringae, was inoculated into the apoplast of Arabidopsis plants grown in Resilience soil compared to Sunshine soil (Figure 1). We also tested plants grown in Resilience or Sunshine soil by spray inoculation, which requires that the bacteria swim into the leaf to establish infection. We found that resistance to P. syringae in plants grown on Resilience soil could be overcome by high bacterial titers (i.e., PtoDC3000 or PmaES4326, Figure 2), while moderately virulent strains such as PmaM6C∆E (Gimenez‐Ibanez et al., 2014; Rohmer et al., 2003) exhibited restricted growth (reduction of ~0.5–1 log) in plants grown in Resilience soil (Figure 2).
Pathogen‐associated molecular patterns such as flg22 and Ef‐Tu are very common targets for the plant immune system (Zipfel et al., 2004, 2006). PAMPs are typically recognized by receptor‐like kinase (RLK) PRRs (Monaghan & Zipfel, 2012; Zipfel et al., 2004, 2006). RLKs are part of multigene families that function in many aspects of plant development and physiology and act as PRRs (Shiu et al., 2004). Only a handful of PAMPs and their cognate PRRs have been identified. We employed a type III secretion system mutant (PtoDC3000∆hrcC) that is unable to secrete effector proteins into its host, to test whether PTI is impacted by the soil that the plants were grown in. We tested Arabidopsis Col‐0 and the fls2 mutant, which lacks the FLS2 PRR required for flg22 recognition (Gomez‐Gomez & Boller, 2000; Gomez‐Gomez et al., 1999). We found that the virulence of PtoDC3000∆hrcC was reduced by ~0.5 log in Arabidopsis Col‐0 or fls2 grown in Resilience soil compared to plants grown in Sunshine soil (Figure 3), indicating that PTI does not contribute to the enhanced resistance. As expected, PtoDC3000∆hrcC also grew to higher bacterial titers in fls2 compared to Col‐0 in both soil types, although these data were only statistically significant in Resilience soil (Figure 3). These data indicate that the resistance observed in plants grown in Resilience soil does not depend on PTI.
Silicon has been hypothesized to impair the function or perhaps secretion of effector proteins in the apoplast (Vivancos et al., 2015). To test these hypotheses, we infiltrated Arabidopsis with PtoDC3000 carrying HopZ1a as HopZ1a elicits a strong HR in Arabidopsis that is dependent on an intact acetyltransferase catalytic triad (Lewis et al., 2008). HopZ1a's acetyltransferase activity is necessary for its recognition by the ZED1 pseudokinase and the ZAR1 resistance protein (Lewis et al., 2008, 2010, 2013; Schreiber et al., 2016). We found no differences in HopZ1a ETI in plants grown in Resilience or Sunshine soil, suggesting that the acetyltransferase activity of HopZ1a is not affected (Figure 4a). We directly tested for the ability of silicon to impact HopZ1a expression, by growing P. syringae carrying HopZ1a in minimal media containing silicon. We found that effector induction was not affected by the addition of potassium silicate to the media (Figure 4c). These data suggest that silicon is unable to interfere with the activity or expression of P. syringae type III effector proteins. Resistance conferred by silicon has been primarily observed toward fungal or oomycete pathogens that form a haustorium inside the plant cell. Fungal effectors are believed to be delivered to the host cell across the fungal–host membrane interface within the plant cell (Yi & Valent, 2013), suggesting that fungal or oomycete effector proteins would not contact silicon in the apoplast. In addition, the broad‐spectrum nature of silicon resistance against bacterial, oomycete, and fungal pathogens argues against a specific effect on diverse effector proteins.
Silicon improves host resistance to many fungal and oomycete diseases, including rice blast caused by Magnaporthe grisea (Rodrigues et al., 2004), powdery mildew of wheat, cucumber and rose (Belanger, Benhamou, & Menzies, 2003; Fawe, Abou‐Zaid, Menzies, & Belanger, 1998; Remus‐Borel, Menzies, & Belanger, 2005; Samuels, Glass, Ehret, & Menzies, 1991; Shetty et al., 2011), and some bacterial diseases, including bacterial wilt of tomato caused by Ralstonia solanacearum (Chen et al., 2015; Ghareeb et al., 2011) and bacterial speck of tomato caused by Pseudomonas syringae (Andrade et al., 2013). Silicon, in the form of uncharged monosilicic acid, is absorbed through the roots, moves through the transpiration stream, and eventually is deposited in the cell walls, intercellular spaces, and cell lumens of leaves and other tissues (Epstein, 1994; Guerriero et al., 2016; Kim, Kim, Park, & Choi, 2002; Samuels et al., 1991). We found that Arabidopsis plants grown in Sunshine or Resilience soil contained ~0.5 mg/g of silicon (Figure 5a), indicating that plants grown in Resilience soil did not take up more silicon than plants grown in Sunshine soil. We observed a similar amount of silicon accumulation as previously reported in nontransgenic Arabidopsis seedlings (0.6–1.5 mg/g of silicon) when they were hydroponically grown in silicon‐supplemented media (Montpetit et al., 2012; Vivancos et al., 2015). Our data indicate that silicon concentrations within the plant are not responsible for the resistance observed in plants grown in Resilience soil. Plants grown on Resilience soil exhibited greater biomass than those grown on Sunshine soil (Figure 5b). As this could be due to nutrient or soil composition, we also analyzed the soil properties and elemental content for both Sunshine and Resilience soils (Tables 1 and 2). Resilience soil contained more bioavailable silicon, as well as more bioavailable potassium and sulfur, and more total calcium, magnesium, and sodium compared to Sunshine soil (Table 2). The pH of Sunshine soil was slightly higher compared to Resilience soil, which can also affect the bioavailability of elements (Barber, 1995). The moisture content of Resilience soil was also slightly higher than that of Sunshine soil. These data suggest that additional differences between Resilience and Sunshine soil help promote resistance to P. syringae when Arabidopsis plants are grown in Resilience soil. It is also possible that plants grown in Resilience soil are more resistant because they have more vegetative resources to devote to defenses, similar to the resource reallocation that has been proposed to occur during the growth–defense trade‐off (Huot, Yao, Montgomery, & He, 2014).
Plant immunity can be primed for rapid defenses to a later infection by applying chemicals such as SA (Conrath et al., 2006), azelaic acid (Jung, Tschaplinski, Wang, Glazebrook, & Greenberg, 2009), and pipecolic acid (Bernsdorff et al., 2016). There is a cost to inducing defenses as this redirects resources away from growth, leading to smaller plants and reduced photosynthetic capacity (Bernsdorff et al., 2016; Bowling, Clarke, Liu, Klessig, & Dong, 1997; Mateo et al., 2006; Mauch et al., 2001). As SA is a major contributor to systemic broad‐spectrum resistance in plants, we tested whether mutants impaired in the production of salicylic acid or its signaling displayed greater resistance when grown in Resilience soil versus Sunshine soil. We observed higher levels of bacterial growth in ics1, npr1, and nahG lines compared to Col‐0, consistent with previous reports (Cao et al., 1994; Delaney et al., 1994; Lewis et al., 2010; Nawrath & Metraux, 1999). ics1, npr1, and nahG lines grown in Resilience soil supported less bacterial growth than plants grown in Sunshine soil, and the difference in bacterial growth on plants grown in Sunshine or Resilience soil was similar between Col‐0 and ics1, npr1, or nahG lines (Figure 6). This suggests that enhanced resistance associated with plant growth in Resilience soil is SA‐independent.
Historically, different soil types were known to have disease suppressive properties and to inhibit disease by root‐colonizing pathogens (Chet & Baker, 1981; Lifshitz, Sneh, & Baker, 1984; Martin & Hancock, 1986; Mazzola, 2002; Schroth & Hancock, 1982). Metagenomic analysis of the soil and rhizosphere microbiome suggests that microbial communities may compete with root‐colonizing pathogens to prevent disease, actively inhibit pathogens, and/or promote plant health or immune responses (De Coninck et al., 2015; Hadar & Papadopoulou, 2012; Mendes, Garbeva, & Raaijmakers, 2013). In addition, chemical and physical attributes of the soil can influence the soil microbiome (Hadar & Papadopoulou, 2012). Interestingly, our data show that the plants' soil environment can influence the resistance of plants to foliar pathogens, and the resistance is SA‐independent (Figures 1, 2 and 6). Plant growth‐promoting bacteria in the rhizosphere have previously been shown to trigger ISR in aerial portions of the plant that are inoculated with a different pathogen (Alstrom, 1991; Gang, Kloepper, & Tuzun, 1991; Pieterse et al., 2014; Van Peer, Niemann, & Schippers, 1991). Although ISR is SA‐independent (Pieterse, vanWees, Hoffland, vanPelt, & vanLoon, 1996), NPR1 appears to have a unique role in ISR compared to SAR (Pieterse et al., 2014; Spoel et al., 2003; Stein, Molitor, Kogel, & Waller, 2008). Our data showed that npr1 plants grown in Resilience soil still exhibited significantly enhanced bacterial resistance compared to plants grown in Sunshine soil (Figure 6). Thus, it is not clear whether Resilience soil may contain plant growth‐promoting bacteria that trigger ISR. We cannot exclude the possibility that silicon or other elements in the soil (Table 2) may affect the microbiome, which may then promote plant resistance. Regardless, our work demonstrates the importance of environmental factors such as soil properties, in contributing to plant resistance to pathogens. In addition, our work highlights the significance of careful documentation of the environmental conditions under which plant–microbe interactions are studied, as soil properties can have substantial effects on resistance to pathogenic bacteria.
AUTHOR CONTRIBUTIONS
JAH and JDL conceived and designed the experiments. JAH, RT‐R, and JDL performed the experiments. JAH, RT‐R, JCW, and JDL analyzed the data. JAH, JCW, and JDL wrote the manuscript.
Supporting information
ACKNOWLEDGMENTS
We thank Rolin Sauceda for his help with plant growth and maintenance; Dr. Mary Wildermuth for the kind gift of the ics1, nahG, and npr1 seeds; and Dr. Karl Schreiber for constructive feedback on the manuscript. Research on plant immunity in the Lewis laboratory was supported by United States Department of Agriculture Agricultural Research Service 5335‐21000‐040‐00D (J.D.L).
Hassan JA, de la Torre‐Roche R, White JC, Lewis JD. Soil mixture composition alters Arabidopsis susceptibility to Pseudomonas syringae infection. Plant Direct. 2018;2:1–13. 10.1002/pld3.44
REFERENCES
- Agrios, G. N. (2005). Plant pathology, 5th ed. Amsterdam, the Netherlands: Elsevier Academic Press. [Google Scholar]
- Alstrom, S. (1991). Induction of disease resistance in common bean susceptible to halo blight bacterial pathogen after seed bacterization with rhizosphere Pseudomonads. Journal of General and Applied Microbiology, 37, 495–501. 10.2323/jgam.37.495 [DOI] [Google Scholar]
- Andrade, C. C. L. , Resende, R. S. , Rodrigues, F. A. , Ferraz, H. G. M. , Moreira, W. R. , Oliveira, J. R. , & Mariano, R. L. R. (2013). Silicon reduces bacterial speck development on tomato leaves. Tropical Plant Pathology, 38, 436–442. 10.1590/S1982-56762013005000021 [DOI] [Google Scholar]
- Bakker, P. , Doornbos, R. F. , Zamioudis, C. , Berendsen, R. L. , & Pieterse, C. M. J. (2013). Induced systemic resistance and the rhizosphere microbiome. Plant Pathology Journal, 29, 136–143. 10.5423/PPJ.SI.07.2012.0111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber, S. A. (1995). Soil nutrient bioavailability: A mechanistic approach, 2nd ed. New York: Wiley. [Google Scholar]
- Barros, J. , de Souza, P. F. , Schiavo, D. , & Nobrega, J. A. (2016). Microwave‐assisted digestion using diluted acid and base solutions for plant analysis by ICP OES. Journal of Analytical Atomic Spectrometry, 31, 337–343. 10.1039/C5JA00294J [DOI] [Google Scholar]
- Belanger, R. R. , Benhamou, N. , & Menzies, J. G. (2003). Cytological evidence of an active role of silicon in wheat resistance to powdery mildew (Blumeria graminis f. sp tritici). Phytopathology, 93, 402–412. 10.1094/PHYTO.2003.93.4.402 [DOI] [PubMed] [Google Scholar]
- Bernsdorff, F. , Doring, A. C. , Gruner, K. , Schuck, S. , Brautigam, A. , & Zeier, J. (2016). Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid‐dependent and ‐independent pathways. Plant Cell, 28, 102–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, V. , Meier, S. , Petersen, L. N. , Ingle, R. A. , & Roden, L. C. (2011). Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS ONE, 6, e26968 10.1371/journal.pone.0026968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block, A. , & Alfano, J. R. (2011). Plant targets for Pseudomonas syringae type III effectors: Virulence targets or guarded decoys? Current Opinion in Microbiology, 14, 39–46. 10.1016/j.mib.2010.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock, R. M. , Pye, M. F. , & Roubtsova, T. V. (2014). Predisposition in plant disease: Exploiting the nexus in abiotic and biotic stress perception and response In VanAlfen N. K. (Ed.), Annual review of phytopathology, vol. 52 (pp. 517–549). Palo Alto, CA: Annual Reviews. [DOI] [PubMed] [Google Scholar]
- Bowling, S. A. , Clarke, J. D. , Liu, Y. D. , Klessig, D. F. , & Dong, X. N. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1‐dependent and NPR1‐independent resistance. Plant Cell, 9, 1573–1584. 10.1105/tpc.9.9.1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H. , Bowling, S. A. , Gordon, A. S. , & Dong, X. N. (1994). Characterization of an Arabidopsis mutant that is non‐responsive to inducers of systemic acquired resistance. Plant Cell, 6, 1583–1592. 10.1105/tpc.6.11.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra‐Shekara, A. C. , Gupte, M. , Navarre, D. , Raina, S. , Raina, R. , Klessig, D. , & Kachroo, P. (2006). Light‐dependent hypersensitive response and resistance signaling against Turnip crinkle virus in Arabidopsis. Plant Journal, 45, 320–334. 10.1111/j.1365-313X.2005.02618.x [DOI] [PubMed] [Google Scholar]
- Chen, Y. T. , Liu, M. , Wang, L. , Lin, W. P. , Fan, X. Y. , & Cai, K. Z. (2015). Proteomic characterization of silicon‐mediated resistance against Ralstonia solanacearum in tomato. Plant and Soil, 387, 425–440. 10.1007/s11104-014-2293-4 [DOI] [Google Scholar]
- Cheng, C. , Gao, X. , Feng, B. , Sheen, J. , Shan, L. , & He, P. (2013). Plant immune response to pathogens differs with changing temperatures. Nature Communications, 4, 2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chet, I. , & Baker, R. (1981). Isolation and biocontrol potential of Trichoderma hamatum from soil naturally suppressive to Rhizoctonia solani . Phytopathology, 71, 286–290. 10.1094/Phyto-71-286 [DOI] [Google Scholar]
- Conrath, U. , Beckers, G. J. M. , Flors, V. , Garcia‐Agustin, P. , Jakab, G. , Mauch, F. , … Mauch‐Mani, B. (2006). Priming: Getting ready for battle. Molecular Plant‐Microbe Interactions, 19, 1062–1071. 10.1094/MPMI-19-1062 [DOI] [PubMed] [Google Scholar]
- Dangl, J. L. , Horvath, D. M. , & Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science, 341, 746–751. 10.1126/science.1236011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J. L. , & Jones, J. D. G. (2001). Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. 10.1038/35081161 [DOI] [PubMed] [Google Scholar]
- Datnoff, L. E. , Deren, C. W. , & Snyder, G. H. (1997). Silicon fertilization for disease management of rice in Florida. Crop Protection, 16, 525–531. 10.1016/S0261-2194(97)00033-1 [DOI] [Google Scholar]
- De Coninck, B. , Timmermans, P. , Vos, C. , Cammue, B. P. A. , & Kazan, K. (2015). What lies beneath: Belowground defense strategies in plants. Trends in Plant Science, 20, 91–101. 10.1016/j.tplants.2014.09.007 [DOI] [PubMed] [Google Scholar]
- Delaney, T. P. , Uknes, S. , Vernooij, B. , Friedrich, L. , Weymann, K. , Negrotto, D. , … Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science, 266, 1247–1250. 10.1126/science.266.5188.1247 [DOI] [PubMed] [Google Scholar]
- Dewdney, J. , Reuber, T. L. , Wildermuth, M. C. , Devoto, A. , Cui, J. P. , Stutius, L. M. , … Ausubel, F. M. (2000). Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant Journal, 24, 205–218. 10.1046/j.1365-313x.2000.00870.x [DOI] [PubMed] [Google Scholar]
- van Dijk, K. , Fouts, D. E. , Rehm, A. H. , Hill, A. R. , Collmer, A. , & Alfano, J. R. (1999). The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (type III) protein secretion system in a temperature‐ and pH‐sensitive manner. Journal of Bacteriology, 181, 4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, E. (1994). The anomaly of silicon in plant biology. Proceedings of the National Academy of Sciences of the United States of America, 91, 11–17. 10.1073/pnas.91.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, E. (1999). Silicon. Annual Review of Plant Physiology and Plant Molecular Biology, 50, 641–664. 10.1146/annurev.arplant.50.1.641 [DOI] [PubMed] [Google Scholar]
- Fawe, A. , Abou‐Zaid, M. , Menzies, J. G. , & Belanger, R. R. (1998). Silicon‐mediated accumulation of flavonoid phytoalexins in cucumber. Phytopathology, 88, 396–401. 10.1094/PHYTO.1998.88.5.396 [DOI] [PubMed] [Google Scholar]
- Feng, F. , & Zhou, J. M. (2012). Plant‐bacterial pathogen interactions mediated by type III effectors. Current Opinion in Plant Biology, 15, 469–476. 10.1016/j.pbi.2012.03.004 [DOI] [PubMed] [Google Scholar]
- Fu, Z. Q. , & Dong, X. N. (2013). Systemic acquired resistance: Turning local infection into global defense In Merchant S. S. (Ed.), Annual review of plant biology, vol. 64 (pp. 839–863). Palo Alto, CA: Annual Reviews. [DOI] [PubMed] [Google Scholar]
- Gang, W. , Kloepper, J. W. , & Tuzun, S. (1991). Induction of systemic resistance of cucumber to Colletotrichum orbiculare by select strains of plant growth‐promoting rhizobacteria. Phytopathology, 81, 1508–1512. [Google Scholar]
- Gao, Q. M. , Kachroo, A. , & Kachroo, P. (2014). Chemical inducers of systemic immunity in plants. Journal of Experimental Botany, 65, 1849–1855. 10.1093/jxb/eru010 [DOI] [PubMed] [Google Scholar]
- Gao, Q.‐M. , Zhu, S. , Kachroo, P. , & Kachroo, A. (2015). Signal regulators of systemic acquired resistance. Frontiers in Plant Science, 6, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanmi, D. , McNally, D. J. , Benhamou, N. , Menzies, J. G. , & Belanger, R. R. (2004). Powdery mildew of Arabidopsis thaliana: A pathosystem for exploring the role of silicon in plant‐microbe interactions. Physiological and Molecular Plant Pathology, 64, 189–199. 10.1016/j.pmpp.2004.07.005 [DOI] [Google Scholar]
- Ghareeb, H. , Bozso, Z. , Ott, P. G. , Repenning, C. , Stahl, F. , & Wydra, K. (2011). Transcriptome of silicon‐induced resistance against Ralstonia solanacearum in the silicon non‐accumulator tomato implicates priming effect. Physiological and Molecular Plant Pathology, 75, 83–89. 10.1016/j.pmpp.2010.11.004 [DOI] [Google Scholar]
- Gimenez‐Ibanez, S. , Boter, M. , Fernandez‐Barbero, G. , Chini, A. , Rathjen, J. P. , & Solano, R. (2014). The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biology, 12, e1001792 10.1371/journal.pbio.1001792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohre, V. , & Robatzek, S. (2008). Breaking the barriers: Microbial effector molecules subvert plant immunity. Annual Review of Phytopathology, 46, 189–215. 10.1146/annurev.phyto.46.120407.110050 [DOI] [PubMed] [Google Scholar]
- Gomez‐Gomez, L. , & Boller, T. (2000). FLS2: An LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Molecular Cell, 5, 1003–1011. 10.1016/S1097-2765(00)80265-8 [DOI] [PubMed] [Google Scholar]
- Gomez‐Gomez, L. , Felix, G. , & Boller, T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana . Plant Journal, 18, 277–284. 10.1046/j.1365-313X.1999.00451.x [DOI] [PubMed] [Google Scholar]
- Grant, S. R. , Fisher, E. J. , Chang, J. H. , Mole, B. M. , & Dangl, J. L. (2006). Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annual Review of Microbiology, 60, 425–449. 10.1146/annurev.micro.60.080805.142251 [DOI] [PubMed] [Google Scholar]
- Griebel, T. , & Zeier, J. (2008). Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: Phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiology, 147, 790–801. 10.1104/pp.108.119503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero, G. , Hausman, J. F. , & Legay, S. (2016). Silicon and the plant extracellular matrix. Frontiers in Plant Science, 7, 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadar, Y. , & Papadopoulou, K. K. (2012). Suppressive composts: Microbial ecology links between abiotic environments and healthy plants In VanAlfen N. K., Leach J. E. & Lindow S. (Eds.), Annual review of phytopathology, vol. 50 (pp. 133–153). Palo Alto, CA: Annual Reviews. [DOI] [PubMed] [Google Scholar]
- Heath, M. C. (2000). Hypersensitive response‐related death. Plant Molecular Biology, 44, 321–334. 10.1023/A:1026592509060 [DOI] [PubMed] [Google Scholar]
- Hirano, S. S. , & Upper, C. D. (2000). Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae – a pathogen, ice nucleus, and epiphyte. Microbiology and Molecular Biology Reviews, 64, 624–653. 10.1128/MMBR.64.3.624-653.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, J. (2013). Modulation of plant immunity by light, circadian rhythm, and temperature. Current Opinion in Plant Biology, 16, 406–413. 10.1016/j.pbi.2013.06.017 [DOI] [PubMed] [Google Scholar]
- Huot, B. , Yao, J. , Montgomery, B. L. , & He, S. Y. (2014). Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Molecular Plant, 7, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, T. V. , Dahlbeck, D. , & Staskawicz, B. J. (1989). Bacterial blight of soybean: Regulation of a pathogen gene determining host cultivar specificity. Science, 245, 1374–1377. 10.1126/science.2781284 [DOI] [PubMed] [Google Scholar]
- Jones, J. D. G. , Vance, R. E. , & Dangl, J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science, 354, 1117. [DOI] [PubMed] [Google Scholar]
- Jung, H. W. , Tschaplinski, T. J. , Wang, L. , Glazebrook, J. , & Greenberg, J. T. (2009). Priming in systemic plant immunity. Science, 324, 89–91. 10.1126/science.1170025 [DOI] [PubMed] [Google Scholar]
- Katagiri, F. , Thilmony, R. , & He, S. Y. (2002). The Arabidopsis thaliana‐Pseudomonas syringae interaction In Somerville C. R., & Meyerowitz E. M. (Eds.), The Arabidopsis book doi/101199/tab 0039 (pp. 1–35). Rockville, MA: American Society of Plant Biologists. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. G. , Kim, K. W. , Park, E. W. , & Choi, D. (2002). Silicon‐induced cell wall fortification of rice leaves: A possible cellular mechanism of enhanced host resistance to blast. Phytopathology, 92, 1095–1103. 10.1094/PHYTO.2002.92.10.1095 [DOI] [PubMed] [Google Scholar]
- Kroukamp, E. M. , Wondimu, T. , & Forbes, P. B. C. (2016). Metal and metalloid speciation in plants: Overview, instrumentation, approaches and commonly assessed elements. TrAC Trends in Analytical Chemistry, 77, 87–99. 10.1016/j.trac.2015.10.007 [DOI] [Google Scholar]
- Lewis, J. D. , Abada, W. , Ma, W. B. , Guttman, D. S. , & Desveaux, D. (2008). The HopZ family of Pseudomonas syringae type III effectors require myristoylation for virulence and avirulence functions in Arabidopsis thaliana . Journal of Bacteriology, 190, 2880–2891. 10.1128/JB.01702-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J. D. , Desveaux, D. , & Guttman, D. S. (2009). The targeting of plant cellular systems by injected type III effector proteins. Seminars in Cell & Developmental Biology, 20, 1055–1063. 10.1016/j.semcdb.2009.06.003 [DOI] [PubMed] [Google Scholar]
- Lewis, J. D. , Lee, A. H.‐Y. , Hassan, J. A. , Wan, J. , Hurley, B. , Jhingree, J. R. , … Desveaux, D. (2013). The Arabidopsis ZED1 pseudokinase is required for ZAR1‐mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proceedings of the National Academy of Sciences of the United States of America, 110, 18722–18727. 10.1073/pnas.1315520110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J. D. , Wu, R. , Guttman, D. S. , & Desveaux, D. (2010). Allele‐specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genetics, 6, e1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. , Nikolic, M. , Belanger, R. R. , Gong, H. , & Song, A. (2015a). History and introduction of silicon research, p 1‐18, Silicon in agriculture: From theory to practice. Dordrecht, the Netherlands: Springer Netherlands. [Google Scholar]
- Liang, Y. , Nikolic, M. , Belanger, R. R. , Gong, H. , & Song, A. (2015b). Silicon and plant‐pathogen interactions, p 181‐196, Silicon in agriculture: From theory to practice. Dordrecht, the Netherlands: Springer Netherlands. [Google Scholar]
- Lifshitz, R. , Sneh, B. , & Baker, R. (1984). Soil suppressiveness to a plant pathogenic Pythium species. Phytopathology, 74, 1054–1061. 10.1094/Phyto-74-1054 [DOI] [Google Scholar]
- Lozano, J. C. , & Sequeira, L. (1970). Differentiation of races of Pseudomonas solanacearum by a leaf infiltration technique. Phytopathology, 60, 833–838. 10.1094/Phyto-60-833 [DOI] [Google Scholar]
- Lu, H. , McClung, C. R. , & Zhang, C. (2017). Tick tock: Circadian regulation of plant innate immunity In Leach J. E. & Lindow S. E. (Ed.), Annual review of phytopathology, vol. 55 (pp. 287–311). Palo Alto, CA: Annual Reviews. [DOI] [PubMed] [Google Scholar]
- Macho, A. P. , & Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Molecular Cell, 54, 263–272. 10.1016/j.molcel.2014.03.028 [DOI] [PubMed] [Google Scholar]
- Martin, F. N. , & Hancock, J. G. (1986). Association of chemical and biological factors in soils suppressive to Pythium ultimum . Phytopathology, 76, 1221–1231. 10.1094/Phyto-76-1221 [DOI] [Google Scholar]
- Mateo, A. , Funck, D. , Muhlenbock, P. , Kular, B. , Mullineaux, P. M. , & Karpinski, S. (2006). Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. Journal of Experimental Botany, 57, 1795–1807. 10.1093/jxb/erj196 [DOI] [PubMed] [Google Scholar]
- Mauch, F. , Mauch‐Mani, B. , Gaille, C. , Kull, B. , Haas, D. , & Reimmann, C. (2001). Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. Plant Journal, 25, 67–77. 10.1046/j.1365-313x.2001.00940.x [DOI] [PubMed] [Google Scholar]
- Mazzola, M. (2002). Mechanisms of natural soil suppressiveness to soilborne diseases. Antonie van Leeuwenhoek, 81, 557–564. 10.1023/A:1020557523557 [DOI] [PubMed] [Google Scholar]
- Mendes, R. , Garbeva, P. , & Raaijmakers, J. M. (2013). The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiology Reviews, 37, 634–663. 10.1111/1574-6976.12028 [DOI] [PubMed] [Google Scholar]
- Monaghan, J. , & Zipfel, C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Current Opinion in Plant Biology, 15, 349–357. 10.1016/j.pbi.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Montpetit, J. , Vivancos, J. , Mitani‐Ueno, N. , Yamaji, N. , Remus‐Borel, W. , Belzile, F. , … Belanger, R. R. (2012). Cloning, functional characterization and heterologous expression of TaLsi1, a wheat silicon transporter gene. Plant Molecular Biology, 79, 35–46. 10.1007/s11103-012-9892-3 [DOI] [PubMed] [Google Scholar]
- Mudgett, M. B. (2005). New insights to the function of phytopathogenic bacterial type III effectors in plants. Annual Review of Plant Biology, 56, 509–531. 10.1146/annurev.arplant.56.032604.144218 [DOI] [PubMed] [Google Scholar]
- Nawrath, C. , & Metraux, J. P. (1999). Salicylic acid induction‐deficient mutants of Arabidopsis express PR‐2 and PR‐5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell, 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D. W. , & Sommers, L. E. (1996). Total carbon, organic carbon, and organic matter In Sparks Dl. (Ed.), Methods of soil analysis, vol Part 3 (pp. 961–1010). SSSA Book Series 5. Madison, WI: Soil Science Society of America. [Google Scholar]
- Pieterse, C. M. J. , vanWees, S. C. M. , Hoffland, E. , vanPelt, J. A. , & vanLoon, L. C. (1996). Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis‐related gene expression. Plant Cell, 8, 1225–1237. 10.1105/tpc.8.8.1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C. M. J. , Zamioudis, C. , Berendsen, R. L. , Weller, D. M. , Van Wees, S. C. M. , & Bakker, P. (2014). Induced systemic resistance by beneficial microbes In VanAlfen N. K. (Ed.), Annual review of phytopathology, vol. 52 (pp. 347–375). Palo Alto, CA: Annual Reviews. [DOI] [PubMed] [Google Scholar]
- Remus‐Borel, W. , Menzies, J. G. , & Belanger, R. R. (2005). Silicon induces antifungal compounds in powdery mildew‐infected wheat. Physiological and Molecular Plant Pathology, 66, 108–115. 10.1016/j.pmpp.2005.05.006 [DOI] [Google Scholar]
- Roden, L. C. , & Ingle, R. A. (2009). Lights, rhythms, infection: The role of light and the circadian clock in determining the outcome of plant‐pathogen interactions. Plant Cell, 21, 2546–2552. 10.1105/tpc.109.069922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, F. A. , McNally, D. J. , Datnoff, L. E. , Jones, J. B. , Labbe, C. , Benhamou, N. , … Belanger, R. R. (2004). Silicon enhances the accumulation of diterpenoid phytoalexins in rice: A potential mechanism for blast resistance. Phytopathology, 94, 177–183. 10.1094/PHYTO.2004.94.2.177 [DOI] [PubMed] [Google Scholar]
- Rohmer, L. , Kjemtrup, S. , Marchesini, P. , & Dangl, J. L. (2003). Nucleotide sequence, functional characterization and evolution of pFKN, a virulence plasmid in Pseudomonas syringae pathovar maculicola . Molecular Microbiology, 47, 1545–1562. 10.1046/j.1365-2958.2003.03402.x [DOI] [PubMed] [Google Scholar]
- Roine, E. , Wei, W. S. , Yuan, J. , Nurmiaho‐Lassila, E. L. , Kalkkinen, N. , Romantschuk, M. , & He, S. Y. (1997). Hrp pilus: an hrp‐dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proceedings of the National Academy of Sciences of the United States of America, 94, 3459–3464. 10.1073/pnas.94.7.3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels, A. L. , Glass, A. D. M. , Ehret, D. L. , & Menzies, J. G. (1991). Mobility and deposition of silicon in cucumber plants. Plant, Cell and Environment, 14, 485–492. 10.1111/j.1365-3040.1991.tb01518.x [DOI] [Google Scholar]
- Schreiber, K. J. , Baudin, M. , Hassan, J. A. , & Lewis, J. D. (2016). Die another day: Molecular mechanisms of effector‐triggered immunity elicited by type III secreted effector proteins. Seminars in Cell & Developmental Biology, 56, 124–133. 10.1016/j.semcdb.2016.05.001 [DOI] [PubMed] [Google Scholar]
- Schroth, M. N. , & Hancock, J. G. (1982). Disease suppressive soil and root‐colonizing bacteria. Science, 216, 1376–1381. 10.1126/science.216.4553.1376 [DOI] [PubMed] [Google Scholar]
- Shetty, R. , Frette, X. , Jensen, B. , Shetty, N. P. , Jensen, J. D. , Jorgensen, H. J. L. , … Christensen, L. P. (2011). Silicon‐induced changes in antifungal phenolic acids, flavonoids, and key phenylpropanoid pathway genes during the interaction between miniature roses and the biotrophic pathogen Podosphaera pannosa . Plant Physiology, 157, 2194–2205. 10.1104/pp.111.185215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenaga, A. M. , & Argueso, C. T. (2016). No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Seminars in Cell & Developmental Biology, 56, 174–189. 10.1016/j.semcdb.2016.06.005 [DOI] [PubMed] [Google Scholar]
- Shiu, S. H. , Karlowski, W. M. , Pan, R. S. , Tzeng, Y. H. , Mayer, K. F. X. , & Li, W. H. (2004). Comparative analysis of the receptor‐like kinase family in Arabidopsis and rice. Plant Cell, 16, 1220–1234. 10.1105/tpc.020834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova, A. , Li, H. Q. , Weingart, H. , Aufhammer, S. , Burse, A. , Finis, K. , … Ullrich, M. S. (2001). Thermoregulated expression of virulence factors in plant‐associated bacteria. Archives of Microbiology, 176, 393–399. 10.1007/s002030100344 [DOI] [PubMed] [Google Scholar]
- Speth, E. B. , Lee, Y. N. , & He, S. Y. (2007). Pathogen virulence factors as molecular probes of basic plant cellular functions. Current Opinion in Plant Biology, 10, 580–586. 10.1016/j.pbi.2007.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel, S. H. , Koornneef, A. , Claessens, S. M. C. , Korzelius, J. P. , Van Pelt, J. A. , Mueller, M. J. , … Pieterse, C. M. (2003). NPR1 modulates cross‐talk between salicylate‐ and jasmonate‐dependent defense pathways through a novel function in the cytosol. Plant Cell, 15, 760–770. 10.1105/tpc.009159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, E. , Molitor, A. , Kogel, K. H. , & Waller, F. (2008). Systemic resistance in Arabidopsis conferred by the mycorrhizal fungus Piriformospora indica requires jasmonic acid signaling and the cytoplasmic function of NPR1. Plant and Cell Physiology, 49, 1747–1751. 10.1093/pcp/pcn147 [DOI] [PubMed] [Google Scholar]
- Suzuki, N. , Rivero, R. M. , Shulaev, V. , Blumwald, E. , & Mittler, R. (2014). Abiotic and biotic stress combinations. New Phytologist, 203, 32–43. 10.1111/nph.12797 [DOI] [PubMed] [Google Scholar]
- Thalineau, E. , Truong, H. N. , Berger, A. , Fournier, C. , Boscari, A. , Wendehenne, D. , & Jeandroz, S. (2016). Cross‐regulation between N metabolism and nitric oxide (NO) signaling during plant immunity. Frontiers in Plant Science, 7, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bockhaven, J. , Steppe, K. , Bauweraerts, I. , Kikuchi, S. , Asano, T. , Hofte, M. , & De Vleesschauwer, D. (2015). Primary metabolism plays a central role in moulding silicon‐inducible brown spot resistance in rice. Molecular Plant Pathology, 16, 811–824. 10.1111/mpp.12236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Peer, R. , Niemann, G. J. , & Schippers, B. (1991). Induced resistance and phytoalexin accumulation in biological control of Fusarium wilt of carnation by Pseudomonas sp. strain WCS417R. Phytopathology, 81, 728–734. 10.1094/Phyto-81-728 [DOI] [Google Scholar]
- Vivancos, J. , Labbe, C. , Menzies, J. G. , & Belanger, R. R. (2015). Silicon‐mediated resistance of Arabidopsis against powdery mildew involves mechanisms other than the salicylic acid (SA)‐dependent defence pathway. Molecular Plant Pathology, 16, 572–582. 10.1111/mpp.12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers, V. , & Oliver, R. P. (2014). Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Molecular Plant‐Microbe Interactions, 27, 196–206. 10.1094/MPMI-10-13-0313-IA [DOI] [PubMed] [Google Scholar]
- Wang, W. , Barnaby, J. Y. , Tada, Y. , Li, H. , Tor, M. , Caldelari, D. , … Dong, X. N. (2011). Timing of plant immune responses by a central circadian regulator. Nature, 470, U110–U126. 10.1038/nature09766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M. C. , Dewdney, J. , Wu, G. , & Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. 10.1038/35107108 [DOI] [PubMed] [Google Scholar]
- Xin, X. F. , & He, S. Y. (2013). Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annual Review of Phytopathology, 51, 473–498. 10.1146/annurev-phyto-082712-102321 [DOI] [PubMed] [Google Scholar]
- Yi, M. , & Valent, B. (2013). Communication between filamentous pathogens and plants at the biotrophic interface In VanAlfen N. K. (Ed.), Annual review of phytopathology, vol. 51 (pp. 587–611). Palo Alto, CA: Annual Reviews. [DOI] [PubMed] [Google Scholar]
- Zhou, J. M. , & Chai, J. (2008). Plant pathogenic bacterial type III effectors subdue host responses. Current Opinion in Microbiology, 11, 179–185. 10.1016/j.mib.2008.02.004 [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Kunze, G. , Chinchilla, D. , Caniard, A. , Jones, J. D. G. , Boller, T. , & Felix, G. (2006). Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell, 125, 749–760. 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E. J. , Jones, J. D. G. , Felix, G. , & Boller, T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. 10.1038/nature02485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


