Abstract
Plant responses to the environment are shaped by external stimuli and internal signaling pathways. In both the model plant Arabidopsis thaliana (Arabidopsis) and crop species, circadian clock factors are critical for growth, flowering, and circadian rhythms. Outside of Arabidopsis, however, little is known about the molecular function of clock gene products. Therefore, we sought to compare the function of Brachypodium distachyon (Brachypodium) and Setaria viridis (Setaria) orthologs of EARLY FLOWERING 3, a key clock gene in Arabidopsis. To identify both cycling genes and putative ELF3 functional orthologs in Setaria, a circadian RNA‐seq dataset and online query tool (Diel Explorer) were generated to explore expression profiles of Setaria genes under circadian conditions. The function of ELF3 orthologs from Arabidopsis, Brachypodium, and Setaria was tested for complementation of an elf3 mutation in Arabidopsis. We find that both monocot orthologs were capable of rescuing hypocotyl elongation, flowering time, and arrhythmic clock phenotypes. Using affinity purification and mass spectrometry, our data indicate that BdELF3 and SvELF3 could be integrated into similar complexes in vivo as AtELF3. Thus, we find that, despite 180 million years of separation, BdELF3 and SvELF3 can functionally complement loss of ELF3 at the molecular and physiological level.
Keywords: circadian clock, circadian RNA‐seq, ELF3, flowering, growth, Setaria
1. INTRODUCTION
Plants have developed sophisticated signaling networks to survive and thrive in diverse environments. Many plant responses are shaped, in part, by an internal timing mechanism known as the circadian clock, which allows for the coordination and anticipation of daily and seasonal variation in the environment (Greenham & McClung, 2015). Circadian clocks, which are endogenous oscillators with a period of approximately 24 hr, are critical for regulating the timing of physiology, development, and metabolism in all domains of life (Bell‐Pedersen et al., 2005; Doherty & Kay, 2010; Edgar et al., 2012; Harmer, 2009; Wijnen & Young, 2006). In plants and blue‐green algae, circadian clocks provide an experimentally observable adaptive advantage by synchronizing internal physiology with external environmental cues (Dodd et al., 2005; Green, Tingay, Wang, & Tobin, 2002; Ouyang, Andersson, Kondo, Golden, & Johnson, 1998; Rubin et al., 2017; Woelfle, Ouyang, Phanvijhitsiri, & Johnson, 2004). Currently, circadian oscillators are best understood in the reference plant Arabidopsis, in which dozens of clock or clock‐associated components have been identified using genetic screens and noninvasive, luciferase‐based oscillating reporters (Hsu & Harmer, 2014; Nagel & Kay, 2012). These morning‐, afternoon‐, and evening‐phased clock oscillators form multiple interconnected transcription–translation feedback loops and compose a complex network (Hsu & Harmer, 2014; Pokhilko et al., 2012). The Arabidopsis circadian clock regulates a significant portion of physiology, including photosynthesis, growth, disease resistance, starch metabolism, and phytohormone pathways (Covington, Maloof, Straume, Kay, & Harmer, 2008; Graf, Schlereth, Stitt, & Smith, 2010; Harmer et al., 2000; Michael, Mockler, Breton, Mcentee, & Byer, 2008; Wang, Barnaby, et al., 2011), with up to 30% of gene expression under circadian control (Covington et al., 2008; Michael et al., 2008).
Within the Arabidopsis clock network, a tripartite protein complex called the evening complex (EC) is an essential component of the evening transcription loop (Huang & Nusinow, 2016b). The EC consists of three distinct proteins, EARLY FLOWERING 3 (ELF3), EARLY FLOWERING 4 (ELF4), and LUX ARRHYTHMO (LUX, also known as PHYTOCLOCK1), with transcript and protein levels peaking in the evening (Doyle et al., 2002; Hazen et al., 2005; Hicks, Albertson, & Wagner, 2001; Nusinow et al., 2011; Onai & Ishiura, 2005). The EC plays a critical role in maintaining circadian rhythms, by repressing expression of key clock genes (Dixon et al., 2011; Helfer et al., 2011; Herrero et al., 2012; Kolmos et al., 2011; Mizuno et al., 2014). Loss‐of‐function mutation of any EC component in Arabidopsis results in arrhythmicity of the circadian clock and causes excessive cellular elongation and early flowering regardless of environmental photoperiod (Doyle et al., 2002; Hazen et al., 2005; Hicks et al., 2001; Khanna, Kikis, & Quail, 2003; Kim, Hicks, & Somers, 2005; Nozue et al., 2007; Nusinow et al., 2011; Onai & Ishiura, 2005).
In Arabidopsis, ELF3 directly interacts with ELF4 and LUX, functioning as a scaffold to bring ELF4 and LUX together (Herrero et al., 2012; Nusinow et al., 2011). Additional protein–protein interaction studies and tandem affinity purification coupled with mass spectrometry (AP‐MS) have identified many ELF3‐associating proteins and established ELF3 as a hub of a complex protein–protein interaction network, which consists of key components from the circadian clock pathway and light signaling pathways (Huang & Nusinow, 2016b; Huang, Alvarez, Bindbeutel, et al., 2016; Liu, Covington, Fankhauser, Chory, & Wagner, 2001; Yu et al., 2008). In this network, ELF3 directly interacts with the major red light photoreceptor phytochrome B (phyB), and CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), which is an E3 ubiquitin ligase required for proper regulation of photomorphogenesis and also interacts with phyB (Liu et al., 2001; Yu et al., 2008). The physical interaction among ELF3, phyB, and COP1, together with recruitment of direct interacting proteins to the network, provides biochemical evidence for cross talk between circadian clock and light signaling pathways (Huang & Nusinow, 2016b). Although much work does translate from Arabidopsis to other plant species, interaction between ELF3 and other proteins has yet to be tested in species outside Arabidopsis. Whether evening complex‐like protein assemblages or a similar ELF3‐containing protein‐protein interaction network exists in species outside Arabidopsis is an interesting question to ask.
Identification and characterization of clock genes in diverse plant species have revealed that many clock components are broadly conserved (Filichkin et al., 2011; Khan, Rowe, & Harmon, 2010; Lou et al., 2012; Song, Ito, & Imaizumi, 2010). Furthermore, comparative genomics analysis has found that circadian clock components are selectively retained after genome duplication events, suggestive of the importance of their role in maintaining fitness (Lou et al., 2012). Recently, mutant alleles of ELF3 were identified associated with the selection of favorable photoperiodism phenotypes in several crops, such as pea (Pisum sativum), rice (Oryza sativa), soybean (Glycine max), and barley (Hordeum vulgare L.) (Faure et al., 2012; Lu et al., 2017; Matsubara et al., 2012; Saito et al., 2012; Weller et al., 2012; Zakhrabekova et al., 2012). These findings are consistent with the reported functions of Arabidopsis ELF3 in regulating the photoperiodic control of growth and flowering (Hicks et al., 2001; Huang & Nusinow, 2016b; Nozue et al., 2007; Nusinow et al., 2011). However, opposed to the early flowering phenotype caused by elf3 mutants in Arabidopsis, pea, and barley (Faure et al., 2012; Hicks et al., 2001; Weller et al., 2012), loss‐of‐function mutation of the rice or soybean ELF3 ortholog results in delayed flowering (Lu et al., 2017; Saito et al., 2012), suggesting ELF3‐mediated regulation of flowering varies in different plant species. The molecular mechanisms underlying this difference have not been thoroughly elucidated.
Brachypodium distachyon is a C3 model grass closely related to wheat (Triticum aestivum), barley, oats (Avena sativa), and rice. Setaria viridis is a C4 model grass closely related to maize (Zea mays), sorghum (Sorghum bicolor), sugarcane (Saccharum officinarum), and other bioenergy grasses. Both grasses are small, transformable, rapid‐cycling plants with recently sequenced genomes, making them ideal model monocots for comparative analysis with Arabidopsis (Bennetzen et al., 2012; Brutnell et al., 2010). Computational analysis of Brachypodium has identified putative circadian clock orthologs (Higgins, Bailey, & Laurie, 2010), including BdELF3. However, no such comparative analysis has been carried out systematically in Setaria to identify putative orthologs of circadian clock genes. Therefore, we generated a RNA‐seq time‐course dataset to analyze the circadian transcriptome of Setaria after either photo‐ or thermo‐entrainment and developed an online gene‐expression query tool (Diel Explorer) for the community. We found that the magnitude of circadian‐regulated genes in Setaria is similar to other monocots after photo‐entrainment, but much less after thermal entrainment. We further analyzed the functional conservation of SvELF3, together with previously reported BdELF3, by introducing both ELF3 orthologs into Arabidopsis elf3 mutant for physiological and biochemical characterization. We found that Brachypodium and Setaria ELF3 can complement the hypocotyl elongation, flowering time and circadian arrhythmia phenotypes caused by the elf3 mutation in Arabidopsis. Furthermore, AP‐MS analyses found that Brachypodium and Setaria ELF3 were integrated into a similar protein–protein interaction network in vivo as their Arabidopsis counterpart. Our data collectively demonstrated the functional conservation of ELF3 among Arabidopsis, Brachypodium, and Setaria are likely due to the association with same protein partners, providing insights of how ELF3 orthologs potentially function in grasses.
2. METHODS
2.1. Plant materials and growth conditions
For Arabidopsis, wild‐type (Columbia‐0) and elf3‐2 plants carrying the CCA1::LUC reporter were described previously (Nusinow et al., 2011; Pruneda‐Paz, Breton, Para, & Kay, 2009). Seeds were surface sterilized and plated on 1/2× Murashige and Skoog (MS) basal salt medium with 0.8% agar + 1% (w/v) sucrose. After 3 days of stratification, plates were placed horizontally in a Percival incubator (Percival Scientific, Perry, IA), supplied with 80 μmol m−2 s−1 white light and set to a constant temperature of 22°C. Plants were grown under 12‐h light/12‐h dark cycles (12L:12D) for 4 days (for physiological experiments) or for 10 days (for AP‐MS) before assays.
For Setaria circadian expression profiling by RNA‐seq, seeds were stratified for 5 days at 4°C before being moved to entrainment conditions. Plants were grown under either LDHH or LLHC (L: light, D: dark, H: hot, C: cold) entrainment condition, and then sampled for RNA‐seq in constant light and constant temperature (32°C) conditions (F, for free‐running) every 2 hr for 48 hr. Light intensity was set to 400 μmol m−2 s−1 white light. In LDHH‐F, stratified Setaria seeds were grown for 10 days under 12L:12D and constant temperature (32°C) before sampling in constant light and constant temperature. In LLHC‐F, stratified Setaria seeds were grown for 10 days under constant light conditions and cycling temperature conditions 12 h at 32°C (subjective day)/12 h at 22°C (subjective night) before sampling in constant conditions. Two experimental replicates were collected for each entrainment condition.
2.2. Setaria circadian RNA‐seq
The second leaf from the top of seventeen S. viridis plants was selected for RNA‐seq sampling at each time point for each sampling condition. Five replicate samples were pooled after being ground in liquid nitrogen and resuspended in lithium chloride lysis binding buffer (Wang, Si, et al., 2011). RNA‐seq libraries from leaf samples were constructed according to the previous literature (Wang, Si, et al., 2011) with one major modification. Rather than extracting RNA then mRNA from ground leaf samples (Wang, Si, et al., 2011), mRNA was extracted directly from frozen ground leaf samples similar to the method described in Kumar et al. (2012), except that two additional rounds of wash, binding, and elution steps after treatment with EDTA were necessary to remove rRNA from samples. mRNA quantity was assessed using a Qubit with a Qubit RNA HS Kit, and mRNA quality was assessed using a Bioanalyzer and Plant RNA PiCO chip. Ninety‐six library samples were multiplexed 12 per lane, for a total of eight lanes of Illumina Hiseq 2000 sequencing. Paired‐end 101‐bp sequencing was carried out at MOgene (St. Louis, MO). Raw data and processed data can be found on NCBI's Gene Expression Omnibus (GEO; (Barrett et al., 2013; Edgar, Domrachev, & Lash, 2002)) and are accessible with identification number GSE97739 ( https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97739).
RNA‐seq data were trimmed with BBTools (v36.20) using parameters: ktrim=r k=23 mink=11 hdist=1 tpe tbo ktrim=l k=23 mink=11 hdist=1 tpe tbo qtrim=rl trimq=20 minlen=20 (Bushnell, 2016). Any parameters not specified were run as default. Before trimming, we had 1,814,939,650 reads with a mean of 18,905,621 reads per sample and a standard deviation of 2,875,187. After trimming, we have 1,646,019,593 reads with a mean of 17,146,037 reads per sample and a standard deviation of 2,411,061. Kallisto (v 0.42.4; (Bray, Pimentel, Melsted, & Pachter, 2016)) was used to index the transcripts with the default parameters and the S. viridis transcripts fasta file (Sviridis_311_v1.1) from Phytozome (Goodstein et al., 2012). The reads were quantified with parameters:‐t 40 ‐b 100. Any parameters not specified were run as default. Kallisto output was formatted for compatibility with JTK‐Cycle (v3.1; (Hughes, Hogenesch, & Kornacker, 2010)), and circadian cycles were detected. To query the Setaria expression data, we developed Diel Explorer. The tool can be found at http://shiny.bioinformatics.danforthcenter.org/diel-explorer/. Underlying code for Diel Explorer is available on Github ( https://github.com/danforthcenter/diel-explorer).
2.3. Plasmid constructs and generation of transgenic plants
Coding sequences (without the stop codon) of AtELF3 (AT2G25930) and SvELF3a (Sevir.5G206400.1) were cloned into the pENTR/D‐TOPO vector (ThermoFisher Scientific, Waltham, MA), verified by sequencing, and were recombined into the pB7HFC vector (Huang, Alvarez, Bindbeutel, et al., 2016) using LR Clonease (ThermoFisher Scientific). Coding sequence of BdELF3 (Bradi2g14290.1) was submitted to the U.S. Department of Energy Joint Genome Institute (DOE‐JGI), synthesized by the DNA Synthesis Science group, and cloned into the pENTR/D‐TOPO vector. Sequence validated clones were then recombined into pB7HFC as described above. The pB7HFC‐At/Bd/SvELF3 constructs were then transformed into elf3‐2 [CCA1::LUC] plants by the floral dip method (Zhang, Henriques, Lin, Niu, & Chua, 2006). Homozygous transgenic plants were validated by testing luciferase bioluminescence, drug resistance, and by PCR‐based genotyping. All primers used in this article were listed in Table S1.
2.4. Hypocotyl and flowering time measurement
Twenty seedlings of each genotype were arrayed and photographed with a ruler for measuring hypocotyl length using the ImageJ software (NIH) (Schneider, Rasband, & Eliceiri, 2012). The procedure was repeated three times. For measuring flowering time, 12 plants of each genotype were placed in a random order and were grown under the long‐day condition (light:dark = 16:8 hr). The seedlings were then observed every day at 12:00 PM; the date on which each seedling began flowering, indicated by the growth of a ~1 cm inflorescence stem, was recorded along with the number of rosette leaves produced up to that date. ANOVA with Bonferroni correction was measured using GraphPad Prism version 6.00 (GraphPad Software, La Jolla California USA, www.graphpad.com).
2.5. Circadian assays in Arabidopsis
Seedlings were transferred to fresh 1/2× MS plates after 5 days of entrainment under the 12L:12D condition and sprayed with sterile 5 mM luciferin (Gold Biotechnology, St. Louis, MO) prepared in 0.1% (v/v) Triton X‐100 solution. Sprayed seedlings were then imaged in constant light (70 μmol m−2 sec−1, wavelengths 400, 430, 450, 530, 630, and 660 set at intensity 350 (Heliospectra LED lights, Göteborg, Sweden)). Bioluminescence was recorded after a 120–180s delay to diminish delayed fluorescence (Gould et al., 2009) over 5 days using an ultra‐cooled CCD camera (Pixis 1024B, Princeton Instruments) driven by Micro‐Manager software (Edelstein, Amodaj, Hoover, Vale, & Stuurman, 2010; Edelstein et al., 2014). The images were processed in stacks by Metamorph software (Molecular Devices, Sunnyvale, CA), and rhythms determined by fast Fourier‐transformed nonlinear least squares (FFT‐NLLS) (Plautz et al., 1997) after background subtraction using the interface provided by the Biological Rhythms Analysis Software System 3.0 (BRASS) available at http://www.amillar.org.
2.6. Yeast two‐hybrid analysis
Yeast two‐hybrid assays were carried out as previously described (Huang, Alvarez, Bindbeutel, et al., 2016). In brief, the DNA binding domain (DBD) or activating domain (AD)‐fused constructs were transformed using the Li‐Ac transformation protocol (Clontech) into Saccharomyces cerevisiae strain Y187 (MATα) and the AH109 (MATa), respectively. Two strains of yeast were then mated to generate diploid with both DBD and AD constructs. Protein–protein interaction was tested in diploid yeast by replica plating on CSM –Leu –Trp –His media supplemented with extra adenine (30 mg/L final concentration) and 2 mM 3‐amino‐1,2,4‐triazole (3AT). Pictures were taken after 4‐day incubation at 30°C. All primers used for cloning plasmid constructs were listed in Table S1.
2.7. Protein extraction and western blotting
Protein extracts were made from 10‐day‐old seedlings as previously described (Huang, Alvarez, Bindbeutel, et al., 2016) and loaded 50 μg to run 10% SDS‐PAGE. For Western blots, all of the following primary and secondary antibodies were diluted into PBS + 0.1% Tween and incubated at room temperature for 1 hr: anti‐FLAG®M2‐HRP (Sigma, A8592, diluted at 1:10,000) and anti‐Rpt5‐rabbit (ENZO Life Science, BML‐PW8245‐0025, diluted at 1:5000), and anti‐rabbit‐HRP secondary antibodies (Sigma, A0545, diluted at 1:10,000).
2.8. Affinity purification and mass spectrometry (AP‐MS)
Protein extraction methods and protocols for AP‐MS were described previously (Huang, Alvarez, & Nusinow, 2016; Huang & Nusinow, 2016a; Huang, Alvarez, Bindbeutel, et al., 2016; Huang, Yoo, et al., 2016). In brief, transgenic seedlings carrying the At/Bd/SvELF3‐HFC constructs were grown under 12L:12D conditions for 10 days and were harvested at dusk (ZT12). Five grams of seedlings was needed per replicate to make protein extracts, which underwent tandem affinity purification utilizing the FLAG and His epitopes of the fusion protein. Purified samples were reduced, alkylated, and digested by trypsin. The tryptic peptides were then injected to an LTQ‐Orbitrap Velos Pro (ThermoFisher Scientific) coupled with a U3000 RSLCnano HPLC (ThermoFisher Scientific) with settings described previously (Huang, Alvarez, Bindbeutel, et al., 2016).
2.9. AP‐MS data analysis
Data analysis was carried out as previously described (Huang, Alvarez, Bindbeutel, et al., 2016). The databases searched were TAIR10 database (20101214, 35,386 entries) and the cRAP database ( http://www.thegpm.org/cRAP/). Peptide identifications were accepted if they could be established at greater than 95.0% probability and the Scaffold Local FDR was <1%. Protein identifications were accepted if they could be established at greater than 99.0% probability as assigned by the Protein Prophet algorithm (Keller, Nesvizhskii, Kolker, & Aebersold, 2002; Nesvizhskii, Keller, Kolker, & Aebersold, 2003). A full list of all identified proteins (reporting total/exclusive unique peptide count and percent coverage) can be found in Table S2. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (Vizcaino et al., 2014) via the PRIDE partner repository with the dataset identifier PXD006352 and https://doi.org/10.6019/PXD006352.
2.10. Accession numbers
AtELF3 (AT2G25930), BdELF3 (Bradi2g14290.1), SvELF3a (Sevir.5G206400.1) and SvELF3b (Sevir.3G123200.1) are used in this article. Accession numbers for all the At/Bd/Sv/ELF3‐associated proteins can be found in Table 1 and Table S2.
Table 1.
Proteins co‐purified with ELF3 orthologs from AP‐MS
| AGI number | Protein name | Molecular weight | Exclusive unique peptide count/percent coveragea (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AtELF3 | SvELF3 #2 | SvELF3 #3 | BdELF3 #2 | BdELF3 #3 | ||||||||
| rep1 | rep2 | rep1 | rep2 | rep1 | rep2 | rep1 | rep2 | rep1 | rep2 | |||
| n/a | AtELF3‐HFC | 84 kDa | 20/36 | 19/31 | – | – | – | – | – | – | – | – |
| n/a | SvELF3‐HFC | 87 kDa | – | – | 19/40 | 29/48 | 22/42 | 33/54 | – | – | – | – |
| n/a | BdELF3‐HFC | 86 kDa | – | – | – | – | – | – | 25/42 | 26/43 | 21/35 | 19/33 |
| AT2G18790 | phyB | 129 kDa | 23/37 | 24/37 | 31/54 | 28/42 | 32/50 | 30/45 | 22/34 | 22/33 | 24/40 | 24/37 |
| AT5G35840 | phyC | 124 kDa | 22/29 | 23/29 | 27/37 | 32/39 | 27/37 | 33/42 | 20/24 | 18/22 | 19/23 | 22/27 |
| AT5G43630 | TZP | 91 kDa | 14/21 | 12/17 | 13/23 | 20/33 | 13/22 | 24/36 | 12/18 | 13/19 | 14/20 | 14/22 |
| AT4G18130 | phyE | 123 kDa | 11/16 | 19/27 | 12/18 | 18/23 | 11/19 | 17/24 | 6/7 | 10/14 | 14/19 | 13/18 |
| AT2G16365.2 | PCH1b | 51 kDa | 9/25 | 9/25 | 9/30 | 14/43 | 11/36 | 16/49 | 9/26 | 9/28 | 11/32 | 11/32 |
| AT2G46340 | SPA1 | 115 kDa | 8/14 | 7/9 | 2/2 | 4/8 | 2/3 | 4/9 | – | – | 6/8 | 5/6 |
| AT3G42170c | DAYSLEEPER | 79 kDa | 8/19 | 5/11 | – | 7/16 | – | 12/27 | 3/7 | 4/8 | 2/4 | – |
| AT2G32950 | COP1 | 76 kDa | 8/16 | 9/17 | 6/17 | 4/8 | 3/6 | 5/12 | 1/2 | 2/4 | 6/11 | 4/8 |
| AT3G22380 | TIC | 165 kDa | 5/5 | 5/5 | 4/4 | 12/12 | 3/3 | 15/15 | 4/4 | 3/3 | 1/1 | 1/1 |
| AT4G11110 | SPA2 | 115 kDa | 5/11 | 6/12 | – | – | – | – | – | – | – | – |
| AT2G40080 | ELF4 | 12 kDa | 4/50 | 4/50 | 3/42 | 5/68 | 4/60 | 4/60 | 4/50 | 4/50 | 5/68 | 5/68 |
| AT1G09340c | CRB | 43 kDa | 4/16 | 3/12 | 1/4 | 1/4 | 1/4 | 4/18 | 1/4 | 1/4 | – | – |
| AT3G13670 | MLK4 | 79 kDa | 3/10 | 3/13 | – | – | – | 1/2 | 6/21 | 3/10 | 3/11 | 5/13 |
| AT5G61380 | TOC1 | 69 kDa | 2/4 | 2/4 | 2/5 | – | 3/7 | – | – | – | 1/2 | 1/2 |
| AT1G53090 | SPA4 | 89 kDa | 2/6 | 5/14 | – | – | – | – | – | – | – | – |
| AT5G18190 | MLK1 | 77 kDa | 2/12 | 2/11 | – | – | – | – | 2/14 | 2/14 | 2/11 | 2/11 |
| AT3G26640 | LWD2 | 39 kDa | 2/21 | 1/21 | – | 1/16 | – | 2/22 | 3/27 | 1/16 | – | – |
| AT1G12910 | LWD1 | 39 kDa | 2/21 | 3/30 | 2/21 | 4/31 | 1/17 | 2/21 | 2/22 | 2/19 | – | – |
| AT4G16250 | phyD | 129 kDa | 1/10 | 1/11 | 2/12 | 1/12 | 3/15 | 2/12 | 2/11 | 1/10 | 2/14 | 2/12 |
| AT1G09570 | phyA | 125 kDa | 1/1 | 1/1 | 7/11 | 5/5 | 7/11 | 2/2 | – | 2/2 | 6/7 | 3/4 |
| AT2G25760 | MLK3 | 76 kDa | 1/5 | 1/6 | – | 1/4 | – | 1/4 | 3/13 | 2/9 | 2/9 | 2/9 |
| AT3G03940 | MLK2 | 78 kDa | 1/8 | 1/8 | – | 1/7 | – | – | 3/20 | 2/13 | 3/13 | 1/9 |
| AT3G46640 | LUX | 35 kDa | 1/3 | 1/3 | – | 3/19 | – | 3/23 | 1/3 | 1/3 | 1/3 | 2/11 |
| AT1G17455 | ELF4‐L4 | 13 kDa | – | – | – | – | – | – | 1/23 | 1/23 | – | – |
| AT1G72630 | ELF4‐L2 | 13 kDa | – | 1/11 | – | 1/13 | – | – | 1/22 | 1/22 | 1/22 | 1/11 |
Proteins co‐purified with ELF3 orthologs (AtELF3, SvELF3 and BdELF3, with C‐terminal His6‐3xFLAG tag) were identified from affinity purification coupled with mass spectrometry (AP‐MS) analyses using 12L:12D grown, 10‐day‐old transgenic plants (in elf3‐2 null mutant backgrounds) harvested at ZT12.
All listed proteins match 99% protein threshold, minimum number peptides of two distinct peptides across all purifications and peptide threshold as 95%. Proteins not matching the criteria were marked with “–”.
Percent coverage for PCH1 is calculated using protein encoded by At2g16365.2.
These proteins have been noted as frequently identified proteins in AP‐MS experiments (see van Leene et al., 2015).
3. RESULTS
3.1. Identifying and cloning ELF3 orthologs from Brachypodium and Setaria
ELF3 is a plant‐specific nuclear protein with conserved roles in flowering and the circadian clock in multiple plant species (Faure et al., 2012; Herrero et al., 2012; Liu et al., 2001; Lu et al., 2017; Matsubara et al., 2012; Saito et al., 2012; Weller et al., 2012; Zakhrabekova et al., 2012). To identify ELF3 orthologs in monocots, we used the protein sequence of Arabidopsis ELF3 (AtELF3) to search the proteomes of two model monocots Brachypodium and Setaria using BLAST (Altschul, Gish, Miller, Myers, & Lipman, 1990). Among the top hits, we identified a previously reported ELF3 homolog in Brachypodium (Bradi2g14290.1, BdELF3) (Calixto, Waugh, & Brown, 2015; Higgins et al., 2010) and two putative ELF3 homologous genes Sevir.5G206400.1 (referred as SvELF3a) and Sevir.3G123200.1 (referred as SvELF3b) in Setaria. We used Clustal Omega ( http://www.ebi.ac.uk/Tools/msa/clustalo/) to conduct multiple sequence alignments of comparing protein sequences of ELF3 orthologs with that of AtELF3 (Sievers et al., 2011). BdELF3, SvELF3a, and SvELF3b encode proteins with similar identity compared to AtELF3 (34.7–36.8%) (Fig. S1). When compared to BdELF3, SvELF3b was 74.3% identical while SvELF3a was 57.4% identical (Fig. S1). Therefore, to maximize the diversity of ELF3 sequences used in this study, we cloned full‐length cDNAs encoding BdELF3 and SvELF3a.
3.2. Diel explorer of Setaria circadian data
In Arabidopsis, ELF3 cycles under diel and circadian conditions (constant condition after entrainment) with a peak phase in the evening (Covington et al., 2001; Hicks et al., 2001; Nusinow et al., 2011). We queried an available diurnal time‐course expression dataset for Brachypodium from the DIURNAL website and found that BdELF3 expression cycles under diel conditions (LDHH, 12‐h light/12‐h dark cycles with constant temperature, Fig. S2a), but not under circadian conditions in available data (LDHC‐F or LDHH‐F, Fig. S2b) (Filichkin et al., 2011; Mockler et al., 2007). Also different from AtELF3, transcript levels of BdELF3 accumulate at dawn rather than peak in the evening (Fig. S2a) when grown under diel conditions, suggesting different regulations on ELF3 expression between monocot and dicot plants. Neither diel nor circadian expression data for Setaria were available. Therefore, we generated RNA‐seq time‐course data to examine SvELF3 expression as well as the circadian expression of other clock orthologs after both photocycle and thermocycle entrainment. In addition, we developed the Diel Explorer tool ( http://shiny.bioinformatics.danforthcenter.org/diel-explorer/) to query and visualize Setaria circadian‐regulated gene expression (Fig. S3). 48,594 Setaria transcripts are represented in the two datasets entrained under either photocycles (LDHH‐F) or thermocycles (LLHC‐F). With Diel Explorer, users can manually enter a list of transcript identifiers, gene ontology (GO) terms, or gene orthologs, plot gene expression, and download data. Alternatively, users can upload files of transcript identifiers or gene orthologs, and/or filter the datasets by entrainment, phase, or significance cutoffs. Data and graphs can be downloaded directly using Diel Explorer. The tool serves as a community resource that can be expanded to include other circadian or diurnal data in the future. The underlying code is available on Github ( https://github.com/danforthcenter/diel-explorer).
For our analysis, JTK‐Cycle was used to detect cycling Setaria genes at cutoff that had a false‐positive rate of 0 with simulated data (Bonferroni‐adjusted p‐value < .001) (Hughes et al., 2010). Under photoperiod entrainment (LDHH‐F), 5,585 of the 48,594 Setaria transcripts are circadian‐regulated (Bonferroni‐adjusted p‐value < .001). This proportion of photoperiod‐entrained circadian genes (~11.5%) is similar to maize (10.8%), rice (12.6%), and poplar (11.2%) data sets, but much smaller than the approximately 30% reported for Arabidopsis (Covington et al., 2008; Filichkin et al., 2011; Khan et al., 2010). Under thermocycle entrainment (LLHC‐F), 582 of the 48,594 Setaria transcripts are circadian‐regulated (Bonferroni‐adjusted p‐value < .001). Therefore, only ~1.2% of Setaria transcripts are circadian cycling under thermocycle entrainment. The ~10‐fold reduction in circadian cycling genes between photocycle and thermocycle entrainment (Fig. S4) is interesting considering that there was less than 1% difference in the number of genes with a circadian period between photocycle and thermocycle entrainment in C3 monocot rice (Oryza japonica) (Filichkin et al., 2011). The reduction in cycling genes between the two entrainment conditions in Setaria compared to rice is an indication that circadian regulation could vary greatly among monocots. Also, the difference in number of cycling genes between monocots and dicots may represent a significant reduction of the role of the circadian clock between these lineages. Wang et al., 2014 found that different metabolites peak along a leaf developmental gradient (Wang et al., 2014) and when we query that expression dataset on Maize eFG browser (Li et al., 2010) for expression of clock genes GRMZM2G175265 and GRMZM2G014902 (zmCCA1), there is an increasing gradient of absolute expression from base to tip. If developmental zones were asynchronous, the overall reduction in cycling genes between monocots and dicots would indicate a stronger role of developmental gradients in modulating the monocot circadian clock along with environment. However, gradients in expression of clock genes are not necessarily evidence that developmental zones are asynchronous, as that gradient could also result in reduction in cycling gene amplitude between developmental zones, but not asynchronicity. Therefore, to examine whether the overall reduction in cycling genes is due to asynchronicity between developmental zones, a circadian time‐course along development would be necessary.
In addition to the overall reduction in circadian genes, the phase with the most number of cycling genes was ZT18 after light entrainment (LDHH‐F; Fig. S4), but ZT12 with temperature entrainment (LLHC‐F; Fig. S4), which is consistent with previous studies that have found significant differences in temperature and light entrainment of the circadian clock (Boikoglou et al., 2011; Michael, Salome, & McClung, 2003; Michael et al., 2008). There are 269 genes that are considered circadian‐regulated and are cycling under both LDHH‐F and LLHC‐F conditions (Bonferroni‐adjusted p‐value < .001). The list of 269 genes that overlap between photocycle and thermocycle entrainment includes best matches for Arabidopsis core clock components TIMING OF CAB EXPRESSION 1 (TOC1, AT5G61380.1; Sevir.1G241000.1), LATE ELONGATED HYPOCOTYL (LHY, AT1G01060; Sevir.6G053100.1), and CCA1‐like gene REVEILLE1 (RVE1, AT5G17300.1; Sevir.1G280700.1). However, putative Setaria orthologs of TOC1, LHY, and RVE1 all have different circadian phases under LDHH entrainment compared to LLHC entrainment (Figure 1). In fact, the majority (233/269) of overlapping circadian genes in Setaria have a distinct circadian phase under thermocycle compared to photocycle entrainment (Fig. S4). We also found that putative orthologs of PSEUDO‐RESPONSE REGULATOR 7 (PRR7, AT5G02810; Sevir.2G456400.1; related to OsPRR73 (Murakami, Ashikari, Miura, Yamashino, & Mizuno, 2003)) and LUX ARRHYTHMO (LUX, AT3G46640; Sevir.5G474200.1) cycle significantly under LDHH‐F but not LLHC‐F conditions (Figure 1). Neither SvELF3a nor SvELF3b cycles under circadian conditions after photo‐ or thermo‐entrainment (Figure 1), similar to ELF3 orthologs in Brachypodium (Fig. S2) (Mockler et al., 2007) and rice (Filichkin et al., 2011). This is different from AtELF3, which continues to cycle under constant condition after either photo‐ or thermos‐entrainment (Fig. S5) (Mockler et al., 2007). The difference in expression of these putative orthologs between Arabidopsis and monocots Setaria, Brachypodium, and rice suggests that the architecture of the circadian clock may have significant differences in response to environmental cues in these two species.
Figure 1.
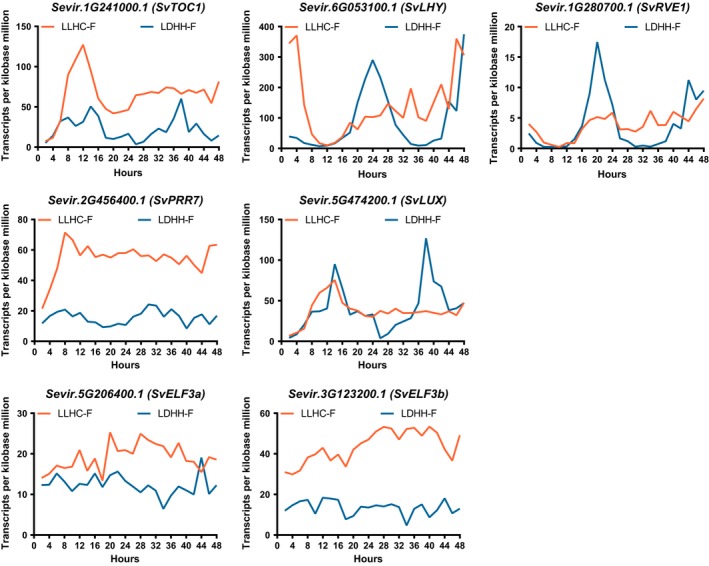
Circadian expression profiles of putative Setaria clock components from Diel Explorer using time‐course RNA‐seq data. Setaria plants were entrained by either photocycle (LDHH) or thermocycle (LLHC), followed by being sampled every 2 hr for 48 hr under constant temperature and light conditions (Free‐Running; F) to generate time‐course RNA‐seq data. Mean values of transcripts per kilobase million (TPM) from two experimental replicates for each timepoints per gene were plotted
3.3. BdELF3 and SvELF3 rescue growth and flowering defects in Arabidopsis elf3 mutant
Although the circadian expression pattern of Brachypodium ELF3 and Setaria ELF3 is different from that of Arabidopsis ELF3, it is still possible that the ELF3 orthologs have conserved biological functions. To test this, we sought to determine whether BdELF3 or SvELF3a could complement the major phenotypic defects of the elf3 mutant in Arabidopsis, namely hypocotyl elongation, time to flowering, or circadian rhythmicity. To this end, we constitutively expressed BdELF3, SvELF3a (hereafter referred as SvELF3), and AtELF3 cDNAs by the 35S Cauliflower mosaic virus promoter in the Arabidopsis elf3‐2 mutant expressing a LUCIFERASE reporter driven by the promoter of CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) (elf3‐2 [CCA1:LUC]) (Pruneda‐Paz et al., 2009). All three ELF3 coding sequences were fused to a C‐terminal His6‐3xFlag affinity tag (HFC), which enables detection by Western blotting and identification of protein‐protein interaction by affinity purification and mass spectrometry (AP‐MS) (Huang, Alvarez, Bindbeutel, et al., 2016). After transforming these constructs, we identified and selected two biologically independent transgenic lines with a single insertion of each At/Bd/SvELF3‐HFC construct. Western blot analysis using FLAG antibodies detected the expression of all ELF3‐HFC fusion proteins (Fig. S6).
Next, we asked whether expressing At/Bd/SvELF3‐HFC fusion proteins could rescue the mutation defects caused by elf3‐2. When plants are grown under light/dark cycles (12‐hr light:12‐hr dark), elf3‐2 mutant plants elongate their hypocotyls much more than wild‐type plants (4.75 ± 0.48 mm vs. 1.95 ± 0.27 mm, respectively. ± = standard deviation) (Figure 2). The long hypocotyl defect in elf3‐2 was effectively suppressed by expressing either AtELF3, or ELF3 orthologs (BdELF3 or SvELF3) (Figure 2). These data show that the monocot ELF3 orthologs function similarly to Arabidopsis ELF3 in the regulation of hypocotyl elongation in seedlings.
Figure 2.
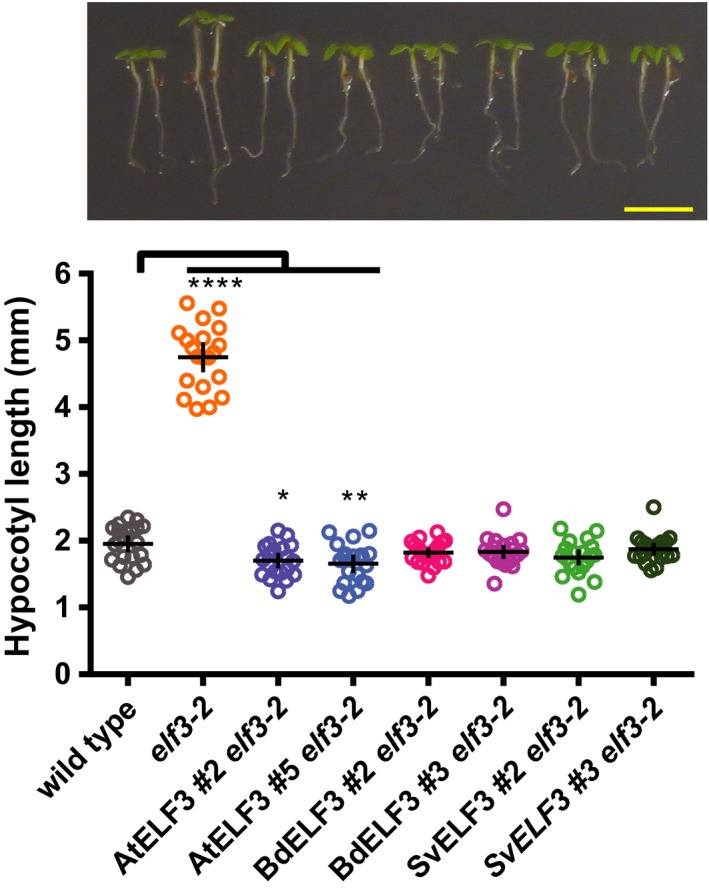
ELF3 orthologs suppress hypocotyl elongation defects in elf3‐2. The hypocotyls of 20 seedlings of wild type, elf3‐2 mutant, AtELF3 elf3‐2, BdELF3 elf3‐2, and SvELF3 elf3‐2 (two independent transgenic lines for each ELF3 ortholog) were measured at 4 days after germination under 12‐hr light:12‐hr dark growth conditions at 22°C. Upper panel shows representative seedlings of each genotype, with scale bar equal to 5 mm. Mean and 95% confidence intervals are plotted as crosshairs. This experiment was repeated three times with similar results. ANOVA with Bonferroni correction was used to generate adjusted p values, * <.05, ** <.01, **** <.0001
In addition to regulating phenotypes in seedlings, ELF3 also functions in adult plants to suppress the floral transition. Loss of function in Arabidopsis ELF3 results in an early flowering phenotype regardless of day length (Hicks et al., 2001; Liu et al., 2001; Zagotta, Shannon, Jacobs, & Meeks‐Wagner, 1992). To determine how monocot ELF3 orthologs compared to Arabidopsis ELF3 in flowering time regulation, we compared flowering responses under long‐day conditions among wild‐type, elf3‐2, and elf3‐2 transgenic lines expressing AtELF3, BdELF3, or SvELF3 (At/Bd/SvELF3‐HFC). Constitutive over‐expression of AtELF3 led to a delay in flowering in long days (Figure 3) as previously observed (Liu et al., 2001). Similarly, constitutive expression of BdELF3 or SvELF3 caused plants to flower significantly later than the elf3 mutants. These data show that all ELF3 orthologs can function to repress the rapid transition to flowering of the elf3 mutation when constitutively expressed in adult plants.
Figure 3.
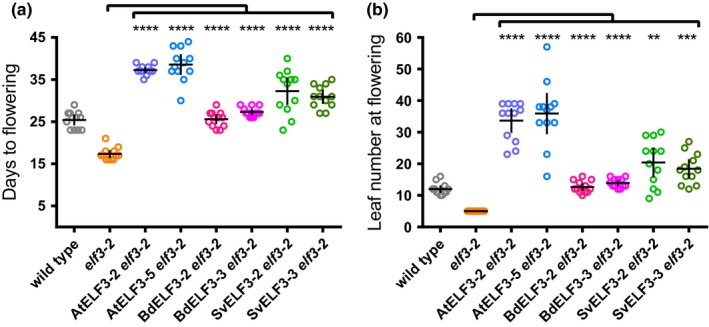
ELF3 orthologs suppress time to flowering of elf3‐2. 12 wild‐type, elf3‐2 mutant, AtELF3 elf3‐2, BdELF3 elf3‐2, and SvELF3 elf3‐2 seedlings from two independent transformations were measured for days (a) and number of rosette leaves (b) at flowering (1 cm inflorescence). Mean and 95% confidence intervals are plotted as crosshairs. This experiment was repeated twice with similar results. ANOVA with Bonferroni correction was used to generate adjusted p values, **<.01, ***<.001, ****<.0001, of measurements when compared to the elf3‐2 mutant line
3.4. BdELF3 and SvELF3 restore the circadian rhythmicity in Arabidopsis elf3 mutant
ELF3 is a key component of the Arabidopsis circadian clock and is critical for maintaining the periodicity and amplitude of rhythms as shown using the CCA1 promoter‐driven luciferase reporter (CCA:LUC) (Covington et al., 2001; Hicks et al., 1996; Nusinow et al., 2011). To determine whether BdELF3 or SvELF3 could rescue the arrhythmic phenotype of the elf3 mutation, we analyzed the rhythms of the CCA1::LUC reporter under constant light conditions after diel entrainment (12‐hr light: 12‐hr dark at constant 22°C). Relative amplitude error (RAE) analysis found that 100% of wild type and nearly all of the three elf3‐2 transgenic lines expressing AtELF3, BdELF3 (BdELF3 #3 was 87.5% rhythmic), and SvELF3 were rhythmic, while only 37.5% of the elf3‐2 lines had measurable rhythms (RAE < 0.5) (Fig. S7). Comparison of average period length found that the AtELF3 expressing lines completely rescued the period and amplitude defects in the elf3 mutant (Figure 4a,d). The SvELF3 and BdELF3 lines also rescued the amplitude defect (Figure 4b,c), but their period was longer than wild type (compare 23.21 ± 0.59 hr for wild type to BdELF3 #2 = 27.68 ± 0.86 hr, BdELF3 #3 = 26.39 ± 1.01 hr, SvELF3 #2 = 24.47 ± 0.50 hr, SvELF3 #3 = 24.32 ± 0.34 hr, and elf3‐2 = 31.27 ± 1.93 hr, ± = standard deviation, Figure 4d). In summary, these data show that expression of any of the ELF3 orthologs is sufficient to recover the amplitude and restore rhythms of the CCA1::LUC reporter.
Figure 4.
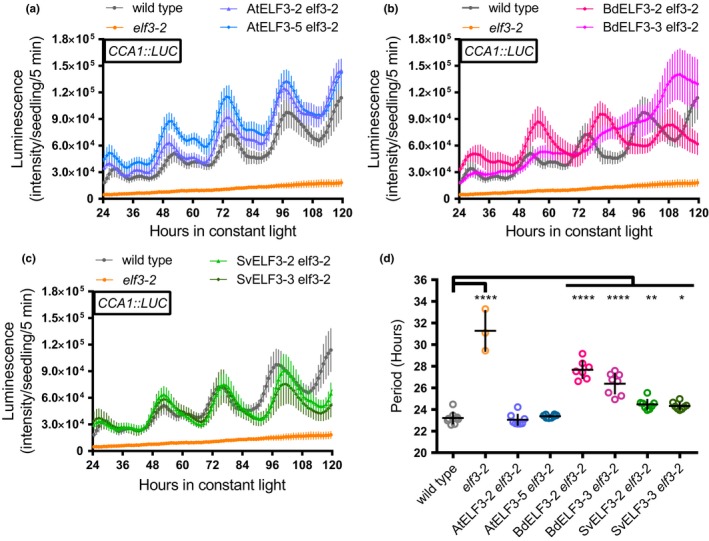
ELF3 orthologs can recover CCA1::LUC rhythms and amplitude in elf3‐2 mutants. Eight seedlings of wild type, elf3‐2 mutant, AtELF3 elf3‐2 (a), BdELF3 elf3‐2 (b), and SvELF3 elf3‐2 (c) from two independent transformations were imaged for bioluminescence under constant light after entrainment in 12‐hr light:12‐hr dark growth conditions at 22°C. Each plot shows average bioluminescence of all seedlings along with 95% confidence interval (error bars). This experiment was repeated four times with similar results. Note that wild‐type and elf3‐2 mutant data were plotted on all graphs for comparison. (d) Periods of seedlings. Only periods with a relative amplitude error below 0.5 (see also Fig. S7) were plotted. Mean and 95% confidence intervals are plotted as crosshairs. ANOVA with Bonferroni correction was used to generate adjusted p values, * <.05, ** <.01, *** <.001, **** <.0001, of measurements when compared to the wild type
3.5. BdELF3 and SvELF3 are integrated into a similar protein‐protein interaction network in Arabidopsis
Despite relatively low sequence conservation at the protein level, the ELF3 orthologs can complement a wide array of elf3 phenotypes (Figures 2–4). As ELF3 functions within the evening complex (EC) in Arabidopsis, which also contains the transcription factor LUX and the DUF‐1313 domain containing protein ELF4 (Herrero et al., 2012; Nusinow et al., 2011), we reasoned that the monocot ELF3 orthologs may also be able to bind to these proteins when expressed in Arabidopsis. To determine whether a composite EC could be formed, we tested whether BdELF3 or SvELF3 could directly interact with AtLUX or AtELF4 in a yeast two‐hybrid assay. Similar to AtELF3 (Nusinow et al., 2011), both BdELF3 and SvELF3 directly interact with both AtELF4 and the C‐terminal portion of AtLUX (Figure 5). We cannot conclude that whether monocot ELF3 orthologs are also able to interact with the N‐terminal AtLUX, as this fragment auto‐activated the reporter gene in the yeast two‐hybrid assay (Figure 5).
Figure 5.
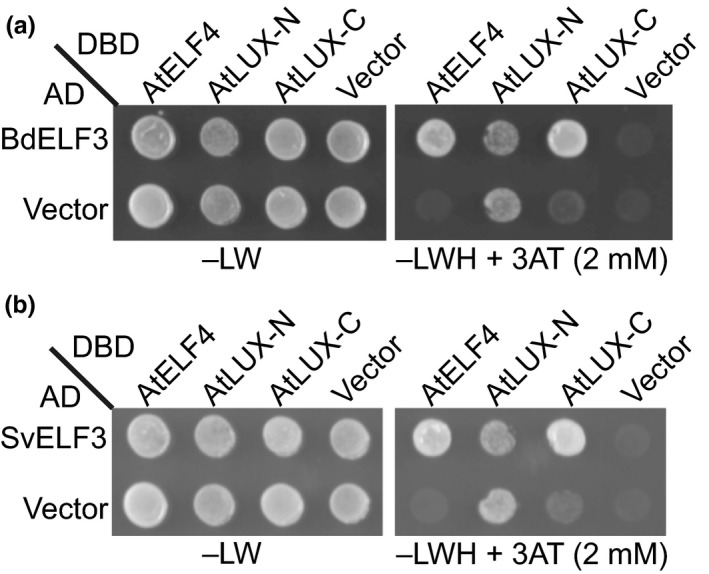
Both BdELF3 and SvELF3 can directly bind to AtELF4 and AtLUX. Yeast two‐hybrid analysis of testing whether either BdELF3 (a) or SvELF3 (b) can directly interact with either AtELF4, the N‐terminal half of AtLUX (AtLUX‐N, a.a. 1‐143) or the C‐terminal half of AtLUX (AtLUX‐C, a.a. 144‐324). –LW tests for the presence of both bait (DBD) and pray (AD) vectors, while the −LWH + 3AT tests for interaction. Vector alone serves as interaction control. This experiment was repeated twice with similar results
ELF3 functions not only as the scaffold of the EC, but also as a hub protein in a protein‐protein interaction network containing multiple key regulators in both the circadian clock and light signaling pathways (Huang & Nusinow, 2016b; Huang, Alvarez, Bindbeutel, et al., 2016). We hypothesize that BdELF3 and SvELF3 could rescue many of the defects of the elf3 mutant because both monocot versions were integrated into the same protein‐protein interaction network. To test this hypothesis, we used affinity purification and mass spectrometry (AP‐MS) to identify the proteins that co‐precipitate with monocot ELF3s when expressed in Arabidopsis. AP‐MS on two biological replicates for each sample with the above‐mentioned independent insertion lines were included for each ELF3 ortholog. For comparison, the same AP‐MS experiment was performed with one of the 35S promoter‐driven AtELF3‐HFC transgenic lines (AtELF3‐2). To detect specific co‐precipitating proteins, we manually removed commonly identified contaminant proteins from plant affinity purifications and mass spectrometry experiments (van Leene et al., 2015), and proteins identified from a control transgenic line expressing GFP‐His6‐3xFlag described previously (Huang, Alvarez, Bindbeutel, et al., 2016) (Table 1, the full list of identified proteins can be found in Table S2).
We have previously reported proteins that co‐precipitated with ELF3 driven from its native promoter using a similar AP‐MS methodology (Huang, Alvarez, Bindbeutel, et al., 2016). When using the 35S promoter‐driven AtELF3 transgenic line, we were able to generate a curated list of 22 proteins that specifically co‐precipitate with AtELF3, including all previously identified proteins, such as all five phytochromes, PHOTOPERIODIC CONTROL OF HYPOCOTYL1 (PCH1) (Huang, Yoo, et al., 2016), and COP1 (Table 1). In addition, we also identified LIGHT‐REGULATED WD 2 (LWD2) and SPA1‐RELATED 4 (SPA4) as now co‐precipitating with AtELF3. These additional interactions may be a result of a combination of altered seedling age, expression level of the ELF3 bait, or tissue‐specificity of expression due to these purifications are from tissues where the epitope‐tagged transgene is constitutively over‐expressed. However, as LWD2 is a known component of the circadian clock (Wu, Wang, & Wu, 2008) and SPA4 is a known component of the COP1‐SPA complex (Zhu et al., 2008), these interactions are likely to be relevant.
In comparing the list of BdELF3 and SvELF3 co‐precipitated proteins with that of AtELF3, we found that neither SvELF3 nor BdELF3 co‐precipitated SPA2 and SPA4, components of the COP1‐SPA complex. In addition, SvELF3 did not co‐precipitate MUT9‐LIKE KINASE1, a kinase with roles in chromatin modification and circadian rhythms as AtELF3 did (Huang, Alvarez, Bindbeutel, et al., 2016; Wang et al., 2015). However, BdELF3 and SvELF3 associated with most of the proteins found in AtELF3 AP‐MS (20 of 22 for BdELF3, 19 of 22 for SvELF3), in at least one of the replicate purifications from each monocot ortholog AP‐MS. Therefore, our data suggest that BdELF3 and SvELF3 are integrated into a similar protein‐protein interaction network as AtELF3, which likely underlies their ability of broadly complementing elf3 mutants.
4. DISCUSSION
Recent work in diverse plant species has found that the circadian clock plays critical roles in regulating metabolism, growth, photoperiodism, and other agriculturally important traits (Bendix, Marshall, & Harmon, 2015; McClung, 2013; Rubin et al., 2017; Shor & Green, 2016). While the relevance of the circadian clock to plant physiology is recognized, it is unclear whether the circadian clock components have conserved function among different plant species. This is particularly true for the majority of clock proteins, whose biological functions are currently poorly understood at the molecular level (Hsu & Harmer, 2014). Also, the divergent modes of growth regulation and photoperiodism between monocots and dicots suggest that the clock evolved to have altered roles in regulating these physiological responses between lineages (Matos et al., 2014; Poire et al., 2010; Song, Shim, Kinmonth‐Schultz, & Imaizumi, 2014). Here, we asked whether orthologs of ELF3 from two monocots could complement any of the loss‐of‐function phenotypes in the model dicot plant Arabidopsis. In this study we found that ELF3 from either Brachypodium or Setaria could complement the hypocotyl elongation, early flowering, and arrhythmic clock phenotype of the elf3 mutant in Arabidopsis, despite the variations in protein sequences and evolutionary divergence between monocot and dicot plants. These data suggest that monocot ELF3s can functionally substitute for Arabidopsis ELF3, albeit with varying efficacy. As monocot and dicot ELF3 are largely different in the protein sequences, functional conservation of ELF3 orthologs also leads to the next open question of identifying the functional domains within ELF3.
Previously, comparison of ELF3 homologs has identified at least five conserved regions that may be important for function (Fig. S1) (Liu et al., 2001; Saito et al., 2012; Weller et al., 2012). Our multiple sequence alignments also show that at least two regions of AtELF3, namely the N‐terminus (AA 1~49) and one middle region (AA 317~389) share many conserved residues with ELF3 orthologs in grasses (Fig. S1). These regions fall within known fragments that are sufficient for binding to phyB (Liu et al., 2001), COP1 (Yu et al., 2008), or ELF4 (Herrero et al., 2012). Consistent with the hypothesis that these conserved regions are critical for proper ELF3 function, a single amino acid substitution (A362V) within this middle region results in defects of ELF3 nuclear localization and changes in the circadian clock period (Anwer et al., 2014). In addition, our protein–protein interaction study and AP‐MS analysis show that both monocot ELF3 can form composite ECs (Figure 5) and that all three ELF3 homologs interact with an almost identical set of proteins in vivo (Table 1), further suggesting that one or more of the conserved regions may mediate the binding between ELF3 and its known interacting proteins. Furthermore, the similar pool of ELF3 interacting proteins identified by Bd/SvELF3 AP‐MS suggests that the overall conformation of ELF3 ortholog proteins is conserved and that similar complexes and interactions with ELF3 orthologs may form in monocot species. However, whether these interactions form in planta and have the same effect on physiology is unclear. For example, Setaria data generated here and public data for Brachypodium and rice showed that ELF3 does not cycle under circadian conditions, which differs from Arabidopsis. Further, different from the fact that the clock plays a key role in regulating elongation in Arabidopsis (Nozue et al., 2007), the circadian clock has no influence on growth in C3 model grass Brachypodium, despite robust oscillating expression of putative clock components (Matos et al., 2014). Similarly, ELF3 from rice and soybean promotes flowering and senescence (Lu et al., 2017; Saito et al., 2012; Sakuraba, Han, Yang, Piao, & Paek, 2016; Yang, Peng, Chen, Li, & Wu, 2013; Zhao et al., 2012), while in Arabidopsis, ELF3 represses these responses (Liu et al., 2001; Sakuraba et al., 2014; Zagotta et al., 1992), which suggests significant rewiring of ELF3 regulated photoperiodic responses of flowering between short‐day (rice/soybean) and long‐day (Arabidopsis) plants. Alternatively, ELF3 may form distinct interactions and complexes in monocot species that were not identified in our trans‐species complementation analysis. Clearly, further work is required to understand ELF3 function in monocots beyond the studies presented here.
In addition to the molecular characterization of ELF3, our analysis of circadian‐regulated genes in Setaria after photo‐ and thermo‐entrainment found significant differences in the behavior of the clock when compared to other monocots. Although the number of circadian‐regulated genes is comparable to studies carried out in maize and rice after photo‐entrainment (between 10% and 12%) (Filichkin et al., 2011; Khan et al., 2010), we found that very few genes (~1%) continue to cycle after release from temperature entrainment in Setaria (Fig. S4) when compared to rice (~11%) (Filichkin et al., 2011). This may reflect a fundamental difference in how the clock interfaces with temperature between these monocot species. Furthermore, proportions of circadian‐regulated genes upon photo‐entrainment in all three monocot plants (Fig. S4) (Filichkin et al., 2011; Khan et al., 2010) are much smaller than the approximately 30% reported for Arabidopsis (Covington et al., 2008), suggesting the divergence of clock functions through evolution or domestication. Further comparisons of circadian responses among monocots or between monocots and dicots will help to determine the molecular underpinning of these differences.
In summary, we find that BdELF3 and SvELF3 form similar protein complexes in vivo as AtELF3, which likely allows for functional complementation of loss of function of elf3 despite relatively low sequence conservation. We also present an online query tool, Diel Explorer that allows for exploration of circadian gene expression in Setaria, which illustrate fundamental differences in clock function among monocots and between monocots and dicots. Collectively, this work is a first step toward functional understanding of the circadian clock in two model monocots, Brachypodium and Setaria.
AUTHOR CONTRIBUTIONS
H.H., M.A.G., T.C.M., and D.A.N. conceived the project; T.C.M., B.S.E., and D.A.N. supervised the experiments; H.H., M.A.G., S.E.H., S.A., C.L., E.L.G., J.G., M.J.N., R.K.B., and D.A.N. performed the experiments; H.H., M.A.G., S.E.H., S.A., C.L., M.J.N., and D.A.N. analyzed the data; H.H., M.A.G., and D.A.N. wrote the manuscript. All authors edited the manuscript.
Supporting information
ACKNOWLEDGEMENTS
We would like to thank all the funding agencies. In addition, we would like to thank the reviewers of this manuscript for the thoughtful comments that improved this work.
Huang H, Gehan MA, Huss SE, et al. Cross‐species complementation reveals conserved functions for EARLY FLOWERING 3 between monocots and dicots. Plant Direct. 2017;00:1–14. 10.1002/pld3.18
Funding information
This work was supported by the National Science Foundation (NSF) grant (IOS‐1456796) to D.A.N., the NSF (DBI‐0922879) grant for acquisition of the LTQ‐Velos Pro Orbitrap LC‐MS/MS, the U.S. Department of Energy grants (DE‐SC0006627, DE‐SC0012639, and DE‐SC0008769) to T.C.M., and the NSF‐Plant Genome grant (IOS‐1202682) to M.A.G.
This manuscript was previously deposited as a preprint at doi: ( https://doi.org/10.1101/131185).
REFERENCES
- Altschul, S. F. , Gish, W. , Miller, W. , Myers, E. W. , & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Anwer, M. U. , Boikoglou, E. , Herrero, E. , Hallstein, M. , Davis, A. M. , Velikkakam James, G. , … Davis, S. J. (2014). Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. eLife, 3, e02206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, T. , Wilhite, S. E. , Ledoux, P. , Evangelista, C. , Kim, I. F. , Tomashevsky, M. , … Soboleva, A. (2013). NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Research, 41, D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell‐Pedersen, D. , Cassone, V. M. , Earnest, D. J. , Golden, S. S. , Hardin, P. E. , Thomas, T. L. , & Zoran, M. J. (2005). Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nature Reviews Genetics, 6, 544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendix, C. , Marshall, C. M. , & Harmon, F. G. (2015). Circadian clock genes universally control key agricultural traits. Molecular Plant, 8, 1135–1152. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L. , Schmutz, J. , Wang, H. , Percifield, R. , Hawkins, J. , Pontaroli, A. C. , … Devos, K. M. (2012). Reference genome sequence of the model plant Setaria. Nature Biotechnology, 30, 555–561. [DOI] [PubMed] [Google Scholar]
- Boikoglou, E. , Ma, Z. , von Korff, M. , Davis, A. M. , Nagy, F. , & Davis, S. J. (2011). Environmental memory from a circadian oscillator: The Arabidopsis thaliana clock differentially integrates perception of photic vs. thermal entrainment. Genetics, 189, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, N. L. , Pimentel, H. , Melsted, P. , & Pachter, L. (2016). Near‐optimal probabilistic RNA‐seq quantification. Nature Biotechnology, 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Brutnell, T. P. , Wang, L. , Swartwood, K. , Goldschmidt, A. , Jackson, D. , Zhu, X. G. , … van Eck, J. (2010). Setaria viridis: A model for C4 photosynthesis. Plant Cell, 22, 2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell, B. (2016). BBMap short read aligner. University of California, Berkeley, California URL http://sourceforge.net/projects/bbmap.
- Calixto, C. P. , Waugh, R. , & Brown, J. W. (2015). Evolutionary relationships among barley and Arabidopsis core circadian clock and clock‐associated genes. Journal of Molecular Evolution, 80, 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington, M. F. , Maloof, J. N. , Straume, M. , Kay, S. A. , & Harmer, S. L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biology, 9, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington, M. F. , Panda, S. , Liu, X. L. , Strayer, C. A. , Wagner, D. R. , & Kay, S. A. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell, 13, 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, L. E. , Knox, K. , Kozma‐Bognar, L. , Southern, M. M. , Pokhilko, A. , & Millar, A. J. (2011). Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Current Biology, 21, 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd, A. N. , Salathia, N. , Hall, A. , Kevei, E. , Toth, R. , Nagy, F. , … Webb, A. A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science, 309, 630–633. [DOI] [PubMed] [Google Scholar]
- Doherty, C. J. , & Kay, S. A. (2010). Circadian control of global gene expression patterns. Annual Review of Genetics, 44, 419–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, M. R. , Davis, S. J. , Bastow, R. M. , McWatters, H. G. , Kozma‐Bognar, L. , Nagy, F. , … Amasino, R. M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature, 419, 74–77. [DOI] [PubMed] [Google Scholar]
- Edelstein, A. , Amodaj, N. , Hoover, K. , Vale, R. , & Stuurman, N. (2010). Computer control of microscopes using microManager. Current Protocols in Molecular Biology, 14(Unit14), 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein, A. D. , Tsuchida, M. A. , Amodaj, N. , Pinkard, H. , Vale, R. D. , & Stuurman, N. (2014). Advanced methods of microscope control using muManager software. Journal of Biological Methods, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. , Domrachev, M. , & Lash, A. E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research, 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. S. , Green, E. W. , Zhao, Y. , van Ooijen, G. , Olmedo, M. , Qin, X. , … Reddy, A. B. (2012). Peroxiredoxins are conserved markers of circadian rhythms. Nature, 485, 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure, S. , Turner, A. S. , Gruszka, D. , Christodoulou, V. , Davis, S. J. , von Korff, M. , & Laurie, D. A. (2012). Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proceedings of the National Academy of Sciences, 109, 8328–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin, S. A. , Breton, G. , Priest, H. D. , Dharmawardhana, P. , Jaiswal, P. , Fox, S. E. , … Mockler, T. C. (2011). Global profiling of rice and poplar transcriptomes highlights key conserved circadian‐controlled pathways and cis‐regulatory modules. PLoS ONE, 6, e16907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein, D. M. , Shu, S. , Howson, R. , Neupane, R. , Hayes, R. D. , Fazo, J. , … Rokhsar, D. S. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Research, 40, D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, P. D. , Diaz, P. , Hogben, C. , Kusakina, J. , Salem, R. , Hartwell, J. , & Hall, A. (2009). Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant Journal, 58, 893–901. [DOI] [PubMed] [Google Scholar]
- Graf, A. , Schlereth, A. , Stitt, M. , & Smith, A. M. (2010). Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proceedings of the National Academy of Sciences, 107, 9458–9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, R. M. , Tingay, S. , Wang, Z. Y. , & Tobin, E. M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiology, 129, 576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenham, K. , & McClung, C. R. (2015). Integrating circadian dynamics with physiological processes in plants. Nature Reviews Genetics, 16, 598–610. [DOI] [PubMed] [Google Scholar]
- Harmer, S. L. (2009). The circadian system in higher plants. Annual Review of Plant Biology, 60, 357–377. [DOI] [PubMed] [Google Scholar]
- Harmer, S. L. , Hogenesch, J. B. , Straume, M. , Chang, H. S. , Han, B. , Zhu, T. , … Kay, S. A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science, 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Hazen, S. P. , Schultz, T. F. , Pruneda‐Paz, J. L. , Borevitz, J. O. , Ecker, J. R. , & Kay, S. A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proceedings of the National Academy of Sciences, 102, 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer, A. , Nusinow, D. A. , Chow, B. Y. , Gehrke, A. R. , Bulyk, M. L. , & Kay, S. A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Current Biology, 21, 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero, E. , Kolmos, E. , Bujdoso, N. , Yuan, Y. , Wang, M. , Berns, M. C. , … Davis, S. J. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell, 24, 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, K. A. , Albertson, T. M. , & Wagner, D. R. (2001). EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell, 13, 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, K. A. , Millar, A. J. , Carre, I. A. , Somers, D. E. , Straume, M. , Meeks‐Wagner, D. R. , & Kay, S. A. (1996). Conditional circadian dysfunction of the Arabidopsis early‐flowering 3 mutant. Science, 274, 790–792. [DOI] [PubMed] [Google Scholar]
- Higgins, J. A. , Bailey, P. C. , & Laurie, D. A. (2010). Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS ONE, 5, e10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P. Y. , & Harmer, S. L. (2014). Wheels within wheels: The plant circadian system. Trends in Plant Science, 19, 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , Alvarez, S. , Bindbeutel, R. , Shen, Z. , Naldrett, M. J. , Evans, B. S. , … Nusinow, D. A. (2016). Identification of evening complex associated proteins in arabidopsis by affinity purification and mass spectrometry. Molecular & Cellular Proteomics: MCP, 15, 201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , Alvarez, S. , & Nusinow, D. A. (2016). Data on the identification of protein interactors with the Evening Complex and PCH1 in Arabidopsis using tandem affinity purification and mass spectrometry (TAP‐MS). Data Brief, 8, 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , & Nusinow, D. (2016a). Tandem purification of His6‐3x FLAG tagged proteins for mass spectrometry from arabidopsis. Bio‐Protocol, 6. [Google Scholar]
- Huang, H. , & Nusinow, D. A. (2016b). Into the evening: complex interactions in the arabidopsis circadian clock. Trends in Genetics, 32, 674–686. [DOI] [PubMed] [Google Scholar]
- Huang, H. , Yoo, C. Y. , Bindbeutel, R. , Goldsworthy, J. , Tielking, A. , Alvarez, S. , … Nusinow, D. A. (2016). CH1 integrates circadian and light‐signaling pathways to control photoperiod‐responsive growth in Arabidopsis. eLife, 5, e13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, M. E. , Hogenesch, J. B. , & Kornacker, K. (2010). JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome‐scale data sets. Journal of Biological Rhythms, 25, 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, A. , Nesvizhskii, A. I. , Kolker, E. , & Aebersold, R. (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical Chemistry, 74, 5383–5392. [DOI] [PubMed] [Google Scholar]
- Khan, S. , Rowe, S. C. , & Harmon, F. G. (2010). Coordination of the maize transcriptome by a conserved circadian clock. BMC Plant Biology, 10, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna, R. , Kikis, E. A. , & Quail, P. H. (2003). EARLY FLOWERING 4 functions in phytochrome B‐regulated seedling de‐etiolation. Plant Physiology, 133, 1530–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, W. Y. , Hicks, K. A. , & Somers, D. E. (2005). Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiology, 139, 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos, E. , Herrero, E. , Bujdoso, N. , Millar, A. J. , Toth, R. , Gyula, P. , … Davis, S. J. (2011). A reduced‐function allele reveals that EARLY FLOWERING3 repressive action on the circadian clock is modulated by phytochrome signals in Arabidopsis. Plant Cell, 23, 3230–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, R. , Ichihashi, Y. , Kimura, S. , Chitwood, D. H. , Headland, L. R. , Peng, J. , … Sinha, N. R. (2012). A high‐throughput method for illumina RNA‐Seq library preparation. Frontiers in Plant Science, 3, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leene, J. , Eeckhout, D. , Cannoot, B. , de Winne, N. , Persiau, G. , van de Slijke, E. , … de Jaeger, G. (2015). An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nature Protocols, 10, 169–187. [DOI] [PubMed] [Google Scholar]
- Li, P. , Ponnala, L. , Gandotra, N. , Wang, L. , Si, Y. , Tausta, S. L. , … Brutnell, T. P. (2010). The developmental dynamics of the maize leaf transcriptome. Nature Genetics, 42, 1060–1067. [DOI] [PubMed] [Google Scholar]
- Liu, X. L. , Covington, M. F. , Fankhauser, C. , Chory, J. , & Wagner, D. R. (2001). ELF3 encodes a circadian clock‐regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell, 13, 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, P. , Wu, J. , Cheng, F. , Cressman, L. G. , Wang, X. , & McClung, C. R. (2012). Preferential retention of circadian clock genes during diploidization following whole genome triplication in Brassica rapa . Plant Cell, 24, 2415–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S. , Zhao, X. , Hu, Y. , Liu, S. , Nan, H. , Li, X. , … Kong, F. (2017). Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nature Genetics, 49, 773–779. [DOI] [PubMed] [Google Scholar]
- Matos, D. A. , Cole, B. J. , Whitney, I. P. , Mackinnon, K. J. , Kay, S. A. , & Hazen, S. P. (2014). Daily changes in temperature, not the circadian clock, regulate growth rate in Brachypodium distachyon. PLoS ONE, 9, e100072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara, K. , Ogiso‐Tanaka, E. , Hori, K. , Ebana, K. , Ando, T. , & Yano, M. (2012). Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant and Cell Physiology, 53, 709–716. [DOI] [PubMed] [Google Scholar]
- McClung, C. R. (2013). Beyond Arabidopsis: The circadian clock in non‐model plant species. Seminars in Cell & Developmental Biology, 24, 430–436. [DOI] [PubMed] [Google Scholar]
- Michael, T. P. , Mockler, T. C. , Breton, G. , Mcentee, C. , & Byer, A. (2008). Network discovery pipeline elucidates conserved time‐of‐day‐specific cis‐regulatory modules. PLoS Genetics, 4, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, T. P. , Salome, P. A. , & McClung, C. R. (2003). Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proceedings of the National Academy of Sciences, 100, 6878–6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, T. , Nomoto, Y. , Oka, H. , Kitayama, M. , Takeuchi, A. , Tsubouchi, M. , & Yamashino, T. (2014). Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night‐time repressor in Arabidopsis thaliana . Plant and Cell Physiology, 55, 958–976. [DOI] [PubMed] [Google Scholar]
- Mockler, T. C. , Michael, T. P. , Priest, H. D. , Shen, R. , Sullivan, C. M. , Givan, S. A. , … Chory, J. (2007). The DIURNAL project: DIURNAL and circadian expression profiling, model‐based pattern matching, and promoter analysis. Cold Spring Harbor Symposia on Quantitative Biology, 72, 353–363. [DOI] [PubMed] [Google Scholar]
- Murakami, M. , Ashikari, M. , Miura, K. , Yamashino, T. , & Mizuno, T. (2003). The evolutionarily conserved OsPRR quintet: Rice pseudo‐response regulators implicated in circadian rhythm. Plant and Cell Physiology, 44, 1229–1236. [DOI] [PubMed] [Google Scholar]
- Nagel, D. H. , & Kay, S. A. (2012). Complexity in the wiring and regulation of plant circadian networks. Current Biology, 22, R648–R657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii, A. I. , Keller, A. , Kolker, E. , & Aebersold, R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chemistry, 75, 4646–4658. [DOI] [PubMed] [Google Scholar]
- Nozue, K. , Covington, M. F. , Duek, P. D. , Lorrain, S. , Fankhauser, C. , Harmer, S. L. , & Maloof, J. N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature, 448, 358–361. [DOI] [PubMed] [Google Scholar]
- Nusinow, D. A. , Helfer, A. , Hamilton, E. E. , King, J. J. , Imaizumi, T. , Schultz, T. F. , … Kay, S. A. (2011). The ELF4‐ELF3‐LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature, 475, 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai, K. , & Ishiura, M. (2005). PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes to Cells, 10, 963–972. [DOI] [PubMed] [Google Scholar]
- Ouyang, Y. , Andersson, C. R. , Kondo, T. , Golden, S. S. , & Johnson, C. H. (1998). Resonating circadian clocks enhance fitness in cyanobacteria. Proceedings of the National Academy of Sciences, 95, 8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz, J. D. , Straume, M. , Stanewsky, R. , Jamison, C. F. , Brandes, C. , Dowse, H. B. , … Kay, S. A. (1997). Quantitative analysis of drosophila period gene transcription in living animals. Journal of Biological Rhythms, 12, 204–217. [DOI] [PubMed] [Google Scholar]
- Poire, R. , Wiese‐Klinkenberg, A. , Parent, B. , Mielewczik, M. , Schurr, U. , Tardieu, F. , & Walter, A. (2010). Diel time‐courses of leaf growth in monocot and dicot species: Endogenous rhythms and temperature effects. Journal of Experimental Botany, 61, 1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko, A. , Fernandez, A. P. , Edwards, K. D. , Southern, M. M. , Halliday, K. J. , & Millar, A. J. (2012). The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Molecular Systems Biology, 8, 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda‐Paz, J. L. , Breton, G. , Para, A. , & Kay, S. A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science, 323, 1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, M. J. , Brock, M. T. , Davis, A. M. , German, Z. M. , Knapp, M. , Welch, S. M. , … Weinig, C. (2017). Circadian rhythms vary over the growing season and correlate with fitness components. Molecular Ecology. [DOI] [PubMed] [Google Scholar]
- Saito, H. , Ogiso‐Tanaka, E. , Okumoto, Y. , Yoshitake, Y. , Izumi, H. , Yokoo, T. , … Tanisaka, T. (2012). Ef7 encodes an ELF3‐like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short‐ and long‐day conditions. Plant and Cell Physiology, 53, 717–728. [DOI] [PubMed] [Google Scholar]
- Sakuraba, Y. , Han, S. H. , Yang, H. J. , Piao, W. , & Paek, N. C. (2016). Mutation of rice early flowering3.1 (OsELF3.1) delays leaf senescence in rice. Plant Molecular Biology, 92, 223–234. [DOI] [PubMed] [Google Scholar]
- Sakuraba, Y. , Jeong, J. , Kang, M. Y. , Kim, J. , Paek, N. C. , & Choi, G. (2014). Phytochrome‐interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nature Communications, 5, 4636. [DOI] [PubMed] [Google Scholar]
- Schneider, C. A. , Rasband, W. S. , & Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor, E. , & Green, R. M. (2016). The impact of domestication on the circadian clock. Trends in Plant Science, 21, 281–283. [DOI] [PubMed] [Google Scholar]
- Sievers, F. , Wilm, A. , Dineen, D. , Gibson, T. J. , Karplus, K. , Li, W. , … Higgins, D. G. (2011). Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology, 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. H. , Ito, S. , & Imaizumi, T. (2010). Similarities in the circadian clock and photoperiodism in plants. Current Opinion in Plant Biology, 13, 594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. H. , Shim, J. S. , Kinmonth‐Schultz, H. A. , & Imaizumi, T. (2014). Photoperiodic flowering: Time measurement mechanisms in leaves. Annual Review of Plant Biology, 66, 441–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino, J. A. , Deutsch, E. W. , Wang, R. , Csordas, A. , Reisinger, F. , Rios, D. , … Hermjakob, H. (2014). ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nature Biotechnology, 32, 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Barnaby, J. Y. , Tada, Y. , Li, H. , Tor, M. , Caldelari, D. , … Dong, X. (2011). Timing of plant immune responses by a central circadian regulator. Nature, 470, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Casas‐Mollano, J. A. , Xu, J. , Riethoven, J. J. , Zhang, C. , & Cerutti, H. (2015). Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana. Proceedings of the National Academy of Sciences, 112, 8487–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Czedik‐Eysenberg, A. , Mertz, R. A. , Si, Y. , Tohge, T. , Nunes‐Nesi, A. , … Brutnell, T. P. (2014). Comparative analyses of C4 and C3 photosynthesis in developing leaves of maize and rice. Nature Biotechnology, 32, 1158–1165. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Si, Y. , Dedow, L. K. , Shao, Y. , Liu, P. , & Brutnell, T. P. (2011). A low‐cost library construction protocol and data analysis pipeline for Illumina‐based strand‐specific multiplex RNA‐seq. PLoS ONE, 6, e26426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller, J. L. , Liew, L. C. , Hecht, V. F. G. , Rajandran, V. , Laurie, R. E. , Ridge, S. , … Lejeune‐Hénaut, I. (2012). A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proceedings of the National Academy of Sciences, 109, 21158–21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen, H. , & Young, M. W. (2006). Interplay of circadian clocks and metabolic rhythms. Annual Review of Genetics, 40, 409–448. [DOI] [PubMed] [Google Scholar]
- Woelfle, M. A. , Ouyang, Y. , Phanvijhitsiri, K. , & Johnson, C. H. (2004). The adaptive value of circadian clocks: An experimental assessment in cyanobacteria. Current Biology, 14, 1481–1486. [DOI] [PubMed] [Google Scholar]
- Wu, J. F. , Wang, Y. , & Wu, S. H. (2008). Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiology, 148, 948–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Peng, Q. , Chen, G.‐X. , Li, X.‐H. , & Wu, C.‐Y. (2013). OsELF3 is involved in circadian clock regulation for promoting flowering under long‐day conditions in rice. Molecular Plant, 6, 202–215. [DOI] [PubMed] [Google Scholar]
- Yu, J. W. , Rubio, V. , Lee, N. Y. , Bai, S. , Lee, S. Y. , Kim, S. S. , … Deng, X. W. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Molecular Cell, 32, 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta, M. , Shannon, S. , Jacobs, C. , & Meeks‐Wagner, D. (1992). Early‐flowering mutants of Arabidopsis thaliana . Functional Plant Biology, 19, 411–418. [Google Scholar]
- Zakhrabekova, S. , Gough, S. P. , Braumann, I. , Muller, A. H. , Lundqvist, J. , Ahmann, K. , … Hansson, M. (2012). Induced mutations in circadian clock regulator Mat‐a facilitated short‐season adaptation and range extension in cultivated barley. Proceedings of the National Academy of Sciences, 109, 4326–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Henriques, R. , Lin, S.‐S. , Niu, Q.‐W. , & Chua, N.‐H. (2006). Agrobacterium‐mediated transformation of Arabidopsis thaliana using the floral dip method. Nature Protocols, 1, 641–646. [DOI] [PubMed] [Google Scholar]
- Zhao, J. , Huang, X. , Ouyang, X. , Chen, W. , Du, A. , Zhu, L. , … Li, S. (2012). OsELF3‐1, an ortholog of Arabidopsis early flowering 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS ONE, 7, e43705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, D. , Maier, A. , Lee, J. H. , Laubinger, S. , Saijo, Y. , Wang, H. , … Deng, X. W. (2008). Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell, 20, 2307–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


