SUMMARY
Stress granule (SG) formation is frequently accompanied by ubiquitin proteasome system (UPS) impairment and ubiquitylated protein accumulation. SGs, ubiquitin, and UPS components co-localize, but the relationship between the ubiquitin pathway and SGs has not been systematically characterized. We utilize pharmacological inhibition of either the ubiquitin- or NEDD8-activating enzyme (UAE or NAE) to probe whether active ubiquitylation or neddylation modulate SG dynamics. We show that UAE inhibition results in rapid loss of global protein ubiquitylation using ubiquitin-specific proteomics. Critically, inhibiting neither UAE nor NAE significantly affected SG formation or disassembly, indicating that active protein ubiquitylation or neddylation is dispensable for SG dynamics. Using antibodies with varying preference for free ubiquitin or polyubiquitin and fluorescently tagged ubiquitin variants in combination with UAE inhibition, we show that SGs co-localize primarily with unconjugated ubiquitin rather than polyubiquitylated proteins. These findings clarify the role of ubiquitin in SG biology and suggest that free ubiquitin may alter SG protein interactions.
Graphical Abstract
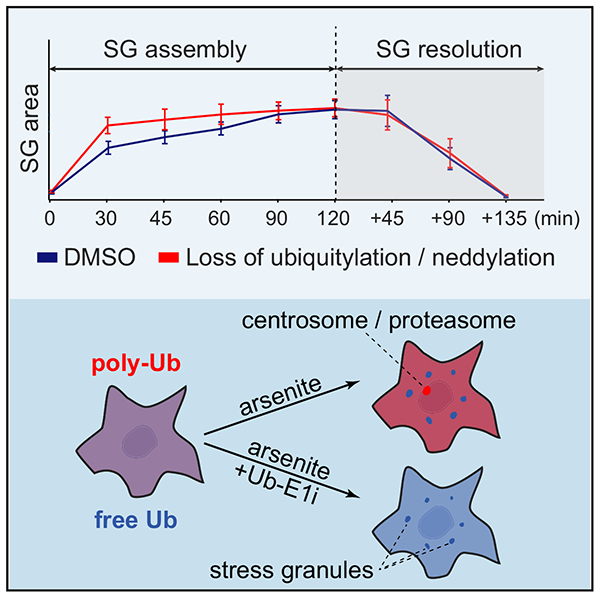
In Brief
Protein ubiquitylation has been implicated in pathways by which cellular stress induces the formation of stress granules (SGs) and affects protein homeostasis through the ubiquitin proteasome system. Markmiller et al. show that ubiquitylation is dispensable for SG dynamics and that SGs co-localize primarily with free ubiquitin rather than polyubiquitylated proteins.
INTRODUCTION
Cellular insults such as oxidative and heat stress that globally disrupt protein folding result in both the accumulation of ubiquitylated proteins and the induction of membrane-less stress granules (SGs) (Kim et al., 2015; Protter and Parker, 2016). SGs are enigmatic cellular structures that comprise translationally repressed mRNAs associated with a variety of RNA-binding proteins (Buchan, 2014). While the cellular function of SGs remains unclear, SG formation and SG resident proteins have been linked to human neurological disorders, including amyotrophic lateral sclerosis (ALS) and frontotemporal degeneration (FTD) (Buchan, 2014; Dewey et al., 2012; Li et al., 2013). Genomic and proteomic characterization of both the SG RNA and protein constituents have revealed a marked compositional diversity in both SG proteins and RNAs (Jain et al., 2016; Khong et al., 2017; Markmiller et al., 2018). Examination of SG proteomes has revealed that proteins involved in regulating distinct post-translational modifications (PTMs) are often enriched within SGs. These findings suggest that PTMs may regulate either global SG dynamics or the recruitment of individual proteins into SGs and that targeting PTMs may be an effective strategy to alter SG dynamics (Ohn and Anderson, 2010).
Numerous lines of evidence have implicated protein ubiquitylation or other ubiquitin-like modification systems, like neddylation, as potential regulators of SG dynamics. First, components of the ubiquitin-proteasome system (UPS), including ubiquitin itself, have been shown to co-localize with SGs induced by a variety of protein homeostasis stressors (Kwon et al., 2007; Mateju et al., 2017; Xie et al., 2018). Second, proteasome inhibition and the concomitant increase in polyubiquitylated proteins results in SG formation (Mateju et al., 2017; Mazroui et al., 2007; Seguin et al., 2014). Third, genetic disruption or pharmacological inhibition of ubiquitin or neddylation components can disrupt SG dynamics in both S. cerevisiae and mammalian cells (Buchan et al., 2013; Jayabalan et al., 2016; Kwon et al., 2007; Ohn et al., 2008; Seguin et al., 2014; Takahashi et al., 2013; Turakhiya et al., 2018; Xie et al., 2018). Despite this evidence, several key questions regarding the role of ubiquitylation in regulating SG dynamics remain unanswered. While ubiquitin has been shown to co-localize with SGs, whether polyubiquitylated proteins them-selves or proteins modified with specific ubiquitin linkages are recruited to SGs is unknown. It is also unknown how many of the ubiquitin-system components that co-localize with SGs require ubiquitin within SGs for their localization. The deubiquitylating enzyme USP10 is a well-characterized SG-localized protein (Ohn et al., 2008; Soncini et al., 2001). However, USP10 SG localization is determined by binding to another SG protein, G3BP1; and mutation of the UPS10 active site, which renders it incapable of removing ubiquitin from substrates, had little impact on its localization or overall SG dynamics (Kedersha et al., 2016; Takahashi et al., 2013). Despite the many links between the UPS and SGs, there has yet to be a demonstration that ubiquitylation of a specific SG protein is required for its SG localization or that overall protein ubiquitylation or other ubiquitin-like protein modification pathways are needed to form or dissolve SGs.
Here, we directly examine the relationship between protein ubiquitylation and SG dynamics. Interrogation of global protein ubiquitylation using ubiquitin proteomics approaches revealed widespread alterations to the ubiquitin-modified proteome upon arsenite-induced stress. Despite clear changes to some SG protein ubiquitylation, arsenite treatment did not result in global changes to known SG-resident protein ubiquitylation. Utilizing potent and specific inhibitors of either the ubiquitin-activating enzyme (UAE) or the NEDD8-activating enzyme (NAE), we demonstrate that active protein ubiquitylation or neddylation is dispensable for arsenite-induced SG formation or dissolution. We demonstrate that free, unconjugated ubiquitin localizes to SGs in a UAE-independent manner. Further, the SG localization of many ubiquitin system components is similarly unperturbed by UAE inhibition and the concomitant ablation of protein ubiquitylation. Taken together, our results clearly demonstrate that active protein neddylation or ubiquitylation is not required for SG dynamics and that unconjugated ubiquitin is the primary form of ubiquitin that localizes to SGs.
RESULTS
Identification of Resident SG Protein Ubiquitylation upon Arsenite Treatment
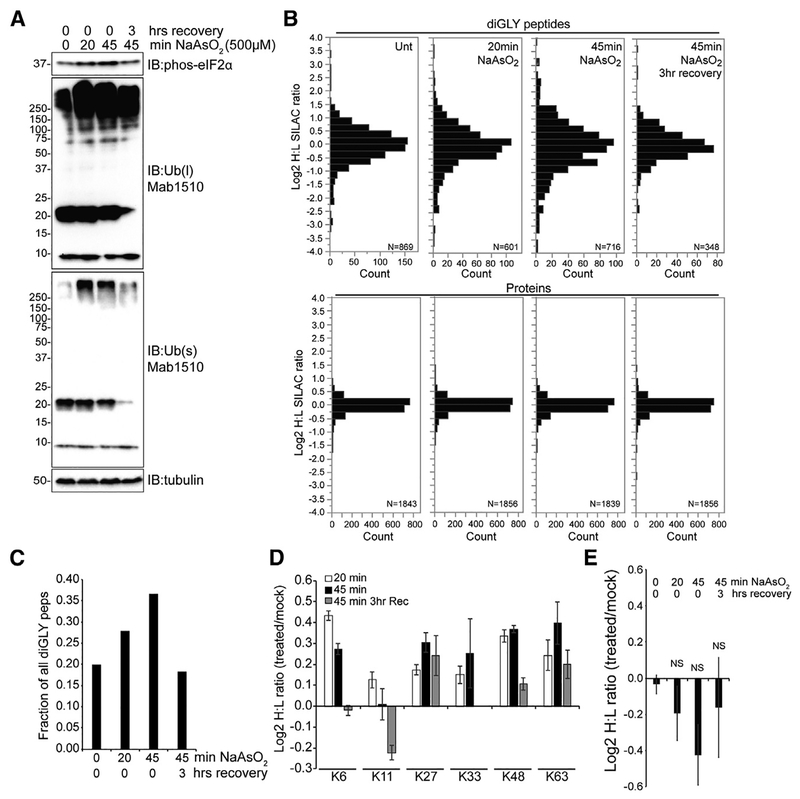
Sodium arsenite is one of the most robust known inducers of transient SGs that form rapidly and disperse within 2–3 h upon washout (Figures S1A and S1B). Arsenite-induced SG formation coincides with eIF2a phosphorylation and a global increase in ubiquitylated proteins (Figures 1A, S1C, and S1D). These results demonstrate that the timing of alterations to protein ubiquitylation largely mirrors SG dynamics, indicating a possible role for protein ubiquitylation during SG for mation or dissolution. To identify proteins whose ubiquitylation may be critical for their SG localization, we utilized quantitative ubiquitin site-specific proteomic approaches to identify proteins whose ubiquitylation status was altered upon arsenite treatment (Gendron et al., 2016; Kim et al., 2011). HeLa cells grown in media containing 13C15N-labeled lysine were either untreated or treated with arsenite for 20 or 45 min or were treated with arsenite for 45 min, followed by washout and recovery for 3 h. Arsenite treatment did not result in overt changes to protein abundance, at least for the depth of proteome coverage achieved in this experiment. However, consistent with our previous results, arsenite treatment resulted in a global increase in protein ubiquitylation, which was reduced upon arsenite removal (Figures 1A and 1B; Table S1). In cells treated with arsenite for 45 min, greater than 35% of all quantified ubiquitin-modified peptides increased or decreased in abundance more than 2-fold, indicating that a significant fraction of protein ubiquitylation is impacted by arsenite treatment (Figure 1C). Modest, but consistent abundance increases in all but lysine 11 (K11)-linked ubiquitin chains were observed upon 45-min arsenite treatment (Figure 1D). To directly examine whether known SG proteins were selectively ubiquitylated upon arsenite treatment (Table S2), we quantified a total of 102 ubiquitylated peptides from 43 proteins from a curated list of validated SG resident proteins. We found no significant difference in the abundance of these peptides upon arsenite treatment or washout compared to untreated samples, indicating that, at a global level, SG resident protein ubiquitylation was not specifically altered during conditions that induce SG formation (Figure 1E; Table S2). While our data suggest that SG protein ubiquitylation is not globally impacted by arsenite treatment, it does not rule out the possibility that the ubiquitylation of specific SG proteins governs their SG localization.
Figure 1. SG Protein Ubiquitylation Is Largely Unaffected by NaAsO2 Treatment.
(A) Immunoblot of phospho-eIF2a, ubiquitin, and tubulin in whole-cell extracts from HeLa cells treated with NaAsO2 as indicated. s and l denote short and long exposures, respectively.
(B) Log2 heavy-to-light ratios (log2 H/L) for all diGLY-modified peptides (top) and total proteins (bottom). Heavy-labeled cells were treated with NaAsO2 as indicated.
(C) The fraction of all quantified diGLY peptides with >2-fold change in abundance upon NaAsO2 treatment or washout.
(D) Log2 H/L corresponding to diGLY-modified peptides from ubiquitin. The individual ubiquitin modified lysine that was quantified is indicated. Error bars denote SEM of multiple quantification events for a given peptide.
(E) The median log2 H/L of all diGLY-modified peptides quantified from known SG proteins. Error bars denote SEM for all diGLY quantification events observed on known SG proteins. NS, not significant.
Acute Pharmacological UAE Inhibition Results in a Rapid Loss of Protein Ubiquitylation
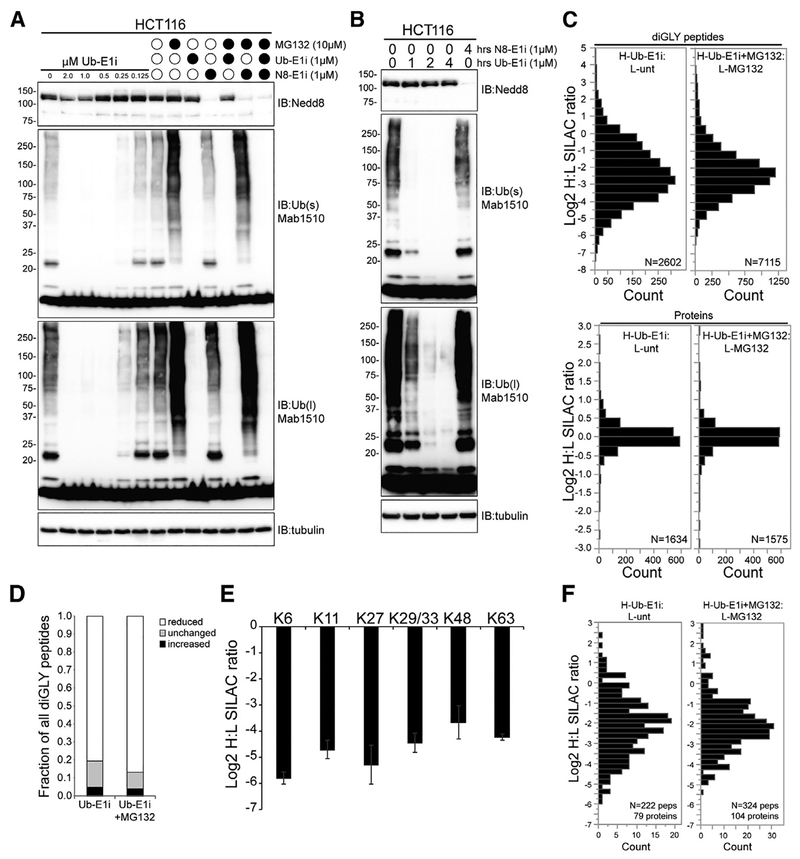
Ubiquitin has been observed to co-localize with SGs, and ubiquitin system components have been demonstrated to both localize to SGs and regulate SG dynamics, implicating a role for protein ubiquitylation during SG formation or dissolution (Buchan et al., 2013; Kwon et al., 2007). However, a direct evaluation of whether active protein ubiquitylation is required to either form or resolve SGs has not been reported. To perform such an examination, we utilized a specific and potent ubiquitin E1 activating enzyme (UAE) inhibitor (TAK-243, also known as MLN7243, which we refer to as Ub-E1i) to acutely inhibit protein ubiquitylation (Hyer et al., 2018). The addition of increasing amounts of UbE1i to HCT116 cells for 4 h resulted in a dose-dependent decrease in polyubiquitylated proteins, with complete abrogation of observable polyubiquitylated material with Ub-E1i treatment above 0.5 μM (Figure 2A). Proteasome inhibition by MG132 resulted in the well-characterized increase in total protein ubiquitylation, which was completely blocked upon co-treatment with Ub-E1i. The addition of Ub-E1i resulted in a time-dependent decrease in polyubiquitylated material. Consistent with previous reports, Ub-E1i treatment was selective for UAE, as cullin neddylation was unaffected after 4 h of Ub-E1i treatment but was completely blocked by addition of the NAE inhibitor MLN4924 (TAK-924, which we refer to as N8-E1i) (Figures 2A and 2B) (Soucy et al., 2009). We next set out to establish the impact of Ub-E1i inhibition on individual protein ubiquitylation using quantitative, ubiquitin site-specific proteomics. Heavy (13C15N) lysine-labeled HCT116 cells were treated with Ub-E1i alone or in combination with MG132 for 4 h and then mixed with unlabeled HCT116 cells that were either untreated or treated with MG132. As expected, Ub-E1i treatment resulted in a robust reduction in the abundance of the clear majority of ubiquitylated peptides identified (Table S3). More than 80% of all quantified ubiquitin-modified peptides were reduced in abundance by more than 1.7-fold with or without MG132 treatment (Figures 2C and 2D). As had previously been observed upon treatment with proteasome inhibitors (Gendron et al., 2016; Kim et al., 2011), UAE inactivation had little impact on overall protein abundance after 4 h of treatment (Figure 2C; Table S3). However, a more complete characterization of low abundance proteins would likely reveal robust protein abundance alterations. As would be predicted upon UAE inhibition, all detected ubiquitin-linkage peptides were reduced more than 8-fold, indicating that Ub-E1i treatment reduces total protein polyubiquitylation, regardless of chain type (Figure 2E). A direct examination of the ubiquitylation status of known SG proteins revealed that Ub-E1i treatment resulted in a clear reduction of SG protein ubiquitylation across a diversity of individual ubiquitylation sites (Figure 2F; Table S2). Our results demonstrate that Ub-E1i treatment results in the rapid loss of more than 80% of all ubiquitylation events.
Figure 2. The Ubiquitin-Activating Enzyme Inhibitor TAK-243 Rapidly Ablates Ubiquitin Conjugates in Cells.
(A and B) Immunoblot of Nedd8, ubiquitin, and tubulin in whole-cell extracts from HCT116 cells treated with Ub-E1i or N8-E1i alone (A and B) or in combination with MG132 (A) as indicated. s and l denote short and long exposures, respectively.
(C) Log2 H/L for all diGLY-modified peptides (top) and total proteins (bottom). Heavy-labeled cells were treated with Ub-E1i (1 μM) with or without MG132 (10 μM) for 4 h.
(D) The fraction of all quantified diGLY-modified peptides that increased, decreased, or remained unchanged upon Ub-E1i treatment with or without proteasome inhibition.
(E) Log2 H/L corresponding to diGLY-modified peptides from ubiquitin after Ub-E1i treatment alone. The individual ubiquitin-modified lysine that was quantified is indicated. Error bars denote SEM of multiple quantification events for a given peptide.
(F) Log2 H/L of all diGLY-modified peptides quantified from known SG proteins. Heavy-labeled cells were treated with Ub-E1i (1 μM) with or without MG132 (10 μM) for 4 h.
Active Protein Ubiquitylation or Neddylation Is Not Required for SG Formation or Dissolution
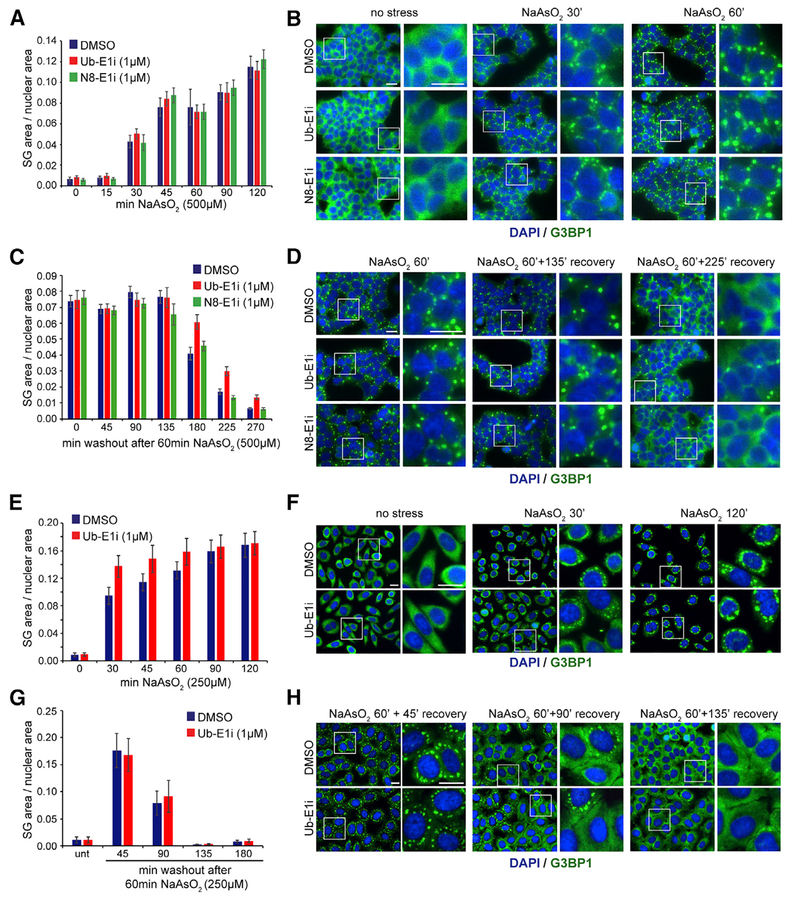
Having established UAE inactivation via Ub-E1i treatment as a powerful tool to rapidly and robustly ablate protein ubiquitylation, we set out to test whether SG formation or dissolution requires protein ubiquitylation. We validated that Ub-E1i treatment ablated the arsenite-induced increase in protein polyubiquitylation in three cell lines (Figure S2A). We utilized a previously characterized 293T cell line expressing the well-established SG protein, G3BP1, tagged with GFP at its endogenous C terminus using CRISPR/Cas9-based approaches (Markmiller et al., 2018) to track SG dynamics. Consistent with previous results, arsenite addition resulted in a rapid increase in SG formation, as determined by G3BP1-GFP coalescence (Figures 3A and 3B). Pre-treatment with Ub-E1i followed by treatment with two different concentrations of arsenite did not delay the kinetics of SG formation over a 2-h time course (Figures 3A, 3B, S2B, and S2C). Although Ub-E1i treatment has been reported to induce the unfolded protein response (UPR) (Best et al., 2019; Hyer et al., 2018; Zhuang et al., 2019), the addition of either Ub-E1i or N8-E1i alone did not induce SG formation (Figure S2B). However, Ub-E1i pre-treatment resulted in a small enhancement of SG formation at early time points, with lower concentrations of arsenite (100 μM), indicating that the added stress of UAE inhibition may accelerate SG formation under these conditions. Pre-treatment with the NAE inhibitor MLN4924 (N8-E1i), followed by arsenite treatment, did not affect SG formation at either of the tested arsenite concentrations (Figures 3A, 3B, S2B, and S2C). Taken together, these observations indicate that neither active protein ubiquitylation nor neddylation is required for initial arsenite-induced SG formation. We then determined whether protein ubiquitylation or neddylation was required for SG dissolution following arsenite removal. 293T G3BP1-GFP cells were pre-treated with DMSO, Ub-E1i, or N8-E1i for 1 h, and SGs were formed by treatment with arsenite in the presence of inhibitors for 1 h. SG dissolution was monitored after arsenite washout in media that lacked Ub-E1i or N8-E1i. While NAE inhibition has no measurable impact on SG dissolution, the addition of Ub-E1i resulted in a minor delay in SG dissolution (Figures 3C and 3D). We validated these results in HeLa cells using G3BP1 antibodies to monitor SG formation and clearance. As observed in 293T cells, UAE inhibition did not delay the kinetics of arsenite-induced SG formation using two different arsenite concentrations, with a slight acceleration of SG formation at early time points (Figures 3E, 3F, and S2D). We were unable to validate the small delay in SG dissolution upon the Ub-E1i treatment seen in 293T cells, as UAE inactivation had no impact on SG clearance in HeLa cells (Figures 3G, 3H, and S2E). It is possible that SG composition may change during longer exposures to stress and that features including the content and function of ubiquitin may be more relevant upon longer stress treatments. Therefore, we analyzed SG dynamics following longer arsenite treatment in HeLa cells (3 h at 250 μM) and found disassembly kinetics to be similarly unaffected by UAE inhibition (Figure S2F). Since UAE and NAE inhibition by Ub-E1i or N8-E1i is reversible in nature, it is possible that gradual enzyme reactivation could occur during SG dissolution. To address this issue, we examined the recovery of protein neddylation and ubiquitylation after pre-treatment of the E1 inhibitors and subsequent arsenite treatment followed by washout conditions with or without E1 inhibitors. In 293T cells, protein ubiquitylation and cullin neddylation were restored to ~20% of pre-treatment levels 5 h after E1 inhibitor removal (Figures S3A–S3D). By contrast, in HeLa and HCT116 cells, the extent of ubiquitylation and neddylation recovery after washout was greatly reduced, with no observable recovery of protein ubiquitylation in HeLa cells 5 h after Ub-E1i washout (Figures S3E and S3F). Notably, SG disassembly was complete in HeLa cells after 5 h, indicating that SG disassembly is not dependent on recovery of UAE activity during the washout phase. Consistent with this finding, the addition of Ub-E1i during the washout period did not delay SG dissolution following arsenite removal (Figure S3G). Taken together, our results clearly indicate that acute inhibition of protein ubiquitylation or neddylation has little to no impact on SG dynamics.
Figure 3. SG Dynamics Are Unaffected by Inhibition of Protein Ubiquitylation or Neddylation.
(A, C, E, and G) Quantification of stress granule (SG) dynamics in 293T G3BP1-GFP (A and C) and HeLa (E and G) cells during SG assembly (A and E) and disassembly (C and G). Cells in multi-well plates were treated with inhibitors and NaAsO2 as indicated, with or without subsequent washout. Following fixation at the indicated time points, HeLa cells were immunostained with an antibody against G3BP1, and plates were imaged using an automated image acquisition system. SG and nuclear area were quantified using custom image analysis scripts, and error bars denote SD. A minimum of ~25,000 cells were analyzed for each cell type, condition, or time point.
(B, D, F, and H) Representative images for indicated time points and treatments quantified in (A),(C), (E), and (G), respectively. Scale bars, 20 μm.
See also Figures S2 and S3.
Unconjugated Ubiquitin Co-localizes with SGs in a UAE-Independent Manner
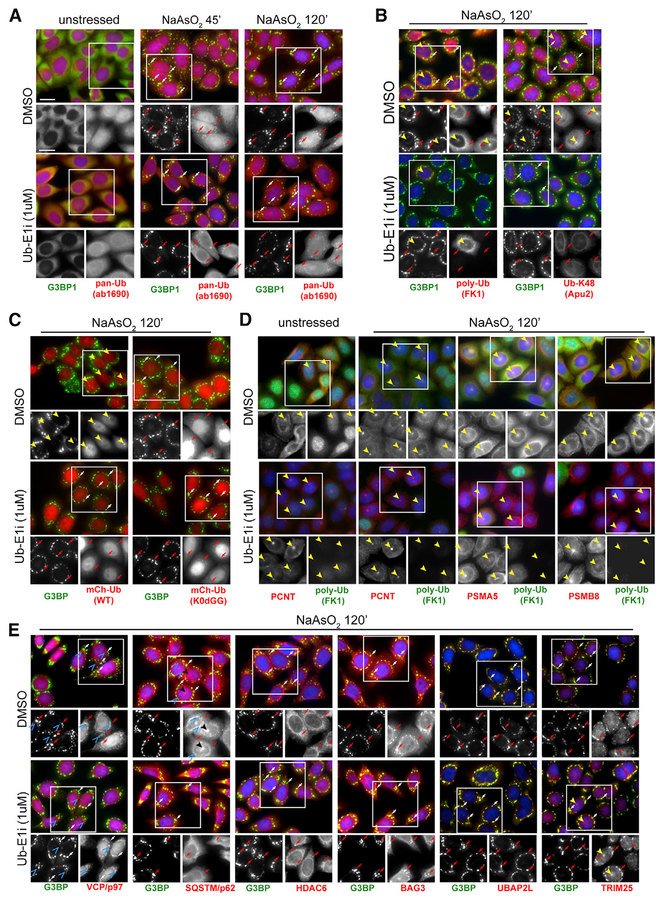
In accordance with previous studies, we were able to show that ubiquitin co-localizes with SGs by immunofluorescence (IF), using an antibody that recognizes both free and conjugated ubiquitin (Figures 4A and S2A). Because ubiquitin exists in a variety of bio-chemically distinct pools within cells (e.g., unconjugated, free ubiquitin, and lysine-48 linked polyubiquitin chains) (Clague et al., 2015), we set out to carefully characterize which forms of ubiquitin specifically co-localize with SGs. Staining with two different ubiquitin antibodies that preferentially recognize polyubiquitin chains under denaturing PAGE-western blotting conditions did not reveal any significant co-localization of polyubiquitin with SGs in HeLa cells (Figures 4B and S4A–S4C) (Fujimuro and Yokosawa, 2005), indicating that polyubiquitylated proteins are not enriched within SGs. This hypothesis is further supported by the finding that K48- or K63-linkage-specific antibodies (Apu2 and Apu3, respectively) also do not recognize antigens that co-localize with arsenite-induced SGs (Figures 4B and S4C). Instead, the IF staining signals from both K48- and K63-specific antibodies, as well as from the polyubiquitin-specific FK1 and FK2 antibodies, all appear enriched at a single perinuclear focus per cell, suggesting a partial compartmentalization of polyubiquitylated proteins outside of SGs during stress (Figures 4B and S4C). Underscoring the specificity of the IF signals obtained with the FK1, FK2, Apu2, and Apu3 antibodies, overall staining intensity was drastically reduced upon Ub-E1i treatment, consistent with the observed depletion of polyubiquitin chains after Ub-E1i treatment by western blotting (Figures 4B and S4A–S4C). By contrast, IF staining with the pan-ubiquitin antibody did not prominently label the same foci as observed with the poly-ubiquitin antibodies (Figures 4A and S4C), and overall signal intensity was slightly increased, while SG staining was unchanged upon Ub-E1i treatment. These results indicate that primarily free, unconjugated ubiquitin localizes to arsenite-induced SGs.
Figure 4. Unconjugated Ubiquitin Localizes to SGs in a UAE-Independent Manner.
(A–E) Immunofluorescence staining of HeLa cells treated with NaAsO2 (250 μM) for 0’, 45’, or 120’ prior to fixation (A) or pretreated with DMSO or UbE1i (1 μ M) for 90’, followed by treatment with NaAsO2 (250 mM) for 120’ prior to fixation (B–E). Cells were stained with antibodies against G3BP1 (A–E), pan-ubiquitin (A), polyubiquitin (B and D), centrosome marker pericentrin (D), proteasome subunits (D), VCP/p97 (E), SQSTM/p62 (E), HDAC6 (E), UBAP2L (E), or TRIM 25 (E). (C) Cells stably expressing either mCherry-tagged wild-type ubiquitin (mCh-Ub-WT) or mCherry-tagged ubiquitin, in which all internal lysine residues were mutated to arginine and the C-terminal diglycine residues were removed (mCh-Ub-K0DGG [K0dGG]) were stained with antibodies against G3BP1. G3BP1-positive SGs are indicated by arrows (both red and white). G3BP1-negative perinuclear foci are indicated by solid yellow arrowheads. G3BP1-negative punctate signal for VCP/p97 and SQSTM/p62 is indicated by blue open arrowheads. Images from Ub-E1i-treated cells in all panels were taken at the same exposure time and acquisition settings in the ubiquitin channel as those for images from DMSO-treated cells. Nuclei were stained using DAPI. Scale bars, 20 μ m in all panels.
See also Figure S4.
To validate these findings, we generated HeLa cell lines that stably expressed either mCherry-tagged wild-type ubiquitin (mCh-Ub-WT) or mCherry-tagged ubiquitin in which all internal lysine residues were mutated to arginine and the C-terminal di-glycine residue was removed (mCh-Ub-K0ΔGG). The K0DGG variant is unable to be incorporated into chains or be utilized by the ubiquitin conjugation machinery, thus serving as a marker for free ubiquitin (Dantuma et al., 2006). Recapitulating the results obtained by IF staining, mCh-Ub-WT strongly localized to a single perinuclear focus per cell, with no observable SG co-localization upon arsenite treatment (Figure 4C). When cells were treated with Ub-E1i prior to and during stress as described earlier, these large foci were largely ablated, and some co-localization with SGs was observed (Figure 4C). By contrast, mCh-Ub-K0ΔGG localized to SGs in a Ub-E1i-independent manner and did not accumulate in larger foci distinct from SGs. (Figure 4C). We obtained the same results in 293T cells stably expressing mCherry-tagged ubiquitin variants (Figure S4D). In summary, our data demonstrate that free ubiquitin is the primary form of ubiquitin that localizes to arsenite-induced SGs.
Polyubiquitin Accumulates at Centrosomes in Response to Stress
We next sought to characterize the structures at which we observed the accumulation of polyubiquitylated proteins in response to arsenite stress. Their subcellular localization was highly reminiscent of the centrosome, which has been implicated in the proteasomal processing of ubiquitylated substrates (Fabunmi et al., 2000; Wigley et al., 1999). Indeed, IF staining showed clear co-localization of the polyubiquitin signal with the centrosomal marker pericentrin (PCNT), as well as proteasome subunits (Figure 4D). Treatment of cells with Ub-E1i did not affect localization of PCNT or the proteasome, indicating that polyubiquitin is not essential for scaffolding or maintaining the centrosome-associated proteasome.
Localization of UPS Proteins Does Not Depend on Polyubiquitin
In light of these findings, we further characterized the effect of ablating polyubiquitylation on the subcellular distribution of several proteins associated either with stress-induced protein homeostasis or with otherwise known or proposed functions within the UPS. Strikingly, of the proteins we analyzed, only the localization of the autophagy-related protein sequestosome-1 (SQSTM/p62) was affected upon Ub-E1i treatment, losing its punctate appearance in what are likely to be previously described p62-positive deposition sites (Ganassi et al., 2016; Minoia et al., 2014) (Figure 4E). By contrast, localization of VCP/p97 was unchanged upon UAE inhibition, as was the SG localization of previously characterized SG-localized UPS components HDAC6, BAG3, TRIM25, and UBAP2L (Ganassi et al., 2016; Kwon et al., 2007; Markmiller et al., 2018) (Figure 4E). These findings are consistent with our hypothesis that protein ubiquitylation is dispensable for SG formation by showing that even proteins with demonstrated functions within the UPS associate with SGs in a UAE-independent manner.
DISCUSSION
Accumulating evidence suggests that multivalent protein-protein, protein-RNA, and RNA-RNA interactions are required to nucleate SGs and that SGs form as a result of liquid-liquid phase separation (LLPS) among SG components (Van Treeck and Parker, 2018; Wheeler et al., 2016). Indeed, many identified SG proteins contain intrinsically disordered domains or domains of low complexity that are critical not only for their SG localization but also for their ability to undergo phase transitions in vitro (Boeynaems et al., 2017; Lin et al., 2015; Mackenzie et al., 2017; Martinez et al., 2016; Molliex et al., 2015; Murakami et al., 2015; Patel et al., 2015; Riback et al., 2017; Xiang et al., 2015). Post-translational modifications on SG proteins could serve to either disrupt critical multivalent interactions or provide new contact surfaces that drive LLPS (Ohn and Anderson, 2010). Because ubiquitin can form polymeric chains and ubiquitin-inter-acting proteins are present in SGs, it was conceivable that protein ubiquitylation might regulate higher order protein-protein interactions critical for SG dynamics. Combined with previous studies, our results demonstrate that ubiquitin co-localizes with SGs nucleated in response to oxidative stress caused by sodium arsenite treatment. However, our results also clearly establish that acute inhibition of active protein ubiquitylation or neddylation does not directly impact SG formation or dissolution, despite the near ablation of overall protein ubiquitylation.
We show that the SG localization of several proteins associated with the UPS is equally unaffected by UAE inhibition, supporting the notion that polyubiquitin plays, at most, a minor role in SG biology. Nevertheless, a systematic characterization of SG composition under Ub-E1i treatment conditions would determine whether any proteins require active ubiquitylation for their SG localization. Our observation that ubiquitin remains detectable within SGs even after acute UAE inhibition could simply be the result of free ubiquitin localizing to SGs in a passive manner. Alternatively, it is possible that free, unconjugated ubiquitin may play a role in nucleating or modulating SGs. Consistent with this hypothesis is the demonstration that the ubiquitin-binding protein UBQLN2 undergoes LLPS in vitro and that this transition can be inhibited by adding unconjugated ubiquitin to the reaction (Dao et al., 2018). It will be difficult to demonstrate that free ubiquitin is critical for SG dynamics, given the inability to ablate cellular ubiquitin without catastrophic consequences to overall cellular function.
Nearly all chemical SG inducers disrupt protein folding and quality control pathways, resulting in the accumulation of polyubiquitylated material within cells. While our data show that polyubiquitylation is not a critical factor in the acute formation and disassembly of SGs, it is likely to be important in determining how the SG response is integrated with the larger proteostasis system during stress. However, the type and extent of cellular stress will need to be carefully controlled to make definitive links between proteostasis dysfunction and SG dynamics. For example, previous studies demonstrated that long-term (18-h) NAE inhibition or knockdown of neddylation components inhibits SG formation (Jayabalan et al., 2016; Ohn et al., 2008). Near-complete NAE inhibition using N8-E1i takes place on minute timescales when added to mammalian cells (Liu et al., 2018). As such, it is critical to separate observations using acute NAE or UAE inhibition from those using prolonged inhibition that will result in widespread cellular dysfunction. Clear temporal studies investigating SG compositional dynamics over long timescales during conditions that promote proteostasis dysfunction are needed to provide insight into how the proteostasis machinery, like the UPS, interact and cooperate with SGs during disease pathogenesis.
STAR⋆METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Eric J. Bennett (e1bennett@ucsd.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Immortalized human cell lines were utilized in this study. The Lenti-X HEK293T cell line is derived from human female tissue, the HCT116 cell line is derived from human colorectal carcinoma tissue, and HeLa S3 cells are derived from human female cervical adenocarcinoma tissue. All cell lines were grown in complete DMEM media (GIBCO) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a humidified incubator under 5% CO2.
METHOD DETAILS
SILAC labeling
For stable isotope labeling by amino acids in cell culture (SILAC) experiments, cells were cultured in custom DMEM without arginine or lysine (Mediatech) supplemented with 10% dialyzed FBS (Life Technologies), 1% penicillin/streptomycin, L-Arginine hydrochloride (85mg/ml Sigma) and either “light” L-Lysine hydrochloride (50μg/ml Sigma) or heavy 13C6,15N2 L-Lysine-hydrochloride (50mg/ml Cambridge Isotopes) and 292 mg/mL L-Glutamine (Mediatech).
Mass Spectrometry
Heavy and light labeled cells were mixed 1:1 and processed for proteomics and diGLY-immuno-affinity enrichment as described previously(Gendron et al., 2016; Kim et al., 2011). Samples were analyzed by LC-MS/MS using a Q-Exactive mass spectrometer (Thermo Scientific, San Jose, CA) and all data was processed as described previously (Gendron et al., 2016). All RAW mass spectrometry data files have been deposited at the MassIVE repository using the accession identifier MassIVE: MSV000082933.
Generation of cell lines stably expressing mCherry-tagged ubiquitin
For creation of stable cell lines, we generated a MSCV-N-mCherry-IRES-PURO gateway destination vector by replacing the Flag-HA cassette from MSCV-N-Flag-HA-IRES-PURO (Addgene #41033) with mCherry. Wild-type ubiquitin (Ub-WT) and ubiquitin in which all internal lysine residues were mutated to arginine and the c-terminal diglycine residues were removed (Ub-K0ΔGG) were cloned into the MSCV-N-Flag-HA-IRES-PURO retroviral vector by gateway cloning. Retrovirus was packaged in 293 Lenti-X cells (Clontech/TaKaRa) according to standard procedures. HeLa and 293T cells were transduced with retroviral particles and polyclonal cell lines with stable expression of mCherry-Ub variants established by puromycin selection.
Immunofluorescence, imaging and image analysis
For immunofluorescence experiments, cells were plates into 96- or 384-well optical bottom plates and left to adhere for 24–28 hours. After inhibitor and stress treatments, cells were fixed in 4% paraformaldehyde in PBS or PHEM buffer (60mM PIPES, 25mM HEPES, 4mM MgCl2, 10mM EGTA, pH 7.4) for 15–20’ at room temperature, permeabilized in 0.25% Triton-X in PBS for 15’ at RT, followed by blocking in 2% BSA + 10% normal goat serum in PBS + 0.01% Tween-20 (PBST) and 0.3M glycine for at least 60min at RT. Primary antibodies were diluted in 1× PHEM buffer + 1% BSA + 5% normal goat serum and incubated either at RT for 1–4 hours or overnight at 4°C. After 2 washes in PBST, cells were incubated with secondary antibodies in PBST + DAPI for 30–60min at RT, followed by two washes in PBST and final resuspension in 50% glycerol in PBS. Cells were then imaged either at 20× or 40× magnification on a ZEISS Axio Vert.A1 inverted microscope with equipped with a Colibri LED light source and Apotome optical sectioning module. Images were processed in ImageJ.
High-content imaging of SG time course plates
96- or 384-well plates were imaged using a Vala Sciences IC200-KIC high-content screening system. Either four or nine fields at the center of each well were imaged with a 20× objective through 460 nm and 535 nm emission filters for DAPI and G3BP1-GFP, respectively. Exposure times were optimized for each set of plates and applied uniformly to all wells.
QUANTIFICATION AND STATISTICAL ANALYSIS
Automated image segmentation and feature quantification
Images from SG assembly and disassembly time courses were segmented and image features identified and quantified using a custom CellProfiler pipeline (Carpenter et al., 2006). Briefly, nuclei were segmented and identified in the DAPI fluorescence channel images using a diameter cutoff of 9–80 pixels for HEK293xT and HeLa cells. Both nuclei count and total nuclear area were calculated for each image. For quantification of SGs, we first identified cell bodies by overlaying the GFP fluorescence channel images and tracing radially outward from the nuclei to the limits of the cytoplasmic G3BP1-GFP signal. The cell bodies were used as masks for the subsequent SG identification to eliminate imaging artifacts outside of cell boundaries, such as background fluorescence or dead cells. After masking, punctate structures within the cell body area were selected by enhancing speckle-like features that were 10 pixels in diameter for HEK293xT and HeLa cells, and these punctate structures were then annotated as stress granules. As for nuclei, both SG count and total SG area were calculated for each image. We chose normalized SG area as the most robust metric for SG dynamics. This measure was obtained by calculating the total area covered by SGs in each image and normalizing this by dividing SG area by the total area covered by nuclei in the same image. Each experimental condition was represented by between 12 and 16 replicate wells, with between 4 and 9 images acquired per well. The mean and standard deviation were calculated across all replicate wells and fields of view acquired per well, with a minimum of ~25,000 cells analyzed per condition.
DATA AND SOFTWARE AVAILABILITY
The proteomics data reported in this paper are available for download from the MassIVE mass spectrometry data repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp). The accession for this dataset is MassIVE: MSV000082933.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-G3BP1 | MBL International | Cat#RN048PW; RRID:AB_10794608 |
| Mouse monoclonal anti-G3BP1 | EMD Millipore | Cat#05–1938; RRID:AB_10561767 |
| Rabbit polyclonal anti-Ubiquitin | EMD Millipore | Cat#AB1690; RRID:AB_2180744) |
| Mouse monoclonal anti-Ubiquitin | EMD Millipore | Cat#MAB1510; RRID:AB_461752 |
| Mouse monoclonal anti-Ubiquitinylated proteins (FK1) | EMD Millipore | Cat# 04–262; RRID:AB_11213557 |
| Mouse monoclonal anti-Ubiquitin (FK2) | EMD Millipore | Cat# ST1200–100UG; RRID:AB_10681625 |
| Rabbit polyclonal anti-K48 (Apu2) | EMD Millipore | Cat# 05–1307; RRID:AB_1587578 |
| Rabbit polyclonal anti-K63 (Apu3) | EMD Millipore | Cat# 05–1308; RRID:AB_1587580 |
| Rabbit polyclonal anti-pericentrin | EMD Millipore | Cat# ABT59; RRID:AB_10947564 |
| Rabbit polyclonal anti-PSMA5 | Thermo Fisher | Cat# PA1–1962; RRID:AB_2171717 |
| Rabbit polyclonal anti-PSMB8 | Thermo Fisher | Cat# PA1–972; RRID:AB_2172350 |
| Rabbit polyclonal anti-VCP | Proteintech | Cat# 10736–1-AP; RRID:AB_2214635 |
| Rabbit polyclonal anti-SQSTM | Bethyl | Cat# A302–856A; RRID:AB_10631138 |
| Rabbit polyclonal anti-HDAC6 | Proteintech | Cat# 12834–1-AP; RRID:AB_10597094 |
| Rabbit polyclonal anti-BAG3 | Bethyl | Cat# A302–806A; RRID:AB_10631035 |
| Rabbit polyclonal anti-UBAP2L | Bethyl | Cat# A300–533A; RRID:AB_477953 |
| Rabbit polyclonal anti-TRIM25 | Bethyl | Cat# A301–856A; RRID:AB_1279507 |
| Phospho-eIF2alpha (Ser51) (D9G8) XP Rabbit mAb antibody | Cell Signaling Technology | Cat# 3398; RRID:AB_2096481 |
| Mouse Anti-alpha-Tubulin Monoclonal Antibody, Unconjugated, Clone DM1A | Cell Signaling Technology | Cat# 3873; RRID:AB_1904178 |
| Goat anti-Rabbit IgG (H+L) Superclonal, Alexa Fluor 647 | Thermo Fisher | Cat# A27040; RRID:AB_2536101 |
| Goat anti-Rabbit IgG (H+L) Superclonal, Alexa Fluor 488 | Thermo Fisher | Cat# A27034; RRID:AB_2536097 |
| Goat anti-Mouse IgG (H+L) Superclonal, Alexa Fluor 647 | Thermo Fisher | Cat# A28181; RRID:AB_2536165 |
| Goat anti-Mouse IgG (H+L) Superclonal, Alexa Fluor 488 | Thermo Fisher | Cat# A28175; RRID:AB_2536161 |
| Goat anti-Mouse IgG / IgM (H+L) Secondary Antibody, Alexa Fluor 488 | Thermo Fisher | Cat# A-10680; RRID:AB_2534062 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Sodium arsenite | Sigma | N/A |
| MG132 | Tocris | Cat# 1748 |
| TAK-243 / MLN7243 / Ub-E1i | Chemietek | Cat# CT-M7243 |
| TAK924 / MLN4924 / N8-E1i | Cayman Chemical | Cat# 15217 |
| L-LYSINE:2HCL (13C6, 99%; 15N2, 99%) | Cambrige Isotope Labs | Cat# CNLM-291 |
| Deposited Data | ||
| Quantitative mass spectrometry data | This paper | MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) accession MassIVE: MSV000082933 |
| Experimental Models: Cell Lines | ||
| Human Lenti-X 293T cells | Clontech | Cat#632180 |
| Human HCT116 | ENCODE Project | N/A |
| Human HeLa-S3 cells | Bennett Lab | N/A |
| HEK293T G3BP1-GFP | Generated in-house (Markmilleretal., 2018) | N/A |
| Software and Algorithms | ||
| CellProfiler | Carpenter et al., 2006 | https://cellprofiler.org/ |
Highlights.
Ubiquitin-activating enzyme inhibition rapidly ablates global protein ubiquitylation
Stress granule (SG) dynamics are unaffected by acute UAE or NAE inhibition
SGs co-localize with unconjugated ubiquitin but not polyubiquitylated proteins
SG-association of ubiquitin-related proteins is independent of polyubiquitylation
ACKNOWLEDGMENTS
We thank Julia Nussbacher and members of the Bennett and Yeo labs for critical comments. We thank the Sanford-Burnham High Content Screening Facility in La Jolla, as well as Cleber Trujillo and Alysson Muotri for use of imaging equipment. This work is partially funded by grants HG004659 and NS103172 (to G.W.Y.) and GM119132 (to E.J.B.) from the NIH; Target ALS (to G.W.Y.); the ALS Association (to G.W.Y.); the Larry L. Hillblom Foundation (to G.W.Y.); and the Lawrence Ellison Foundation (to E.J.B.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.04.015.
DECLARATION OF INTERESTS
G.W.Y. is a co-founder, member of the board of directors, equity holder, and paid consultant for Locana and Eclipse BioInnovations. G.W.Y. is a co-founder of Enzerna and ProteoNA. G.W.Y. is a paid consultant for Aquinnah Pharmaceuticals and Ionis Pharmaceuticals and a scientific advisory board member of Ribometrix and the Allen Institute of Immunology. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies. All other authors declare no competing interests.
REFERENCES
- Best S, Hashiguchi T, Kittai A, Bruss N, Paiva C, Okada C, Liu T, Berger A, and Danilov AV (2019). Targeting ubiquitin-activating enzyme induces ER stress-mediated apoptosis in B-cell lymphoma cells. Blood Adv 3, 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De Decker M, Jaspers T, Ryan VH, et al. (2017). Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR (2014). mRNP granules. Assembly, function, and connections with disease. RNA Biol 11, 1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis RM, Taylor JP, and Parker R (2013). Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell pheno-types. Genome Biol 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague MJ, Heride C, and Urbé S (2015). The demographics of the ubiquitin system. Trends Cell Biol 25, 417–426. [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Groothuis TA, Salomons FA, and Neefjes J (2006). A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J. Cell Biol 173, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao TP, Kolaitis RM, Kim HJ, O’Donovan K, Martyniak B, Colicino E, Hehnly H, Taylor JP, and Castaneda CA (2018). Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell 69, 965–978.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CM, Cenik B, Sephton CF, Johnson BA, Herz J, and Yu G (2012). TDP-43 aggregation in neurodegeneration: are stress granules the key? Brain Res 1462, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabunmi RP, Wigley WC, Thomas PJ, and DeMartino GN (2000). Activity and regulation of the centrosome-associated proteasome. J. Biol. Chem 275, 409–413. [DOI] [PubMed] [Google Scholar]
- Fujimuro M, and Yokosawa H (2005). Production of antipolyubiquitin mono-clonal antibodies and their use for characterization and isolation of polyubiqui-tinated proteins. Methods Enzymol 399, 75–86. [DOI] [PubMed] [Google Scholar]
- Ganassi M, Mateju D, Bigi I, Mediani L, Poser I, Lee HO, Seguin SJ, Morelli FF, Vinet J, Leo G, et al. (2016). A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol. Cell 63, 796–810. [DOI] [PubMed] [Google Scholar]
- Gendron JM, Webb K, Yang B, Rising L, Zuzow N, and Bennett EJ (2016). Using the ubiquitin-modified proteome to monitor distinct and spatially restricted protein homeostasis dysfunction. Mol. Cell. Proteomics 15, 2576–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyer ML, Milhollen MA, Ciavarri J, Fleming P, Traore T, Sappal D, Huck J, Shi J, Gavin J, Brownell J, et al. (2018). A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med 24, 186–193. [DOI] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, and Parker R (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayabalan AK, Sanchez A, Park RY, Yoon SP, Kang GY, Baek JH, Anderson P, Kee Y, and Ohn T (2016). NEDDylation promotes stress granule assembly. Nat. Commun 7, 12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, and Anderson P (2016). G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol 212, 845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, and Parker R (2017). The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808–820.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. (2011). Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MN, Choi J, Ryu HW, and Ryu KY (2015). Disruption of polyubiquitin gene Ubc leads to attenuated resistance against arsenite-induced toxicity in mouse embryonic fibroblasts. Biochim. Biophys. Acta 1853, 996–1009. [DOI] [PubMed] [Google Scholar]
- Kwon S, Zhang Y, and Matthias P (2007). The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev 21, 3381–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, and Gitler AD (2013). Stress granules as crucibles of ALS pathogenesis. J. Cell Biol 201, 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, and Parker R (2015). Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Reitsma JM, Mamrosh JL, Zhang Y, Straube R, and Deshaies RJ (2018). Cand1-mediated adaptive exchange mechanism enables variation in F-box protein expression. Mol. Cell 69, 773–786.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, Pottier C, Annu K, Baker M, Perkerson RB, Kurti A, et al. (2017). TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron 95, 808–816.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, et al. (2018). Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FJ, Pratt GA, Van Nostrand EL, Batra R, Huelga SC, Kapeli K, Freese P, Chun SJ, Ling K, Gelboin-Burkhart C, et al. (2016). Protein-RNA networks regulated by normal and ALS-associated mutant HNRNPA2B1 in the nervous system. Neuron 92, 780–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju D, Franzmann TM, Patel A, Kopach A, Boczek EE, Maharana S, Lee HO, Carra S, Hyman AA, and Alberti S (2017). An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J 36, 1669–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Di Marco S, Kaufman RJ, and Gallouzi IE (2007). Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol. Biol. Cell 18, 2603–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoia M, Boncoraglio A, Vinet J, Morelli FF, Brunsting JF, Poletti A, Krom S, Reits E, Kampinga HH, and Carra S (2014). BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: implications for a proteasome-to-autophagy switch. Autophagy 10, 1603–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FT, Michel CH, et al. (2015). ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 88, 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, and Anderson P (2010). The role of posttranslational modifications in the assembly of stress granules. Wiley Interdiscip. Rev. RNA 1, 486–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, and Anderson P (2008). A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol 10, 1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077. [DOI] [PubMed] [Google Scholar]
- Protter DSW, and Parker R (2016). Principles and properties of stress granules. Trends Cell Biol 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, and Drummond DA (2017). Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168, 1028–1040.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin SJ, Morelli FF, Vinet J, Amore D, De Biasi S, Poletti A, Rubinsztein DC, and Carra S (2014). Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ 21, 1838–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncini C, Berdo I, and Draetta G (2001). Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene 20, 3869–3879. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. (2009). An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Higuchi M, Matsuki H, Yoshita M, Ohsawa T, Oie M, and Fujii M (2013). Stress granules inhibit apoptosis by reducing reactive oxygen species production. Mol. Cell. Biol 33, 815–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turakhiya A, Meyer SR, Marincola G, Bohm S, Vanselow JT, Schlosser A, Hofmann K, and Buchberger A (2018). ZFAND1 recruits p97 and the 26S proteasome to promote the clearance of arsenite-induced stress granules. Mol. Cell 70, 906–919.e7. [DOI] [PubMed] [Google Scholar]
- Van Treeck B, and Parker R (2018). Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell 174, 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JR, Matheny T, Jain S, Abrisch R, and Parker R (2016). Distinct stages in stress granule assembly and disassembly. eLife 5, e18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley WC, Fabunmi RP, Lee MG, Marino CR, Muallem S, DeMartino GN, and Thomas PJ (1999). Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol 145, 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, and McKnight SL (2015). The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell 163, 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Matsumoto S, Endo A, Fukushima T, Kawahara H, Saeki Y, and Komada M (2018). Deubiquitinases USP5 and USP13 are recruited to and regulate heat-induced stress granules by deubiquitinating activities. J. Cell Sci 131, jcs210856. [DOI] [PubMed] [Google Scholar]
- Zhuang J, Shirazi F, Singh RK, Kuiatse I, Wang H, Lee HC, Berkova Z, Berger A, Hyer M, Chattopadhyay N, et al. (2019). Ubiquitin-activating enzyme inhibition induces an unfolded protein response and overcomes drug resistance in myeloma. Blood 133, 1572–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomics data reported in this paper are available for download from the MassIVE mass spectrometry data repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp). The accession for this dataset is MassIVE: MSV000082933.