Abstract
Many of mRNAs synthesized during pollen development are translated after germination, and we hypothesize that they are stored in cytoplasmic granules. We analyzed the cellular localization of the SKS14 and AT59 Arabidopsis mRNAs, which are orthologues of the tobacco NTP303 and tomato LAT59 pollen mRNAs, respectively, by artificially labeling the transcripts with a MS2‐GFP chimera. A MATLAB‐automated image analysis helped to identify the presence of cytoplasmic SKS14 and AT59 mRNA granules in mature pollen grains. These mRNA granules partially colocalized with VCS and DCP1, two processing body (PB) proteins. Finally, we found a temporal correlation between SKS14 protein accumulation and the disappearance of SKS14 mRNA granules during pollen germination. These results contribute to unveil a mechanism for translational regulation in Arabidopsis thaliana pollen.
Keywords: MATLAB, MS2, pollen, processing body, translational regulation
1. INTRODUCTION
Arabidopsis pollen development involves two stages: an early stage that includes microspores and bicellular pollen followed by a late stage of tricellular and mature pollen. These two stages differ in their transcriptional profiles: early genes expressed during the microspore stage decrease their abundance before pollen maturation. In turn, late genes are expressed after asymmetric mitosis and accumulate during pollen maturation, bringing about a stable pool of mRNAs that govern pollen germination and pollen tube early growth. Thereby, at anthesis, all proteins or mRNAs required for germination and pollen tube growth would be present (Boavida, Becker, & Feijó, 2005). Consistent with this, in many species, pollen germination is independent of transcription but dependent on translation (Twell, 1994).
Pollen tube growth is a process that occurs in an explosive way, and tip extension requires a rapid increase in pectin amount. Some of the late pollen genes encode proteins homologous to enzymes linked to pectin metabolism, including polygalacturonases (Brown & Crouch, 1990; Niogret, Dubald, Mandaron, & Mache, 1991; Rogers & Lonsdale, 1992), pectin methylesterases (Mu, Stains, & Kao, 1994; Wakeley, Rogers, Rozycka, Greenland, & Hussey, 1998), and pectate lyases (Rogers, Harvey, & Lonsdale, 1992; Wing et al., 1989). The tomato late gene LAT59 codifies a protein related to the pectate lyase family, potentially involved in cell wall degradation by pectin cleavage. The translation of LAT59 mRNA is highly regulated and occurs since final stages of pollen development (Curie & McCormick, 1997). In turn, the tomato LAT52 is a pollen gene that codifies a cysteine‐rich extracellular protein involved in pollen hydration and pollen germination (Muschietti, Dircks, Vancanneyt, & McCormick, 1994). LAT52 transcript levels gradually increase during pollen development reaching its maximum at pollen maturity (Twell, Klein, Fromm, & McCormick, 1989).
Another example is the tobacco NTP303 gene, which is transcribed through pollen development from the early binucleate stages (Weterings et al., 1992) and translated once germination occurs (Wittink et al., 2000). NTP303 has homology with ascorbate oxidases, and according to its time of expression, it would be linked to pollination or fertilization (Schrauwen et al., 1999).
Processing body (PB) are highly conserved cytoplasmic organelles involved in translation inhibition, mRNA degradation, and storage. PB formation includes translationally repressed messenger ribonucleoproteins (mRNPs) that aggregate into larger structures through protein–protein interactions. mRNPs localized in PBs can be degraded or undergo a rearrangement where translation initiation factors are recruited, thus allowing mRNAs to reenter polysomes (Decker & Parker, 2012).
As in yeast and animals, it has been shown that Arabidopsis thaliana PBs include decapping factors and coactivators, such as decapping protein one (DCP1), decapping protein two (DCP2), decapping protein five (DCP5), and varicose (VCS) (Xu & Chua, 2009; Xu, Yang, Niu, & Chua, 2006). The knockout of these genes affects the growth of vascular and epidermal cells, stomata, and root hairs, suggesting that decapping and/or PBs have a fundamental role during plant development (Maldonado‐Bonilla, 2014; Xu et al., 2006).
Here, we focus in the Arabidopsis mature pollen SKS14 and AT59 mRNAs (Loraine, McCormick, Estrada, Patel, & Qin, 2013) which are putative orthologues of the tobacco NTP303 and tomato LAT59 mRNAs, respectively. We found SKS14 and AT59 mRNAs in cytoplasmic granules that colocalize with the PB markers VCS and DCP1. Finally, we show that SKS14 protein accumulates during pollen germination while the number of SKS14 mRNA granules decreases. These observations are compatible with the notion that the SKS14 mRNA is released from PBs to allow translation in a controlled manner.
2. MATERIAL AND METHODS
2.1. Plant material and growth conditions
Sterilized seeds from Arabidopsis thaliana (ecotype Columbia‐0) wild‐type, single mutants, and transgenic plants were plated on 0.5X Murashige and Skoog (1962) medium with 1% sucrose and selective agent (50 mg/l kanamycin) if necessary and cold stratified 4 days in dark at 4°C. Seeds were germinated and grown under continuous light at 22°C for 7 days. Seedlings were then transferred to soil or peat, mixed with vermiculite and perlite (2:1:1), and grown in a chamber at 22°C under long‐day (16/8 hr light/dark) photoperiod and 60% relative humidity.
2.2. Plasmid constructs
MS2 system is based on the strong association between bacteriophage MS2 capsid protein (MCP) and six repeat loops of a 19‐nucleotide fragment containing bacteriophage's replicase start codon (Bertrand et al., 1998). The pMS2‐GFP and pSL‐MS2‐12X plasmids were donated by Robert Singer (Addgene plasmid #27121 and #27119, respectively) (Bertrand et al., 1998; Fusco et al., 2003).
MS2 control vector was generated through an LR recombination system (Invitrogen) using the binary vector pK7WG2D. After PCR amplification from the pMS2‐GFP plasmid, the GFP‐MCP fragment was inserted into the pZD05 vector under the control of LAT52 promoter, obtaining the pLAT52::GFP‐MCP. Then, the pLAT52::GFP‐MCP fragment was inserted in the pENTR1a entry vector and then recombined in the binary vector pK7WG2D. All plasmids were confirmed by sequencing.
SKS14 and AT59 5′UTRs and coding regions were cloned by PCR from Arabidopsis mature pollen cDNA in the pSL‐MS2‐12X plasmid. The SKS14‐MS2‐12X and AT59‐MS2‐12X fragments were PCR‐cloned into pZD05. pLAT52::SKS14‐MS2‐12X and pLAT52::AT59‐MS2‐12X fragments were PCR‐cloned and inserted into the binary vector pK7WG2D‐pLAT52::GFP‐MCP obtaining pLAT52::GFP‐MCP/pLAT52::SKS14‐MS2‐12X and pLAT52::GFP‐MCP/pLAT52::AT59‐MS2‐12X. All plasmids were confirmed by sequencing.
pLAT52::RFP‐VCS and pLAT52::RFP‐DCP1 vectors were generated using the binary vector pK7WG2D. The VCS or DCP1 fragments were amplified from the pda09249 DNA stock (ABRC) or Arabidopsis mature pollen cDNA, respectively. They were inserted in the pENTR1a entry vector and then recombined in pK7WG2D. The pLAT52‐RFP fragment was amplified from the pZD05 vector, digested, and cloned in pK7WG2D containing VCS or DCP1. All plasmids were confirmed by sequencing.
pSKS14::SKS14‐RFP vector was generated using binary vector pGWB408. The SKS14 coding region and the RFP fragment were amplified from Arabidopsis cDNA and from the pZD05 vector, respectively, and cloned in the pENTR1a vector. The 35S promoter of the pGWB408 vector was replaced by a 1091‐bp fragment corresponding to the SKS14 promoter that was cloned by PCR from Arabidopsis cDNA. All plasmids were confirmed by sequencing.
EHA105 Agrobacterium strain was used for transformation. Plant transformation was carried on by floral dip method (Clough & Bent, 1998).
2.3. Microscopy analysis
Mature pollen grains of twenty‐day‐old plants were collected and stored in Tris–EDTA buffer in the presence or absence of puromycin (100 μg/ml) for 1 hr at 22°C. Images were taken with a fluorescence microscope Olympus BX41 or in a confocal microscope Olympus IX81 FV1000 (Laser 488 nm, filter BP 505‐525, or Laser 543 nm, filter BP 560‐620) and analyzed by MATLAB scripts.
2.4. MATLAB scripts
2.4.1. mRNA granules detection
In order to analyze the cytoplasmic granules by MATLAB (Fig. S2), initially a mask was applied to eliminate vegetative nucleus and facilitate the cytoplasm visualization. Then, the image was split into 49 boxes and the average fluorescence of each box was determined (Fig. S2C). Those groups of pixels that were between 5 and 30 pixels in size and had an average fluorescence higher than three standard deviations from the average fluorescence of the corresponding box were marked with a yellow point. The grid was moved 20 pixels to the right and down, and a new round of analysis was carried out. Those groups of pixels that followed similar parameters were marked with a light blue circle (Fig. S2D). Pixels detected with both analysis (yellow point and light blue circle) were processed in a new round of the script to confirm the cytoplasmic granules (Fig. S2E). The validated cytoplasmic granules were marked with a red point and a green circle, indicating the center and the approximate area of the granule, respectively (Fig. S2F). Determination of the number of cytoplasmic granules was carried out by taking one image of the central plane of each pollen grain; by this approach, we obtained an approximated value of the number of cytoplasmic granules for each pollen grain analyzed. All the analysis was carried out in the presence of puromycin 50 μg/ml.
2.4.2. PBs detection
A similar MATLAB script used for the mRNA granules was applied for PBs detection except that positive PB marker protein foci were defined between 5 and 80 pixels.
2.4.3. Colocalization analysis
To address statistical significance of the proximity between the mRNAs granules and PB marker protein foci (i.e., whether or not they may colocalize by chance given the specific layout of mRNAs and PBs within an image), a shuffling analysis was performed using a custom‐made MATLAB routine. After confirming the cytoplasmic granules and the PB marker protein foci, both were modeled as circles in the 2D plane using the center of mass and area obtained from previously described analysis. The random distribution of the distance between the mRNA granule and the nearest PB foci was obtained by randomly repositioning the mRNA granule 10,000 times while keeping the PB's foci layout fixed. From that random distribution, we proceeded to obtain the p‐value for the experimentally obtained distance. The sum of the radii of the mRNA granule and the nearest PB marker protein foci was used to determine whether they were colocalizing, proximal, or distant. All the analyses were carried out in the presence of puromycin 50 μg/ml.
2.5. Statistical analysis
Results are expressed as scattered plots. Points represent data dispersion for n = 7‐10 quantifications of cytoplasmic granules per pollen grain (each n corresponds to the media of the number of cytoplasmic granules of 10‐20 pollen grains) and n = 10‐25 for determination of fluorescence intensity (each n corresponds to one vegetative nucleus fluorescence intensity measurement); probability values of <.1 were considered statistically significant. Statistical analysis of the data and further processing were performed using GraphPad Prism version 5.00 for Windows (GraphPad Software).
3. RESULTS
3.1. SKS14 and AT59 mRNAs form cytoplasmic granules in mature pollen
LAT59 protein is present at final stages of pollen development and its amount increases after pollen germination (Curie & McCormick, 1997) while NTP303 is synthesized only once pollen germinates (Wittink et al., 2000). We sought to investigate the post‐transcriptional regulation of the Arabidopsis LAT59 and NTP303 orthologues. Arabidopsis AT59 (At1 g14420) is the LAT59 ortholog (Kulikauskas & McCormick, 1997). Among the putative NTP303 orthologues, we focused on the SKU5 similar (SKS) family that has 19 members in Arabidopsis (Sedbrook et al., 2002). Among them, SKS11, SKS12, SKS13, and SKS14 are expressed in pollen (Honys & Twell, 2000). We then considered the length and free energies of their 5′UTRs regions and compared these parameters to those of NTP303 5′UTR. Furthermore, SKS14 is the pollen SKS gene that shows the highest increment in expression levels as pollen development proceeds, showing a maximum in mature pollen (Honys & Twell, 2000). This makes SKS14 a good candidate to test our hypothesis and therefore was chosen for this study.
Paralleling the fate of maternal mRNA in animal oocytes, we hypothesize that SKS14 and AT59 mRNAs are stored in cytoplasmic granules during pollen development. To analyze the subcellular localization of these transcripts in vivo, we artificially labeled the mRNA using the MS2‐MCP system, which is based on the strong binding of the bacteriophage MS2 coat protein (MCP) to specific RNA loops termed MCP‐binding site (Lampasona & Czaplinski, 2016). The GFP‐MCP protein was fused to a nuclear localization signal (NLS) so that only the GFP‐MCP bound to the target mRNAs is found in the cytoplasm. Fig. S1 shows that the free GFP‐MCP protein is largely accumulated in the vegetative nuclei identified by DAPI staining. We obtained independent Arabidopsis transgenic lines containing the GFP‐MCP construct together with either the SKS14 or the AT59 mRNA fused to the MCP‐binding site, termed SKS14‐MS2 and AT59‐MS2, respectively (Fig. 1). All constructs are under the control of the tomato pollen‐specific promoter LAT52 (Twell, Yamaguchi, Wing, Ushiba, & McCormick, 1991). For comparison, we generated control transgenic plants termed MS2 lines carrying only the GFP‐MCP construct (Fig. 1). With these tools, we found that both SKS14 and AT59 messengers form granules in the cytoplasm of mature pollen grains (Fig. 2; Table 1).
Figure 1.
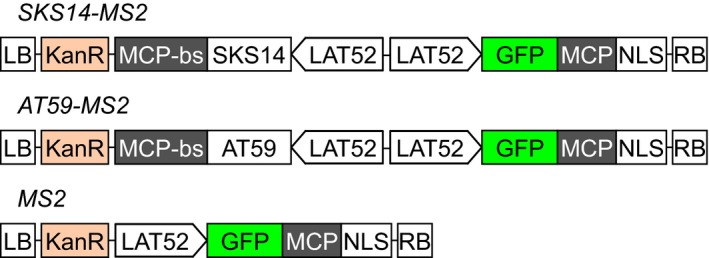
mRNA detection by the MS2 system. The two depicted constructs were inserted in the same vector: GFP fused to the MS2 coat protein (MCP) with a nuclear localization signal (NLS), and the SKS14 or AT59 transcripts fused to MCP‐binding site (MCP‐bs). A control construct termed MS2 encodes the GFP‐MCP chimera and no target mRNA. The three constructs SKS14‐MS2,AT59‐MS2, and MS2 were under the control of the pollen‐specific promoter LAT52. LB and RB, left and right borders of the translocation cassette, respectively
Figure 2.
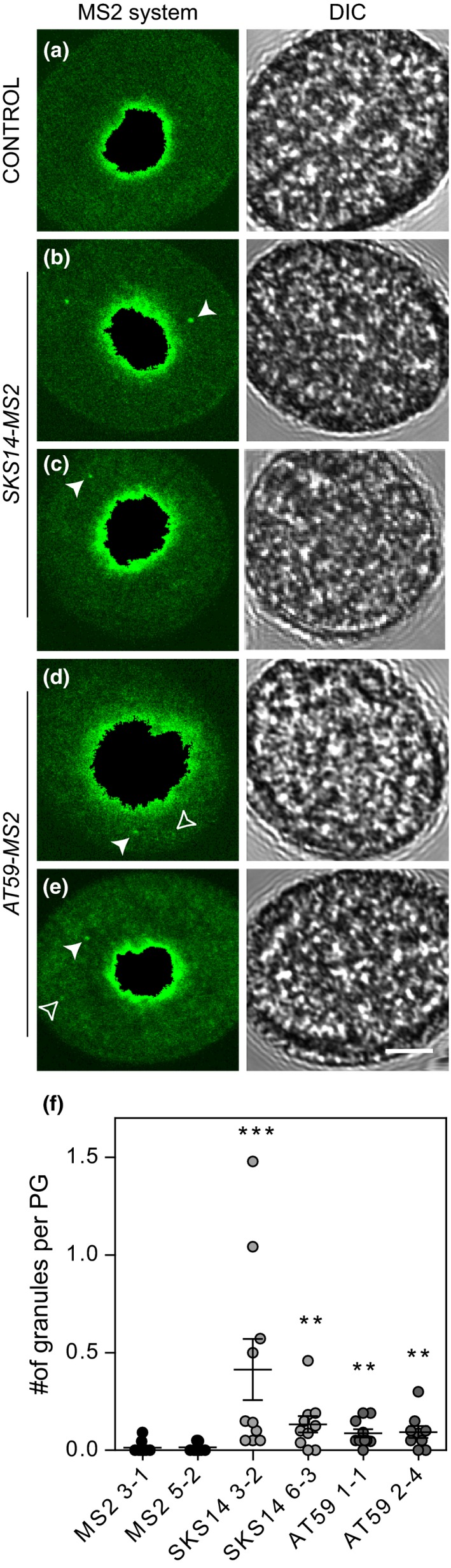
Arabidopsis SKS14 and AT59 mRNAs form cytoplasmic granules in mature pollen grains. (a) Confocal images of a representative pollen grain from the control MS2 line. A mask was applied to eliminate vegetative nucleus and facilitate cytoplasm visualization. (b‐e) Representative images of two independent SKS14‐MS2 (b and c) and two AT59‐MS2 lines (d and e). In the left panels, white arrowheads show examples of cytoplasmic granules identified by the MATLAB script while empty arrowheads show cytoplasmic aggregates that were not detected by the MATLAB script. Right panels, DIC images. Size bar, 5 μm. (f) Quantification of cytoplasmic granules. Each point corresponds to the mean value of an independent sample that included 20 pollen grains. The media and standard error for each transgenic line are shown. Statistical significance (Mann–Whitney test) relative to the control line MS2 3‐1 is indicated (***p < .001 and **p < .01)
Table 1.
Quantification of cytoplasmic granules per pollen grain in WT pollen. Mean values from 20 pollen grains in independent samples are shown, and the media and standard error for each transgenic line are indicated. N: number of pollen grains analyzed. ND: Not determined (Mann–Whitney test)
| Line | Media | Standard error | N | p‐value | p‐value summary |
|---|---|---|---|---|---|
| MS2 3‐1 | 0.014 | 0.010 | 10 | ND | ND |
| SKS14 3‐2 | 0.413 | 0.156 | 10 | .0003 | *** |
| SKS14 6‐3 | 0.133 | 0.042 | 10 | .0053 | ** |
| AT59 1‐1 | 0.087 | 0.021 | 10 | .0029 | ** |
| AT59 2‐4 | 0.093 | 0.031 | 9 | .0092 | ** |
Representative images of pollen grains showing cytoplasmic granules of SKS14‐MS2 mRNA and AT59‐MS2 mRNA are depicted in Fig. 2b‐e. A representative control MS2 pollen grain is shown in Fig. 2a.
To perform an automated unbiased analysis of these images, we implemented a MATLAB script that detects granules in the cytoplasm, applying a mask to the nucleus and measuring pixel size and fluorescence intensity (see Material and Methods and Fig. S2). Fig. 2f shows that the number of cytoplasmic granules of fluorescent protein in the SKS14‐MS2 and AT59‐MS2 lines was 10‐40 times higher than in the control MS2 line (Table 1).
The number of mRNA granules per pollen grain is somehow lower than expected considering that the strong LAT52 promoter was used. Given that a whole scan of the entire pollen grain along Z‐axis rapidly quenched the fluorescent GFP signal, a maximum of five confocal sections of each pollen grain were analyzed, with a final size of 2.5 um thick. This represents about 10% of the volume of an Arabidopsis pollen grain, and thus, we speculate that the total number of RNA granules would be 10 times larger than the experimental value. In addition, the presence submicroscopic RNA granules of less than the resolution of the confocal microscope (~150 nm) should also be considered. A second factor that may be introducing a systematic error is that granules close to the perinuclear region were not included, due to the strong GFP nuclear signal and because most of the potential positives close to the perinuclear region were extensions of the vegetative cell nuclei indentations. Thus, a significant proportion of mRNA granules was lost in our analysis, but at the same time, we largely reduced the chances of including false positives.
To test that the differences observed in the number of granules were not due to dissimilar expression levels of the GFP‐MCP chimera in the different Arabidopsis lines, we measured the fluorescence intensity in the vegetative nuclei, where nonbound GFP‐MCP accumulates due to the NLS included in the construct. Although GFP‐MCP intensity varies between lines, we found that there is no correlation between the nuclear fluorescence intensity and the number of cytoplasmic granules, strongly supporting that these granules are specific and are not due to the overexpression of heterologous GFP‐MCP protein (Fig. S3).
3.2. SKS14 and AT59 mRNA cytoplasmic granules colocalize with PB proteins
After demonstrating that the SKS14 and AT59 mRNAs accumulate in cytoplasmic granules in mature pollen, we sought to determine whether these granules contain PBs marker proteins. We first investigated the localization of VCS and DCP1 in mature pollen by expressing RFP‐VCS and RFP‐DCP1 under the LAT52 promoter. Fig. 3 shows that RFP‐VCS and RFP‐DCP1 appeared as cytoplasmic foci. As expected, the size of these bodies increased upon incubation with puromycin, a drug that inhibits translation elongation‐releasing mRNAs from ribosomes, thus allowing mRNA recruitment to PBs (Thomas, Loschi, Desbats, & Boccaccio, 2011). The effect was stronger for the RFP‐VCS construct where PBs increased both in size and number upon exposure to puromycin (Fig. 3).
Figure 3.
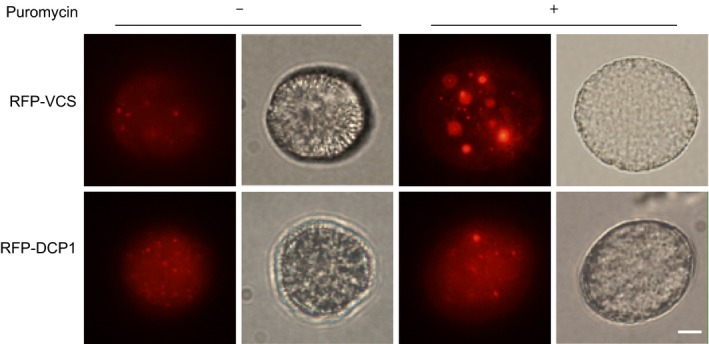
The PB proteins VCS and DCP1 form cytoplasmic foci in mature pollen. Representative images of mature pollen grains in the absence (‐) or presence (+) of puromycin 50 μg/ml. DIC images are shown in the right panels. Size bar, 5 μm
Next, we generated four different Arabidopsis lines by crossing the SKS14‐MS2 or AT59‐MS2 with either the RFP‐VCS or RFP‐DCP1 lines. Automated PBs and mRNA granule detection was performed by a MATLAB script as above (for details see “Materials and Methods”). To investigate colocalization, the two structures were modeled as circles using the area and center obtained with the MATLAB algorithm, as detailed in Materials and Methods. Colocalization was considered to occur when the distance between the centers of the structures was equal or lower than the sum of their radii. Proximity was defined when the distance was larger but less than twice the sum of their radii. Finally, no relationship was assumed if the mRNA granule and the PB were separated by more than twice the sum of their radii (Fig. S4). The position and size of the mRNA granules and PBs were determined by specific MATLAB scripts (for details see “Materials and Methods”), merged the results and determined whether the mRNA granules colocalized, were proximal, or showed no relationship to the PBs protein markers. Fig. 4 a, b, and c shows representative SKS14‐MS2 RFP‐VCS pollen grains depicting the three types of spatial colocalization relationships. We performed the same study for three additional lines, SKS14‐MS2 RFP‐DCP1 (Fig. S5), AT59‐MS2 RFP‐VCS (Fig. S6), and AT59‐MS2 RFP‐DCP1 (Fig. S7), with similar results.
Figure 4.
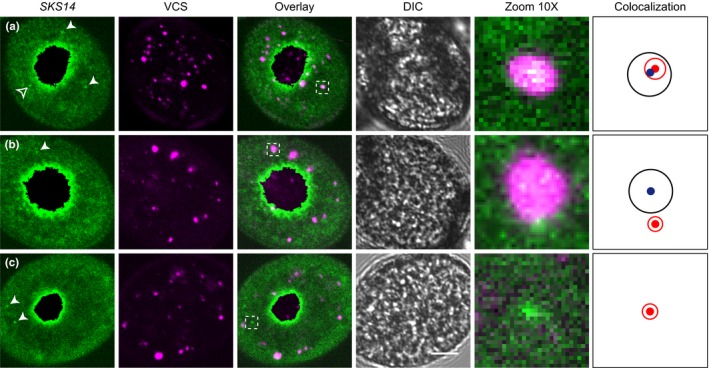
Colocalization of SKS14 mRNA with RFP‐VCS. (a) Confocal image of a representative mature pollen grain showing high colocalization between SKS14 mRNA and a RFP‐VCS body. (b) Confocal image of a representative mature pollen grain showing a SKS14 mRNA cytoplasmic granule contiguous to a RFP‐VCS body. (c) Confocal image of a representative cytoplasmic granule with no relationship with any RFP‐VCS body. In the left panels, white arrowheads show examples of cytoplasmic granules identified by MATLAB while empty arrowheads show cytoplasmic aggregates not detected by MATLAB. The insets in the merged (“Overlay”) column are enlarged on the 10X panels. In the “Colocalization” column, the blue point and black circle indicate the localization and size of the VCS body, respectively, and the red point and circle correspond to the SKS14 mRNA granule. DIC images are shown. Size bar, 5 μm
To address the extent of random colocalization in these images, we performed a shuffling analysis (for description of the script, see “Materials and Methods”). Fig. S4 shows examples of shuffling analysis for each one of the colocalization groups. At an alpha value of 0.05, the experimental distance observed for colocalized and proximal granule pairs, but not for distant granules, was significantly different than the values expected by chance.
Then, we quantified the number of mRNA granules that colocalized or were nearby PBs (Table 2). We found that around 20% of the SKS14 mRNA bodies colocalized with either VCS or DCP1, while approximately 10% of them were found proximal. Similarly, 10% of the AT59 mRNA granules colocalized with the VCS or DCP1 bodies and a similar fraction was found nearby them (Table 2). These results suggest that both SKS14 and AT59 mRNAs showed partial localization with PBs identified by VCS or DCP1.
Table 2.
SKS14 and AT59 mRNA cytoplasmic granules colocalize with PB proteins. Percentages of granules showing colocalization or proximity for each mRNA and PB protein pair are indicated. N: number of pollen grains analyzed
| mRNA/protein | Colocalization (%) | Proximal (%) | N |
|---|---|---|---|
| SKS14‐VCS | 19.6 | 6.5 | 46 |
| 21.4 | 16.7 | 42 | |
| SKS14‐DCP1 | 23.8 | 0 | 21 |
| 20 | 10 | 30 | |
| AT59‐VCS | 13 | 9.5 | 63 |
| 14.3 | 11.4 | 35 | |
| AT59‐DCP1 | 11.4 | 11.4 | 35 |
| 8.8 | 0 | 34 |
3.3. SKS14‐RFP protein is expressed at early stages during pollen germination
Next, we asked when the SKS14 mRNA is translated. We generated two independent Arabidopsis lines (S2 and S13) that express both pLAT52::GFP‐MCP/pLAT52::SKS14‐MS2 and pSKS14::SKS14‐RFP simultaneously. Fig. 5 shows that the SKS14‐RFP protein started to be detected during pollen germination, being localized to the region where the pollen tube will emerge (Fig. 5a‐b). Later, SKS14‐RFP is accumulated at the margins of pollen tubes as long as they grow (Fig. 5c‐d), suggesting that there is an increase in the total amount of SKS14‐RFP. These observations agree with previous results that show that the tobacco NTP303 is present at the cell wall and at callose plugs of growing pollen tubes (Wittink et al., 2000).
Figure 5.

SKS14‐RFP protein is expressed from early stages of germination and localizes at the margins of pollen tubes. Representative images of S2 line pollen grains expressing SKS14‐RFP protein (left panels). Right panels, DIC images. Size bars, 5 μm for early and late stages (a and b) and 15 μm for the pollen tube (c). (d) A 5X magnification shows localization at the tip (Size bar, 5 μm). (e) Quantification of SKS14 mRNA cytoplasmic granules per pollen grain compared to a MS2 control (MS2 3‐1). ES and LS, early and late stages of pollen germination, respectively. Each point represents the mean value of independent samples including 9‐10 pollen grains. Lines link data from paired samples (same experiment). Statistically different values are indicated (***p < .001 and **p < .01). The p‐value for the ES‐MS2/LS‐MS2 pair was 0.99, and for the LS‐MS2/LS‐S13 pair, 0.48 (two‐way ANOVA randomized block, Bonferroni post‐test)
In general, mRNA translation correlates with silencing foci remodeling or dissolution (reviewed in Thomas et al., 2011; Pascual, Maschi, Luchelli, & Boccaccio, 2014). We wonder whether SKS14 expression involves changes in the number of SKS14 mRNA granules. We analyzed in the double‐transgenic lines (S2 and S13) the presence of SKS14 mRNA granules at the early (ES) and late (LS) stages of pollen germination (Fig. 5a‐b). We found a statistically significant decrease in the number of granules at the late stage of pollen germination when compared to the corresponding early stage in the S13 line (Fig. 5e). We also found for both S2 and S13 lines that the number of fluorescent mRNA granules at the early stage, but not at the late stage, was significantly higher in comparison with the control MS2 line (ES‐MS2/ES‐S2 pair p = .019 (*p < .1) and LS‐MS2/LS‐S2 pair p = .25; ES‐MS2/ES‐S13 pair p = .0002 (***p < .001) and LS‐MS2/LS‐S13 pair p = .48). These results suggest that during pollen germination, when the SKS14‐RFP protein is being synthetized, there is a simultaneous decrease in the amount of SKS14 mRNA granules, suggesting that the SKS14 mRNA granules release their content to allow a controlled production of the SKS14 protein.
4. DISCUSSION
Both in plants and animals, gamete development depends on the translation of stored mRNAs. The post‐transcriptional control of genes expressed during the spermatogenesis in fly and mouse is well known (Lasko, 2012; Nguyen‐Chi & Morello, 2011). In plants, the translational inhibitor cycloheximide, but not the transcriptional inhibitor actinomycin D, inhibits early pollen tube growth, suggesting that translation of preexisting mRNAs is required (Hao, Li, Hu, & Lin, 2005). Studies on the regulation of the tobacco NTP303 and tomato LAT59 genes demonstrated that they are both transcribed during pollen development while their proteins are mostly or exclusively synthesized after germination, respectively (Curie & McCormick, 1997; Hulzink et al., 2002). Antisense NTP303 plants are male sterile due to the arrest of pollen tubes within the style (de Groot et al., 2004). Fig. 5 shows a negative correlation between the presence of SKS14 mRNA granules and RFP‐SKS14 expression, which is compatible with a role for the granules in mRNA storage. We found that upon initiation of pollen germination, the related SKS14 protein localizes where the pollen tube emerges, and later in the pollen tube margins. Tobacco NTP303 is related to ascorbate oxidases and localizes at the plasma membrane (Wittink et al., 2000), and we propose that SKS14 could have a similar role in Arabidopsis pollen.
Here, we visualized the mRNAs of the Arabidopsis SKS14 and AT59 mRNAs in mature pollen and during germination. We identified SKS14 and AT59 mRNA in cytoplasmic granules related to PBs in mature pollen and propose that these bodies would function in mRNA storage while waiting for being translated. In addition, the presence of PB components in these RNA granules may be linked to mRNA degradation. However, we favor the notion of mRNA storage given that SKS‐RFP accumulates latter during pollen tube growth (Fig. 5c).
It is generally accepted that mRNAs translationally inactive are stored in stress granules (SGs), processing body (PB), or related organelles (Decker & Parker, 2012; Thomas et al., 2014). We propose that an active exchange of mRNAs between PBs and polysomal mRNPs finely tune gene expression during the late stages of pollen development and during germination. Supporting this, in a pioneer work, the NTP303 mRNA was found in polysomal and in a fraction of particles resistant to EDTA/puromycin treatment referred to as EPPs (Honys, Combe, Twell, & Capková, 2009). These particles have been proposed as long‐term storage complexes. EPPs include a set of mRNAs that are stored and translationally repressed at early stages of pollen development. Some of the stored mRNAs are massively translated at late stages of pollen development and/or transported to the pollen tube where they are translated. Relevantly, most of the translationally inactive NTP303 mRNA was present both in the polysomal and EPP fractions. Whether the EPPs and the granules identified here represent the same entity remains open. Against this possibility, VCS and DCP1 are absent from tobacco EPPs (Honys et al., 2003).
We also described the presence of processing body in Arabidopsis mature pollen. As in other cell types and organisms, RFP‐VCS and RFP‐DCP1 localized to discrete cytoplasmic bodies that become larger and more abundant upon treatment with puromycin, suggesting that pollen PBs recruit transcripts that are released from active polysomes. Accumulation of translationally repressed mRNAs in PBs has been previously reported in yeast, Drosophila, and mammals (Decker & Parker, 2012; Thomas et al., 2011). However, when analyzing the colocalization of SKS14 and AT59 mRNAs with VCS or DCP1, we found partial overlapping with about 30% of the granules in close contact. We speculate that pollen grains may contain several types of PBs, and only a fraction of them would contain VCS and DCP1. Several animal cell types display a variety of PB containing subsets of PB components, and in addition, it has been proposed that mRNAs are differentially located on PBs depending on their translational requirements (Weil et al., 2012). In mammalian neurons, only 50% of dendritic DCP1a bodies contain Hedls, the VCS ortholog, and vice versa, 50% of Hedls‐positive dendritic puncta contain DCP1a (Luchelli, Thomas, & Boccaccio, 2015). Likewise, in untreated mammalian cells, only 18% of the beta‐actin and Cerulean‐mini‐dystrophin mRNAs colocalized with PB using Hedls as a protein marker (Aizer et al., 2014). Further analysis including super‐resolution microscopy and the study of additional PB proteins and RNA regulation pathways will shed light on the dynamic relationship between pollen mRNAs and PBs and will allow a more complete understanding of their translational regulation.
In several examples, translation is repressed by proteins that bind to the UTRs and mRNA silencing is necessary for the proper localization and function of the encoded proteins (Chartrand et al., 2002; Jambor, Brunel, & Ephrussi, 2011). Several common elements in the 5′UTR of pollen‐expressed genes have been identified (Hulzink et al., 2003). Some of these consensus sequences are present in the NTP303 5′UTR and affect translation efficiency (Hulzink et al., 2002), thus opening the possibility that these pollen‐specific 5′UTR sequences play a regulatory role during development and germination. While the repression mechanism remains poorly understood, it has been proposed that translation of the NTP303 mRNA is activated by factors that bind to the 5′UTR after pollen germination (Hulzink et al., 2002). In turn, binding of regulatory factors to the 5′UTR of the LAT59 mRNA would inhibit translation at early stages and the release of the repressors upon pollen maturation would allow LAT59 protein synthesis (Curie & McCormick, 1997). Whether these 5′UTRs sequences mediate the targeting of these transcripts to PBs remain to be investigated. In this regard, it has been recently reported that CGG repeats in the 5′UTR of the Fragile X Mental Retardation 1 (FMR1) RNA mediate RNA localization into cytoplasmic granules (Rovozzo et al., 2016).
To securely define which pollen mRNAs are stored or translated, a robust and tight post‐transcriptional regulation is expected to occur. The presence of cytoplasmic granules that contain the SKS14 and AT59 mRNAs in mature pollen suggests a mechanism for translational regulation. Our results suggest that granules related to PBs store mRNAs to postpone their translation until necessary or that they regulate mRNA levels. An appealing speculation is whether the observations here reported for SKS14 and AT59 mRNAs can be extrapolated to other late pollen genes.
AUTHOR CONTRIBUTIONS
MRS and JPM conceived the experiments; MRS, LS, SGT, GLB, LIP, and JPM designed the research; MRS performed the experiments; LS, SGT, and LIP provided technical assistance; MRS, LS, SGT, GLB, LIP, and JPM analyzed and interpreted the data; MRS and JPM wrote the manuscript with contributions of all the authors; GLB complemented the writing.
Supporting information
ACKNOWLEDGMENTS
We thank Dr. Federico Fuentes for the excellent confocal microscopy technical assistance.
Scarpin MR, Sigaut L, Temprana SG, Boccaccio GL, Pietrasanta LI, Muschietti JP. Two Arabidopsis late pollen transcripts are detected in cytoplasmic granules. Plant Direct. 2017;00:1–10. 10.1002/pld3.12
Funding information
JPM, GLB, LS, and LIP are investigators of the National Research Council (CONICET) from Argentina. MRS and SGT are recipients of postdoctoral fellowships from the CONICET. This work was supported by grants from ANPCyT (PICT 2014‐0423 and PICT 2015‐0078) and from the Universidad de Buenos Aires to JPM.
REFERENCES
- Aizer, A. , Kalo, A. , Kafri, P. , Shraga, A. , Ben‐Yishay, R. , Jacob, A. , … Shav‐Tal, Y. (2014). Quantifying mRNA targeting to P bodies in living human cells reveals a dual role in mRNA decay and storage. Journal of Cell Science, 127, 4443–4456. 10.1242/jcs.152975 [DOI] [PubMed] [Google Scholar]
- Bertrand, E. , Chartrand, P. , Schaefer, M. , Shenoy, S. M. , Singer, R. H. , & Long, R. M. (1998). Localization of ASH1 mRNA particles in living yeast. Molecular Cell, 2(4), 437–445. 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- Boavida, L. C. , Becker, J. D. , & Feijó, J. A. (2005). The making of gametes in higher plants. International Journal of Developmental Biology, 49(5–6), 595–614. 10.1387/ijdb.052019lb [DOI] [PubMed] [Google Scholar]
- Brown, S. M. , & Crouch, M. L. (1990). Characterization of a gene family abundantly expressed in Oenothera organensis pollen that shows sequence similarity to polygalacturonase. The Plant Cell, 2(3), 263–274. 10.1105/tpc.2.3.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand, P. , Meng, X. H. , Huttelmaier, S. , Donato, D. , & Singer, R. H. (2002). Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Molecular Cell, 10(6), 1319–1330. [DOI] [PubMed] [Google Scholar]
- Clough, S. , & Bent, A. (1998). Floral dip: A simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana. The Plant Journal, 16(6), 735–743. [DOI] [PubMed] [Google Scholar]
- Curie, C. , & McCormick, S. (1997). A strong inhibitor of gene expression in the 5′ untranslated region of the pollen‐specific LAT59 gene of tomato. The Plant Cell, 9(11), 2025–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C. J. , & Parker, R. (2012). P‐bodies and stress granules : Possible roles in the control of translation and mRNA degradation. Cold Spring Harbor Perspectives in Biology, 4(9), 1–16. 10.1101/cshperspect.a012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco, D. , Accornero, N. , Lavoie, B. , Shenoy, S. M. , Blanchard, J. M. , Singer, R. H. , & Bertrand, E. (2003). Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Current Biology, 13(2), 161–167. 10.1016/S0960-9822(02)01436-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot, P. , Weterings, K. , de Been, M. , Wittink, F. , Hulzink, R. , Custers, J. , … Wullems, G. (2004). Silencing of the pollen‐specific gene NTP303 and its family members in tobacco affects in vivo pollen tube growth and results in male sterile plants. Plant Molecular Biology, 55(5), 715–726. 10.1007/s11103-004-1964-6 [DOI] [PubMed] [Google Scholar]
- Hao, H. , Li, Y. , Hu, Y. , & Lin, J. (2005). Inhibition of RNA and protein synthesis in pollen tube development of Pinus bungeana by actinomycin D and cycloheximide. New Phytologist, 165(3), 721–729. 10.1111/j.1469-8137.2004.01290.x [DOI] [PubMed] [Google Scholar]
- Honys, D. , Combe, J. P. , Twell, D. , & Capková, V. (2000). The translationally repressed pollen‐specific ntp303 mRNA is stored in non‐polysomal mRNPs during pollen maturation. Sexual Plant Reproduction, 13(3), 135–144. 10.1007/s004970000047 [DOI] [Google Scholar]
- Honys, D. , Ren, D. , Fecikova, J. , Jedelsky, P. L. , Nebesa, J. , & Dobrev, P. (2009). Cytoskeleton‐associated large RNP complexes in tobacco male gametophyte (EPPs) are associated with ribosomes and are involved in protein synthesis, processing, and localization. Journal of Proteome Research, 8, 2015–2031. [DOI] [PubMed] [Google Scholar]
- Honys, D. , & Twell, D. (2003). Comparative analysis of the arabidopsis pollen transcriptome 1. Plant Physiology, 132(2), 640–652. 10.1104/pp.103.020925.fitness [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulzink, R. J. M. , de Groot, P. F. M. , Croes, A. F. , Quaedvlieg, W. , Twell, D. , Wullems, G. J. , & van Herpen, M. M. A. (2002). The 5 ‐untranslated region of the ntp303 gene strongly enhances translation during pollen tube growth, but not during pollen maturation. Plant Physiology, 129(1), 342–353. 10.1104/pp.001701.342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulzink, R. J. M. , Weerdesteyn, H. , Croes, A. F. , Gerats, T. , van Herpen, M. M. A. , & van Helden, J. (2003). In silico identification of putative regulatory sequence elements in the 5 ‐untranslated region of genes that are expressed during male gametogenesis. Plant Physiology, 132(1), 75–83. 10.1104/pp.102.014894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor, H. , Brunel, C. , & Ephrussi, A. (2011). Dimerization of oskar 3′ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA, 17(12), 2049–2057. 10.1261/rna.2686411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikauskas, R. , & McCormick, S. (1997). Identification of the tobacco and Arabidopsis homologues of the pollen‐expressed LAT59 gene of tomato. Plant Molecular Biology, 34(5), 809–814. [DOI] [PubMed] [Google Scholar]
- Lampasona, A. A. , & Czaplinski, K. (2016). RNA voyeurism: A coming of age story, Methods, 98, (pp. 10–17). London: Elsevier Inc. 10.1016/j.ymeth.2015.11.024 [DOI] [PubMed] [Google Scholar]
- Lasko, P. (2012). mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harbor Perspectives in Biology, 4(10), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loraine, A. E. , McCormick, S. , Estrada, A. , Patel, K. , & Qin, P. (2013). RNA‐seq of arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiology, 162(2), 1092–1109. 10.1104/pp.112.211441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchelli, L. , Thomas, M. G. , & Boccaccio, G. L. (2015). Synaptic control of mRNA translation by reversible assembly of XRN1 bodies. Journal of Cell Science, 128(8), 1542–1554. 10.1242/jcs.163295 [DOI] [PubMed] [Google Scholar]
- Maldonado‐Bonilla, L. D. (2014). Composition and function of P bodies in Arabidopsis thaliana. Frontiers In Plant Science, 5, 201 10.3389/fpls.2014.00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu, J. H. , Stains, J. P. , & Kao, T. (1994). Characterization of a pollen‐expressed gene encoding a putative pectin esterase of Petunia inflata. Plant Molecular Biology, 25(3), 539–544. 10.1007/bf00043881 [DOI] [PubMed] [Google Scholar]
- Muschietti, J. , Dircks, L. , Vancanneyt, G. , & McCormick, S. (1994). LAT52 protein is essential for tomato pollen development: Pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant Journal, 6(3), 321–328. 10.1046/j.1365-313X.1994.06030321.x [DOI] [PubMed] [Google Scholar]
- Nguyen‐Chi, M. , & Morello, D. (2011). RNA‐binding proteins, RNA granules, and gametes: Is unity strength? Reproduction, 142(6), 803–817. 10.1530/REP-11-0257 [DOI] [PubMed] [Google Scholar]
- Niogret, M. F. , Dubald, M. , Mandaron, P. , & Mache, R. (1991). Characterization of pollen polygalacturonase encoded by several cDNA clones in maize. Plant Molecular Biology, 17(6), 1155–1164. 10.1007/bf00028732 [DOI] [PubMed] [Google Scholar]
- Rogers, H. J. , Harvey, A. , & Lonsdale, D. M. (1992). Isolation and characterization of a tobacco gene with homology to pectate lyase which is specifically expressed during microsporogenesis. Plant Molecular Biology, 20(3), 493–502. 10.1007/bf00040608 [DOI] [PubMed] [Google Scholar]
- Rogers, H. J. , & Lonsdale, D. M. (1992). Genetic manipulation of male sterility for the production of hybrid seed. Plant Growth Regulation, 11(1), 21–26. [Google Scholar]
- Rovozzo, R. , Korza, G. , Baker, M. W. , Li, M. , Bhattacharyya, A. , Barbarese, E. , & Carson, J. H. (2016). CGG repeats in the 5′ UTR of FMR1 RNA regulate translation of other RNAs localized in the same RNA granules. PLoS ONE, 11(12), 1–23. 10.1371/journal.pone.0168204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen, J. A. M. , Derksen, J. , Knuiman, B. , & Wullems, G. J. (1999). The location of the pollen‐specific NTP303 protein In Clément C., Pacini E. & Audran J. C. (Eds.), Anther and pollen (pp. 113–118). Berlin, Heidelberg: Springer; 10.1007/978-3-642-59985-9_10 [DOI] [Google Scholar]
- Sedbrook, J. C. , Carroll, K. L. , Hung, K. F. , Masson, P. H. , & Somerville, C. R. (2002). The arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol – anchored glycoprotein involved in directional root growth. The Plant Cell, 14(7), 1635–1648. 10.1105/tpc.002360.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, M. G. , Loschi, M. , Desbats, M. A. , & Boccaccio, G. L. (2011). RNA granules: The good, the bad and the ugly. Cellular Signalling, 23(2), 324–334. 10.1016/j.cellsig.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, M. G. , Pascual, M. L. , Maschi, D. , Luchelli, L. , & Boccaccio, G. L. (2014). Synaptic control of local translation: The plot thickens with new characters. Cellular and Molecular Life Sciences, 71(12), 2219–2239. 10.1007/s00018-013-1506-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell, D. (1994). The diversity and regulation of gene expression in the pathway of male gametophyte development In Scott R. & Stead A. (Eds.), Molecular and cellular aspects of plant reproduction (Society for Experimental Biology Seminar Series)(pp. 83–136). Cambridge: Cambridge University Press; 10.1017/cbo9780511752339.007 [DOI] [Google Scholar]
- Twell, D. , Klein, T. M. , Fromm, M. E. , & McCormick, S. (1989). Transient expression of chimeric genes delivered into pollen by microprojectile bombardment. Plant Physiology, 91(4), 1270–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell, D. , Yamaguchi, J. , Wing, R. A. , Ushiba, J. , & McCormick, S. (1991). Promoter analysis of genes that are coordinately expressed during pollen development reveals pollen‐specific enhancer sequences and shared regulatory elements. Genes and Development, 5(3), 496–507. 10.1101/gad.5.3.496 [DOI] [PubMed] [Google Scholar]
- Wakeley, P. R. , Rogers, H. J. , Rozycka, M. , Greenland, A. J. , & Hussey, P. J. (1998). A maize pectin methylesterase‐like gene, ZmC5, specifically expressed in pollen. Plant Molecular Biology, 37(1), 187–192. 10.1023/a:1005954621558 [DOI] [PubMed] [Google Scholar]
- Weil, T. T. , Parton, R. M. , Herpers, B. , Soetaert, J. , Veenendaal, T. , Xanthakis, D. , … Davis, I. (2012). Drosophila patterning is established by differential association of mRNAs with P bodies. Nature Cell Biology, 14(12), 1305–1313. 10.1038/ncb2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings, K. , Reijnen, W. , van Aarssen, R. , Kortstee, A. , Spijkers, J. , van Herpen, M. , … Wullems, G. (1992). Characterization of a pollen‐specific cDNA clone from Nicotiana tabacum expressed during microgametogenesis and germination. Plant Molecular Biology, 18(6), 1101–1111. 10.1007/BF00047713 [DOI] [PubMed] [Google Scholar]
- Wing, R. A. , Yamaguchi, J. , Larabell, S. K. , Ursin, V. M. , & McCormick, S. (1989). Molecular and genetic characterization of two pollen‐expressed genes that have sequence similarity to pectate lyases of the plant pathogen Erwinia. Plant Molecular Biology. Springer, 14(1), 17–28. [DOI] [PubMed] [Google Scholar]
- Wittink, F. R. A. , Knuiman, B. , Derksen, J. , Vera, Č. , Twell, D. , Schrauwen, J. A. M. , … Čapková, V. (2000). The pollen‐specific gene Ntp303 encodes a 69‐kDa glycoprotein associated with the vegetative membranes and the cell wall. Sexual Plant Reproduction, 12(5), 276–284. 10.1007/s004970050195 [DOI] [Google Scholar]
- Xu, J. , & Chua, N.‐H. (2009). Arabidopsis decapping 5 is required for mRNA decapping, P‐body formation, and translational repression during postembryonic development. The Plant Cell, 21(10), 3270–3279. 10.1105/tpc.109.070078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Yang, J.‐Y. , Niu, Q.‐W. , & Chua, N.‐H. (2006). Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. The Plant Cell, 18(12), 3386–3398. 10.1105/tpc.106.047605 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


