Abstract
Multiple genetic mutations within melanoma not only cause lesion-specific responses to targeted therapy but also alter the molecular route of resistance to that therapy. Inactivation of PTEN occurs in up to 30% of melanomas, frequently with a concurrent activating BRAF mutation. PTEN loss regulates both acquired and intrinsic drug resistance. Here we show that AXL/AKT axis mediated-resistance to BRAF inhibitor (BRAFi) depends upon PTEN status in melanoma. Hyperactivation of both ERK and AKT pathways was associated with BRAFi resistance in melanoma with wildtype PTEN. The PTEN-impaired melanoma cells required only the ERK resistance mechanism. Moreover, we identified AXL as a key upstream effector of AKT pathway-associated resistance to BRAFi in melanoma with wildtype PTEN, but not in melanoma with impaired PTEN. Notably, we confirmed that blocking AXL by shRNA and a small molecular inhibitor could rescue the sensitivity of resistant melanoma cells with wildtype PTEN to BRAFi and inhibit their growth in vitro and in vivo. Our study has uncovered a mechanism by which PTEN status contributes to acquired resistance to BRAFi and offers a rational strategy to guide clinical testing in pre-identified subsets of patients who relapse during treatment with BRAFi. The identified protein AXL represents a promising therapeutic target for BRAF mutant melanoma patients with wildtype PTEN.
Keywords: PTEN, BRAF inhibitor, resistance, AXL, BRAF mutant melanoma
Introduction
Malignant melanoma is the deadliest form of skin cancer. More than 50% of melanomas have mutations in BRAF, 90% of which are BRAFV600E substitutions. This oncogenic mutation activates the downstream kinase MEK/ERK within the MAPK pathway, promoting melanogenesis.1 Targeting this BRAF mutant kinase by inhibitors demonstrates a striking response in melanomas bearing BRAFV600E mutations.2–4 However, acquired resistance rapidly develops, hindering its durable efficacy. Recently identified mechanisms of acquired resistance to BRAFV600E inhibitors (BRAFi), including mutations in NRAS or MEK1, and overexpression of COT, EGFR, PDGFRβ, IGF1R or MET,5–9 have encouraged clinicians to employ a combination of inhibitors to more effectively block the MAPK signaling pathway.10,11 However, due to multiple genetic, epigenetic and heterogeneous alternations in melanomas, combinations have had less than the desired effect.12 Therefore, innovative strategies are still necessary to improve the precision and durability of anti-melanoma therapies.13
The tumor suppressor phosphatase and tensin homolog deleted on chromosome 10 (PTEN) is mutated or epigenetically silenced in more than 30% of melanomas.14–16 Interestingly, the PTEN mutations are coincident with BRAF mutations.17,18 PTEN effectively antagonizes PI3K and AKT, thereby inhibiting cell proliferation and promoting apoptosis.19,20 The loss of PTEN has been reported to contribute to intrinsic resistance to BRAFi via the suppression of BIM-mediated apoptosis.21 However, only ~̴ 10% of BRAFV600E/PTEN-null melanomas exhibit de novo resistance to BRAFi, and others have been reported to be sensitive,22–24 implying that the contribution of PTEN status to acquired resistance to BRAFi in melanoma may be complex and context dependent.
In this study, we created BRAFi resistant mutant melanoma cell lines with/without wildtype PTEN as a model for assessing the role of PTEN in BRAFi resistance. We found that the hyperactivation of both ERK and AKT pathways was associated with BRAFi resistance in melanoma with wildtype PTEN. The PTEN-impaired melanoma cells required only the ERK resistance mechanism. Moreover, we demonstrate that AXL is a key upstream effector of AKT pathway-associated resistance to BRAFi in melanomas with wildtype PTEN, but not in melanomas with impaired PTEN. Our study has uncovered a mechanism by which PTEN status contributes to acquired BRAFi resistance, and identified a promising new target for the subset of patients bearing BRAF mutant/PTEN wildtype melanomas.
Results
Establishing models of acquired resistance to BRAF inhibition in human melanomas with/without wildtype PTEN
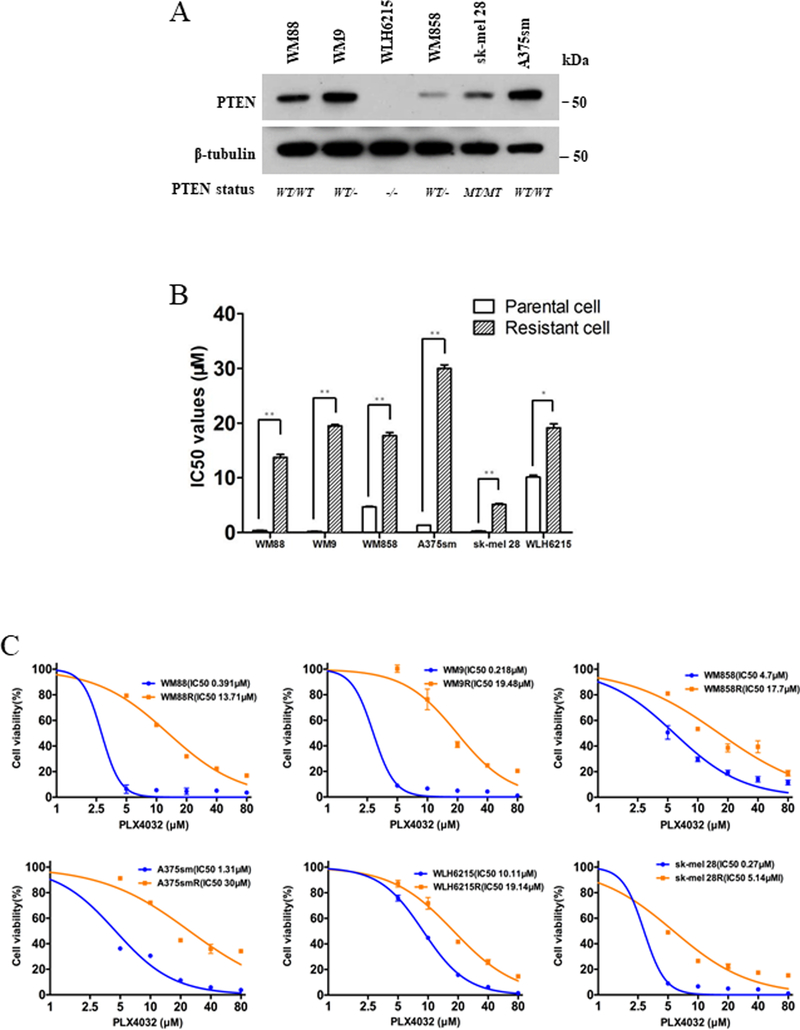
To study the molecular mechanisms of acquired resistance to BRAFi in melanoma bearing wildtype vs. impaired PTEN, we established 6 PLX4032-resistant human melanoma (PRM) models for BRAFV600E cell lines with varying PTEN status. Four cell lines have at least one allele of wildtype PTEN (WM88, A375sm, WM9, and WM858), while two other cell lines have impaired PTEN (sk-mel 28 with PTEN mutation, WLH6215 with PTEN loss) (Supplementary Table 1). The PTEN protein levels in these cell lines were confirmed using western blot assay (Figure 1A, Supplementary Table 1). Interestingly, the PTEN protein levels did not correlate with the number of alleles.
Figure 1. The melanoma models of acquired resistance to BRAF inhibitor in human melanoma with/without wildtype PTEN.
(A) The Western blotting showed the protein level of PTEN in 6 BRAFV600E human melanoma cell lines with various PTEN genetic background, the protein level of tubulin as loading control. (B) The values of 50%-inhibitory-concentration (IC50) of 6 pairs parental and resistant human melanoma cells were determined using CCK8 assays. Error bars denote s.d. for biological three repeats. Results are statistically significant between parental and resistant groups by Student’s t-test (* p<0.05, **p<0.01). (C) The cell viability curves of 6 pairs parental and resistant human melanoma cells were determined using CCK8 assays. Error bars denote s.d. for biological three repeats.
After six months of stepwise exposure to increasing concentration of PLX4032, we successfully generated 6 PRM cell lines (WM88R, WM9R, WM858R, A375smR, sk-mel 28R and WLH6215R) (Figure 1B, 1C; Supplementary Table 2). Compared with the parental cells, all PRM cells exhibited higher IC50 values to PLX4032 with resistance indexes (RI) of 35.1(WM88R), 89.4 (WM9R), 3.8 (WM858R), 22.9 (A375smR), 1.9 (WLH6215R), and 19.0 (sk-mel 28R) (Figure 1B, 1C; Supplementary Table 2).
BRAF inhibitor resistance via hyperactivated ERK and AKT pathways is dependent on PTEN status
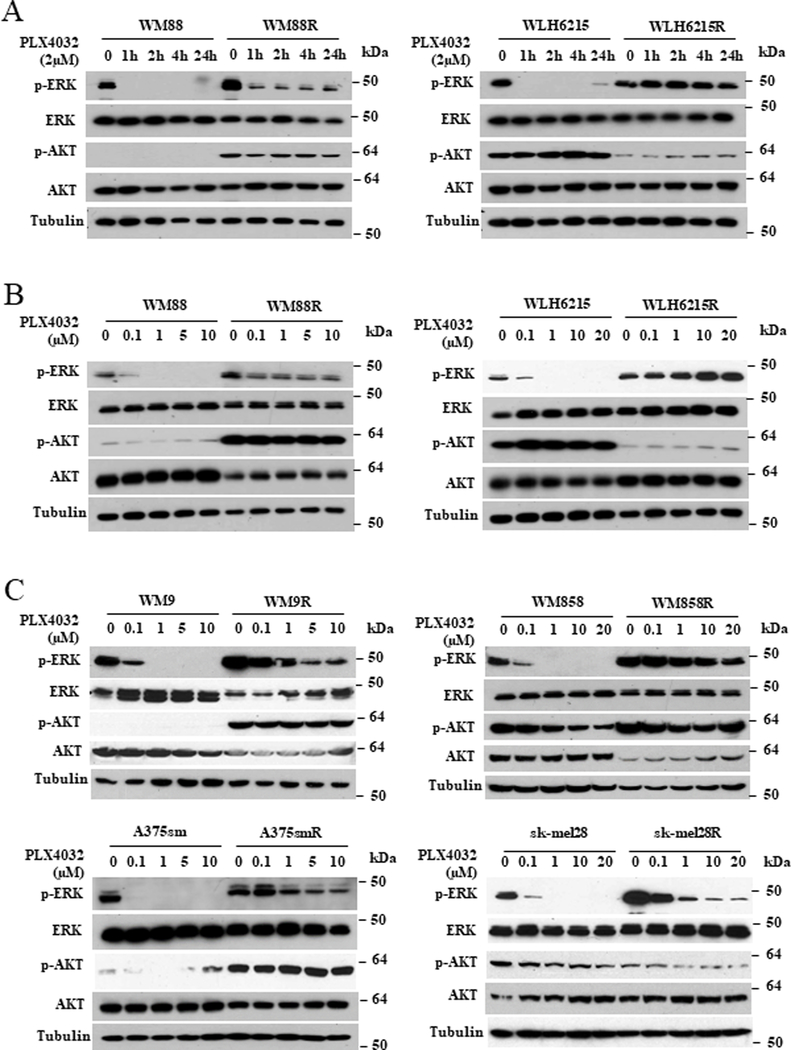
To identify the molecular signaling pathways of acquired resistance to BRAFi in melanoma cells with altered PTEN, we first examined the activation of MAPK and PI3K/AKT signalings in parental and resistant cells with and without the BRAFi PLX4032 (Figure 2). As expected, PLX4032 could rapidly and significantly reduce the phosphorylation of ERK½ in a dose or time dependent-manner in all parental cells, whereas the phosphorylation of ERK½ in all resistant cells was maintained at higher and significantly longer levels compared to parental cells (Figure 2), consistent with MAPK reactivation in resistant cells. Notably, all resistant cells with wildtype PTEN maintained a significantly higher level of activated AKT compared to their parental counterparts (Figure 2), correlating hyperactivated AKT pathways with resistance in wildtype PTEN melanomas. In contrast with resistant melanoma cells carrying wildtype PTEN, activation of AKT in resistant cell lines with PTEN loss (WLH6215) or PTEN mutant (sk-mel 28) was overtly or slightly decreased, suggesting that the activation of the AKT pathway is only associated with resistance in melanomas with wildtype PTEN. More importantly, our data raise the possibility that mechanisms of resistance to BRAFi may be different in wildtype and PTEN-deficient melanomas.
Figure 2. Oncogene addiction resulted by both hyperactivated ERK and AKT pathways is dependents on PTEN status in BRAF inhibitor-resistant melanoma.
(A and B) WM88 cells with wildtype PTEN or WLH6215 cells with impaired PTEN and their PLX4032-resistant counterparts were treated with 2μM PLX4032 for the indicated time (A) or indicated the concentration of PLX4032 for 2 hours (B), and the effects on the activation of ERK½ and AKT were determined by immunoblotting for p-ERK and p-AKT. The protein level of total ERK, total AKT and tubulin are as loading controls. (C) WM9, WM858, sk-mel 28 and A375sm cells and their PLX4032-resistant counterparts were treated with indicated concentration of PLX4032 for 2 hours, and the effects on the activation of ERK½ and AKT were determined by immunoblotting for p-ERK and protein p-AKT levels. The protein level of total ERK, total AKT and tubulin are as loading controls.
Synergistic growth inhibition by combining AKT, MEK, and BRAF suppression depends on PTEN status in BRAF inhibitor-resistant melanoma
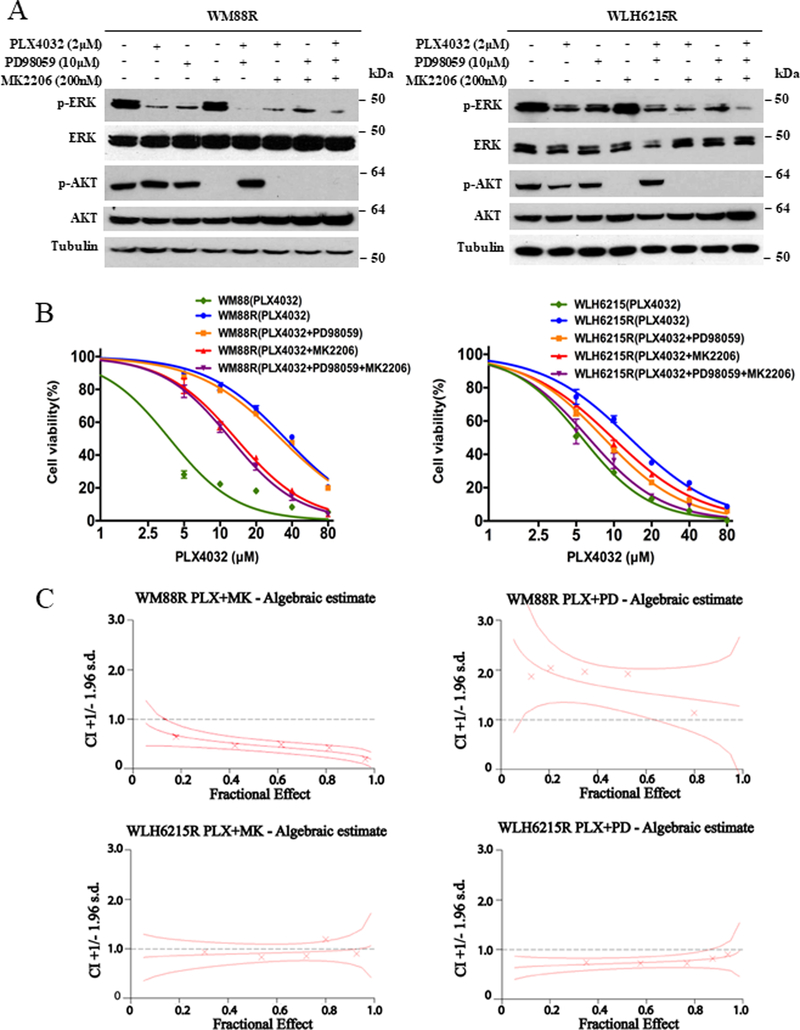
To further assess the distinct roles of the MAPK and AKT signaling pathways in BRAFi-resistant melanoma with/without wildtype PTEN, we tested the ability of the AKT inhibitor MK2206 and the MEK inhibitor PD98059 on inhibiting the growth of PRM cells. As shown in Figure 3A, although both PLX4032 and PD98059 were able to reduce the activation of ERK independently, neither PLX4032 nor PD98059 was able to inhibit cell growth (Figure S1) in BRAFi-resistant/wildtype PTEN melanoma cells. Moreover, a combination PLX4032 and PD98059 could significantly reduce the activation of ERK½ (Figure 3A, S2A, and S3A); yet still do not affect the cell growth (Figure 3B, S2B, and S3B). This data suggests that inhibition of MAPK cannot suppress the proliferation of BRAFi-resistant melanoma cells with wildtype PTEN; however, it could also indicate the existence of reactivation of MAPK signaling. In fact, PD98059 could not enhance the efficacy of PLX4032, and there was no synergism in the combination of PD98059 and PLX4032 (combination index (CI) greater than 1) (Figure 3C, Supplementary Table 3). However, the AKT inhibitor MK2206 alone or combination with PLX4032 significantly suppressed the activation of AKT and inhibited the WM88R cells proliferation with high synergistic inhibition (Figure 3B, 3C, and S1B; Supplementary Table 3), suggesting that the suppression of AKT can rescue the sensitivity to BRAFi in resistant melanoma WM88R with wildtype PTEN.
Figure 3. Synergistic growth inhibition of combination with AKT, MEK, and BRAF inhibitors also is dependents on PTEN status in BRAF inhibitor-resistant melanoma.
(A) Western blotting showed indicated protein levels from WM88R and WLH6215R human BRAF inhibitor-resistant melanoma cells treated for 2 hours with 2.0μmol/L PLX4032 (+), 10μmol/L MEK inhibitor PD98059 (+) or 200 nmol/L AKT inhibitor MK2206 (+) and DMSO (−) control. The protein level of total ERK, total AKT and tubulin are as loading controls. (B) Cell viability curves of WM88R or WLH6215R human BRAF inhibitor-resistant melanoma cells in response to PLX4032 or combination of PLX4032 and PD98059, MK2206 or both were determined using CCK8 assay. Error bars denote s.d. for biological three repeats. (C) Graphs of combination indexes (CI) for WM88R or WLH6215R human BRAF inhibitor-resistant melanoma cells treated with the combination of PLX4032 and MK2206 or PD98059 were determined using the Chou and Talalay method.
Similar results were noted in the other three BRAFi-resistant melanoma cell lines with wildtype PTEN (WM9R, WM858R, and A375smR). That is, treatments with the BRAFi PLX4032 and/or the MEK inhibitor PD98059 inhibited the activation of ERK½, but not cell growth, while the AKT inhibitor MK2206 alone or in combination with PLX4032 significantly inhibited both AKT activation and cell growth in all BRAFi-resistant/wildtype PTEN melanoma cell lines, confirming that activated AKT confers the resistance to BRAFi in melanoma with wildtype PTEN (Figure S2 and S3, Supplementary Table 3).
Interestingly, PD98059 or MK2206 not only suppressed phosphorylation of ERK½ or AKT respectively but either in combinations with PLX4032 inhibited the cell growth with moderate to high synergy in the BRAFi-resistant/PTEN-impaired melanoma cell lines WLH6215R and sk-mel 28R (Figure 3, S3C; Supplementary Table 3). These data suggest that suppression of p-ERK level mediated by the combination of PLX4032 and PD98059 is critical for this synergistic growth inhibition in BRAFi-resistant melanoma with impaired PTEN.
We also found that the combined inhibition of BRAF, MEK, and AKT, through the triplet-combination of PLX4032 with PD98059 and MK2206, was more effective in growth inhibition of PTEN-impaired PRM cells than BRAFi combined with either AKT or MEK inhibitor alone. In contrast, there was no significant difference in the growth of PTEN wildtype PRM cells when the triple combination was compared to PLX4032 plus MK2206 (Figure 3B, S2B, and S3B).
Impaired PTEN in BRAFV600E mutant melanoma influences resistance to BRAF inhibition
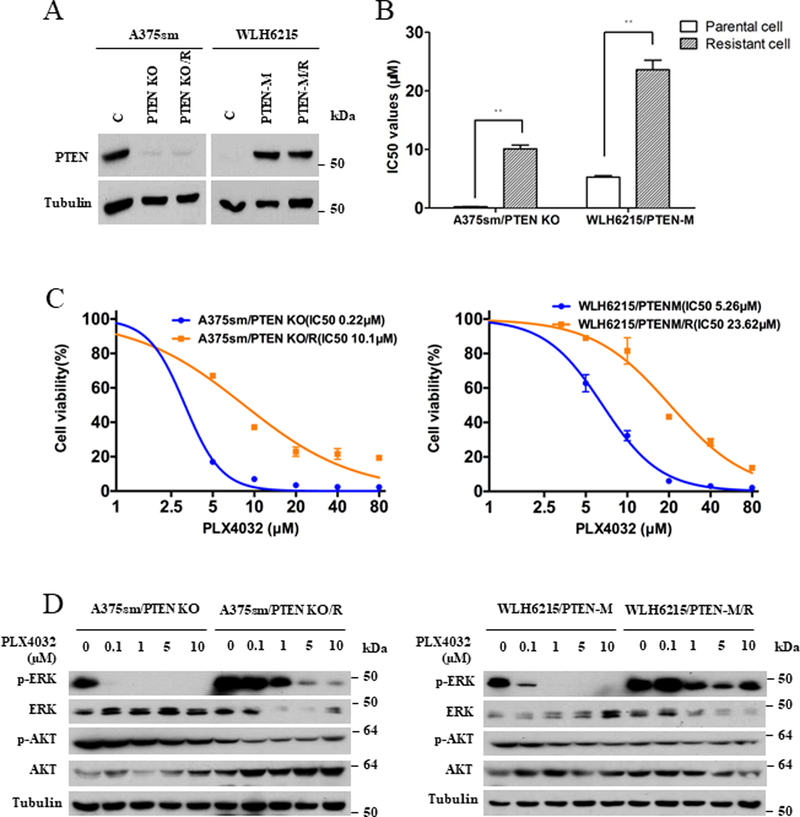
To further confirm that the roles of the MAPK or PI3K/AKT signaling pathways in BRAFi resistance are dependents on the PTEN status in melanoma cells, we used CRISPR/Cas 9 technology to create both a PTEN deficient cell line (A375sm/PTEN KO) and a PTEN mutant cell line (WLH6215/PTEN-M) in which the mutation PTEN C124A was introduced into WLH6215 cells, which do not express PTEN. Figure 4A shows that PTEN protein was absent in A375sm/PTEN KO cells and re-expressed in WLH6215/PTEN-M cells using western blot assays. We then employed the A375sm/PTEN KO and WLH6215/PTEN-M cells successfully to establish the BRAFi-resistant cell lines A375sm/PTEN-KO/R and WLH6215/PTEN-M/R with RI 46.0 and 4.5 respectively (Figure 4B, 4C; Supplementary Table 2). The activation of MAPK or PI3K/AKT signaling in these cells was examined with and without PLX4032 treatment. As expected, PLX4032 reduced phosphorylated Erk½ in a dose-dependent fashion in all parental cells, whereas all resistant cells showed strong resistance to PLX4032-induced ERK inhibition (Figure 4D). However, similar to the results of WLH6215R and sk-mel 28R cells with impaired PTEN (Figure 2A, 2C), p-AKT was decreased in A375sm/PTEN KO/R and WLH6215/PTEN-M/R cells (Figure 4D), verifying that AKT does not contribute to the resistance to BRAFi in melanoma with impaired PTEN. Moreover, similar to WLH6215R and sk-mel 28R cells, treatment of either PD98059 or MK2206 in combination with PLX4032 could significantly suppress the phosphorylation of ERK½ or AKT, respectively, in A375sm/PTEN KO/R and WLH6215/PTEN-M/R cells (Figure S4A). Both inhibitors could enhance the sensitivity to PLX4032 and inhibit cell growth with moderate synergistic to highly synergistic inhibition in BRAFi-resistant A375sm/PTEN KO/R and WLH6215/PTEN-M/R melanoma cells (Figure S4B, S4C; Supplementary Table 3). These data confirm, in contrast to PTEN wildtype cells, PTEN-impaired melanoma cells require only MAPK-based resistance mechanism for BRAF inhibition.
Figure 4. Impaired PTEN in BRAF V600E mutated melanoma influences the resistance to BRAF inhibitor.
(A) The protein level of PTEN in A375sm (C), A375sm/PTEN KO (PTEN KO) and its BRAF inhibitor-resistant counterpart A375sm/PTEN KO/R (PTEN KO/R) or WLH6215 (C), WLH6215/PTEN-M (PTEN-M) cells and its BRAF inhibitor-resistant counterpart WLH6215/PTEN-M/R (PTEN-M/R) was determined using western blot. The protein level of tubulin is as loading controls. (B) IC50 values of A375sm/PTEN KO and WLH6215/PTEN-M parental and it’s BRAF inhibitor-resistant cells. Error bars denote s.d. for biological three repeats. Results are statistically significant between parental and resistant groups by Student’s t-test (*p<0.05, **p<0.01). (C) Cell viability curve of A375sm/PTEN KO and WLH6215/PTEN-M parental and their BRAF inhibitor-resistant cells. Error bars denote s.d. for biological three repeats. (D) A375sm/PTEN KO and WLH6215/PTEN-M cells and their BRAF inhibitor-resistant counterparts were treated with indicated concentration of PLX4032 for 2 hours, and the effects on MAPK or PI3K/AKT signaling pathway were determined by immunoblotting for p-ERK and p-AKT levels. The protein level of total ERK, total AKT and tubulin are as loading controls.
The activated AXL kinase is required for acquired resistance to BRAF inhibitors in melanoma with wildtype PTEN
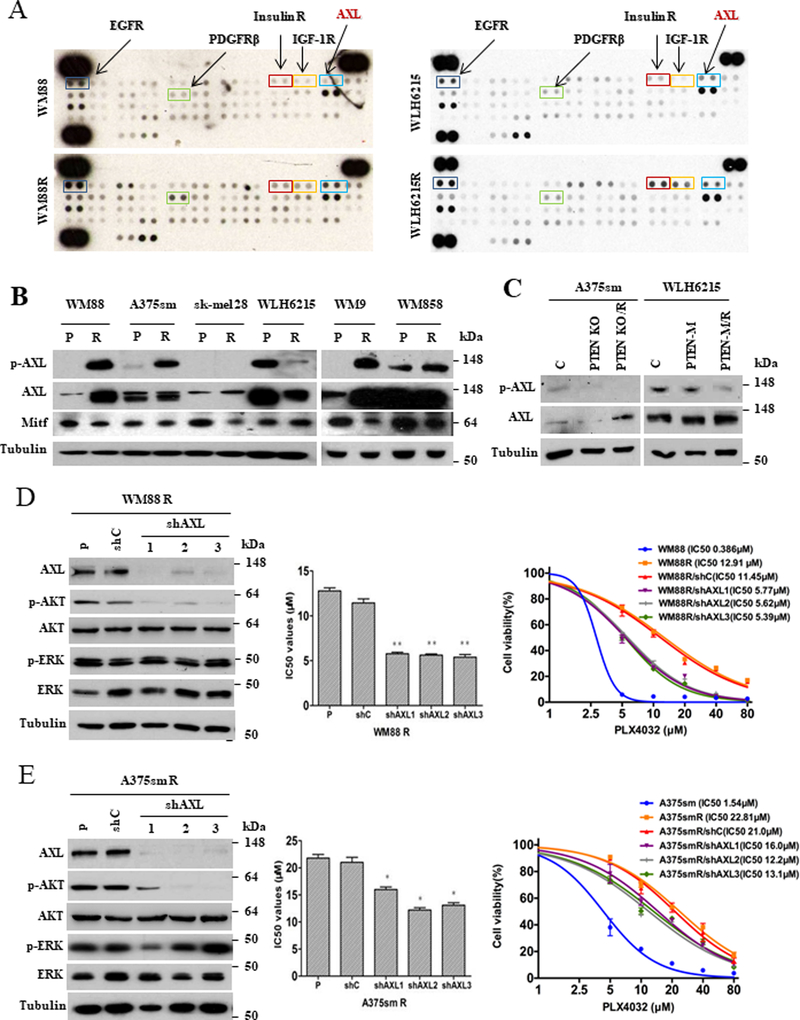
To explore the molecular mechanism by which the role of AKT in BRAFi resistance is dependent on the status of PTEN, we parallelly compared phosphorylation levels of receptor tyrosine kinases (RTKs) in WM88R with wildtype PTEN and WLH6215R with impaired PTEN using human phospho-RTKs array analysis. Consistent with previous studies,5 the activation of EGFR, PDGFRβ, Insulin R, and IGF-1R were increased in both BRAFi-resistant WM88R and WLH6215R cells (Figure 5A, S5A). Interestingly, the phosphorylation of AXL, an RTK of the TAM (TYRO3, AXL, and MERTK) family,25 was increased only in WM88R cells (Figure 5A). Moreover, we confirmed that AXL phosphorylation was elevated significantly in all PTEN wildtype PRM cells, including WM88R, WM9R, WM858R and A375smR cells (Figure 5B). However, the phosphorylation of AXL was decreased or unchanged in PTEN-impaired WLH6215R and sk-mel 28R PRM cells, as well as A375sm/PTEN KO/R and WLH6215/ PTEN-M/R cells (Figure 5C). These data suggest that AXL may help to regulate resistance to BRAF inhibitor in melanoma with wildtype PTEN. Because the high AXL and low MITF (low MITF/AXL ratio) were proposed to play an important role in the early resistance to multiple targeted drugs,26 we also examined MITF expression in all parental cells and their BRAFi-resistant counterparts. However, we found no association between low MITF/high AXL and resistance to BRAFi in our models (Figure 5B, S5A).
Figure 5. AXL protein mediates acquired-resistance to PLX4032 via activating PI3K/AKT pathway in melanoma with wild-type PTEN.
(A) Whole-cell extracts from WM88 and WM88R or WLH6215 and WLH6215R were incubated with the RTK antibody arrays, and phosphorylated proteins were determined by subsequent incubation with anti-phosphotyrosine horseradish peroxidase (each RTK spotted in duplicate, positive controls in corners). (B) The expression of AXL and p-AXL were confirmed using western blot with indicated parental (P), BRAF inhibitor-resistant (R) human melanoma cells. The protein level of tubulin is a loading control. (C) Western blotting (stripping the same western blots in figure 4A) showed the expression level of AXL and p-AXL in cells with PTEN knockout (PTEN KO) or mutant PTEN (PTEN-M) and their counterpart resistant cells (PTEN KO/R or PTEN-M/R). The protein level of tubulin is a loading control. C, the cells with empty vector as control. (D) Knockdown of AXL by shRNAs rescued the sensitivity of BRAF inhibitor-resistant WM88R cells to BRAF inhibitor and decreased the activation of AKT. The left panel showed the expression level of AXL, pAKT and indicated proteins using western blot. The middle panel showed the IC50 values of BRAF inhibitor for the WM88R with/without shRNA for AXL. The right panel showed the cell viability of cells stable expressing shRNAs for AXL in the treatment of BRAF inhibitor determined by CCK8 assay. WM88R: p, parent cells; shC, stable expressing shRNA control; shAXL1, shAXL2, and shAXL3, stable expressing different three shRNA constructs. Error bars denote s.d. for biological repeats. Results are statistically significant for all shAXL groups vs control by one-way ANOVA (*p < 0.05, **p < 0.01). (E) Knockdown of AXL by shRNA rescued the sensitivity of BRAF inhibitor-resistant A375smR cells to BRAF inhibitor and decreased the activation of AKT. The left panel showed the expression level of AXL, pAKT and indicated proteins using western blot. The middle panel showed the IC50 values of BRAF inhibitor for the A375smR with/without shRNA for AXL. The right panel showed the cell viability of cells stable expressing shRNAs for AXL in the treatment of BRAF inhibitor determined by CCK8 assay. A375smR: p, parent cells; shC, stable expressing shRNA control; shAXL1, shAXL2, and shAXL3, stable expressing different three shRNA constructs. Error bars denote s.d. for biological repeats. Results are statistically significant for all shAXL groups vs control by one-way ANOVA (*p < 0.05, **p < 0.01).
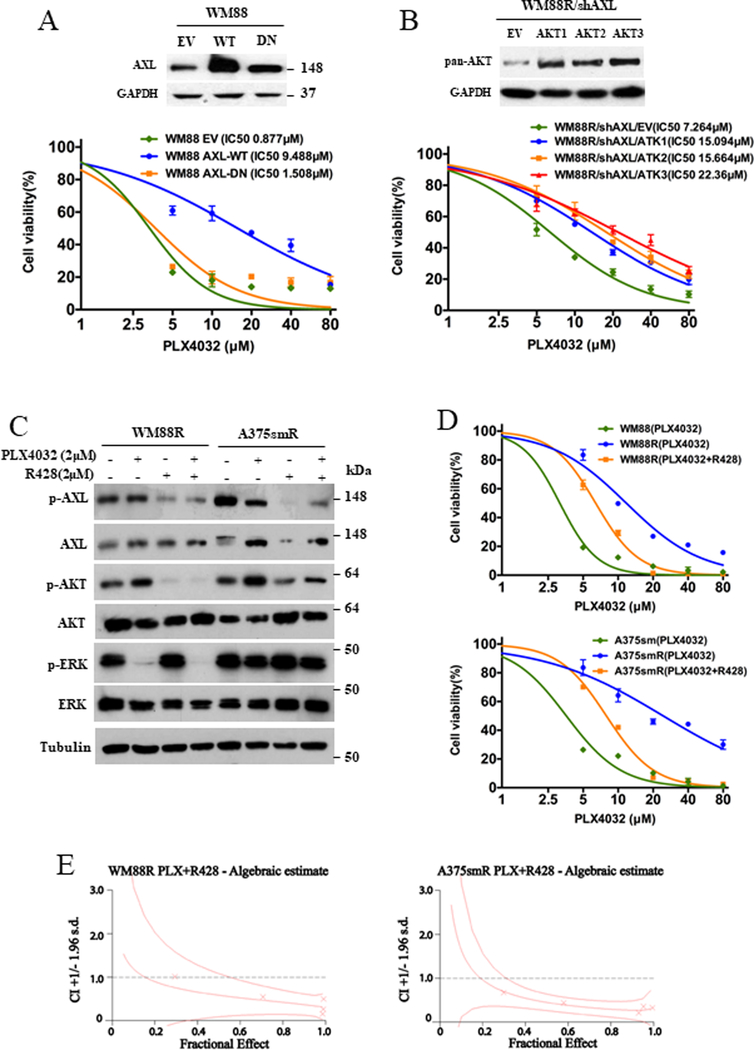
To further evaluate the importance of AXL in the regulation of resistance in BRAFV600E-mutant melanoma with wildtype PTEN, we examined whether downregulation of AXL in BRAFi-resistant PTEN-wildtype melanoma cells with elevated AXL could rescue the sensitivity of resistant-melanoma to BRAFi. Three shRNA targeting AXL successfully downregulated the protein level of AXL in both WM88R and A375smR cells (Figure 5D and 5E, left panels). Interestingly, knockdown of AXL in both resistant cell lines with wildtype PTEN enhanced the sensitivity to PLX4032 (Figure 5D and 5E, middle and right panels). In contrast, the overexpression of a wild-type AXL inhibited the sensitivity to PLX4032 in PTEN wildtype WM88, but the kinase-dead form of AXL did not (Figure 6A). However, neither wildtype nor mutant AXL could affect the ability of BRAFi resistance in PTEN loss WLH6215 cells (Figure S5E). Our data indicate that AXL is involved in the regulation of resistance in PTEN wildtype melanoma. Moreover, knockdown of AXL decreased the phosphorylation of AKT (Figure 5D and 5E, left panel), suggesting that the activation of AKT may involve the AXL-mediated BRAFi resistance in PTEN wildtype melanoma. To confirm the importance of AKT in AXL mediated BRAFi resistance, we introduced the expressing vector of Myr-AKT1, Myr-AKT2, and Myr-AKT3 into WM88R/shAXL cells, and found that all constitutively active AKT isoforms could enhance the ability of PLX4032 resistance and abrogate the shAXL-mediated inhibition of PLX4032 resistance (Figure 6B). We noted that the activation of AKT is required for BRAFi resistance in PTEN wildtype melanomas (Figure 2, 3, S1B, S2 and S3). Together, our data suggest that AXL activates the AKT pathway, thereby contributing to BRAFi resistance in melanoma with wildtype PTEN.
Figure 6. (A) AXL promotes resistance to BRAF inhibitor in PTEN wildtype melanoma cells.
(Top panel) The western blotting showed that the protein level of AXL in WM88 cells transfected with empty vector (EV), ALX wildtype (WT) or AXL kinase-dead mutant form (DN). (bottom panel) Cell viability curves of WM88 cells transfected with empty vector (EV), expressing AXL wildtype (WT) or AXL kinase-dead mutant form (DN) plasmid in response to PLX4032 were determined using CCK8 assay. Error bars denote s.d. for biological three repeats. (B) The constitutively active Akt abrogates the inhibition of PLX-4032 resistance mediated by shAXL. (Top panel) The western blotting showed that the protein level of AKT in WM88R/shAXL cells transfected with empty vector (EV), expressing Myr-AKT1 (AKT1), Myr-AKT2 (AKT2) or Myr-AKT3 (AKT3) plasmid. (bottom panel) Cell viability curves of WM88R/shAXL cells transfected with empty vector (EV), expressing Myr-AKT1 (AKT1), Myr-AKT2 (AKT2) or Myr-AKT3 (AKT3) plasmid in response to PLX4032 were determined using CCK8 assay. Error bars denote s.d. for biological three repeats. (C, D, and E) A pharmacological inhibitor R428 of AXL decreases the phosphorylation of AKT and restores the sensitivity of WM88R and A375smR cells to BRAF inhibitor with synergistic inhibition. (C) The expression of indicated proteins in WM88R and A375smR cells treated with 2 μM AXL inhibitor R428 and 2 μM BRAF inhibitor PLX4032 for 2 hours was determined by western blot. (D) The cell viability of WM88R and A375smR treated with BRAF inhibitor, or combination of BRAF inhibitor and AXL inhibitor R428 was determined by CCK8 assay. Error bars denote s.d. for biological three repeats. (E) Combination indexes (CI) for WM88R or A375smR cells treated with the combination of PLX4032 and R428 were determined using the Chou and Talalay method.
We next sought to validate our findings using a pharmacological inhibitor of AXL. Figure 6C and D show that the AXL inhibitor R428 could decrease AXL and AKT phosphorylation and restore the sensitivity to BRAFi in WM88R and A375smR cells with synergistic inhibition (CI<1) (Figure 6E, Supplementary Table 4). Taken together, our results demonstrate that AXL activation is necessary for the acquisition of resistance to PLX4032 in melanoma cells with wildtype PTEN via regulating the activation of AKT pathway. Moreover, AXL inhibitor along or combination with PLX4032 upregulated the protein level of cleaved-PARP, cleaved- caspase 3, cleaved-caspase 7 and cleaved-caspase 9 in BRAFi resistant melanoma cells (Figure S6A), suggesting that the inhibition of AXL by small molecular inhibitor could reduce tumor cell apoptosis.
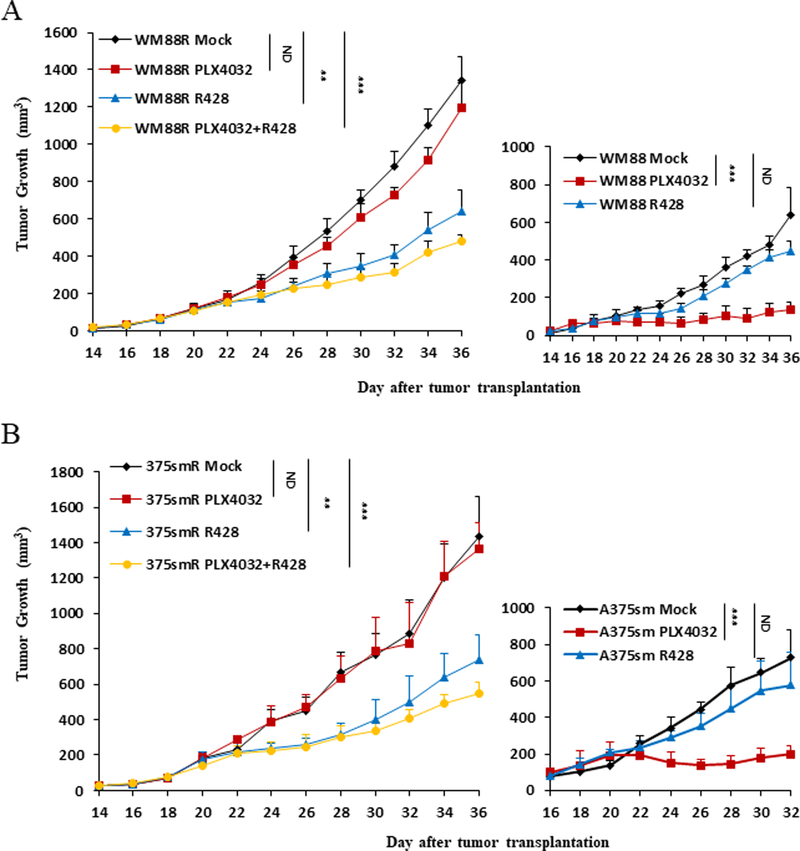
To further examine the consequences of AXL inhibition in a more relevant preclinical mouse model in vivo, we designed and performed animal studies using xenograft models. Fourteen or 16 days after transplantation of WM88 and WM88R or A375sm and A375smR cells by subcutaneous injection into NSG mice, PLX4032 and R428 were administered to mice orally twice daily for more than two weeks. As expected, the tumors arising from parent WM88 and A375sm cells in the mice treated with PLX4032 grew significantly more slowly than those in mice treated with vehicle control (Figure 7A and 7B, right panel). In contrast, the tumors arising from resistant WM88R and A375smR cells treated with PLX4032 grew similarly to those treated with vehicle control (Figure 7A and 7B, left panel). However, an AXL inhibitor R428 or combination of R428 with PLX4032 significantly inhibited melanoma growth in mice compared with their vehicle control (Figure 7A and 7B, left panel ), and reduced the growth rate of resistant WM88R and A375smR melanomas to a level comparable to parent WM88 and A375sm melanomas, although the resistant WM88R and A375smR melanomas grew significantly faster than WM88 and A375sm parent tumors in vivo due to constitutively activated Erk½ and AKT in resistant cells (Figure 2). Consistent with the results from treated cells in vivo, the tumor treated with R428 showed weeker AKT and AXL activation by immunohistochemical staining (Figure S6B). Our data demonstrate that blocking AXL activation inhibits BRAFi-resistant melanoma growth and suggest that AXL is a promising diagnostic marker and therapeutic target for BRAFi-resistant melanoma with wildtype PTEN.
Figure 7. Inhibition of AXL by a pharmacological inhibitor R428 blocks tumor growth in vivo.
(A, left panel) The growth curves of resistant WM88R tumor cells treated with mock (WM88R Mock, n=10), BRAF inhibitor (WM88R PLX 4032, n=7), AXL inhibitor R428 (WMR R428, n=8) or combination of AXL inhibitor R428 with PLX4032 (WM88R PLX4032+R428, n=7). (A, right panel) The growth curves of parent WM88 tumor cells treated with mock (WM88 Mock, n=6), BRAF inhibitor (WM88 PLX 4032, n=6), AXL inhibitor R428 (WM88 R428, n=7). Each point represents mean tumor volume ± s.e.m. ND, no statistical difference; *p<0.05; ** p<0.01; *** p<0.001. (B, left panel) The growth curves of resistant A375smR tumor cells treated with mock (375smR Mock, n=10), BRAF inhibitor (375smR PLX 4032, n=8), AXL inhibitor R428 (375smR R428, n=10) or combination of AXL inhibitor R428 with PLX4032 (375smR PLX4032+R428, n=8). (B, right panel) The growth curves of parent A375sm tumor cells treated with mock (A375sm Mock, n=6), BRAF inhibitor (A375sm PLX 4032, n=6), AXL inhibitor R428 (A375sm R428, n=7). Each point represents mean tumor volume ± s.e.m. ND, no statistical difference; *p<0.05; ** p<0.01; *** p<0.001.
Discussion
Multiple genetic mutations within tumors not only cause lesion-specific responses to targeted therapy but also alter the molecular route to resistance to the targeted therapy. Understanding the alternative molecular route that causes resistance would improve the clinical application of targeted therapies through improved patient selection. Malignant melanoma is recently characterized as widely mutated in multiple genes and with poor response to current treatment. The revolutionary discovery of BRAF mutation in most melanoma led to the development of targeted inhibition by inhibitors in patients with BRAF mutation, but the strength of efficacy is short and limited by acquired drug resistance, resulting in disease progression due to multiple genetic mutations. Loss or mutation of PTEN occurs in up to 30% of melanomas, frequently with a concurrent activating BRAF mutation18 and advanced malignancy.27,28 Clinically, inactivation of PTEN in patients with melanoma with BRAF mutations is associated with worse outcomes in late stages patients with BRAFi compared with in wild-type PTEN melanoma.29,30 Recent studies reported that melanoma cell lines with inactivated PTEN can be growth arrested by BRAF and MEK inhibitors, but that they are resistant to apoptosis induction.31,32 These studies support that PTEN inactivation identifies a distinct clinically significant subset of melanomas, implying that the status of PTEN may affect the molecular mechanism of late acquired resistance to BRAFi.
Through comparative study, we here report that the MAPK signaling pathway was reactivated in all BRAFi resistant melanoma cells as a major pathway of acquired BRAFi resistance independent of PTEN status, but the PI3K/AKT signaling pathway was only reactivated in BRAFi resistant melanoma cells with wildtype PTEN. Interestingly, neither treatment with MEK inhibitor alone nor combination with PLX4032 could inhibit the growth of resistant melanoma cells with wildtype PTEN, whereas an AKT inhibitor could significantly inhibit their growth. However, both MEK inhibitor and AKT inhibitor combination with BRAFi PLX4032 or triple combinations could inhibit proliferation of the resistant melanoma cells with impaired PTEN. Notably, we found that activated AXL played a significant role in the regulation of acquired BRAFi resistance in melanoma cells with wildtype PTEN through activating AKT, but not in melanoma with impaired PTEN. Our data demonstrated that AXL/AKT axis mediated-resistance to BRAFi depends on PTEN status.
Consistent with previous studies,5 the activation of several receptor tyrosine kinases (RTK) such as EGFR, PDGFR, IGF1R and Insulin receptor was enhanced in BRAF inhibitor-resistant melanomas, supporting MAPK reactivation as a pathway of acquired BRAF inhibitor resistance in melanoma. Although the combination of BRAF and MEK inhibitors is the backbone of targeted therapy in BRAF-mutated melanoma,33 treatment failure still occurs with the resistance mechanisms observed in the BRAF/MEK inhibitor combination being analogous to those seen in patients on BRAF inhibitor monotherapy.34 Indeed, our study shows that both the MEK inhibitor alone or the BRAF/MEK inhibitor combination fail to abrogate the growth of melanomas with wildtype PTEN due to activated AKT of the second core PI3K pathway of acquired BRAF inhibitor resistance.23
The activated AKT/PI3K core pathway of acquired BRAFi resistance in melanoma is highly dependent on PTEN status in our models. In BRAF mutant melanoma with wildtype PTEN, the PI3K/AKT pathway is a second bona fide core pathway of late drug resistance.23 In contrast, in BRAF mutant melanoma with impaired PTEN, the activation of AKT was decreased, while the MAPK/ERK pathway was reactivated as the core pathway of drug resistance35 because of the intrinsic higher level of activated AKT due to PTEN inactivation. Indeed, the combination of either an AKT or MEK inhibitor with PLX4032 or the three-inhibitor combination could abrogate the growth of BRAFi resistant melanoma with impaired PTEN.
As reported, BRAF inhibition leads to increase activation of multiple receptor tyrosine kinase signalings, which may then promote PI3K signaling.5,9,36 In our study, although EGFR, PDGFR, IGF1R, and Insulin receptor were elevated in BRAFi-resistant melanomas, only BRAFi resistant melanomas with wildtype PTEN exhibited elevated activated AKT, suggesting the activated AKT signaling does not associate with these RTKs, and an alternative factor exists. Indeed, we showed that activated AXL, an RTK of the UFO and Ark family,25 is dependent on PTEN status in BRAFi-resistant melanomas, and we confirmed that AXL could activate AKT and confer resistance to BRAFi in melanoma with wildtype PTEN.
AXL can promote aggressiveness of various types of tumors including melanoma through PI3K/AKT and MAPK signaling pathways, and its expression can be transcriptionally regulated by multiple factors.37,38 Consistent with Brand et al.’s study of activation of AXL by EGFR-MAPK signaling via crosstalk 39 we also found an elevated EGFR in most of BRAFi resistant cells (Figure 5SA). The knockdown of AXL in both PTEN wildtype WM88R and A375smR cells could slightly block phosphorylation of EGFR (Figure S5B & S5C). Moreover, downregulation of EGFR by siRNA could significantly reduce the AXL expression, suggesting that there is a crosstalk between AXL and EGFR (Figure S5D). However, we found that level of EGFR was increased in both PTEN impaired sk-mel 28R and WLH6215R cells; in contrast, activated AXL was decreased or undetectable compared with their parent-sensitive cells (Figure S5A). Moreover, both PTEN wildtype allele WM9R and WM858R with a elevated AXL expressed a reduced or undetectable EGFR compared with their BRAFi sensitive cells (Figure S5A), suggesting that there is no correlation between AXL and EGFR in those cells, and others mechanisms may involve the regulation of AXL activation such lipid interaction or crosstalk with other factors. 40 AXL has been identified to play a role in resistance to EGFR tyrosine kinase inhibitors (TKIs)39 or other MAPK pathway inhibitors in non-small cell lung cancer (NSCLC),41 head and neck cancer (HNC),42 and triple-negative breast cancer (TNBC),43 as well as melanoma.26,37,38 Similar to our study, Yao et al proposed that AXL activity confers resistance to BRAF inhibitor in BRAFV600E mutant gliomas. 44 A recent study by Miller et al. 45 also reported that inhibition of MAPK signaling by MEK inhibitor increased total and phosphorylated AXL and that co-treatment with AXL inhibitor synergistically reduced tumor growth in vivo.
In contrast to previous studies of MITF low and AXL high resistant phenotypes,26 our models all express MITF and do not clearly associate MITF low/AXL high with the resistant phenotype. The identification of activated AXL is associated with the resistance to BRAFi in melanoma with wildtype PTEN, but not in melanoma with impaired PTEN, suggesting that activated AXL is not the mechanism for resistance in the subsets of melanoma with BRAF mutant and impaired PTEN.
We showed that inhibition of AXL by both shRNA and inhibitor not only inhibited the activation of AKT but also blocked tumor growth in vitro and in vivo in melanoma with wildtype PTEN. The constitutively active AKTs could abrogate the inhibition of BRAF inhibitor resistance mediated by silencing AXL. Our data demonstrate for the first time that AXL plays a major role in the regulation of acquired BRAFi resistance by activating the AKT/PI3K core signaling pathway, raising the possibility that AXL is a new prognostic marker and therapeutic target for BRAFi resistance in BRAFV600E mutant melanoma with wildtype PTEN. Our findings have significant implications for personalized medicine strategies and therapies for acquired BRAFi resistance in melanomas with different PTEN status.
Materials and Methods
Plasmids, antibodies, cell lines, cell culture, and reagents
Plasmids: AXL shRNAs expressing plasmids, siAXL and siEGFR were purchased from Open Biosystems (GE Dharmacon, Lafayette, CO). CRISPR/cas9 knockout vectors for PTEN were constructed using lentiCrispr V2 vector (Addgene, Cambridge MA). PTEN mutant (C124A) expressing plasmid, AXL wildtype expressing plasmid, Myr-AKT1, Myr-AKT2, and Myr-AKT3 were provided by Addgene (Cambridge, MA). Antibodies: anti-phospho-AKT (Ser473), anti-AKT, anti-phospho-ERK ½ (Thr202/Tyr204), anti-ERK½, anti-tubulin, anti-PTEN, anti-Axl, anti-phospho-Axl, anti-GAPDH, anti-EGFR, anti-PDGFRβ, anti-Her2/ErbB2, anti-cleaved Caspase −3 (Asp175), anti-Caspase-3, anti-PARP, anti-cleaved PARP (Asp214), anti-Caspase-9, anti-cleaved Caspase-9 (Asp330), anti-Caspase-7 and anti-cleaved Caspase-7 (Asp198) were purchased from Cell Signaling (Danvers, MA, USA); anti-phospho-Axl was purchased from LifeSpan BioScience, Inc (Seattle, WA); anti-β-actin antibody was purchased from Santa Cruz (Dallas, TX). Anti-Mitf antibodies were purchased from Abcam (Cambridge, MA). Human melanoma cell lines: A375sm was a gift from Dr. Isaiah Fidler (M.D. Anderson Medical Center, Houston, TX); WM88, WM9, WLH6215, WM858 cell lines were a gift from Dr. Meenhard Herlyn (The Wistar Institute, Philadelphia, PA); sk-mel 28 was obtained from American Type Culture Collection (ATCC, Manassas, VA). The stable expressing cell lines were established by transfection using lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) and selected by antibiotics G418 or puromycin (Sigma, St. Louis, MO). All cell lines were tested negative with mycoplasma and authenticated using short tandem repeat (STR) DNA profiling routinely. The cell lines for in vivo animal studies were negative for infectious microbes by molecular testing of biological materials (MTBM) test [the animal proposal LCBG023, approval by NCI-Bethesda Animal Care and Use Committee (ACUC)]. A BRAF inhibitor (PLX4032), a MEK inhibitor (PD98059) and an AKT inhibitor (MK2206) were purchased from Selleckchem (Houston, TX) or APExBIO (An Apoptosis and Epigenetics Company, Houston, TX).
Establishment of PLX4032-resistant melanoma cells
Aliquots of melanoma cells in the exponential growth phase were seeded into 75 cm2 cell culture flasks. PLX4032 (1μmol/ml) was added for 48 h during the mitotic phase, and then the cells were transferred into drug-free culture medium until the next mitotic phase (around 7–10 d), after which PLX4032 was added for the next 48 h at twice the previous concentration. We continued this process in a stepwise increasing concentration of PLX4032 while observing cell death every day, changing to the fresh complete culture medium, and performing the CCK8 assay regularly. This process was continued for six months until the melanoma cells grew stably in the PLX4032-containing medium.
Cellular proliferation and drug treatments
Cell proliferation experiments were carried out in 96-well plates and measured using CCK8 assay.46,47 Cells in the exponential phase of growth were inoculated into each well at a density of 3×103 cells per well, with five wells for each set of conditions. Drug treatments initiated at 24 h and lasted for 72 h. After then, 10μl CCK8 was added to each well, and the cells were incubated at 37°C under 5% CO2 for 4 h. The optical density (OD) of each well at a wavelength of 450 nm was determined using a microplate reader. Cell viability was calculated according to the following equation: (drug-supplemented OD-blank control OD)/ (normal control OD-blank control OD) ×100%. Origin 6.1 or GraphPad prism 6 software was utilized to plot the survival versus drug concentration curve and calculate the 50% inhibitory concentration (IC50). The resistance index (RI) was calculated as the ratio between the IC50 value of resistant cells and that of parental cells.
Western blot analysis
Immunoblots were performed on lysates generated from cultured cells and tissues solubilized in RIPA buffer.46,47
Phospho-protein array
The profiling of phosphorylated proteins was determined using the proteome profiler human phospho-kinase array (R&D systems, ARY003B, Minneapolis, MN). Briefly, whole-cell extracts were incubated on the human phospho-kinase antibody arrays, and phosphorylation status was determined by subsequent incubation with anti-phosphotyrosine horseradish peroxidase as described by the manufacturer.
Immunohistochemistry
Tumor tissues were fixed in 10% buffered formalin solution (pH7.2) for 16 h, and/or frozen in OCT compound and serially sectioned to 15 μm at 20 °C. Immunohistochemistry was performed as described.46,47 Immunoreactivity scores were analyzed using ImageScope V 10.0 software from Aperio Technologies (Vista, CA).
Preclinical drugs treatments
Parents and resistant melanoma cell lines WM88 or WM88R cells (200 μl, 1×10e6) were transplanted into NSG male mice (purchased from The Jackson Laboratory) between 4 and 6 weeks of age subcutaneously (SQ). Mice harboring the inoculated tumor cells 14 days after transplantation with similar tumor size were randomly assigned to different treatment groups using a parallel group design and treated with either 50 mg/kg or 75 mg/kg of the PLX4032 or 7 mg/kg or 75 mg/kg of AXL inhibitor R428 twice daily by oral gavage for 21 days. The cage of mouse and treatments were coded with a number. The tumor size was monitored by measuring the length (L) and width (W) using caliper every 2 days, and the volumes were calculated via the formula: (L ×W2) × 0.5. Each group was initialed with 10 mice [based on the typical power of 80% to 90%, 6 animals is considered adequate for hypothesis testing. 48, 49 All mouse procedures were performed according to NIH guidelines [the animal proposal LCBG023, approval by NCI-Bethesda Animal Care and Use Committee (ACUC)].
Calculation of combination index
Using the CalcuSyn Version 2.11 (Copyright Biosoft, USA) software, the combination index (CI) was calculated for cells receiving combination therapy according to the Chou and Talalay mathematical model for drug interactions. The resulting CI theorem of Chou-Talalay offers a quantitative definition for an additive effect (CI =1), synergism (CI < 1), and antagonism (CI > 1) in drug combinations.50
Statistical analysis
All data were represented as the mean of at least triplicate samples ± standard deviation. All statistical analyses in this study were performed using SPSS 13.0 and GraphPad Prism 6. A two-tailed Student’s t-test or one-way ANOVA was used to test the differences in sample means for data with normally distributed means. The significance of mean values between two groups was analyzed by Student’s t-test or multiple t-tests. When three or more groups were compared, the one-way ANOVA test and post hoc Bonferroni analysis were used. Mann–Whitney U-test was used for non-parametric data. P values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported in part by funding from the NIH intramural research program. Thanks, are due to Dr. Miriam Anver for assistance with immunohistochemistry.
Footnotes
Conflict of interest
The authors have no competing financial interest to declare.
The authors have no conflict of interest to declare.
REFERENCES
- 1.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature 2002; 417(6892):949–954. [DOI] [PubMed] [Google Scholar]
- 2.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011; 364(26):2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 2010; 363(9):809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med 2012; 366(8):707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010; 468(7326):973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 2010; 468(7326):968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, Haanen J, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 2014; 508(7494):118–122. [DOI] [PubMed] [Google Scholar]
- 8.Girotti MR, Pedersen M, Sanchez-Laorden B, Viros A, Turajlic S, Niculescu-Duvaz D, et al. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer Discov 2013; 3(2):158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 2010; 18(6):683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012; 367(18):1694–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G, Davies MA. Targeted therapy resistance mechanisms and therapeutic implications in melanoma. Hematol Oncol Clin North Am 2014; 28(3):523–536. [DOI] [PubMed] [Google Scholar]
- 12.Ribas A, Gonzalez R, Pavlick A, Hamid O, Gajewski TF, Daud A, et al. Combination of vemurafenib and cobimetinib in patients with advanced BRAF(V600)-mutated melanoma: a phase 1b study. Lancet Oncol 2014; 15(9):954–965. [DOI] [PubMed] [Google Scholar]
- 13.Sullivan RJ, Fisher DE. Understanding the biology of melanoma and therapeutic implications. Hematol Oncol Clin North Am 2014; 28(3):437–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene 2003; 22(20):3113–3122. [DOI] [PubMed] [Google Scholar]
- 15.Healy E, Rehman I, Angus B, Rees JL. Loss of heterozygosity in sporadic primary cutaneous melanoma. Genes Chromosomes Cancer 1995; 12(2):152–156. [DOI] [PubMed] [Google Scholar]
- 16.Herbst RA, Weiss J, Ehnis A, Cavenee WK, Arden KC. Loss of heterozygosity for 10q22–10qter in malignant melanoma progression. Cancer Res 1994; 54(12):3111–3114. [PubMed] [Google Scholar]
- 17.Tsao H, Zhang X, Benoit E, Haluska FG. Identification of PTEN/MMAC1 alterations in uncultured melanomas and melanoma cell lines. Oncogene 1998; 16(26):3397–3402. [DOI] [PubMed] [Google Scholar]
- 18.Haluska FG, Tsao H, Wu H, Haluska FS, Lazar A, Goel V. Genetic alterations in signaling pathways in melanoma. Clin Cancer Res 2006; 12(7 Pt 2):2301s–2307s. [DOI] [PubMed] [Google Scholar]
- 19.Marsh Durban V, Deuker MM, Bosenberg MW, Phillips W, McMahon M. Differential AKT dependency displayed by mouse models of BRAFV600E-initiated melanoma. J Clin Invest 2013; 123(12): 5104–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med 2004; 10:789–799. [DOI] [PubMed] [Google Scholar]
- 21.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res 2011; 71(7):2750–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov 2014; 4(1):94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 2014; 4:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poulikakos PI, Rosen N. Mutant BRAF melanomas--dependence and resistance. Cancer Cell 2011;19(1):11–15. [DOI] [PubMed] [Google Scholar]
- 25.Linger RM, Keating AK, Earp HS, Graham DK. TAM receptor tyrosine kinases: biologic functions, signaling, and potential therapeutic targeting in human cancer. Adv Cancer Res 2008; 100:35–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C, et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun 2014; 5:5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet 2009; 41:544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stahl JM, Cheung M, Sharma A, Trivedi NR, Shanmugam S, Robertson GP. Loss of PTEN promotes tumor development in malignant melanoma. Cancer Res 2003; 63:2881–2890. [PubMed] [Google Scholar]
- 29.Nathanson KL, Martin AM, Wubbenhorst B, Greshock J, Letrero R, D’Andrea K, et al. Tumor genetic analyses of patients with metastatic melanoma treated with the BRAF inhibitor dabrafenib (GSK2118436). Clin Cancer Res 2013; 19:4868–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bucheit AD, Chen G, Siroy A, Tetzlaff M, Broaddus R, Milton D, et al. Complete loss of PTEN protein expression correlates with shorter time to brain metastasis and survival in stage IIIB/C melanoma patients with BRAFV600 mutations. Clin Cancer Res 2014; 20:5527–5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gopal YN, Deng W, Woodman SE, Komurov K, Ram P, Smith PD, et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res 2010; 70:8736–8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng W, Gopal YN, Scott A, Chen G, Woodman SE, Davies MA. Role and therapeutic potential of PI3K-mTOR signaling in de novo resistance to BRAF inhibition. Pigment Cell Melanoma Res 2012; 25:248–258. [DOI] [PubMed] [Google Scholar]
- 33.Sullivan R, LoRusso P, Boerner S, Dummer R. Achievements and challenges of molecular targeted therapy in melanoma. Am Soc Clin Oncol Educ Book 2015; 177–186. [DOI] [PubMed] [Google Scholar]
- 34.Wagle N1, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol 2011; 29(22): 3085–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byron SA, Loch DC, Wellens CL, Wortmann A, Wu J, Wang J, et al. Sensitivity to the MEK inhibitor E6201 in melanoma cells is associated with mutant BRAF and wildtype PTEN status. Mol Cancer 2012; 11:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turke AB, Song Y, Costa C, Cook R, Arteaga CL, Asara JM, et al. MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer Res 2012; 72:3228–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Ye X, Tan C, Hongo JA, Zha J, Liu J, et al. AXL as a potential therapeutic target in cancer: role of AXL in tumor growth, metastasis and angiogenesis. Oncogene 2009; 28(39):3442–3455. [DOI] [PubMed] [Google Scholar]
- 38.Sensi M, Catani M, Castellano G, Nicolini G, Alciato F, Tragni G, et al. Human cutaneous melanomas lacking MITF and melanocyte differentiation antigens express a functional Axl receptor kinase. J Invest Dermatol 2011; 131(12):2448–2457. [DOI] [PubMed] [Google Scholar]
- 39.Brand TM, Iida M, Stein AP, Corrigan KL, Braverman CM, Luthar N, et al. AXL mediates resistance to cetuximab therapy. Cancer Res. 2014; 74(18):5152–64. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 40.Meyer AS, Zweemer AJ, Lauffenburger DA. The AXL Receptor is a Sensor of Ligand Spatial Heterogeneity. Cell Syst. 2015; 1(1):25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet 2012; 44(8):852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giles KM, Kalinowski FC, Candy PA, Epis MR, Zhang PM, Redfern AD, et al. Axl mediates acquired resistance of head and neck cancer cells to the epidermal growth factor receptor inhibitor erlotinib. Mol Cancer Ther 2013; 12(11):2541–2558. [DOI] [PubMed] [Google Scholar]
- 43.Meyer AS, Miller MA, Gertler FB, Lauffenburger DA. The receptor AXL diversifies EGFR signaling and limits the response to EGFR-targeted inhibitors in triple-negative breast cancer cells. Sci Signal 2013; 6(287): ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao TW, Zhang J, Prados M, Weiss WA, James CD, Nicolaides T. Acquired resistance to BRAF inhibition in BRAFV600E mutant gliomas. Oncotarget. 2017; 8(1):583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller MA, Oudin MJ, Sullivan RJ, Wang SJ, Meyer AS, Im H, et al. Reduced Proteolytic Shedding of Receptor Tyrosine Kinases Is a Post-Translational Mechanism of Kinase Inhibitor Resistance. Cancer Discov 2016; 6(4):382–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Y, Davicioni E, Triche TJ, Merlino G. The homeoprotein six1 transcriptionally activates multiple protumorigenic genes but requires ezrin to promote metastasis. Cancer Res 2006; 66(4):1982–1989. [DOI] [PubMed] [Google Scholar]
- 47.Yu Y, Khan J, Khanna C, Helman L, Meltzer PS, Merlino G. Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nat Med 2004; 10(2):175–181. [DOI] [PubMed] [Google Scholar]
- 48.Sheskin DJ. Handbook of parametric and nonparametric statistical procedures. 3rd ed. Boca Raton:CRC Press; 2003. [Google Scholar]
- 49.Hather G, Liu R, Bandi S, Mettetal J, Manfredi M, Shyu WC, et al. Growth rate analysis and efficient experimental design for tumor xenograft studies. Cancer Informatics 2014:13(Suppl 4)65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer research 2010; 70(2):440–446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.