Abstract
Despite the importance of proper cell death regulation across broad evolutionary distances, an understanding of the molecular machinery underpinning this fundamental process in plants remains largely elusive. This is despite its critical importance to development, homeostasis, and proper responses to stress. The identification of endogenous plant regulators of cell death has been hindered by the fact that many core regulators of cell death in animals are absent in plant genomes. Remarkably, numerous studies have shown that the ectopic expression of animal prosurvival genes in plants can suppress cell death imposed by many stresses. In this study, we capitalize on the ectopic expression of one of these animal prosurvival genes, an inhibitor of apoptosis from Spodoptera frugiperda (SfIAP), to identify novel cell death regulators in plants. A yeast two‐hybrid assay was conducted using SfIAP as bait to screen a tomato cDNA library. This screen identified several transcription factors of the SQUAMOSA promoter‐binding protein (SBP) family as potential SfIAP binding partners. We confirmed this interaction in vivo for our top two interactors, SlySBP8b and SlySBP12a, using coimmunoprecipitation. Interestingly, overexpression of SlySBP8b and SlySBP12a induced cell death in Nicotiana benthamiana leaves. Overexpression of these two transcription factors also induced the accumulation of reactive oxygen species and enhanced the growth of the necrotrophic pathogen Alternaria alternata. Fluorescence microscopy confirmed the nuclear localization of both SlySBP8b and SlySBP12a, while SlySBP12a was also localized to the ER membrane. These results suggest a prodeath role for SlySBP8b and SlySBP12a and implicate ER membrane tethering as a means of regulating SlySBP12a activity.
Keywords: cell death, fumonisin B1, inhibitor of apoptosis, necrotrophic, SQUAMOSA promoter‐binding protein‐like, SQUAMOSA promoter‐binding protein
1. INTRODUCTION
Proper cell death regulation is a fundamental aspect of development and stress response that is conserved throughout all kingdoms of life (Allocati, Masulli, Di Ilio, & De Laurenzi, 2015). This process of genetically regulated cellular suicide is referred to as programmed cell death (PCD). Programmed cell death has been studied extensively in animal systems, and the results of these research efforts have led to major treatment advances for many human diseases (Fuchs & Steller, 2011). In contrast, our understanding of the biochemical pathways underlying PCD in plants is severely lacking. This is largely due to the absence of obvious orthologs of core regulators of apoptosis, a well‐studied form of PCD in animals (Kabbage, Kessens, Bartholomay, & Williams, 2017). While this has undoubtedly slowed progress on plant PCD research, it has also presented a unique opportunity for the discovery of novel cell death regulators in plant systems.
Apoptosis is a specific type of PCD characterized by distinct morphological and biochemical features (Kroemer et al., 2009). Apoptotic cell death in animals is executed through the activation of cysteine‐dependent aspartate‐specific proteases termed caspases. Caspases exist as inactive proenzymes that can be activated by external or internal cellular cues. Once activated, caspases execute an orderly demise of the cell by targeting negative regulators of apoptosis, cytoskeletal components, and other caspases (Parrish, Freel, & Kornbluth, 2013). Due to the terminal nature of apoptosis, caspases must be kept under multiple layers of regulation. The inhibitor of apoptosis (IAP) family is an important group of proteins that negatively regulate caspase activity. The defining feature of all IAPs is the presence of one or more baculovirus IAP repeat (BIR) domains, which are used by IAP proteins for substrate binding (Verhagen, Coulson, & Vaux, 2001). Additionally, some IAPs contain a really interesting new gene (RING) domain that serves as a functional E3 ubiquitin ligase domain. Inhibitor of apoptosis proteins can inhibit caspase activity by preventing procaspases from becoming active or by suppressing active caspases. This can be accomplished by simply blocking the active site pocket of a caspase or by utilizing the RING domain to ubiquitinate a caspase and mark it for proteasome‐mediated degradation (Feltham, Khan, & Silke, 2012; Gyrd‐Hansen & Meier, 2010).
Despite the fact that obvious orthologs of IAPs and caspases are absent in plant genomes, the ectopic expression of animal and viral apoptotic regulators in tobacco (Nicotiana spp.) and tomato (Solanum lycopersicum) modulate plant cell death. This was first reported nearly two decades ago when the expression of Bax, a mammalian proapoptotic gene absent in plant genomes, induced localized tissue collapse and cell death in Nicotiana benthamiana (Lacomme & Santa, 1999). Shortly thereafter, Dickman et al. (2001) demonstrated that expression of a viral IAP (OpIAP), as well as anti‐apoptotic members of the Bcl‐2 family, conferred resistance to a suite of necrotrophic fungal pathogens in Nicotiana tabacum. Pathogens with a necrotrophic lifestyle require dead host tissue for nutrient acquisition, and studies on Cochliobolus victoriae, Sclerotinia sclerotiorum, and Fusarium spp. revealed that these necrotrophic fungal pathogens hijack host cell death machinery to kill cells (Asai et al., 2000; Glenn et al., 2008; Kabbage, Williams, & Dickman, 2013; Lorang et al., 2012; Williams, Kabbage, Kim, Britt, & Dickman, 2011).
More recently, we showed that overexpression of an IAP from Spodoptera frugiperda (fall armyworm; SfIAP) in tobacco and tomato prevented cell death associated with a wide range of abiotic and biotic stresses (Kabbage, Li, Chen, & Dickman, 2010; Li, Kabbage, & Dickman, 2010). Tobacco and tomato lines expressing SfIAP had increased heat and salt stress tolerance, two abiotic stresses that induce cell death. These transgenic lines were also resistant to the fungal necrotroph Alternaria alternata and the mycotoxin fumonisin B1 (FB1) (Li et al., 2010). Fumonisin B1 is produced by some species of Fusarium and is a potent inducer of apoptosis in animal cells and apoptotic‐like PCD in plant cells (Gilchrist, 1997).
It has been over 15 years since it was first reported that overexpression of animal anti‐apoptotic regulators in plants conferred enhanced resistance against a wide assortment of necrotrophic pathogens. During this time, numerous studies have confirmed the efficacy of animal apoptotic regulators in plants without identifying the means by which these regulators function. In this study, we used an unbiased approach to identify in planta binding partners of SfIAP in tomato to better understand how this insect IAP is able to inhibit cell death and confer stress tolerance in plants. Yeast two‐hybrid and coimmunoprecipitation (CoIP) assays show that SfIAP interacts with members of the SQUAMOSA promoter‐binding protein (abbreviated SBP in tomato or SQUAMOSA promoter‐binding protein‐like in some other species) transcription factor family. Overexpression of two tomato SBPs, SlySBP8b and SlySBP12a, induced cell death in tobacco leaves accompanied by enhanced production of reactive oxygen species (ROS). Overexpression of SlySBP8b and SlySBP12a also created an environment that was more conducive to the growth of the necrotrophic fungal pathogen A. alternata. In summary, our findings uncover SlySBP8b and SlySBP12a as novel SfIAP binding partners that exhibit prodeath attributes.
2. MATERIALS AND METHODS
2.1. Plant material and growth conditions
Nicotiana benthamiana plants were grown on a 16‐hr light cycle (~50 microeinsteins m−2 s−1) at 26°C and ~60% humidity. Nicotiana glutinosa (PI 555510) and tomato (Solanum lycopersicum cv. Bonny Best) plants were grown on a 16‐hr light cycle (~100 microeinsteins m−2 s−2) at 22°C and ~60% humidity. The soil composition for all plants consisted of SunGro® propagation mix and Sunshine® coarse vermiculite in a 3:1 ratio. Plants were watered with deionized water supplemented with Miracle‐Gro® all‐purpose fertilizer (1 g/L) as needed.
2.2. Plasmid construction
The full‐length open reading frames of SlySBP‐like (Solyc07g062980), SlySBP4 (Solyc07g053810), SlySBP6a (Solyc03g114850), SlySBP6c (Solyc12g038520), SlySBP8b (Solyc01g090730), and SlySBP12a (Solyc01g068100) were amplified by PCR from cDNA collected from tomato inflorescence tissue (Supporting information Table S1). AttB1 and attB2 adapters were added to forward and reverse primers, respectively, to generate attB‐flanked amplicons suitable for Gateway™ Recombination Cloning (Invitrogen).
Amplicons were recombined into the entry vector pDONR™/Zeo using BP clonase II (Invitrogen). SlySBP8b(NLS mt ) and SlySBP12a(NLS mt ) constructs were generated using the Q5® Site‐Directed Mutagenesis Kit (New England Biolabs). SlySBP12a(ΔTMD) and TMD SlySBP12a were amplified from SlySBP12a in pDONR™/Zeo using the primers indicated in Supporting information Table S1 and recombined into pDONR™/Zeo. For overexpression in N. benthamiana leaves and tomato protoplasts, entry vectors were mixed with the desired pEarleyGate destination vectors (Earley et al., 2006) and recombined using LR clonase II (Invitrogen). pEarleyGate vectors drive transgene expression using a cauliflower mosaic virus 35S (35S) promoter and were used to generate N‐terminal yellow fluorescent protein (YFP; pEarleyGate104) or N‐terminal influenza hemagglutinin (HA; pEarleyGate201) fusions. All constructs were verified using Sangar sequencing before being transformed into Agrobacterium tumefaciens GV3101.
Plasmids for the yeast two‐hybrid screen were prepared as follows. SfIAP, SfIAP BIR1, and luciferase cDNAs were cloned into the bait vector pGilda under control of the GAL1 promoter and in‐frame with an N‐terminal fusion of the E. coli LexA DNA binding protein (Takara Bio USA, Inc.). Luciferase (firefly luciferase from Photinus pyralis) was cut from an existing plasmid using a 5′‐Nco1 restriction site in the START codon and a 3′‐Not1 restriction site outside of the ORF and ligated into pGilda. Primers for SfIAP (GenBank: AF186378.1) and SfIAP BIR1 amplification were designed to place an EcoR1 site at the 5′ end and a BamH1 site at the 3′ end of the ORF. Primers used for amplification can be found in Supporting information Table S1. Amplicons were cut using these restriction enzymes and ligated into pGilda. Tomato cDNAs were expressed from the GAL1 promoter with an N‐terminal fusion of the B42 activation protein in the pB42AD plasmid (Takara Bio USA, Inc.). Bait and prey library were sequentially transformed into EGY48 yeast using standard protocols.
2.3. Yeast two‐hybrid screening
Yeast containing bait and plasmid were plated on SD galactose (‐His/‐Trp/‐Leu) to induce gene expression and select for bait–prey interactions. After incubating at 28°C for ~5 days, colonies were pooled in 10 ml of sorbitol/phosphate buffer (1.2 M sorbitol, 0.1 M NaPO4, pH 7.5) per plate, pelleted, and resuspended in 2 ml of sorbitol/phosphate buffer supplemented with 500 U of lyticase (Sigma: L2524‐25KU) and 250 μg of RNase A. Yeast cells were incubated in the lyticase buffer for 3 hr at 37°C prior to plasmid recovery. Plasmid DNA was extracted using a Wizard Plus SV Miniprep kit (Promega) and a modified protocol. Briefly, 2.5 ml of lysis solution and 80 μl of alkaline protease solution were added to yeast protoplasts and incubated at room temperature for 10 min. Next, 3.5 ml of neutralization solution was added and cellular debris was pelleted by centrifugation. Supernatant was run through the provided columns and plasmid DNA eluted according to the manufacturer's instructions. Low‐cycle PCR was performed to amplify cDNA's from the prey library. Briefly, MyFi™ proofreading DNA polymerase (Bioline) and pB42AD forward and reverse primers (flanking the cDNA insertion site of pB42AD) were used to amplify cDNA's (Supporting information Table S1). A QIAquick PCR purification kit (Qiagen) was used to clean PCR products before sequencing.
2.4. Illumina sequencing and data analysis
Sequencing was performed by the Biotechnology Center at UW‐Madison using Illumina next‐generation sequencing with 100‐bp paired‐end reads. The sequencing data were uploaded to the Galaxy web platform, and we used the public server at usegalaxy.org to analyze the data (Afgan et al., 2016). Reads were groomed and trimmed to remove low‐quality bases and adapter sequences before alignment (Bolger, Lohse, & Usadel, 2014). Bait (pGilda) and prey (pB42AD) plasmid sequences were concatenated with the Saccharomyces cerevisiae reference genome (S288C_reference_sequence_R64‐2‐1_20150113) to create a FASTA file containing sources of plasmid and gDNA contamination. Reads were aligned to this file using Bowtie 2 (Langmead & Salzberg, 2012). Aligned reads (plasmid and gDNA) were discarded while unaligned reads were aligned to the tomato reference genome (Solgenomics: ITAG2.4) with Bowtie 2. Cufflinks (v2.2.1) was used to assemble transcripts from these aligned reads and calculate FPKM values for each locus (Trapnell et al., 2012). Enrichment scores for each locus were calculated using R Studio and scripts written in‐house (RStudio Team, 2016). Details of Galaxy pipeline, in‐house scripts, and complete dataset are available upon request.
2.5. Transient expression in N. benthamiana and N. glutinosa
Agrobacterium strain GV3101 was grown overnight in liquid LB supplemented with gentamycin and kanamycin (50 μg/ml) at 28°C with shaking. Cells were harvested by centrifugation, washed once with sterile deionized water, and resuspended in infiltration medium (10 mM MgSO4, 9 mM MES, 10 mM MgCl2, 300 μM acetosyringone, pH 5.7) to a final concentration of OD600 = 0.9. Cultures were incubated at room temperature for 4 hr before infiltration. Nicotiana benthamiana plants were infiltrated with a 1‐ml needleless syringe at 4–5 weeks of age with the two youngest and easily infiltratable leaves being used. Nicotiana glutinosa plants were infiltrated at 5–6 weeks of age with a single leaf being used on each plant, typically corresponding to the 4th or 5th true leaf. Plants were transformed at different ages due to differences in rate of growth between the two species.
For total protein extraction, leaf tissue was frozen in liquid nitrogen and ground in 3× Laemmli buffer (10% β‐mercaptoethanol). Samples were boiled for 10 min followed by centrifugation at 10,000 g for 5 min. Supernatants were removed and transferred to new tubes. Total proteins were separated by electrophoresis on a 12% Tris‐Glycine‐SDS polyacrylamide gel (Bio‐Rad). Proteins were transferred to a nitrocellulose membrane. Total protein was detected using Ponceau S stain. Epitope‐tagged proteins were detected by probing with α‐GFP (Cell Signaling 2955S) or α‐HA (Cell Signaling 3724S) primary antibodies. The α‐GFP antibody was detected using goat α‐mouse IgG conjugated to horseradish peroxidase (HRP) (Cell Signaling 7076P2) while the α‐HA antibody was detected using goat α‐rabbit IgG conjugated to HRP (Cell Signaling 7074P2). Amersham™ ECL™ reagent (GE Life Sciences) was used to detect HRP‐conjugated antibodies.
2.6. Transient transfection of tomato protoplasts
Mesophyll protoplasts form tomato cotyledons were isolated from 10‐day‐old plants using the Tape Sandwich method (Wu et al., 2009). A total of 6 μg of plasmid was used for each transfection with an equal ratio used for cotransfections. Transfections were performed using polyethylene glycol (PEG) as described previously (Yoo, Cho, & Sheen, 2007). Protoplasts were used for imaging the day after transfection.
2.7. Coimmunoprecipitation assays
Agrobacterium strains harboring free 35S:YFP or 35S:YFP‐SfIAP M4 (I332A) were coinfiltrated with strains harboring 35S:HA‐SlySBP8b or 35S:HA‐SlySBP12a. A 7:2 ratio of YFP strains to HA strains was used due to relatively low accumulation of YFP‐SfIAPM4(I332A) protein compared to HA‐SlySBP8b and HA‐SlySBP12a. Approximately 40 hr post‐agroinfiltration, transformed leaves were collected and ground in liquid nitrogen to a fine powder. Extraction buffer (50 mM Tris‐HCl, 150 mM NaCl, 5 mM EDTA, 10% glycerol, 0.2% IGEPAL, and 1% plant protease inhibitor cocktail [Sigma]) was added at a concentration of 2 ml/g of leaf tissue. YFP‐tagged proteins were immunoprecipitated by incubating the lysate with α‐GFP magnetic agarose beads (GFP‐Trap_MA; Chromotek) for 2 hr at 4°C. Beads were washed three times in extraction buffer (w/o IGEPAL) and boiled in 30 μl of 2× SDS loading buffer before loading on duplicate 12% Tris‐Glycine‐SDS polyacrylamide gels (Bio‐Rad). Proteins were transferred to duplicate nitrocellulose membranes and probed with α‐GFP (Cell Signaling 2955S) or α‐HA (Cell Signaling 3724S) primary antibodies. The α‐GFP antibody was detected using goat α‐mouse IgG conjugated to HRP (Cell Signaling 7076P2) while the α‐HA antibody was detected using goat α‐rabbit IgG conjugated to HRP (Cell Signaling 7074P2). Amersham™ ECL™ reagent (GE Life Sciences) was used to detect HRP‐conjugated antibodies.
2.8. Confocal laser scanning microscopy
Confocal laser scanning microscopy was performed on a Zeiss ELYRA LSM780 inverted confocal microscope using a 40×, 1.1‐numerical aperture, water objective. Protoplasts were stained with DHE at a concentration of 5 μM by adding DHE directly to protoplast solution and imaging approximately 10 min after DHE addition. YFP fusions, chlorophyll autofluorescence, and DHE were excited with a 488 nm argon laser. YFP emission was detected between 502 and 542 nm, chlorophyll emission was detected between 657 and 724 nm, and DHE was detected between 606 and 659 nm. mCherry was excited with a 561 nm He–Ne laser, and emission was detected between 606 and 651 nm. Colocalization analysis was performed using the Coloc 2 package in ImageJ. A region of interest was selected on the image, and the analysis was performed using a PSF of 3.0 with 100 Costes randomizations.
2.9. Electrolyte leakage analysis
Cell death progression in N. benthamiana leaves was assessed by measuring ion leakage. Approximately 24 hr post‐agroinfiltration, eight leaf disks were collected from two leaves on the same plant and pooled into a single well of a 12‐well plate, representing a single biological replicate. Leaf disks were washed for 30 min in 4 ml of deionized water by rotating plates at 50 rpm at room temperature. Wash water was removed and replaced with 4 ml of fresh deionized water. Immediately after adding freshwater, the conductivity of the solution was recorded, representing the 24 hr post‐agroinfiltration measurement. The conductivity of the water was measured using an ECTestr 11+ MultiRange conductivity meter (Oakton) at the indicated time points.
2.10. DAB staining of N. benthamiana leaves
Staining solution was prepared by dissolving 3,3′‐diaminobenzidine (DAB; Sigma) in HCl at pH 2. Once dissolved, this solution was added to Na2HPO4 buffer (10 mM) for a final DAB concentration of 1 mg/ml. Tween 20 (0.05% v/v) was added, and the final pH was adjusted to 7.2. Whole leaves were collected, placed in petri dishes, submerged in DAB staining solution, and vacuum infiltrated. Plates were covered in aluminum foil and incubated at room temperature with shaking. After 4 hr, DAB staining solution was removed and replaced with clearing solution A (25% acetic acid, 75% ethanol). Leaves were heated at 80°C for 10 min to remove chlorophyll. Clearing solution A was removed and replaced with clearing solution B (15% acetic acid, 15% glycerol, 70% ethanol). Leaves were incubated in clearing solution B overnight at room temperature to remove residual chlorophyll.
2.11. A. alternata inoculation of N. glutinosa leaves
Alternaria alternata isolated from potato was provided by Dr. Amanda Gevens (University of Wisconsin, Madison, WI). For FB1 treatments, leaves were coinfiltrated with an Agrobacterium suspension containing the 35S:YFP construct and 5 μM FB1 (Cayman Chemicals). Leaves were harvested from N. glutinosa plants one day after agroinfiltration. Detached leaves were placed adaxial‐side up in petri dishes (100 mm × 20 mm) containing three layers of wet filter paper. Five‐mm‐diameter agar plugs were collected from the edge of an actively growing fungal colony on potato dextrose agar. Leaves were wounded with a 1‐ml pipette tip along the midrib, and agar plugs were placed fungal‐side‐down on top of the wound. Inoculated leaves were kept at room temperature (~23°C) for the duration of the experiment. Lesion areas were recorded 3 days after fungal inoculation.
2.12. Image acquisition and analysis
All leaf images were taken using a Nikon D5500 camera with a Nikon AF‐S NIKKOR 18‐55 mm lens. Quantification of DAB staining intensity and fungal growth was performed using the Fiji package for ImageJ (Schindelin et al., 2012). For quantification of DAB staining intensity, the Colour Deconvolution package was used to isolate the DAB color channel for each DAB‐stained leaf (Ruifrok & Johnston, 2001). Staining intensity caused by 35S:YFP expression on the left half of each leaf was subtracted from the staining intensity caused by 35S:HA‐SlySBP8b or 35S:HA‐SlySBP12a expression on the right half of the same leaf. Fungal lesions were quantified by tracing the periphery of the lesion and calculating the area within the periphery using ImageJ. Statistical analyses were performed using a one‐way analysis of variance (ANOVA) with Tukey's honest significant difference (HSD) test in R Studio (RStudio Team, 2016).
2.13. Identification of SBP cis‐elements in Arabidopsis genes
The online bioinformatic tool FindM was used to search the eukaryotic promoter database (EPD) for promoters containing the 5′‐CCGTAC(A/G)‐3′ cis‐element bound by the SBP domain of SBP transcription factors. Promoter regions were defined as the genomic region 1,000‐bp upstream of the transcription start site, and a bidirectional search mode was used. Genomic sequences used were from the TAIR 10 version of the Arabidopsis genome.
3. RESULTS
3.1. Identification of SfIAP binding partners in tomato
To identify putative binding partners of SfIAP from tomato, we performed a yeast two‐hybrid assay coupled with next‐generation sequencing using a method developed by Lewis et al. (2012) termed quantitative interactor screen sequencing (QIS‐Seq). This method enables the entire pool of interactors to be sequenced by pooling all yeast colonies together instead of individually sequencing each colony using Sanger sequencing. The high‐throughput nature of QIS‐Seq proved useful for screening multiple baits, including a negative control, against the library as well as sequencing the entire cDNA library itself (Supporting information Figure S1A).
SfIAP contains two BIR domains and a C‐terminal RING domain. The BIR1 domain and the RING domain are essential for complete SfIAP function in plants while the BIR2 domain is dispensable (Kabbage et al., 2010). Full‐length SfIAP and the BIR1 domain alone (SfIAPBIR1) were used as bait to screen a tomato cDNA library produced under stressed conditions. The SfIAPBIR1 construct was used to prolong transient interactions that can occur upon ubiquitination of substrates by the RING domain of full‐length SfIAP. Luciferase served as a negative control to account for non‐specific protein interactions and potential autoactivation of the selectable marker. The cDNA library itself was also sequenced to account for biases in transcript abundance.
Enrichment scores were calculated for each locus using the equation in Supporting information Figure S1B. A total of 13 putative interactors with enrichment scores of 50 or higher were identified in our screen (Table 1). Interestingly, this list contained six members of the SQUAMOSA promoter‐binding protein (SBP) family of transcription factors. Based on enrichment scores, the top interactor with full‐length SfIAP was SlySBP8b (95.7) while the top interactor with SfIAPBIR1 was SlySBP12a (98.7). Also present at lower enrichments were SlySBP4, ‐6a, ‐6c, and an unannotated homolog referred to as “SlySBP‐like” (Table 1).
Table 1.
Enriched genes identified from QIS‐Seq using full‐length SfIAP or the BIR1 domain alone as bait
| Locus ID | Annotation | FPKM values | Enrichment | ||
|---|---|---|---|---|---|
| SfIAP | Luciferase | Library | |||
| Solyc01g090730 | SlySBP8b | 66124.2 | 1434.5 | 13.2 | 95.7 |
| Solyc02g071010 | Chlorophyll a/b binding | 52964.4 | 765.5 | 4551.3 | 89.6 |
| Solyc05g005560 | BURP‐domain containing | 94.9 | 2.5 | 9.3 | 86.6 |
| Solyc03g114850 | SlySBP6a | 626.3 | 44.6 | 1.1 | 86.6 |
| Solyc07g062980 | SlySBP‐like | 1489.9 | 294.3 | 6.9 | 66.8 |
| Solyc12g038520 | SlySBP6c | 43.2 | 8.9 | 2.3 | 63.2 |
| Solyc07g053810 | SlySBP4 | 576.7 | 171.2 | 11.0 | 53.4 |
| Locus ID | Annotation | FPKM values | Enrichment | ||
|---|---|---|---|---|---|
| SfIAPBIR1 | Luciferase | Library | |||
| Solyc01g068100 | SlySBP12a | 7378.1 | 42.8 | 12.1 | 98.7 |
| Solyc06g073090 | Ribosomal sub. interface | 5669.4 | 196.7 | 61.1 | 92.3 |
| Solyc01g080020 | Xylanase inhibitor | 85.7 | 4.4 | 7.0 | 83.7 |
| Solyc01g090690 | Elongation factor G | 28.4 | 1.3 | 3.2 | 82.4 |
| Solyc01g090730 | SlySBP8b | 14122.1 | 1434.5 | 13.2 | 81.5 |
| Solyc01g094200 | NAD‐dep. malic enzyme | 54.5 | 2.0 | 9.4 | 79.7 |
| Solyc12g038520 | SlySBP6c | 69.2 | 8.9 | 2.3 | 75.0 |
| Solyc07g053810 | SlySBP4 | 1031.9 | 171.2 | 11.0 | 70.9 |
| Solyc07g062980 | SlySBP‐like | 1257.7 | 294.3 | 6.9 | 61.8 |
| Solyc01g009750 | Unknown Protein | 69.1 | 10.2 | 33.2 | 52.4 |
FPKM: fragments per kilobase per million reads.
3.2. Induction of tissue death by SlySBP8b and SlySBP12a
SfIAP is known to inhibit apoptosis in S. frugiperda and suppress cell death when ectopically expressed in tomato and tobacco (Li et al., 2010; Muro, Hay, and Clem, 2002). Thus, we anticipated that SfIAP‐interacting partners in plants may be positive regulators of cell death. To narrow our list of candidate genes, we transiently overexpressed full‐length cDNA clones of the six tomato SBPs identified from our yeast two‐hybrid screen in N. benthamiana leaves and monitored these leaves for signs of tissue death. The generated cassettes contained an N‐terminal hemagglutinin (HA) tag and were driven by a cauliflower mosaic virus 35S promoter (35S). The structures of these SBPs are quite diverse; the SBP domain being the only domain conserved among all six members (Supporting information Figure S2).
The left half of each leaf was transformed with free YFP as a negative control for cell death induction while the right half was transformed with the corresponding SBP transcription factor. Cell death was recorded if tissue collapse or lesion formation was present on the right half (SBP) but absent on the left half (YFP) of the leaf. A total of 10 leaves from five plants were scored at 5 days post‐transformation. The results are displayed alongside a representative image of the phenotype in Figure 1a. Tissue death induced by 35S:HA‐SlySBP8b and 35S:HA‐SlySBP12a expression occurred in 9/10 and 10/10 leaves, respectively (Figure 1a). Overexpression of 35S:HA‐SlySBP‐like induced weak cell death in 1/10 leaves while the other SBPs failed to produce any visible signs of tissue death (Figure 1a). Immunoblots using an α‐HA antibody confirmed protein accumulation for all constructs (Figure 1b). These results show that at least two SfIAP interactors induce clear signs of cell death upon overexpression in N. benthamiana.
Figure 1.
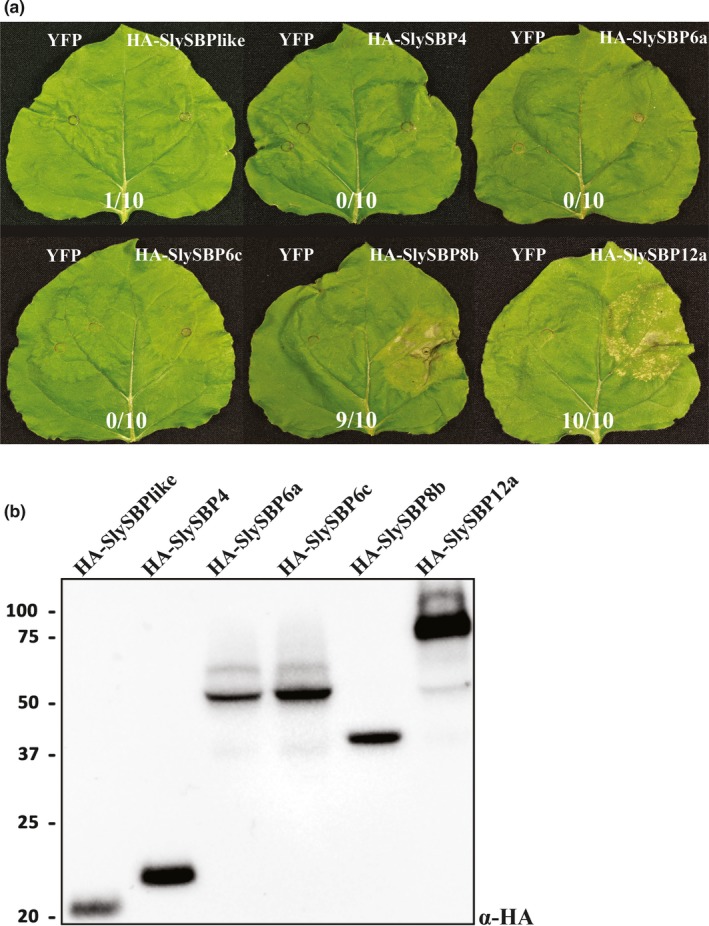
Cell death induced by overexpression of SlySBP transcription factors in N. benthamiana. Full‐length cDNA clones of enriched SlySBP transcription factors from the yeast two‐hybrid assay were transiently overexpressed in N. benthamiana. (a) The left half of each leaf was transformed with free YFP as a negative control while the right half was transformed with the corresponding SlySBP gene containing an N‐terminal HA tag and 35S promoter. Images were taken 5 days post‐transformation. Cell death was recorded if tissue collapse or lesion formation was present on the right half (SBP) but absent on the left half (YFP) of the leaf. A total of 10 leaves from five plants were scored for cell death with the results displayed below each leaf. (b) An immunoblot was performed on tissue collected 2 days post‐transformation to confirm accumulation of SlySBP proteins. Proteins were detected using an α‐HA antibody
3.3. SlySBP8b and SlySBP12a interact with SfIAPM4(I332A) in planta
Due to the strong tissue death phenotype associated with the overexpression of SlySBP8b and SlySBP12a, we focused our subsequent efforts on these two SBP variants. For in vivo confirmation of the yeast two‐hybrid results, we performed coimmunoprecipitation (CoIP) assays in N. benthamiana leaves. A truncated version of SfIAP beginning at the 4th methionine residue was used as our bait. This version maintains its function in S. frugiperda cells but lacks a caspase recognition site that is typically cleaved in S. frugiperda (Cerio, Vandergaast, & Friesen, 2010). This is particularly important as we show that cleavage at the N‐terminus of the full‐length protein occurs in N. benthamiana, thus removing N‐terminal epitope tags (Supporting information Figure S3).
We noticed that SlySBP12a was not enriched in our yeast two‐hybrid when full‐length SfIAP was used as bait but only appeared when the SfIAPBIR1 truncation was used (Table 1). To account for the possibility that SfIAP may interact transiently with SlySBP12a, an E3 ligase mutant of the truncated SfIAP protein was generated by mutating a conserved residue in the RING domain (Cerio et al., 2010). This construct, referred to as SfIAPM4(I332A), is resistant to N‐terminal cleavage in N. benthamiana (Supporting information Figure S3).
Two days after coexpression of 35S:YFP‐SfIAP M4 (I332A) with 35S:HA‐SlySBP8b or 35S:HA‐SlySBP12a, total proteins were extracted from leaves and incubated with GFP‐Trap_MA beads (Chromotek, Germany). All proteins were detected in the input fraction, and HA‐SlySBP8b and HA‐SlySBP12a were successfully pulled‐down by YFP‐SfIAPM4(I332A) but not by free YFP (Figure 2). These data confirm the yeast two‐hybrid results and demonstrate that SfIAPM4(I332A) interacts with SlySBP8b and SlySBP12a in plant cells.
Figure 2.
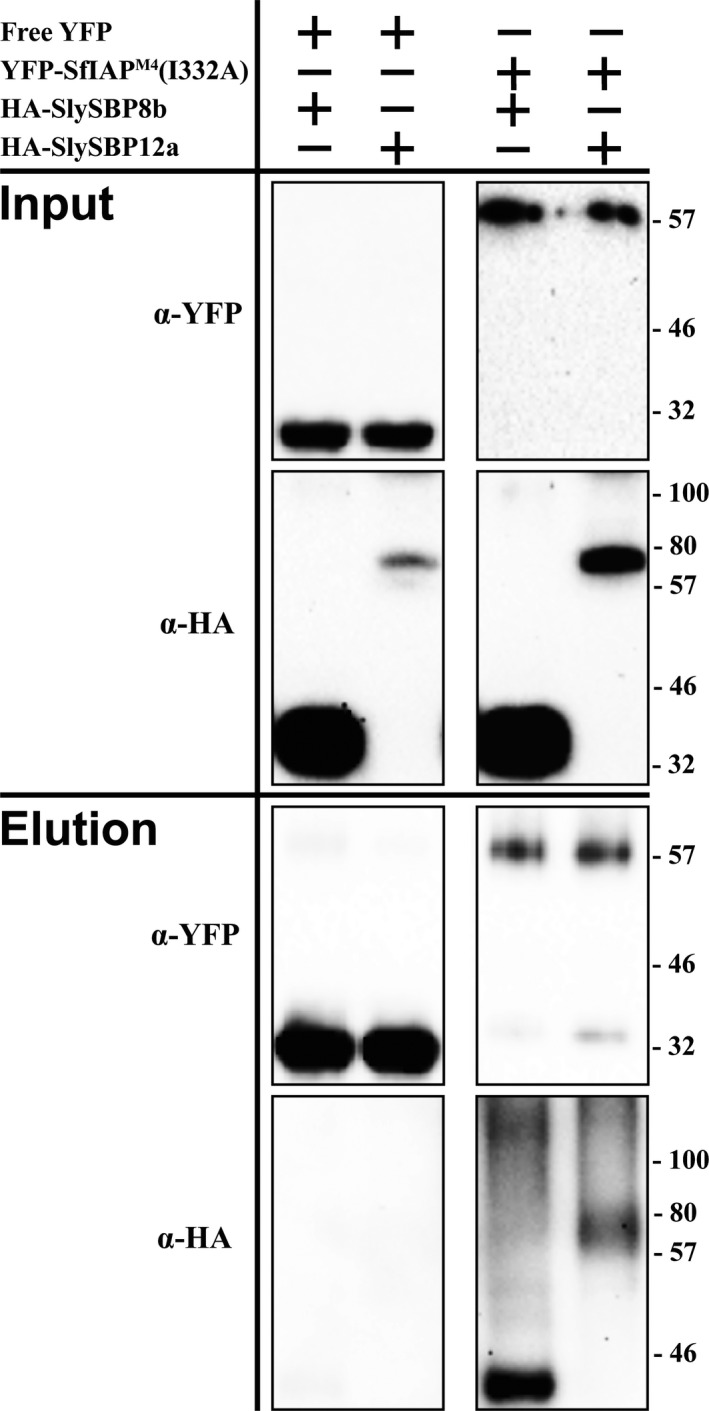
Coimmunoprecipitation of SfIAPM 4(I332A) with SlySBP8b and SlySBP12a in N. benthamiana. 35S:YFP‐Sf IAPM 4 (I332A) or free YFP was transiently coexpressed with 35S:HA‐SlySBP8b or 35S:HA‐SlySBP12a in N. benthamiana leaves. Proteins were immunoprecipitated with an α‐YFP affinity matrix. A portion of each sample was taken before immunoprecipitation to serve as the input control. An immunoblot was performed on input and elution fractions using the indicated antibodies to detect the epitope‐tagged proteins
3.4. SlySBP8b and SlySBP12a localize to the nucleus
As putative transcription factors, we reasoned that SlySBP8b and SlySBP12a function in the nucleus through regulation of genes involved in cell death induction. Additionally, a predicted bi‐partite nuclear localization signal (NLS) is present in the SBP domain of all tomato SBP transcription factors (Salinas, Xing, Hohmann, Berndtgen, & Huijser, 2012). Localization was assessed by expressing 35S:YFP‐SlySBP8b and 35S:YFP‐SlySBP12a in tomato mesophyll protoplasts. Confocal laser scanning microscopy (CLSM) revealed that both YFP‐SlySBP8b and YFP‐SlySBP12a colocalized with the nuclear marker dihydroethidium (DHE) (Figure 3).
Figure 3.
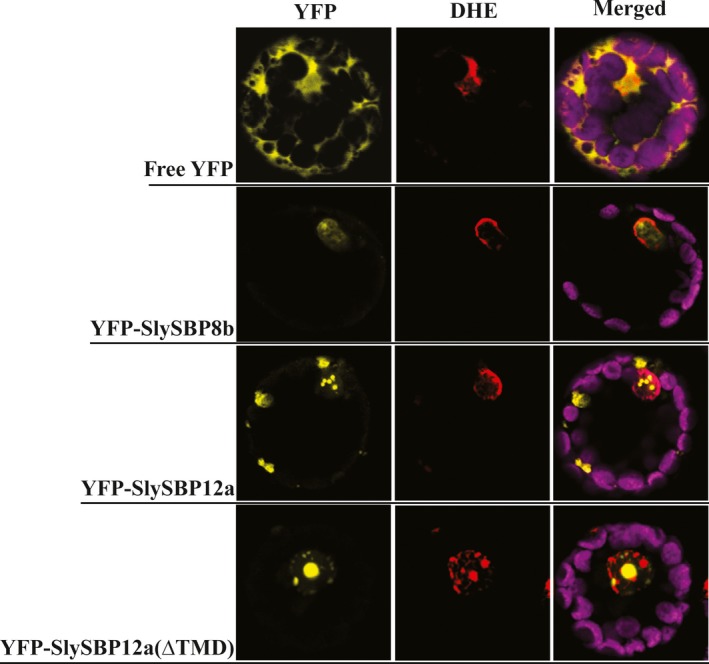
Nuclear localization of free YFP, SlySBP8b, SlySBP12a, and SlySBP12a(ΔTMD) in tomato protoplasts. Tomato protoplasts were transfected with plasmids encoding 35S:YFP, 35S:YFP‐SlySBP8b, 35S:YFP‐SlySBP12a, or 35S:YFP‐SlySBP12a(ΔTMD) and imaged by CLSM. Dihydroethidium (red) was used as a nuclear counterstain while the magenta signal represents chloroplast autofluorescence
3.5. Disruption of the NLS in SlySBP8b and SlySBP12a abolishes cell death induction
In addition to its role in nuclear targeting, the NLS of SBP proteins forms a positively charged surface that is required for DNA binding (Birkenbihl, Jach, Saedler, & Huijser, 2005). Site‐directed mutagenesis was used to substitute conserved lysine and arginine residues in the NLS with leucine to disrupt this positive charge (Supporting information Figure S4). Overexpression of the two NLS mutants, 35S:HA‐SlySBP8b(NLS mt ) and 35S:HA‐SlySBP12a(NLS mt ), in N. benthamiana leaves did not induce visible signs of cell death (Figure 4a). Electrolyte leakage induced by overexpression of HA‐SlySBP8b(NLSmt) and HA‐SlySBP12a(NLSmt) was similar to overexpression of the negative control YFP (Figure 4b and c). Immunoblots performed on tissues overexpressing both the wild‐type and NLS mutants confirmed that protein accumulation was not greatly affected by mutations in the NLS (Figure 4d).
Figure 4.
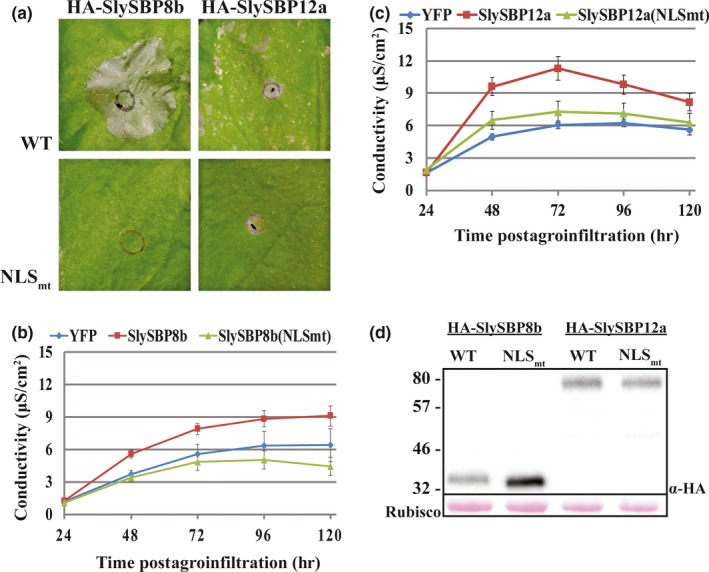
Disruption of the NLS in SlySBP8b and SlySBP12a prevents cell death in N. benthamiana upon overexpression. 35S:HA‐SlySBP8b, 35S:HA‐SlySBP8b(NLS mt ), 35S:HA‐SlySBP12a, or 35S:HA‐SlySBP12a(NLS mt ) were transiently transformed in N. benthamiana. (a) Images of leaves taken 5 days post‐transformation. (b) Electrolyte leakage assay used to quantify cell death. 35S:YFP—blue diamond; 35S:HA‐SlySBP8b—red square; 35S:HA‐SlySBP8b(NLSmt)—green triangle. Two independent experiments were pooled together for a total of 13 biological replicates for each gene. Error bars represent a 95% confidence interval. (c) Electrolyte leakage assay used to quantify cell death. 35S:YFP—blue diamond; 35S:HA‐SlySBP12a —red square; 35S:HA‐SlySBP12a(NLSmt)—green triangle. Three independent experiments were pooled together for a total of 22 biological replicates for each gene. Error bars represent a 95% confidence interval. (d) Immunoblot performed on tissue collected 2 days post‐transformation. An α‐HA antibody was used to detect SlySBP proteins, and Ponceau S stain was used to detect Rubisco as a loading control
Nuclear localization of YFP‐SlySBP8b(NLSmt) and YFP‐SlySBP12a(NLSmt) in N. benthamiana epidermal cells was still observed (Figure 5). Localization of YFP‐SlySBP8b(NLSmt) was also observed outside of the nucleus around the periphery of the cell, which was not observed with YFP‐SlySBP8b, indicating nuclear import is impaired but not abolished in this mutant (Figure 5). The cis‐regulatory element for SBP transcription factor binding to promoters (5′‐CCGTAC(A/G)‐3′) was previously described (Franco‐Zorrilla et al., 2014; Liang, Nazarenus, & Stone, 2008). We searched for this motif in the promoter region of Arabidopsis genes and found 523 genes containing this motif within 1,000‐bp upstream of the transcription start site. These genes are involved in many diverse biological processes (Supporting information Figure S5). A closer look revealed WRKY transcription factors, NBS‐LRR genes, and an SBP homolog (Supporting information Table S2). Two of these genes, RPP4 and RRS1, are known lesion‐mimic mutants in Arabidopsis (Huang, Li, Bao, Zhang, & Yang, 2010; Noutoshi et al., 2005). These data suggest a functional NLS within the SBP domain is required for cell death induction caused by SlySBP8b and SlySBP12a overexpression.
Figure 5.
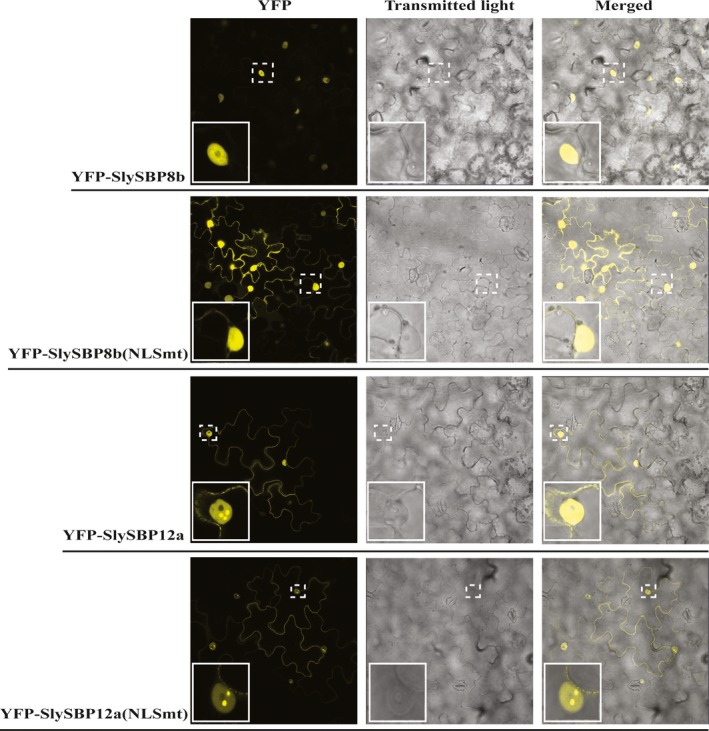
Localization of SlySBP8b, SlySBP8b(NLSmt), SlySBP12a, and SlySBP12a(NLSmt) in N. benthamiana epidermal cells. Leaves were transiently transformed with 35S:YFP‐SlySBP8b, 35S:YFP‐SlySBP8b(NLSmt), 35S:YFP‐SlySBP12a, or 35S:YFP‐SlySBP12a(NLSmt) and imaged using CLSM 2 days later. The dashed‐line box in each panel is magnified and displayed in the lower‐left corner of each panel. A transmitted light image was included to highlight the periphery of epidermal cells and nuclei
3.6. SlySBP12a is anchored to the ER membrane by a C‐terminal transmembrane domain
While YFP‐SlySBP8b was found to be strictly nuclear localized, YFP‐SlySBP12a was also localized to diffuse pockets outside of the nucleus (Figure 3). The presence of a putative C‐terminal transmembrane domain (TMD) in SlySBP12a (Supporting information Figure S2) suggested that this localization pattern could be due to the anchoring of SlySBP12a to a cellular membrane. Removal of the last 73 amino acids of SlySBP12a eliminated the putative TMD and resulted in complete localization of YFP‐SlySBP12a(ΔTMD) to the nucleus (Figure 3; Figure 6). Additionally, overexpression of 35S:HA‐SlySBP12a(ΔTMD) in N. benthamiana caused enhanced cell death characterized by extensive tissue collapse at the site of transgene expression and increased electrolyte leakage compared to the full‐length construct (Figure 7a and b). The TMD of SlySBP12a may thus regulate its access to the nucleus and the subsequent induction of cell death.
Figure 6.
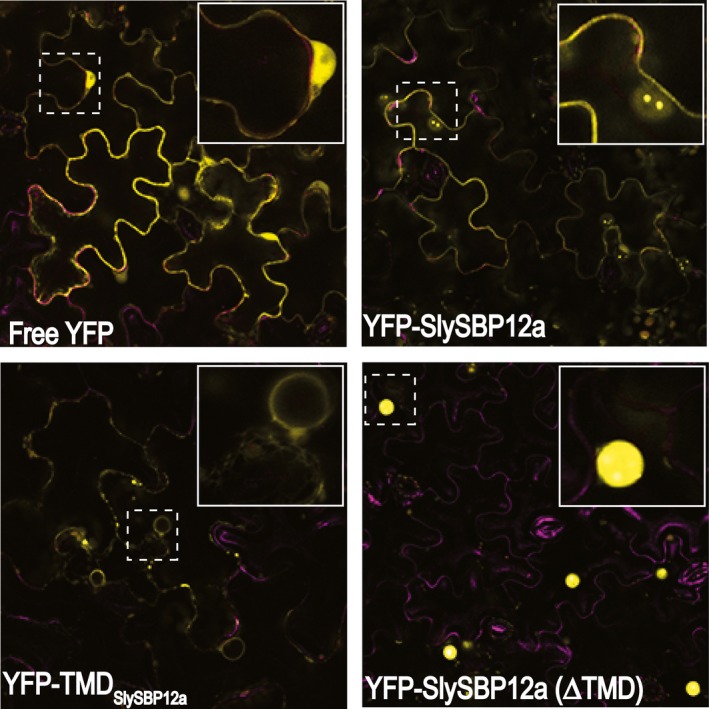
Localization of SlySBP12a, TMDS ly SBP 12a, and SlySBP12a(ΔTMD) in N. benthamiana epidermal cells. Leaves were transiently transformed with 35S:YFP‐SlySBP12a, 35S:YFP‐SlySBP12a(ΔTMD), 35S:YFP‐ TMDS ly SBP 12a , or 35S:YFP and imaged using CLSM 2 days post‐transformation. The dashed‐line box in each panel is magnified and displayed in the upper‐right corner of each panel. Chlorophyll autofluorescence is shown in magenta
Figure 7.
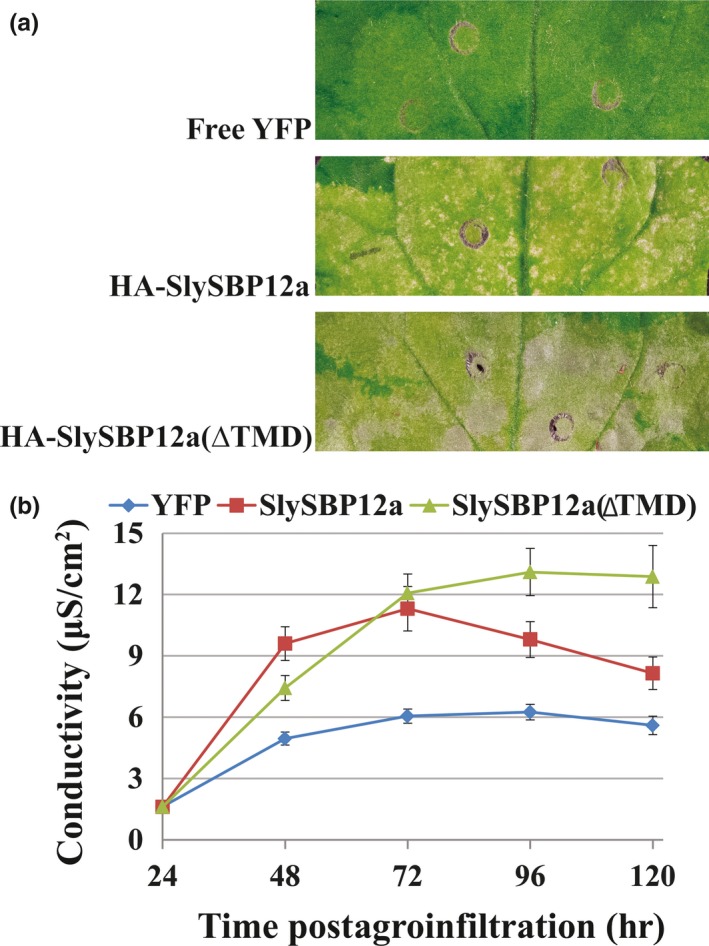
Removal of the TMD from SlySBP12a results in enhanced cell death upon overexpression. 35S:HA‐SlySBP12a, 35S:HA‐SlySBP12a(ΔTMD), or 35S:YFP was transiently transformed in N. benthamiana. (a) Images of leaves taken 5 days post‐transformation. (b) Electrolyte leakage assay used to quantify cell death. 35S:YFP—blue diamond; 35S:HA‐SlySBP12a —red square; and 35S:HA‐SlySBP12a(ΔTMD)—green triangle. Three independent experiments with similar results were pooled together for a total of 22 biological replicates for each gene. Error bars represent a 95% confidence interval
To determine the membrane localization of SlySBP12a, the last 73 amino acids of the protein containing the putative TMD were fused to the C‐terminal end of YFP (YFP‐TMDSlySBP12a) (Supporting information Figure S4). This construct was expressed in N. benthamiana where it localized to the periphery of epidermal cells and a ring‐like structure around the nucleus that resembled endoplasmic reticulum (ER) localization (Figure 6). Endoplasmic reticulum localization was confirmed in tomato protoplasts, where the YFP‐TMDSlySBP12a fusion colocalized with the ER marker SP‐mCherry‐HDEL (Figure 8a). This ER marker consists of the fluorescent protein mCherry with a signal peptide at its N‐terminus and an ER retention motif at its C‐terminus (Nelson, Cai, & Nebenfuhr, 2007). We were also able to show colocalization between the full‐length YFP‐SlySBP12a construct and the ER marker in tomato protoplasts (Figure 8a). Intensity histograms were generated, and Pearson's R values and Costes p‐values were calculated for four regions. Regions around and outside of the nucleus, where we see YFP‐SlySBP12a and YFP‐TMDSlySBP12a colocalization with the ER marker, produced Pearson's R values of 0.77, 0.72, and 0.67 with a Costes p‐Value of 1.00 (Figure 8b). With Pearson's R values close to 1.00 and a Costes p value of 1.00, there is strong evidence that the YFP and mCherry signals overlap. However, inside of the nucleus where only YFP‐SlySBP12a appears to accumulate, the Pearson's R value was −0.02 with a Costes p‐value of 0.83 (Figure 8b). A Pearson's R value close to 0.00 and a Costes p value below 0.95 provide no statistically significant evidence of overlap between YFP and mCherry signals in the nucleus. Taken together, these results confirm that SlySBP12a contains a functional TMD that integrates the full‐length protein into the ER membrane.
Figure 8.
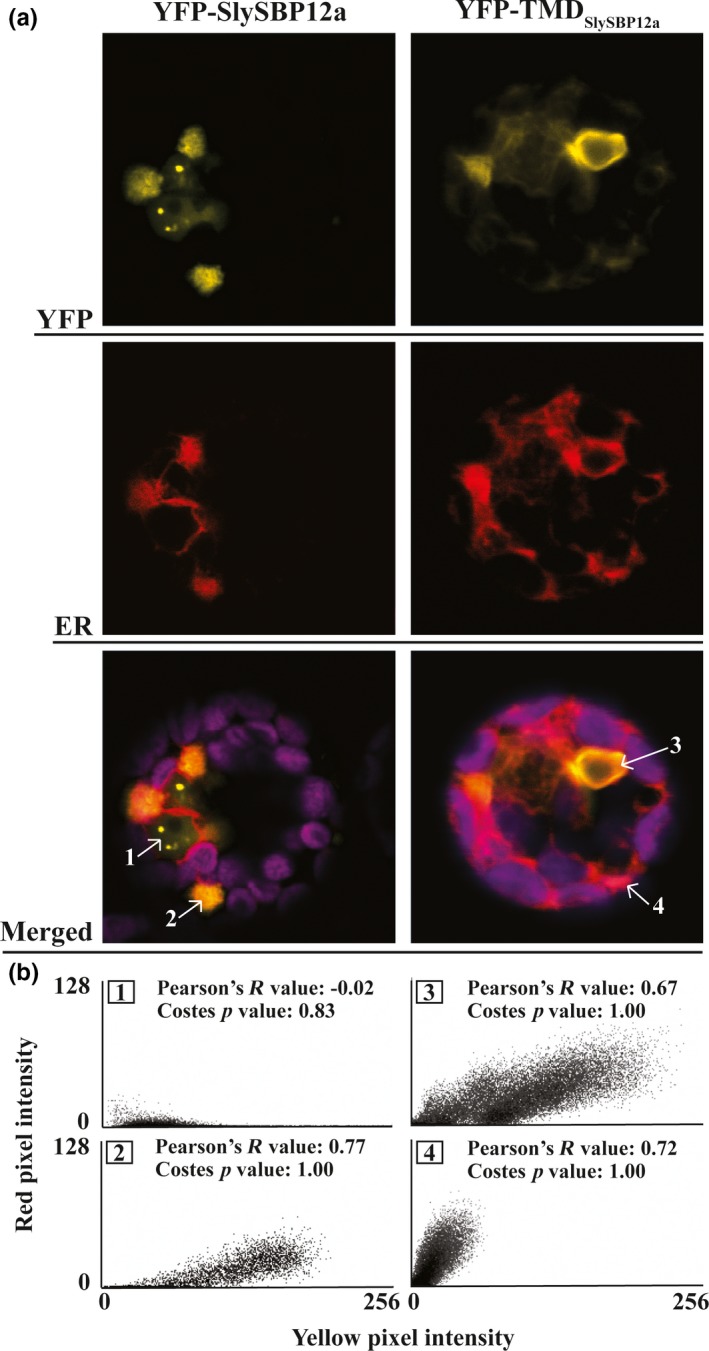
Endoplasmic reticulum localization of SlySBP12a and TMDS ly SBP 12a in tomato protoplasts. Tomato protoplasts were transfected with plasmids encoding 35S:YFP‐SlySBP12a or 35S:YFP‐ TMDS ly SBP 12a and imaged by CLSM. An SP‐mCherry‐HDEL construct was cotransfected to serve as an ER marker (red). The magenta signal represents chloroplast autofluorescence. (a) Representative images of tomato protoplasts expressing 35S:YFP‐SlySBP12a or 35S: TMDS ly SBP 12a with the ER marker. Numbered regions indicated by arrows were used for colocalization analysis. (b) Intensity histograms of the four regions selected for colocalization analysis. Pearson's R values and Costes p‐values are displayed for each region
3.7. ROS production and fungal growth in leaves overexpressing SlySBP8b and SlySBP12a
Reactive oxygen species (ROS) are important cell death intermediaries, and their accumulation is a key feature of cell death imposed by necrotrophic fungal pathogens and the death‐inducing toxins they produce (Heller & Tudzynski, 2011; Kim, Min, & Dickman, 2008; Sakamoto, Tada, Nakayashiki, Tosa, & Mayama, 2005; Shi et al., 2007). Following transient expression in N. benthamiana leaves, we monitored the accumulation of hydrogen peroxide (H2O2) for 4 days using 3′3‐diaminobenzidine (DAB) staining. Leaves expressing 35S:HA‐SlySBP8b and 35S:HA‐SlySBP12a displayed enhanced DAB staining intensity relative to expression of the 35S:YFP control on the same leaf (Figure 9a). Accumulation of H2O2 occurred as early as 2 and 3 days post‐transformation for 35S:HA‐SlySBP8b and 35S:HA‐SlySBP12a, respectively (Figure 9b). ImageJ software was used to measure DAB staining intensity (Schindelin et al., 2012).
Figure 9.
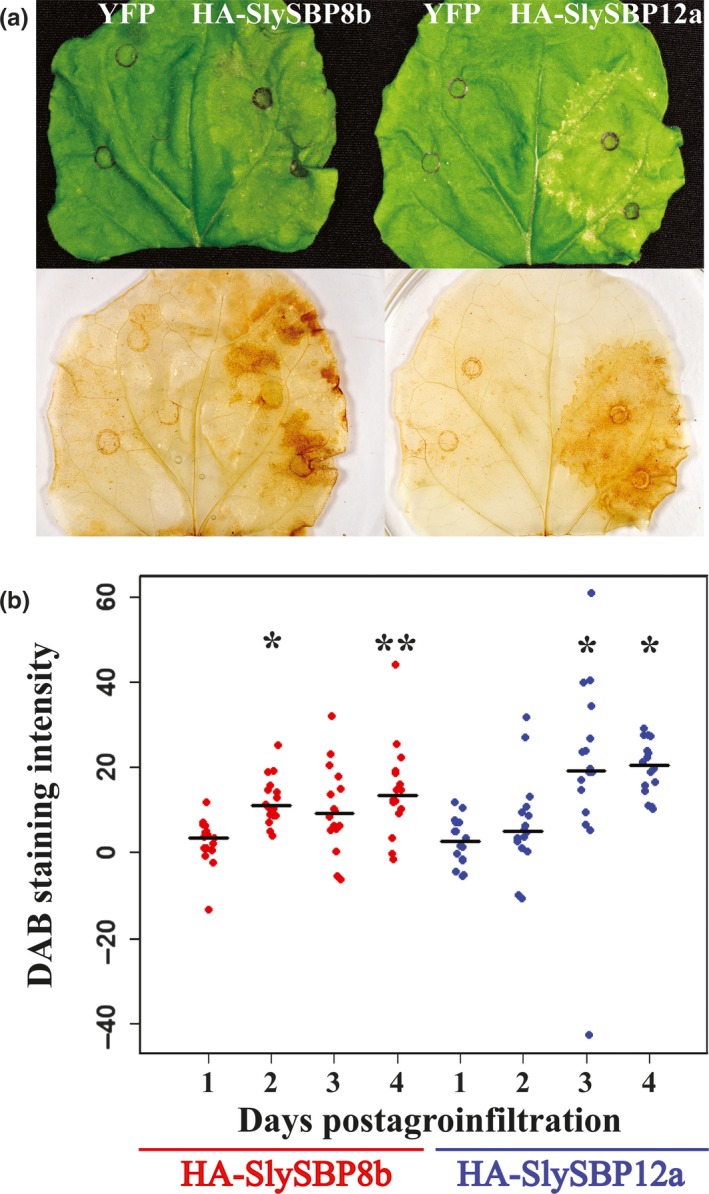
Overexpression of SlySBP8b and SlySBP12a in N. benthamiana induces H2O2 accumulation. 35S:HA‐SlySBP8b and 35S:HA‐SlySBP12a were transiently transformed in N. benthamiana. Leaves were cleared and stained with DAB to detect H2O2. (a) Images of leaves before and after DAB staining taken 4 days post‐agroinfiltration. (b) Quantification of DAB‐stained area for each SlySBP relative to YFP expression on the same leaf. ImageJ was used to analyze 16 leaves for each gene at each time point. Data are displayed as a dotplot with the medians represented as black horizontal lines. Statistical significance for each gene compared to its Day 1 time point was determined using a one‐way ANOVA with Tukey's HSD post hoc test (*p < 0.01; **p < 0.001)
Transgenic SfIAP plants were reported to accumulate lower levels of ROS under stress conditions compared to wild‐type plants (Li et al., 2010). Necrotrophic fungal pathogens are known to exploit host ROS production as means to kill host cells for their own benefit (Govrin & Levine, 2000; Heller & Tudzynski, 2011; Kabbage et al., 2013; Ranjan et al., 2017). In addition to reduced ROS accumulation, transgenic SfIAP plants are also resistant to the necrotrophic fungal pathogen A. alternata (Li et al., 2010). Therefore, we reasoned that leaves overexpressing SlySBP8b and SlySBP12a would support enhanced growth of this pathogen. Unfortunately, N. benthamiana is not susceptible to this pathogen, so we screened Nicotiana germplasm for susceptible species. We found that Nicotiana glutinosa was susceptible to A. alternata and previous work confirmed that transgenes could be expressed effectively in this species using Agrobacterium‐mediated transient transformation (Kessens, Ashfield, Kim, & Innes, 2014).
While the differences were small, a total of 54 biological replicates from four randomized and blind experiments showed that leaves expressing 35S:YFP‐SlySBP8b or 35S:YFP‐SlySBP12a had increased A. alternata lesion areas compared to leaves expressing 35S:YFP alone (Figure 10a and b). This effect was more pronounced with 35S:YFP‐SlySBP12a than with 35S:YFP‐SlySBP8b expression. As a positive control, leaves were treated with 5 μM FB1 to simulate cell death induction by a fungal toxin. Lesion development in 35S:YFP‐SlySBP12a‐expressing tissue and FB1‐treated tissue was comparable (Figure 10b). Fluorescence microscopy was used to confirm protein accumulation in each leaf before fungal inoculation, and 35S:YFP‐SlySBP8b and 35S:YFP‐SlySBP12a were able to induce tissue death in N. glutinosa (Supporting information Figure S6). Lesion areas were measured using ImageJ software (Schindelin et al., 2012). Overall, we show that these two transcription factors are able to increase ROS levels and promote A. alternata growth, phenotypes that are dampened in plants expressing SfIAP.
Figure 10.
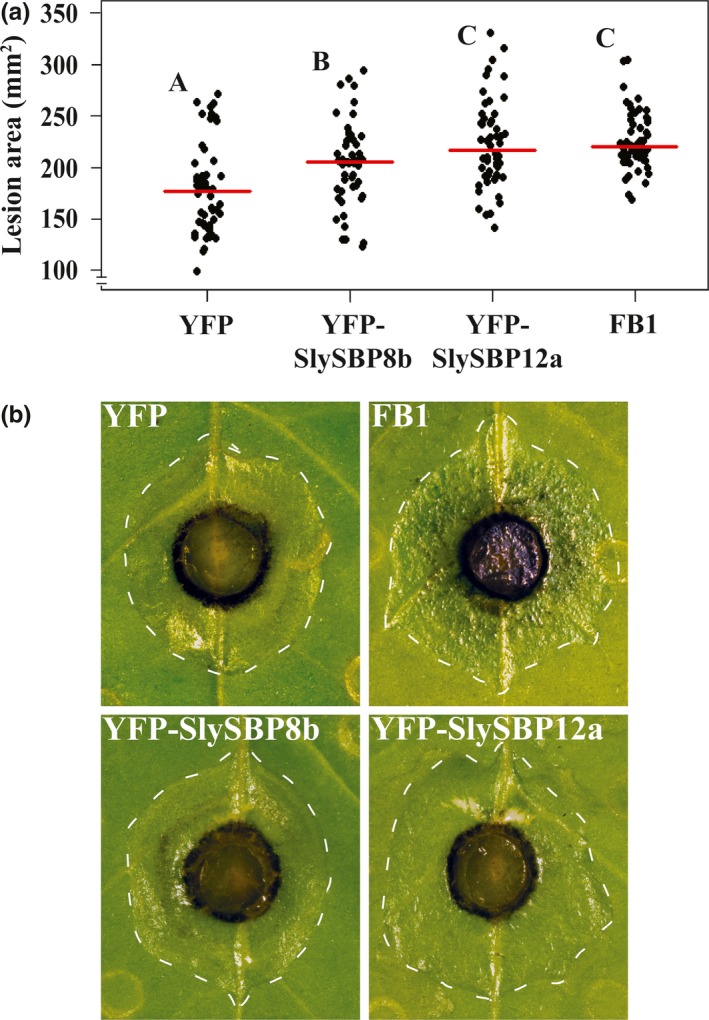
Overexpression of SlySBP8b and SlySBP12a enhances A. alternata growth on N. glutinosa. 35S:YFP‐SlySBP8b, 35S:YFP‐SlySBP12a, or 35S:YFP was transiently transformed in N. glutinosa. As a positive control for cell death induction, leaves were treated with 5 μM FB1. Agar plugs containing actively growing A. alternata mycelium were placed fungal‐side‐down on leaves. Images were taken, and lesion areas were recorded, 3 days after fungal inoculation. (a) Quantification of lesion area using ImageJ. The results of four randomized and blind experiments were pooled representing 54 leaves for each treatment. Data are displayed as a dotplot with the medians represented by red horizontal lines. Treatments with the same letter are not statistically significant as determined by a one‐way ANOVA with Tukey's HSD post hoc test (YFP/SBP8b, p = 0.02; YFP/SBP12a, p = 2.0E‐7; YFP/FB1, p = 5.0 E‐7; SBP8b/SBP12a, p = 0.02; SBP8b/FB1, p = 0.04). (b) Images of inoculated leaves with lesions outlined by a dotted white line
4. DISCUSSION
Over 15 years ago, it was first reported that heterologous expression of a viral IAP (OpIAP) in tobacco suppressed cell death induced by the necrotrophic fungal pathogen S. sclerotiorum and the necrosis‐inducing viral pathogen tomato spotted wilt virus (Dickman et al., 2001). Subsequent studies revealed that an IAP from Spodoptera frugiperda (SfIAP) suppressed cell death imposed by numerous abiotic and biotic stresses (Hoang, Williams, Khanna, Dale, & Mundree, 2014; Kabbage et al., 2010; Li et al., 2010). However, the biochemical mechanism by which these IAPs suppress cell death in plant systems remains unknown. In this study, we utilize SfIAP as a biochemical tool to identify novel prodeath regulators and shed light on how SfIAP functions in plants.
4.1. SlySBP8b and SlySBP12a associate with SfIAPM4(I332A) and exhibit characteristics of prodeath regulators
The yeast two‐hybrid and CoIP data presented clearly show that SlySBP8b and SlySBP12a associate with SfIAPM4(I332A) (Table 1 & Figure 2). Remarkably, SlySBP8b and SlySBP12a exhibit attributes of prodeath regulators, demonstrated by cell death induction and ROS accumulation upon overexpression (Figures 1 and 9). We anticipated that coexpression of SfIAP with SlySBP8b or SlySBP12a would suppress cell death induction. However, numerous attempts to suppress cell death induced by SlySBP8b and SlySBP12a overexpression were unsuccessful when SfIAP or SfIAP M4 was coexpressed. One possible explanation is the fact that SfIAP and SfIAPM4 protein accumulation is significantly less than SlySBP8b and SlySBP12a when expressed in N. benthamiana. In fact, SfIAP and SfIAPM4 accumulation was only detected when these proteins were fused to YFP. While an N‐terminal YFP tag did not impact binding to SlySBP8b and SlySBP12a, we cannot rule out the possibility that this large tag could affect the ability of SfIAP or SfIAPM4 to suppress the activity of these transcription factors. An excess of either transcription factor could allow enough to enter the nucleus and influence cell death gene expression.
SlySBP8b and SlySBP12a belong to a family of plant‐specific transcription factors known as SQUAMOSA promoter‐binding proteins (SBPs), of which 15 members are present in tomato (Salinas et al., 2012). Members of this family are defined by a highly conserved SBP box DNA binding domain and can be further divided into nine phylogenetically distinct clades (Preston & Hileman, 2013; Yamasaki, Kigawa, Seki, Shinozaki, & Yokoyama, 2013). SBP genes are known to regulate diverse developmental processes such as flowering time, branching, trichome development, apical dominance, and pollen sac development to name a few (Wang & Wang, 2015; Yamasaki et al., 2013). Interestingly, silencing of the SBP gene Colorless non‐ripening (Cnr) in tomato results in fruit with delayed ripening, a phenotype observed in tomatoes overexpressing SfIAP (Li et al., 2010; Manning et al., 2006).
While much is known about the role of SBP transcription factors in plant development, only a few studies to date have associated SBPs with stress responses. The deletion of Arabidopsis SPL14 (AtSPL14) conferred enhanced tolerance to FB1, thus implicating this SBP transcription factor in the cellular response to this mycotoxin (Stone, Liang, Nekl, & Stiers, 2005). Tolerance to FB1 is a phenotype that we have also observed in SfIAP‐overexpressing tomato seedlings (Li et al., 2010). Interestingly, AtSPL14 and SlySBP12a both reside in clade‐II and display similar structural characteristics with large SBP proteins that contain a predicted C‐terminal transmembrane domain (Preston & Hileman, 2013).
Another clade‐II member, GmSPL12l from soybean, was shown to be a target of the Phakopsora pachyrhizi (Asian soybean rust) effector PpEC23 (Qi et al., 2016). This effector suppressed the hypersensitive response (HR) in soybean and tobacco and also interacted with other clade‐II members from N. benthamiana and Arabidopsis: NbSPL1 and AtSPL1 (Qi et al., 2016). In another study, the N immune receptor of N. benthamiana was found to associate with the SBP transcription factor NbSPL6 upon activation of HR. This interaction only occurred when plants were challenged with an HR‐eliciting strain of tobacco mosaic virus (TMV‐U1) but not a non‐eliciting strain (TMV‐Ob) (Padmanabhan et al., 2013). Taken together, our results and the findings of previous studies clearly show that SBP transcription factors are critical regulators of plant stress responses that result in cell death.
Fungal pathogens with a necrotrophic lifestyle are known to exploit host ROS production for cell death induction and successful pathogenesis (Govrin & Levine, 2000; Heller & Tudzynski, 2011). As positive regulators of cell death and ROS production, we hypothesized that overexpression of SlySBP8b and SlySBP12a would support enhanced growth of necrotrophic fungal pathogens. Additionally, SfIAP transgenic plants are resistant to cell death induced by the necrotrophic fungal pathogen A. alternata (Li et al., 2010). The results of four randomized and blind experiments clearly show that while the contribution of SlySBP8b or SlySBP12a overexpression to A. alternata lesion areas was small, it was significantly greater than leaves expressing the negative control 35S:YFP (Figure 10). The small differences in growth could be explained by the fact that A. alternata is already an aggressive pathogen and the benefits of priming its host for death would be small. To test this, we also treated leaves with FB1, which is a structural analog of the AAL toxin produced by A. alternata f. sp. lycopersici that induces cell death in tomato (Mirocha et al., 1992). Pre‐treatment of N. glutinosa leaves with FB1 led to enhanced growth of A. alternata comparable to SlySBP12a overexpression (Figure 10). These results provide further evidence that SlySBP8b and SlySBP12a are positive regulators of cell death, which in this case, contribute to pathogenic development of A. alternata.
4.2. SlySBP8b and SlySBP12a require a functional SBP domain for cell death induction
As members of a transcription factor family, we hypothesize that SlySBP8b and SlySBP12a exert their prodeath activity through the regulation of genes involved in cell death. The NLS of SBP transcription factors serves a dual role in nuclear import and DNA binding, making this an essential motif for SBP function (Birkenbihl et al., 2005). We show that these transcription factors are clearly localized to the nucleus of tomato protoplasts and N. benthamiana epidermal cells (Figures 3 and 5) and mutation of the bi‐partite NLS of both transcription factors abolishes cell death (Figure 4). While the NLS mutation was unable to abolish nuclear localization of SlySBP8b(NLSmt) or SlySBP12a(NLSmt), it did result in accumulation of some SlySBP8b(NLSmt) protein in the cytoplasm, likely due to partial impairment of nuclear import (Figure 5). These results highlight the importance of a functional SBP domain for cell death induction caused by SlySBP8b and SlySBP12a overexpression, possibly through the regulation of genes involved in cell death.
To identify genes involved in cell death that may be regulated by SBP transcription factors, we searched the Arabidopsis genome for promoters that contain the SBP cis‐element (Supporting information Figure S5). We identified 523 genes involved in a diverse array of biological processes, which is expected of a large transcription factor family known to be involved in diverse developmental and stress‐related processes. Further investigation of these genes revealed a subset with known roles in stress responses (Supporting information Table S2). Four genes encode nucleotide‐binding site leucine‐rich repeat (NBS‐LRR) proteins, which are known to play important roles in plant immunity through activation of hypersensitive‐programmed cell death (HR‐PCD) (McHale, Tan, Koehl, & Michelmore, 2006). Additionally, several WRKY transcription factors with known regulatory roles in plant immunity to both biotrophic and necrotrophic pathogens were identified (Bhattarai, Atamian, Kaloshian, & Eulgem, 2010; Hsu et al., 2013; Lai, Vinod, Zheng, Fan, & Chen, 2008). Perhaps the most interesting finding is that two genes, RPP4 and RRS1, are known lesion‐mimic mutants (Huang et al., 2010; Noutoshi et al., 2005). Due to the large number of genes with predicted SBP‐binding sites in their promoters, future studies will need to utilize chromatin immunoprecipitation sequencing (ChIP‐Seq) to determine genes regulated by SlySBP8b and SlySBP12a in vivo. These studies will provide more concrete information on the downstream components responsible for cell death execution in plants.
4.3. SlySBP12a localizes to the ER
Unlike SlySBP8b, which we found to be strictly nuclear localized, SlySBP12a was also present outside of the nucleus (Figures 3 and 6). By fusing the putative C‐terminal TMD of SlySBP12a to YFP, we were able to show that the TMD of SlySBP12a localized YFP around the nucleus and at the periphery of N. benthamiana epidermal cells (Figure 6). We hypothesized that this pattern was due to ER localization. This was confirmed in tomato protoplasts, where both YFP‐SlySBP12a and YFP‐TMDSlySBP12a colocalize with the ER marker SP‐mCherry‐HDEL (Figure 8a and b).
In response to environmental stress, plant cells increase production of secreted proteins, which in turn can cause ER stress due to the sudden influx of proteins that must be properly folded before moving through the rest of the secretory pathway (Eichmann & Schafer, 2012). This makes the ER an important sensor of cellular stress as the accumulation of unfolded proteins is first detected by the ER. Membrane‐tethered transcription factors (MTTFs) residing at the ER membrane play important roles in ER stress perception and regulation of genes involved in stress relief and cell death in mammalian and plant systems (Slabaugh & Brandizzi, 2011). Membrane tethering provides spatial regulation of transcription factor activity, as MTTFs must be removed from the membrane before the transcription factor domain can translocate to the nucleus (Slabaugh & Brandizzi, 2011). This type of regulation allows these transcription factors to act quickly in response to cellular stress.
In this study, we show that SlySBP12a exhibits a localization pattern similar to previously described ER‐MTTFs from Arabidopsis: NAC089, bZIP28, and bZIP60. These transcription factors are activated upon perception of ER stress and activate cell death (NAC089), heat stress (bZIP28), and ER stress (bZIP60) responses through transcriptional regulation of genes involved in these processes (Gao, Brandizzi, Benning, & Larkin, 2008; Iwata & Koizumi, 2005; Liu, Srivastava, Che, & Howell, 2007; Yang et al., 2014). Removal of the TMD from these transcription factors results in their complete localization to the nucleus and constitutive activation of the processes they regulate (Gao et al., 2008; Iwata & Koizumi, 2005; Liu et al., 2007; Yang et al., 2014). This mirrors what we have observed with SlySBP12a. Removal of the TMD results in complete nuclear localization in N. benthamiana and tomato cells and enhanced cell death induction compared to full‐length SlySBP12a (Figures 3, 6, and 7).
With our data and previous studies of ER‐MTTFs, we can speculate that SlySBP12a is cleaved from the ER membrane upon stress perception and translocates to the nucleus where it regulates genes involved in cell death. However, we must keep in mind that our experiments were performed with a cDNA copy of SlySBP12a, preventing the detection of splice isoforms that could lack the TMD. This is important to consider as bZIP60 was originally thought to be proteolytically cleaved from the ER membrane upon stress induced by tunicamycin treatment (Iwata, Fedoroff, & Koizumi, 2008). A follow‐up study by the same group showed that in addition to being proteolytically cleaved, bZIP60 is also alternatively spliced in response to tunicamycin treatment, resulting in a truncated protein lacking the C‐terminal TMD (Nagashima et al., 2011). Future experiments looking at the translocation of SlySBP12a upon stress induction must consider the possibility of alternative splice isoforms.
5. CONCLUSION
While the expression of IAP and other anti‐apoptotic genes in plants confers enhanced stress tolerance, the animal‐derived nature of these genes will likely prevent their broad commercial use. Thus, the identification of endogenous plant cell death regulators, such as SBP transcription factors, that can be targeted to ameliorate stress tolerance is appealing. This is exemplified by recent interest in exploiting SBP genes for crop improvement due to the many developmental traits they regulate (Liu, Harberd Nicholas, & Fu, 2016; Wang & Wang, 2015). Efforts are underway in our laboratory to determine whether the disruption of these transcription factors impact tolerance to a range of abiotic and biotic insults.
AUTHOR CONTRIBUTIONS
R.K. performed and designed experiments, analyzed data, and wrote the manuscript. N.S. performed experiments and analyzed data. M.K. performed and designed experiments, analyzed data, and wrote the manuscript. All authors reviewed the manuscript.
Supporting information
ACKNOWLEDGMENTS
We would like to thank Andrew Bent and members of his laboratory for their valuable insight and shared laboratory resources. The Alternaria alternata isolate used was provided by Shunping Ding of Amanda Gevens’ Lab (University of Wisconsin‐Madison). Next‐generation sequencing for the yeast two‐hybrid assay was performed by the Biotechnology Center at UW‐Madison. Tomato protoplasts were generated with assistance from Stacy Anderson of Donna Fernandez's Lab (University of Wisconsin‐Madison). Confocal microscopy was performed at the Newcomb Imaging Center at UW‐Madison. Nicotiana glutinosa seeds were obtained from the U.S. Nicotiana Germplasm Collection donated by the University of California‐Berkeley. R.K. is supported by a National Institutes of Health National Research Service Award T32 GM007215. This work was supported by a United States Department of Agriculture (USDA) Hatch award (WIS01818) to M.K.
Kessens R, Sorensen N, Kabbage M. An inhibitor of apoptosis (SfIAP) interacts with SQUAMOSA promoter‐binding protein (SBP) transcription factors that exhibit pro‐cell death characteristics. Plant Direct. 2018;2:1–17. 10.1002/pld3.81
This manuscript was previously deposited as a preprint at https://doi.org/10.1101/267435
REFERENCES
- Afgan, E. , Baker, D. , van den Beek, M. , Blankenberg, D. , Bouvier, D. , Cech, M. , … Goecks, J. (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Research, 44, W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allocati, N. , Masulli, M. , Di Ilio, C. , & De Laurenzi, V. (2015). Die for the community: An overview of programmed cell death in bacteria. Cell Death & Disease, 6, e1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, T. , Stone, J. M. , Heard, J. E. , Kovtun, Y. , Yorgey, P. , Sheen, J. , & Ausubel, F. M. (2000). Fumonisin B1‐induced cell death in arabidopsis protoplasts requires jasmonate‐, ethylene‐, and salicylate‐dependent signaling pathways. Plant Cell, 12, 1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai, K. K. , Atamian, H. S. , Kaloshian, I. , & Eulgem, T. (2010). WRKY72‐type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene‐for‐gene resistance mediated by the tomato R gene Mi‐1. The Plant Journal, 63, 229–240. [DOI] [PubMed] [Google Scholar]
- Birkenbihl, R. P. , Jach, G. , Saedler, H. , & Huijser, P. (2005). Functional dissection of the plant‐specific SBP‐domain: Overlap of the DNA‐binding and nuclear localization domains. Journal of Molecular Biology, 352, 585–596. [DOI] [PubMed] [Google Scholar]
- Bolger, A. M. , Lohse, M. , & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerio, R. J. , Vandergaast, R. , & Friesen, P. D. (2010). Host insect inhibitor‐of‐apoptosis SfIAP functionally replaces baculovirus IAP but is differentially regulated by Its N‐terminal leader. Journal of Virology, 84, 11448–11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman, M. B. , Park, Y. K. , Oltersdorf, T. , Li, W. , Clemente, T. , & French, R. (2001). Abrogation of disease development in plants expressing animal antiapoptotic genes. Proceedings of the National Academy of Sciences of the United States of America, 98, 6957–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K. W. , Haag, J. R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K. , & Pikaard, C. S. (2006). Gateway‐compatible vectors for plant functional genomics and proteomics. The Plant Journal, 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Eichmann, R. , & Schafer, P. (2012). The endoplasmic reticulum in plant immunity and cell death. Frontiers in Plant Science, 3, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltham, R. , Khan, N. , & Silke, J. (2012). IAPS and ubiquitylation. IUBMB Life, 64, 411–418. [DOI] [PubMed] [Google Scholar]
- Franco‐Zorrilla, J. M. , López‐Vidriero, I. , Carrasco, J. L. , Godoy, M. , Vera, P. , & Solano, R. (2014). DNA‐binding specificities of plant transcription factors and their potential to define target genes. Proceedings of the National Academy of Sciences, 111, 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, Y. , & Steller, H. (2011). Programmed cell death in animal development and disease. Cell, 147, 742–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, H. , Brandizzi, F. , Benning, C. , & Larkin, R. M. (2008). A membrane‐tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 105, 16398–16403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist, D. G. (1997). Mycotoxins reveal connections between plants and animals in apoptosis and ceramide signaling. Cell Death and Differentiation, 4, 689–698. [DOI] [PubMed] [Google Scholar]
- Glenn, A. E. , Zitomer, N. C. , Zimeri, A. M. , Williams, L. D. , Riley, R. T. , & Proctor, R. H. (2008). Transformation‐mediated complementation of a FUM gene cluster deletion in Fusarium verticillioides restores both fumonisin production and pathogenicity on maize seedlings. Molecular Plant‐Microbe Interactions, 21, 87–97. [DOI] [PubMed] [Google Scholar]
- Govrin, E. M. , & Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Current Biology, 10, 751–757. [DOI] [PubMed] [Google Scholar]
- Gyrd‐Hansen, M. , & Meier, P. (2010). IAPs: From caspase inhibitors to modulators of NF‐kappaB, inflammation and cancer. Nature Reviews Cancer, 10, 561–574. [DOI] [PubMed] [Google Scholar]
- Heller, J. , & Tudzynski, P. (2011). Reactive oxygen species in phytopathogenic fungi: Signaling, development, and disease. Annual Review of Phytopathology, 49, 369–390. [DOI] [PubMed] [Google Scholar]
- Hoang, T. M. L. , Williams, B. , Khanna, H. , Dale, J. , & Mundree, S. G. (2014). Physiological basis of salt stress tolerance in rice expressing the antiapoptotic gene SfIAP. Functional Plant Biology, 41, 1168. [DOI] [PubMed] [Google Scholar]
- Hsu, F. C. , Chou, M. Y. , Chou, S. J. , Li, Y. R. , Peng, H. P. , & Shih, M. C. (2013). Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. Plant Cell, 25, 2699–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Li, J. , Bao, F. , Zhang, X. , & Yang, S. (2010). A gain‐of‐function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature. Plant Physiology, 154, 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, Y. , Fedoroff, N. V. , & Koizumi, N. (2008). Arabidopsis bZIP60 is a proteolysis‐activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell, 20, 3107–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, Y. , & Koizumi, N. (2005). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proceedings of the National Academy of Sciences of the United States of America, 102, 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage, M. , Kessens, R. , Bartholomay, L. C. , & Williams, B. (2017). The life and death of a plant cell. Annual Review of Plant Biology, 68, 375–404. [DOI] [PubMed] [Google Scholar]
- Kabbage, M. , Li, W. , Chen, S. , & Dickman, M. B. (2010). The E3 ubiquitin ligase activity of an insect anti‐apoptotic gene (SfIAP) is required for plant stress tolerance. Physiological and Molecular Plant Pathology, 74, 351–362. [Google Scholar]
- Kabbage, M. , Williams, B. , & Dickman, M. B. (2013). Cell death control: The interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLOS Pathogens, 9, e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessens, R. , Ashfield, T. , Kim, S. H. , & Innes, R. W. (2014). Determining the GmRIN4 requirements of the soybean disease resistance proteins Rpg1b and Rpg1r using a nicotiana glutinosa‐based agroinfiltration system. PLoS ONE, 9, e108159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K. S. , Min, J. Y. , & Dickman, M. B. (2008). Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Molecular Plant‐Microbe Interactions, 21, 605–612. [DOI] [PubMed] [Google Scholar]
- Kroemer, G. , Galluzzi, L. , Vandenabeele, P. , Abrams, J. , Alnemri, E. S. , Baehrecke, E. H. , … Melino, G. (2009). Classification of cell death: Recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death and Differentiation, 16, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme, C. , & Santa, Cruz. S. (1999). Bax‐induced cell death in tobacco is similar to the hypersensitive response. Proceedings of the National Academy of Sciences of the United States of America, 96, 7956–7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, Z. , Vinod, K. , Zheng, Z. , Fan, B. , & Chen, Z. (2008). Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biology, 8, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. , & Salzberg, S. L. (2012). Fast gapped‐read alignment with Bowtie 2. Nature Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J. D. , Wan, J. , Ford, R. , Gong, Y. , Fung, P. , Nahal, H. , … Guttman, D. S. (2012). Quantitative interactor screening with next‐generation sequencing (QIS‐Seq) identifies Arabidopsis thaliana MLO2 as a target of the Pseudomonas syringae type III effector HopZ2. BMC Genomics, 13, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Kabbage, M. , & Dickman, M. B. (2010). Transgenic expression of an insect inhibitor of apoptosis gene, SfIAP, confers abiotic and biotic stress tolerance and delays tomato fruit ripening. Physiological and Molecular Plant Pathology, 74, 363–375. [Google Scholar]
- Liang, X. , Nazarenus, T. J. , & Stone, J. M. (2008). Identification of a consensus DNA‐binding site for the Arabidopsis thaliana SBP domain transcription factor, AtSPL14, and binding kinetics by surface plasmon resonance. Biochemistry, 47, 3645–3653. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Harberd Nicholas, P. , & Fu, X. (2016). SQUAMOSA promoter binding protein‐like transcription factors: Targets for improving cereal grain yield. Molecular Plant, 9, 765–767. [DOI] [PubMed] [Google Scholar]
- Liu, J. X. , Srivastava, R. , Che, P. , & Howell, S. H. (2007). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane‐associated transcription factor, bZIP28. Plant Cell, 19, 4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang, J. , Kidarsa, T. , Bradford, C. S. , Gilbert, B. , Curtis, M. , Tzeng, S. C. , … Wolpert, T. J. (2012). Tricking the guard: Exploiting plant defense for disease susceptibility. Science, 338, 659–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, K. , Tor, M. , Poole, M. , Hong, Y. , Thompson, A. J. , King, G. J. , … Seymour, G. B. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP‐box transcription factor inhibits tomato fruit ripening. Nature Genetics, 38, 948–952. [DOI] [PubMed] [Google Scholar]
- McHale, L. , Tan, X. , Koehl, P. , & Michelmore, R. W. (2006). Plant NBS‐LRR proteins: Adaptable guards. Genome Biology, 7, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirocha, C. J. , Gilchrist, D. G. , Shier, W. T. , Abbas, H. K. , Wen, Y. , & Vesonder, R. F. (1992). AAL toxins, fumonisins (biology and chemistry) and host‐specificity concepts. Mycopathologia, 117, 47–56. [DOI] [PubMed] [Google Scholar]
- Muro, I. , Hay, B. A. , & Clem, R. J. (2002). The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. Journal of Biological Chemistry, 277, 49644–49650. [DOI] [PubMed] [Google Scholar]
- Nagashima, Y. , Mishiba, K. , Suzuki, E. , Shimada, Y. , Iwata, Y. , & Koizumi, N. (2011). Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Scientific Reports, 1, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B. K. , Cai, X. , & Nebenfuhr, A. (2007). A multicolored set of in vivo organelle markers for co‐localization studies in Arabidopsis and other plants. The Plant Journal, 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Noutoshi, Y. , Ito, T. , Seki, M. , Nakashita, H. , Yoshida, S. , Marco, Y. , … Shinozaki, K. (2005). A single amino acid insertion in the WRKY domain of the Arabidopsis TIR‐NBS‐LRR‐WRKY‐type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. The Plant Journal, 43, 873–888. [DOI] [PubMed] [Google Scholar]
- Padmanabhan, M. S. , Ma, S. , Burch‐Smith, T. M. , Czymmek, K. , Huijser, P. , & Dinesh‐Kumar, S. P. (2013). Novel positive regulatory role for the SPL6 transcription factor in the N TIR‐NB‐LRR receptor‐mediated plant innate immunity. PLoS Pathogens, 9, e1003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish, A. B. , Freel, C. D. , & Kornbluth, S. (2013). Cellular mechanisms controlling caspase activation and function. Cold Spring Harbor Perspectives in Biology, 5, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, J. C. , & Hileman, L. C. (2013). Functional evolution in the plant SQUAMOSA‐promoter binding protein‐like (SPL) gene family. Frontiers in Plant Science, 4, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, M. , Link, T. I. , Muller, M. , Hirschburger, D. , Pudake, R. N. , Pedley, K. F. , … Whitham, S. A. (2016). A small cysteine‐rich protein from the Asian soybean rust fungus, phakopsora pachyrhizi. Suppresses plant immunity. PLoS Pathogens, 12, e1005827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan, A. , Jayaraman, D. , Grau, C. , Hill, J. H. , Whitham, S. A. , Ane, J. M. , … Kabbage, M. (2017). The pathogenic development of Sclerotinia sclerotiorum in soybean requires specific host NADPH oxidases. Molecular Plant Pathology, 19, 700–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team (2016). RStudio: Integrated development for R. Boston, MA: RStudio, Inc. [Google Scholar]
- Ruifrok, A. C. , & Johnston, D. A. (2001). Quantification of histochemical staining by color deconvolution. Analytical and Quantitative Cytology and Histology, 23, 291–299. [PubMed] [Google Scholar]
- Sakamoto, M. , Tada, Y. , Nakayashiki, H. , Tosa, Y. , & Mayama, S. (2005). Two phases of intracellular reactive oxygen species production during victorin‐induced cell death in oats. Journal of General Plant Pathology, 71, 387–394. [Google Scholar]
- Salinas, M. , Xing, S. , Hohmann, S. , Berndtgen, R. , & Huijser, P. (2012). Genomic organization, phylogenetic comparison and differential expression of the SBP‐box family of transcription factors in tomato. Planta, 235, 1171–1184. [DOI] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , … Cardona, A. (2012). Fiji: An open‐source platform for biological‐image analysis. Nature Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L. , Bielawski, J. , Mu, J. , Dong, H. , Teng, C. , Zhang, J. , … Zuo, J. (2007). Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Research, 17, 1030–1040. [DOI] [PubMed] [Google Scholar]
- Slabaugh, E. , & Brandizzi, F. (2011). Membrane‐tethered transcription factors provide a connection between stress response and developmental pathways. Plant Signaling & Behavior, 6, 1210–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, J. M. , Liang, X. , Nekl, E. R. , & Stiers, J. J. (2005). Arabidopsis AtSPL14, a plant‐specific SBP‐domain transcription factor, participates in plant development and sensitivity to fumonisin B1. The Plant Journal, 41, 744–754. [DOI] [PubMed] [Google Scholar]
- Trapnell, C. , Roberts, A. , Goff, L. , Pertea, G. , Kim, D. , Kelley, D. R. , … Pachter, L. (2012). Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nature Protocols, 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen, A. M. , Coulson, E. J. , & Vaux, D. L. (2001). Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biology, 2, REVIEWS3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , & Wang, H. (2015). The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Molecular Plant, 8, 677–688. [DOI] [PubMed] [Google Scholar]
- Williams, B. , Kabbage, M. , Kim, H.‐J. , Britt, R. , & Dickman, M. B. (2011). Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLOS Pathogens, 7, e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F.‐H. , Shen, S.‐C. , Lee, L.‐Y. , Lee, S.‐H. , Chan, M.‐T. , & Lin, C.‐S. (2009). Tape‐Arabidopsis Sandwich – a simpler Arabidopsis protoplast isolation method. Plant Methods, 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki, K. , Kigawa, T. , Seki, M. , Shinozaki, K. , & Yokoyama, S. (2013). DNA‐binding domains of plant‐specific transcription factors: Structure, function, and evolution. Trends in Plant Science, 18, 267–276. [DOI] [PubMed] [Google Scholar]
- Yang, Z. T. , Wang, M. J. , Sun, L. , Lu, S. J. , Bi, D. L. , Sun, L. , … Liu, J. X. (2014). The membrane‐associated transcription factor NAC089 controls ER‐stress‐induced programmed cell death in plants. PLoS Genetics, 10, e1004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S. D. , Cho, Y. H. , & Sheen, J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nature Protocols, 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


