Abstract
Tef [Eragrostis tef (Zucc.) Trotter], an allotetraploid cereal that is a staple food to over 60 million people in the Horn of Africa, has a high nutritional content and is resistant to many biotic and abiotic stresses such as waterlogging and drought. Three tef genotypes, Alba, Tsedey, and Quncho, were subjected to waterlogging conditions and their growth, physiology, and change in transcript expression were measured with the goal of identifying targets for breeding cultivars with improved waterlogging tolerance. Root and shoot growth and dry weight were observed over 22 days. Stomatal conductance and chlorophyll and carotenoid contents were quantified. Microscopy was used to monitor changes in the stem cross sections. Illumina RNA sequencing was used to obtain the expression profiles of tef under flooding and control conditions and was verified using qPCR. Results indicated differences in growth between the three genotypes. Waterlogged Tsedey plants grew higher and had more root biomass than normally watered Tsedey plants. Quncho and Alba genotypes were more susceptible to the excess moisture stress. The effects of these changes were observed on the plant physiology. Among the three tested tef genotypes, Tsedey formed more aerenchyma than Alba and had accelerated growth under waterlogging. Tsedey and Quncho had constitutive aerenchyma. Genes affecting carbohydrate metabolism, cell growth, response to reactive oxygen species, transport, signaling, and stress responses were found to change under excess moisture stress. In general, these results show the presence of substantial anatomical and physiological differences among tef genotypes when waterlogged during the early growth stage.
Keywords: adventitious roots, aerenchyma, differential expression, Eragrostis tef, flooding, grass family, RNA‐Seq, tef, transcriptome, waterlogging
1. INTRODUCTION
Climate change is one of the most challenging problems facing global food security. Along with changes in temperature, the frequency and severity of both extreme droughts and extreme precipitation events are expected to increase (Hartmann, Tank, & Rusticucci, 2013) causing billions of dollars of crop losses (Bailey‐Serres, Lee, & Brinton, 2012). Understanding of the genetic basis of the plant response to abiotic constraints is critical for the development of cultivars that are resilient to stress and to decrease the gap between yield potential and actual yield under stressed conditions (Setter & Waters, 2003).
Tef [Eragrostis tef (Zucc.) Trotter], an allotetraploid cereal crop, is the major crop in the Horn of Africa, especially in Ethiopia where it is annually cultivated on over three million hectares of land and is staple food to over 60 million people (CSA 2015). Of the approximately 350 members of the Eragrostis genus, tef is the only one cultivated for use as a human food although several species are used as livestock fodder or forage grasses (Ketema, 1993). Tef grows in wide agroecological conditions ranging from semiarid areas with low rainfall to areas with high rainfall. It is also a dominant crop in poorly drained soils commonly known as vertisols which cover about 10% of the total area of Ethiopia, much of which are in high‐rainfall areas (Mekonen, Tesfaye, & Bayu, 2013). The combination of poorly drained soil and high rainfall is conducive to waterlogging and is responsible for both low yields and underutilization of this land for farming (Asamenew, Beyene, Negatu, & Ayele, 1993).
Waterlogging is a soil condition in which excess water causes inadequate gas exchange between the soil and the atmosphere. The effects on the plant are manifold. Oxygen diffuses approximately 10,000 times slower in water than in air, and the flow of oxygen into waterlogged soil is around 320,000 times less (Watanabe, Nishiuchi, Kulichikhin, & Nakazono, 2013). Therefore, the amount of oxygen available to the roots decreases (Jackson & Colmer, 2005; Lee et al., 2011), and photosynthesis and respiration are limited. Waterlogging can decrease a cell's resistance to pathogens (Hsu et al., 2013). Additionally, phytotoxic compounds such as sulfides and the reduced forms of minerals (e.g., Mn2+ and Fe2+) accumulate in waterlogged soil (Laanbroek, 1990; Nishiuchi, Yamauchi, Takahashi, Kotula, & Nakazono, 2012), creating oxidative stress (Fukao, Yeung, & Bailey‐Serres, 2011).
Waterlogging‐tolerant plants have many morphological responses to these stresses. A key response of many plants to flooding is the formation of adventitious roots, roots that form from non‐root tissue (Steffens & Rasmussen, 2016). In rice, excess water triggers a signal cascade involving many hormones including ethylene, abscisic acid (ABA), gibberellic acid (GA), auxin (Pacurar, Perrone, & Bellini, 2014), and cytokinin (Dat, Capelli, Folzer, Bourgeade, & Badot, 2004). Ethylene is unable to escape, and the consequent buildup of ethylene triggers adventitious root growth (Lorbiecke & Sauter, 1999). The newly formed adventitious roots often contain aerenchyma, air‐filled spaces that provide an internal pathway for the movement of oxygen from the well‐aerated shoots to the roots. Wetland plant species often possess constitutive aerenchyma or can form aerenchyma upon flooding (Colmer & Pedersen, 2008; Yamauchi, Shimamura, Nakazono, & Mochizuki, 2013), while in many dryland flooding‐tolerant species, waterlogging induces aerenchyma formation (Colmer & Voesenek, 2009). Several studies have correlated the formation of aerenchyma with waterlogging tolerance (Abiko et al., 2012; Herzog, Striker, Colmer, & Pedersen, 2016; Setter & Waters, 2003; Zhang et al., 2015).
Another morphological adaptation to flooding is the formation of barriers to radial oxygen loss (ROL) in the basal part of the root to minimize oxygen loss to the environment and keep the oxygen moving toward the root apex (Abiko et al., 2012; Colmer, Cox, & Voesenek, 2006; Nishiuchi et al., 2012; Watanabe et al., 2013). These barriers may also impede the movement of soil toxins and gases into the root (Armstrong, 1979; Colmer, 2003; Watanabe et al., 2013). Both suberin (Watanabe et al., 2013) and lignin (Kotula, Ranathunge, Schreiber, & Steudle, 2009) may be components of this apoplastic barrier. A hybrid between wheat (Triticum aestevum) and a wild waterlogging‐tolerant barley with constitutive aerenchyma and an inducible ROL barrier was found to be more waterlogging tolerant than the parental wheat line (Malik, Islam, & Colmer, 2011). Similarly, aerenchyma formation combined with an inducible barrier to ROL contributed to the waterlogging tolerance of a newly discovered teosinte (Zea nicaraguensis) compared to cultivated maize (Zea mays) (Abiko et al., 2012).
The production of reactive oxygen species (ROS) is a consequence of aerobic metabolism and may be a part of the signaling that stimulates the growth of adventitious roots (Steffens & Rasmussen, 2016). Detoxification of ROS is mediated by both enzymatic (superoxide dismutase, ascorbate peroxidase, etc.) and non‐enzymatic (carotenoids, ascorbic acid, glutathione) antioxidants (Lekshmy, Jha, & Sairam, 2015). In crop plants, upregulation of antioxidants has been found to be correlated with abiotic stress tolerance (Rivera‐Contreras et al., 2016; Ushimaru et al., 1997; Zhang et al., 2015) and has even been suggested to be the basis for waterlogging tolerance (Thirunavukkarasu et al., 2013).
Two strategies employed by rice to combat flood are (i) escape, rapid growth of the shoots that allows the plant to reach the surface as quickly as possible, and (ii) quiescence, entering a state of inactivity until the flooding conditions have passed (Nishiuchi et al., 2012). Both strategies are mediated by ethylene. Escape is a useful response in the case of deep and long‐lasting flooding, while quiescence is employed for shallower floods of short duration. Submergence‐tolerant rice cultivars employing the quiescence strategy restrict the consumption of carbohydrates, retain chlorophyll to maintain limited photosynthesis, and express an ethylene‐responsive factor (ERF) gene, Submergence 1A (SUB1A), which confers submergence tolerance by dampening ethylene production and GA responsiveness (Fukao, Xu, Ronald, & Bailey‐Serres, 2006; Xu et al., 2006). In deepwater rice, the escape strategy is employed. When ethylene accumulates in tissues, it stimulates internode elongation (Hattori et al., 2009; Kende, van der Knaap, & Cho, 1998; Raskin & Kende, 1983), via two ethylene‐responsive factors, Snorkel1 (SK1) and Snorkel2 (SK2) (Fukao et al., 2006). To allow rapid shoot elongation, proteins such as expansins are expressed to loosen the cell wall (Li, Jones, & McQueen‐Mason, 2003). Rapid shoot elongation under hypoxic conditions (escape strategy) has also been reported for several other plants including arrowhead (Sagittaria pygmaea Miq.), pondweed (Potamogeton distinctus A. Benn) (Ishizawa, Murakami, Kawakami, & Kuramochi, 1999; Summers & Jackson, 1994), and two rumex species (Rumex Palustris and Rumex acetosa) (van Veen et al., 2013) although it has been suggested that rapid shoot growth is detrimental to plant growth after the floodwaters recede (Kawano, Ito, & Sakagami, 2009).
The physiological response of plants subjected to waterlogging stress involves a switch from aerobic respiration to fermentative metabolism, and consequently, genes that are involved in starch breakdown such as the glycolytic and fermentative pathways are affected (Parent, Capelli, Berger, Crèvecoeur, & Dat, 2008). Tolerant plants can also photosynthesize and respire while flooded (Caudle & Maricle, 2012). Thus, physiological measurements such as stomatal conductance and leaf chlorophyll content are valuable probes of the plant response.
The Tef Improvement Project was established to increase tef productivity using modern plant breeding methods and has undertaken genome and transcriptome sequencing for this purpose (Cannarozzi et al., 2018). Here, we report a study on the response to waterlogging of three tef genotypes—a landrace, Alba, and two improved varieties Tsedey and Quncho. Quncho, a cross between Dukem (maternal) and Magna (paternal), is a popular variety as it combines the high yield of Dukem with the high seed quality of Magna. Tsedey, an improved variety on the market since 1984, can grow in a wide range of soil, water, and air conditions. Here, growth and biomass have been measured for tef subjected to early (at 4 days) waterlogging. In addition, we combined an expression study using RNA‐Seq and qPCR with physiological studies and microscopy to understand the response of tef to waterlogging at the tillering stage. The results show that there are substantial differences in growth, waterlogging tolerance, anatomy, and physiology between the three genotypes studied in the early and tillering stages of waterlogging, diversity that is necessary for breeding the trait into new germplasm and identification of the responsible genes using association mapping. The most tolerant genotype, Tsedey, has greater growth of both the shoot and the root, more constitutive aerenchyma, and aerenchyma production. Genes affecting carbohydrate metabolism, cell growth, response to reactive oxygen species, transport, signaling, and stress responses were found to change under stress in the Tsedey improved variety, consistent with the physiological and anatomical changes observed.
2. MATERIALS AND METHODS
Experiments designed to monitor root and shoot growth, aerenchyma size, physiology, and gene expression are outlined in Table 1.
Table 1.
Summary of growth, expression, and physiology experiments conducted
| Experiment type | Genotypes | Number of plants of each genotype | Tissue | Age of plant during treatment | Evaluation methods | Parameters measured | Stage Figure or Table | |
|---|---|---|---|---|---|---|---|---|
| 1 | Growth 1 | Alba, Quncho, Tsedey | 18 | Roots, shoots | From 4 to 28 days | Measure shoot length in pots | Shoot length |
Early stage Figure S1 |
| 2 | Growth 2 | Quncho, Tsedey | 66 | Roots, shoots | From 4 to 28 days | Harvest 4 controls and 4 waterlogged at 2, 4, 7, 9, 11, 16, and 24 days after treatment | Root and shoot length, dry weight, cross section |
Early stage Figures 1 and S6 Tables 2, S1, S2 |
| 3 | Root growth 1 | Alba, Quncho, Tsedey | 2 | Roots | From 4 to 22 days | Visualization with rhizotron | Root growth |
Early stage Figure S2 |
| 4 | Root growth 2 | Wheat, maize, Alba, Tsedey | 2 | Roots, shoots | From 7 to 35 days | Visualization with rhizotron | Root growth |
Early stage Figures S3 (wheat) and S4 (maize) |
| 5 | Physiology | Tsedey | Shoots | From 19 to 28 days |
Porometer SPAD |
Chlorophyll a and b, carotenoid, stomatal conductance |
Tillering stage Figure 2 |
|
| 6 | Cross sections | Alba, Quncho, Tsedey | 21 | Roots | From 19 to 28 days | Count adventitious roots, cross sections |
Number of adventitious roots Aerenchyma size |
Tillering stage Figures 3, 4 and S5 Table S3 |
| 7 | qPCR | Alba, Quncho, Tsedey | Shoots | From 19 to 28 days | Microscopy, qPCR | Differential gene expression |
Tillering stage Figure 5 Table S6 |
|
| 8 | RNA‐Seq | Tsedey | Shoots | From 19 to 28 days | RNA‐Seq | Differential gene expression |
Tillering stage Figures S7, S8, S9, S10, S11, and S12 Tables 3, 4, 5, S4, and S5 |
2.1. Tef genotypes used
Three Eragrostis tef genotypes used in these experiments were as follows: (i) Tsedey (DZ‐Cr‐37), an improved variety whose genome and transcriptome have been sequenced and which is thought to be waterlogging resistant, (ii) Alba, a landrace obtained from the US Department of Agriculture (USDA), and (iii) Quncho (DZ‐Cr‐387 RIL‐355), a popular high‐yielding and white‐seeded variety. Tsedey and Quncho were obtained from the Ethiopian Institute of Agricultural Research (EIAR).
2.2. Growing conditions for Experiment 1 (Growth 1): plant height
Three seeds of tef genotypes (Alba, Tsedey, and Quncho) were sown in pots and grown under long‐day conditions (16‐h light at 22°C and 65% relative humidity; 8‐h dark at 18°C and 65% relative humidity). For the waterlogging treatment, the pots were put in a plastic tray in which the water level was maintained at 1 cm below the surface of the soil. The control plants were put in an identical tray and watered from below every 3 days followed by drainage of the tray. The waterlogging treatment was applied to 18 (9 controls, 9 waterlogged) 4‐day‐old plants. Day 0 marked the first day of treatment. Plant height (from the base of the root to the end of the longest leaf) was recorded for the next 24 days, namely at 0, 1, 3, 4, 7, 8, 14, 17, and 22 days after waterlogging.
2.3. Growing conditions for Experiment 2 (Growth 2): Early waterlogging time series of plant height, root length, and root/shoot dry weight
Experiment 2 is a repetition of Experiment 1 using 66 seedlings of Tsedey and Quncho sown in pots and grown under long‐day conditions as indicated above. For the waterlogging treatment, the pots were put in a plastic tray in which the water level was maintained at 1 cm below the surface of the soil. The control plants were put in an identical tray and watered from below every 3 days followed by drainage of the tray. Four days after germination, treatment started and the plants were grown for the next 24 days under control or waterlogging conditions. At 2, 4, 7, 9, 11, 16, and 24 days after treatment, four plants were harvested, one root was reserved for cross‐section, the remaining roots washed under tap water, the separated roots and shoots were dried at 60°C overnight, and the root and shoot dry weight and length were measured.
2.4. Growing conditions for Experiment 3: tef root visualization using rhizotrons
In experiment 3, seeds of Alba, Quncho, and Tsedey were grown in Plexiglass rhizotrons with dimensions of 20 cm wide, 30 cm high, and 1.5 cm deep. After germination, all plantlets except the one with the most visible root were culled. Water was available from the bottom of the rhizotron or could leak in at the seals on the sides. For the waterlogging treatment, the water level was maintained at 1 cm below the soil surface. The control plants were watered from the bottom and moistened at the top. When not being photographed, the rhizotrons were kept in a box with a lid that protected them from the light.
2.5. Growing conditions for Experiment 4: wheat and maize root visualization using rhizotrons
In experiment 4, bobwhite wheat and Akku maize varieties were obtained from the group of Matthias Erb at the Institute of Plant Sciences at the University of Bern. The seeds of wheat and maize were soaked in 2.5% bleach solution for 3–4 min and then repeatedly rinsed while agitating. They were then planted on wet paper towels in Petri dishes and then transferred to the rhizotrons after 4 days. Treatment was started one week after transplantation into the rhizotron. The waterlogging conditions were the same as for Experiment 3.
2.6. Growing conditions for Experiment 5 (Physiology) and Experiment 8 (RNA‐Seq)
Seedlings of the improved tef variety Tsedey were grown under long‐day conditions (16 h of light at 24°C and 8 h of dark at 18°C, 65% relative humidity) in pots. The seedlings were grown for 19 days and then exposed for 9 days to two conditions (waterlogging and normal watering). Waterlogging was achieved by maintaining the water level at 1 cm below the soil surface. After 9 days of treatment, the stomatal conductance, chlorophyll a and b, and carotenoid content were measured, the plants were harvested, and RNA was extracted from leaf tissue (described below).
2.7. Growing conditions for cross sections at the tillering stage Experiment 6 and Experiment 7 (qPCR)
Alba, Tsedey and Quncho varieties were sown in 7.5‐cm‐diameter pots and grown under long‐day conditions in the growth room as indicated above. Waterlogging treatment (water level maintained at 1 cm below the soil surface) was applied for 9 days to 19‐day‐old plants. After 9 days of growth under control or treatment conditions, the plants were harvested, the wet weight of the root and shoot was measured, the number of adventitious roots per plant was counted, plant material was collected for qPCR, and one or two adventitious roots were taken for cross sections. This procedure was performed in duplicate, both times with 10 or 11 plants (observation 1 and observation 2). For the cross sections, sometimes more than one cross section from the same plant and root location were taken which were considered as technical replicates and hence were not counted as independent observations. At the middle of the root, it was often difficult to get an unblemished cross section. The filenames for the cross sections are named for the plant (T: for Tsedey, Q: for Quncho, or A: for Alba), the number of the plants (1‐17), the location on the root (oben (root base), mitte (middle), or unten (tip)), the replicate (1‐5), and the magnification.
2.8. Physiological measurements
Stomatal conductance of the flag leaf for the adaxial side was determined using an AP4 diffusion porometer (Delta T, Cambridge Life Sciences, Cambridge, UK) at the end of the water stress period. Chlorophyll a and b, as well as carotenoids which comprise carotenes and xanthophylls, were extracted using 95% ethanol and measured with UV–Vis spectroscopy as previously described (Lichtenthaler, 1987). The amount of these pigments was normalized by fresh weight. For the physiological measurements, 10 measurements from 10 different plants were used.
2.9. Statistical analysis for the physiology and growth measurements
Data are presented as means ± standard deviation. Statistical tests were made using R version 3.0.1 using the built‐in function wilcox.test. First, an ANOVA test was used to determine whether significant differences existed. If so, outliers were removed using a modified Thompson tau test. Nonparametric tests (Mann–Whitney U‐test or Kruskal–Wallis H tests) with a p‐value of ≤.05 were used to determine statistical significance between treatment means.
2.10. RNA extraction for RNA‐Seq
RNA was extracted from samples of leaf tissue using the TRIzol kit (ThermoFisher Scientific) according to the supplier's protocol. The quality and quantity of RNA were quantified using ND‐1000 spectrophotometer for which the average 260/280 ratio was 2.0, indicating good‐quality RNA.
2.11. Transcriptome library construction and sequencing for RNA‐Seq
The RNA extracted from plants grown under waterlogging and normal watering conditions was sent to Fasteris (Geneva, Switzerland, www.fasteris.com) for further quality testing and sequencing using Illumina HiSeq2000 with the goal of analysis for differential expression.
Two biologic replicates were made leading to two libraries for control (GNY1 and GNY10) and two from the waterlogged plants (GNY3 and GNY12). The GNY10 and GNY12 libraries were prepared using the AccuPrime polymerase (Invitrogen, Carlsbad, CA) and following the protocol for high GC content. The GNY1a and GNY3a samples were prepared with the TruSeq SBS version 5 kit and the data analysis pipeline consisting of HiSeq Control Software version 1.1.37.8, RTA 1.7.48, and CASAVA 1.7. GNY1b, GNY3b, GNY10, and GNY12 also used the TruSeq SBS version 5 kit and flow cell version 3 with software: HiSeq Control Software version 1.4.8, RTA 1.12.4.2, and CASAVA 1.8.2. The six cDNA libraries were sequenced to generate a total of 205 million single‐end reads. Before assembly, the reads were trimmed such that the Phred quality scores were above 28. In addition, all primer and adaptor sequences detected by FastQC were removed.
2.12. Quantification of gene expression levels and differential expression experiments
The reads from each condition were mapped onto the 14,057 scaffolds of size 1000 bp or greater obtained from the recently sequenced tef genome (Cannarozzi et al., 2014) using STAR 2.3.0 with the default parameters. These were converted to BAM format with SAMtools (Li et al., 2009).
A count table was obtained using the HTSeq‐count program with options stranded=no, type=gene, and attribute=ID (Anders, Pyl, & Huber, 2015) and using the Maker gene predictions provided by the Tef Genome Project (Et_genome_1.0.fasta.gz) (Cannarozzi et al., 2014). HTSeq tabulates the percentage of uniquely mapping reads, reads that map to no feature in the predicted transcriptome, ambiguous reads (map to more than one gene simultaneously), and reads that do not map uniquely to one location. Only uniquely mapping reads were used for counting. The newCountDataSetFromHTSeqCount function of the DESeq package was used to generate the count table used as input into DESeq, a Bioconductor package that estimates the variance–mean dependence of high‐throughput sequence data and tests for differential expression using the negative binomial model (Anders & Huber, 2010).
2.13. Annotation and enrichment analysis
2.13.1. Background set
The transcriptome used in these experiments was from the file Et_all.maker.transcripts.shortids.fasta.gz provided by the Tef Improvement Project. This file contains genes predicted in the genome by Maker and described in (Cannarozzi et al., 2014). To define a background set for the enrichment analysis, the reads were mapped onto the set of 42,052 genes predicted from the tef genomic scaffolds, creating a background set consisting of all 34,761 genes detected in the experiment. The background set was annotated using Blast2GO with the default parameters (E‐value 1.0 e‐03, blastp, non‐redundant database at NCBI). Differentially regulated genes were defined as those found by DESeq to have an adjusted p‐value of less than 0.05 and a fold change of 2 or more. Assignment and clustering of GO terms produced biologic process, molecular function, and cellular component of these differentially regulated terms. Enrichment analysis using Fisher's exact test at α = 0.05 was used to determine genes differentially expressed.
2.13.2. Mercator
Sequences were also placed into MapMan functional categories based on the sequence similarity using the Web interface to Mercator (http://mapman.gabipd.org/web/guest/mercator) (Lohse et al., 2014) using annotated sequences from the following: the Arabidopsis Information Resource (TAIR) (Berardini et al., 2015), SwissProt/UniProt plant proteins (Bateman et al., 2017), TIGR5 rice proteins (Kawahara et al., 2013) and the COG database (Tatusov et al., 2003) and BLAST cutoff 80. Multiple bin assignments were allowed. Enrichment analyses of various pathways using a Wilcoxon rank‐sum test with Benjamini–Hochberg corrections for multiple testing were conducted using a local installation of the MapMan software.
2.14. Verification of expression levels using qPCR
Plant material was collected as described above and was then ground in liquid nitrogen. Total RNA was extracted using the Total RNA Isolation System (Promega, USA) and treated with the DNA‐free kit from Life Technologies, USA. RNA was quantified using a NanoDrop ND‐1000 spectrophotometer (Thermo Scientific, USA). For calculating the relative gene expression, the 2‐ΔΔCq method was used (Livak & Schmittgen, 2001). CYP and PP2A were used as reference genes. Each biologic replicate was sampled three times, and the measured Cq values were averaged.
2.15. Preparation and microscopy of cross sections of adventitious roots
The adventitious roots were cut into three sections of equal length, referred as “base” (closest to the shoot), “middle,” and “tip” (closest to the root tip or apex). These were embedded in 5% agarose and sliced into 100‐ to 200‐μm slices using a Vibratome. No staining or dyes were used. The cross sections were visualized in a Zeiss Axioskop 2 microscope with an AxioCam color (412–312) camera using 5× to 20× magnification. The microscope software was AxioVision Release 4.8.2 SP2 (06‐2012), and ImageJ (Duarte et al., 2016) was also used in the image processing.
2.16. Quantification of aerenchyma
Processing of the cross sections with Photoshop included manually selecting the cortex and aerenchyma.
2.17. Phylogeny
Sequences were aligned using Mafft (L‐INS‐I) (Katoh & Standley, 2013) with the default settings. PhyML (Guindon et al., 2010) was used to obtain a maximum‐likelihood tree using the default model of HKY85 + G. Trees were visualized using FigTree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
The expansin sequences from (Li et al., 2003) were downloaded from the National Center for Biotechnology Information (NCBI) Web site (Coordinators, 2017). The accession numbers are as follows: AF247163.1, AF247164.1, AF247165.1, AF394545.1, AF394546.1, AF394548.1, AF394549.1, AF394550.1, AF394551.1, AF394552.1, AF394553.1, AF394554.1, AF394555.1, AF394556.1, AF394557.1, AF394558.1, AF394559.1, AF394560.1, AF394561.1, AF394562.1, U30477.1, U30479.1, U85246.1, Y07782.1, AF261270.1, AF391105.1, AF391106.1, AF391108.1, AF391110.1, AF391111.1, AF261271.1, AF261272.1, AF261273.1, AF261274.1, AF261275.1, AF261276.2, AF261277.1, AF261278.1, AF466188.1, AY039023.1, AY046928.1, AY046929.1, AY100692.1, AY100693.1, AY100694.1, and U95968.1.
The ribonuclease accession numbers are as follows: AAF82615.1, AAG09465.1, gi|15149819, ref|XP_002311302.1, ref|XP_002311303.1, ref|XP_002316136.1, ref|XP_002321228.1, gb|EEE95823.2, gb|AAM18521.1, gb|AAS01727.1, gb|AAS07016.1, gi|68563425, gi|3860325, ref|XP_001691379.1, gi|113374061, gb|AAB58718.1, gb|AAB58719.1, gb|AAF45043.1, gi|71611076, gb|AAF45022.1, gb|AAM80567.1, gi|133173, gi|31620998, gi|31621000, gi|31621002, gi|116058653, gb|AAA21135.1, gi|976231, gi|976233, gi|18394083, gi|18394085, gi|18396065, gi|18405157, gi|1710615, gi|1710616, gi|116634825, gi|50513542, gi|7707689, gi|1698670, gi|195628852, gi|642956, gb|AAC49326.1, Et_s3159‐0.24‐1, LOC_Os07g43670.1, LOC_Os01g67180.1, LOC_Os08g33710.1, LOC_Os09g36680.1, LOC_Os09g36700.1, LOC_Os01g67190.1, Os07g0629900, Obart07g23070, Orgla07g0180700, Oglum07g23130, Oniva07g22690, Bgiosga026191, Orufi07g24240, Os07g0629300, Opunc07g21760, Obart07g23080, Oglum07g23140, Oniva11g11010, Bgiosga026192, and Orufi07g24250.
2.18. Accession numbers
This project has been archived at GenBank under BioProject PRJNA413657 with BioSamples: GNY1: SAMN07764637, GNY3:SAMN07764639, GNY10:SAMN07764640, and GNY12: SAMN07764642. The reads have been deposited in the GenBank Sequence Read Archive as study SRP119988 with accession numbers: SRR6175533 (GNY1‐1), SRR6175534 (GNY2‐1), SRR6175535 (GNY3‐1), SRR6175536 (GNY1‐2), SRR6175529 (GNY2‐2), SRR6175530 (GNY3‐2), SRR6175531 (GNY10), SRR6175532 (GNY11), and SRR6175528 (GNY12).
3. RESULTS
3.1. Time series on growth of tef shoots under early waterlogging
The time course Experiment 1 was undertaken to measure shoot growth of Alba, Quncho, and Tsedey genotypes under waterlogging and normal watering conditions where plant height was measured at the interval of 2–4 days. The difference between the height of the control plants and those treated with waterlogging varied between the three cultivars in response to waterlogging (Fig. S1). For Alba, the difference between the height of the control plants and those that were waterlogged was not significantly different at any time (Mann–Whitney U‐test, p ≤ .05). The waterlogged Quncho plants were significantly shorter than the control plants at 3 and 7 days after watering, while the waterlogged Tsedey plants were significantly taller than the control plants 1, 3, 4, and 7 days after the onset of stress. At 8 days after waterlogging treatment, the heights of the waterlogged and control plants were no longer significantly different for any cultivar.
3.2. Time series studies on plant development under early waterlogging
Experiment 2 is a repetition of Experiment 1 but with four waterlogged and four control plants harvested at 2, 4, 7, 9, 11, 16, and 24 days after the waterlogging stress was applied. This enabled visualization of root characteristics as well as measurements of root and shoot biomass. The Quncho and Tsedey varieties were chosen for this experiment as their responses to waterlogging in Experiment 1 differed, namely Tsedey grew taller during the onset of waterlogging, while Quncho's growth was suppressed.
At each time point, shoot dry weight, root dry weight, root length and shoot length, and the number of leaves were measured (Figure 1 for all days, Table 2 for day 24, and all raw data in Tables S1 (Quncho) and S2 (Tsedey)). As in Experiment 1, Tsedey grew more robustly than Quncho in the waterlogged environment. The number of replicates in Experiment 2 (three or four replicates) was smaller than that of Experiment 1 (nine replicates) because of the destructive sampling. The small number of replicates combined with the large variance between observations resulted in fewer statistically significant measurements in Experiment 2. On the last day, day 24, all remaining plants were harvested, and their parameters measured. For this reason, there were nine replicates of each condition and genotype on day 24.
Figure 1.
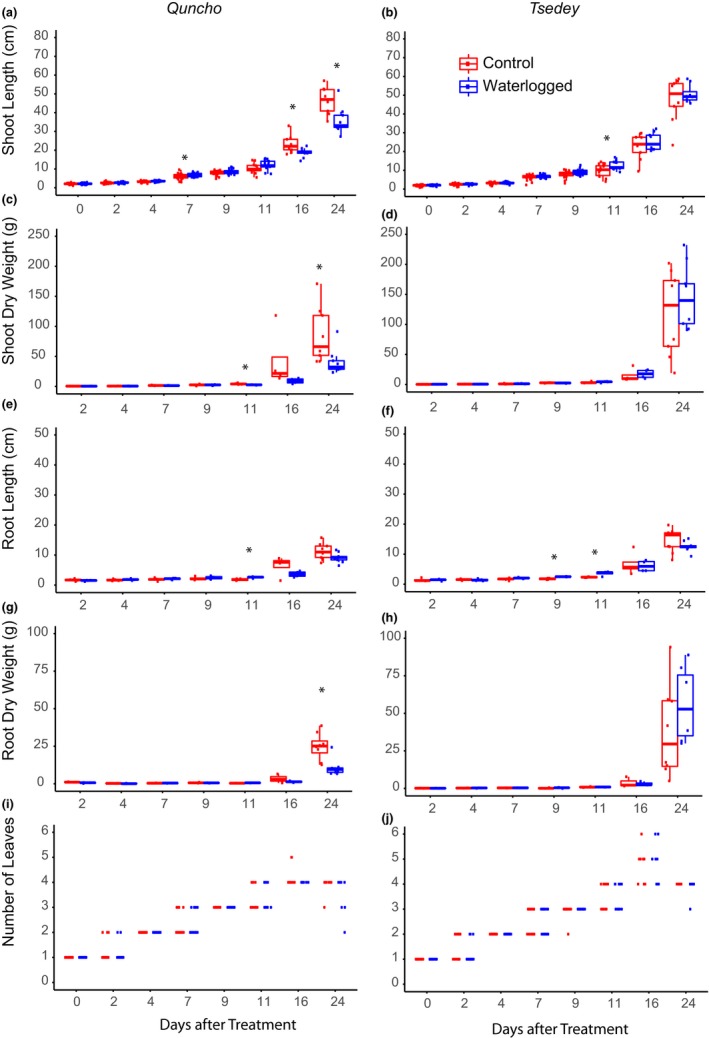
Growth of 4‐day‐old seedlings of two tef genotypes (Quncho and Tsedey) subjected to waterlogging and normal watering. Shoot length (a, b), shoot dry weight per plant (c, d), root length (e, f), root dry weight per plant (g, h), and number of leaves per plant (i, j). Means marked with an asterisk are significantly different from each other (Mann–Whitney: *p ≤ .05)
Table 2.
Growth characteristics of Alba, Quncho, and Tsedey after 24 days of waterlogging at the early growth stage (4 days)
| Parameter | Quncho | Tsedey | ||||
|---|---|---|---|---|---|---|
| Control | Waterlog | % of control | Control | Waterlog | % of control | |
| Shoot length | 46.5 ± 7 | 35.8 ± 7.1 | 76.9* | 47.6 ± 11.6 | 50.6 ± 4.8 | 106 |
| Shoot dry weight | 82.6 ± 48 | 40.4 ± 20 | 48.9* | 118 ± 68 | 145.2 ± 52 | 123 |
| Root length | 11.2 ± 2.7 | 9.1 ± 1.6 | 81.3 | 14.5 ± 3.8 | 12.6 ± 1.7 | 87 |
| Root dry weight | 24.8 ± 9 | 10.8 ± 5.7 | 43.5* | 38.0 ± 31 | 56.1 ± 24 | 148 |
| Number of leaves | 3.9 ± 0.4 | 3.4 ± 0.8 | 87.2 | 4.0 ± 0 | 3.9 ± 0.4 | 97.5 |
Four‐day‐old plants were grown in soil either with normal watering or with water maintained at 1 cm below the soil surface. All values are means (n = 3 or 4) ± SD. Significance differences at p ≤ .05 are denoted with a * (Mann–Whitney).
3.2.1. Shoot growth
In the Tsedey cultivar, the shoot length, measured from the base of the plant to the end of the longest leaf, was in most cases similar in the control and waterlogged plants. However, the waterlogged plants were significantly longer than those of the control plants at 11 days after stress (Figure 1). The shoot dry weight was similar for Tsedey plants under control and waterlogged conditions.
For Quncho, the length of the shoot of the waterlogged plants was significantly smaller than that of the control plants at 7, 16, and 24 days after stress. The waterlogged plants started showing signs of stress (smaller plants with browning and dying leaves) at 16 days after the onset of stress. This resulted in the shoot dry weight of Quncho being significantly lower at 11 and 24 days after stress.
The summary of growth statistics at 24 days after waterlogging is shown in Table 2. After 24 days of waterlogging, Quncho subjected to waterlogging had 49% less shoot dry weight, while the shoot dry weight of Tsedey increased by 23%. In addition, the shoot length of Quncho was negatively affected, while Tsedey consistently grew larger under waterlogged conditions. The number of leaves did not change significantly for either cultivar.
3.2.2. Root growth
The root structure of tef plants contains many fine root hairs. As the plants were grown in soil, it was necessary to wash the root ball before drying and measurement. To remove all soil without damaging, the fine root structure was difficult and added uncertainty to the measurements of root weight. In terms of root length, the roots of Tsedey subjected to early waterlogging were significantly longer at 9 and 11 days after stress, while the roots of Quncho subjected to early waterlogging were significantly shorter at 11 days after treatment (Figure 1). Similarly, Quncho roots had significantly less dry weight at 24 days after waterlogging where they were reduced by 56% compared to that of Tsedey which had increased in weight by 48% (Table 2). The length of the longest root of both Tsedey and Quncho decreased under waterlogging by about 20% (roots taken from Experiment 2). However, it was observed that under waterlogging, the fine roots of the control plants were replaced with significantly shorter, thicker, and whiter adventitious roots in all cultivars studied.
3.3. Root growth of cereal plants waterlogged from 4 days
For better visualization of the roots, in Experiment 3, single plants of Alba, Quncho, and Tsedey were grown in rhizotrons to view the root growth, and indeed, after harvesting, the root ball of Tsedey was sometimes much larger under waterlogging than under control conditions. However, the variation in root growth between plants and experiments was high.
For comparison, the experiment was repeated with Alba, Tsedey, wheat, and maize (Experiment 4). In Tsedey, the control roots were longer, but the waterlogged plants had thicker roots and more adventitious roots than normally watered plants (Fig. S2). The aboveground portion of the control and waterlogged Tsedey plants looked similar (not shown). The root plate of the waterlogged Tsedey plants had a wider root plate, while the roots of the control plants tended to grow longer and toward the bottom of the pot. While the Alba control plants had more roots at every stage than the waterlogged roots, clear adventitious root growth was not visible (not shown). The waterlogged plants were always smaller and had fewer leaves and tillers.
In wheat, after one day of waterlogging, the root systems of the control and waterlogging plants were similar (Fig. S3). However, after 3 days of waterlogging, the control plants had distinctly more roots than the waterlogged plants, and this phenomenon became more pronounced with time. Both control and waterlogged plants grew adventitious roots, but they were more pronounced in the control plants. By 25 days, the control roots filled the rhizotron and the plants looked much healthier than the waterlogged.
In maize, waterlogging was started on plants 7 days after germinated seeds were transplanted into rhizotrons. The difference in root growth becomes apparent one week after waterlogging where the roots of the control plants were longer (Fig. S4) and the roots of the waterlogged plants thicker. After 10 days, the waterlogged roots formed a fine root structure but remained in the upper part of the rhizotron, while the roots of the control plant reached the bottom. At 25 days, the maize roots of the control plant filled the rhizotron.
3.4. Physiological response of tef to waterlogging at the tillering stage
Nineteen‐day‐old seedlings of improved tef variety Tsedey (DZ‐Cr‐37) were exposed to either normal watering or waterlogging for 9 days. Physiological measurements of stomatal conductance, as well as carotenoid, chlorophyll a, and chlorophyll b, were made before the plants were harvested and the tissue sent for sequencing (Experiment 5). Only stomatal conductance was altered significantly under waterlogging conditions (Figure 2). The difference was large, however, as stomatal conductance was roughly three times higher under waterlogging conditions than controls.
Figure 2.
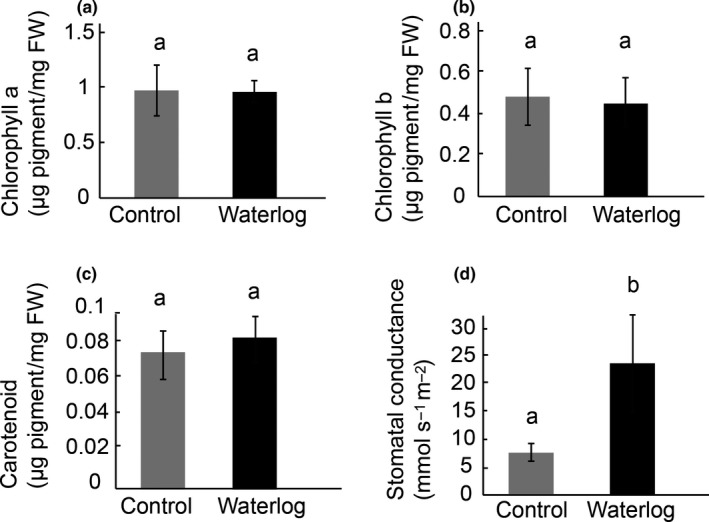
Physiological response of tef plants to waterlogging. The amounts of chlorophyll a (a), chlorophyll b (b), and carotenoid (c), as well as the stomatal conductance (d), were measured for waterlogged and control plants. Treatments marked with different letters have values that are significantly different (Mann–Whitney, p ≤ .05)
3.5. Aerenchyma formation in roots under waterlogging at the tillering stage
Experiment 6 was conducted to observe aerenchyma formation at the tillering stage, 19‐day‐old seedling subjected to 9 days of waterlogging or normal watering. Cross sections of extracted adventitious roots were taken to observe whether aerenchyma were being formed (Figure 3: summary, Fig. S5: all photographs). The number of roots successfully cross‐sectioned ranged from 1 to 3 per genotype and condition. The percentage of cortex area containing aerenchyma in the base, middle, and tip of the root is summarized in Figure 4. All three cultivars had some aerenchyma under normal watering conditions with the number of aerenchyma increasing under waterlogging. The middle of the roots contained more aerenchyma than the base of the root, while no aerenchyma were observed in the root tips of any cultivar under either waterlogged or control condition. Quncho and Tsedey exhibited constitutive aerenchyma, the size of which increased dramatically upon waterlogging. Alba had little constitutive aerenchyma at the base but was able to form them upon excessive moisture stress.
Figure 3.
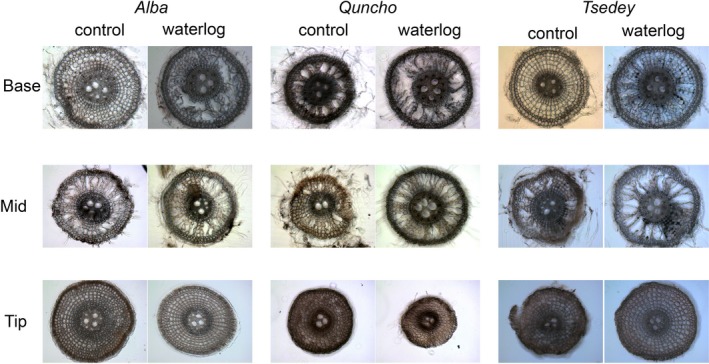
Formation of aerenchyma in adventitious roots of three tef genotypes at the tillering stage. Unstained cross sections of base, middle, and tip of roots of Alba, Tsedey, and Quncho genotypes subjected to 9 days of waterlogging at the tillering stage. All pictures are on the same scale. Scale bar: 100 μm
Figure 4.
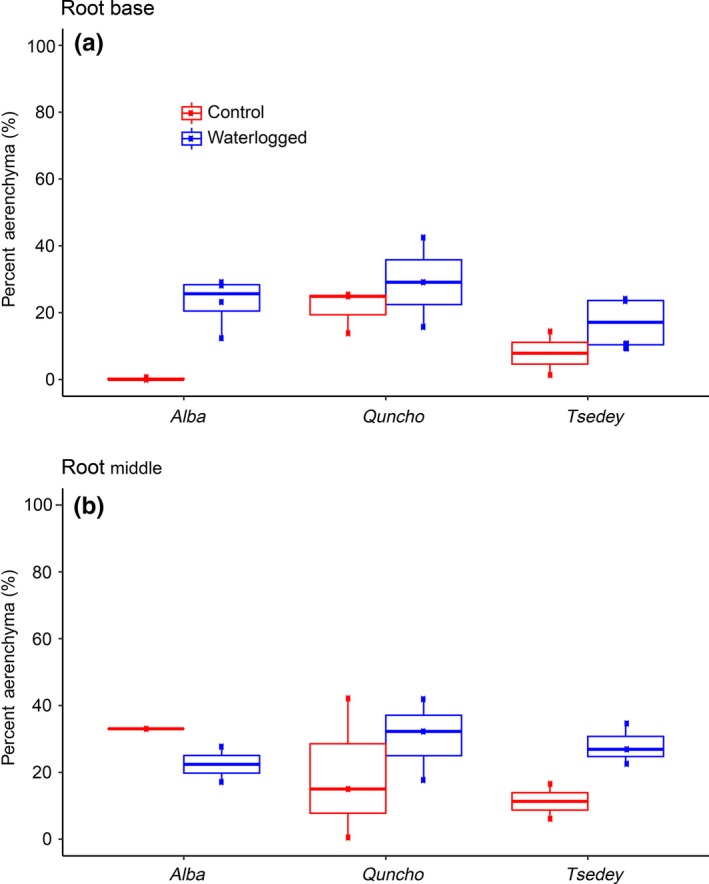
Percentage of cortex occupied by aerenchyma in three cultivars under control and waterlogged conditions. The percentage of aerenchyma in Alba, Quncho, and Tsedey was measured at the root base (a) and middle (b) under control and waterlogged conditions. No aerenchyma were present in the root tip
In addition, the number of adventitious roots was counted for replicates of this experiment. For all three genotypes, the number of adventitious roots increased with waterlogging. The variance was high, however, from 109% in Alba to 336% in Tsedey (Table S3).
The roots of one replicate from each day of Experiment 2 (waterlogging of 4‐day‐old plants for 22 days) were used to visualize the formation of aerenchyma of Quncho and Tsedey with samples taken at 2, 4, 7, 9, 16, and 24 days after waterlogging (Fig. S6). Quncho was the earliest to show aerenchyma formation which appeared only 4 days after waterlogging, while in Tsedey they were first observed 7 days after stress.
3.6. RNA‐Seq study of differential expression at the tillering stage
Changes in gene expression in leaves of the Tsedey genotype (Experiment 7) were measured using RNA‐Seq (pipeline is shown in Fig. S7). For transcriptome sequencing, a total of 132,416,127 sequence reads were generated with Illumina sequencing (Table 3). These reads were filtered for adaptor and vector sequences and trimmed such that the Phred score was greater than 30 across the sequence.
Table 3.
RNA‐Seq data collected and mapping statistics
| Treatments | Library | Length after trimming (bp) | Number of reads | Number of bases after trimming (bp) | Percentage uniquely mapped (%) | Percentage of reads mapped to multiple loci | Percentage unmapped |
|---|---|---|---|---|---|---|---|
| Normal watering | GNY1 | 70 | 6,817,359 | 477,215,120 | 67.5 | 18.5 | 14.0 |
| GNY1b | 90 | 18,854,229 | 1,696,880,610 | 70.6 | 15.2 | 14.2 | |
| GNY10 | 90 | 35,853,151 | 3,226,783,590 | 75.3 | 19.2 | 5.5 | |
| Subtotal | 61,524,739 | 5,400,879,320 | |||||
| Waterlogging | GNY3 | 70 | 9,413,900 | 658,973,000 | 68.3 | 15.6 | 14.6 |
| GNY3b | 90 | 23,625,313 | 2,126,278,170 | 71.7 | 15.4 | 12.9 | |
| GNY12 | 90 | 37,852,175 | 3,406,695,750 | 75.1 | 19.3 | 5.5 | |
| Subtotal | 70,891,388 | 6,191,946,920 | |||||
| Total | 132,416,127 | 11,592,826,240 | |||||
3.6.1. Annotation of the background set using Blast2GOand Mercator
Although the Tef Genome Project provided annotations, only a small percentage of sequences had gene ontology (GO) annotations, which are helpful for enrichment analysis. Therefore, the background genes were reannotated with descriptions and GO terms, InterPro protein signatures (Finn et al., 2017), and KEGG pathways (Kanehisa, Furumichi, Tanabe, Sato, & Morishima, 2017) using Blast2GO (Conesa & Götz, 2008). These annotations are available in Table S4 and at http://www.tef-research.org/genome.html. For the background set, the top hit was from Setaria italica for 14,777 proteins followed by Sorghum bicolor (5,436 proteins), Zea mays (4,571 proteins), and Oryza sativa (2,771 proteins).
GO terms were assigned via Blast2GO using the GOSlim plant annotations resulting in 27,338 proteins annotated with GO terms in the background set (Fig. S8). For the biologic process, the GO terms “metabolic process,” “cellular process,” “single‐organism process,” “biologic regulation,” and “response to stimulus” were the five most represented categories. The molecular function category was dominated by “catalytic activity” and “binding” with “transporter activity” also being well represented. The cellular components most often found were “cell,” “organelle,” and “membrane”.
In addition, the background set was annotated with pathways using the KEGG annotation system in Blast2GO. Prediction of the pathways containing the background proteins resulted in 127 pathways containing 8,768 proteins (25.2% of the genes detected in the experiment were classified). The pathways including the most transcripts were “purine metabolism,” “starch and sucrose metabolism,” “phenylpropanoid biosynthesis,” “phenylalanine metabolism,” “pyrimidine metabolism,” and “glycolysis and glucogenesis.”
3.6.2. Annotation of the background set using Mercator
Mercator, a Web application that assigns DNA or protein sequences to one of 35 MapMan bins (Lohse et al., 2014), was also used for annotation of the background set. MapMan categorization provides an overview of metabolism and cellular process and is tailored for functional annotation of plants (Thimm et al., 2004). Many of these categories deal with metabolic pathways and enzyme functions, providing an overview of the metabolic networks involved in the response. The results of the mapping of the background set revealed the categories: transport (11.7%), cell wall (10.0%), protein (8.3%), and cell (8.3%) to be the most represented functional categories (Fig. S9).
3.6.3. Differential expression
Differential expression analysis was performed to determine the change in transcript expression from the waterlogging treatment to the control (normal watering) (Fig. S7). First, the reads were mapped to the genome using STAR aligner (Dobin et al., 2013). A count table was generated for each predicted gene from the tef genome for each sequencing dataset using HTSeq (Anders et al., 2015), which was given the genome and the predicted locations of the genes provided by the tef genome sequencing (Cannarozzi et al., 2014). Only the roughly 70% of the reads that map uniquely to one location in the genome were used for counting (Table 3). Count tables generated by HTSeq were then used as input into DESeq (Anders & Huber, 2010). Differentially expressed genes were defined as those with an adjusted p‐value of ≤.05 and with a fold change in expression greater than two and are tabulated in Tables 4, 5, and S5. Downregulation was defined as fewer transcripts in the treatment compared to control and the opposite for upregulated. Differential expression analysis identified 37 genes significantly upregulated and 19 genes significantly downregulated under waterlogging conditions. Tables 4 and 5 containing the differentially regulated genes are listed in the MapMan category number.
Table 4.
Genes upregulated under flooding, their Blast2GO and MapMan annotations, their fold change, and p‐value organized using the MapMan categories
| Name | Blast2GO description | Mercator function | Fold change | Log2 fold change | Adj. p‐valuea |
|---|---|---|---|---|---|
| Photosynthesis | |||||
| Et_s1919‐1.19 | Ribulose bisphosphate carboxylase oxygenase activase chloroplast‐like RuBisCO | 1.3.13 PS.calvin cycle.rubisco interacting | 8.33 | 3.06 | .0178 |
| Major carbohydrate metabolism | |||||
| Et_s6672‐0.28 | Granule‐bound starch synthase ii | 2.1.2.2 major CHO metabolism.synthesis.starch.starch synthase | 11.34 | 3.50 | .0001 |
| Et_s66‐0.13 | Granule‐bound starch synthase ii | 2.1.2.2 major CHO metabolism.synthesis.starch.starch synthase | 5.23 | 2.39 | .0231 |
| Et_s2233‐0.29 | Glucose‐6‐phosphate phosphate translocator precursor | 2.2.2.1.2 major CHO metabolism.degradation.starch.starch cleavage.β amylase | 10.57 | 3.40 | .0002 |
| Et_s2217‐0.39 | β‐amylase | 2.2.2.1.2 major CHO metabolism.degradation.starch.starch cleavage.Β‐ amylase | 15.70 | 3.97 | |
| Et_s7847‐0.25 | NADP‐dependent malic enzyme | 8.2.10 TCA/org transformation.other organic acid transformatons.malic’ | 6.46 | 2.69 | .0042 |
| Growth | |||||
| Et_s2486‐1.7 | β‐expansin 1a precursor | 10.7 cell wall.modification | 11.60 | 3.54 | .0001 |
| Et_s5792‐0.0 | β‐expansin 1a precursor | 10.7 cell wall.modification | 8.58 | 3.10 | .0231 |
| Et_s5823‐0.14 | Xyloglucan endotransglucosylase hydrolase protein 8 precursor |
10.6.2 cell wall.degradation.mannan‐xylose‐arabinose‐fucose 10.7 cell wall.modification |
6.01 | 2.59 | .0171 |
| Et_s9915‐0.7 | β‐expansin 1a isoform ×1 | 10.7 cell wall.modification | 5.90 | 2.56 | .0304 |
| Et_s11804‐0.16 | Probable xyloglucan endotransglucosylase hydrolase‐like | 10.7 cell wall.modification | 6.42 | 2.68 | .0108 |
| Metal handling and acquisition | |||||
| Et_s12869‐0.19 | Ferric reduction oxidase chloroplast‐like | 15.1 metal handling.acquisition | 6.46 | 2.69 | .0068 |
| Et_s13065‐0.28 | Ferric reduction oxidase chloroplast‐like | 15.1 metal handling.acquisition | 6.24 | 2.64 | .0444 |
| Et_s1056‐0.47 | Heavy metal‐associated domain‐containing expressed | “35.2” “not assigned.unknown” | 12.43 | 3.64 | .0009 |
| Secondary metabolism | |||||
| Et_C8513699‐0.0 | Transresveratrol di‐o‐methyltransferase‐like | 16.2 secondary metabolism.phenylpropanoids | 8.81 | 3.14 | .0311 |
| Et_s20148‐0.9 | Cinnamoyl reductase 1‐like | 16.2.1.7 secondary metabolism.phenylpropanoids.lignin biosynthesis.CCR1 | 17.5176 | 4.13 | .0132 |
| Et_s517‐0.16 | Phytoene synthase 2 | 16.1.4.1 secondary metabolism.isoprenoids.carotenoids.phytoene synthase | 6.73 | 2.75 | .0311 |
| Et_s4037‐1.43 | GDP‐l‐galactose phosphorylase 2‐like | “21.2.1.2” “redox.ascorbate and glutathione.ascorbate.GDP‐L‐galactose‐hexose‐1‐phosphate guanyltransferase” | 5.94 | 2.57 | .0077 |
| Et_s786‐1.34 | PAP‐specific phosphatase mitochondrial‐like | “23.2” “nucleotide metabolism.degradation” “et_s786‐1.34‐1” | 7.08 | 2.82 | .0328 |
| Et_s4382‐0.38 | Short‐chain dehydrogenase tic chloroplast‐like | “26.22” “misc.short chain dehydrogenase/reductase (SDR)” | 11.18 | 3.48 | .0077 |
| RNA regulation | |||||
| Et_s7183‐0.17 | Salt tolerance‐like protein | “27.3.7” “RNA.regulation of transcription.C2C2(Zn) CO‐like, Constans‐like zinc finger family” | 7.52 | 2.91 | .0011 |
| Et_s6551‐0.36 | Salt tolerance‐like protein | “27.3.7” “RNA.regulation of transcription.C2C2(Zn) CO‐like, Constans‐like zinc finger family” | 5.61 | 2.49 | .0180 |
| Protein functions | |||||
| Et_s3682‐2.26 | Pentatricopeptide repeat‐containing protein at5 g25630‐like | “29.4” “protein.postranslational modification” | 7.77 | 2.96 | .0055 |
| Et_s409‐2.7 | Carboxyl terminal‐processing peptidase chloroplastic | “29.5.5” “protein.degradation.serine protease” | 8.72 | 3.12 | .0062 |
| Et_s2871‐0.32 | C‐terminal processing chloroplastic‐like | “29.5.5” “protein.degradation.serine protease” | 6.09 | 2.61 | .0399 |
| Et_s1636‐1.49 | BTB POZ and TAZ domain‐containing protein 3 isoform ×1 | “29.5.11.4.5.2” “protein.degradation.ubiquitin.E3.BTB/POZ Cullin3.BTB/POZ” | 7.17 | 2.84 | .0399 |
| Signaling | |||||
| Et_s781‐0.29 | Early light‐induced protein | “30.11” “signalling.light” | 10.54 | 3.40 | .0077 |
| Cell division | |||||
| Et_s10318‐0.21 | “31.2” “cell.division” | 10.56 | 3.41 | .0018 | |
| Et_s7686‐1.48 | “31.2” “cell.division” Regulator of chromosome condensation (RCC1) family protein; | 8.67 | 3.12 | .0075 | |
| Transport | |||||
| Et_s7784‐0.11 | Magnesium proton exchanger isoform 1 | “34.12” “transport.metal” | 21.45 | 4.42 | .0055 |
| Et_s6198‐0.9 | High‐affinity nitrate transporter‐like | “34.4” “transport.nitrate” “et_s6198‐0.9‐1” | 12.43 | 3.64 | .0149 |
| Et_s594‐1.9 | High‐affinity nitrate transporter‐like | “34.4” “transport.nitrate” | 10.09 | 3.33 | .0221 |
| Unknown | |||||
| Et_s90‐1.51 | “35.2” “not assigned.unknown” | 29.46 | 4.88 | .0098 | |
| Et_s304‐1.36 | Late embryogenesis abundant protein lea5‐d‐like | “35.2” “not assigned.unknown” | 6.70 | 2.74 | .0106 |
| Et_s1487‐0.5 | “35.2” “not assigned.unknown” | 8.55 | 3.10 | .0355 | |
| Et_s2486‐1.2 | β‐expansin 1a precursor | “not assigned.unknown” “et_s2486‐1.2‐1” | 6.4651 | 2.69 | .0373 |
| Et_s1663‐0.25 | “35.2” “not assigned.unknown” | 5.33 | 2.41 | .0378 | |
Adjusted p‐value from HTSeq.
Table 5.
Genes downregulated under flooding, their Blast2GO and MapMan annotations, their fold change, and p‐value organized using the MapMan categories
| Name | Blast2GO description | Mercator function | Fold change | Log2 fold change | Adj. p‐valuea |
|---|---|---|---|---|---|
| Et_s2617‐0.51 | Aldo‐keto reductase family 4 member c9 | “3.5” “minor CHO metabolism.others” | 0.190 | −2.39 | .0493 |
| Et_s1215‐0.33 | Malate glyoxysomal | “6.2” “gluconeogenesis/glyoxylate cycle.malate synthase” | 0.106 | −3.23 | .0089 |
| Cofactor and vitamin metabolism | |||||
| Et_s4931‐1.11 | Phosphomethylpyrimidine chloroplastic‐like isoform ×1 | “18.2” “Co‐factor and vitamine metabolism.thiamine” | 0.078 | −3.67 | .0082 |
| Et_s780‐1.30 | Thiazole biosynthetic enzyme thi4 family | “18.2” “Co‐factor and vitamine metabolism.thiamine” | 0.124 | −3.01 | .0231 |
| Abiotic stress | |||||
| Et_s346‐4.13 | Bifunctional nuclease 2‐like isoform ×1 | “20.2.4” “stress.abiotic.touch/wounding” | 0.022 | −5.49 | .0000 |
| Et_s11116‐0.0 | Chaperone protein 1‐like | “20.2.1” “stress.abiotic.heat” | 0.016 | −5.90 | .0000 |
| Et_s3654‐1.34 | Bifunctional nuclease 2‐like isoform ×1 | “20.2.4’ “stress.abiotic.touch/wounding” | 0.072 | −3.79 | .0007 |
| Et_s3572‐0.37 | Thioredoxin‐like 3‐chloroplastic | “21.1” “redox.thioredoxin” | 0.080 | −3.63 | .0004 |
| Et_s190‐0.32 | Class III peroxidase | “26.9” “misc.glutathione S transferases” | 0.116 | −3.11 | .0011 |
| Regulation/signaling | |||||
| Et_s2666‐0.25 | TUBBY‐like f‐box protein 7‐like | “27.3” “RNA.regulation of transcription” | 0.133 | −2.91 | .0020 |
| Et_s3159‐0.24 | RNase s‐like protein precursor | “27.1.19” “RNA.processing.:qucleases” | 0.090 | −3.47 | .0399 |
| Et_s521‐0.23 | GTP‐binding protein hflx | “30.5” “signalling.G‐proteins” | 0.084 | −3.57 | .0009 |
| Et_s788‐0.23 | “31.1” “cell.organisation” | 0.032 | −4.94 | .0000 | |
| Et_s4931‐1.10 | “31.1” “cell.organisation” | 0.029 | −5.09 | .0009 | |
| Transport | |||||
| Et_s819‐0.0 | Aquaporin tip4‐2 | “34.19.2” “transport.Major Intrinsic Proteins.TIP” | 0.133 | −2.90 | .0088 |
| Et_s4179‐0.34 | Aquaporin tip4‐1 | “34.19.2” “transport.Major Intrinsic Proteins.TIP” | 0.146 | −2.77 | .0053 |
| Unknown | |||||
| Et_s5368‐0.25 | Single‐strand binding protein | “35.2” “not assigned.unknown” | 0.084 | −3.56 | .0328 |
| Et_s482‐0.32 | “35.2” “not assigned.unknown” | 0.085 | −3.55 | .0387 | |
Adjusted p‐value from HTSeq.
3.6.4. Functional classification of genes differentially expressed under waterlogging stress
Mercator was also used to classify the differentially regulated sequences into the 35 MapMan functional plant categories (Fig. S10) (Thimm et al., 2004). Under waterlogging, the upregulated categories included bin 10: cell wall (15.8%), bin 29: protein (10.5%), bin 34: transport (10.5%), bin 2: major carbohydrate metabolism (7.9%), and bin 16: secondary metabolism (7.9%) with 15.8% unassigned. The downregulated categories included bin 20: stress (13.64%), bin 31: cell (13.64%), bin 34: transport (13.64%), bin 27: RNA (9.1%), bin 18: cofactor and vitamin metabolism (9.1%), bin 6: glyconeogenesis (4.6%), bin 3: minor CHO metabolism (4.55%), bin 21: redox (4.55%), and bin 30: signaling (4.55%) with the classification of 9.1% as unassigned.
3.7. Validation of the RNA‐Seq expression values
From the genes determined to be significantly differentially expressed under waterlogging conditions using RNAS‐Seq on the Tsedey genotype, four genes were chosen for experimental validation with qPCR using Alba, Quncho, and Tsedey tef genotypes. These include two genes that were upregulated, a cinnamoyl reductase‐like gene and a granule‐bound starch synthase gene, as well as two that were downregulated, a bifunctional nuclease 2‐like gene and an aquaporin. Two genes, serine/threonine–protein phosphatase (PP2A) and cyclophilin/peptidyl‐prolyl Isomerase (CYP), have been identified as the most stable for use as reference genes in qPCR of abiotic stress response genes in sorghum (Reddy et al., 2016). All measurements were taken using both reference genes. Comparisons between the RNA‐Seq expression measurements in Tsedey and the qPCR expression in Tsedey, Alba, and Quncho are shown in Figure 5 and Table S6. All four genes were regulated in the same direction with similar magnitudes. The use of the two reference genes resulted in similar behavior except in the Alba genotype for the aquaporin gene and for granule‐bound starch synthase (Figure 5).
Figure 5.
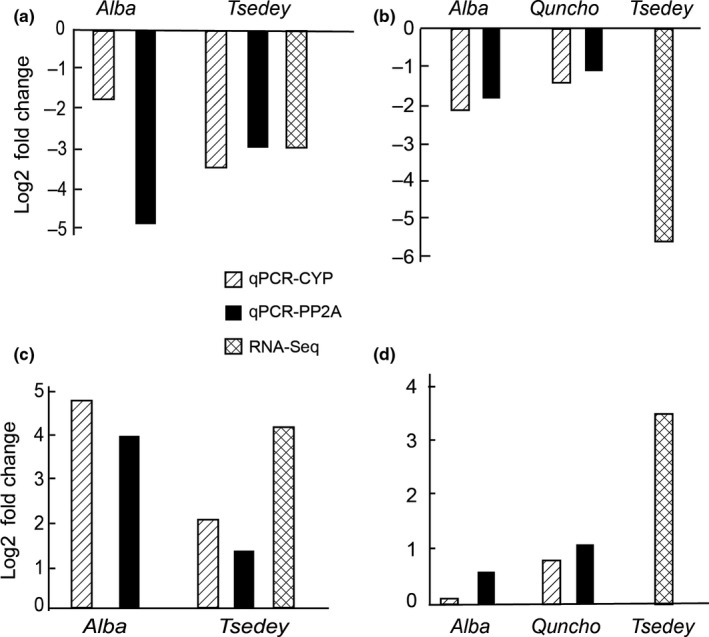
Validation of RNA‐Seq expression measurements by qPCR in three tef genotypes. Aquaporin (a), bifunctional nuclease 2‐like gene (b), cinnamoyl reductase‐like gene (c), and granule‐bound starch synthase (d) were regulated under waterlogging conditions using RNA‐Seq. Expression measurements for these four transcripts were verified with qPCR, using either CYP or PP2A as reference genes. Three replicates were used for each genotype, condition, and transcript
4. DISCUSSION
Eight experiments were carried out to probe the morphology of roots, growth of shoots and roots, and gene expression. The experiments were carried out either on 4‐day‐old seedlings with parameter measurement every 2–4 days for up to 24 days (early waterlogging) or on plants at the tillering stage (19‐day‐old plants which were then waterlogged for 9 days).
4.1. Growth
The Tsedey genotype grew faster under early waterlogging conditions, while the growth of Quncho was suppressed (Figure 1 and S1). Measurements of Tsedey seedling growth over time showed that significant differences in growth occurred in the first week of waterlogging. After 22 days of waterlogging, the shoot dry weight of Tsedey was 123% larger under waterlogging, while for Quncho it was reduced to 49% of the control length (Table 2). The largest difference between Tsedey and Quncho tef genotypes was seen in the roots, where Tsedey consistently grew a larger root mass when waterlogged. The root length decreased for both genotypes, but the number of adventitious roots increased for both, resulting in a denser root mass. The root dry weight was significantly increased in Tsedey by 148%, while in Quncho it was reduced to 48%. A part of the difference may be attributed to the fact that thicker roots are less likely to be damaged or removed when washing the root ball. In a similar study, the dry weight of the flooding‐tolerant Zea nicaraguensis decreased by 54%, while in the flooding‐susceptible Zea mays, the dry weight was reduced by 60% (Abiko et al., 2012). A global increase in biomass under waterlogging conditions is often associated with waterlogging adaptation (Naidoo & Naidoo, 2015) as is the ability to produce adventitious roots (Steffens, Wang, & Sauter, 2006). The number of leaves was not affected, but the leaves of Quncho started browning after 16 days, a sign of waterlogging stress.
4.2. Aerenchyma formation at the tillering stage
Cross sections of three tef genotypes (Alba, Quncho, and Tsedey) revealed the amount of aerenchyma in the root tip, middle, and base of the roots under conditions of waterlogging and normal watering (Figure 3 and Fig. S5). No genotype formed aerenchyma in the root tips under any condition, as was shown for Z. mays (Abiko et al., 2012). Alba had aerenchyma only in the middle of the root under control conditions where after 9 days of waterlogging, aerenchyma were induced in the base and the number and size of aerenchyma in the middle of the root increased. Quncho had more and larger constitutive aerenchyma than Tsedey, especially at the base of the root. However, both cultivars developed larger aerenchyma upon waterlogging in both the middle and the base of the root. In a similar study, Zea mays roots formed a negligible amount of aerenchyma in drained soil which increased to 15% by area under waterlogging. The roots of Z. nicaraguensis, a waterlogging‐tolerant species, had 22% aerenchyma by area, which increased to 29% under waterlogging (Abiko et al., 2012). Aerenchyma are induced by waterlogging but are found constitutively in wetland or waterlogging‐adapted species (Abiko et al., 2012; Justin & Armstrong, 1987). A QTL for aerenchyma formation has been identified in barley, and the newly developed markers explain 44% of the phenotypic variance (Zhang et al., 2016), while a recently discovered allele in wild barley is said to account for 76.8% of the phenotypic variance (Zhang et al., 2017).
4.3. Physiological response at the tillering stage
Measurements of the seedling growth over time show that the largest changes in growth occurred directly after the application of the stress, while in this experiment the tissue was collected 9 days after the application of the stress. For the physiological measurements, no significant difference was found in chlorophyll a, chlorophyll b, or carotenoid which may indicate that the plants were not stressed to a large extent or had already adapted to the stress (Figure 2). Stomatal conductance, however, increased threefold under flooding. Caudle and Maricle suggested that the characteristics of plants that correlated best with tolerance to flooding were photosynthesis rate, respiration upon flooding, and the ability to avoid oxygen shortage (Caudle & Maricle, 2012). Leaf‐level stomatal conductance was also suggested to be an indicator of flooding tolerance in comparison with the responses of the flooding‐sensitive Johnson grass (Sorghum halepense) and the flooding‐tolerant common reed (Phragmites australis). In the sensitive Sorghum halepense, transpiration decreased in response to flooding, while in the tolerant Phragmites australis, the opposite occurred (Waring & Maricle, 2013). A study in tolerant versus sensitive wheat concluded that a significant reduction in grain and straw yield happened only if the waterlogging was prolonged for more than 20 days (Arduini, Orlandi, Pampana, & Masoni, 2016).
4.4. Differentially expressed genes under waterlogging
A total of 56 genes were found to be regulated under waterlogging at the tillering stage (Tables 4, 5 and S5). The small number of genes for which significant changes in expression were found between normal and waterlogging conditions in Tsedey contrasts with studies in Brassica napus L. roots where 4432 genes changed expression after 12 h of waterlogging (Zou et al., 2013), while in Jatropha 1968 mRNA transcripts had a significant change in abundance (Juntawong et al., 2014). Similarly, in cypress tree (Taxodium “Zhongshansa 406”), 2090 differentially expressed genes were found in roots, while only 394 were found in shoots (with 174 shared between the two groups). The RNA used in the current study was extracted from seedlings (not including the roots) 9 days after waterlogging at the tillering stage, which may explain the relatively small number of genes detected.
The differentially regulated genes are ordered by their MapMan annotation bins in Tables 4 and 5 and are discussed in this order. The distribution of the functional categories of the regulated genes shows that after 19 days of waterlogging, the MapMan bins with the most upregulated genes were 15.8% cell wall, 11% transport, 11% protein (amino acid activation, synthesis, targeting, modification, and degradation), major carbohydrate metabolism, and secondary metabolism. The most downregulated bins were as follows: 14% cell (cell organization, division, cycle, and vesicle transport), 14% stress, 14% transport, 9% miscellaneous, 9% RNA, and 9% cofactor/vitamin metabolism (Fig. S10).
4.4.1. Photosynthesis
In the expression experiments, RuBisCO activase (ribulose bisphosphate carboxylase oxygenase activase), Et_s919‐1.19, was upregulated under flooding conditions in Tsedey. RuBisCO is important in the first step of fixing carbon dioxide into organic carbon molecules in the Calvin cycle. The principal role of RuBisCO activase is to release inhibitory sugar phosphates from the active sites of RuBisCO (Jordan & Chollet, 1983) and it also acts as a chaperone during stress (Rokka, Zhang, & Aro, 2001). In tomato, RuBisCO activase has been found to decrease under flooding stress and is susceptible to degradation from reactive oxygen species (Ahsan et al., 2007).
4.4.2. Cell wall
Both β‐expansins (Et_s2486‐1.7, Et_2486‐1.2, Et_s5792‐0.0, Et_s9915‐0.7) and xyloglucan endotransglucosylase/hydrolases (XTHs) (Et_s5823‐0.14, Et_s11804‐0.16) were upregulated in Tsedey. Scaffold2486 contains two tandem genes annotated by Blast2GO as expansin genes, in which only one was identified as an expansin by MapMan. With only two exons, the ambiguous gene looks incomplete, supporting the annotation of MapMan which annotates it as a gene of unknown function (Table 4). Under flooding conditions, rapid elongation of internodes and petioles is often observed as plants use an escape strategy to try to reach an environment with a better oxygen supply. Expansins and XTHs are involved in breaking down the cell walls to allow rapid expansion and growth (Li et al., 2002; Rose, Braam, Fry, & Nishitani, 2002; Sampedro & Cosgrove, 2005; Tsuchiya, Satoh, & Iwai, 2015) and have been shown to contribute to rapid coleoptile growth in rice (Miro & Ismail, 2013). One β‐expansin has been shown to induce the extension of maize coleoptiles (Cosgrove, Bedinger, & Durachko, 1997). Expansins have also been implicated in the formation of adventitious roots in the loblolly pine (Pinus taeda) (Hutchison, Singer, McInnis, Diaz‐Salaz, & Greenwood, 1999) and deepwater rice (Cho & Kende, 1997). The action of XTHs involves breaking linkages in the xyloglucan–cellulose network and then reforming them in a new position, allowing expansion without impairing overall wall integrity. XTH transcripts have been shown to increase in maize roots after 12 h of hypoxia due to flooding (Saab & Sachs, 1996).
Lysigenous aerenchyma result from the death of certain cells in the root cortex and are found in crops such as wheat, rice, barley, and maize (Shiono, Takahashi, Colmer, & Nakazono, 2008). Ethylene accumulation has been implicated as the primary signal in the formation of lysigenous aerenchyma (Shiono et al., 2008). This triggers a signal transduction pathway involving phosphoinositides and Ca2+ (Drew, He, & Morgan, 2000). Although no genes in this pathway have been detected to have differential expression under waterlogging, an XTH is upregulated (Table 4). An XTH has also been found to be associated with aerenchyma formation (Saab & Sachs, 1996). In maize, a mechanism for cortical cell‐specific programmed cell death has been proposed that includes the generation of ROS in combination with suppression of a metallothionein ROS scavenger. The consequent buildup of ROS leads to cell death and aerenchyma formation (Yamauchi, Rajhi, & Nakazono, 2011).
Addition of the regulated tef expansin sequences to the phylogenetic tree of Li et al. (2003) shows that the regulated tef sequence is of type β1, the most closely related to the Li sequences EXPβ1.10 which forms a clade with EXPβ1.7, EXPβ1.8, EXPβ1.9, and EXPβ1.11 (Fig. S11). Expression of β‐expansins has been correlated with internodal elongation in deepwater rice (Lee & Kende, 2001).
Peroxidases also affect cell wall extensibility. Correlations between the decrease in cell wall‐bound peroxidase activity and coleoptile elongation have been observed (Ismail, Ella, Vergara, & Mackill, 2009; Lee & Lin, 1995). Peroxidase activity has been found to be significantly higher in flooding‐sensitive genotypes of rice. Indeed, in the current study, a class III peroxidase, Et_s190.0.32, is also downregulated.
4.4.3. Changes in carbohydrate metabolism
The MapMan category primary metabolism includes the metabolism of amino acids, nitrogen metabolism, sugar and its derivatives, and energy. Plant roots subjected to flood stress switch from aerobic respiration to fermentative metabolism, requiring soluble sugar, which is produced from carbohydrates stored as starches. Starch breakdown is the result of the action of the hydrolytic enzymes alpha amylase, β‐amylase, debranching enzyme, and alpha glucosidase (Guglielminetti, Yamaguchi, Perata, & Alpi, 1995). Several genes that function in the metabolism of sugar and its derivatives were upregulated under water stress. One is granule‐bound starch synthase (Et_s66‐0.13‐1 and Et_s6672‐0.28‐1), an enzyme that converts ADP‐glucose to amylose. This gene is more upregulated in Tsedey than in Quncho or Alba (Figure 5). Another upregulated gene is Et_2217‐0.39‐1, β‐amylase, which is active in the conversion of starch to maltose. Amylase activity has a positive correlation with both shoot and root elongation and with plant survival when comparing tolerant and non‐tolerant rice genotypes (Ismail et al., 2009). Upregulation of β‐amylase transcripts under waterlogging stress has also been observed in Rumex (van Veen et al., 2013).
The glucose‐6‐phosphate translocator precursor (Et_s2233‐0.29) is homologous to rice Os10g33920 and Os08g01410 and is also upregulated under waterlogging stress in tef. The expression of Glc6P/phosphate translocator 2 (GPT2) has been found to have a crucial role in the seedling response to exogenous sugars. Sugar transporters tend to be upregulated in shoots but downregulated in roots under flooding in model plant Arabidopsis (Dyson, Webster, & Johnson, 2014). Under the category minor carbohydrate metabolism, Et_s2617‐0.51 (aldo‐keto reductase family 4 member c9) was found to be downregulated. This is a member of a family of proteins with roles in metabolism and stress response.
4.4.4. Metal handling
Excess water and oxygen depletion in the soil cause a sharp drop in the soil redox potential, creating reducing conditions and resulting in a buildup of Fe II and Mn II (Shabala, 2011). Et_s12869‐0.19 and Et_s13065‐0.28, annotated as chloroplast‐like ferric reduction oxidases, involved in the reduction of Fe III to Fe II, and maintaining iron homeostasis, were found to be upregulated. Other upregulated genes involving metals include Et_s1056‐0.47, a heavy metal‐associated domain‐containing protein often found in proteins involved in heavy metal transport or detoxification, Et_s7784‐0.11, annotated as isoform 1 of a magnesium proton exchanger, and Et_s786‐1.34, a pap‐specific phosphatase, which has the GO molecular function of magnesium ion binding and the MapMan function of degradation.
4.4.5. Secondary metabolism (response to reactive oxygen species)
Both the pathway for the synthesis of phenylpropanoids, specifically lignin, and the pathway for the synthesis of carotenoids have enzymes that are upregulated under flooding stress. In the volatile carotenoid pathway of MapMan, isoprenoids.carotenoids.phytoene synthase, Et_s517‐0.16‐1 is upregulated 2.75 times. Its function is to convert GGPP into phytoene. In the volatile phenyl propanoid pathway (secondary metabolism phenylpropanoid lignan biosynthesis), two genes are upregulated. The first is Et_s20148‐0.9‐1 [cinnamoyl CoA reductase 1 (CCR1)], which is involved in lignin biosynthesis and probably involved in the formation of lignin in defense responses in rice (Kawasaki et al., 2006).
Lignin, a component of the plant cell wall, has functions associated with mechanical support, water transport, and defense against pests. Both biotic and abiotic stresses are known to invoke changes in lignin content and composition in plants (Moura, Bonine, Viana, Dornelas, & Mazzafera, 2010). The development barrier to ROL is often associated with suberization and sometimes also lignification of the outer part of the roots (Abiko et al., 2012).
The second gene upregulated in this pathway is Et_c8513699‐0.0‐1 (isoflavone‐7‐O‐methyltransferase), implicated in the biosynthesis of phenylpropanoids. Alfalfa transformants overexpressing isoflavone O‐methyltransferase have increased induction of phenylpropanoid/isoflavonoid pathway gene transcripts after infection with P. medicaginis and also have increased resistance (He & Dixon, 2000).
Another upregulated antioxidant is Et_s4037‐1.43, a GDP‐1‐galactose phosphorylase 2‐like protein which catalyzes a step in the biosynthetic pathway of vitamin C, an important antioxidant. Recently, oxidative stress was suggested to be a major tolerance factor in submergence‐tolerant Brachypodium distachyon, and ascorbate oxidases and ascorbate peroxides were among the upregulated antioxidants (Rivera‐Contreras et al., 2016; Ushimaru et al., 1997; Zhang et al., 2015).
4.4.6. Stress tolerance
Proteins known to be involved in the abiotic stress response had mixed changes in expression. Two salt tolerance‐like proteins, Et_s7183‐0.17 and Et_s6551‐0.36, were upregulated as was Et_s304‐1.36, a late embryogenesis abundant protein (LEA5‐like) that was annotated by Blast2GO but not by MapMan. Salt stress and waterlogging stress often occur together on irrigated land as waterlogged soils prevent leaching of the salts brought by the irrigation water. Adequate drainage is effective in reducing both problems. However, as tef is not usually irrigated, the likelihood that breeding has selected for both traits is small.
Downregulated genes under waterlogging were Et_s346‐4.13 and Et_s3572‐0.37, annotated as bifunctional nuclease 2‐like isoform ×1 and annotated by MapMan as being involved in touch/wounding stress. Expression measurements indicated that the bifunctional nuclease 2‐like gene is downregulated more in Tsedey than in Alba or Quncho (Figure 5). A chaperone protein 1‐like (Et_s11116‐0.0) involved in heat stress was also downregulated.
4.4.7. RNA regulation
Three proteins are categorized into MapMan category 27, RNA regulation. The first is Et_s3159‐0.24, a ribonuclease s‐like protein precursor, downregulated 3.5 times. This sequence was added to the phylogenetic tree of ribonuclease sequences produced by MacIntosh, Hillwig, Meyer, & Flagel (2010) in Fig. S12, which shows that the tef ribonuclease groups with the class 1 ribonucleases. In general, class I ribonucleases show tissue specificity and are associated with stress regulation. Interestingly, it appears in a monocot‐specific class I S‐like clade, which includes proteins that have lost their ribonuclease activity due to the loss of two catalytic histidines. Although they are not active ribonucleases, they are expressed and the rice homolog is expressed under drought conditions (Salekdeh, Siopongco, Wade, Ghareyazie, & Bennett, 2002) indicating a change in function. Indeed, both of these histidines have undergone substitution in the tef homolog.
In addition, a TUBBY‐like f‐box protein type 7, Et_s2666‐0.25 is downregulated approximately by a factor of 3. F‐box proteins are characterized by a conserved F‐box domain, which has a length of about 40 amino acids. A subfamily of these proteins contains a TUBBY domain. The Arabidopsis TUBBY‐like protein type 7 may play a role in regulating phytohormone signaling and appears to be regulated by a homeobox transcription factor, KN1. In addition, it was found to be downregulated in Arabidopsis under conditions of auxin induction (naphthylacetic acid‐treated roots) and an excess of potassium (Lai et al., 2004).
4.4.8. Signaling and transport
Aquaporins form a family of membrane proteins that allow control of water flow through a membrane either by changing the abundance of these proteins in the membrane or by changing the rate of flow through the pores. Aquaporin activity may be key in influencing water transport through waterlogged roots (Bramley, Turner, Tyerman, & Turner, 2007) and may be a critical part of determining a plant's isohydric characteristics, that is, how much they maintain the same water potential during variations in water availability. In the current work, two aquaporins of the TIP4 subtype, Et_s819‐0.0 (TIP4‐2) and Et_s4179‐0.34 (TIP4‐1), were downregulated. Downregulation of the PIP and TIP aquaporin subtypes under waterlogging stress has been found in Arabidopsis (Liu et al., 2005). In sorghum, waterlogging‐tolerant genotypes showed differential regulation of PIP2‐6, PIP2‐7, TIP2‐2, TIP4‐4, and TIP5‐1. However, SbTIP4‐4 was downregulated in the sensitive genotypes (Kadam et al., 2017). TIP2 regulates the response to abiotic stresses (salt and drought) in bread wheat (Triticum aestivum) (Kayum et al., 2017) and tomato (Solanum lycopersicum) (Sade et al., 2009). In addition to the aquaporins, some proteins involved in metal and nitrate transport (Et_s7784‐0.11, Et_s6198‐0.9, Et_s594‐1.9) were upregulated.
4.4.9. Not found
Surprisingly, none of the stress‐induced hormones in the ABA, GA, or JA pathways had detectable changes in expression after 9 days of waterlogging stress. Ethylene has been shown to play a role in both the induction of the GA pathway (Voesenek et al., 2003) and aerenchyma formation in maize and rice (Nishiuchi et al., 2012). Gene regulation in Arabidopsis shoots under flooding was also affected in a mutant with an ethylene signaling mutation. In addition, genes associated with ABA biosynthesis were upregulated in shoots and downregulated in roots, indicating a role for ethylene in the response. An ABA signaling mutation, abi4‐1, affected the expression of several systemic responsive genes, suggesting that ABA biosynthesis may be part of the systemic response to flood response (Hsu, Chou, Peng, Chou, & Shih, 2011).
5. CONCLUSIONS
We report here on the morphology, growth, physiology, and differential gene expression in tef under conditions of long‐term water stress starting at the tillering stage and early in development. Although tef has been reported to be waterlogging tolerant, this is the first quantification of changes to gene expression, morphology, and physiology under waterlogging. Three tef genotypes, two improved varieties and one landrace, were investigated and differences in waterlogging tolerance among the three genotypes were found. The improved variety Tsedey was found to have increased root and shoot biomass under waterlogging conditions when waterlogged in early development. It formed more adventitious roots and aerenchyma than the other two genotypes and is a promising candidate for further study. Differences in plant growth characteristics peaked at 9–11 days after the onset of stress for early waterlogging. Differential expression of 19‐day‐old seedlings waterlogged at the tillering stage shows changes in the cell wall, carbohydrate metabolism, and upregulation of genes involved in the response to ROS. In addition, genes affecting transport and lignification were affected. The identification of lines more tolerant to waterlogging conditions will aid the cultivation of cereal crops in poorly drained soils as tolerant waterlogging lines will be introgressed to elite tef genotypes, and the relevant genetic loci may be introduced to other cultivated crops. As this study shows that there is significant genetic diversity in tef genotypes with respect to waterlogging tolerance, a large‐scale screening is underway to identify and quantify the performance of 500 tef genotypes under waterlogging stress.
AUTHOR CONTRIBUTIONS
G.C., A.W., and Z.T. conceived experiments; G.C., M.S., C.R., A.W., R.B., S.B., S.P.W., and Z.T. performed experiments; G.C., M.S., and C.R. analyzed the data; G.C. wrote the manuscript; all authors contributed to manuscript revision; S.C. and K.A. contributed to the original research plans; Z.T. conceived the original research plans.
Supporting information
ACKNOWLEDGMENTS
We thank Syngenta Foundation for Sustainable Agriculture and the University of Bern for financial and technical support. We also thank Christopher Ball and Jasmin Sekulovski for taking care of our plants.
Cannarozzi G, Weichert A, Schnell M, et al. Waterlogging affects plant morphology and the expression of key genes in tef (Eragrostis tef). Plant Direct. 2018;2:1–22. 10.1002/pld3.56
REFERENCES
- Abiko, T. , Kotula, L. , Shiono, K. , Malik, A. I. , Colmer, T. D. , & Nakazono, M. (2012). Enhanced formation of aerenchyma and induction of a barrier to radial oxygen loss in adventitious roots of Zea nicaraguensis contribute to its waterlogging tolerance as compared with maize (Zea mays ssp mays). Plant Cell and Environment, 35, 1618–1630. 10.1111/j.1365-3040.2012.02513.x [DOI] [PubMed] [Google Scholar]
- Ahsan, N. , Lee, D. G. , Lee, S. H. , Kang, K. Y. , Bahk, J. D. , Choi, M. S. , … Lee, B. H. (2007). A comparative proteomic analysis of tomato leaves in response to waterlogging stress. Physiologia Plantarum, 131, 555–570. 10.1111/j.1399-3054.2007.00980.x [DOI] [PubMed] [Google Scholar]
- Anders, S. , & Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biology, 11, R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P. T. , & Huber, W. (2015). HTSeq‐a Python framework to work with high‐throughput sequencing data. Bioinformatics, 31, 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduini, I. , Orlandi, C. , Pampana, S. , & Masoni, A. (2016). Waterlogging at tillering affects spike and spikelet formation in wheat. Crop & Pasture Science, 67, 703–711. [Google Scholar]
- Armstrong, A. C. (1979). Aeration in higher plants. Advances in Botanical Research, 7, 225–332. [Google Scholar]
- Asamenew, G. , Beyene, H. , Negatu, W. , & Ayele, G. (1993). A survey of the farming systems of Vertisol areas of the Ethiopian highlands In Mamo T., Astatke A., Srivastava K. L., & Dibabe A. (Eds.), Improved management of Vertisols for sustainable crop – Livestock production in the Ethiopian highlands: Synthesis report. Addis Ababa, Ethiopia: Technical Committee of the Joint Vertisol Project. [Google Scholar]
- Bailey‐Serres, J. , Lee, S. C. , & Brinton, E. (2012). Waterproofing crops: Effective flooding survival strategies. Plant Physiology, 160, 1698–1709. 10.1104/pp.112.208173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A. , Martin, M. J. , O'Donovan, C. , Magrane, M. , Alpi, E. , Antunes, R. , … Zhang, J. (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Research, 45, D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini, T. Z. , Reiser, L. , Li, D. H. , Mezheritsky, Y. , Muller, R. , Strait, E. , & Huala, E. (2015). The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis, 53, 474–485. 10.1002/dvg.22877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramley, H. , Turner, D. W. , Tyerman, S. D. , & Turner, N. C. (2007). Water flow in the roots of crop species: The influence of root structure, aquaporin activity, and waterlogging. Advances in Agronomy, 96, 133–196. 10.1016/S0065-2113(07)96002-2 [DOI] [Google Scholar]
- Cannarozzi, G. , Chanyalew, S. , Assefa, K. , Bekele, A. , Blösch, R. , Weichert, A. , … Tadele, Z. (2018). Technology generation to dissemination: Lessons learned from the tef improvement project. Euphytica, 214, 31 10.1007/s10681-018-2115-5 [DOI] [Google Scholar]
- Cannarozzi, G. , Plaza‐Wuthrich, S. , Esfeld, K. , Larti, S. , Wilson, Y. S. , Girma, D. , … Tadele, Z. (2014). Genome and transcriptome sequencing identifies breeding targets in the orphan crop tef (Eragrostis tef). BMC Genomics, 15, 581 10.1186/1471-2164-15-581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudle, K. L. , & Maricle, B. R. (2012). Effects of flooding on photosynthesis, chlorophyll fluorescence, and oxygen stress in plants of varying flooding tolerance. Transactions of the Kansas Academy of Science, 115, 5–18. 10.1660/062.115.0102 [DOI] [Google Scholar]
- Cho, H. T. , & Kende, H. (1997). Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell, 9, 1661–1671. 10.1105/tpc.9.9.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmer, T. D. (2003). Long‐distance transport of gases in plants: A perspective on internal aeration and radial oxygen loss from roots. Plant Cell and Environment, 26, 17–36. 10.1046/j.1365-3040.2003.00846.x [DOI] [Google Scholar]
- Colmer, T. D. , Cox, M. C. H. , & Voesenek, L. A. C. J. (2006). Root aeration in rice (Oryza sativa): Evaluation of oxygen, carbon dioxide, and ethylene as possible regulators of root acclimatizations. New Phytologist, 170, 767–777. 10.1111/j.1469-8137.2006.01725.x [DOI] [PubMed] [Google Scholar]
- Colmer, T. D. , & Pedersen, O. (2008). Oxygen dynamics in submerged rice (Oryza sativa). New Phytologist, 178, 326–334. 10.1111/j.1469-8137.2007.02364.x [DOI] [PubMed] [Google Scholar]
- Colmer, T. D. , & Voesenek, L. A. C. J. (2009). Flooding tolerance: Suites of plant traits in variable environments. Functional Plant Biology, 36, 665–681. 10.1071/FP09144 [DOI] [PubMed] [Google Scholar]
- Conesa, A. , & Götz, S. (2008). Blast2GO: A comprehensive suite for functional analysis in plant genomics. International Journal of Plant Genomics, 2008, 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coordinators, N. R. (2017). Database resources of the national center for biotechnology information. Nucleic Acids Research, 45, D12–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, D. J. , Bedinger, P. , & Durachko, D. M. (1997). Group I allergens of grass pollen as cell wall‐loosening agents. Proceedings of the National Academy of Sciences of the United States of America, 94, 6559–6564. 10.1073/pnas.94.12.6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CSA (2015) Agricultural sample survey 2014/2015. Volume I. Report on area and production of major crops. Statistical Bulletin 578. Addis Ababa, Ethiopia: Central Statistical Agency (CSA). [Google Scholar]
- Dat, J. F. , Capelli, N. , Folzer, H. , Bourgeade, P. , & Badot, P. M. (2004). Sensing and signalling during plant flooding. Plant Physiology and Biochemistry, 42, 273–282. 10.1016/j.plaphy.2004.02.003 [DOI] [PubMed] [Google Scholar]
- Dobin, A. , Davis, C. A. , Schlesinger, F. , Drenkow, J. , Zaleski, C. , Jha, S. , … Gingeras, T. R. (2013). STAR: Ultrafast universal RNA‐seq aligner. Bioinformatics, 29, 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew, M. C. , He, C. J. , & Morgan, P. W. (2000). Programmed cell death and aerenchyma formation in roots. Trends in Plant Science, 5, 123–127. 10.1016/S1360-1385(00)01570-3 [DOI] [PubMed] [Google Scholar]
- Duarte, R. F. , Prom‐u‐Thai, C. , Amaral, D. C. , Faquin, V. , Guilherme, L. R. G. , Reis, A. R. , & Alves, E. (2016). Determination of zinc in rice grains using DTZ staining and ImageJ software. Journal of Cereal Science, 68, 53–58. 10.1016/j.jcs.2015.11.006 [DOI] [Google Scholar]
- Dyson, B. C. , Webster, R. E. , & Johnson, G. N. (2014). GPT2: A glucose 6‐phosphate/phosphate translocator with a novel role in the regulation of sugar signalling during seedling development. Annals of Botany, 113, 643–652. 10.1093/aob/mct298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R. D. , Attwood, T. K. , Babbitt, P. C. , Bateman, A. , Bork, P. , Bridge, A. J. , … Mitchell, A. L. (2017). InterPro in 2017‐beyond protein family and domain annotations. Nucleic Acids Research, 45, D190–D199. 10.1093/nar/gkw1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao, T. , Xu, K. N. , Ronald, P. C. , & Bailey‐Serres, J. (2006). A variable cluster of ethylene response factor‐like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell, 18, 2021–2034. 10.1105/tpc.106.043000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao, T. , Yeung, E. , & Bailey‐Serres, J. (2011). The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell, 23, 412–427. 10.1105/tpc.110.080325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielminetti, L. , Yamaguchi, J. , Perata, P. , & Alpi, A. (1995). Amylolytic activities in cereal seeds under aerobic and anaerobic conditions. Plant Physiology, 109, 1069–1076. 10.1104/pp.109.3.1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon, S. , Dufayard, J. F. , Lefort, V. , Anisimova, M. , Hordijk, W. , & Gascuel, O. (2010). New algorithms and methods to estimate maximum‐likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology, 59, 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hartmann, D. L. , Tank, A. M. G. K. , & Rusticucci, M. (2013) IPCC Fifth Assessment Report, Climate Change 2013: The Physical Science Basis. In IPCC, AR5, pp. 31–39.
- Hattori, Y. , Nagai, K. , Furukawa, S. , Song, X. J. , Kawano, R. , Sakakibara, H. , … Ashikari, M. (2009). The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature, 460, 1026–U1116. 10.1038/nature08258 [DOI] [PubMed] [Google Scholar]
- He, X. Z. , & Dixon, R. A. (2000). Genetic manipulation of isoflavone 7‐O‐methyltransferase enhances biosynthesis of 4 ‘‐O‐methylated isoflavonoid phytoalexins and disease resistance in alfalfa. Plant Cell, 12, 1689–1702. 10.1105/tpc.12.9.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog, M. , Striker, G. G. , Colmer, T. D. , & Pedersen, O. (2016). Mechanisms of waterlogging tolerance in wheat – A review of root and shoot physiology. Plant Cell and Environment, 39, 1068–1086. 10.1111/pce.12676 [DOI] [PubMed] [Google Scholar]
- Hsu, F. C. , Chou, M. Y. , Chou, S. J. , Li, Y. R. , Peng, H. P. , & Shih, M. C. (2013). Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis . Plant Cell, 25, 2699–2713. 10.1105/tpc.113.114447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, F. C. , Chou, M. Y. , Peng, H. P. , Chou, S. J. , & Shih, M. C. (2011). Insights into hypoxic systemic responses based on analyses of transcriptional regulation in Arabidopsis . PLoS ONE, 6, e28888 10.1371/journal.pone.0028888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, K. W. , Singer, P. B. , McInnis, S. , Diaz‐Salaz, C. , & Greenwood, M. S. (1999). Expansins are conserved in conifers and expressed in hypocotyls in response to exogenous auxin. Plant Physiology, 120, 827–831. 10.1104/pp.120.3.827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizawa, K. , Murakami, S. , Kawakami, Y. , & Kuramochi, H. (1999). Growth and energy status of arrowhead tubers, pondweed turions and rice seedlings under anoxic conditions. Plant Cell and Environment, 22, 505–514. 10.1046/j.1365-3040.1999.00439.x [DOI] [Google Scholar]
- Ismail, A. M. , Ella, E. S. , Vergara, G. V. , & Mackill, D. J. (2009). Mechanisms associated with tolerance to flooding during germination and early seedling growth in rice (Oryza sativa). Annals of Botany, 103, 197–209. 10.1093/aob/mcn211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, M. B. , & Colmer, T. D. (2005). Response and adaptation by plants to flooding stress – Preface. Annals of Botany, 96, 501–505. 10.1093/aob/mci205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, D. B. , & Chollet, R. (1983). Inhibition of ribulose bisphosphate carboxylase by substrate ribulose 1,5‐bisphosphate. Journal of Biological Chemistry, 258, 13752–13758. [PubMed] [Google Scholar]
- Juntawong, P. , Sirikhachornkit, A. , Pimjan, R. , Sonthirod, C. , Sangsrakru, D. , Yoocha, T. , … Srinives, P. (2014). Elucidation of the molecular responses to waterlogging in Jatropha roots by transcriptome profiling. Frontiers in Plant Science, 5, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justin, S. H. F. W. , & Armstrong, W. (1987). The anatomical characteristics of roots and plant‐response to soil flooding. New Phytologist, 106, 465–495. 10.1111/j.1469-8137.1987.tb00153.x [DOI] [Google Scholar]
- Kadam, S. , Abril, A. , Dhanapal, A. P. , Koester, R. P. , Vermerris, W. , Jose, S. , & Fritschi, F. B. (2017). Characterization and regulation of aquaporin genes of sorghum [Sorghum bicolor (L.) Moench] in response to waterlogging stress. Frontiers Plant Science, 8, 862 10.3389/fpls.2017.00862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. , Furumichi, M. , Tanabe, M. , Sato, Y. , & Morishima, K. (2017). KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research, 45, D353–D361. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. , & Standley, D. M. (2013). MAFFT multiple sequence alignment software Version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara, Y. , de la Bastide, M. , Hamilton, J. P. , Kanamori, H. , McCombie, W. R. , Ouyang, S. , … Matsumoto, T. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice, 6, 4 10.1186/1939-8433-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano, N. , Ito, O. , & Sakagami, J. I. (2009). Morphological and physiological responses of rice seedlings to complete submergence (flash flooding). Annals of Botany, 103, 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, T. , Koita, H. , Nakatsubo, T. , Hasegawa, K. , Wakabayashi, K. , Takahashi, H. , … Shimamoto, K. (2006). Cinnamoyl‐CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proceedings of the National Academy of Sciences of the United States of America, 103(1), 230–235. 10.1073/pnas.0509875103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayum, M. A. , Park, J. I. , Nath, U. K. , Biswas, M. K. , Kim, H. T. , & Nou, I. S. (2017). Genome‐wide expression profiling of aquaporin genes confer responses to abiotic and biotic stresses in Brassica rapa . BMC Plant Biology, 17, 23 10.1186/s12870-017-0979-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende, H. , van der Knaap, E. , & Cho, H. T. (1998). Deepwater rice: A model plant to study stem elongation. Plant Physiology, 118, 1105–1110. 10.1104/pp.118.4.1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketema, S. (1993). Tef (Eragrostis tef): Breeding, agronomy, genetic resources, utilization, and role in Ethiopian agriculture. Addis Ababa, Ethiopia: Institute of Agricultural Research. [Google Scholar]
- Kotula, L. , Ranathunge, K. , Schreiber, L. , & Steudle, E. (2009). Functional and chemical comparison of apoplastic barriers to radial oxygen loss in roots of rice (Oryza sativa L.) grown in aerated or deoxygenated solution. Journal of Experimental Botany, 60, 2155–2167. 10.1093/jxb/erp089 [DOI] [PubMed] [Google Scholar]
- Laanbroek, H. J. (1990). Bacterial cycling of minerals that affect plant‐growth in waterlogged soils – A review. Aquatic Botany, 38, 109–125. 10.1016/0304-3770(90)90101-P [DOI] [Google Scholar]
- Lai, C. P. , Lee, C. L. , Chen, P. H. , Wu, S. H. , Yang, C. C. , & Shaw, J. F. (2004). Molecular analyses of the Arabidopsis TUBBY‐like protein gene family. Plant Physiology, 134, 1586–1597. 10.1104/pp.103.037820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. , & Kende, H. (2001). Expression of beta‐expansins is correlated with internodal elongation in deepwater rice. Plant Physiology, 127, 645–654. 10.1104/pp.010345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T. M. , & Lin, Y. H. (1995). Changes in soluble and cell wall‐bound peroxidase‐activities with growth in anoxia‐treated rice (Oryza sSativa L) coleoptiles and roots. Plant Science, 106, 1–7. 10.1016/0168-9452(94)04053-J [DOI] [Google Scholar]
- Lee, S. C. , Mustroph, A. , Sasidharan, R. , Vashisht, D. , Pedersen, O. , Oosumi, T. , … Bailey‐Serres, J. (2011). Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytologist, 190, 457–471. 10.1111/j.1469-8137.2010.03590.x [DOI] [PubMed] [Google Scholar]
- Lekshmy, S. , Jha, S. K. , & Sairam, R. K. (2015). Physiological and molecular mechanisms of flooding tolerance in plants In Pandey G. (Ed.), Elucidation of Abiotic stress signaling in plants. New York: Springer. [Google Scholar]
- Li, Y. , Darley, C. P. , Ongaro, V. , Fleming, A. , Schipper, O. , Baldauf, S. L. , & McQueen‐Mason, S. J. (2002). Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiology, 128, 854–864. 10.1104/pp.010658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , … 1000 Genome Project Data Processing Subgroup . (2009). The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Jones, L. , & McQueen‐Mason, S. (2003). Expansins and cell growth. Current Opinion in Plant Biology, 6, 603–610. 10.1016/j.pbi.2003.09.003 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler, H. K. (1987). Chlorophyll fluorescence signatures of leaves during the autumnal chlorophyll breakdown. Journal of Plant Physiology, 131, 101–110. 10.1016/S0176-1617(87)80271-7 [DOI] [Google Scholar]
- Liu, F. L. , VanToai, T. , Moy, L. P. , Bock, G. , Linford, L. D. , & Quackenbush, J. (2005). Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiology, 137, 1115–1129. 10.1104/pp.104.055475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K. J. , & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2(T)(‐Delta Delta C) method. Methods, 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lohse, M. , Nagel, A. , Herter, T. , May, P. , Schroda, M. , Zrenner, R. , … Usadel, B. (2014). Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell and Environment, 37, 1250–1258. 10.1111/pce.12231 [DOI] [PubMed] [Google Scholar]
- Lorbiecke, R. , & Sauter, M. (1999). Adventitious root growth and cell‐cycle induction in deepwater rice. Plant Physiology, 119, 21–29. 10.1104/pp.119.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntosh, G. C. , Hillwig, M. S. , Meyer, A. , & Flagel, L. (2010). RNase T2 genes from rice and the evolution of secretory ribonucleases in plants. Molecular Genetics and Genomics, 283, 381–396. 10.1007/s00438-010-0524-9 [DOI] [PubMed] [Google Scholar]
- Malik, A. I. , Islam, A. K. M. R. , & Colmer, T. D. (2011). Transfer of the barrier to radial oxygen loss in roots of Hordeum marinum to wheat (Triticum aestivum): Evaluation of four H‐marinum‐wheat amphiploids. New Phytologist, 190, 499–508. 10.1111/j.1469-8137.2010.03519.x [DOI] [PubMed] [Google Scholar]
- Mekonen, M. , Tesfaye, K. , & Bayu, W. (2013). Soil drainage and nutrient management to improve productivity of waterlogged Vertisols for small‐scale farmers. Engineering International Journal, 1, 27–39. 10.18034/ei.v1i2.122 [DOI] [Google Scholar]
- Miro, B. , & Ismail, A. M. (2013). Tolerance of anaerobic conditions caused by flooding during germination and early growth in rice (Oryza sativa L.). Frontiers Plant Science, 4, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura, J. C. M. S. , Bonine, C. A. V. , Viana, J. D. F. , Dornelas, M. C. , & Mazzafera, P. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. Journal of Integrative Plant Biology, 52, 360–376. 10.1111/j.1744-7909.2010.00892.x [DOI] [PubMed] [Google Scholar]
- Naidoo, G. , & Naidoo, Y. (2015). Waterlogging responses of Schoenoplectus scirpoides (Schrad) Browning (Cyperaceae). African Journal of Ecology, 53, 36–43. 10.1111/aje.12144 [DOI] [Google Scholar]
- Nishiuchi, S. , Yamauchi, T. , Takahashi, H. , Kotula, L. , & Nakazono, M. (2012). Mechanisms for coping with submergence and waterlogging in rice. Rice, 5, 2 10.1186/1939-8433-5-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacurar, D. I. , Perrone, I. , & Bellini, C. (2014). Auxin is a central player in the hormone cross‐talks that control adventitious rooting. Physiologia Plantarum, 151, 83–96. 10.1111/ppl.12171 [DOI] [PubMed] [Google Scholar]
- Parent, C. , Capelli, N. , Berger, A. , Crèvecoeur, M. , & Dat, J. F. (2008). An overview of plant responses to soil waterlogging. Plant Stress, 2, 20–27. [Google Scholar]
- Raskin, I. , & Kende, H. (1983). How does deep‐water rice solve its aeration problem. Plant Physiology, 72, 447–454. 10.1104/pp.72.2.447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, P. S. , Reddy, D. S. , Sivasakthi, K. , Bhatnagar‐Mathur, P. , Vadez, V. , & Sharma, K. K. (2016). Evaluation of sorghum [Sorghum bicolor (L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real‐time PCR data normalization. Frontiers Plant Science, 7, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera‐Contreras, I. K. , Zamora‐Hernandez, T. , Huerta‐Heredia, A. A. , Capataz‐Tafur, J. , Barrera‐Figueroa, B. E. , Juntawong, P. , & Pena‐Castro, J. M. (2016). Transcriptomic analysis of submergence‐tolerant and sensitive Brachypodium distachyon ecotypes reveals oxidative stress as a major tolerance factor. Scientific Reports, 6, 27686 10.1038/srep27686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokka, A. , Zhang, L. X. , & Aro, E. M. (2001). Rubisco activase: An enzyme with a temperature‐dependent dual function? Plant Journal, 25, 463–471. 10.1046/j.1365-313x.2001.00981.x [DOI] [PubMed] [Google Scholar]
- Rose, J. K. C. , Braam, J. , Fry, S. C. , & Nishitani, K. (2002). The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant and Cell Physiology, 43, 1421–1435. 10.1093/pcp/pcf171 [DOI] [PubMed] [Google Scholar]
- Saab, I. N. , & Sachs, M. M. (1996). A flooding‐induced xyloglucan endo‐transglycosylase homolog in maize is responsive to ethylene and associated with aerenchyma. Plant Physiology, 112, 385–391. 10.1104/pp.112.1.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade, N. , Vinocur, B. J. , Diber, A. , Shatil, A. , Ronen, G. , Nissan, H. , … Moshelion, M. (2009). Improving plant stress tolerance and yield production: Is the tonoplast aquaporin SlTIP2;2 a key to isohydric to anisohydric conversion? New Phytologist, 181, 651–661. 10.1111/j.1469-8137.2008.02689.x [DOI] [PubMed] [Google Scholar]
- Salekdeh, G. H. , Siopongco, J. , Wade, L. J. , Ghareyazie, B. , & Bennett, J. (2002). Proteomic analysis of rice leaves during drought stress and recovery. Proteomics, 2, 1131–1145. [DOI] [PubMed] [Google Scholar]
- Sampedro, J. , & Cosgrove, D. J. (2005). The expansin superfamily. Genome Biology, 6, 242 10.1186/gb-2005-6-12-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setter, T. L. , & Waters, I. (2003). Review of prospects for germplasm improvement for waterlogging tolerance in wheat, barley and oats. Plant and Soil, 253, 1–34. 10.1023/A:1024573305997 [DOI] [Google Scholar]
- Shabala, S. (2011). Physiological and cellular aspects of phytotoxicity tolerance in plants: The role of membrane transporters and implications for crop breeding for waterlogging tolerance. New Phytologist, 190, 289–298. 10.1111/j.1469-8137.2010.03575.x [DOI] [PubMed] [Google Scholar]
- Shiono, K. , Takahashi, H. , Colmer, T. D. , & Nakazono, M. (2008). Role of ethylene in acclimations to promote oxygen transport in roots of plants in waterlogged soils. Plant Science, 175, 52–58. 10.1016/j.plantsci.2008.03.002 [DOI] [Google Scholar]
- Steffens, B. , & Rasmussen, A. (2016). The physiology of adventitious roots. Plant Physiology, 170, 603–617. 10.1104/pp.15.01360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens, B. , Wang, J. X. , & Sauter, M. (2006). Interactions between ethylene, gibberellin and abscisic acid regulate emergence and growth rate of adventitious roots in deepwater rice. Planta, 223, 604–612. 10.1007/s00425-005-0111-1 [DOI] [PubMed] [Google Scholar]
- Summers, J. E. , & Jackson, M. B. (1994). Growth and energy status of arrowhead tubers, pondweed turions and rice seedlings under anoxic conditions. Journal of Experimental Botany, 45, 1309–1318. 10.1093/jxb/45.9.1309 [DOI] [Google Scholar]
- Tatusov, R. L. , Fedorova, N. D. , Jackson, J. D. , Jacobs, A. R. , Kiryutin, B. , Koonin, E. V. , … Natale, D. A. (2003). The COG database: An updated version includes eukaryotes. BMC Bioinformatics, 4, 41 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm, O. , Blasing, O. , Gibon, Y. , Nagel, A. , Meyer, S. , Kruger, P. , … Stitt, M. (2004). MAPMAN: A user‐driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant Journal, 37, 914–939. 10.1111/j.1365-313X.2004.02016.x [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu, N. , Hossain, F. , Mohan, S. , Shiriga, K. , Mittal, S. , Sharma, R. , … Gupta, H. S. (2013). Genome‐wide expression of transcriptomes and their co‐expression pattern in subtropical maize (Zea mays L.) under waterlogging stress. PLoS ONE, 8, e70433 10.1371/journal.pone.0070433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, M. , Satoh, S. , & Iwai, H. (2015). Distribution of XTH, expansin, and secondary‐wall‐related CesA in floral and fruit abscission zones during fruit development in tomato (Solanum lycopersicum). Frontiers in Plant Science, 6, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushimaru, T. , Maki, Y. , Sano, S. , Koshiba, K. , Asada, K. , & Tsuji, H. (1997). Induction of enzymes involved in the ascorbate‐dependent antioxidative system, namely, ascorbate peroxidase, monodehydroascorbate reductase and dehydroascorbate reductase, after exposure to air of rice (Oryza sativa) seedlings germinated under water. Plant and Cell Physiology, 38, 541–549. 10.1093/oxfordjournals.pcp.a029203 [DOI] [Google Scholar]
- van Veen, H. , Mustroph, A. , Barding, G. A. , Vergeer‐van Eijk, M. , Welschen‐Evertman, R. A. M. , Pedersen, O. , … Sasidharan, R. (2013). Two Rumex species from contrasting hydrological niches regulate flooding tolerance through distinct mechanisms. Plant Cell, 25, 4691–4707. 10.1105/tpc.113.119016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek, L. A. C. J. , Benschop, J. J. , Bou, J. , Cox, M. C. H. , Groeneveld, H. W. , Millenaar, F. F. , … Peeters, A. J. M. (2003). Interactions between plant hormones regulate submergence‐induced shoot elongation in the flooding‐tolerant dicot Rumex palustris . Annals of Botany, 91, 205–211. 10.1093/aob/mcf116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring, E. F. , & Maricle, B. R. (2013). Stomatal conductance correlates with flooding tolerance in Phragmites australis and Sorghum halepense . Transactions of the Kansas Academy of Science, 115, 161–166. 10.1660/062.115.0310 [DOI] [Google Scholar]
- Watanabe, K. , Nishiuchi, S. , Kulichikhin, K. , & Nakazono, M. (2013). Does suberin accumulation in plant roots contribute to waterlogging tolerance? Frontiers in Plant Science, 4, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, K. , Xu, X. , Fukao, T. , Canlas, P. , Maghirang‐Rodriguez, R. , Heuer, S. , … Mackill, D. J. (2006). Sub1A is an ethylene‐response‐factor‐like gene that confers submergence tolerance to rice. Nature, 442, 705–708. 10.1038/nature04920 [DOI] [PubMed] [Google Scholar]
- Yamauchi, T. , Rajhi, I. , & Nakazono, M. (2011). Lysigenous aerenchyma formation in maize root is confined to cortical cells by regulation of genes related to generation and scavenging of reactive oxygen species. Plant Signaling & Behavior, 6, 759–761. 10.4161/psb.6.5.15417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, T. , Shimamura, S. , Nakazono, M. , & Mochizuki, T. (2013). Aerenchyma formation in crop species: A review. Field Crops Research, 152, 8–16. 10.1016/j.fcr.2012.12.008 [DOI] [Google Scholar]
- Zhang, X. C. , Fan, Y. , Shabala, S. , Koutoulis, A. , Shabala, L. , Johnson, P. , … Zhou, M. X. (2017). A new major‐effect QTL for waterlogging tolerance in wild barley (H. spontaneum). Theoretical and Applied Genetics, 130, 1559–1568. 10.1007/s00122-017-2910-8 [DOI] [PubMed] [Google Scholar]
- Zhang, X. C. , Shabala, S. , Koutoulis, A. , Shabala, L. , Johnson, P. , Hayes, D. , … Zhou, M. X. (2015). Waterlogging tolerance in barley is associated with faster aerenchyma formation in adventitious roots. Plant and Soil, 394, 355–372. 10.1007/s11104-015-2536-z [DOI] [Google Scholar]
- Zhang, X. C. , Zhou, G. F. , Shabala, S. , Koutoulis, A. , Shabala, L. , Johnson, P. , … Zhou, M. X. (2016). Identification of aerenchyma formation‐related QTL in barley that can be effective in breeding for waterlogging tolerance. Theoretical and Applied Genetics, 129, 1167–1177. 10.1007/s00122-016-2693-3 [DOI] [PubMed] [Google Scholar]
- Zou, X. L. , Tan, X. Y. , Hu, C. W. , Zeng, L. , Lu, G. Y. , Fu, G. P. , … Zhang, X. K. (2013). The transcriptome of Brassica napus L. roots under waterlogging at the seedling stage. International Journal of Molecular Sciences, 14, 2637–2651. 10.3390/ijms14022637 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


