Abstract
The recruitment of TATA binding protein (TBP) to gene promoters is a critical rate-limiting step in transcriptional regulation for all three eukaryotic RNA polymerases. However, little is known regarding the dynamics of TBP in live mammalian cells. In this report, we examined the distribution and dynamic behavior of green fluorescence protein (GFP)-tagged TBP in live HeLa cells using fluorescence recovery after photobleaching (FRAP) analyses. We observed that GFP-TBP associates with condensed chromosomes throughout mitosis without any FRAP. These results suggest that TBP stably associates with the condensed chromosomes during mitosis. In addition, endogenous TBP and TBP-associated factors (TAFs), specific for RNA polymerase II and III transcription, cofractionated with mitotic chromatin, suggesting that TBP is retained as a TBP-TAF complex on transcriptionally silent chromatin throughout mitosis. In interphase cells, GFP-TBP distributes throughout the nucleoplasm and shows a FRAP that is 100-fold slower than the general transcription factor GFP-TFIIB. This difference supports the idea that TBP and, most likely, TBP-TAF complexes, remain promoter- bound for multiple rounds of transcription. Altogether, our observations demonstrate that there are cell cycle specific characteristics in the dynamic behavior of TBP. We propose a novel model in which the association of TBP-TAF complexes with chromatin during mitosis marks genes for rapid transcriptional activation as cells emerge from mitosis.
INTRODUCTION
In eukaryotic cells, the three RNA polymerases I, II, and III are dedicated to the transcription of distinct classes of genes. Distinct promoter architectures and the assembly of polymerase-specific initiation complexes at gene promoters are keys that dictate the recruitment of the particular class of polymerases. TATA binding protein (TBP) interacts with a variety of TBP-associated factors (TAFs) to form the selectivity factor-1 (SL1), transcription factor TFIID, and TFIIIB complexes that are important for specifying RNA polymerase I, II, and III transcription, respectively (Hernandez, 1993). TBP-TAF complexes are critical players in determining levels of transcription initiation. Thus, the formation of specific TBP-TAF complexes potentially regulates transcription of specific genes under different growth conditions. Increasing the recruitment of these complexes to gene promoters by regulatory proteins is one mechanism for transcriptional activation (Albright and Tjian, 2000; Hampsey and Reinberg, 1999; Hernandez, 1993; Lee and Young, 1998). Once recruited to a promoter, TBP-TAF complexes can perform additional functions that are important for transcriptional regulation, including recruitment of additional members of the general transcriptional machinery to the promoter, induction of conformational changes in DNA topology, and recruitment of coactivator or corepressor proteins that influence gene transcription (reviewed in Tansey and Herr, 1997).
The various TBP-TAF complexes have been purified and characterized extensively in vitro and in vivo; however, little is known regarding the dynamics of TBP in live mammalian cells. In this report, we describe the dynamics of TBP throughout the cell cycle in live HeLa cells by examining the distribution and mobility of green fluorescent protein (GFP)-tagged TBP as measured by fluorescence recovery after photobleaching (FRAP) analysis. FRAP analysis involves photobleaching an area containing fluorescently-tagged molecules and measuring the level and rate of fluorescence recovery as fluorescent molecules outside the photobleached zone migrate into this area. In this way, a measure of the ability of a fluorescent molecule to move over time can be determined. Using GFP-TBP and GFP-TFIIB as surrogate markers for endogenous TBP and TFIIB respectively, we observed that GFP-TBP and GFP-TFIIB are located in transcriptionally active interphase nuclei and that the rate of fluorescence recovery after photobleaching for GFP-TBP is 100-fold slower than that for GFP-TFIIB, suggesting that TBP remains chromatin-bound for multiple rounds of transcription, while TFIIB cycles on and off promoters. In addition, GFP-TBP specifically associates with condensed chromosomes throughout all stages of mitosis. There is no fluorescence recovery after photobleaching, suggesting that active recruitment of GFP- TBP to gene promoters does not occur after cells enter mitosis. Our data indicate that a significant number of genes may preload TBP-TAF complexes before entering mitosis, allowing these genes to be primed for transcriptional initiation as cells enter the next cell cycle.
MATERIALS AND METHODS
Plasmids
Open reading frames encoding TBP and TFIIB were cloned into pEGFP-C1 (Clontech, Palo Alto, CA) to generate the expression vectors pEGFP-TBP and pEGFP-TFIIB, respectively. Expression constructs were sequenced to confirm that they contained the correct cDNA sequence.
Protein Extractions
GFP-TBP and GFP-TFIIB were expressed by transient transfection of pEGFP-TBP or pEGFP-TFIIB into HeLa cells as described previously (Chen and Huang, 2001). Proteins were extracted 24–30 h posttransfection by incubating cells in CSK buffer (100 mM NaCl, 300 mM sucrose, 10 mM PIPES (pH 6.8), 3 mM MgCl2, 1 mM PMSF) on ice for 5 min. Supernatants from this low salt extraction were collected, and the cell pellet was further extracted with CSK buffer containing 250 mM NaCl to generate the higher salt extraction fraction. The remaining pellet was resuspended in Laemmli sample buffer and used as the extraction-resistant fraction. Equivalent amounts of total protein from each fraction were analyzed by Western blot analysis.
Coimmunoprecipitations
Whole-cell extracts from HeLa cells transiently transfected with GFP-TBP were prepared by sonication of cells in lysis buffer containing 10 mM Tris-HCl (pH 8), 150 mM NaCl, 20% glycerol, 1 mM EDTA, 0.1% NP40, 1 mM DTT, 0.5 mM PMSF, 1 mM benzamidine, and 1 mM sodium metabisulfite. Fifty microliters HeLa whole-cell extract (6 μg protein/ul) were incubated for 1 h at 4°C with either 2 μl anti-BRF polyclonal (Mital et al., 1996) or 2 μg normal rabbit (Santa Cruz) antibodies in 500 μl total volume of lysis buffer. Twenty microliters protein G agarose (Upstate Biotechnology, Lake Placid, NY) were then added, and reactions were incubated 1 h at 4°C. The beads were washed four times with 500 μl PBS and resuspended in 25 μl Laemmli sample buffer. Proteins were separated by SDS- PAGE, transferred to Hybond ECL nitrocellulose membrane (Amersham, Arlington Heights, IL), and analyzed by Western blot analysis using monoclonal antibodies directed against GFP (Clontech).
Characterization of Chromatin Fractions
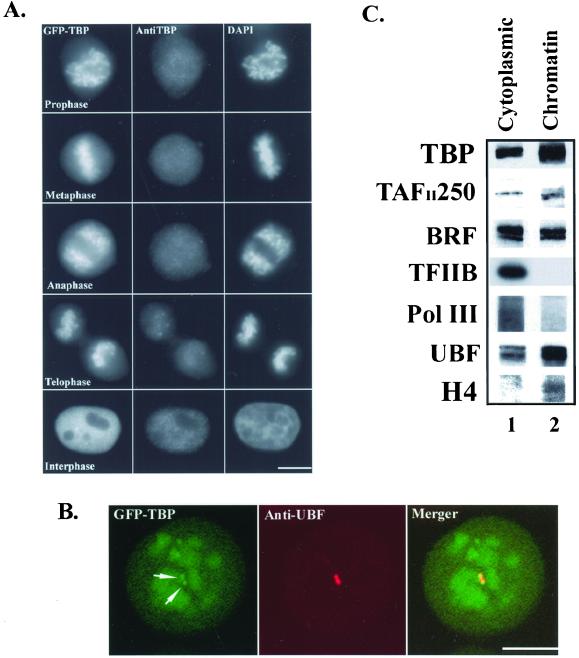
The isolation and purification of mitotic chromatin were performed as described (Valdivia, 1998). HeLa cells were treated with nocodazole (500 ng/ml, Sigma-Aldrich, St. Louis, MO) for ∼ 16 h. Mitotic cells were then incubated in 75 mM KCl on ice for 20 min and subsequently in disruption buffer (10 mM Tris-HCl (pH 7.4), 120 mM KCl, 20 mM NaCl, 0.1% Triton X-100, 2 mM CaCl2, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 0.5 μg/ml pepstatin A and 0.1 mM PMFS) for 10 min. The cells were homogenized and the resultant extract centrifuged at 3000g for 15 min at 4°C. The supernatant was used as the cytoplasmic fraction. The pellet, which is enriched in crude chromosomes, was further purified by centrifugation in a 36-ml linear gradient consisting of 20–60% wt/vol sucrose in disruption buffer. Fractions containing chromatin were pooled and centrifuged at 2500g for 10 min at 4°C. The chromatin pellet was washed, sonicated, and resuspended in 1X Laemmli buffer. Proteins present in the chromatin and cytoplasmic fractions were separated by SDS-PAGE for Western blot analyses using antibodies directed against TBP, TAFII250, TFIIB, UBF, BRF, and the RNA polymerase III large subunit. The results shown in Figure 2 were generated by reprobing the same nitrocellulose membrane with the different primary antibodies as indicated. Three chromosome purification experiments were performed, which all produced similar results.
Figure 2.
TBP-TAF complexes involved in RNA polymerase II and III transcription are associated with mitotic chromatin. (A) GFP-TBP fluorescence (left panels) exhibits a labeling pattern similar to that for DAPI staining (right panels) throughout mitosis in GFP-TBP transfected cells. Immunolabeling of the same cells using anti-TBP antibodies (middle panels) does not reveal an obvious chromatin-labeling pattern. The bar represents 10 μm. (B) Confocal microscopic analyses of prophase cells show a heterogenous fluorescence pattern of GFP-TBP that is signficantly more concentrated at NORs (left panel, arrows), identified by anti-UBF immunolabeling (middle panel), then other regions of chromosomes. The bar represents 10 μm. (C) Western blot analyses of mitotic cytoplasmic (lane 1) and purified mitotic chromatin (lane 2) fractions demonstrate that endogenous TBP, TAFII250, and BRF are associated with mitotic chromatin. Both histone H4 and UBF1 are also enriched in the mitotic chromatin fractions, as expected. However, both TFIIB and the RNA polymerase III large subunit are overwhelmingly enriched in the mitotic cytoplasmic fractions.
FRAP Calculation
The relative fluorescence intensity (RFI) at each time point was calculated similarly as described by Misteli and Phair (Phair and Misteli, 2000). RFI = (It/TNt)/(I0/TN0), where It= the average fluorescence intensity of the photobleached region at various time points after photobleaching, TNt= the average fluorescence intensity of the entire nucleus at the corresponding time point, I0= the average fluorescence intensity of the photobleached region before photobleaching, and TN0= the average fluorescence intensity of the entire nucleus before photobleaching. When It/TNt = I0/TN0, namely, when RFI = 1, the fluorescence recovery of the photobleached region reaches 100%.
RESULTS
GFP-TBP and Endogenous TBP Behave Similarly
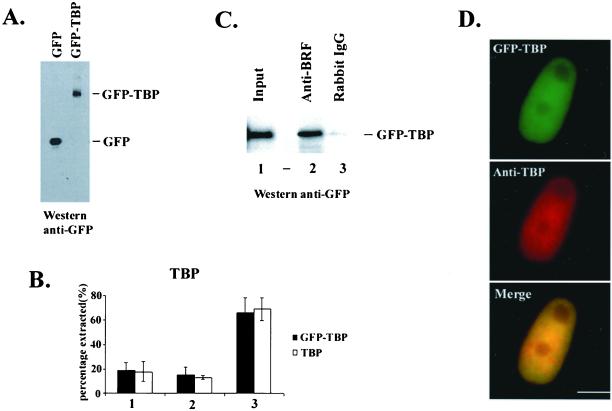
To determine whether the behavior of GFP-tagged TBP is similar to that of endogenous TBP in transiently transfected HeLa cells, we performed the following experiments. First, the expression of GFP-TBP was analyzed by Western blot analyses using anti-GFP antibodies 24–30 h posttransfection. A single band with the expected mobility of the full-length fusion protein was detected in extracts from GFP-TBP transfected cells, but not from GFP transfected cells (Figure 1A). The ability of GFP-TBP to be extracted from transfected cells treated with Triton X-100 and two concentrations of salt was also compared with endogenous TBP. Results from these experiments demonstrate that the proportion of GFP-TBP and TBP extracted at each salt condition is nearly identical (Figure 1B), suggesting that GFP-TBP and TBP are similar in their association with chromatin or other cellular complexes that contribute to their extractability in these assays. Furthermore, we determined whether GFP-TBP is assembled into TBP-TAF complexes in transfected cells using coimmunoprecipitation assays. As shown in Figure 1C, using antibodies directed against BRF, a component of the TBP-containing TFIIIB complex that is required for polymerase III transcription, GFP-TBP was coimmunoprecipitated from nuclear extracts. This association was specific because GFP-TBP was not detected in immunoprecipitations using nonspecific rabbit antibodies. Thus, the N-terminal GFP tag did not inhibit the assembly of GFP-TBP into a TBP-TAF complex. The subcellular localization of GFP-TBP in transfected cells was also examined by immunolabeling using anti-TBP antibodies. The overlapping of fluorescent signals from anti-TBP antibodies (Figure 1D, middle panel) and GFP-TBP (Figure 1D, top panel) demonstrates that GFP-TBP and TBP localize to the same nucleoplasmic region (Figure 1D, bottom panel). Furthermore, the intensity of anti-TBP immunolabeling was indistinguishable between cells that express relatively low concentrations of GFP-TBP and nontransfected cells, indicating that cellular TBP levels were not grossly perturbed by exogenous GFP-TBP expression (unpublished data). These low levels of GFP-TBP expression mimic native physiological conditions, and these conditions were used for all FRAP experiments described below. Altogether, the above experiments demonstrated that GFP-TBP and endogenous TBP behave similarly, and therefore, GFP-TBP can faithfully serve as a marker for the endogenous TBP in live cells.
Figure 1.
GFP-TBP behaves similarly as endogenous TBP in transiently transfected HeLa cells. (A) GFP (left lane) or GFP-PTB (right lane) proteins are expressed as full-length polypeptides with the expected mobility as detected by Western blot analysis using anti- GFP antibodies (Clontech). (B) Similar proportions of GFP- TBP and endogenous TBP are extracted from GFP-TBP transfected HeLa cells when treated with 0.1% Triton X-100 and either 150 mM NaCl (1) or 250 mM KCl (2) and are present in the insoluble cell pellet after extraction (3). The amounts of each extracted protein were analyzed by Western blot using antibodies directed against TBP, and the proportions extracted were calculated by comparing the level of extracted protein to the level of each protein present before extraction. (C) GFP-TBP is functional for complex assembly into TFIIIB. Whole cell extracts were prepared from HeLa cells transfected with GFP- TBP, and immunoprecipitations were performed using anti- TFIIIB (anti-BRF, Mital and Hernandez, 1996) or nonspecific antibodies (lanes 2 and 3, respectively). Western blot analysis was performed using antibodies directed against GFP. (D) GFP-TBP (top) and total cellular TBP (middle) localize to similar nuclear regions in GFP-TBP transfected cells, as indicated in the merged image (bottom). Endogenous TBP and GFP-TBP were detected by immunolabeling using anti-TBP antibodies. The bar represents 10 μm.
TBP-TAF Complexes Are Associated with Mitotic Chromatin
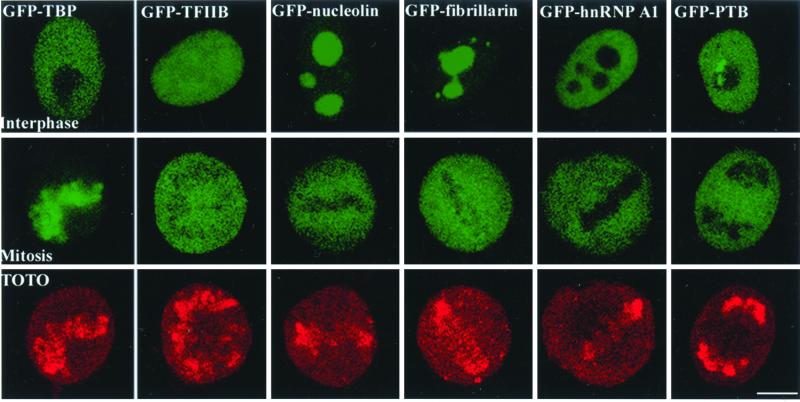
Active transcription of eukaryotic genes during interphase is rapidly silenced as cells enter mitosis. Several models explaining this transcriptional inhibition include phosphorylation of general transcription factors and polymerases, activation of general repressor proteins, and prevention of factor access to DNA by chromosome condensation (Gottesfeld and Forbes, 1997; Kornberg and Lorch, 1999). Any of these activities could change the behavior of TBP-TAFs. Thus, using fluorescence microscopy, we were interested in examining the behavior of GFP-TBP as cells transited from a transcriptionally active to inactive state during mitosis. When cells entered prophase, the fluorescence pattern of GFP-TBP (Figure 2A, left top panel) was remarkably similar to the DAPI-staining pattern in the same cells (Figure 2A, right top panel). This finding indicates that a significant proportion of GFP-TBP remains chromosome- bound as cells enter mitosis. Surprisingly, very little GFP- TBP or endogenous TBP was detected on mitotic chromosomes when the same cells were immunolabeled using anti-TBP antibodies (Figure 2A, middle top panel), which is consistent with a previously reported observation (Segil et al., 1996). In fact, there is no difference in the anti-TBP immunofluorescence pattern seen in GFP-TBP transfected cells and nontransfected cells during mitosis (unpublished data). This observation suggests that the reactive epitope in TBP is compacted into condensed chromosomes precluding detection by antibodies. The association of GFP-TBP with chromatin is specific to TBP, as neither GFP alone nor other GFP-tagged nucleic acid binding proteins such as GFP-TFIIB, GFP-nucleolin, GFP- fibrillarin, GFP-hnRNP A1, and GFP-hnRNP I (PTB) were detected on mitotic chromosomes (Figure 3). Chromatin association of GFP-TBP persists throughout anaphase and telophase. Altogether, our findings implicate that a subpopulation of cellular TBP remains chromatin-bound during mitosis and that chromatin compaction does not displace TBP from DNA. Furthermore, upon close examination using confocal microscopy, we observed a heterogeneous fluorescence pattern of GFP-TBP on mitotic chromatin (Figure 2B). Interestingly, the fluorescence of GFP-TBP is stronger at the nuclear organization regions (NORs) that contain rDNA repeats (Figure 2B, arrows) compared with other chromosomal regions. Each NOR contains ∼ 200 tandem copies of rDNA and is, therefore, one of the more gene dense chromosome segments. Because the SL1 complex associates with NORs throughout mitosis (Jordan et al., 1996; Roussel et al., 1996; Grummt, 1999), the higher concentration of GFP-TBP at NORs suggests that GFP- TBP binds DNA specifically at promoter sequences rather than associating with DNA nonspecifically.
Figure 3.
GFP-TBP is associated with mitotic chromatin whereas other DNA or RNA binding proteins, including GFP-TFIIB, -nucleolin, -fibrillarin, -hnRNP A1, and –hnRNP I (PTB), are not associated with mitotic chromatin, as examined by confocal laser scanning microscopy. In fact, they are excluded from regions where mitotic chromatin localized. Top panels demonstrate the localization pattern of corresponding GFP tagged proteins in interphase nuclei. Middle panels show the localization pattern of these proteins in mitotic cells, and bottom panels show the TOTO staining of the same mitotic cells to demonstrate the location of mitotic chromatin. The bar represents 10 um.
To determine whether endogenous TBP and other transcription factors are also associated with mitotic chromatin, cell extracts prepared from nocodozole-synchronized metaphase HeLa cells were examined by Western blot analyses. Cell extracts were fractionated into mitotic cytoplasmic and chromatin fractions, and the chromatin fraction was further purified using sucrose gradient centrifugation. To evaluate the integrity of the chromatin fraction, we tested for the presence of UBF, a polymerase I specific transcription factor, and for histone H4, a core histone. Previous studies have shown that the polymerase I transcription machinery, including SL1, UBF, and RNA polymerase I, associates with chromatin at NORs during mitosis (Jordan et al., 1996; Roussel et al., 1996; Grummt, 1999), and that H4 is a constituent of chromatin (Aalfs and Kingston, 2000). As shown in Figure 2C, a subpopulation of UBF and the majority of H4 were detected in the chromatin fraction, confirming that the chromatin fraction is indeed chromatin- enriched. The cytoplasmic and chromatin fractions were then examined for TBP and TBP-associated proteins using antibodies directed against TBP and two TBP- TAFs, TAFII250 (polymerase II-specific) and BRF (polymerase III-specific). Both TAFs and TBP were detected in the chromatin fraction suggesting that a significant proportion of TFIID and TFIIIB are chromatin-associated during mitosis. We also tested for the presence of TFIIB and RNA polymerase III in the chromatin fraction, since DNA binding by TFIID allows subsequent recruitment of TFIIB (Zawel et al., 1995). For RNA polymerase III transcription, the polymerase is recruited to promoters containing prebound TFIIIB (Kassavetis et al., 1990). In contrast to the enrichment of TBP, TAFII250, and BRF in the chromatin fractions, both TFIIB and the RNA polymerase III large subunits were overwhelmingly enriched in the mitotic cytoplasmic fraction. Thus, as opposed to the polymerase I transcription system where transcription factors and the polymerase were associated with NORs, other general transcription factors and RNA polymerases did not necessarily associate with condensed chromosomes. Altogether, the above experiments demonstrate that TBP-TAF complexes involved in Pol II and Pol III transcriptions remain bound to condensed chromatin throughout mitosis.
GFP-TBP Does Not Exchange On and Off Mitotic Chromosomes
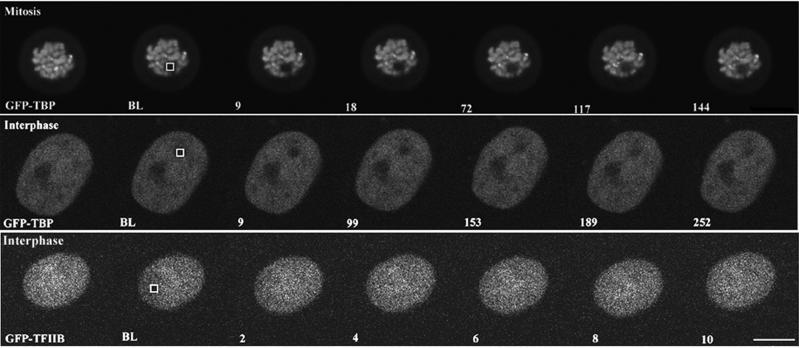
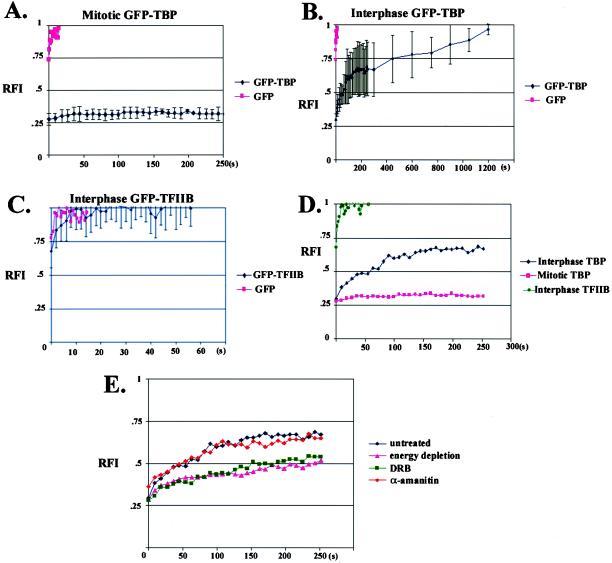
The recruitment of TBP- containing complexes to gene promoters is a crucial early step in preinitiation complex assembly, and controlling the rate of promoter occupancy by TBP is an integral component of transcriptional regulation (Albright and Tjian, 2000; Hampsey and Reinberg, 1999; Hernandez, 1993; Lee and Young, 1998). We were interested, therefore, in determining whether GFP-TBP exchanges on and off or remains statically associated with transcriptionally silent chromatin throughout mitosis. To do this, we used FRAP analysis (White and Stelzer, 1999), which involves photobleaching an area containing fluorescent-tagged molecules and measuring the level and rate of the fluorescence recovery as fluorescent molecules outside the photobleached zone migrate into this area. In this way, a measure of the ability of a fluorescent molecule to be replaced over time can be determined. FRAP analysis has been used to analyze the kinetics of chromatin binding proteins such as histones and has been shown to be a good approach for understanding the dynamics of chromatin- binding proteins in vivo (Dey et al., 2000; Lever et al., 2000; Misteli et al., 2000). HeLa cells transiently transfected with GFP-TBP were grown on glass-bottom dishes and mounted onto a Zeiss 510 scanning laser microscope (Oberkochen, Germany). A 2- μm2 area over mitotic chromosomes was photobleached, and a series of images were acquired at 9-s intervals immediately after bleaching. Subsequently, the relative intensity of fluorescence within the photobleached area was measured using the area density measurement tool of the Metamorph software (Universal Imaging, Media, PA). The fluorescence recovery, represented as RFI, was calculated similarly as previously described (Phair and Misteli, 2000). As shown in Figures 4 (top panel) and 5A, the mitotic chromosome-associated GFP-TBP did not show fluorescence recovery even at 20 min after bleaching, demonstrating that the photobleached GFP-TBP was not replaced by emission-competent GFP-TBP from within the mitotic cytoplasm throughout mitosis. The little fluorescence recovery for mitotic chromosome-associated GFP- TBP is in contrast to the rapid recovery observed in the mitotic cytoplasm for either GFP or GFP-TFIIB, both of which approach 100% within 2 s (unpublished data). These findings demonstrate that TBP does not exchange on and off chromatin and, therefore, is stable in its association with chromatin throughout mitosis.
Figure 4.
FRAP analyses of GFP-TBP and GFP- TFIIB during mitosis and interphase in live HeLa cells. HeLa cells transfected with GFP-TBP were photobleached either during mitosis (top panel) or during interphase (middle panel). HeLa cells were also transfected with GFP- TFIIB and then photobleached during interphase (bottom panel). The bar represents 10 μm.
GFP-TBP Exchanges On and Off Chromatin More Slowly than GFP-TFIIB in Interphase Cells
To determine the behavior of GFP-TBP in transcriptionally active cells, interphase nuclei were examined. GFP-TBP distributes throughout the nucleoplasm, with lower concentrations observed in the nucleolus. FRAP analyses revealed that the fluorescence recovery of GFP-TBP in interphase nuclei has a t1/2 of ∼ 1 min, with nearly 100% recovery observed at ∼ 20 min (Figure 4B). Thus, in contrast to what is observed in mitotic cells, the bleached GFP-TBP is replaceable with unbleached GFP-TBP in the nucleoplasm of interphase cells. The recovered fluorescence was not due to newly synthesized GFP-TBP because fluorescence recovery was not affected when protein synthesis was inhibited by cycloheximide treatment (unpublished data). In addition, the fluorescence recovery of GFP-TBP approached 100%, demonstrating that almost all GFP-TBP within the bleached region was replaceable and suggesting, therefore, that nearly all TBP exchanges on and off chromatin in interphase nuclei. The significantly slower fluorescence recovery for GFP-TBP, compared with GFP alone, is likely indicative of a slow TBP dissociation rate off chromatin (Figure 5B).
Figure 5.
Quantitative analyses of fluorescence recovery after photobleaching for GFP-TBP and GFP-TFIIB. The fluorescence recovery (y-axis, RFI, 100% recovery = 1) as a function of time (x-axis, seconds) is shown for GFP-TBP during mitosis (A) and interphase (B), and GFP-TFIIB during interphase (C). The fluorescence recovery within the 2 μm2 bleached zone was calculated (Materials and Methods) at various time points after photobleaching. Little fluorescence recovery is observed during mitosis for GFP-TBP (panel A). In interphase, the nucleoplasmic-distributed GFP-TBP shows fluorescence recovery approaching 100% within 20 min (panel B), which is 100-fold slower than that for GFP-TFIIB (panels C and D). The effects of transcriptional inhibition by α-amanitin, DRB, and energy depletion on fluorescence recovery of GFP-TBP (E) during interphase were examined.
To compare the dynamics of TBP with another general transcription factor involved in initiation complex formation for RNA polymerase II transcription, FRAP analyses was also performed for GFP-tagged TFIIB. We first determined whether GFP-TFIIB behaved similarly to endogenous TFIIB. Both localized to the nucleus and showed similar salt extraction properties from transfected cells (unpublished data). Furthermore, GFP-TFIIB coimmunoprecipitated with TFIIF from HeLa cells that were transfected with GFP-TFIIB, but not with GFP alone, demonstrating that GFP-TFIIB assembles into a complex containing TFIIF (unpublished data). These results suggest that the behavior of GFP-TFIIB and endogenous TFIIB are similar. Using FRAP analysis, we observed that, in marked contrast to GFP-TBP, the fluorescent recovery of GFP-TFIIB in the nucleoplasm is ∼100-fold more rapid, and fluorescence recovery reaches 100% within a few seconds after bleaching (Figure 4, and Figure 5, C and D). The significant difference in the rate of fluorescence recovery between GFP-TBP and GFP-TFIIB in live cells suggests that GFP-TBP and GFP-TFIIB may have different chromatin binding kinetics.
To address whether the fluorescence recovery of GFP-TBP or GFP-TFIIB is dependent on transcriptional activity, FRAP analyses were performed on transfected HeLa cells that were treated with transcription inhibitors including DRB, α- amanitin, and glucose analogues plus sodium azide. DRB is a kinase inhibitor that blocks TFIIH kinase activity, and prevents phosphorylation of the CTD from the large subunit of RNA polymerase II (Sehgal et al., 1976; Dubois et al., 1994), and affects a number of kinases (Mittleman et al., 1983; Zandomeni et al., 1986; Hidaka et al., 1991). α-Amanitin binds specifically to the RNA polymerase II large subunit and, at a higher concentration, to the polymerase III large subunit (Bartolomei and Corden, 1987). Treatment with glucose analogues plus sodium azide reduces ATP levels in cells. Each treatment was performed on transfected cells expressing GFP-TBP. The efficacy of transcription inhibition of the drugs was ensured by examining the redistribution of splicing factors that typically occurs during transcription inhibition (unpublished data). As shown in Figure 5E, both DRB and energy depletion significantly reduced the rate of fluorescence recovery for GFP-TBP. The reduction of fluorescence recovery for GFP-TBP in ATP-depleted cells suggests that one or more ATP- dependent processes influences the kinetics of TBP binding to chromatin. It is possible that phosphorylation, an ATP- dependent process, may regulate TBP cycling on and off chromatin, as supported by the reduction of GFP-TBP replacement in the presence of DRB, a kinase inhibitor. These observations are consistent with the model in which phosphorylation of transcription factors regulate preinitiation complex assembly. Interestingly, treatment with α-amanitin at levels that inhibit RNA polymerase II and III transcription did not affect the dynamics of GFP- TBP in the nucleoplasm (Figure 5E). This finding suggests that the exchange of GFP-TBP on and off chromatin is independent of RNA polymerization in cells.
DISCUSSION
TBP-TAFs Are Associated with Condensed Chromatin During Mitosis
Through direct visualization of GFP- TBP by immunofluorescence in live cells as well as the analysis of fractions biochemically enriched in chromatin, we demonstrate that TBP-TAFs are associated with condensed mitotic chromosomes. The association is likely specific, since the concentration of GFP-TBP is greater on the gene- dense NORs, compared to other chromosomal regions. In addition, the association is stable, as demonstrated by the absence of fluorescence recovery of GFP-TBP on mitotic chromosomes after photobleaching. These findings indicate that TBP-TAFs bind not only to transcriptionally active chromatin during interphase but also to highly condensed and transcriptionally inactive chromatin during mitosis. These findings are consistent with a recent study demonstrating that TBP-TAF complexes are associated with transcriptionally suppressed chromatin in yeast cells (Sekinger and Gross, 2001). However, in our study we did not observe active recruitment of TBP to chromatin during mitosis. Based on our findings, we propose a model addressing the relationship between TBP-TAFs and DNA at a transcriptionally silent stage of the cell cycle, in which TBP-TAF complexes remain bound to promoters after transcriptional silencing and are incorporated into the higher order chromatin structure during mitosis. This is consistent with previous observations that certain TAFs contain histone-like domains (Hoffmann et al., 1996; Xie et al., 1996), and that the central cavity of TFIID could structurally accommodate an entire nucleosome (Andel et al., 1999; Jacobson et al., 2000). When cells emerge from mitosis, the presence of prebound TBP-TAF complexes may allow the rapid activation of these promoters. However, we believe that the initial activation of promoters preoccupied by TBP- TAFs likely involves regulation at a step that is distinct from TBP-TAF recruitment. This model may help explain the apparent paradox regarding the ordered recruitment of histone acetyl transferases to promoters that may be inaccessible because of chromatin structure. An intriguing possibility is that the TAFII250 subunit of a prebound TFIID complex is readily available to modify chromatin via histone acetylation or phosphorylation, which induces structural changes in chromatin, thus facilitating the recruitment of other chromatin modifying proteins or ATP- dependent chromatin remodeling machines. This hypothesis does not contradict the conventional model where TFIID and TFIIIB are recruited to promoters in the early stages of initiation complex formation since GFP-TBP does exchange on and off chromatin in interphase nuclei.
GFP-TBP Exchanges On and Off Chromatin More Slowly than GFP-TFIIB in Interphase Cells
FRAP analyses of GFP-TBP in interphase nuclei demonstrate a complete fluorescence recovery in ∼ 20 min after photobleaching a 2-μm2 nuclear area. The time required for full recovery of GFP- TBP is at least 1000 times slower than GFP, demonstrating that the fluorescence recovery of GFP-TBP is most likely not the result of protein diffusion alone. In addition, the fluorescence recovery within the bleached area approached 100%, suggesting that nearly all the GFP-TBP within the 2- μm2 bleached area was replaced by emission-competent GFP-TBP from unbleached nuclear regions. In comparison with another basal transcription factor GFP- TFIIB, the complete fluorescence recovery of GFP-TBP is ∼100 times slower. Although the nonchromatin-associated proportion may not be the same for both GFP-TBP and GFP- TFIIB, it could not be the sole explanation for the 100- fold difference of FRAP between the two proteins. We interpret that this significant difference of FRAP contains information reflective of the difference in the on and off rate of chromatin-binding by by the GFP-TBP and GFP-TFIIB. This slower mobility of GFP-TBP is consistent with other kinetic studies demonstrating that TFIID dissociates from chromatin slowly (Burley, 1996). The significant difference in the rate of fluorescence recovery between GFP-TBP and GFP-TFIIB provides evidence in live cells, supporting that TFIID remains promoter-associated during transcription, whereas TFIIB dissociates during the transition from initiation to elongation (Van Dyke et al., 1988; Van Dyke et al., 1989; Zawel et al., 1995), and that TFIIB reassociates with TFIID, individually, to reform the RNA polymerase II docking site or as part of a holoenzyme (Zawel et al., 1995).
Summarily, we have analyzed the dynamic behavior of TBP in live mammalian cells for the first time, using GFP-TBP and FRAP analyses. We have shown that TBP-TAF complexes involved in RNA polymerase II and III transcription are associated with transcriptionally silent mitotic chromatin. This association may allow some promoters to be activated rapidly as cells emerge from mitosis.
ACKNOWLEDGMENTS
We thank Nouria Hernandez for antibodies to TFIIB, BRF, and RNA polymerase III large subunit; and Edward K.L. Cheng for anti-UBF antibody. We also thank Steve Adam, Andrew Belmont, Grace Chen, Joseph Gall, Tom Misteli, Tim Spann, and David Spector for their discussion and helpful comments during the preparation of this manuscript. This work was supported by grants from the National Cancer Institute of the National Institutes of Health (NIH) to S. Huang (1-R01-CA77560–01A1 and 5-K01-CA74988–03) and from NIH to R.W. Henry (1-R01-GM59805–01A2).
Abbreviations used:
- DRB
5,6-Dichloro- β-d- ribofuranosylbenzimidazole, FRAP, fluorescence recovery after photobleaching, GFP, green fluorescent protein, hnRNP I, heterogeneous nuclear ribonucleoprotein type I, NOR, nucleolar-organizing region, Pol I, RNA polymerase I, Pol II, RNA polymerase II, Pol III, RNA polymerase III, RFI, relative fluorescence intensity, SL1, selectivity factor, TAF, TBP-associated factor, TAFII250, TBP-associated factor 250, TBP, TATA- binding protein, TFIIB, transcription factor IIB, TFIID, transcription factor IID, TFIIIB, transcription factor IIIB, UBF, upstream binding factor
Footnotes
DOI:10.1091/mbc.01–10-0523.
Corresponding author. E- mail address: s-huang2@northwestern.edu
REFERENCES
- Aalfs JD, Kingston RE. What does “chromatin remodeling”mean? Trends Biochem Sci. 2000;25:548–555. doi: 10.1016/s0968-0004(00)01689-3. [DOI] [PubMed] [Google Scholar]
- Albright SR, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- Andel F, Ladurner AG, Inouye C, Tjian R, Nogales E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science. 1999;286:2153–2156. doi: 10.1126/science.286.5447.2153. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Corden JL. Localization of an alpha-amanitin resistance mutation in the gene encoding the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1987;7:586–594. doi: 10.1128/mcb.7.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK. The TATA box binding protein. Curr Opin Struct Biol. 1996;6:69–75. doi: 10.1016/s0959-440x(96)80097-2. [DOI] [PubMed] [Google Scholar]
- Chen D, Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol. 2001;153:169–176. doi: 10.1083/jcb.153.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois MF, Nguyen VT, Bellier S, Bensaude O. Inhibitors of transcription such as 5,6-dichloro-1- beta-D-ribofuranosylbenzimidazole and isoquinoline sulfonamide derivatives (H- 8 and H-7) promote dephosphorylation of the carboxyl-terminal domain of RNA polymerase II largest subunit. J Biol Chem. 1994;269:13331–13336. [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Grummt I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog Nucleic Acid Res Mol Biol. 1999;62:109–154. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. RNA polymerase II as a control panel for multiple coactivator complexes. Curr Opin Genet Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- Hidaka H, Watanabe M, Kobayashi R. Properties and use of H-series compounds as protein kinase inhibitors. Methods Enzymol. 1991;201:328–339. doi: 10.1016/0076-6879(91)01029-2. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Chiang CM, Oelgeschlager T, Xie X, Burley SK, Nakatani Y, Roeder RG. A histone octamer-like structure within TFIID. Nature. 1996;380:356–359. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–5142. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Jordan P, Mannervik M, Tora L, Carmo-Fonseca M. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Braun BR, Nguyen LH, Geiduschek EP. S. cerevisiaeTFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA. Regulation of gene expression by TBP-associated proteins. Genes Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- Lever MA, Th'ng JP, Sun X, Hendzel MJ. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- Mital R, Kobayashi R, Hernandez N. RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol Cell Biol. 1996;16:7031–7042. doi: 10.1128/mcb.16.12.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittleman B, Zandomeni R, Weinmann R. Mechanism of action of 5,6-dichloro-1-beta-D- ribofuranosylbenzimidazole. II. A resistant human cell mutant with an altered transcriptional machinery. J Mol Biol. 1983;165:461–473. doi: 10.1016/s0022-2836(83)80213-7. [DOI] [PubMed] [Google Scholar]
- Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Roussel P, Andre C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs. J Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segil N, Guermah M, Hoffmann A, Roeder RG, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- Sehgal PB, Darnell JE, Tamm I. The inhibition by DRB (5,6-Dichloro-1-β-D- ribofuranosylbenzimidazole) of hnRNA and mRNA production in HeLa cells. Cell. 1976;9:473–480. doi: 10.1016/0092-8674(76)90092-1. [DOI] [PubMed] [Google Scholar]
- Sekinger EA, Gross DS. Silenced chromatin is permissive to activator binding and pic recruitment. Cell. 2001;105:403–414. doi: 10.1016/s0092-8674(01)00329-4. [DOI] [PubMed] [Google Scholar]
- Tansy WP, Herr W. TAFs: guilt by association? Cell. 1997;88:729–732. doi: 10.1016/s0092-8674(00)81916-9. [DOI] [PubMed] [Google Scholar]
- Valdivia MM. Chromosome isolation for biochemical and morphological analysis. In: Spector DL, Goldman RD, Leinwand LA, editors. Cells: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. pp. 49.1–12. [Google Scholar]
- Van Dyke MW, Roeder RG, Sawadogo M. Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science. 1988;241:1335–1338. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]
- Van Dyke MW, Sawadogo M, Roeder RG. Stability of transcription complexes on class II genes. Mol Cell Biol. 1989;9:342–344. doi: 10.1128/mcb.9.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Stelzer E. Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol. 1999;9:61–65. doi: 10.1016/s0962-8924(98)01433-0. [DOI] [PubMed] [Google Scholar]
- Xie X, Kokubo T, Cohen SL, Mirza UA, Hoffmann A, Chait BT, Roeder RG, Nakatani Y, Burley SK. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- Zandomeni R, Zandomeni MC, Shugar D, Weinmann R. Casein kinase type II is involved in the inhibition by 5,6-dichloro-1-beta-D- ribofuranosylbenzimidazole of specific RNA polymerase II transcription. J Biol Chem. 1986;261:3414–3419. [PubMed] [Google Scholar]
- Zawel L, Kumar KP, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]