Abstract
Over the last decade, redundant entry of data in electronic medical records (EMR) for health care and electronic data capture (EDC) systems for research has been the typical medical research methodology. The corresponding data transcription this methodology requires not only increases the burden for clinician investigators and clinical research coordinators (CRCs), but it also decreases the quality of data. We designed and developed a new standards‐based and platform‐independent system to use data in the EMR to directly populate clinical data management systems in the EDC to eliminate the need for data transcription, streamline the clinical research process, and reduce clinician burden. Standardized structured medical information eXchange2 (SS‐MIX2) was implemented along with the Integrating the Healthcare Enterprise (IHE) Retrieve Form for Data Capture (RFD) Integration Profile. Standards from Clinical Data Interchange Standards Consortium (CDISC) were used to define metadata for research data collection forms and as a means to standardize data exchange semantics. These standards and the associated methodology were applied to observational research in patients with diabetes mellitus. The system we developed complies with global requirements for regulated research. It provides a standard‐based and platform‐independent method that can serve to accelerate the cycle of a learning health system.
Keywords: 21 CFR Part 11, CDISC, CSV, EMR, IHE RFD, SS‐MIX2
1. INTRODUCTION
Redundant entry of data from electronic medical records (EMR) to electronic data capture (EDC) systems is the typical methodology when EMR and EDC systems are separately operated in clinical research. This process of transcription increases the burden for investigators and clinical research coordinators (CRCs) at research sites and decreases the quality of data.
Reuse of clinical data directly from EMR for clinical research has already been done in many universities and organizations,1 but is still limited to specific EMR/EDC systems combinations. Typically, when changes are necessary in the data items to be collected or in the systems design, the burden falls on the EMR vendors to make the changes because the EDC entry form is usually incorporated as a function of EMR system. These methods are not practical for regulated research on investigational new drugs because the entry form is built into the EMR system. It is impossible to validate the total system to make it compliant with applicable regulatory requirements and regulations, including 21 CFR Part 11.
To address these barriers, we have designed a system to simultaneously use EMR and EDC without “building in” the EDC form yet enabling auto‐population of this form from EMR data without transcription. This can be done by using the Integrating the Healthcare Enterprise (IHE) Integration Profile, Retrieve Form for Data Capture (RFD).2 This is a global standard that enables data collection from EMR with seamless data transfer to EDC systems. Separating the entry form from EMR systems by using RFD makes it possible to conduct computerized system validation (CSV) that is required in 21 CFR Part 11. Vendor neutral and platform‐independent EMR/EDC systems paired with international standards have been highly recommended to resolve the current barriers to streamlining global research and formed the basis for this project.
Standardized structured medical information eXchange2 (SS‐MIX2) Standardized Storage and SS‐MIX2 Annex Storage were used as the export data from EMR (Figure 1).3 , 4 The SS‐MIX project was promoted by Japan's Ministry of Health, Labor and Welfare (MHLW) and was initially part of the Shizuoka Style EMR project in 2006FY.4 According to investigations completed by MHLW in 2015FY,5 EMR systems were operating in 2542 hospitals (34%) out of 7426 hospitals in Japan, and SS‐MIX2 Standardized Storage was being implemented in 865 hospitals (34% of the hospitals with operational EMR systems). Confining these metrics to 710 hospitals with more than 400 beds, EMR systems were operating in 550 hospitals (78%), and SS‐MIX Standardized Storage in 237 hospitals (43% of hospitals with EMR). “SS‐MIX2 Standardized Storage: explanation of the structure and guidelines for implementation Ver. 1.2”6 was authorized as the standard specification of MHLW7 on March 28, 2016. Six hundred and thirty hospitals in Japan are storing prescription orders and laboratory examination results in HL7 v2.5 format using SS‐MIX2 Standardized Storage (Figure 2). Thus, the SS‐MIX2 specification is the de facto standard for the export from EMR in Japan.
Figure 1.

Description of the schematic structure of standardized structured medical information eXchange2 (SS‐MIX2) Standardized Storage and Annex Storage
Figure 2.

Geographical distribution of 630 hospitals (red circles) in Japan, which store prescription orders, laboratory examination results in HL7 v2.5 format in standardized structured medical information eXchange2 (SS‐MIX2) Standardized Storage
SS‐MIX2 Standardized Storage stores data such as patient demographics, prescription orders, laboratory examination results, and diagnostic disease classification in ICD‐10. Hamamatsu University Hospital then developed and implemented an EMR “Stamp” on EMR and used this for data entry by investigators to record clinical data and also for clinical research data, which are stored in SS‐MIX2 Annex Storage in case card form.
In this study, we designed and developed a new system to use data in EMR for data within the clinical data management system (CDMS) in an EDC system by using SS‐MIX2 Standardized Storage, Annex Storage using IHE RFD Profile. We also used Clinical Data Interchange Standards Consortium (CDISC) Standards to define metadata used for entry forms and to standardize data exchange,8, 9, 10 Specifically, we designed this system for observational clinical research on patients with type 2 diabetes mellitus. We adopted metadata from the Diabetes Therapeutic Area User Guide v1.0, which CDISC developed through the Coalition For Accelerating Standards and Therapies (CFAST) as a Therapeutic Area (TA) Standard in 2014. We also collected cardiovascular events defined in the CDISC Cardiovascular Studies Therapeutic Area User Guide v1.0.
We converted data collected from the EMR “Stamp” and SS‐MIX2 Standardized Storage into CDISC Clinical Data Acquisition Standards Harmonization (CDASH) format and also employed the CDISC Operational Data Model (ODM) transport format to apply to various types of clinical research such as disease registries and data to be included in submissions to regulatory authorities (eg, US FDA and Japan's PMDA) for approval of new therapies.
Our goal was to develop an ecosystem for clinical research that would eliminate the aforementioned technical challenges and transcription burden. Our ecosystem would increase the efficiency of data entry, eliminate transcription, facilitate source data verification, and improve the quality of data collected (reused from EMR) for research for the purpose of accelerating a learning health system cycle.
2. METHODS
A diagram of the total system developed for this observational clinical research study of patients with type 2 diabetes mellitus (UMIN‐CTR Number: UMIN000031987) is shown in Figure 3.
Figure 3.

Diagram of the total system. Data flow from the electronic medical record (EMR) “Stamp” in the EMR network in hospital to the Translational Research Center for Medical Innovation (TRI) System in the TRI network in hospital via router is shown. Data flow is also shown among the four actors of the Healthcare Enterprise (IHE) Retrieve Form for Data Capture (RFD) profile, ie, form filler, form archiver, form manager, and form receiver in TRI network in hospital and TRI Data Center. The scope for computerized system validation (CSV) is indicated within the yellow dashed line
EMR “Stamp” (Figure 4) has been implemented on the EMR at Hamamatsu University Hospital and used for data entry by investigators to input data such as findings and events of patients during each clinical session. Data entered in EMR “Stamp” were sent and stored in SS‐MIX2 Annex Storage in the EMR Network in hospital. Patient demographics, prescription orders, and laboratory examination results data were sent from EMR to SS‐MIX2 Standardized Storage. The Translational Research Center for Medical Innovation (TRI) System in the TRI Network in the hospital received the data stored in SS‐MIX2 Standardized Storage and Annex storage via a router, which connected and separated the EMR Network and the TRI Network in the hospital. Data in the TRI System were sent electronically to eClinical Base (eCB), an EDC system operating in the TRI Data Center. The IHE RFD Profile was implemented, ensuring that the scope for computerized system validation (CSV) would not include the EMR itself. Four actors in IHE RFD Profile—Form Filler, Form Archiver, Form Manager, and Form Receiver—communicated with each other in accordance with the profile to facilitate the data exchange between the TRI Network in the hospital and the TRI Data Center.
Figure 4.
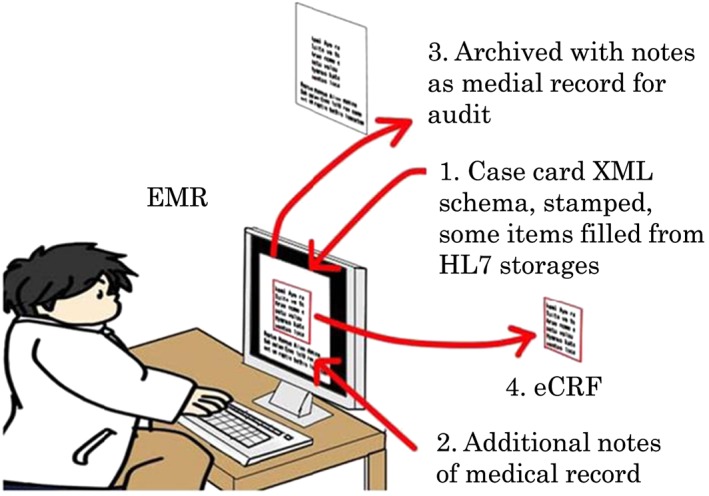
Schematic method of electronic medical record (EMR) “Stamp” on the EMR and how this methodology saves investigators from redundant entry of data
We developed three EMR “Stamps” and used them to enter the following:
Minimal items in the ordinary clinical sessions for patients with type 2 diabetes mellitus such as weight, blood pressure, total cholesterol, blood glucose, HbA1C, urinary albumin, and urinary creatinine; this was called as “Minimum Data Set” (Figure 5);11
Cardiovascular events such as death, transient ischemic attack and stroke, myocardial infarction, percutaneous coronary intervention, and peripheral vascular intervention, which are defined as cardiovascular endpoints in the CDISC Cardiovascular Studies Therapeutic Area User Guide v1.0 (Figure 6);
Side effects or adverse events that occurred in patients with type 2 diabetes mellitus (Figure 7).
Figure 5.

Electronic medical record (EMR) “Stamp” for “Minimum Data Set” from patients with type 2 diabetes mellitus on the EMR. An investigator is required to complete data entry at every patients clinical visits
Figure 6.

Electronic medical record (EMR) “Stamp” for cardiovascular events of patients with type 2 diabetes mellitus on the EMR; this is used only when cardiovascular event occurs
Figure 7.

Electronic medical record (EMR) “Stamp” for side effects experienced by patients with type 2 diabetes mellitus on the EMR; this is used only when a side effect occurs
To minimize duplicate entries by investigators, EMR data were used simultaneously for medical chart completion and for investigational purposes such as case card contents. The data collected via the EMR “Stamp” was sent and stored in SS‐MIX2 Annex Storage and was stored in EMR concurrently after data entry; confirmation of the data was done by investigators. The TRI System was specified to receive the data stored in SS‐MIX2 Annex Storage via the router. Thus, the TRI System acted as the EDC in this case.
SS‐MIX2 Standardized Storage and Annex Storage were constructed in accordance with “SS‐MIX2 Standardized Storage explanation of the structure and guidelines for implementation Ver. 1.2” and data such as patient demographics, disease classifications, prescription orders, and laboratory examination results were replicated in real time in Hamamatsu University Hospital.
The schematic structure of SS‐MIX2 Standardized Storage and Annex Storage is shown in Figure 1. Standardized Storage and Annex Storage use the same directory structures of hierarchized folders and files by the file management system that is generally utilized in a computer operating system to perform filing and storing of the medical care information. In subordinate of the Standardized Storage root folder (shown in red) is the “patient ID folder” (shown in blue). (To prevent response deterioration due to storage of multiple folders in the root folder data from a large number of patients registered in the medical institution is stored in Standardized Storage.) The patient ID is separated by three digits to be hierarchized in three levels. In subordinate of “patient ID folder,” the following two types of folders are arranged:
Folder that contains patient's basic information (folder name is shown to be “‐”(hyphen));
Medical treatment date folder (folder name is shown to be medical treatment date), which falls under medical information to be stored
In subordinate of the “Medical treatment date folder” is the “Data type folder.” Under the “Data type folder,” the data file that stores various standardized medical information messages described in HL7 Ver 2.5 is stored in “Standardized Storage” and nonstandardized medical information, such as JPEG and CDA files, is stored in “Annex Storage.”
The SS‐MIX2 Standardized Storage and Annex Storage were implemented in the EMR Network in the hospital. The TRI System received patient demographics, prescription orders, and laboratory examination results data stored in SS‐MIX2 Standardized Storage and data in EMR “Stamp,” which were sent and stored in Annex Storage via the router, which connected and separated the EMR Network and the TRI Network in the hospital.
The TRI System acquired data from SS‐MIX2 Standardized Storage and Annex Storage and created new forms to display via the EMR screen. TRI System utilized forms previously defined in eCB, the EDC system developed by TRI leveraging the CDISC ODM + (Style) format. Data were converted to CDISC ODM format, a zip file was created, and its HASH value was shown on the screen. The zip file was stored in “Form Archiver” and sent to eCB in the TRI Data Center electronically after confirmation by investigators. An optional system structure was specified to strengthen the security of EMR when the TRI System sent data to eCB in TRI Data Center electronically. Both the online and offline data transfer could be accommodated, between the TRI Network in the hospital and electronically as shown in Figure 3. The HASH value was shown in eCB and source data verification was simplified by confirming the HASH values shown in the TRI System and eCB.
The system developed in this study is comprises three networks as shown in Figure 3. EMR and EMR “Stamp”, SS‐MIX2 Standardized Storage and Annex Storage in EMR on the left were set up in the closed EMR Network in the hospital. “Form Filler” (whose role is to receive data from EMR “Stamp” via SS‐MIX2 Annex Storage and send them to “Form Receiver” and “Form Archiver”) was set up in the central TRI Network in the hospital. These two networks were connected and separated by a router. “Form Manager” (which sent forms to “Form Filler” and “Form Receiver”) was set up in the TRI Data Center.
We were able to exclude the EMR from the scope of computerized system validation (CSV) required by the regulation, 21 CFR Part 11, by using IHE RFD Integration Profile and by making Form Archiver the eSource. The scope of the CSV is shown in Figure 3 as the area defined by the yellow dotted line.
3. RESULTS
The principal theme of the system developed in this study is to use data in the EMR directly for research (EDC) to decrease duplication of data input by investigators and to increase the quality of data by eliminating the need for transcription. We used CDISC standards and the IHE RFD Integration Profile and adhered to the CSV requirements in compliance with regulations, specifically 21 CFR Part 11. Thus, the system developed in the present study could be used for studies to be included in submissions to regulatory authorities. We used the SS‐MIX2 Standardized Storage and Annex Storage, which are standard specifications acknowledged by MHLW and popularly used for exporting data from EMR in Japan.
In this study, we specifically used a newly developed system for an observational clinical research study of adult patients with type 2 diabetes mellitus to prove the feasibility. Patient data stored in the EMR were collected by the use of EMR “Stamp,” SS‐MIX2 Standardized Storage, and Annex Storage. We used the Diabetes Therapeutic Area User Guide v1.0 as a reference to define clinical elements to be collected for this study.
EMR “Stamps” as described in the previous section were developed and implemented in Hamamatsu University Hospital. It was confirmed that the required clinical information could be entered and provided in the format of a research electronic case report form (eCRF) via the EMR without reentry. EMR “Stamp” was able to store not only clinical information but also data for clinical research in EMR along with the audit trail to ensure data provenance using the SS‐MIX2 Annex Storage. Provenance (ie, an audit trail) is required in order to adhere to 21CFR11.
In this study, we were able to use EMR “Stamps” when investigators entered data for clinical research in EMR. At the initial registry of a patient, the investigator performed eligibility evaluation based on the description in the research protocol, and informed consent was obtained from the patient evaluated to be eligible. The TRI System registered the patient, and a patient identification number for the study was issued when the informed consent was agreed. This was done to pseudonymize the data for research purposes since patient identifiers are not brought into the EDC database. Investigators completed data entry when the patient visited Hamamatsu University Hospital for a clinical session.
Figure 5 shows the EMR “Stamp,” which investigators are required to complete by entering appropriate data when a type 2 diabetes mellitus patient presents for a clinical visit. Investigators entered the minimal data items at the ordinary visits for patients with type 2 diabetes mellitus, these included weight and blood pressure. Other data items from laboratory research results, such as total cholesterol, blood glucose, HbA1C, urinary albumin, and urinary creatinine were directly imported from SS‐MIX2 Standardized Storage. The number of data items was minimized to reduce the burden of investigators by designing two other research‐relevant EMR “Stamps” for cardiovascular events and side effects or adverse events, which are important indications of the change of condition of patients. Investigators entered data for these two EMR “Stamps” only when these events occurred. Figure 6 shows the EMR “Stamp” to enter cardiovascular events. Investigators used this EMR “Stamp” to enter cardiovascular events such as death, transient ischemic attack and stroke, myocardial infarction, percutaneous coronary intervention, and peripheral vascular intervention, which are defined as cardiovascular endpoints in CDISC Cardiovascular Studies Therapeutic Area User Guide v1.0. This EMR “Stamp” was also used to indicate dialysis introduction.
Figure 7 shows the EMR “Stamp” used when side effects occurred. We used a dropdown list for the selection of side effects previously foreseen and described in the protocol and also prepared for free text input for other side effects as a precaution.
Data of these EMR “Stamps” were stored in the EMR and sent to the TRI System via SS‐MIX2 Annex Storage. Investigators were able to use these data for their own research by exporting them in CSV format. Figure 8 shows an excel sheet of the time series of the “Minimum Data Set” of a specific patient with type 2 diabetes mellitus. Investigators were able to use this sheet both for medical practice and clinical research, which removes the burden of reentry of data for research. This, in turn, should generate motivation for investigators to participate in clinical research by integrating research into their clinical care workflow.
Figure 8.

Excel sheet of the time series data collected by electronic medical record (EMR) “Stamp” for “Minimum Data Set” of a specific patient with type 2 diabetes mellitus on EMR. An investigator is able to see this excel sheet anytime in the EMR for the clinical practice
Figure 9 shows an example of SS‐MIX2 Standardized Storage output received by the TRI System. These data were messages of clinical information described in HL7 Version 2.5 format that adhere to “SS‐MIX2 Standardized Storage: explanation of the structure and guidelines implementation Ver. 1.2.”
Figure 9.

Example of stored data in standardized structured medical information eXchange2 (SS‐MIX2) Standardized Storage, which was acquired by Translational Research Center for Medical Innovation (TRI) System. These messages in HL7 Version 2.5 were stored under the “Data type folder” in standardized structured medical information eXchange2 (SS‐MIX2) Standardized Storage
Patient demographics, medical history and laboratory research results, prescription orders, hypoglycemic event, and cardiovascular events/outcomes and adverse events stored in EMR were collected via the EMR “Stamps” and SS‐MIX2 Standardized Storage and Annex Storage, and the TRI System was able to receive these data.
Figures 10, 11, and 12 show forms used for confirmation of items in the “Minimum Data Set.” Specifically, Figure 10 shows confirmation of items in the “Minimum Data Set” in the TRI system collected by the EMR “Stamp” for Minimum Data Set, with the exception of laboratory results, which were directly imported into the system. These data were stored in SS‐MIX2 Annex Storage. Figures 11 and 12 are confirmation for data used from that stored data in SS‐MIX2 Standardized Storage during the last clinical session and the current session. The Form Filler of IHE RFD Profile was composed of these three forms, and the two additional forms for cardiovascular event and side effects.
Figure 10.

Confirmation form for items in the “Minimum Data Set” in TRI System collected by electronic medical record (EMR) “Stamp” for “Minimum Data Set”; note that laboratory results are not included
Figure 11.

Confirmation form for prescription orders in Translational Research Center for Medical Innovation (TRI) System collected from standardized structured medical information eXchange2 (SS‐MIX2) Standardized Storage
Figure 12.

Confirmation form for laboratory examination results in Translational Research Center for Medical Innovation (TRI) System collected from standardized structured medical information eXchange2 (SS‐MIX2) Standardized Storage
The forms in Figures 10, 11, and 12 were used only for data confirmation. If an investigator found incorrect data and made a decision that revision of data was necessary, the investigator had the capability within this system to revise the data by using EMR “Stamp” and/or enter order(s) again to export prescription orders and laboratory examination results to SS‐MIX2 Standardized Storage.
Once an investigator confirmed the accuracy of the collected data through the confirmation forms in Figures 10, 11, and 12, the data were converted to CDISC ODM format. Their zip file was created, and its HASH value was calculated by HASH function of JAVA using the Secure Hash Algorithm (SHA)‐256 developed by the National Security Agency (NSA). The HASH value of the zip file was displayed on the screen of the TRI System. The zip file was stored in “Form Archiver” and sent to eCB in the TRI Data Center electronically after confirmation by the investigators. We confirmed that the zip file was sent successfully from TRI System in the hospital to eCB in the TRI Data Center. The HASH value of the zip file, which eCB received, was calculated by the same HASH function used in the TRI System and shown on the screen of eCB. We confirmed that the HASH values shown on the screens of TRI System in the hospital and eCB in TRI Data Center were identical. This showed that data in TRI System and eCB were the same and simplified the source data verification (SDV) by confirming the HASH values shown in TRI System and eCB.
This process, which was vendor neutral and platform‐independent, enabled Form Archiver to keep the audit trails since all revisions of data by the investigator were stored in Form Archiver.
4. DISCUSSION
Collection of clinical data from the EMR for reuse in clinical research is one of the key capabilities that will enable learning health systems.12
We developed a vendor neutral, standards‐based, and platform‐independent system that meets regulatory requirements and enables seamless data transfer from EMR to EDC systems. Our system was developed specifically for use in observational research of patients with type 2 diabetes mellitus; however, it is readily applicable to other areas of research by designing EMR “Stamps” based upon the research data required for those therapeutic areas. In this project, forms were designed to be populated with minimum data points from EMR systems and additional research data related to cardiovascular events and side effects. The standards implemented were the IHE RFD Integration Profile, SS‐MIX2, SS‐MIX Annex and CDISC CDASH, and ODM. We were able to comply with global regulatory requirements for clinical research, excluding the EMR from the scope of CSV required by 21 CFR Part 11 by implementing the vendor‐neutral and platform‐independent IHE RFD Integration Profile, which is a global standard for data collection from EMR and seamless data transfer to EDC.
As data are directly imported from SS‐MIX2 Standardized Storage and are automatically transferred to the EDC‐CDMS, redundant data entry is no longer necessary. Further, data quality is maintained by eliminating the need for transcription. There is no need to reenter the source data and no source data verification (SDV) is required as it is automatically conducted by comparison of HASH values in the TRI System and eCB, the EDC system. These process improvements are accompanied by reduced burden on the clinical investigator or site personnel. This should, in turn, increase interest in clinical research and expand the patient resource pool for research studies. Elimination of SDV should also help to minimize manual data‐cleaning work and thus contribute to cost reduction for clinical research.
Nordo et al13 demonstrated that the use of the IHE RFD Integration Profile as a part of the Epic EHR research model along with the REDCap EDC system and middleware (RADaptor) developed by the Duke University Office of Research Informatics produced significant time and resource savings and improved data quality. Specifically, this eSource pilot for a registry study produced a 37% time savings and required one fewer full‐time employee while the error rate was reduced from 9% to 0%. For the Duke study, data elements mapped with RADaptor included those contained within the EHR's continuity of care document (CCD), a standard used to comply with initial US meaningful use requirements. Data elements not in CCD could be entered anew into the eCRF.
The system developed for this project cannot be applied to every study because there is a limit in the data from SS‐MIX2 Standardized Storage; namely, they are limited to patient demographics, prescription orders, laboratory examination results, and disease classifications. However, it is immediately applicable to simpler protocols and certainly holds the potential to give better cost‐benefit performances, particularly in large cohort studies or postmarketing surveillance. Because this system is standards‐based and platform‐independent, hospitals with such standards in place can quickly participate in a new clinical study. Use of the CDISC ODM transport standard with CDASH metadata also readily supports multicenter research studies around the globe, even if the data are based upon CCD vs SS‐MIX2., An enabling global ecosystem for multicenter collaborative research will be created as the number of hospitals with the ability to produce research data in a standard format grows.
In addition, wider adoption of standards‐based systems will improve the quality of clinical practice and help maintain the quantity and quality of clinical research as the burden of investigators is reduced and more clinicians participate in research. This will reduce cost, and the time necessary to conduct clinical research and promote the development of new therapies, medical devices, and therapeutic methods. Further, this readily supports the acceleration of learning health cycles and learning health systems.
Understanding the efficiencies and efficacy of this system will promote the implementation of SS‐MIX2 Standardized Storage and Annex Storage with IHE RFD Integration Profile and data standards in more hospitals. This could contribute to regional health information organizations, restoration of data in a disaster, and actualize the stated vision of CDISC in 2011: “Informing patient care and safety through higher quality medical research.”
We hope that the implementation of SS‐MIX2 Standardized Storage and Annex Storage in hospitals will become more popular, not only in Japan but also globally, in the US, Europe, and Asia. Harmonization of storage standards for EMR data and global use of the standards‐based system as described in this report is feasible and could accelerate learning health systems.
5. CONCLUSION
We developed a vendor neutral, standards‐based, and platform‐independent EMR‐EDC system for use in an observational study on type 2 diabetes mellitus. The system leverages a number of available standards: SS‐MIX2 Standardized Storage, SS‐MIX2 Annex Storage, IHE RFD Integration Profile, and CDISC standards (CDASH, SDTM, ODM, and TA‐specific standards). We developed the design of the EMR‐EDC system to make it possible to eliminate redundant entry of data in the EMR and the EDC system and to exclude EMR from the scope of CSV required by 21 CFR Part 11. Thus, it adheres to global regulatory requirements for research while fitting nicely into the workflow of a busy clinician. The system will make it possible to improve the quality of clinical research while maintaining the quality of clinical care, enabling clinicians to more easily participate in important research that can provide additional learning from a larger pool of research participants. This, in turn, can accelerate learning health cycles supporting global learning health systems.
CONFLICT OF INTEREST
The following individuals have no conflicts of interest to declare:
Dr Takenouchi, Mr Yuasa, Mr Shioya, Prof Hiroshi Watanabe, Dr Oki, Dr Akio Hakamata, Dr Hiroshi Watanabe, and Dr Fukushima. Prof Kimura declares that he receives research funds from NEC Corporation and Fujifilm Corporation.
Takenouchi K, Yuasa K, Shioya M, et al. Development of a new seamless data stream from EMR to EDC system using SS‐MIX2 standards applied for observational research in diabetes mellitus. Learn Health Sys. 2019;3:e10072 10.1002/lrh2.10072
REFERENCES
- 1. De Moor G, Sundgren M, Karla D, et al. Using electronic health records for clinical research: The case of the EHR4CR project. J Biomed Inform. 2015;53:162‐173. [DOI] [PubMed] [Google Scholar]
- 2. Integrating the Healthcare Enterprise (IHE): IT infrastructure technical framework . Retrieve Form for Data Capture (RFD). 2017. https://wiki.ihe.net/index.php/Retrieve_Form_for_Data_Capture
- 3. Kimura M, Tani S, Sakusabe T. Towards Japanese EHR; Shizuoka Style EMR Project, Deployment Stage, CJKMI2005, Proceedings 4‐5, Shenzhen,Chaina, Nov. 10, 2005.
- 4. Kimura M, Nakayasu K, Ohshima Y, et al. SS‐MIX: A ministry project to promote standardized healthcare information exchange. Methods Inf Med. 2011;50(2):131‐139. [DOI] [PubMed] [Google Scholar]
- 5. Ministry of Health, Labor and Welfare: Investigations of Medical Sites . 2016. http://www.mhlw.go.jp/toukei/list/79‐1.html
- 6. Japan Association for Medical Informatics . Oct. 2014. https://www.jami.jp/english/about/
- 7. Ministry of Health, Labor and Welfare: Standard Specifications for Healthcare Information . 2016. http://www.mhlw.go.jp/file/06‐Seisakujouhou‐10800000‐Iseikyoku/0000118987.pdf
- 8. Kush R. CDISC & the world of health information technology. Appl Clin Trials. 2007;16(2):64‐70. [Google Scholar]
- 9. Carol R, Kush R, Helton E. Saving time and money. Appl Clin Trials. 2007;16(6):70‐74. [Google Scholar]
- 10. Kush R, Fukushima M, Takenouchi K, Nagai Y, Jono T, Kojima S. History, current status and future scope of CDISC as the global standards. Clin Eval. 2012;39(3):547‐557. [Google Scholar]
- 11. Nakashima N, Tajima N. Determination and standardization of self‐management items of lifestyle‐related diseases as diabetes mellitus by clinical associations. Endocrinol Diabetes Metab. 2014;38(6):573‐580. [Google Scholar]
- 12. Friedman CP, Wong AK, Blumenthal D. Achieving a nationwide learning health system. Sci Transl Med. 2010;2(57):29. [DOI] [PubMed] [Google Scholar]
- 13. Nordo AH, Eisenstein E, Hawley J, et al. A comparative effectiveness study of eSource used for data capture for a clinical research registry. Int J Med Inform. July 2017;103:89‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]


