Abstract
Objectives
Human papillomavirus-related oropharyngeal squamous cell carcinoma (HPV+ OPSCC) is increasing in incidence. Although HPV+ OPSCC has favorable prognosis, 10 to 25% of HPV+ OPSCCs eventually recur. We sought to evaluate the feasibility of detection of HPV16 E6/E7 expression in Circulating Tumor Cells (CTCs) and its utility as a prognostic tool in HPV16-associated OPSCC.
Materials and methods
We developed a highly sensitive RT-qPCR assay for HPV mRNA expression in EpCAM(+) CTCs. In 22 patients with early stage and locally advanced OPSCC we evaluated HPV16 E6/E7 expression in the EpCAM(+) CTC fraction at baseline and at the end of concurrent chemoradiotherapy. HPV status in pre-therapy formalin-fixed paraffin-embedded (FFPE) tumor biopsies was assessed by p16 immunohistochemistry and polymerase chain reaction (PCR) and double positives were subjected to Real-time qPCR assay for detection of HPV16, 18 and 31 types.
Results
Fourteen of 22 OPSCC (63.6%) were HPV DNA+/p16+. Among HPV+/p16+ patients, 10 patients (71.4%) were HPV16 DNA+. HPV16 E6/E7(+) CTCs were detected in 3 of 10 patients (30%) at baseline and 4 of 9 patients (44.4%) at the end-of-treatment, all of which were p16+/HPV16 DNA+. Survival analysis showed a significantly higher risk for disease relapse (p = 0.001) and death (p = 0.005) in patients with HPV16 E6/E7(+) baseline CTCs.
Conclusion
Detection of HPV E6/E7(+) CTCs might be a useful noninvasive test in liquid biopsy samples for determination of a clinically relevant HPV infection in HPV+ OPSCC. Combined interpretation of HPV E6/E7(+) CTCs with UICC staging data may lead to alteration of risk definition of patient subsets, with improved risk discrimination in early-stage disease.
Introduction
Human papillomavirus (HPV) is a well-recognized etiologic factor in oropharyngeal squamous cell carcinoma (OPSCC), and it has been demonstrated that seropositivity for antibodies against HPV16 oncogenic proteins E6 /E7 is present in prediagnostic samples of patients with OPSCC many years before diagnosis [1]. HPV16 is the subtype most commonly implicated in the pathogenesis of OPSCC, with a prevalence over 90%, followed by HPV18 (3%) [2]. During the past few years, the global burden of the disease has steadily increased, and it has been predicted to surpass cervical cancer in some developed countries [3]. It is commonly accepted that HPV-associated oropharyngeal tumors (HPV+ OPSCC) comprise a distinct disease entity, showing a distinct clinical behavior and better survival outcomes [1]. However, approximately 10–25% of HPV+ OPSCCs have bad prognosis and eventually recur [4]; this population needs to be identified.
The capacity to measure tumor cells circulating through the vasculature (circulating tumor cells-CTCs) might provide evidence of the aggressiveness of a tumor prior to detection of identifiable metastases [5]. Studies have shown that CTCs in the peripheral blood could be a valuable tool for real-time monitoring of tumor status, predicting potential metastasis and recurrence, monitoring treatment efficacy and predicting survival of cancer patients [6]. In addition, carcinogenesis in HPV+ OPSCC is driven by sustained expression of viral E6 and E7 oncogenes. E6 inhibits p53, while E7 binds to retinoblastoma protein (pRb), promotes its degradation and the release of the E2F transcription factor, resulting in deregulation of the G1/S cell cycle check point and the activation of S-phase re-entry and viral replication [7]. Therefore, HPV-E6/E7 oncogene expressing CTCs could be chosen as a unique biomarker for detection of viable tumor cells in the peripheral blood of OPSCC patients with transcriptionally active HPV infection.
Herein, we sought to assess whether CTCs expressing HPV16 E6/E7 oncogenes (HPV16 E6/E7(+)) could be detected before initiation of treatment in OPSCC patients and correlate with treatment outcome. Thus, we aimed to evaluate HPV16 E6/E7(+) CTCs as a non-invasive prognostic tool in HPV16+ OPSCC.
Patients and methods
Study design
Twenty-two (n = 22) patients with early stage and locally advanced (LA) OPSCC were included in this analysis. Written informed consent was obtained from all patients before participating in the study. The present study was approved by the Medical Ethical Committee of Attikon University hospital (Athens, Greece) and complies with the principles laid down in the Declaration of Helsinki.
Pre-therapy tumor biopsies (FFPE tissue) were assessed for high risk (HR) HPV infection by p16 immunohistochemistry (IHC) and PCR (GP5+/6+ and MY systems), as well as for the detection of HPV16, 18 and 31 by qPCR. HPV DNA+/p16+ tumors were subjected to E6/E7 oncogene expression evaluation by RT-qPCR.
OPSCC patients were treated with radical radiotherapy (early stage, n = 3) or cisplatin chemoradiotherapy +/- TPF (docetaxel, cisplatin, 5-fluorouracil) induction chemotherapy (IC) (locally advanced, n = 19). Nine (47.3%) of the nineteen patients with LA disease received IC. Patients were evaluated with head and neck MRI and chest CT scan +/- PET/CT scan at baseline and 3 months post radiotherapy completion. Liquid biopsy samples were obtained before treatment initiation. For 15 out of 22 patients, liquid biopsy samples were also available at the end of treatment. EpCAM(+) CTCs were immunomagnetically selected and subjected to HPV16 E6/E7 oncogene expression analysis by RT-qPCR.
Immunohistochemical (IHC) staining for p16
IHC was performed to determine p16 expression using a p16 mouse monoclonal antibody (predilute, mtm-CINtech, E6H4) as previously described [8]. p16 was considered to be positive when defined as strong and diffuse nuclear and cytoplasmic staining in ≥70% of the tumor cells, which is the same scoring criteria used by Ang et al [8].
DNA extraction from paraffin sections
Two 10 μm-paraffin sections from OPSCC specimens were used for DNA extraction by the QIAamp DNA FFPE Tissue kit (QIAGEN, Germany), according to manufacturer’s instructions. The extracted DNA was stored at -20°C until analysis.
Detection of high-risk HPV DNA by PCR
High-risk HPV DNA detection was performed using the two most popular worldwide consensus PCR assays: the MY system [9] and the GP5+/6+ system [10] both amplifying regions of HPV L1 gene. DNA integrity was assessed by PCR amplification of β-globin with PC04 and GH020 primers [9].
PCR reactions were performed in the GENEAmp PCR System 9600 (Applied Biosystems, USA). The 25 μl total reaction volume, contains 50–100 ng genomic DNA, 2.5 μl of 10X PCR buffer (w/o MgCl2), 3 μl of 10 mM dNTPs mix, 0.75 μl of 50 mM MgCl2, 1.75 μl of 10 μM forward primer, 1.75 μl of 10 μM reverse primer and 0.25 μl of 5 U/μl Platinum Taq DNA polymerase (Invitrogen, Life Technologies). Amplification for β-globin was performed at 94°C for 5 min followed by 36 cycles of 45 sec denaturation at 94°C, 45 sec annealing at 58°C and 45 sec elongation at 72°C. The last cycle was followed by a final extension of 5 min at 72°C. The thermal protocol for MY and GP5+/6+ systems consists of incubation at 94°C for 5 min followed by 40 cycles of 2 min denaturation at 94°C, 2 min annealing at 55°C for MY and 40°C for GP5+/6+ and 2 min elongation at 72°C. The last cycle was followed by a final extension of 5 min at 72°C. Appropriate controls including DEPC-H2O (blank), DNA-negative for HPV and DNA-positive for HPV16 from SiHa cervical carcinoma cells, were used. All necessary standard precautions were observed in order to avoid contamination through PCR carry-over. PCR products were analyzed in 1.5% w/v agarose gels.
HPV RFLP typing
In case of HPV DNA+/p16+ samples, reactions were performed again in quadruplicate, mixed and their product was subjected to restriction fragment polymorphism (RFLP) analysis using the BamHI, DdeI, HaeIII, HinfI, PstI and RsaI restriction enzymes (New England Biolabs, USA). In precisely, 13 μl PCR product, 1.5 μl restriction buffer NEB 2 and 0.5 μl of each of the abovementioned restriction enzymes, in separate tubes, were incubated for 4h at 37°C. Analysis performed in 2% Nusieve 1:1 agarose gel as previously reported [11–12]. Assignment of an HPV type to a particular risk category was done according to Munoz et al [13].
Real time qPCR for detection of HPV16, 18 and 31
Real-time qPCR assays were applied in HPV DNA+/p16+ FFPE samples in order to provide a more sensitive detection for HPV16, 18 and 31 types, most commonly found in OPSCC. The qPCR assays amplify a 93bp HPV16 E6 region [14], a 185bp HPV18 E1 region [15] and a 350bp HPV31 E6 region [12].
The qPCR reactions were performed in the 7500 Real-Time PCR System (Applied Biosystems). The 10 μl reactions consisted of 50–100 ng genomic DNA, 5 μl of Kapa SYBR Fast Universal 2X qPCR Master Mix (Kapa Biosystems, Inc., Woburn, MA, USA) and 300 nM of each qPCR primer. The thermal protocol includes 3 min polymerase activation step at 95°C, followed by 40 cycles of denaturation at 95°C for 15 sec and the primer annealing and extension step at 60°C for 1 min. Duplicate reactions were performed for each tested sample. Melting curve analysis and agarose gel electrophoresis were performed following the amplification in order to distinguish the accumulation of the specific reaction products from non-specific ones or primer-dimers.
Isolation of EpCAM(+) CTCs
For the isolation of EpCAM(+) CTCs from peripheral blood (30mL) we followed our previously described protocols [16–18].
RNA extraction
Total RNA was extracted from two 10 μm-paraffin sections using Nucleospin totalRNA FFPE XS (MACHEREY-NAGEL, Germany), with on-column rDNase digestion, according to manufacturer’s instructions. 1μg of the extracted RNA was further digested with DNAase I (Life technologies, USA) before cDNA synthesis. From the EpCAM(+) CTCs fraction, total RNA was isolated using the miRNeasy micro kit (QIAGEN, Germany), according to manufacturer’s instructions.
cDNA synthesis
First-strand cDNA synthesis was carried out by the SuperScript First-Strand Synthesis System for RT-PCR (Life technologies, USA) according to manufacturer’s protocol, using 7μl of isolated total RNA as starting template.
Detection of HPV16 E6/E7 expression in CTCs and paraffin embedded tissues
Expression analysis of HPV16 E6/E7 oncogenes in CTCs and HPV16(+) tumor samples was performed by RT-qPCR, using specific qPCR primers for HPV16 E6 and β-actin genes, amplifying a 109 bp HPV16 E6-specific and 105 bp β-actin-specific regions, previously described [12, 19].
The qPCR assay was performed in the 7500 Real-Time PCR System (Applied Biosystems). The 10 μl reaction mixture consists of Kapa SYBR Fast Universal 2X qPCR Master Mix (Kapa Biosystems), 300 nM of each qPCR primer and 2.0 μl cDNA, both for HPV16 E6 and β-actin reactions. The thermal protocol consists of a 3 min polymerase activation step at 95°C, followed by 40 cycles of denaturation at 95°C for 15 sec and the primer annealing and extension step at 60°C for 1 min. Duplicate reactions were performed for each tested sample. Melting curve analysis and agarose gel electrophoresis were performed following the amplification in order to distinguish the accumulation of the specific reaction products from non-specific ones or primer-dimers.
Statistical analysis
Patients’ recurrence and death were used as clinical endpoint events for progression-free survival (PFS) and overall survival (OS) of patients. PFS and OS were defined as the time from disease diagnosis to documented disease progression or patients’ death. Alive patients without documented events were censored at the time of the last disease evaluation. Co-primary objectives of the study were the associations between E6/E7 expression at baseline and the end of treatment CTCs with PFS and OS intervals.
Chi-square test for categorical data used to assess the univariate differences of study parameters according to the gene expression. Survival analysis was performed by Kaplan-Meier curves, tested for significance by Mantel-Cox log-rank test. A level of p≤0.05 is considered statistically significant unless specified otherwise.
Results
A total of 22 FFPE samples from patients with OPSCC were obtained and assessed for p16/HPV markers. Median follow-up was 31 months. Stage was determined according to American Joint Committee on Cancer (AJCC) 7th edition TNM staging system. Baseline clinicopathological characteristics of patients are shown in Table 1.
Table 1. Clinicopathological features of OSCC patients.
| Variable | No. of patients, n = 22 | p-valuea |
|---|---|---|
| Tumor (T) classification | ||
| T1 | 2 (9.1%) | 0.45 |
| T2 | 7 (31.8%) | |
| T3 | 4 (18.2%) | |
| T4 | 9 (40.9%) | |
| Nodal (N) classification | ||
| N0 | 6 (27.3%) | 1.00 |
| N+ | 16 (72.7%) | |
| Stage | ||
| II | 3 (13.7%) | 1.00 |
| III | 1 (4.5%) | |
| IVa | 14 (63.6%) | |
| IVb | 4 (18.2%) | |
| Grade | ||
| 1 | 1 (4.5%) | 0.12 |
| 2 | 11 (50.0%) | |
| 3 | 8 (36.4%) | |
| unknown | 2 (9.1%) | |
| Tobacco consumption | ||
| No | 4 (18.2%) | 0.61 |
| Ex Smoker | 2 (9.1%) | |
| Light Smoker | 1 (4.5%) | |
| Heavy Smoker | 15 (68.2%) | |
| Alcohol consumption | ||
| No | 8 (36.4%) | 0.53 |
| Social | 9 (40.9%) | |
| Heavy | 5 (22.7%) | |
| Gender | ||
| Male | 16 (72.7%) | 1.00 |
| Female | 6 (27.3%) |
a Chi-square test for the correlation of HPV16 E6/E7 oncogene expression in baseline CTCs with HPV16(+) patients clinicopathological features
p16 Immunohistochemistry
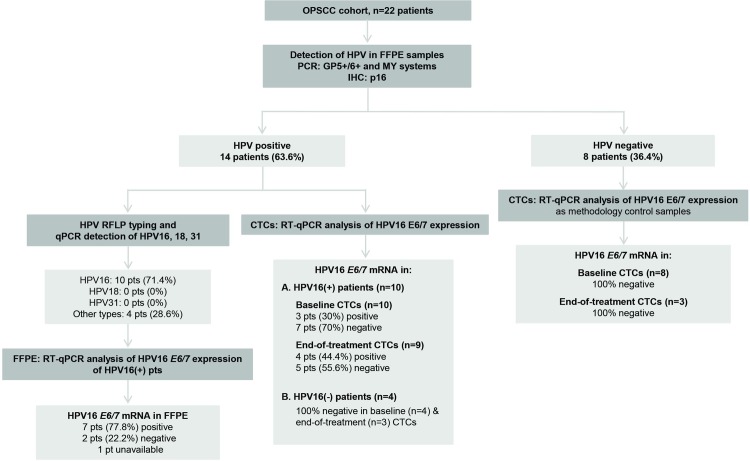
Twenty-two specimens were evaluable for p16 by immunohistochemistry. Fourteen of the twenty-two patients (63.6%) were positive for p16 (Fig 1).
Fig 1. REMARK diagram of the study.
Detection of high-risk HPV DNA by PCR
Twenty-two FFPE samples were evaluable for HPV DNA by PCR. All samples were subjected to histopathological evaluation, DNA quality control, and HPV DNA detection. HPV infection was detected in 14 of 22 OPSCC (63.6%) samples by PCR (GP5+/6+, MY systems) (Fig 1). Ten of 14 patients (71.4%) were HPV16+ by qPCR evaluation. Subsequently, HPV16 DNA+ tumors were subjected to HPV16 E6/E7 oncogene expression analysis by RT-qPCR. Nine samples were evaluable for high-risk HPV16 mRNA analysis by RT-qPCR and HPV16 E6/E7 expression was detected in 7 of them (77.8%) (Fig 1).
E6/E7 oncogene expression analysis in CTCs
HPV16 E6/E7 oncogene expression analysis was performed in immunomagnetically positive selected CTCs isolated before treatment initiation, as well as the end of treatment for 15 patients of the cohort. HPV16 E6/E7(+) CTCs were detected in 3 of 10 HPV16+ patients (30%) at baseline, and in 4 of 9 HPV16+ patients (44.4%) at the end of treatment, all of which were p16+/HPV DNA+; however only 2 patients were HPV16 E6/E7(+) in both pre- and end-of-treatment CTCs. We found no HPV16 E6/E7 mRNA expression in CTCs isolated from p16-/HPV DNA- or HPV16- patients. Clinicopathological characteristics of HPV16+ cases and the correlation with HPV16 E6/E7 positive CTCs are shown in Table 2.
Table 2. Clinicopathological characteristics of HPV16+ cases and correlation with HPV16 E6/E7 positive CTCs.
| Case | Gender | Age at diagnosis | Stage | Tobacco | Alcohol | Treatment | CTCs at baseline | CTCs at the end of treatment |
|---|---|---|---|---|---|---|---|---|
| 1 | Μ | 74 | IVa | Heavy | Heavy | CRT | (+) | (+) |
| 2 | Μ | 41 | IVa | Νο | Νο | CRT | (-) | (-) |
| 3 | Μ | 46 | IVa | Heavy | Social | IC, CRT | (+) | (-) |
| 4 | F | 55 | IVa | Heavy | Social | IC, CRT | (-) | NA |
| 5 | Μ | 42 | IVb | Heavy | Social | CRT | (-) | (-) |
| 6 | Μ | 45 | II | Light | Social | CRT | (-) | (+) |
| 7 | F | 44 | IVa | Heavy | Social | CRT | (-) | (+) |
| 8 | M | 76 | IVa | Heavy | No | CRT | (-) | (-) |
| 9 | F | 50 | IVa | Heavy | Social | IC, CRT | (+) | (+) |
| 10 | Μ | 52 | IVa | Heavy | Heavy | CRT | (-) | (-) |
Abbreviations: CRT = Chemoradiotherapy, F = Female, IC = Induction Chemotherapy, M = Male
Association with clinical outcome
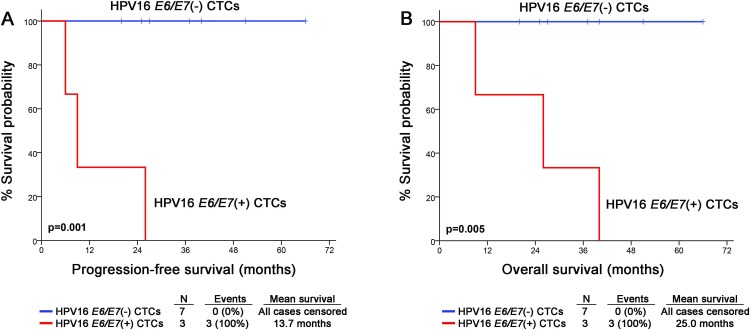
Twenty-one patients were successfully followed-up, whereas one patient was excluded due to unclear monitoring data. During a median follow-up time (reverse Kaplan–Meier method) of 31 months (95% CI: 26.42–35.58) patients’ progression and death was detected in 7 (33.3%) patients. Among them, 4 patients were HPV16+ and 3 patients were HPV16-. Among HPV16+ cases, 3 patients had HPV16 E6/E7 positive CTCs at baseline and one had not. The mean PFS and OS was 45.41 months (95% CI: 33.15–57.68) and 44.39 months (95%CI: 31.86–56.92), respectively. The preplanned survival analysis of the HPV16+ patients showed a statistically significant higher risk for disease relapse (p = 0.001) and worse OS outcome (p = 0.005) in patients with HPV16 E6/E7(+) baseline CTCs. Kaplan-Meier curves are shown in Fig 2. Finally, Kaplan-Meier analysis did not show any significant correlation between E6/E7 expression in CTCs isolated at the end of treatment and patients’ PFS (p = 0.596) or OS (p = 0.504) (S1 Fig). As expected, HPV+ OPSCC patients had improved prognosis in terms of PFS (p = 0.008) and OS (p = 0.049) compared to HPV- OPSCC. No statistically significant difference in prognosis was noted between HPV16 and HPV- other genotypes probably due to small numbers.
Fig 2. Survival analysis of OPSCC patients according to HPV16 E6/E7 expression in CTCs.
Kaplan-Meier survival curves for (A) the progression-free survival (PFS), and (B) the overall survival (OS) of patients. P-values were calculated by log-rank test.
Discussion
In the present report, we sought to evaluate whether HPV16 E6/E7 oncogene expressing CTCs could be detected before and after treatment in a cohort of patients with OPSCC. Importantly, we show for the first time a significant association between HPV16 E6/E7(+) CTCs at baseline and both PFS (p = 0.001) and OS (p = 0.005). Although HPV+ OPSCC has favorable prognosis, 10% to 25% of HPV+ OPSCCs eventually recur. The identification of poor prognosis HPV16+ OPSCC has important treatment implications. The favorable prognosis of HPV+ OPSCC has fueled the development of deintensification strategies aiming to spare these patients the devastating consequences of aggressive treatment. HPV16 E6/E7(+) CTC detection could be used for better selection of deintensification candidates.
Although HPV16 is known to be responsible for more than 90% of HPV-related OPSCCs, in our cohort, only 71% of patients were found to be HPV16 positive. This discrepancy might be related to national differences, since Greece lacks official data registries regarding prevalence of HPV in head and neck cancer. Nevertheless, sample size is small to draw definite conclusions.
Several studies have emphasized the magnitude of molecular assays for CTC molecular characterization [16–18, 20]. Indeed, our group has demonstrated that real-time PCR-based methods performed in RNA or genomic DNA extracted from EpCAM(+) CTCs can provide useful information for the molecular characterization of CTCs. HPV E6/E7 oncogene expression provides the most specific genetic marker distinguishing tumor cells bearing transcriptionally active HPV in peripheral blood [21]. In the present study, HPV16 E6/E7(+) CTCs were detected in 3 of 10 patients with HPV16(+) LA OPSCC (30%). Taking into account the paradigm of HPV-related cervical cancer, Tseng et al. found that the presence of HPV16 and HPV18 E6 oncogene mRNA expression in the peripheral blood of patients with locally advanced disease was associated with bulky tumor volume and pelvic lymph node metastasis [22]. Furthermore, after a median follow-up of 22 months, patients with positive E6 oncogene expression had a significantly higher risk of recurrence than those who were E6 oncogene expression negative. Similarly, we found that patients with HPV16 E6/E7(+) CTCs before initiation of treatment had significantly higher risk for relapse and death. Thus, the clinical utility of HPV16 mRNA detection in CTCs isolated from patients with LA OPSCC deserves evaluation in large cohorts.
To date, this is the first study evaluating HPV16 E6/E7 (+) CTCs as a prognostic tool in OPSCC patients. The majority of studies to date have evaluated the utility of circulating HPV DNA as a surveillance tool in patients with head and neck squamous cell carcinoma (HNSCC). A recent meta-analysis including 5 studies (n = 600 patients) reports a high pooled estimated specificity in detecting recurrence but an inferior pooled sensitivity [23]. Recent technical advances in detecting circulating DNA might improve the sensitivity [24]. Mazurek et al. based the diagnosis of HPV+ HNSCC only on detection of plasma HPV DNA using E6/E7 PCR. The detection rate in that study was 14%. Interestingly, patients with more advanced disease had higher levels of circulating-free DNA [25].
Cao et al. detected HPV DNA in 65% of the pretreatment plasma samples from HPV+ OPSCC. Pre-treatment plasma HPV DNA copy number correlated significantly with nodal metabolic tumor volume (assessed on FDG-PET). Serial measurements in 14 patients showed rapid decline in HPV DNA that became undetectable at RT completion. Accordingly, post-treatment HPV DNA was elevated in 3 out of 4 patients with recurrent or residual disease prior to clinically and radiologically detected metastatic disease [26]. Dahlstrom et al. detected circulating HPV DNA in pre-treatment serum and found that the presence of circulating HPV DNA was associated with higher N category and overall stage. Although patients with HPV+ tumors with detectable pre-treatment levels of circulating HPV DNA had worse PFS, this difference did not reach statistical significance [27]. Similarly, Lee et al. recently demonstrated that detection of HPV DNA in sequential samples through and after chemo-radiotherapy predicted response and residual disease at the primary site and in cervical lymph nodes in patients with HPV + LA disease [28].
One important application of our results is the identification of OPSCC patients with poor prognosis. Current clinical trials address separately HPV+ and HPV- OPSCC and focus on treatment deintensification. If validated, the test described in this paper could be easily incorporated into routine clinical practice as a prognostic tool for therapy selection. Importantly, in this small cohort there was no association between high tumor burden and the presence of HPV16 E6/E7(+) CTCs at baseline indicating that the latter might be an independent prognostic marker in HPV+ OPSCC. Due to small sample size we did not find a significant correlation between clinical outcome and HPV16 E6/E7(+) CTCs at the end of treatment; a prospective study evaluating this question in a large number of patients is needed.
A major limitation of our study is that it is a single institution small cohort and our results need to be validated in large cohorts with longer follow-up. In addition, EpCAM-based isolation methods may miss CTCs bearing mesenchymal rather than epithelial phenotype, a phenomenon that is not uncommon in HNSCC.
In conclusion, we sought to assess the feasibility of detection of HPV E6/E7(+) CTCs in OPSCC. We demonstrated a significant association between HPV16 E6/E7 mRNA detection and clinical outcome in this pilot cohort. Combined interpretation of HPV E6/E7(+) CTCs with UICC staging data may lead to alteration of risk definition of patient subsets, with improved risk discrimination in early-stage disease. Validation studies are awaited.
Supporting information
Kaplan-Meier survival curves for (a) Progression Free Survival (PFS) and (b) Overall Survival (OS) of patients. P values were calculated by long-rank test.
(TIF)
Acknowledgments
The authors wish to thank all patients and their families for agreeing to participate in the study.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Nelson HH, Pawlita M, Michaud DS, McClean M, Langevin SM, Eliot MN, et al. Immune Response to HPV16 E6 and E7 Proteins and Patient Outcomes in Head and Neck Cancer. JAMA Oncol. 2016. 10.1001/jamaoncol.2016.4500 . [DOI] [PubMed] [Google Scholar]
- 2.Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14(2):467–75. 10.1158/1055-9965.EPI-04-0551 . [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi AK, Anderson WF, Lortet-Tieulent J, Curado MP, Ferlay J, Franceschi S, et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31(36):4550–9. 10.1200/JCO.2013.50.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taberna M, Mena M, Pavon MA, Alemany L, Gillison ML, Mesia R. Human papillomavirus-related oropharyngeal cancer. Ann Oncol. 2017;28(10):2386–98. 10.1093/annonc/mdx304 . [DOI] [PubMed] [Google Scholar]
- 5.McMullen KP, Chalmers JJ, Lang JC, Kumar P, Jatana KR. Circulating tumor cells in head and neck cancer: A review. World J Otorhinolaryngol Head Neck Surg. 2016;2(2):109–16. 10.1016/j.wjorl.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun T, Zou K, Yuan Z, Yang C, Lin X, Xiong B. Clinicopathological and prognostic significance of circulating tumor cells in patients with head and neck cancer: a meta-analysis. Onco Targets Ther. 2017;10:3907–16. 10.2147/OTT.S136530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bravo IG, Felez-Sanchez M. Papillomaviruses: Viral evolution, cancer and evolutionary medicine. Evol Med Public Health. 2015;2015(1):32–51. 10.1093/emph/eov003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. 10.1056/NEJMoa0912217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ting Y MM. Detection and typing of genital human papillomaviruses. In: Innis MA GD, Sninsky JJ, White TJ editor. PCR Protocols: A Guide to Methods and Applications1990. p. 356–67.
- 10.de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3' ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995;76 (Pt 4):1057–62. 10.1099/0022-1317-76-4-1057 . [DOI] [PubMed] [Google Scholar]
- 11.Bernard HU, Chan SY, Manos MM, Ong CK, Villa LL, Delius H, et al. Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction amplification, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J Infect Dis. 1994;170(5):1077–85. . [DOI] [PubMed] [Google Scholar]
- 12.Psyrri A, Fortpied C, Koutsodontis G, Avgeris M, Kroupis C, Goutas N, et al. Evaluation of the impact of tumor HPV status on outcome in patients with locally advanced unresectable head and neck squamous cell carcinoma (HNSCC) receiving cisplatin, 5-fluorouracil with or without docetaxel: a subset analysis of EORTC 24971 study. Ann Oncol. 2017;28(9):2213–8. Epub 2017/06/28. doi: 3884599 [pii] 10.1093/annonc/mdx320 . [DOI] [PubMed] [Google Scholar]
- 13.Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348(6):518–27. 10.1056/NEJMoa021641 . [DOI] [PubMed] [Google Scholar]
- 14.Kroupis C, Markou A, Vourlidis N, Dionyssiou-Asteriou A, Lianidou ES. Presence of high-risk human papillomavirus sequences in breast cancer tissues and association with histopathological characteristics. Clin Biochem. 2006;39(7):727–31. 10.1016/j.clinbiochem.2006.03.005 . [DOI] [PubMed] [Google Scholar]
- 15.Vincent-Salomon A, de la Rochefordiere A, Salmon R, Validire P, Zafrani B, Sastre-Garau X. Frequent association of human papillomavirus 16 and 18 DNA with anal squamous cell and basaloid carcinoma. Mod Pathol. 1996;9(6):614–20. . [PubMed] [Google Scholar]
- 16.Strati A, Markou A, Parisi C, Politaki E, Mavroudis D, Georgoulias V, et al. Gene expression profile of circulating tumor cells in breast cancer by RT-qPCR. BMC Cancer. 2011;11:422 10.1186/1471-2407-11-422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markou A, Strati A, Malamos N, Georgoulias V, Lianidou ES. Molecular characterization of circulating tumor cells in breast cancer by a liquid bead array hybridization assay. Clin Chem. 2011;57(3):421–30. 10.1373/clinchem.2010.154328 . [DOI] [PubMed] [Google Scholar]
- 18.Strati A, Koutsodontis G, Papaxoinis G, Angelidis I, Zavridou M, Economopoulou P, et al. Prognostic significance of PD-L1 expression on circulating tumor cells in patients with head and neck squamous cell carcinoma. Ann Oncol. 2017;28(8):1923–33. Epub 2017/08/26. doi: 3920775 [pii] 10.1093/annonc/mdx206 . [DOI] [PubMed] [Google Scholar]
- 19.Shi W, Kato H, Perez-Ordonez B, Pintilie M, Huang S, Hui A, et al. Comparative prognostic value of HPV16 E6 mRNA compared with in situ hybridization for human oropharyngeal squamous carcinoma. J Clin Oncol. 2009;27(36):6213–21. Epub 2009/11/04. doi: JCO.2009.23.1670 [pii] 10.1200/JCO.2009.23.1670 . [DOI] [PubMed] [Google Scholar]
- 20.Muller V, Riethdorf S, Rack B, Janni W, Fasching PA, Solomayer E, et al. Prognostic impact of circulating tumor cells assessed with the CellSearch System and AdnaTest Breast in metastatic breast cancer patients: the DETECT study. Breast Cancer Res. 2012;14(4):R118 10.1186/bcr3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cogliano V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, et al. Carcinogenicity of human papillomaviruses. Lancet Oncol. 2005;6(4):204 . [DOI] [PubMed] [Google Scholar]
- 22.Tseng CJ, Pao CC, Lin JD, Soong YK, Hong JH, Hsueh S. Detection of human papillomavirus types 16 and 18 mRNA in peripheral blood of advanced cervical cancer patients and its association with prognosis. J Clin Oncol. 1999;17(5):1391–6. 10.1200/JCO.1999.17.5.1391 . [DOI] [PubMed] [Google Scholar]
- 23.Jensen KK, Gronhoj C, Jensen DH, von Buchwald C. Circulating human papillomavirus DNA as a surveillance tool in head and neck squamous cell carcinoma: A systematic review and meta-analysis. Clin Otolaryngol. 2018. Epub 2018/05/16. 10.1111/coa.13267 [DOI] [PubMed] [Google Scholar]
- 24.Hanna GJ, Supplee JG, Kuang Y, Mahmood U, Lau CJ, Haddad RI, et al. Plasma HPV cell-free DNA monitoring in advanced HPV-associated oropharyngeal cancer. Ann Oncol. 2018. Epub 2018/07/17. doi: 5053586 [pii] 10.1093/annonc/mdy251 . [DOI] [PubMed] [Google Scholar]
- 25.Mazurek AM, Rutkowski T, Fiszer-Kierzkowska A, Malusecka E, Skladowski K. Assessment of the total cfDNA and HPV16/18 detection in plasma samples of head and neck squamous cell carcinoma patients. Oral Oncol. 2016;54:36–41. 10.1016/j.oraloncology.2015.12.002 . [DOI] [PubMed] [Google Scholar]
- 26.Cao H, Banh A, Kwok S, Shi X, Wu S, Krakow T, et al. Quantitation of human papillomavirus DNA in plasma of oropharyngeal carcinoma patients. Int J Radiat Oncol Biol Phys. 2012;82(3):e351–8. 10.1016/j.ijrobp.2011.05.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dahlstrom KR, Li G, Hussey CS, Vo JT, Wei Q, Zhao C, et al. Circulating human papillomavirus DNA as a marker for disease extent and recurrence among patients with oropharyngeal cancer. Cancer. 2015;121(19):3455–64. Epub 2015/06/23. 10.1002/cncr.29538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JY, Garcia-Murillas I, Cutts RJ, De Castro DG, Grove L, Hurley T, et al. Predicting response to radical (chemo)radiotherapy with circulating HPV DNA in locally advanced head and neck squamous carcinoma. Br J Cancer. 2017;117(6):876–83. 10.1038/bjc.2017.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kaplan-Meier survival curves for (a) Progression Free Survival (PFS) and (b) Overall Survival (OS) of patients. P values were calculated by long-rank test.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.