Abstract
The signaling lymphocytic activation molecule (SLAM) family is comprised of nine distinct receptors (SLAMF1 through SLAMF9) that are expressed on hematopoietic cells. All of these receptors, with the exception of SLAMF4, are homotypic by nature as downstream signaling occurs when hematopoietic cells that express the same SLAM receptor interact. The SLAM family receptor function is largely controlled via SLAM associated protein (SAP) family adaptors. The SAP family adaptors consist of SAP, Ewing sarcoma associated transcript (EAT)-2, and EAT-2-related transducer (ERT). These adaptors associate with the cytoplasmic domain of the SLAM family receptors through phosphorylated tyrosines. Defects in SLAM family members and SAP adaptors have been implicated in causing immune deficiencies. This is exemplified in patients with X-linked lymphoproliferative (XLP) disease, where SAP undergoes a loss of function mutation. Furthermore, evidence has been accumulating that SLAM family members are potential targets for inflammatory and autoimmune diseases. This review will discuss the structure and function of the SLAM family receptors and SAP family adaptors, their role in immune regulation, and potential approaches to target this family of receptors therapeutically.
Introduction
The SLAM family of receptors consists of nine distinct members. These members include: SLAMF1 (SLAM or CD150), SLAMF2 (CD48), SLAMF3 (Ly-9 or CD229), SLAMF4 (2B4 or CD244), SLAMF5 (CD84), SLAMF6 (Ly108 in mice, NTB-A or SF2000 in humans), SLAMF7 (CRACC, CD319 or CS1), SLAMF8 (CD353 or BLAME), and SLAMF9 (SF2001 or CD84H). In terms of classification, SLAMF2, SLAMF8 and SLAMF9 are not considered full members of the SLAM family and can be designated as atypical (Table 1) (1). This is due to the fact that SLAMF2, SLAMF8, and SLAMF9 do not share homology in their cytoplasmic domains when compared to the rest of the typical SLAM family (Table 1). All the receptors in this family are assigned to the CD2 superfamily immunoglobulin (Ig) domain-containing molecules and are known to be widely expressed on hematopoietic cells, where most cells express between 3 to 5 individual SLAM family members (2). Interestingly, although SLAM family receptors are considered to be homophilic, it has been reported that they could also bind to several morbilliviruses, such as the measles (3).
Table 1.
Classification and nomenclature of the SLAM family members.
| Typical family members | Atypical family members |
|---|---|
| SLAMF1 (CD150) | SLAMF2 (CD48) |
| SLAMF3 (Ly-9 or CD229) | SLAMF8 (CD353 or BLAME) |
| SLAMF4 (2B4 or CD244) | SLAMF9 (SF2001 orCD84H) |
| SLAMF5 (CD84) | |
| SLAMF6 (Ly108, NTB-A or SF2000) | |
| SLAMF7 (CRACC, CD319orCS1) |
SLAMF1 will form homodimers when examined via surface plasmon resonance (SPR), in a “head to head” fashion (4), meaning that the binding occurs between the membrane distal Ig V-like domains. SLAMF2 does not self-associate, in contrast to SLAMF3 which exhibits a similar self-association mode when studied by introducing point mutations in the Ig V-like domains (5). When studied by crystallography, SLAMF4, SLAMF5, and SLAMF6 bind to their respective self-ligands by their Ig V-like domain as well (1, 6–8). SLAMF7 is also a self-ligand that binds via its Ig V-like domain as determined by antibody binding analyzed by flow cytometry (9). SLAMF8 and SLAMF9 have no known ligands (1).
The SLAM family receptors and SLAM family adaptors are rapidly becoming a target of interest in the understanding and treatment of diseases that range from XLP (10–14) to HIV (15), and several types of cancers (2, 16–17). Moreover, it is becoming increasingly clear that SLAM family receptors play an integral role in the pathogenesis of autoimmune disorders (2, 16–21). Therefore, it is imperative to develop a holistic understanding of the mechanisms by which these proteins function in health and disease.
The structure of the SLAM family receptors
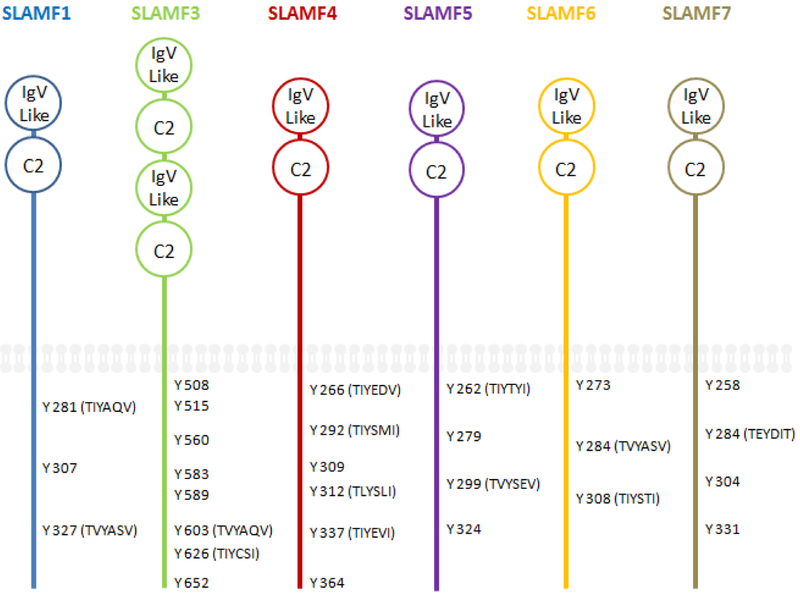
All the SLAM family proteins are considered part of the CD2 superfamily. The typical SLAM family receptors are type I glycoproteins that contain an amino terminal Ig like variable (V) domain which has no canonical disulfide bonds (Figure 1). They also have a membrane proximal constant 2 (C2) domain that houses two disulfide bonds (22). Among the typical members, SLAMF3 has a repeated IgV like domain and a C2 domain and thus has four extra cellular domains (Figure 1) (23–24). This occurs in a simple repeat of the V-like and C2-like domains (25). The cytoplasmic tails of the typical SLAM family members contain multiple immunoreceptor tyrosine-based switch motifs (ITSMs) (Figure 1) (26–27). These ITSMs act as ligands for the SAP family adaptors SAP, EAT-2 and ERT (28). The name ITSM comes from the need to differentiate it from immunoreceptor tyrosine activation motifs (ITAMs), which recruit Syk kinases, and immunoreceptor tyrosine inhibitory motifs (ITIMs), which frequently recruit phosphatases (1, 28). Each SLAM family member contains at least a single ITSMs and the homology across the ITSMs in the cytoplasmic domain is TxYxxV/I/L/ (where x is any amino acid).
Figure 1. The structure of the typical SLAM family receptors.
The SLAM family receptors are type I glycoproteins that contain an amino terminal Ig like variable (IgV) domains, a membrane proximal constant 2 (C2) domains, and phosphorylated tyrosines (Y), some of which are part of immune tyrosine switch motifs (ITSM) sequences (TxYxxV/I/L).
In regards to the atypical SLAM family members, SLAMF2 differs its cytoplasmic region from the other members, although it is analogous in the ecto domain. This receptor is held to the cell surface by means of a glycosylphosphatidylinositol (GPI) anchor (29), has no cytoplasmic domain (30), and thus lacks any ITSMs. However, this receptor is the only of a ligand for SLAMF4 and a potent co-stimulating receptor that is expressed on nearly all hematopoietic cells (30–32). As mentioned, SLAMF8 and SLAMF9 have only a short cytoplasmic domain and lack ITSMs (33–34).
The SAP family adaptors bind to the SLAM family receptors
The SAP family adaptors are comprised almost exclusively of a Src homology 2 (SH2) domain. The SAP adaptor family includes SAP, EAT-2, and ERT (Table 2). All of the SAP family members are cytosolic proteins and are expressed exclusively in hematopoietic cells. SAP is expressed in human and mouse T cells, natural killer (NK) cells, NK T cells as well as platelets (Table 2) (14, 25–26, 35–39). SAP is lowly expressed in B cells, both in humans and mice (40). EAT-2 is also expressed in humans and mice and typically originates in NK cells (37, 41) but can also be found in dendritic cells (DCs) and macrophages (25) where it is lowly expressed (41). ERT is found only in mouse NK cells as it exists as a pseudo gene in humans (25). While SH2D1A SAP is located on chromosome X, SH2D1B (EAT-2) and SH2D1B2 (ERT) are on chromosome 1 (Table 2).
Table 2.
The SAP family adaptor expression and chromosomal location.
| Family member | Expression | Chromosomal location |
|---|---|---|
| SAP | T, NK-T, NK, B cells and platelets | X (Humans and mice) |
| EAT-2 | DC, NK, and macrophage | 1 (Humans and mice) |
| ERT | NK | 1 (Mice and pseudo gene in humans) |
SAP tends to bind to ITSMs (homology to TxYxxV/I/L/T) (42), located in the cytosolic tail of the SLAM family receptors but will bind preferentially to the specific ITSM sequence of TIYxxV/I/L/T (43). EAT-2 is not known to have a preference for specific ITSM sequence (44). This is exemplified by the fact that EAT-2 will bind to SLAMF7, but SAP will not (17). SLAMF7 contains only one ITSM (Figure 1) with the sequence of TEYxxV/I/L/T. Moreover, EAT-2 will associate with SLAMF1, SLAMF3, SLAMF4, SLAMF5, and SLAMF6 (25), none of which contain a TEYxxV/I/L/T ITSM. It is not clear what are the binding preferences of ERT are.
In addition to the SAP family adaptors, several SH2 domain containing phosphatases can bind to the ITSMs of the SLAM family receptors. These include SH2 domain-containing protein tyrosine phosphatases SHP-1 and SHP-2, as well as the SH2 domain-containing inositol phosphatase SHIP-1 (45–46). It is widely accepted that these phosphatases are prevented from binding to ITSMs through competition with SAP on the same binding sites (25, 41). Current literature has extensively exhibited that SLAM family receptor functions in immunity are dependent on their interactions with SAP family adaptors as well as SH2 domain-containing protein tyrosine phosphatases (26, 37–38, 40, 45–48). The next sections will discuss both activating and inhibitory pathways that are differently regulated by these adaptors and phosphatases.
Activating pathways initiated by the recruitment of SAP adaptors to the SLAM family receptors
SLAM receptors can act as inhibitory receptors or as activating receptors, depending on the binding of SAP family adaptors to the ITSMs in the cytoplasmic domains of SLAM family receptors (49–50). This is speculated to occur in both NK-T cells and in follicular helper T cells (TFH) (25). Fyn mediated phosphorylation of the ITSMs occurs after self-association of SLAM family members (50–54), and is required for SAP binding to most SLAM family members (50–51). At first it was not clear how the binding of SH2 domain that has no intrinsic function could alter the function of a receptor. However, this issue became elucidated when it was shown that SAP can compete with other SH2 domain containing proteins such as SHP-2 for binding to the same ITSMs on SLAMF4 (26). This means that one function of SAP is to act as a “cap” and to competitively prevent phosphatases from binding to ITSMs.
This will serve a dual function. First, it will act to block the binding of SH2 domain containing phosphatases such as SHP-1, SHP-2 and SHIP-1, which in some setting, act as inhibitory mediators. Second, SAP can enhance downstream signaling through recruiting of the Src family kinase Fyn (Figure 2). The binding of Fyn to SAP takes place via the arginine 78 residue (R78) of SAP binding to the SH3 domain of Fyn (55–57). The R78 residue is located outside the phosphotyrosine binding pocket, allowing for the simultaneous interaction between the SLAM ITSM and the SH3 domain of Fyn (57). This will trigger additional downstream signaling events leading to cellular activation.
Figure 2. Dual signaling functions of SLAMF6.
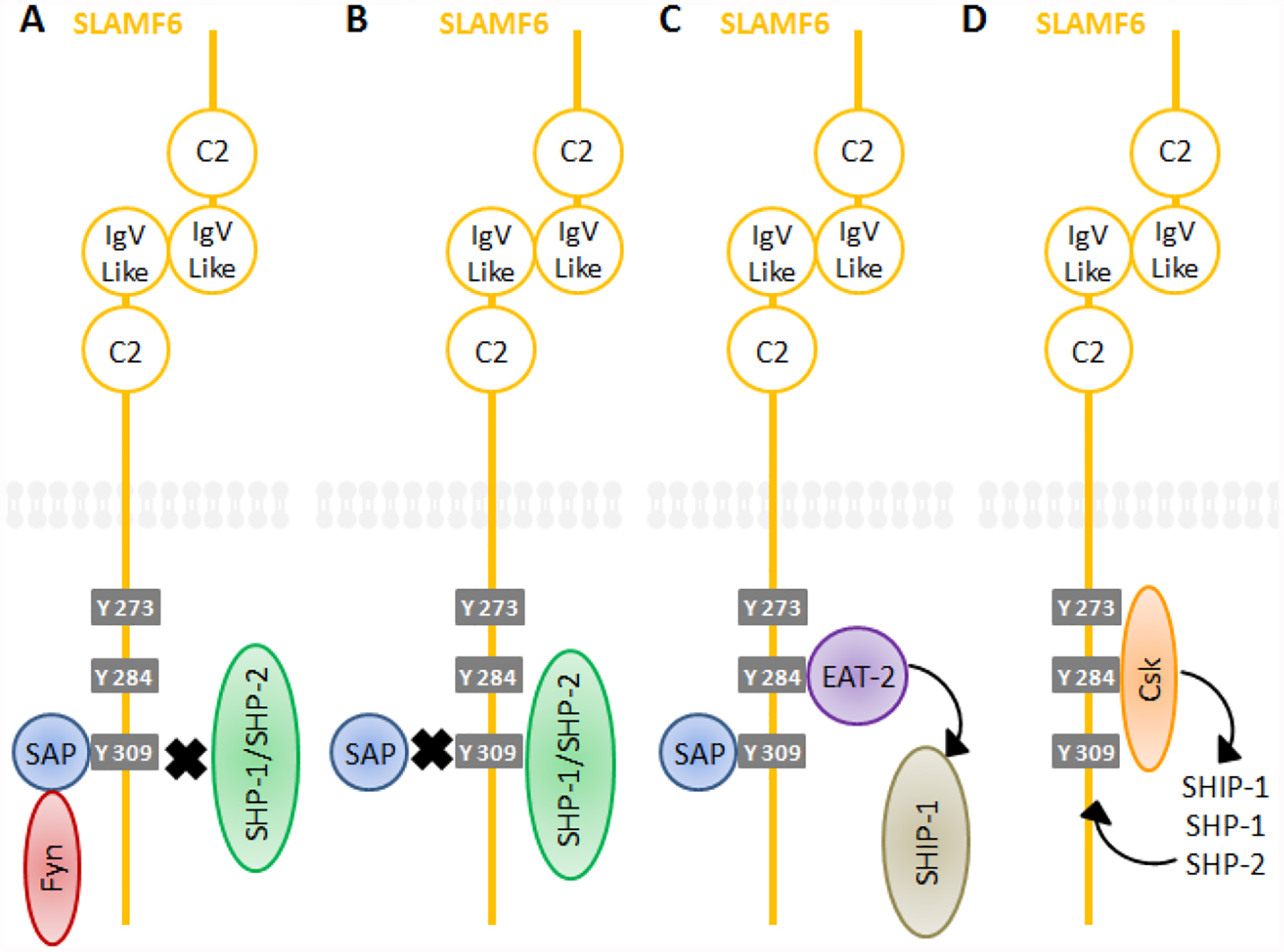
Under SAP recruitment to phosphorylated ITSMs, the receptor behaves as activating receptors (A) while the kinase Fyn will bind to SAP via SAP’s arginine 78 residue. Under SHP-1, SHP-2 and SHIP-1 recruitment, the receptor behaves as inhibitory receptors (B). SLAMF6 can also recruit EAT-2 (C) through a different tyrosines, further leading to cellular activation as a result of SHIP-1 phosphorylation. The inhibitory kinase Csk (D) can recruit additional phosphatases via its phosphorylation of additional ITSMs.
The signaling events following the formation of SLAM-SAP-Fyn complex will differ depending on the type of SLAM family receptor. For example, in the case of SLAMF1 (43, 53), that can bind to SAP at low affinity even without tyrosine phosphorylation, the SLAM-SAP-Fyn formation will lead to recruitment and phosphorylation of SHIP-1 docking protein 1 (Dok1), docking protein 2 (Dok2), and the exchange factor RasGAP (58). Furthermore, it has been reported that in CD4 T cells SLAMF1 engagement will lengthen the conscription of PKCθ to the site of contact between CD4 T cells and antigen presenting cells (APCs) (59). Also, NF-κB, a protein that controls transcription of DNA, cytokine production and cell survival, has enhanced activation during this process (59), leading to IL-4 secretion (1).
Moreover, preventing SAP association with other members such as SLAMF3, SLAMF4, and SLAMF5 act as inhibitory receptors (45), implying that at least in this context, the presence SAP adaptors control the cell activation states. Interestingly, the same SLAM-SAP-Fyn complex in the context of SLAMF6 (60) will lead to the phosphorylation of the small G protein Vav-1 (47). It has been further demonstrated that the removal of SAP from the same complex will alter the cell cytotoxic function, from targeting hematopoietic cells to targeting non-hematopoietic cells in a NKG2D dependent manner (47).
It seems as though the type of SLAM in question is the determining factor for which SH2 domain containing phosphate will bind to the ITSMs. For example, SLAMF4 has a propensity to bind to SHIP-1 when studied in NK cells (61) and SLAMF6 has a tendency to bind to SHP-1 in T cells (62). Furthermore, SAP deficiency has been shown to inhibit polarization of TFH cells, B cells functions, and initiating germinal center (GC) formation (48) via the interaction of SAP with SLAMF5 (63–64). This can lead to the prevention of lupus like symptoms (3). Although the process for how exactly SAP deficiency gives rise to TFH cell dysfunction is not yet fully understood, a study has shown that in the absence of SAP, Ly108 (the mouse analogue to SLAMF6) becomes an inhibitory receptor in TFH cells (62). SAP expression is also required in T cells, and partly in NK cells or NK T cells, to elicit a protective immune response. This was shown when floxed mice that have had SAP specifically deleted from T cells exhibited the same defects in humoral immunity as SAP deleted mice (40). Furthermore, SAP deficient mice show a deficiency in TH2 cytokine production in CD4 T cells (65–66). Another interesting facet to SAP regulation is that it binds directly to CD3ζ via the first membrane proximal ITAM to regulate T cell receptor (TCR) signaling in activated T lymphocytes (67). This may play an important role in disease where SLAMF3 and SLAMF6 are over expressed in T lymphocytes (68) as the over expression of SAP ligands may alter SAP trafficking and cause T cells to become over activated. This implies that SAP plays a more ubiquitous role in T cell activation than previously thought, allowing for the association or non-association of other molecules, such as inhibitory factors to the ITSMs of the SLAM family receptors which will be discussed next.
The recruitment of inhibitory factors to SLAM family receptors
As mentioned above, the SLAM family members SLAMF4 and SLAMF6 are known to recruit the SH2 domain-containing phosphatases, SHP-1, SHP-2 and SHIP-1 as well as the inhibitory kinase Csk (Figure 2) (1). SAP is known to prevent these interactions from occurring (13, 26, 60) by binding to the same phosphorylated ITSMs. Moreover, the kinase Csk is known to recruit additional phosphatases via its phosphorylation of additional ITSMs of SLAMF4 (69). Csk also phosphorylates and inhibits other members of the Src family kinases, further supporting downstream inhibitory signals. One more important layer of complexity is that SAP function differs depending on what tyrosine is binding to. In SLAMF4 it is thought that SAP binding to tyrosine 309 will block the interaction with SHP-2 (70). Moreover, SLAMF2 on target cells has been shown to behave as an inhibitory receptor when binding to SLAMF4 on NK cells (71–73). This behavior has also been reported with SLAMF7 (54). Consequently, and at least in some setting, it is likely that the physiological function of some SLAM family members is inhibitory by nature.
It has been witnessed in NK cells that Vav-1 is the primary substrate of SHP-1 (47). Vav-1 is an important initiator of intracellular Ca2+ flux and cytokine secretion in T cells and NK cells (74–75). Therefore, SLAMF6 as well as other SLAM family receptors may play roles as a docking site for phosphatases that control activation in NK cells and T cells by inhibition of downstream regulators such as Vav-1, suggesting that SLAM receptors themselves do not control activation states, rather they provide a platform for activation regulation by functioning as adaptors.
Differential binding of EAT-2 and ERT to SLAM family receptors
EAT-2 shares around a 50% homology in amino acid sequence with SAP in both human and mice. However, these two proteins differ in several key ways. Unlike SAP, EAT-2 and ERT do not have the arginine residue at position 78 that is required for binding to the kinase Fyn and are not coupled to Vav-1 (2). Rather, EAT-2 and ERT both have additional tyrosines; mouse EAT-2 and ERT have two additional tyrosines and human EAT-2 has only 1 (ERT is not known to be expressed in humans) (37). Furthermore, these tyrosines are located in the carboxy-terminal tail and may participate in the activating or inhibitory signaling of EAT-2 and possibly ERT (37, 54). In studies involving NK cells from mice, it has been shown that EAT-2 acts as an inhibitor to NK cell activation by its association with SLAMF4 (69). Also, it has been shown in EAT-2 deficient mice, NK cells secrete more interferon gamma (IFN-γ) and IL-2 downstream of the receptors CD16, NKG2D, Ly49D and SLAMF4 (37). The same study shows a similar trend in ERT deficient mice, although the increase in killing was seen only toward hematopoietic cells (such as YBDD cells) (37). Conversely, EAT-2 has been shown to take part in the activation of human NK cells when it associates with SLAMF6 (76). This may be due to the fact that it can bind to SLAMF6 simultaneously with SAP (76); EAT-2 binds to the tyrosine 285 in SLAMF6. This perhaps allows it to block SHP-1, SHP-2 and SHIP-1 from binding (27). Moreover, the ITSMs that bind EAT-2 are different in SLAMF4 and SLAMF5 as well as the aforementioned SLAMF6 (27, 77–78). Altogether, the dichotomous effect of EAT-2 and ERT association with SLAM family members warrants further investigation (79–82).
The role of SLAM and SAP family members in human diseases
Conceivably the most discussed human disease in which SAP deficiency is the cause of is XLP. Patients with XLP exhibit an increase in immune response to Epstein Bar virus (EBV) infection (10, 12). This is characterized by an overstated and in many times fatal infectious mononucleosis-like disease secondary to polyclonal B and CD8 T cell proliferation with massive infiltration to the liver and to the bone marrow. This is due to the fact that XLP1 patients fail to clear infected and reactive B cell clones (1, 11). This inevitably leads to hepatic necrosis and bone marrow aplasia and some additional evidence of hemophogocytosis has been suggested (83). Furthermore, B and T cell dysfunction, unrelated to EBV infection, also occurs in XLP1 manifested by hypogammaglobulinemia (84–85). Research has come about to describe the mechanism that causes the improper interactions between CD8 T cells and B cells in XLP1 patients. This uncommon genetic disorder is often fatal as patients develop hemophagocytic lymphohistiocytosis (HLH) syndrome (11). HLH is marked by a hyper activation of immune cells that attack healthy tissue in the bone marrow, liver, spleen or in the lymph nodes (79). Patients with XLP experience swelling of lymph nodes, an enlarged liver and spleen, and hepatitis. Some patients with XLP develop recurrent B-cell non-Hodgkin’s lymphoma even in the absence of EBV infection (80). Technically speaking there are two types of XLP: XLP1 and XLP2. These two types of XLP share homology in the clinical symptoms but not in the genetic root cause. XLP1 is caused by a mutation in the SH2D1A gene (13, 14), the gene that encodes SAP, whereas XLP2 patients exhibit X-linked inhibitor of apoptosis (XIAP) deficiency which is caused by BIRC4 mutations (81–82).
It has been demonstrated in human XLP patients that blocking SLAM family interactions by antibodies restores T cell function against B cell targets that also express SLAM family members. Furthermore, a synergistic effect was witnessed when antibodies were used to block SLAMF4 and SLAMF6 interactions (19). This follows from the fact that SLAMF2, the ligand for SLAMF4, is upregulated on EBV infected B cells (31). Also, it is important to note that XLP1 patients exhibit defects also in the functions of NK-T and NK cells (41). Furthermore, EBV has been discovered to have involvement in rheumatoid synovitis (86–87). Moreover, SLAMF2 is highly up regulated in EBV transformed B cells; which will induce NK cell activation via interaction with SLAMF4 (88). However, rheumatoid arthritis (RA) patients exhibit a lack in SAP association to SLAM. Therefore, this may be the reason for the inability of T cells and NK cells to clear EBV-infected synovial cells and B cells as seen in patients with RA (87–88). A similar mechanism may be at play with regards to patients with XLP. Lastly, patients with RA are at a much higher risk of myocardial infarction (89). For this condition patients are prescribed TNF-α blockers. However, this has the unintended consequence of causing autoimmunity via the lowering of SAP (90). Given the relation that both RA and XLP have to SLAM and SAP this topic warrants further investigation.
Another autoimmune disease in which SLAM takes part in is systemic lupus erythematosus (SLE). The sleb1 locus corresponds to the SLAM genes (SLAMF1 through SLAM) and is located on chromosome 1 (1, 21, 33, 35). This locus takes part in SLE pathogenesis due to polymorphisms in Slamf6 as was shown in 129Sv mice when compared to C57BL/6 mice (21). This results in augmented signaling by the SLAMF6 receptor and changes in B and T cell functions that give rise to inflammatory symptoms (21, 91–94). Furthermore, it is now understood that both SLAMF3 and SLAMF6 are involved in SLE as shown in human T cells collected from lupus patients (68). This occurs via SLAM receptors co-stimulating TCR when both receptors are engaged with their respective ligands simultaneously, resulting in downstream enhanced IL-17 production (68). Furthermore, SLAM co-stimulation with TCR advances Th17 differentiation in naive CD4 T cells. This is a very import finding as patients who exhibit SLE tend to have higher expression levels of SLAMF3 and SLAMF6 on their T cells in correlation with disease severity (68). Evidence has also been elucidated that SAP is involved in SLE. This was shown in a MRL/lpr mouse model that exhibited a loss in the SAP gene leading to defects in auto antibody production (95–96). The contribution and the function of SLAMF6 in NK cells to lupus pathogenies needs further investigation (47).
Clinical advantage of targeting the SLAM family receptors
The kinase inhibitor sorafenib was given FDA approval to treat hepatocellular carcinoma (HCC) in 2007 and since then has shown to increase patient survival by 44% (97–100). It acts as a multi kinase inhibitor of several protein tyrosine kinases such as VEGFR, PDGFR and Raf-family kinases (97–99). Despite the relative successes of this drug there remain multiple patients whose resistance to sorafenib leads to therapeutic failure. Interestingly, SLAMF3 shows reduced expression in cancerous liver tissue and in vitro introduction of SLAMF3 inhibits tumor growth (18), suggesting that SLAMF3 expression might predict resistance to sorafenib. Moreover, it has been attempted to determine if SLAMF3 was inhibiting the ability of sorafenib to treat HCC patients. Future research will have to determine if there is any use to combine this drug with anti-SLAMF3 interventions.
SAP deficient NK cells extracted from mice have been demonstrated to target and kill non-hematopoietic cells (47). This targeting allows NK cells to lyses B16F melanoma cells in a manner that is SAP dependent (47). This study also suggested that the upstream binding partner SLAMF6 is a potential target in cancer immunotherapy, allowing NK cells to act on both hematopoietic and non-hematopoietic transformed cells (47). Moreover, SLAMF6 has promise also as a biomarker for HIV infection. HIV infected cells will exhibit a down regulation of the major histocompatibility complex class I molecules (MHC-I) (101) which typically allows NK cells to sense and kill such cells (102). However, HIV infected cells are not killed by NK cells despite this fact (103). Moreover, it has been shown that this is the case because HIV infected T cells will undergo an up regulation of human leukocyte antigen (HLA)-C and HLA-E, which bind to inhibitory NK cell receptors and inhibit NK cell activity (104). Previous studies have revealed that lymphoblastic leukemia cells are able to avoid NK cell detection because they down modulated the expression of SLAMF4 and SLAMF6 (105). Therefore, it was investigated whether or not SLAMF4 and SLAMF6 play a role in NK cell killing of HIV infected cells. When a 51Cr release assay was conducted on NK cell targets (autologous infected CD4 cells) with and without blocking antibodies for SLAMF4 and SLAMF6 on NK cells, it was demonstrated that NK cell killing of HIV infected cells depends on SLAMF4 and SLAMF6 interactions (15). Furthermore, it was demonstrated that SLAMF4 and SLAMF6 are down regulated during infection of the same cells with HIV (15).
Evidence has also begun to compile regarding targeting SLAMF7 for some types of cancer. This receptor is over expressed on multiple myeloma cells (93), a cancer that is formed by malignant plasma cells (94). In light of these findings the IgG1 non-blocking antibody elotuzumab was developed (16) to recognize specifically SLAMF7 (17). The antibody acts by engaging SLAMF7 on multiple myeloma cells via the Fab portion of the antibody. The Fc portion of elotuzumab will bind to the activating receptor CD16 on NK cells which increases their cytotoxicity towards multiple myeloma cells (2) and has recently been approved in combination with lenalidomide and dexamethasone to treat patients with relapsed or refractory multiple myeloma. SLAMF7 has also exhibited therapeutic potential for patients with RA. Targeting the antigen CD20 with anti-CD20 monoclonal antibodies, which depletes B cells from the circulation (106), has been shown to be insufficient in some patients. These patients have a persistence of CD20 negative plasmablast and plasma cell populations (107). However, these cells strongly express SLAMF7 (16). Therefore, a humanized antibody dubbed PDL241 was developed to target SLAMF7. PDL241 was subsequently shown to inhibit the production of immunoglobulins in a mode that was an Fc-dependent in vitro by killing plasmablasts and plasma cells, but not B cells, in peripheral blood mononuclear cells (PBMC) cultures (108). It was further shown in an in vivo rhesus monkey model that PDL214 treatment reduced the severity of joint-related disease parameters via the reduction of IgG and IgM antibodies (108).
Concluding remarks
Over the past decade it has become increasingly clear that the SLAM family of receptors are important regulators of immune response. This has been demonstrated on most types and subsets of lymphocytes, particularly in NK cells, although SLAM family receptors are also required for T and B cell immunity. At first it has been shown that the receptor functions of SLAM family members such as SLAMF4 are SAP dependent. Furthermore, the function of the receptor can change from inhibitory to activating depending on the context of the interactions and beyond. The context of the interactions can be thought of as function switch that is due to the binding of SH2 domain containing proteins such as SAP or SHP-1 phosphorylated tyrosines in the cytoplasmic domain of the SLAM family proteins. This may be further affected by the concentration of SAP family adaptors as well as the expression levels and sub cellular location of the various SLAM family receptors or the availability of other SAP binding proteins such as CD3ζ. This is further complicated when it is considered that SLAMF2 binding to SLAMF4 has been shown to act as an inhibitory mechanism for NK cells when SLAMF2 is expressed on NK cell targets. As a result, it could be that SAP recruitment to SLAMF4 after the interaction between SLAMF2 and SLAMF4 shut down NK cell activation. Alternatively, phosphatases such as SHP-1, SHP-2, and SHIP-1 may be primarily recruited after the interaction between SLAMF2 and SLAMF4. This potentiates SHP-1, SHP-2, and SHIP-1 as NK cell activators. However, SLAM family members tend to only interact with one another via their IgV like domains, with the exception of SLAMF4, which will interact with SLAMF2.
SLAM family receptors have been identified in many types of disease pathologies. Patients who exhibit XLP1 have a marked inability for their T cells to clear B cells (109). Also, the B cells of XLP1 patients demonstrate a distinct phenotype (CD10+CD24HighCD38HighCD5+Bcl2−) that makes them behave more as immature B cells (110). Therefore, these cells will exhibit a reduction in survival, proliferation, differentiation, chemotaxis, and antibody production compared with mature B cells (110). This will inevitably make them less able to clear infection. Moreover, both T cells and B cells express SLAMF6. When SLAMF6 interactions are blocked in XLP1 cells, normal T cell clearance of B cells can be seen. This is an example of SLAMF6 acting as a receptor that inhibits T cell function; although, B cells express little to no SAP, which can affect the role of SLAMF6 with respect to their cytotoxicity. However, in SAP deleted cells, SLAMF6 deletion will halt NK cell cytotoxicity against non-hematopoietic targets. These targets do not express ligands for SLAMF6. Therefore, SLAMF6 may be acting as an adaptor molecule by altering the trafficking of SHP-1. To further complicate this situation SLAMF4 exhibits similar behavior, but is not only a self-ligand; SLAMF4 is seen to affect B cell clearance in a synergistic manner with SLAMF6 in XLP1, but it does this via interaction with SLAMF2 although it can still interact in a homotypic manner.
To summarize, it is important to understand the nature of the contexts in which SLAM family members switch their function from inhibitory to activating receptors and also the ramifications of this functional alteration. Furthermore, investigation should be done to see if this switch of function is due to SLAM family members behaving as receptors or adaptors. A clear pattern of observations has elucidated the importance SLAM family proteins in immune responses. Now a clear understanding of these mechanisms must be understood in order to design interventional approaches to treat both cancer and other inflammatory conditions.
Table 3.
The SLAM family receptor ligands, expression and interaction with SAP family adaptors.
| Family member | Ligand | Expression | SAP binding | EAT-2 binding | Homology to SLAM family |
|---|---|---|---|---|---|
| SLAMF1 | SLAMF1, Measles virus | Activated T, B, DC, Macrophage, Platelet, GC TFH | + | + | Ecto/cytoplasmic domain |
| SLAMF2 | SLAMF4 | T, Activated T, B, DC | − | − | Ectodomain |
| SLAMF3 | SLAMF3 | Activated T, B, DC, Macrophage, Platelet | + | + | Ecto/cytoplasmic domain |
| SLAMF4 | SLAMF2 (CD48) | CD8T, NK, DC, Macrophage | + | + | Ecto/cytoplasmic domain |
| SLAMF5 | SLAMF5 | T, TFH, B, NK, DC, Macrophage, Platelet | + | + | Ecto/cytoplasmic domain |
| SLAMF6 | SLAMF6 | T, TFH, B, NK, DC, Neutrophil | + | + | Ecto/cytoplasmic domain |
| SLAMF7 | SLAMF7 | Activated T, B, NK, DC, Macrophage | − | + | Ecto/cytoplasmic domain |
| SLAMF8 | Unknown | Macrophage | − | − | Ectodomain |
| SLAMF9 | Unknown | T, B, NK, DC | − | − | Ectodomain |
Acknowledgment
The work was funded by NIH grant (1R01AI125640–01 and 2T32HL007151–36), Rheumatology Research Foundation, Hirschl trust, Colton family scholarship program, and NTB Pharma Ltd.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol. 2011;29:665–705. doi: 10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- 2.Wu N, Veillette A. SLAM family receptors in normal immunity and immune pathologies. Current opinion in immunology. 2016;38:45–51. doi: 10.1016/j.coi.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Tatsuo H, Ono N, Tanaka K, Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406(6798):893–7. [DOI] [PubMed] [Google Scholar]
- 4.Mavaddat N, Mason DW, Atkinson PD, Evans EJ, Gilbert RJC, Stuart DI, Fennelly JA, Barclay AN, Davis SJ, Brown MH. Signaling lymphocytic activation molecule (CDw150) is homophilic but self-associates with very low affinity. Journal of Biological Chemistry. 2000;275(36):28100–9. [DOI] [PubMed] [Google Scholar]
- 5.Romero X, Zapater N, Calvo M, Kalko SG, de la Fuente MA, Tovar V, Ockeloen C, Pizcueta P, Engel P. CD229 (Ly9) lymphocyte cell surface receptor interacts homophilically through its N-terminal domain and relocalizes to the immunological synapse. Journal of immunology. 2005;174(11):7033–42. [DOI] [PubMed] [Google Scholar]
- 6.Cao E, Ramagopal UA, Fedorov A, Fedorov E, Yan QR, Lary JW, Cole JL, Nathenson SG, Almo SC. NTB-A receptor crystal structure: Insights into homophilic interactions in the signaling lymphocytic activation molecule receptor family. Immunity. 2006;25(4):559–70. doi: 10.1016/j.immuni.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 7.Velikovsky CA, Deng L, Chlewicki LK, Fernandez MM, Kumar V, Mariuzza RA. Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family. Immunity. 2007;27(4):572–84. doi: 10.1016/j.immuni.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan QR, Malashkevich VN, Fedorov A, Fedorov E, Cao E, Lary JW, Cole JL, Nathenson SG, Almo SC. Structure of CD84 provides insight into SLAM family function. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(25):10583–8. doi: DOI 10.1073/pnas.0703893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumaresan PR, Lai WC, Chuang SS, Bennett M, Mathew PA. CS1, a novel member of the CD2 family, is homophilic and regulates NK cell function. Mol Immunol. 2002;39(1–2):1–8. [DOI] [PubMed] [Google Scholar]
- 10.Booth C, Gilmour KC, Veys P, Gennery AR, Slatter MA, Chapel H, Heath PT, Steward CG, Smith O, O’Meara A, Kerrigan H, Mahlaoui N, Cavazzana-Calvo M, Fischer A, Moshous D, Blanche S, Pachlopnik Schmid J, Latour S, de Saint-Basile G, Albert M, Notheis G, Rieber N, Strahm B, Ritterbusch H, Lankester A, Hartwig NG, Meyts I, Plebani A, Soresina A, Finocchi A, Pignata C, Cirillo E, Bonanomi S, Peters C, Kalwak K, Pasic S, Sedlacek P, Jazbec J, Kanegane H, Nichols KE, Hanson IC, Kapoor N, Haddad E, Cowan M, Choo S, Smart J, Arkwright PD, Gaspar HB. X-linked lymphoproliferative disease due to SAP/SH2D1A deficiency: a multicenter study on the manifestations, management and outcome of the disease. Blood. 2011;117(1):53–62. doi: 10.1182/blood-2010-06-284935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols KE, Ma CS, Cannons JL, Schwartzberg PL, Tangye SG. Molecular and cellular pathogenesis of X-linked lymphoproliferative disease. Immunological reviews. 2005;203:180–99. doi: 10.1111/j.0105-2896.2005.00230.x. [DOI] [PubMed] [Google Scholar]
- 12.Pachlopnik Schmid J, Canioni D, Moshous D, Touzot F, Mahlaoui N, Hauck F, Kanegane H, Lopez-Granados E, Mejstrikova E, Pellier I, Galicier L, Galambrun C, Barlogis V, Bordigoni P, Fourmaintraux A, Hamidou M, Dabadie A, Le Deist F, Haerynck F, Ouachee-Chardin M, Rohrlich P, Stephan JL, Lenoir C, Rigaud S, Lambert N, Milili M, Schiff C, Chapel H, Picard C, de Saint Basile G, Blanche S, Fischer A, Latour S. Clinical similarities and differences of patients with X-linked lymphoproliferative syndrome type 1 (XLP-1/SAP deficiency) versus type 2 (XLP-2/XIAP deficiency). Blood. 2011;117(5):1522–9. doi: 10.1182/blood-2010-07-298372. [DOI] [PubMed] [Google Scholar]
- 13.Nichols KE, Harkin DP, Levitz S, Krainer M, Kolquist KA, Genovese C, Bernard A, Ferguson M, Zuo L, Snyder E, Buckler AJ, Wise C, Ashley J, Lovett M, Valentine MB, Look AT, Gerald W, Housman DE, Haber DA. Inactivating mutations in an SH2 domain-encoding gene in X-linked lymphoproliferative syndrome. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(23):13765–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffey AJ, Brooksbank RA, Brandau O, Oohashi T, Howell GR, Bye JM, Cahn AP, Durham J, Heath P, Wray P, Pavitt R, Wilkinson J, Leversha M, Huckle E, Shaw-Smith CJ, Dunham A, Rhodes S, Schuster V, Porta G, Yin L, Serafini P, Sylla B, Zollo M, Franco B, Bolino A, Seri M, Lanyi A, Davis JR, Webster D, Harris A, Lenoir G, de St Basile G, Jones A, Behloradsky BH, Achatz H, Murken J, Fassler R, Sumegi J, Romeo G, Vaudin M, Ross MT, Meindl A, Bentley DR. Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nature genetics. 1998;20(2):129–35. doi: 10.1038/2424. [DOI] [PubMed] [Google Scholar]
- 15.Ward J, Bonaparte M, Sacks J, Guterman J, Fogli M, Mavilio D, Barker E. HIV modulates the expression of ligands important in triggering natural killer cell cytotoxic responses on infected primary T-cell blasts. Blood. 2007;110(4):1207–14. doi: 10.1182/blood-2006-06-028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsi ED, Steinle R, Balasa B, Szmania S, Draksharapu A, Shum BP, Huseni M, Powers D, Nanisetti A, Zhang Y, Rice AG, van Abbema A, Wong M, Liu G, Zhan FH, Dillon M, Chen SH, Rhodes S, Fuh F, Tsurushita N, Kumar S, Vexler V, Shaughnessy JD, Barlogie B, van Rhee F, Hussein M, Afar DEH, Williams MB. CS1, a potential new therapeutic antibody target for the treatment of multiple myeloma. Clinical Cancer Research. 2008;14(9):2775–84. doi: 10.1158/1078-0432.CCR-07-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veillette A, Guo H. CS1, a SLAM family receptor involved in immune regulation, is a therapeutic target in multiple myeloma. Critical reviews in oncology/hematology. 2013;88(1):168–77. doi: 10.1016/j.critrevonc.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Bouhlal H, Ouled-Haddou H, Debuysscher V, Singh AR, Ossart C, Reignier A, Hocini H, Fouquet G, Al Baghami M, Eugenio MS, Nguyen-Khac E, Regimbeau JM, Marcq I. RB/PLK1-dependent induced pathway by SLAMF3 expression inhibits mitosis and control hepatocarcinoma cell proliferation. Oncotarget. 2016;7(9):9832–43. doi: 10.18632/oncotarget.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hislop AD, Palendira U, Leese AM, Arkwright PD, Rohrlich PS, Tangye SG, Gaspar HB, Lankester AC, Moretta A, Rickinson AB. Impaired Epstein-Barr virus-specific CD8(+) T-cell function in X-linked lymphoproliferative disease is restricted to SLAM family-positive B-cell targets. Blood. 2010;116(17):3249–57. doi: 10.1182/blood-2009-09-238832. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki A, Yamada R, Kochi Y, Sawada T, Okada Y, Matsuda K, Kamatani Y, Mori M, Shimane K, Hirabayashi Y, Takahashi A, Tsunoda T, Miyatake A, Kubo M, Kamatani N, Nakamura Y, Yamamoto K. Functional SNPs in CD244 increase the risk of rheumatoid arthritis in a Japanese population. Nature genetics. 2008;40(10):1224–9. doi: 10.1038/ng.205. [DOI] [PubMed] [Google Scholar]
- 21.Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, Yim YS, Pertsemlidis A, Garner HR Jr., Morel L, Wakeland EK. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. 2004;21(6):769–80. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Davis SJ, van der Merwe PA. The structure and ligand interactions of CD2: implications for T-cell function. Immunology today. 1996;17(4):177–87. [DOI] [PubMed] [Google Scholar]
- 23.Sandrin MS, Gumley TP, Henning MM, Vaughan HA, Gonez LJ, Trapani JA, McKenzie IF. Isolation and characterization of cDNA clones for mouse Ly-9. Journal of immunology. 1992;149(5):1636–41. [PubMed] [Google Scholar]
- 24.Sandrin MS, Henning MM, Lo MF, Baker E, Sutherland GR, McKenzie IF. Isolation and characterization of cDNA clones for Humly9: the human homologue of mouse Ly9. Immunogenetics. 1996;43(1–2):13–9. [DOI] [PubMed] [Google Scholar]
- 25.Veillette A SLAM-Family Receptors: Immune Regulators with or without SAP-Family Adaptors. Cold Spring Harbor perspectives in biology. 2010;2(3). doi: ARTN a002469 10.1101/cshperspect.a002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, van Schaik S, Notarangelo L, Geha R, Roncarolo MG, Oettgen H, De Vries JE, Aversa G, Terhorst C. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 1998;395(6701):462–9. doi: Doi 10.1038/26683. [DOI] [PubMed] [Google Scholar]
- 27.Morra M, Lu J, Poy F, Martin M, Sayos J, Calpe S, Gullo C, Howie D, Rietdijk S, Thompson A, Coyle AJ, Denny C, Yaffe MB, Engel P, Eck MJ, Terhorst C. Structural basis for the interaction of the free SH2 domain EAT-2 with SLAM receptors in hematopoietic cells. The EMBO journal. 2001;20(21):5840–52. doi: 10.1093/emboj/20.21.5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sidorenko SP, Clark EA. The dual-function CD150 receptor subfamily: the viral attraction. Nature immunology. 2003;4(1):19–24. doi: 10.1038/ni0103-19. [DOI] [PubMed] [Google Scholar]
- 29.Staunton DE, Fisher RC, LeBeau MM, Lawrence JB, Barton DE, Francke U, Dustin M, Thorley-Lawson DA. Blast-1 possesses a glycosyl-phosphatidylinositol (GPI) membrane anchor, is related to LFA-3 and OX-45, and maps to chromosome 1q21–23. The Journal of experimental medicine. 1989;169(3):1087–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elishmereni M, Levi-Schaffer F. CD48: A co-stimulatory receptor of immunity. The international journal of biochemistry & cell biology. 2011;43(1):25–8. doi: 10.1016/j.biocel.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Thorley-Lawson DA, Schooley RT, Bhan AK, Nadler LM. Epstein-Barr virus superinduces a new human B cell differentiation antigen (B-LAST 1) expressed on transformed lymphoblasts. Cell. 1982;30(2):415–25. [DOI] [PubMed] [Google Scholar]
- 32.McArdel SL, Terhorst C, Sharpe AH. Roles of CD48 in regulating immunity and tolerance. Clinical immunology. 2016;164:10–20. doi: 10.1016/j.clim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kingsbury GA, Feeney LA, Nong Y, Calandra SA, Murphy CJ, Corcoran JM, Wang Y, Prabhu Das MR, Busfield SJ, Fraser CC, Villeval JL. Cloning, expression, and function of BLAME, a novel member of the CD2 family. Journal of immunology. 2001;166(9):5675–80. [DOI] [PubMed] [Google Scholar]
- 34.Fraser CC, Howie D, Morra M, Qiu Y, Murphy C, Shen Q, Gutierrez-Ramos JC, Coyle A, Kingsbury GA, Terhorst C. Identification and characterization of SF2000 and SF2001, two new members of the immune receptor SLAM/CD2 family. Immunogenetics. 2002;53(10–11):843–50. doi: 10.1007/s00251-001-0415-7. [DOI] [PubMed] [Google Scholar]
- 35.Morra M, Howie D, Grande MS, Sayos J, Wang N, Wu C, Engel P, Terhorst C. X-linked lymphoproliferative disease: a progressive immunodeficiency. Annu Rev Immunol. 2001;19:657–82. doi: 10.1146/annurev.immunol.19.1.657. [DOI] [PubMed] [Google Scholar]
- 36.Thompson AD, Braun BS, Arvand A, Stewart SD, May WA, Chen E, Korenberg J, Denny C. EAT-2 is a novel SH2 domain containing protein that is up regulated by Ewing’s sarcoma EWS/FLI1 fusion gene. Oncogene. 1996;13(12):2649–58. [PubMed] [Google Scholar]
- 37.Roncagalli R, Taylor JE, Zhang S, Shi X, Chen R, Cruz-Munoz ME, Yin L, Latour S, Veillette A. Negative regulation of natural killer cell function by EAT-2, a SAP-related adaptor. Nature immunology. 2005;6(10):1002–10. doi: 10.1038/ni1242. [DOI] [PubMed] [Google Scholar]
- 38.Calpe S, Erdos E, Liao GX, Wang NH, Rietdijk S, Simarro M, Scholtz B, Mooney J, Lee CH, Shin MS, Rajnavolgyi E, Schatzle J, Morse HC, Terhorst C, Lanyi A. Identification and characterization of two related murine genes, Eat2a and Eat2b, encoding single SH2-domain adapters. Immunogenetics. 2006;58(1):15–25. doi: 10.1007/s00251-005-0056-3. [DOI] [PubMed] [Google Scholar]
- 39.Chung B, Aoukaty A, Dutz J, Terhorst C, Tan RS. Cutting edge: Signaling lymphocytic activation molecule-associated protein controls NKT cell functions. Journal of immunology. 2005;174(6):3153–7. [DOI] [PubMed] [Google Scholar]
- 40.Veillette A, Zhang S, Shi X, Dong Z, Davidson D, Zhong MC. SAP expression in T cells, not in B cells, is required for humoral immunity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(4):1273–8. doi: 10.1073/pnas.0710698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veillette A Immune regulation by SLAM family receptors and SAP-related adaptors. Nature reviews Immunology. 2006;6(1):56–66. doi: 10.1038/nri1761. [DOI] [PubMed] [Google Scholar]
- 42.Shlapatska LM, Mikhalap SV, Berdova AG, Zelensky OM, Yun TJ, Nichols KE, Clark EA, Sidorenko SP. CD150 association with either the SH2-containing inositol phosphatase or the SH2-containing protein tyrosine phosphatase is regulated by the adaptor protein SH2D1A. Journal of immunology. 2001;166(9):5480–7. [DOI] [PubMed] [Google Scholar]
- 43.Poy F, Yaffe MB, Sayos J, Saxena K, Morra M, Sumegi J, Cantley LC, Terhorst C, Eck MJ. Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition. Molecular cell. 1999;4(4):555–61. [DOI] [PubMed] [Google Scholar]
- 44.Latour S, Veillette A. The SAP family of adaptors in immune regulation. Seminars in immunology. 2004;16(6):409–19. doi: 10.1016/j.smim.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 45.Dong Z, Davidson D, Perez-Quintero LA, Kurosaki T, Swat W, Veillette A. The adaptor SAP controls NK cell activation by regulating the enzymes Vav-1 and SHIP-1 and by enhancing conjugates with target cells. Immunity. 2012;36(6):974–85. doi: 10.1016/j.immuni.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 46.Wilson TJ, Garner LI, Metcalfe C, King E, Margraf S, Brown MH. Fine specificity and molecular competition in SLAM family receptor signalling. PloS one. 2014;9(3):e92184. doi: 10.1371/journal.pone.0092184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu N, Zhong MC, Roncagalli R, Perez-Quintero LA, Guo H, Zhang Z, Lenoir C, Dong Z, Latour S, Veillette A. A hematopoietic cell-driven mechanism involving SLAMF6 receptor, SAP adaptors and SHP-1 phosphatase regulates NK cell education. Nature immunology. 2016;17(4):387–96. doi: 10.1038/ni.3369. [DOI] [PubMed] [Google Scholar]
- 48.Zhong MC, Veillette A. Critical role of SAP in progression and reactivation but not maintenance of T cell-dependent humoral immunity. Molecular and cellular biology. 2013;33(6):1223–32. doi: 10.1128/MCB.01591-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong ZJ, Cruz-Munoz ME, Zhong MC, Chen RY, Latour S, Veillette A. Essential function for SAP family adaptors in the surveillance of hematopoietic cells by natural killer cells. Nature immunology. 2009;10(9):973–U62. doi: 10.1038/ni.1763. [DOI] [PubMed] [Google Scholar]
- 50.Bottino C, Falco M, Parolini S, Marcenaro E, Augugliaro R, Sivori S, Landi E, Biassoni R, Notarangelo LD, Moretta L, Moretta A. NTB-A [correction of GNTB-A], a novel SH2D1A-associated surface molecule contributing to the inability of natural killer cells to kill Epstein-Barr virus-infected B cells in X-linked lymphoproliferative disease. The Journal of experimental medicine. 2001;194(3):235–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong MC, Veillette A. Control of T lymphocyte signaling by Ly108, a signaling lymphocytic activation molecule family receptor implicated in autoimmunity. The Journal of biological chemistry. 2008;283(28):19255–64. doi: 10.1074/jbc.M800209200. [DOI] [PubMed] [Google Scholar]
- 52.Simarro M, Lanyi A, Howie D, Poy F, Bruggeman J, Choi M, Sumegi J, Eck MJ, Terhorst C. SAP increases FynT kinase activity and is required for phosphorylation of SLAM and Ly9. International immunology. 2004;16(5):727–36. doi: 10.1093/intimm/dxh074. [DOI] [PubMed] [Google Scholar]
- 53.Li SC, Gish G, Yang D, Coffey AJ, Forman-Kay JD, Ernberg I, Kay LE, Pawson T. Novel mode of ligand binding by the SH2 domain of the human XLP disease gene product SAP/SH2D1A. Current biology : CB. 1999;9(23):1355–62. [DOI] [PubMed] [Google Scholar]
- 54.Cruz-Munoz ME, Dong Z, Shi X, Zhang S, Veillette A. Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function. Nature immunology. 2009;10(3):297–305. doi: 10.1038/ni.1693. [DOI] [PubMed] [Google Scholar]
- 55.Chen RY, Latour S, Shi XC, Veillette A. Association between SAP and FynT: Inducible SH3 domain-mediated interaction controlled by engagement of the SLAM receptor. Molecular and cellular biology. 2006;26(15):5559–68. doi: 10.1128/Mcb.00357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Latour S, Roncagalli R, Chen RY, Bakinowski M, Shi XC, Schwartzberg PL, Davidson D, Veillette A. Binding of SAP SH2 domain to FynT SH3 domain reveals a novel mechanism of receptor signalling in immune regulation. Nat Cell Biol. 2003;5(2):149–54. doi: 10.1038/ncb919. [DOI] [PubMed] [Google Scholar]
- 57.Li CJ, Iosef C, Jia CYH, Han VKM, Li SSC. Dual functional roles for the X-linked lymphoproliferative syndrome gene product SAP/SH2D1A in signaling through the signaling lymphocyte activation molecule (SLAM) family of immune receptors. Journal of Biological Chemistry. 2003;278(6):3852–9. doi: 10.1074/jbc.M206649200. [DOI] [PubMed] [Google Scholar]
- 58.Latour S, Gish G, Helgason CD, Humphries RK, Pawson T, Veillette A. Regulation of SLAM-mediated signal transduction by SAP, the X-linked lymphoproliferative gene product. Nature immunology. 2001;2(8):681–90. doi: 10.1038/90615. [DOI] [PubMed] [Google Scholar]
- 59.Cannons JL, Yu LJ, Hill B, Mijares LA, Dombroski D, Nichols KE, Antonellis A, Koretzky GA, Gardner K, Schwartzberg PL. SAP regulates T(H)2 differentiation and PKC-theta-mediated activation of NF-kappa B1. Immunity. 2004;21(5):693–706. doi: DOI 10.1016/j.immuni.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 60.Snow A, Marsh R, Krummey S, Roehrs P, Zhang KJ, Filipovich L, Su H, Bleesing J, Lenordo M. SAP Augments Proximal T Cell Receptor Signal Strength Necessary for Restimulation-induced Apoptosis of Activated T Cells. Clinical immunology. 2009;131:S160–S. doi: 10.1016/j.clim.2009.03.473. [DOI] [Google Scholar]
- 61.Dong Z, Cruz-Munoz ME, Zhong MC, Chen R, Latour S, Veillette A. Essential function for SAP family adaptors in the surveillance of hematopoietic cells by natural killer cells. Nature immunology. 2009;10(9):973–80. doi: 10.1038/ni.1763. [DOI] [PubMed] [Google Scholar]
- 62.Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, Schwartzberg PL, Crotty S. The Receptor Ly108 Functions as a SAP Adaptor-Dependent On-Off Switch for T Cell Help to B Cells and NKT Cell Development. Immunity. 2012;36(6):986–1002. doi: 10.1016/j.immuni.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gomez-Martin D, Diaz-Zamudio M, Romo-Tena J, Ibarra-Sanchez MJ, Alcocer-Varela J. Follicular helper T cells poise immune responses to the development of autoimmune pathology. Autoimmunity reviews. 2011;10(6):325–30. doi: 10.1016/j.autrev.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 64.Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, Mackay CR. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. Journal of immunology. 2004;173(1):68–78. [DOI] [PubMed] [Google Scholar]
- 65.Wu C, Nguyen KB, Pien GC, Wang N, Gullo C, Howie D, Sosa MR, Edwards MJ, Borrow P, Satoskar AR, Sharpe AH, Biron CA, Terhorst C. SAP controls T cell responses to virus and terminal differentiation of TH2 cells. Nature immunology. 2001;2(5):410–4. doi: 10.1038/87713. [DOI] [PubMed] [Google Scholar]
- 66.Czar MJ, Kersh EN, Mijares LA, Lanier G, Lewis J, Yap G, Chen A, Sher A, Duckett CS, Ahmed R, Schwartzberg PL. Altered lymphocyte responses and cytokine production in mice deficient in the X-linked lymphoproliferative disease gene SH2D1A/DSHP/SAP. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(13):7449–54. doi: 10.1073/pnas.131193098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Proust R, Bertoglio J, Gesbert F. The adaptor protein SAP directly associates with CD3zeta chain and regulates T cell receptor signaling. PloS one. 2012;7(8):e43200. doi: 10.1371/journal.pone.0043200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chatterjee M, Rauen T, Kis-Toth K, Kyttaris VC, Hedrich CM, Terhorst C, Tsokos GC. Increased expression of SLAM receptors SLAMF3 and SLAMF6 in systemic lupus erythematosus T lymphocytes promotes Th17 differentiation. Journal of immunology. 2012;188(3):1206–12. doi: 10.4049/jimmunol.1102773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eissmann P, Beauchamp L, Wooters J, Tilton JC, Long EO, Watzl C. Molecular basis for positive and negative signaling by the natural killer cell receptor 2B4 (CD244). Blood. 2005;105(12):4722–9. doi: 10.1182/blood-2004-09-3796. [DOI] [PubMed] [Google Scholar]
- 70.Tangye SG, Lazetic S, Woollatt E, Sutherland GR, Lanier LL, Phillips JH. Cutting edge: human 2B4, an activating NK cell receptor, recruits the protein tyrosine phosphatase SHP-2 and the adaptor signaling protein SAP. Journal of immunology. 1999;162(12):6981–5. [PubMed] [Google Scholar]
- 71.Mooney JM, Klem J, Wulfing C, Mijares LA, Schwartzberg PL, Bennett M, Schatzle JD. The murine NK receptor 2B4 (CD244) exhibits inhibitory function independent of signaling lymphocytic activation molecule-associated protein expression. Journal of immunology. 2004;173(6):3953–61. [DOI] [PubMed] [Google Scholar]
- 72.Lee KM, McNerney ME, Stepp SE, Mathew PA, Schatzle JD, Bennett M, Kumar V. 2B4 acts as a non-major histocompatibility complex binding inhibitory receptor on mouse natural killer cells. The Journal of experimental medicine. 2004;199(9):1245–54. doi: 10.1084/jem.20031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vacca P, Pietra G, Falco M, Romeo E, Bottino C, Bellora F, Prefumo F, Fulcheri E, Venturini PL, Costa M, Moretta A, Moretta L, Mingari MC. Analysis of natural killer cells isolated from human decidua: Evidence that 2B4 (CD244) functions as an inhibitory receptor and blocks NK-cell function. Blood. 2006;108(13):4078–85. doi: 10.1182/blood-2006-04-017343. [DOI] [PubMed] [Google Scholar]
- 74.Tarakhovsky A, Turner M, Schaal S, Mee PJ, Duddy LP, Rajewsky K, Tybulewicz VL. Defective antigen receptor-mediated proliferation of B and T cells in the absence of Vav. Nature. 1995;374(6521):467–70. doi: 10.1038/374467a0. [DOI] [PubMed] [Google Scholar]
- 75.Chan G, Hanke T, Fischer KD. Vav-1 regulates NK T cell development and NK cell cytotoxicity. European journal of immunology. 2001;31(8):2403–10. doi: . [DOI] [PubMed] [Google Scholar]
- 76.Eissmann P, Watzl C. Molecular analysis of NTB-A signaling: a role for EAT-2 in NTB-A-mediated activation of human NK cells. Journal of immunology. 2006;177(5):3170–7. [DOI] [PubMed] [Google Scholar]
- 77.Tangye SG, Nichols KE, Hare NJ, van de Weerdt BC. Functional requirements for interactions between CD84 and Src homology 2 domain-containing proteins and their contribution to human T cell activation. Journal of immunology. 2003;171(5):2485–95. [DOI] [PubMed] [Google Scholar]
- 78.Dupre L, Andolfi G, Tangye SG, Clementi R, Locatelli F, Arico M, Aiuti A, Roncarolo MG. SAP controls the cytolytic activity of CD8+ T cells against EBV-infected cells. Blood. 2005;105(11):4383–9. doi: 10.1182/blood-2004-08-3269. [DOI] [PubMed] [Google Scholar]
- 79.Wang H, Xiong L, Tang W, Zhou Y, Li F. A systematic review of malignancy-associated hemophagocytic lymphohistiocytosis that needs more attentions. Oncotarget. 2017. doi: 10.18632/oncotarget.19230.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Strahm B, Rittweiler K, Duffner U, Brandau O, Orlowska-Volk M, Karajannis MA, Stadt U, Tiemann M, Reiter A, Brandis M, Meindl A, Niemeyer CM. Recurrent B-cell non-Hodgkin’s lymphoma in two brothers with X-linked lymphoproliferative disease without evidence for Epstein-Barr virus infection. British journal of haematology. 2000;108(2):377–82. [DOI] [PubMed] [Google Scholar]
- 81.Marsh RA, Madden L, Kitchen BJ, Mody R, McClimon B, Jordan MB, Bleesing JJ, Zhang K, Filipovich AH. XIAP deficiency: a unique primary immunodeficiency best classified as X-linked familial hemophagocytic lymphohistiocytosis and not as X-linked lymphoproliferative disease. Blood. 2010;116(7):1079–82. doi: 10.1182/blood-2010-01-256099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda JE, MacKenzie A, Korneluk RG. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature. 1996;379(6563):349–53. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- 83.Sullivan JL, Byron KS, Brewster FE, Baker SM, Ochs HD. X-linked lymphoproliferative syndrome. Natural history of the immunodeficiency. J Clin Invest. 1983;71(6):1765–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Purtilo DT, Sakamoto K, Barnabei V, Seeley J, Bechtold T, Rogers G, Yetz J, Harada S. Epstein-Barr virus-induced diseases in boys with the X-linked lymphoproliferative syndrome (XLP): update on studies of the registry. The American journal of medicine. 1982;73(1):49–56. [DOI] [PubMed] [Google Scholar]
- 85.Ma CS, Hare NJ, Nichols KE, Dupre L, Andolfi G, Roncarolo MG, Adelstein S, Hodgkin PD, Tangye SG. Impaired humoral immunity in X-linked lymphoproliferative disease is associated with defective IL-10 production by CD4+ T cells. J Clin Invest. 2005;115(4):1049–59. doi: Doi 10.1172/Jci23139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takei M, Mitamura K, Fujiwara S, Horie T, Ryu J, Osaka S, Yoshino S, Sawada S. Detection of Epstein-Barr virus-encoded small RNA 1 and latent membrane protein 1 in synovial lining cells from rheumatoid arthritis patients. International immunology. 1997;9(5):739–43. [DOI] [PubMed] [Google Scholar]
- 87.Sawada S, Takei M. Epstein-Barr virus etiology in rheumatoid synovitis. Autoimmunity reviews. 2005;4(2):106–10. doi: 10.1016/j.autrev.2004.08.034. [DOI] [PubMed] [Google Scholar]
- 88.Sawada S, Takei M, Ishiwata T. SAP discovery: the sword edges--beneficial and harmful. Autoimmunity reviews. 2007;6(7):444–9. doi: 10.1016/j.autrev.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 89.Solomon DH, Karlson EW, Rimm EB, Cannuscio CC, Mandl LA, Manson JE, Stampfer MJ, Curhan GC. Cardiovascular morbidity and mortality in women diagnosed with rheumatoid arthritis. Circulation. 2003;107(9):1303–7. [DOI] [PubMed] [Google Scholar]
- 90.Ferraccioli G, Gremese E. Thrombogenicity of TNF alpha in rheumatoid arthritis defined through biological probes: TNF alpha blockers. Autoimmunity reviews. 2004;3(4):261–6. doi: 10.1016/j.autrev.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 91.Kumar KR, Li L, Yan M, Bhaskarabhatla M, Mobley AB, Nguyen C, Mooney JM, Schatzle JD, Wakeland EK, Mohan C. Regulation of B cell tolerance by the lupus susceptibility gene Ly108. Science. 2006;312(5780):1665–9. doi: 10.1126/science.1125893. [DOI] [PubMed] [Google Scholar]
- 92.Tsokos GC. Systemic lupus erythematosus. The New England journal of medicine. 2011;365(22):2110–21. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 93.Veillette A, Guo HJ. CS1, a SLAM family receptor involved in immune regulation, is a therapeutic target in multiple myeloma. Crit Rev Oncol Hemat. 2013;88(1):168–77. doi: 10.1016/j.critrevonc.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 94.Tai YT, Dillon M, Song W, Leiba M, Li XF, Burger P, Lee AI, Podar K, Hideshima T, Rice AG, van Abbema A, Jesaitis L, Caras I, Law D, Weller E, Xie W, Richardson P, Munshi NC, Mathiot C, Avet-Loiseau H, Afar DEH, Anderson KC. Anti-CS1 humanized monoclonal antibody HuLuc63 inhibits myeloma cell adhesion and induces aritibody-dependent cellular cytotoxicity in the bone marrow mitieu. Blood. 2008;112(4):1329–37. doi: 10.1182/blood-2007-08-107292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Komori H, Furukawa H, Mori S, Ito MR, Terada M, Zhang MC, Ishii N, Sakuma N, Nose M, Ono M. A signal adaptor SLAM-associated protein regulates spontaneous autoimmunity and Fas-dependent lymphoproliferation in MRL-Faslpr lupus mice. Journal of immunology. 2006;176(1):395–400. [DOI] [PubMed] [Google Scholar]
- 96.Sawada S Slam-associated protein plays a key role in development of autoimmunity. Autoimmunity reviews. 2012;11(11):804–5. doi: 10.1016/j.autrev.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 97.Keating GM, Santoro A. Sorafenib A Review of its Use in Advanced Hepatocellular Carcinoma. Drugs. 2009;69(2):223–40. [DOI] [PubMed] [Google Scholar]
- 98.Smalley KSM, Xiao M, Villanueva J, Nguyen TK, Flaherty KT, Letrero R, Van Belle P, Elder DE, Wang Y, Nathanson KL, Herlyn M. CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene. 2009;28(1):85–94. doi: 10.1038/onc.2008.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilhelm SM, Adnane L, Newell P, Villanueva A, Llovet JM, Lynch M. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Molecular cancer therapeutics. 2008;7(10):3129–40. doi: 10.1158/1535-7163.MCT-08-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Haussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J, Group SIS. Sorafenib in advanced hepatocellular carcinoma. The New England journal of medicine. 2008;359(4):378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 101.Bonaparte MI, Barker E. Killing of human immunodeficiency virus-infected primary T-cell blasts by autologous natural killer cells is dependent on the ability of the virus to alter the expression of major histocompatibility complex class I molecules. Blood. 2004;104(7):2087–94. doi: 10.1182/blood-2004-02-0696. [DOI] [PubMed] [Google Scholar]
- 102.Huard B, Fruh K. A role for MHC class I down-regulation in NK cell lysis of herpes virus-infected cells. European journal of immunology. 2000;30(2):509–15. doi: . [DOI] [PubMed] [Google Scholar]
- 103.Bonaparte MI, Barker E. Inability of natural killer cells to destroy autologous HIV-infected T lymphocytes. Aids. 2003;17(4):487–94. doi: 10.1097/01.aids.0000050814.06065.23. [DOI] [PubMed] [Google Scholar]
- 104.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10(6):661–71. [DOI] [PubMed] [Google Scholar]
- 105.Pende D, Spaggiari GM, Marcenaro S, Martini S, Rivera P, Capobianco A, Falco M, Lanino E, Pierri I, Zambello R, Bacigalupo A, Mingari MC, Moretta A, Moretta L. Analysis of the receptor-ligand interactions in the natural killer-mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: evidence for the involvement of the Poliovirus receptor (CD155) and Nectin-2 (CD112). Blood. 2005;105(5):2066–73. doi: 10.1182/blood-2004-09-3548. [DOI] [PubMed] [Google Scholar]
- 106.Edwards JC, Szczepanski L, Szechinski J, Filipowicz-Sosnowska A, Emery P, Close DR, Stevens RM, Shaw T. Efficacy of B-cell-targeted therapy with rituximab in patients with rheumatoid arthritis. The New England journal of medicine. 2004;350(25):2572–81. doi: 10.1056/NEJMoa032534. [DOI] [PubMed] [Google Scholar]
- 107.Owczarczyk K, Lal P, Abbas AR, Wolslegel K, Holweg CT, Dummer W, Kelman A, Brunetta P, Lewin-Koh N, Sorani M, Leong D, Fielder P, Yocum D, Ho C, Ortmann W, Townsend MJ, Behrens TW. A plasmablast biomarker for nonresponse to antibody therapy to CD20 in rheumatoid arthritis. Science translational medicine. 2011;3(101):101ra92. doi: 10.1126/scitranslmed.3002432. [DOI] [PubMed] [Google Scholar]
- 108.Woo J, Vierboom MP, Kwon H, Chao D, Ye S, Li J, Lin K, Tang I, Belmar NA, Hartman T, Breedveld E, Vexler V, t Hart BA, Law DA, Starling GC. PDL241, a novel humanized monoclonal antibody, reveals CD319 as a therapeutic target for rheumatoid arthritis. Arthritis research & therapy. 2013;15(6):R207. doi: 10.1186/ar4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hron JD, Caplan L, Gerth AJ, Schwartzberg PL, Peng SL. SH2D1A regulates T-dependent humoral autoimmunity. The Journal of experimental medicine. 2004;200(2):261–6. doi: 10.1084/jem.20040526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cuss AK, Avery DT, Cannons JL, Yu LJ, Nichols KE, Shaw PJ, Tangye SG. Expansion of functionally immature transitional B cells is associated with human-immunodeficient states characterized by impaired humoral immunity. Journal of immunology. 2006;176(3):1506–16. [DOI] [PubMed] [Google Scholar]