Abstract
ARGONAUTES are the central effector proteins of RNA silencing which bind target transcripts in a small RNA‐guided manner. Arabidopsis thaliana has 10 ARGONAUTE (AGO) genes, with specialized roles in RNA‐directed DNA methylation, post‐transcriptional gene silencing, and antiviral defense. To better understand specialization among AGO genes at the level of transcriptional regulation we tested a library of 1497 transcription factors for binding to the promoters of AGO1,AGO10, and AGO7 using yeast 1‐hybrid assays. A ranked list of candidate DNA‐binding TFs revealed binding of the AGO7 promoter by a number of proteins in two families: the miR156‐regulated SPL family and the miR319‐regulated TCP family, both of which have roles in developmental timing and leaf morphology. Possible functions for SPL and TCP binding are unclear: we showed that these binding sites are not required for the polar expression pattern of AGO7, nor for the function of AGO7 in leaf shape. Normal AGO7 transcription levels and function appear to depend instead on an adjacent 124‐bp region. Progress in understanding the structure of this promoter may aid efforts to understand how the conserved AGO7‐triggered TAS3 pathway functions in timing and polarity.
Keywords: Argonaute, leaf development, leaf polarity, post‐transcriptional regulation, transcriptional regulation
1. INTRODUCTION
Small RNAs regulate developmental timing and morphogenesis in a wide range of eukaryotes. Heterochronic (abnormal timing) mutants of the model nematode Caenorhabditis elegans led to the discovery of the first microRNA (miRNA) (Lee, Feinbaum, & Ambros, 1993; Wightman, Ha, & Ruvkun, 1993). Similar screens for Arabidopsis thaliana heterochronic mutants led to the elucidation of a specialized pathway in which trans‐acting small interfering (tasi)RNA are produced from noncoding TAS3 transcripts (Allen & Howell, 2010; Hunter, Sun, & Poethig, 2003; Peragine, Yoshikawa, Wu, Albrecht, & Poethig, 2004; Yoshikawa, Peragine, Park, & Poethig, 2005). Genetic analysis of leaf morphology has also led to the discovery of several other aspects of RNA silencing, including the cloning of the first ARGONAUTE (AGO) gene (Bohmert et al., 1998). AGO proteins bind small RNAs and effect small‐RNA‐guided regulatory changes. Several families of MIRNA genes are conserved in all land plants (Cuperus, Fahlgren, & Carrington, 2011), and miRNA from the majority of these families repress TFs controlling developmental programs, suggesting that AGO‐miRNA‐TF circuits became embedded in the core regulatory networks for the plant body plant early in land plant evolution (Rubio‐Somoza & Weigel, 2011).
The A. thaliana genome contains 10 AGO genes, which function in development, stress resistance, and defense against viruses and transposons (Zhang, Xia, Meyers, & Walbot, 2015). AGO7 and AGO10 are highly specialized: each has limited adaxial and vascular expression (Chitwood et al., 2009; Lynn et al., 1999) and a single main binding partner: miR390 and miR166, respectively Montgomery et al., 2008; Zhu et al., 2011). AGO7 triggers production of phased siRNAs from TAS3 noncoding transcripts (Allen, Xie, Gustafson, & Carrington, 2005; Axtell, Jan, Rajagopalan, & Bartel, 2006; Montgomery et al., 2008; Williams, Carles, Osmont, & Fletcher, 2005). Effects on ARF3, ARF4, and possibly ARF2 are the main downstream output of the AGO7/TAS3/SGS3/RDR6/DCL4 pathway (Adenot et al., 2006; Fahlgren et al., 2006; Garcia, Collier, Byrne, & Martienssen, 2006; Hunter et al., 2006). AGO7 action is thought to limit production of TAS3 tasiRNAs such that tasiRNA movement creates a graded accumulation pattern in developing leaf primordia (Chitwood et al., 2009; Schwab et al., 2009). This gradient contributes to the patterning of ARF target mRNA, establishing either an opposing gradient or a sharp boundary, which may contribute to robust maintenance of polarity (Skopelitis, Husbands, & Timmermans, 2012). The TAS3 pathway has important roles in leaf development in all plants examined thus far, including moss (Plavskin et al., 2016), maize (Dotto et al., 2014; Douglas et al., 2010; Nogueira, Madi, Chitwood, Juarez, & Timmermans, 2007), tomato (Yifhar et al., 2012), lotus (Yan et al., 2010), and alfalfa (Zhou et al., 2013).
Understanding the functions of miRNA such as miR390 and miR166 will require information on the signals controlling tissue‐specificity of their AGO partners. Our objective in this work was to identify upstream regulators of AGO genes and link them to existing genetic knowledge. We capitalized on new yeast‐based tools that provide a fast way to identify upstream regulators. We identified unexpected connections to two other conserved miRNA‐TF circuits that control leaf morphogenesis and defined two other functional regions of the AGO7 promoter.
2. METHODS
2.1. Plasmid construction
Promoter fragments were PCR‐amplified from previously described plasmids (Montgomery et al., 2008), with the primers listed in Table 1. Truncated and modified forms of the AGO7 promoter were made with the oligonucleotides listed in Tables 2 and 3. Gel‐purified PCR products were cloned with the pENTR D‐TOPO kit (Invitrogen) and LR‐recombined into several destination vectors: pGLacZi for Y1H screens (Helfer et al., 2011), pMDC162 for GUS transcriptional reporters, and pMDC99 for transgenic complementation assays (Curtis & Grossniklaus, 2003). The destination vector pY1‐gLUC59(GW) used for the secreted Gaussia luciferase Y1H reporter system has been described (Bonaldi, Li, Kang, Breton, & Pruneda‐Paz, 2017).
Table 1.
Oligonucleotide sequences used for AGO promoter TOPO cloning. Primer names indicate position of 5′‐most genomic base relative to the annotated transcription start site
| Oligo name | Sequence |
|---|---|
| AGO1_‐2308_FWD_cac | CACCCGCTTGTTAAAACTCATAATC |
| AGO1_‐1706_REV | TTAGGTGAAAGAATATCTAGAC |
| AGO1_‐1755_FWD_cacc | CACCATCTAGACAATCTTTTGTTAG |
| AGO1_‐1121_REV | GTTGCTCGTGCGTGAAGA |
| AGO1_‐1170_FWD_cacc | CACCTACTCGTGACATATTCTCTA |
| AGO1_‐536_REV | TATAAAGGATGTTATACAGTTAAG |
| AGO1_‐585_FWD_cacc | CACCACAAGTACCAATTTTAAACTG |
| AGO1_‐1_REV | TGCTACACTTTAAATTCAAGG |
| AGO7_‐1934_FWD_c | CACCTGTCTCTTCTTCTGTACATGC |
| AGO7_‐1436_REV | TAAGTATATTAAAAAATATCAGATGAC |
| AGO7_‐1485_FWD_cacc | CACCTTATAGGTAAATGGATATGACT |
| AGO7_‐941_REV | TGCTAAAACAAAAGATGCTCAA |
| AGO7_‐991_FWD_cac | CACCCAAAGACATACATCTATAATATA |
| AGO7_‐446_REV | AATTATGGGGACCATTCTGT |
| AGO7_‐495_FWD_cacc | CACCAAGAAAATAGTACAAAGAATAAAT |
| AGO7_‐1_REV | AGAAAGGGATTGTCTGAGTTT |
| AGO10_‐2033_FWD_cacc | CACCGATTTCTATAAAAAATACATTCC |
| AGO10_‐1511_REV | AGACCCCATTTCGTGACT |
| AGO10_‐1560_FWD_cacc | CACCGGAAGAAAACAAAATTAATGAG |
| AGO10_‐991_REV | TAGTCTAGGTTAGTTTCCG |
| AGO10_‐1040_FWD_cacc | CACCTATCACAAACTAGACAATCC |
| AGO10_‐471_REV | ACATCATTGTTACAAGATGG |
| AGO10_‐520_FWD_cacc | CACCTTTTTATAATAAGATTAGAGAATTAT |
| AGO10_‐1_REV | ATAGCTTTCCTCTCAATGTG |
Names also list the nucleotides added to create “CACC” sequences for directional TOPO cloning.
Table 2.
Oligonucleotide sequences directly cloned for AGO7 promoter mutation analysis in yeast

Table 3.
Forward primer sequences used for TOPO cloning of truncated versions of the AGO7 promoter
| Oligo name | Sequence |
|---|---|
| AGO7_‐482_FWD_cacc | CACCAAAGAATAAATAATTAAACAGAATGGTCC |
| AGO7_‐453_FWD_cacc | CACCCCATAATTCGATTTAATGAGTGTATTG |
| AGO7_‐422_FWD_cacc | CACCATTTTATAAAACATGTGTAACAACAACAA |
| AGO7_‐298_FWD | CACCAAACATTATCGGTAATCACTA |
| AGO7_‐150_FWD_cacc | CACCTATTTTCTTTTATTATTGCCAACAATT |
| AGO7_1_FWD_cacc | CACCGCCTCTTTTATCTCTCTCTCTCATAAA |
Names follow Table 1. Bases ‐298/‐295 are a natural “CACC” sequence suitable for directional TOPO cloning.
2.2. Y1H screens
Automated lacZ screens were done as previously described (Pruneda‐Paz, Breton, Para, & Kay, 2009; Pruneda‐Paz et al., 2014) using a collection of 1497 TFs and an Agilent BioCel 1200 robotic platform. The TF‐activation domain fusion yeast strain collection (arrayed in 384‐well plates) was mated to bait strains. Diploid cells were selected in media lacking uracil and tryptophan, lysed by freeze‐thaw, and assayed for β‐galactosidase activity. Targeted Y1H assays were done similarly, with the lysis and assay steps replaced, essentially as described (Bonaldi et al., 2017). Briefly, diploid cells were resuspended in phosphate‐buffer saline, 50 μl of cells were transferred to a clear‐bottom plate, and a Synergy H1 plate reader (Biotek) was used to inject 10 μl of 20 μM coelenterazine substrate solution into each well and read luminescence immediately afterward (0.1 s integration time).
2.3. Plant materials and growth conditions
All A. thaliana plants descended from the reference Col‐0 accession. The zippy‐1 mutant allele was isolated by Hunter et al. (2003), and is referred to throughout as “ago7”. Plants were transformed by floral dip using Agrobacterium strain GV3101 (Clough & Bent, 1998; Holsters et al., 1980).
Plants were grown under short day conditions (8 hr light, 16 hr dark) in a Conviron MTR25 reach‐in chamber with PolyLux fluorescent bulbs (200 μmol photons s−1 m2) at 22°C with 50% humidity.
2.4. ago7 mutant complementation tests
Measurement of leaf phenotypes followed previous work (Fahlgren et al., 2006): we scored the index of the earliest leaf with at least one abaxial trichome using a stereomicroscope at 28–30 days post‐stratification, and concurrently measured the blade length and petiole length for the sixth true leaf with digital calipers (Mitutoyo, Japan). At a later timepoint (33 and 35 days post‐stratification), we dissected and scanned the first 10 true leaves from each plant with a Canon Pixma MP190 flatbed scanner. Leaf shape parameters were measured with the LeafJ plug‐in for ImageJ (Maloof, Nozue, Mumbach, & Palmer, 2013). Plants were also photographed from above (per Tovar et al., 2018) from 11 days post‐stratification onward and the time‐lapse image data documented with the rest of the experiment: see Zenodo records 1340636, 439652, and 1256716.
2.5. GUS assays
Histological GUS assays were essentially as described (Bomblies, 2002; Chitwood et al., 2009; Strader et al., 2011). Seedlings were collected into ice‐cold 90% acetone, incubated at −20°C for 20 min and then room temperature for another 20 min. Seedlings were washed twice (5 min each) with staining buffer (100 mM sodium phosphate [pH 7], 20% methanol, 0.1% Triton X‐100, 1.5 mM ferri‐ and ferrocyanide).
Staining buffer with 0.5 mg/mL 5–bromo–4–chloro–3–indolyl–β–d–glucuronic acid (X‐Gluc) was vacuum‐infiltrated into seedlings on ice for two rounds of 15 min each. Samples were then incubated at 37°C for 20 hr, taken through an ethanol/histoclear series, and infiltrated with Paraplast Plus at 60°C, before embedding (Bomblies, 2002). Tissue sections (10 μm thickness) were mounted on Probe‐On Plus slides (Thermo Fisher), deparaffinized with histoclear, and coverslipped. Sections were viewed and photographed with a Leica DM750 microscope and ICC50 HD camera.
2.6. Data and code availability
Data and software code supporting this manuscript have been deposited as supplemental datasets:
Hoyer, S. (2018). jshoyer/y1h‐AGO7‐promoter: Yeast 1‐hybrid screens for upstream regulators of A. thaliana AGO1, AGO7, and AGO10: raw data and R code. Retrieved from https://doi.org/10.5281/zenodo.1472704
Hoyer, J.S. (2018). Yeast 1‐hybrid screens for upstream regulators of A. thaliana AGO1, AGO7, and AGO10: ranked tables of candidate direct upstream TFs. Retrieved from https://doi.org/10.5281/zenodo.1472235
Hoyer, J.S., Holcolm, E. E. (2018). Photomicrographs: transverse sections of GUS‐stained apices for AGO7 promoter analysis. Retrieved from https://doi.org/10.5281/zenodo.1319761
Hoyer, J.S. (2018). Scans of leaves dissected in phyllotactic order: complementation of ago7 mutant A. thaliana plants with truncated promoter transgenes. Retrieved from https://doi.org/10.5281/zenodo.1322799
Hoyer, S. (2018). jshoyer/raspi‐photo‐and‐leaf‐scan‐metadata: Metadata and code for A. thaliana leaf scans and top‐down time‐course photos. Retrieved from https://doi.org/10.5281/zenodo.1472768
Data processing was done with the R Statistical Computing Environment (R Core Team 2017) and the Bioconductor BioStrings package was used for PWM scans (Gentleman et al., 2004; Pagès, Aboyoun, Gentleman, & DebRoy, 2017). Supporting Information Data S1 also include results from “Find Individual Occurences of Motifs” tool (FIMO) scans (Grant et al., 2011) done via the online MEME Suite (Bailey et al., 2009) version 4.12.0, with default settings (p < 10−4 cutoff) and three collections of DNA‐binding specificity models (Franco‐Zorrilla et al., 2014; O'Malley et al., 2016; Weirauch et al., 2014).
3. RESULTS
3.1. Multiple SPLs and TCPs bind the AGO7 promoter
We sought to identify TFs controlling the expression of the three main AGO genes involved in post‐transcriptional control of development (AGO1, AGO10, and AGO7) using high‐throughput yeast 1‐hybrid assays. Our automated strategy, described previously (Pruneda‐Paz et al., 2009; Pruneda‐Paz et al., 2009, 2014), uses a large collection of arrayed A. thaliana TFs (details below) and also short promoter bait sequences, for high resolution and sensitivity. We considered four fragments for each promoter, with ~50 bp of overlap between fragments, to ensure that fragment‐edge binding sites were assayed. For AGO7 these fragments spanned a 1,934 bp region (Figure 1a).
Figure 1.
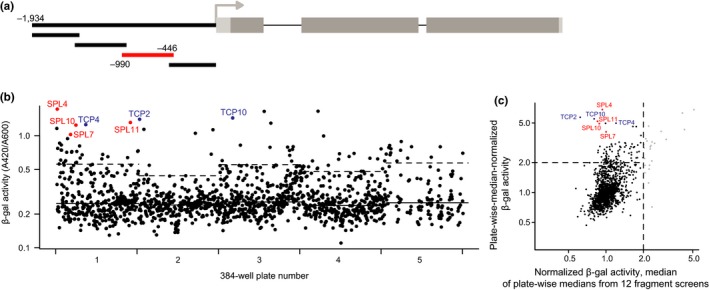
SPL and TCP TFs bind the AGO7 promoter in yeast. (a) Schematic of AGO7 promoter illustrating four fragments screened with Y1H assays. Subsequent panels show results for the fragment indicated in red, which spans the region from 990 bp to 446 bp upstream of the transcription start site. (b) Scatterplot of β‐gal activities for each prey TF constructs screened. Wells are shown in row‐first order for each of the five plates. Median activity for each plate is indicated with solid lines. Dashed lines indicate a cutoff of 6 median absolute deviations above the median for each plate. Hits from SPL and TCP families are highlighted. (c) Diagnostic plot incorporating data from 12 screens. Y‐dimension reflects the same values as panel B, normalized by plate median. X‐dimension results from taking the median of plate‐wise‐median activities from all twelve AGO promoter fragment screens. Vertical dashed line demarcates TFs for which median reporter activity is two‐fold higher than the median for their plate (nonspecific activators, light gray)
Transgenes driven by the collective sequences represented by these fragments are sufficient to complement corresponding ago mutants (Baumberger & Baulcombe, 2005; Montgomery et al., 2008; Tucker et al., 2008), suggesting that they contain the most important upstream regulatory elements. Promoter fragments were screened against a TF‐activation domain fusion library in 384‐well format with one prey TF per well (Pruneda‐Paz et al., 2014), using β‐galactosidase reporter activity from fusion to promoterless uidA coding sequence as a quantitative readout (Figure 1).
A total of 1497 TFs were tested for AGO promoter binding. This collection consisted mainly of sequence‐specific TFs, but also includes transcriptional co‐factors and empty vector control wells (Pruneda‐Paz et al., 2014). Each TF was tested against each promoter fragment a single time. We ranked TF candidates based on normalizing promoter‐fragment‐driven β‐gal activity by the median value for each plate (as illustrated in Figure 1b), to account for systematic differences between plates. We separately plotted signal distributions across all 12 screens (Supporting Information Figures S1–S3) to assess which TFs “hits” act as nonspecific activators in this system, as described below. (Supporting Information Data S1 and S2).
Of the TFs families assayed, only two were represented by multiple hits 6 median absolute deviations or more above the median for their plate (Figure 1b). The first group, Teosinte Branched/Cycloidea/PCF family factors (TCPs), had previously been suggested to directly regulate AGO7 (Koyama, Mitsuda, Seki, Shinozaki, & Ohme‐Takagi, 2010). The three TCP hits identified are miR319 targets (Palatnik et al., 2003) and redundantly control leaf margin development and senescence (Schommer et al., 2008). The second group, SPL factors, are master regulators of heteroblasty in A. thaliana and other plants (Poethig, 2013), the same context in which AGO7 was discovered (Hunter et al., 2003). AGO7 was prioritized over AGO10 and AGO10 for mutagenesis and functional analysis based on interest in these TFs, for which roles in timing but not polarity are well established.
We examined the distribution of reporter activity for other promoter fragments screened, confirming that these SPL and TCPs specifically hit the second proximal region of the AGO7 promoter. Plate‐wise median β‐gal activities for the SPL and TCP hits were close to the median (across all twelve screens) for their plate (Figure 1c), indicating that they do not fall in the group of TFs that are nonspecific reporter gene activators.
We further tested a group of SPL and TCP factors with a second Y1H system, based on a secreted luciferase reporter with an improved dynamic range (Bonaldi et al., 2017); repeated testing reduces statistical false positives and use of alternative reporters can reveal reporter‐gene‐specific technical false positives (Walhout, 2011). This secondary screening confirmed that multiple SPL and TCP TFs bind the second proximal AGO7 promoter fragment tested, despite considerable experimental variability (Supporting Information Figure S4). Some TFs yielded a small degree of activation relative to two different empty vector controls; it is not clear whether these small differences reflect lack of binding (i.e. nonspecific binding only) or indicate binding that is weak but specific.
We assessed possible SPLs and TCPs binding sites using DNA‐binding specificity models determined based on in vitro sequence affinity with protein‐binding microarrays (Weirauch et al., 2014). These position‐weight matrices (PWM, downloaded from the CisBP database) match consensus binding sequences previously determined with in vitro selection for SPLs (Birkenbihl, Jach, Saedler, & Huijser, 2005) and TCP4 (Schommer et al., 2008). These targeted scans complement a wider analysis done with the “Find Individual Occurences of Motifs” tool (FIMO) tool (Grant, Bailey, & Noble, 2011) and three collections of A. thaliana TF PWMs (Franco‐Zorrilla et al., 2014; O'Malley et al., 2016; Weirauch et al., 2014) documented in Supporting Information Data S1. An example sequence logo for one model, for SPL11, is shown in Figure 2a. Because the Y1H bait of interest extends to position ‐990 (Figure 1a), we considered the 1‐kb region adjacent to the annotated AGO7 transcription start site. For SPL11, the highest‐scoring positions (on both strands) were centered on the only two “GTAC” motifs (SPL core binding sites) in that region, at ‐500/‐497 and ‐486/‐483 (Figure 2, panels A and B).
Figure 2.
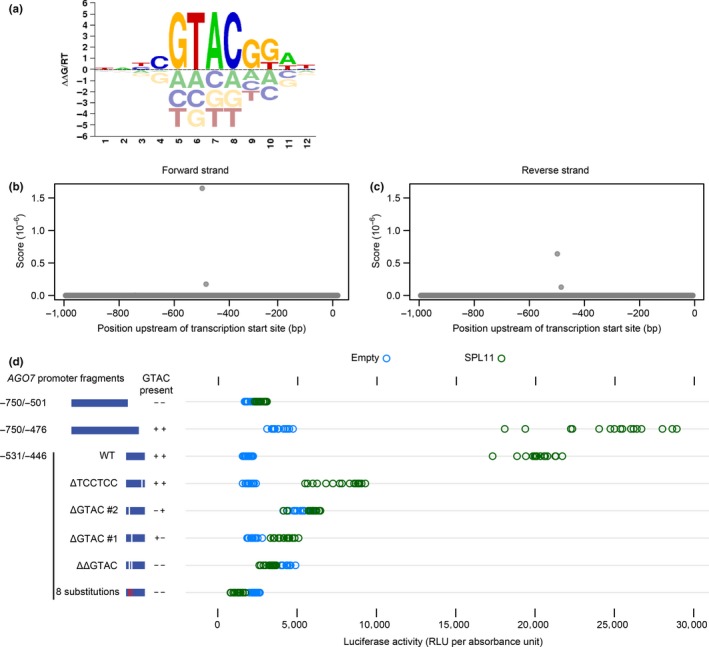
Identification of SPL11 binding sites. (a) Sequence logo for SPL11 PWM, as downloaded from CisBP. Individual position weights can be interpreted as binding specificity contributions (changes in free energy, arbitrary units). (b and c) Scores for SPL11 PWM at each position of the 1‐kb region upstream of the annotated AGO7 transcription start site. (d) Reporter activity (relative luminescence units normalized by A600) for SPL11 and pDEST22 (empty vector) tested in yeast against AGO7 promoter baits including several derivatives of the ‐531/‐446 region. Modifications included one or two 4‐bp deletions, 8 substitutions (TCCG/AAGG), and an unrelated 6‐bp deletion; see Table 2, below
We tested the significance of these “GTAC” sequences using the luciferase reporter gene in yeast. Truncated Y1H bait sequences (‐531/‐446 and ‐750/‐476) containing core binding sequences yielded activation of the reporter when tested against SPL11, but not with the corresponding empty prey vector (Figure 2c). By contrast, activation was not observed for a 3′‐truncated bait lacking “GTAC” sites (‐750/‐501), nor for modified ‐531/‐446 bait sequences with one or both 4‐mers deleted or scrambled (Figure 2c). Deletion of an unrelated 6‐bp region reduced reporter activation (compared to empty vector) but not to the same extent. These results are consistent with direct SPL binding, possibly with some degree of cooperativity, at one or both “GTAC” sites in the yeast system.
We similarly scanned the promoter sequence with empirically determined PWM for five of eight CINCINNATA‐like TCPs, a set that includes four of the five miR319 targets in A. thaliana (Nath, Crawford, Carpenter, & Coen, 2003; Palatnik et al., 2003) but does not include TCP10. The highest scoring positions for four TCPs were centered on a “TGGTCC” motif at ‐459/‐454 (Figure 3, panels E to I). This 6‐mer was the most highly enriched sequence in the promoters of a set of experimentally defined TCP targets (Schommer et al., 2008), and is present in the “most preferred” sequences for TCP3, TCP4, and TCP5 PWMs. A second “TGGTCC” site at ‐428/‐423 was among the four highest‐scoring sequences for all five TCPs considered (Figure 3), but was absent from the ‐990/‐446 region that yielded TCP hits in the initial Y1H screen. High‐scoring positions for the TCP2 PWM included a related “GGGACC” sequence at ‐764/‐770 followed by the ‐459/‐454 “TGGTCC” motif (Figure 3, panels A and F). The second highest scoring position for TCP24 was centered on a nearby “GTTCCC” sequence (Figure 3j).
Figure 3.
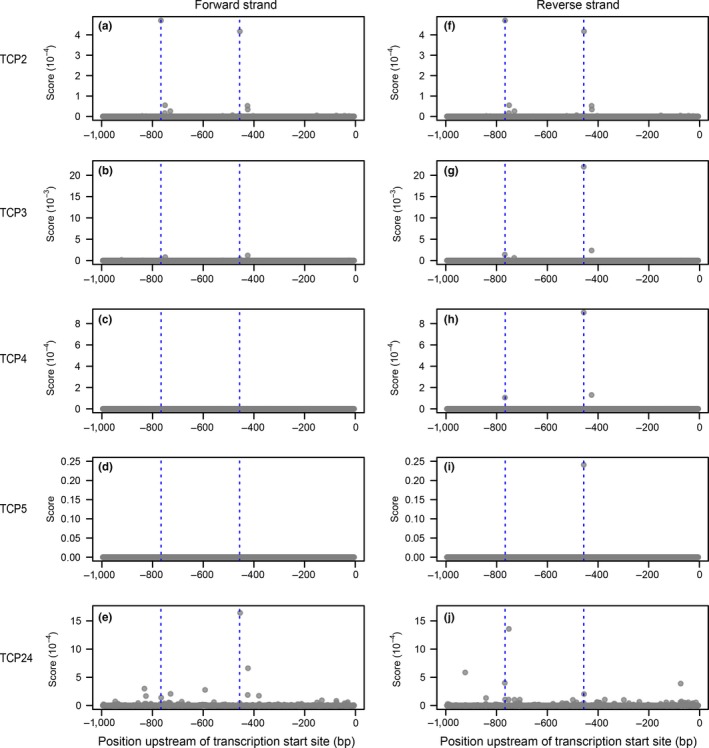
Identification of TCP binding sites. (a to j) PWM scores at each position of the 1‐kb region upstream of the annotated AGO7 transcription start site for the TCPs indicated. Dashed blue lines indicate the two highest scoring positions for TCP2: a “GGGACC“ sequence at ‐764/‐770 and a “TGGTCC“ sequence at ‐459/‐454. Panels A and F are identical, because the CisBP model for TCP2 is perfectly symmetrical
We tested requirements for candidate TCP binding sites with the luciferase Y1H system. Truncated bait sequences (‐750/‐501 and ‐750/‐476) lacking all four sites described above did not drive reporter activation (relative to the empty prey vector control) when tested with TCP2 (Figure 4). The ‐990/‐446 region used in the initial screen yielded reporter induction, as did a 5′‐truncated 86 bp bait region (‐531/‐446) containing the higher scoring “TGGTCC” motif (Figure 4). The same truncated bait sequence with the “TGGTCC” 6‐mer deleted did not yield reporter activation (Figure 4). We conclude that the ‐459/‐454 “TGGTCC” is a high‐affinity TCP binding site that functions in the yeast system and possibly in planta.
Figure 4.
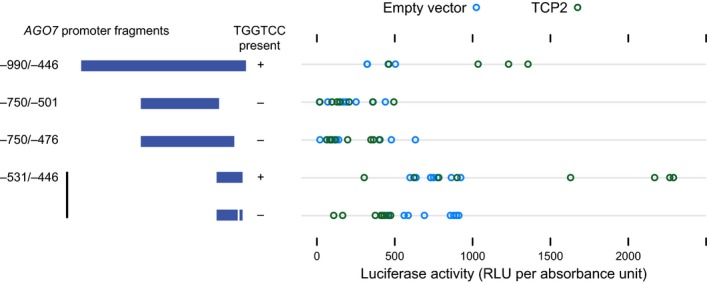
Testing of TCP binding sites in yeast. Reporter activity for TCP2 and pDEST22 (empty vector) tested against Y1H bait from initial screens (‐990/‐446), two truncated versions lacking candidate TCP binding sites, and the ‐531/‐446 region, with candidate TCP binding site (“TGGTCC“) deleted or intact
3.2. SPL binding sites are not required for polar AGO7 transcription
To test the possibility that SPL and/or TCP binding sites contribute to polar AGO7 transcription, we fused a series of truncated versions of the AGO7 promoter to GUS for comparison to previously described transcriptional reporter lines (Chitwood et al., 2009; Montgomery et al., 2008). Consistent with previous results (Chitwood et al., 2009), the 1,934 bp region upstream of the annotated AGO7 transcription start site yielded clear adaxial signal in transverse sections of leaf primordia (Figure 5a). A 482 bp version of the promoter yielded the same pattern in almost all plants tested (Figure 5b), indicating that SPL core binding sites (‐500/‐496 and ‐486/‐483) are not required for this pattern. By contrast, the TSS‐proximal 298 bp region rarely yielded visible blue reporter signal (Figure 5c). Weak adaxial signal was visible for a small proportion of plants, including two of seven plants for one of two transgenic families for the experiment illustrated (Figure 5c; see also Supporting Information Data S3). Given that promoterless‐GUS transformants did not yield visible blue signal (Figure 5d) in any of our experiments, this raises the possibility that cis elements in the proximal 298 bp region or 5′ UTR can confer adaxial polarity to AGO7 transcription. The ‐482/‐299 region, however, is a larger determinant of AGO7 transcription level, as discussed further below.
Figure 5.

Histological analysis of GUS reporter gene activity driven by truncated AGO7 promoter constructs. Core SPL binding sites are indicated in red; the 482 bp promoter construct illustrated in panel B ends immediately adjacent to the second site. Blue tick mark indicates TCP binding motif at ‐459/‐454. For each construct, results are shown for two independent transgenic families (groups of T3 siblings, each descended from a different transformant; each group was stained in a separate scintillation vial). The predominant class for each family (categorized as having distinct abaxial signal or not) is illustrated with a representative transverse section through young leaf primordia. The number of plants in the majority class is indicated as a fraction. Two and one plants yielded a weaker and/or less strongly adaxial pattern than shown here for 1,934 bp and 482 bp promoter constructs, respectively. Between three and nineteen independent lines were tested for all of the constructs shown here, with broadly similar results across multiple experiments
3.3. SPL and TCP binding sites are not strictly required for AGO7 function
We similarly tested cis requirements for transgenic complementation of ago7 mutants. We inserted a series of truncated versions of the AGO7 promoter upstream of the AGO7 coding sequence (including an N‐terminal 3x‐hemagglutinin (HA) tag). Previous results (Montgomery et al., 2008) indicated that the 1,934 bp promoter version of this transgene is functional for complementation of transformed ago7 mutants. For the experiment illustrated in Figure 6, blinded classification of downward leaf curling assigned 100% of empty‐vector‐transformed reference genotype plants (ago7 mutant and wild‐type Col‐0, n = 21 and 20 plants, respectively) to the expected phenotype class. Groups of mutant plants transformed with 3xHA‐AGO7 constructs were predominantly assigned to one or the other class: primary transformants for 422 bp to 1,934 bp promoter constructs were mostly scored as complemented, whereas most transformants for 298 bp and 0 bp promoter construct displayed the downward‐curled‐leaf mutant defect (Figure 6).
Figure 6.
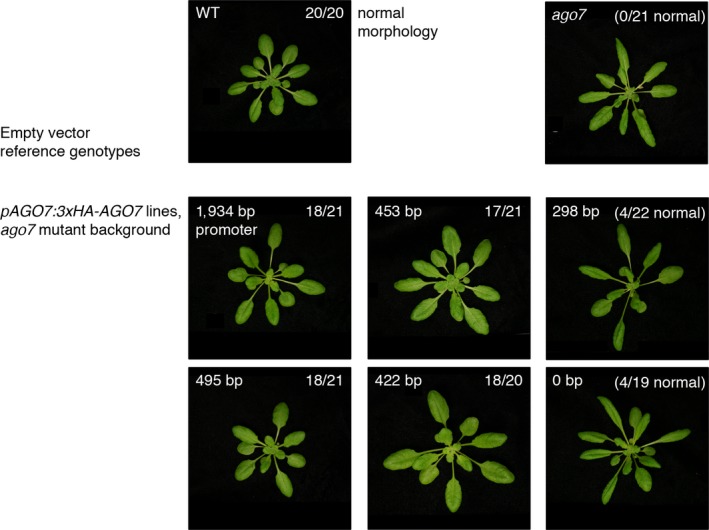
Complementation of ago7 mutant leaf shape phenotype (top right) with 3xHA‐AGO7 transgenes driven by truncated versions of the AGO7 promoter. One representative (major class) primary transformant is shown for each genotype. Upper‐left corner labels for middle and bottom rows indicate the length of upstream AGO7 regulatory sequence used to drive the 3xHA‐AGO7 coding sequence in each construct. Upper‐right corner numbers indicate the fraction of plants blindly assigned to the normal morphology category
We extended this result by quantifying leaf shape for a smaller number of transformants, by dissecting, scanning, and measuring leaves in order (Maloof et al., 2013; see Supporting Information Data S4 and S5 for full details). For the reference genotypes, leaf blade length‐to‐width ratios were higher for wild‐type relative to mutant plants, due to increased curling and/or elongation (Figure 7, panels A and H). Promoterless and 298 bp promoter construct transformants were not distinguishable from empty vector mutant controls (Figure 7, panels F and G). Longer promoter constructs shifted blade length‐to‐width ratios down toward wild‐type levels (Figure 7, panels B to E), which we interpret as partial complementation, consistent with the rosette‐level results in Figure 6. Independently measuring these leaf dimensions at one position (true leaf 6) with calipers yielded similar results (Supporting Information Figure S5).
Figure 7.
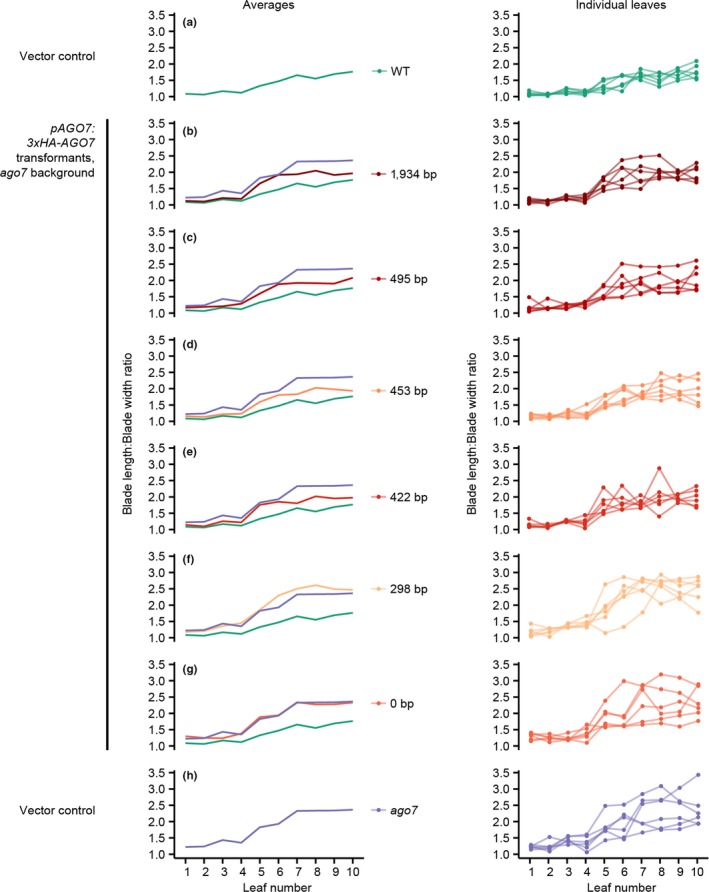
Transgenic complementation of ago7 leaf shape defects, quantified based on leaf blade length to width ratio for true leaves 1 to 10. Values for each individual plant are connected with lines on the right‐hand graphs, and the average of these values is plotted on the left. Averages for empty vector control genotypes (panels a and h) are repeated in each left‐hand panel to facilitate comparison
Results were similar for a related metric that quantifies leaf elongation, the ratio of leaf blade length to petiole length (Supporting Information Figures S5 and S6). The difference between wild‐type and mutant background control plants was smaller for this metric (Supporting Information Figure S6, panels A and H), as was the difference, if any, between means for the 1,934 bp promoter construct lines and wild‐type empty vector control lines (Supporting Information Figure S6B). Means were longer at most leaf positions (i.e. closer to wild‐type) for intermediate‐length promoter constructs (Supporting Information Figure S6, panels C, D, and E) than for short promoter constructs (Supporting Information Figure S6, panels F and G). Exceptions at one position (true leaf 10) were caused by recently emerged leaf “outliers”, the petioles of which were very short and thus disproportionately affected by technical variation (Supporting Information Figure S6C).
The promoter lengths tested end immediately adjacent to core SPL and TCP binding sites (two “TGGTCC” sites and one of two “GTAC” motifs; see Supporting Information Figure S7). We therefore tentatively conclude that SPL and TCP binding is not required for AGO7 transcription at levels that are sufficient for normal leaf morphology. The morphological data described allow us to estimate possible small differences between leaf shape in the complemented lines, but further experimentation would be necessary to relate such differences to cellular parameters or promoter structure.
Finally, we scored appearance on trichomes on abaxial leaf surfaces to assess complementation of the forward shift in ago7 mutants (Hunter et al., 2003). Consistent with results from previous transgenic experiments (Carbonell et al., 2012; Montgomery et al., 2008), abaxial trichomes were visible on an earlier leaf for empty‐vector‐transformed mutant plants relative to corresponding wild‐type plants (Supporting Information Figure S7); abaxial trichomes appeared 1.7 leaf positions earlier on average (95% confidence interval 0.5–2.9, p = 4 × 10−4, Tukey's honest significant difference method). However, there was considerable variability, possibly due to effects from hygromycin selection. No 3xHA‐AGO7 transgenic line showed a detectable increase in earliest abaxial trichome position (relative to empty‐vector‐transformed mutant plants; p > 0.3), indicating that none of the promoter lengths tested were able to drive full complementation of this defect. Alternative strategies may be required to assess ARF‐mediated effects of AGO7 levels on trichome production.
4. DISCUSSION
We characterized the structure of the AGO7 promoter with transgenic analyses and a large‐scale screen for upstream regulators. Figure 8 provides a possible interpretation these results in terms of TF binding events. The most notable result from our Y1H analysis was a direct connection to multiple miR156‐targeted SPL and miR319‐targeted TCP factors. This result appears to reinforce the idea that gradual repression of MIR156 transcription is the key regulatory step controlling heteroblasty in plants (Poethig, 2013). This connection, if verified in future studies, provides an additional example of functional linkage between SPL and TCP TFs (Lu et al., 2013; Rubio‐Somoza et al., 2014; Studer, Wang, & Doebley, 2017) and a link to ARF repressors, the other main regulators of dynamic changes in leaf shape (Figure 9). However, we were not able to assign a clear function to the candidate SPL and TCP binding sites in the AGO7 promoter, particularly because a 422 bp proximal promoter region lacking all these sites is sufficient for substantial transgenic complementation of leaf morphology defects in ago7 mutants (Figures 6 and 7, Supporting Information Figures S5 and S6).
Figure 8.
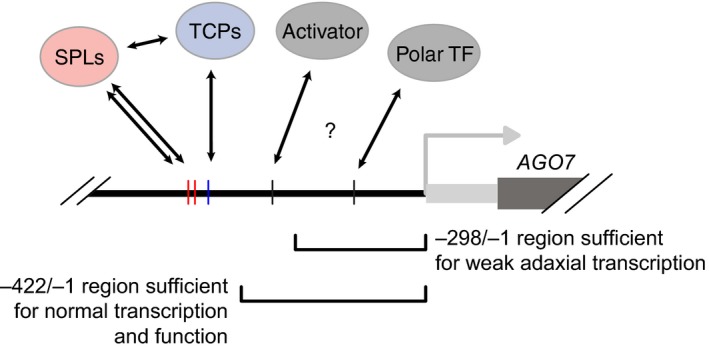
Schematic of the AGO7 proximal promoter region with hypothesized TF binding sites and summary results from transgenic analyses indicated. One or more activator and polarity determinant TFs are proposed to bind at undetermined sites in the regions indicated, as discussed in the text
Figure 9.

Revised model for control of heteroblasty in A. thaliana, incorporating the likely activation of AGO7 by SPLs. SPL levels gradually increase until they reach a hypothetical activity threshold, controlled in part by ARFs, and trigger changes in leaf characteristics (Telfer, Bollman, & Poethig, 1997, Hunter et al., 2006). When the AGO7 pathway is disrupted, ARF levels go up, lowering the threshold for transition such that it is reached earlier. This perturbation results in leaves that are thinner, longer, and more curled in ago7 mutants. Indirect repression of ARFs via AGO7 and TAS3 tasiRNAs may have significance for feedback control of SPL activity
Our truncation analysis provided preliminary evidence for two other functional regions of the AGO7 promoter (Figure 8). We obtained different outcomes for mutant plants tested with 422 bp promoter constructs (largely complemented) versus 298 bp promoter constructs (not complemented). This difference suggests that one or more functionally important binding sites are present in the ‐422/‐299 region. In general agreement with this idea, signal was qualitatively weaker for a 298 bp promoter:GUS reporter than for the next‐longest promoter fragment tested (Figure 4). Multiple experiments suggest that the minimal core promoter and possibly one or more polarizing cis elements are intact in the 298 bp proximal region, but dissecting this further has been technically challenging because of the faintness of the signal.
Despite progress, we did not succeed in identifying TF binding events necessary and/or sufficient for polar expression of AGO7 and AGO10. The YABBY1/FILAMENTOUS FLOWER gene is one promising candidate because of its well‐known role in polarity (Sarojam et al., 2010; Siegfried et al., 1999): as documented in Supporting Information Data S1, YAB1 is predicted to have high affinity for a site in the AGO7 proximal promoter but did not emerge as a hit from the Y1H screens. Surprisingly, we also did not recover the polarity factor REVOLUTA for the AGO10 promoter (Brandt et al., 2013); this likely represents a biological false negative. AGO1 is ubiquitously expressed (Lynn et al., 1999), and therefore expected to be under very robust transcriptional control which may be difficult to dissect. The Y1H results presented here should be a useful resource as further genome‐wide chromatin immunoprecipitation data for A. thaliana become available.
The truncation strategy used for our transgenic assays preserves the distance between cis elements, but also has inherent limitations. We did not test the possibility that SPL and TCP core binding sites are sufficient for specific genetic functions. The apparent enhancer(s) in the ‐422/‐299 region may be functionally redundant with these binding sites, and may therefore have masked any contributions to morphology through AGO7. Redundant clusters of activator binding sites are thought to be common, and may contribute to robustness (Frankel et al., 2010; Hong, Hendrix, & Levine, 2008; Levine, 2010; Perry, Boettiger, Bothma, & Levine, 2010). Effects may be larger in other tissues, given the important functions of ARF repressors in fruits and roots (Marin et al., 2010; Sessions & Zambryski, 1995; Simonini, Bencivenga, Trick, & Østergaard, 2017; Simonini et al., 2016; Su et al., 2017). Alternatively, the sites may simply be nonfunctional, at least in A. thaliana. Testing SPL and TCP binding in multiple tissues would help in assessing these possibilities; such data would aid evaluation of the possibility that the AGO7 promoter integrates both temporal and spatial signals. More broadly, linking such TF binding events to changes at the cellular level in diverse plants should remain a challenging but productive approach (Fouracre & Poethig, 2016; Skopelitis, Benkovics, Husbands, & Timmermans, 2017).
AUTHOR CONTRIBUTIONS
JSH, JLP‐P, GB, SAK, and JCC designed the research. JSH, JLP‐P, GB, MAH, EEH, HF, KMB, and JM performed research. JSH, JLP‐P, MAH, EEH, KMB, and JCC analyzed data. JSH and JCC drafted the paper; all authors commented on and approved the paper.
Supporting information
ACKNOWLEDGMENTS
We thank Danforth Center Plant Growth Facility staff members for excellent plant care, G. Nguyen and R. Allscheid for logistical support, and members of the Carrington lab for helpful discussions. D.H. Chitwood and J.G. Hodge provided essential advice on histological analysis. We thank T.C. Mockler and members of his lab (J. Gierer, D. O'Brien, M. Wiechert) for assistance with their plate readers and liquid‐handling equipment. This work was supported by US National Science Foundation award 1330562 (to JCC) and US National Institute of Health grants AI043288 (to JCC), GM056006 (to SAK and JLP‐P), and GM067837 (to SAK). JSH was supported by an NSF graduate research fellowship (award 1143954).
Hoyer JS, Pruneda‐Paz JL, Breton G, et al. Functional dissection of the ARGONAUTE7 promoter. Plant Direct. 2019;3:1–14. 10.1002/pld3.102
This manuscript was previously deposited as a preprint at bioRxiv.org: https://doi.org/10.1101/392910
REFERENCES
- Adenot, X. , Elmayan, T. , Lauressergues, D. , Boutet, S. , Bouché, N. , Gasciolli, V. , & Vaucheret, H. (2006). DRB4‐dependent TAS3 trans‐acting siRNAs control leaf morphology through AGO7. Current Biology, 16, 927–932. 10.1016/j.cub.2006.03.035 [DOI] [PubMed] [Google Scholar]
- Allen, E. , & Howell, M. D. (2010). miRNAs in the biogenesis of trans‐acting siRNAs in higher plants. Seminars in Cell & Developmental Biology, 21, 798–804. 10.1016/j.semcdb.2010.03.008 [DOI] [PubMed] [Google Scholar]
- Allen, E. , Xie, Z. , Gustafson, A. M. , & Carrington, J. C. (2005). microRNA‐directed phasing during trans‐acting siRNA biogenesis in plants. Cell, 121, 207–221. 10.1016/j.cell.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Axtell, M. J. , Jan, C. , Rajagopalan, R. , & Bartel, D. P. (2006). A two‐hit trigger for siRNA biogenesis in plants. Cell, 127, 565–577. 10.1016/j.cell.2006.09.032 [DOI] [PubMed] [Google Scholar]
- Bailey, T. L. , Boden, M. , Buske, F. A. , Frith, M. , Grant, C. E. , Clementi, L. , … Noble, W. S. (2009). MEME Suite: Tools for motif discovery and searching. Nucleic Acids Research, 37, W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger, N. , & Baulcombe, D. C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proceedings of the National Academy of Sciences United States of America, 102, 11928–11933. 10.1073/pnas.0505461102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl, R. P. , Jach, G. , Saedler, H. , & Huijser, P. (2005). Functional dissection of the plant‐specific SBP‐domain: Overlap of the DNA‐binding and nuclear localization domains. Journal of Molecular Biology, 352, 585–596. 10.1016/j.jmb.2005.07.013 [DOI] [PubMed] [Google Scholar]
- Bohmert, K. , Camus, I. , Bellini, C. , Bouchez, D. , Caboche, M. , & Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO Journal, 17, 170–180. 10.1093/emboj/17.1.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies, K. (2002). Whole mount GUS staining In Weigel D., & Glazebrook J. (Eds.), Arabidopsis: A laboratory manual (pp. 243–245). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Bonaldi, K. , Li, Z. , Kang, S. E. , Breton, G. , & Pruneda‐Paz, J. L. (2017). Novel cell surface luciferase reporter for high‐throughput yeast one‐hybrid screens. Nucleic Acids Research, 45(18), e157 10.1093/nar/gkx682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt, R. , Xie, Y. , Musielak, T. , Graeff, M. , Stierhof, Y. D. , Huang, H. , … Wenkel, S. (2013). Control of stem cell homeostasis via interlocking microRNA and microProtein feedback loops. Mechanisms of Development, 130, 25–33. 10.1016/j.mod.2012.06.007 [DOI] [PubMed] [Google Scholar]
- Carbonell, A. , Fahlgren, N. , Garcia‐Ruiz, H. , Gilbert, K. B. , Montgomery, T. A. , Nguyen, T. , … Carrington, J. C. (2012). Functional analysis of three Arabidopsis ARGONAUTES using slicer‐defective mutants. The Plant Cell, 24, 3613–3629. 10.1105/tpc.112.099945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood, D. H. , Nogueira, F. T. , Howell, M. D. , Montgomery, T. A. , Carrington, J. C. , & Timmermans, M. C. (2009). Pattern formation via small RNA mobility. Genes & Development, 23, 549–554. 10.1101/gad.1770009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S. J. , & Bent, A. F. (1998). Floral dip: A simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . The Plant Journal, 16, 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cuperus, J. T. , Fahlgren, N. , & Carrington, J. C. (2011). Evolution and functional diversification of MIRNA genes. The Plant Cell, 23, 431–442. 10.1105/tpc.110.082784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M. D. , & Grossniklaus, U. (2003). A Gateway cloning vector set for high‐throughput functional analysis of genes in planta . Plant Physiology, 133, 462–469. 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto, M. C. , Petsch, K. A. , Aukerman, M. J. , Beatty, M. , Hammell, M. , & Timmermans, M. C. (2014). Genome‐wide analysis of leafbladeless1‐regulated and phased small RNAs underscores the importance of the TAS3 ta‐siRNA pathway to maize development. PLoS Genetics, 10, e1004826 10.1371/journal.pgen.1004826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, R. N. , Wiley, D. , Sarkar, A. , Springer, N. , Timmermans, M. C. , & Scanlon, M. J. (2010). ragged seedling2 encodes an ARGONAUTE7‐like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. The Plant Cell, 22, 1441–1451. 10.1105/tpc.109.071613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N. , Montgomery, T. A. , Howell, M. D. , Allen, E. , Dvorak, S. K. , Alexander, A. L. , & Carrington, J. C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta‐siRNA affects developmental timing and patterning in Arabidopsis . Current Biology, 16, 939–944. 10.1016/j.cub.2006.03.065 [DOI] [PubMed] [Google Scholar]
- Fouracre, J. P. , & Poethig, R. S. (2016). The role of small RNAs in vegetative shoot development. Current Opinion in Plant Biology, 29, 64–72. 10.1016/j.pbi.2015.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco‐Zorrilla, J. M. , López‐Vidriero, I. , Carrasco, J. L. , Godoy, M. , Vera, P. , & Solano, R. (2014). DNA‐binding specificities of plant transcription factors and their potential to define target genes. Proceedings of the National Academy of Sciences United States of America, 111, 2367–2372. 10.1073/pnas.1316278111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel, N. , Davis, G. K. , Vargas, D. , Wang, S. , Payre, F. , & Stern, D. L. (2010). Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature, 466, 490–493. 10.1038/nature09158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D. , Collier, S. A. , Byrne, M. E. , & Martienssen, R. A. (2006). Specification of leaf polarity in Arabidopsis via the trans‐acting siRNA pathway. Current Biology, 16, 933–938. 10.1016/j.cub.2006.03.064 [DOI] [PubMed] [Google Scholar]
- Gentleman, R. C. , Carey, V. J. , Bates, D. M. , Bolstad, B. , Dettling, M. , Dudoit, S. , … Zhang, J. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biology, 5, R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, C. E. , Bailey, T. L. , & Noble, W. S. (2011). FIMO: Scanning for occurrences of a given motif. Bioinformatics, 27, 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer, A. , Nusinow, D. A. , Chow, B. Y. , Gehrke, A. R. , Bulyk, M. L. , & Kay, S. A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Current Biology, 21, 126–133. 10.1016/j.cub.2010.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters, M. , Silva, B. , Van Vliet, F. , Genetello, C. , De Block, M. , Dhaese, P. , … Schell, J. (1980). The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid, 3, 212–230. 10.1016/0147-619X(80)90110-9 [DOI] [PubMed] [Google Scholar]
- Hong, J. W. , Hendrix, D. A. , & Levine, M. S. (2008). Shadow enhancers as a source of evolutionary novelty. Science, 321, 1314 10.1126/science.1160631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, C. , Sun, H. , & Poethig, R. S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Current Biology, 13, 1734–1739. 10.1016/j.cub.2003.09.004 [DOI] [PubMed] [Google Scholar]
- Hunter, C. , Willmann, M. R. , Wu, G. , Yoshikawa, M. , de la Luz Gutiérrez‐Nava, M. , & Poethig, S. R. (2006). Trans‐acting siRNA‐mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis . Development, 133, 2973–2981. 10.1242/dev.02491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama, T. , Mitsuda, N. , Seki, M. , Shinozaki, K. , & Ohme‐Takagi, M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis . The Plant Cell, 22, 3574–3588. 10.1105/tpc.110.075598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R. C. , Feinbaum, R. L. , & Ambros, V. (1993). The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14 . Cell, 75, 843–854. 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Levine, M. (2010). Transcriptional enhancers in animal development and evolution. Current Biology, 20, R754–R763. 10.1016/j.cub.2010.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Z. , Yu, H. , Xiong, G. , Wang, J. , Jiao, Y. , Liu, G. , … Li, J. (2013). Genome‐wide binding analysis of the transcription activator IDEAL PLANT ARCHITECTURE1 reveals a complex network regulating rice plant architecture. The Plant Cell, 25, 3743–3759. 10.1105/tpc.113.113639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn, K. , Fernandez, A. , Aida, M. , Sedbrook, J. , Tasaka, M. , Masson, P. , & Barton, M. K. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development, 126, 469–481. [DOI] [PubMed] [Google Scholar]
- Maloof, J. N. , Nozue, K. , Mumbach, M. R. , & Palmer, C. M. (2013). LeafJ: An ImageJ plugin for semi‐automated leaf shape measurement. Journal of Visualized Experiments, 71, e50028. 10.3791/50028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin, E. , Jouannet, V. , Herz, A. , Lokerse, A. S. , Weijers, D. , Vaucheret, H. , … Maizel, A. (2010). miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. The Plant Cell, 22, 1104–1117. 10.1105/tpc.109.072553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, T. A. , Howell, M. D. , Cuperus, J. T. , Li, D. , Hansen, J. E. , Alexander, A. L. , … Carrington, J. C. (2008). Specificity of ARGONAUTE7‐miR390 interaction and dual functionality in TAS3 trans‐acting siRNA formation. Cell, 133, 128–141. 10.1016/j.cell.2008.02.033 [DOI] [PubMed] [Google Scholar]
- Nath, U. , Crawford, B. C. , Carpenter, R. , & Coen, E. (2003). Genetic control of surface curvature. Science, 299, 1404–1407. 10.1126/science.1079354 [DOI] [PubMed] [Google Scholar]
- Nogueira, F. T. , Madi, S. , Chitwood, D. H. , Juarez, M. T. , & Timmermans, M. C. (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes & Development, 21, 750–755. 10.1101/gad.1528607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley, R. C. , Huang, S. C. , Song, L. , Lewsey, M. G. , Bartlett, A. , Nery, J. R. , … Ecker, J. R. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell, 166, 1598 10.1016/j.cell.2016.08.063 [DOI] [PubMed] [Google Scholar]
- Pagès, H. , Aboyoun, P. , Gentleman, R. , & DebRoy, S. (2017). Biostrings: Efficient manipulation of biological strings.
- Palatnik, J. F. , Allen, E. , Wu, X. , Schommer, C. , Schwab, R. , Carrington, J. C. , & Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature, 425, 257–263. 10.1038/nature01958 [DOI] [PubMed] [Google Scholar]
- Peragine, A. , Yoshikawa, M. , Wu, G. , Albrecht, H. L. , & Poethig, R. S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans‐acting siRNAs in Arabidopsis . Genes & Development, 18, 2368–2379. 10.1101/gad.1231804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, M. W. , Boettiger, A. N. , Bothma, J. P. , & Levine, M. (2010). Shadow enhancers foster robustness of Drosophila gastrulation. Current Biology, 20, 1562–1567. 10.1016/j.cub.2010.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plavskin, Y. , Nagashima, A. , Perroud, P. F. , Hasebe, M. , Quatrano, R. S. , Atwal, G. S. , & Timmermans, M. C. (2016). Ancient trans‐acting siRNAs confer robustness and sensitivity onto the auxin response. Developmental Cell, 36, 276–289. 10.1016/j.devcel.2016.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig, R. S. (2013). Vegetative phase change and shoot maturation in plants. Current Topics in Developmental Biology, 105, 125–152. 10.1016/B978-0-12-396968-2.00005-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda‐Paz, J. L. , Breton, G. , Nagel, D. H. , Kang, S. E. , Bonaldi, K. , Doherty, C. J. , … Kay, S. A. (2014). A genome‐scale resource for the functional characterization of Arabidopsis transcription factors. Cell Reports, 8, 622–632. 10.1016/j.celrep.2014.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda‐Paz, J. L. , Breton, G. , Para, A. , & Kay, S. A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science, 323, 1481–1485. 10.1126/science.1167206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2017). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rubio‐Somoza, I. , & Weigel, D. (2011). MicroRNA networks and developmental plasticity in plants. Trends in Plant Science, 16, 258–264. 10.1016/j.tplants.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Rubio‐Somoza, I. , Zhou, C. M. , Confraria, A. , Martinho, C. , von Born, P. , Baena‐Gonzalez, E. , … Weigel, D. (2014). Temporal control of leaf complexity by miRNA‐regulated licensing of protein complexes. Current Biology, 24, 2714–2719. 10.1016/j.cub.2014.09.058 [DOI] [PubMed] [Google Scholar]
- Sarojam, R. , Sappl, P. G. , Goldshmidt, A. , Efroni, I. , Floyd, S. K. , Eshed, Y. , & Bowman, J. L. (2010). Differentiating Arabidopsis shoots from leaves by combined YABBY activities. The Plant Cell, 22, 2113–2130. 10.1105/tpc.110.075853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer, C. , Palatnik, J. F. , Aggarwal, P. , Chételat, A. , Cubas, P. , Farmer, E. E. , … Weigel, D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biology, 6, e230 10.1371/journal.pbio.0060230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab, R. , Maizel, A. , Ruiz‐Ferrer, V. , Garcia, D. , Bayer, M. , Crespi, M. , … Martienssen, R. A. (2009). Endogenous tasiRNAs mediate non‐cell autonomous effects on gene regulation in Arabidopsis thaliana. PLoS ONE, 4, e5980 10.1371/journal.pone.0005980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, R. A. , & Zambryski, P. C. (1995). Arabidopsis gynoecium structure in the wild and in ettin mutants. Development, 121, 1519–1532. [DOI] [PubMed] [Google Scholar]
- Siegfried, K. R. , Eshed, Y. , Baum, S. F. , Otsuga, D. , Drews, G. N. , & Bowman, J. L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis . Development, 126, 4117–4128. [DOI] [PubMed] [Google Scholar]
- Simonini, S. , Bencivenga, S. , Trick, M. , & Østergaard, L. (2017). Auxin‐induced modulation of ETTIN activity orchestrates gene expression in Arabidopsis . The Plant Cell, 29(8), 1864–1882. 10.1105/tpc.17.00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonini, S. , Deb, J. , Moubayidin, L. , Stephenson, P. , Valluru, M. , Freire‐Rios, A. , … Østergaard, L. (2016). A noncanonical auxin‐sensing mechanism is required for organ morphogenesis in Arabidopsis . Genes & Development, 30, 2286–2296. 10.1101/gad.285361.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelitis, D. S. , Benkovics, A. H. , Husbands, A. Y. , & Timmermans, M. (2017). Boundary formation through a direct threshold‐based readout of mobile small RNA gradients. Developmental Cell, 43, 265–273. e6. 10.1016/j.devcel.2017.10.003 [DOI] [PubMed] [Google Scholar]
- Skopelitis, D. S. , Husbands, A. Y. , & Timmermans, M. C. (2012). Plant small RNAs as morphogens. Current Opinion in Cell Biology, 24, 217–224. 10.1016/j.ceb.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Strader, L. C. , Wheeler, D. L. , Christensen, S. E. , Berens, J. C. , Cohen, J. D. , Rampey, R. A. , & Bartel, B. (2011). Multiple facets of Arabidopsis seedling development require indole‐3‐butyric acid‐derived auxin. The Plant Cell, 23, 984–999. 10.1105/tpc.111.083071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer, A. J. , Wang, H. , & Doebley, J. F. (2017). Selection during maize domestication targeted a gene network controlling plant and inflorescence architecture. Genetics, 207, 755–765. 10.1534/genetics.117.300071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Z. , Zhao, L. , Zhao, Y. , Li, S. , Won, S. , Cai, H. , … Chen, X. (2017). The THO complex non‐cell‐autonomously represses female germline specification through the TAS3‐ARF3 module. Current Biology, 27, 1597–1609. e2. 10.1016/j.cub.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer, A. , Bollman, K. M. , & Poethig, R. S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana . Development, 124, 645–654. [DOI] [PubMed] [Google Scholar]
- Tovar, J. C. , Hoyer, J. S. , Lin, A. , Tielking, A. , Callen, S. T. , Castillo, E. S. , … Gehan, M. A. (2018). Raspberry Pi‐powered imaging for plant phenotyping. Applications in Plant Sciences, 6, e1031 10.1002/aps3.1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, M. R. , Hinze, A. , Tucker, E. J. , Takada, S. , Jürgens, G. , & Laux, T. (2008). Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development, 135, 2839–2843. 10.1242/dev.023648 [DOI] [PubMed] [Google Scholar]
- Walhout, A. J. (2011). What does biologically meaningful mean? A perspective on gene regulatory network validation. Genome Biology, 12, 109 10.1186/gb-2011-12-4-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch, M. T. , Yang, A. , Albu, M. , Cote, A. G. , Montenegro‐Montero, A. , Drewe, P. , … Hughes, T. R. (2014). Determination and inference of eukaryotic transcription factor sequence specificity. Cell, 158, 1431–1443. 10.1016/j.cell.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman, B. , Ha, I. , & Ruvkun, G. (1993). Posttranscriptional regulation of the heterochronic gene lin‐14 by lin‐4 mediates temporal pattern formation in C. elegans . Cell, 75, 855–862. 10.1038/338313a0 [DOI] [PubMed] [Google Scholar]
- Williams, L. , Carles, C. C. , Osmont, K. S. , & Fletcher, J. C. (2005). A database analysis method identifies an endogenous trans‐acting short‐interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proceedings of the National Academy of Sciences United States of America, 102, 9703–9708. 10.1073/pnas.0504029102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J. , Cai, X. , Luo, J. , Sato, S. , Jiang, Q. , Yang, J. , … Luo, D. (2010). The REDUCED LEAFLET genes encode key components of the trans‐acting small interfering RNA pathway and regulate compound leaf and flower development in Lotus japonicus . Plant Physiology, 152, 797–807. 10.1104/pp.109.140947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yifhar, T. , Pekker, I. , Peled, D. , Friedlander, G. , Pistunov, A. , Sabban, M. , … Eshed, Y. (2012). Failure of the tomato trans‐acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. The Plant Cell, 24, 3575–3589. 10.1105/tpc.112.100222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa, M. , Peragine, A. , Park, M. Y. , & Poethig, R. S. (2005). A pathway for the biogenesis of trans‐acting siRNAs in Arabidopsis . Genes & Development, 19, 2164–2175. 10.1101/gad.1352605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Xia, R. , Meyers, B. C. , & Walbot, V. (2015). Evolution, functions, and mysteries of plant ARGONAUTE proteins. Current Opinion in Plant Biology, 27, 84–90. 10.1016/j.pbi.2015.06.011 [DOI] [PubMed] [Google Scholar]
- Zhou, C. , Han, L. , Fu, C. , Wen, J. , Cheng, X. , Nakashima, J. , … Wang, Z. Y. (2013). The trans‐acting short interfering RNA3 pathway and NO APICAL MERISTEM antagonistically regulate leaf margin development and lateral organ separation, as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula . The Plant Cell, 25, 4845–4862. 10.1105/tpc.113.117788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H. , Hu, F. , Wang, R. , Zhou, X. , Sze, S. H. , Liou, L. W. , … Zhang, X. (2011). Arabidopsis ARGONAUTE10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell, 145, 242–256. 10.1016/j.cell.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and software code supporting this manuscript have been deposited as supplemental datasets:
Hoyer, S. (2018). jshoyer/y1h‐AGO7‐promoter: Yeast 1‐hybrid screens for upstream regulators of A. thaliana AGO1, AGO7, and AGO10: raw data and R code. Retrieved from https://doi.org/10.5281/zenodo.1472704
Hoyer, J.S. (2018). Yeast 1‐hybrid screens for upstream regulators of A. thaliana AGO1, AGO7, and AGO10: ranked tables of candidate direct upstream TFs. Retrieved from https://doi.org/10.5281/zenodo.1472235
Hoyer, J.S., Holcolm, E. E. (2018). Photomicrographs: transverse sections of GUS‐stained apices for AGO7 promoter analysis. Retrieved from https://doi.org/10.5281/zenodo.1319761
Hoyer, J.S. (2018). Scans of leaves dissected in phyllotactic order: complementation of ago7 mutant A. thaliana plants with truncated promoter transgenes. Retrieved from https://doi.org/10.5281/zenodo.1322799
Hoyer, S. (2018). jshoyer/raspi‐photo‐and‐leaf‐scan‐metadata: Metadata and code for A. thaliana leaf scans and top‐down time‐course photos. Retrieved from https://doi.org/10.5281/zenodo.1472768
Data processing was done with the R Statistical Computing Environment (R Core Team 2017) and the Bioconductor BioStrings package was used for PWM scans (Gentleman et al., 2004; Pagès, Aboyoun, Gentleman, & DebRoy, 2017). Supporting Information Data S1 also include results from “Find Individual Occurences of Motifs” tool (FIMO) scans (Grant et al., 2011) done via the online MEME Suite (Bailey et al., 2009) version 4.12.0, with default settings (p < 10−4 cutoff) and three collections of DNA‐binding specificity models (Franco‐Zorrilla et al., 2014; O'Malley et al., 2016; Weirauch et al., 2014).


