Abstract
Intercellular signaling mediated by small peptides is critical to coordinate organ formation in animals, but whether extracellular polypeptides play similar roles in plants is unknown. Here we describe a role in Arabidopsis leaf development for two members of the CLAVATA3/ESR‐RELATED peptide family, CLE5 and CLE6, which lie adjacent to each other on chromosome 2. Uniquely among the CLE genes, CLE5 and CLE6 are expressed specifically at the base of developing leaves and floral organs, adjacent to the boundary with the shoot apical meristem. During vegetative development CLE5 and CLE6 transcription is regulated by the leaf patterning transcription factors BLADE‐ON‐PETIOLE1 (BOP1) and ASYMMETRIC LEAVES2 (AS2), as well as by the WUSCHEL‐RELATED HOMEOBOX (WOX) transcription factors WOX1 and PRESSED FLOWER (PRS). Moreover, CLE5 and CLE6 transcript levels are differentially regulated in various genetic backgrounds by the phytohormone auxin. Analysis of loss‐of‐function mutations generated by genome engineering reveals that CLE5 and CLE6 independently and together have subtle effects on rosette leaf shape. Our study indicates that the CLE5 and CLE6 peptides function downstream of leaf patterning factors and phytohormones to modulate the final leaf morphology.
Keywords: Arabidopsis, CLE, leaf development, signaling, WOX
1. INTRODUCTION
Plants are unique in their ability to generate new organs and tissues throughout their life span, producing intricate structures such as flowers and leaves via complex molecular regulatory mechanisms (Bar & Ori, 2014; Tsukaya, 2013). During vegetative development, leaves initiate as small, regularly spaced primordia on the flanks of the shoot apical meristem. Following leaf initiation, individual primordia develop along three axes of polarity: the adaxial‐abaxial, proximal‐distal, and medial‐lateral axes. Early polarization along these three axes serves to specify the unique cell types within the emergent leaf. In simple‐leaved species, such as Arabidopsis, subsequent cell growth and differentiation then results in a mature three‐dimensional structure with a narrow petiole and a broad lamina, or blade (Kalve, De Vos, & Beemster, 2014). The blade tissue contains an epidermal layer of jigsaw‐shaped pavement cells bounded by several narrow layers of elongated cells at the margin, and is specialized for light capture. Yet despite the importance of leaves as the main sites for photosynthesis, as well as carbon fixation and gas exchange in plants (Tsukaya, 2013), much remains to be understood about the genetic mechanisms that control leaf formation, shape, and function.
The coordination of complex developmental activities such as leaf formation by growing plants is critically dependent on the communication of information between cells. Long‐range intercellular signaling is mediated by phytohormones such as cytokinin, auxin, gibberellin (GA), abscisic acid (ABA), and brassinosteroids (BL). Among these hormones, auxin, GA and BL have well‐characterized roles in orchestrating leaf development (Kalve et al., 2014). In addition to setting the positions of newly arising leaf primordia, auxin contributes to the establishment of leaf adaxial‐abaxial polarity as well as the coordinated transition from cell proliferation to cell expansion. GA and also BL regulate cell division and expansion during the leaf maturation process. These phytohormone‐mediated effects on leaf morphogenesis can occur via changes in hormone biosynthesis, as in the case of GA and BL, and/or in hormone transport or response, as in the case of auxin (Kalve et al., 2014; Tsukaya, 2013).
In addition to phytohormone signaling pathways, families of secreted signaling peptides are involved in regulating plant developmental events (Grienenberger & Fletcher, 2015; Matsubayashi, 2014). The CLAVATA3/EMBRYO SURROUNDING REGION‐related (CLE) family is one of the largest and best‐studied secreted signaling peptide families in plants. CLE family members are found throughout the plant kingdom as well as in some plant‐parasitic nematodes (Miyawaki, Tabata, & Sawa, 2013). The Arabidopsis genome encodes 32 CLE gene family members, which are expressed in a wide variety of tissues and developmental stages (Jun, Fiume, et al., 2010). The genes encode secreted 12‐ to 13‐amino‐acid mature polypeptides, derived from a conserved C‐terminal CLE domain, which undergo various posttranslational modifications (Matsubayashi, 2014).
Although intercellular signaling molecules are crucial for orchestrating plant growth and development, determining their biological activities has proven challenging. To date only a handful of Arabidopsis CLE family members have defined functions. The founding CLE family member CLAVATA3 (CLV3) acts in a negative feedback loop that regulates stem cell homeostasis in the shoot apical meristem (Brand, Fletcher, Hobe, Meyerowitz, & Simon, 2000; Schoof et al., 2000). In addition CLE40 plays a role in root stem cell homeostasis, and CLE41 and CLE44 function in vascular development and lateral root formation (Matsubayashi, 2014; Wang, Zhang, & Wu, 2016). Although a comprehensive library of CLE loss‐of‐function alleles now exists (Yamaguchi et al., 2017), plants carrying most single cle null alleles show no obvious developmental or physiological phenotypes (Jun, Fiume, et al., 2010). One possible explanation is that due to a high degree of sequence homology, many CLE genes play largely redundant roles in plant biology (Strabala et al., 2006).
Key components of CLE‐mediated signaling pathways are members of the WUSCHEL‐RELATED HOMEOBOX (WOX) family of homeodomain‐containing transcription factors (Haecker et al., 2004). Expression of the founding Arabidopsis WOX gene family member, WUSCHEL (WUS), is limited by CLV3 signaling to the most central region of the shoot apical meristem (Laux, Mayer, Berger, & Jurgens, 1996), where it functions in a non‐cell‐autonomous manner to maintain stem cell activity in the overlying cells (Brand, Grunewald, Hobe, & Simon, 2002; Schoof et al., 2000; Yadav et al., 2011). In roots, CLE40 likewise restricts the expression domain of WOX5 (Stahl, Wink, Ingram, & Simon, 2009), which acts non‐cell‐autonomously to promote columella stem cell maintenance (Sarkar et al., 2007). The identical CLE41 and CLE44 peptides induce WOX4 and WOX14 expression to promote vascular cell division (Etchells, Provost, Mishra, & Turner, 2013; Hirakawa, Kondo, & Fukuda, 2010). These examples suggest that the use of CLE‐WOX signaling modules during plant development is widespread.
Evidence is accumulating that CLE polypeptide signaling pathways intersect with classical phytohormone signaling pathways to direct various plant developmental processes (Wang et al., 2016). For example, the CLV3 pathway target WUS maintains shoot and floral meristem activity by directly regulating components of cytokinin response pathways (Leibfried et al., 2005). CLE40 controls the expression of auxin, cytokinin, and ABA signaling genes to inhibit cell differentiation in the root apical meristem (Pallakies & Simon, 2014). CLE6 and CLE41/44 peptide‐stimulated vascular cell proliferation is positively regulated by auxin, and exogenous CLE6 peptide application induces the expression of auxin signaling‐related promoters such as proPIN1:GUS in the hypocotyl stele (Whitford, Fernandez, De Groodt, Ortega, & Hilson, 2008). In addition, GA promotes CLE6 expression in the root stele, and CLE6 over‐expression can partly suppress the phenotypes of GA‐deficient plants (Bidadi et al., 2014). Based on such observations it has been proposed that CLE peptides may play general roles in controlling stem cell fate via their communication with plant hormone‐regulated signaling networks (Whitford et al., 2008).
Yet although there is increasing evidence that CLE peptide and phytohormone signaling pathways connect to regulate Arabidopsis growth and development, most studies to date have been conducted using root or vascular tissues (Wang et al., 2016). In contrast, beyond the role of CLV3 in shoot apical meristem maintenance, very little is known about the regulation or activity of CLE genes in shoot or shoot‐derived tissues. However, a good deal of work has focused on the regulation of leaf development, including the adaxial side adjacent to the SAM boundary as well as at the base of the developing floral organs (Ha, Jun, Nam, & Fletcher, 2004; Hepworth, Zhang, McKim, Li, & Haughn, 2005; Norberg, Holmlund, & Nilsson, 2005). Two closely related genes that encode transcriptional regulatory proteins, BLADE‐ON‐PETIOLE1 (BOP1) and BOP2, are expressed at the base of developing rosette leaves and have largely redundant functions in developing lateral organs, including suppressing ectopic blade outgrowth from the leaf petiole (Ha, Jun, Nam, & Fletcher, 2007; Ha et al., 2003). The BOP proteins activate the transcription of ASYMMETRIC LEAVES2 (AS2) in the proximal region of developing leaf primordia. AS2 encodes a member of the LATERAL ORGAN BOUNDARIES (LOB) family of leucine‐zipper proteins (Iwakawa et al., 2002; Lin, Shuai, & Springer, 2003; Xu et al., 2003) that physically interacts with the ARP domain transcription factor AS1 (Xu et al., 2003) to promote lateral organ identity and adaxial leaf polarity (Machida, Nakagawa, Kojima, Takahashi, & Machida, 2015). AS2 transcription occurs in developing leaf and floral primordia in a broad domain (Byrne et al., 2000; Iwakawa et al., 2007) that overlaps with those of BOP1 and BOP2.
Here we investigate the expression and function of the closely related CLE5 and CLE6 genes, which lie adjacent to one another on chromosome 2 and encode identical CLE polypeptides (Cock & McCormick, 2001). We show that CLE5 and CLE6 have highly specific, overlapping expression patterns at the base of lateral organ primordia. We find that, despite having a high degree of overall sequence similarity, they are differentially regulated during vegetative development by the BOP and AS2 genes, the WOX transcription factors PRS and WOX1, and by auxin. Using null alleles of CLE5 and CLE6 generated by genome editing, we demonstrate that although neither single nor double mutations in CLE5 and CLE6 have a detectable impact on Arabidopsis organ initiation or patterning, they have subtle effects on overall leaf shape. Our studies indicate that CLE5 and CLE6 act downstream of leaf patterning factors and phytohormones to direct formation of the final leaf morphology.
2. EXPERIMENTAL PROCEDURES
2.1. Plant material and growth conditions
Seeds were imbibed at 4°C for 5 days before sowing and plants were grown in Percival growth chambers at 21°C under long day conditions (16 hr light, 8 hr dark) with a light fluence rate of approximately 110 μmol m−2 s−1. Transgenic plants carrying the pCLE5:GUS or pCLE6:GUS constructs were generated as described (Jun, Fiume, et al., 2010), using 2,570 bp upstream of the CLE5 translation start site or 1,713 bp upstream of the CLE6 translation start site. Promoter alignments were performed using the EMBOSS Needle program for pairwise sequence alignment (Rice, Longden, & Bleasby, 2000). For the generation of the pBOP1:CLE6 bop1‐4 bop2‐11 lines, a 5797 bp BOP1 promoter fragment was PCR‐amplified using the primers pBOP1(‐5797) FW and pBOP1 RV (primers listed in Supporting Information Table S1). The CLE6 coding sequence was PCR‐amplified using the primers pBOP1:CLE6 FW and CLE6_CDS+Stop RV (Supporting Information Table S1). Both PCR products were gel‐purified and used as a template for a third PCR with the primers pBOP1(‐5797) FW and CLE6_CDS+Stop RV to generate a pBOP1:CLE6 product. The product was subcloned into the TOPO pCR8‐GW vector (ThermoFisher) and recombined using Gateway technology into the pEarley Gate 300 destination vector. The recombined vector was used for floral dip transformation with Agrobacterium tumefasciens GV3101. T1 transgenic plants were analyzed for CLE6 expression. All lines were in the Arabidopsis Columbia‐0 (Col‐0) background unless otherwise stated.
2.2. Histochemical assays
GUS staining of 10 to 14‐day‐old seedlings was performed as described (Jun, Fiume, et al., 2010). For Dex treatments, seedlings were transferred into half strength liquid Murashige and Skoog (MS) media and then either mock‐treated with 70% ethanol (as a solvent for Dex) or treated with Dex at a final concentration of 10 μM for 4 hr. RNA in situ hybridization was performed on 7‐day‐old seedlings as described (Jun, Ha, & Fletcher, 2010). Non‐radioactively labeled probes were generated from the full cDNA sequences of CLE5 and CLE6 transcripts using primers listed in Supporting Information Table S1.
2.3. p35S:BOP1‐GR and p35S:AS2‐GR Activation Assays
Eleven‐day‐old p35S:BOP1‐GR bop1‐1 or p35S:AS2‐GR as2‐1 seedlings were transferred into half strength liquid MS media. Seedlings were then mock‐treated with 70% ethanol or treated with Dex at a final concentration of 10 μM, incubated under slight agitation, and harvested between 30 min and 4 hr for RT‐qPCR analysis. For the hormone assays, a final concentration of 10 μM IAA, 10 μM NAA, 20–30 μM GA4, or 2 μM BL was added to the wild‐type and p35S:BOP1‐GR bop1‐1 seedlings at the same time as the Dex.
2.4. RT‐qPCR analyses
For RT‐qPCR studies, total RNA was extracted using TriReagent (Sigma‐Aldrich), treated with DNase I and purified with the RNeasy Mini Kit (Qiagen). Four to five μg of total RNA were used for reverse transcription with SuperScript III and oligo(dT15) (ThermoFisher Scientific). Quantitative real‐time Polymerase Chain Reaction (PCR) was performed using the iTaq Universal Sybr Green Supermix (Bio‐Rad) with the primers listed in Supporting Information Table S1. PCR reactions were run and analyzed using a MyiQTM Single‐Color real‐time PCR detection system (Bio‐Rad). Two‐step PCR conditions were as follows: initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Quantification of relative gene expression was performed using the ΔΔCt method (Livak & Schmittgen, 2001), and calculated based on at least three biological replicates with three technical replicates each. Expression values were normalized to TUBULIN2 (TUB2) or MON1 (Czechowski, Stitt, Altmann, Udvardi, & Scheible, 2005).
2.5. ChIP‐qPCR analyses
ChIP was performed as described (Yamaguchi et al., 2014) using an anti‐GR antibody (SC‐1004, Santa Cruz Biotechnology). Seedlings were mock‐treated in 70% ethanol or Dex‐treated for 4 hr. Quantification of immunoprecipitated DNA was performed by semi‐quantitative PCR using the primers listed in Supporting Information Table S1.
2.6. CRISPR/Cas9 cloning and analysis
The Cas9 and sgRNA cassettes were PCR‐amplified from the At‐psgR‐Cas9 vectors (obtained from Dr. Jian‐Kang Zhu, Purdue University) using M13 FW and M13 RV primers and subcloned into the pCR8/GW/TOPO vector (ThermoFisher Scientific) to add the attL1 and attL2 Gateway sequences. The CRISPR/Cas9 Gateway‐compatible cassette was then PCR‐amplified using primers T7_promoter and pCR8_FW (Supporting Information Table S1) and cloned into the GEM‐T vector (Promega) for subsequent experiments (named thereafter At‐psgR/GW). The genomic target sequences for CLE5 and CLE6 were 5′‐AGTTCCGACAGGGTTTCACCCGG‐3′ and 5′‐TACATATCGCCCCACAACCATGG‐3′, respectively. The At‐psgR/GW plasmid was digested with BbsI restriction enzyme and used for ligation with the annealed primers CLE5 P1 and P2 or CLE6 P1 and P2 (Supporting Information Table S1). At‐psgR/GW plasmids containing the CLE5 or CLE6 genomic target sequences were transferred into the pEarleyGate 301 vector using the LR enzyme mix (ThermoFisher Scientific). The recombinant pEarleyGate 301 constructs were transferred into Agrobacterium tumefaciens GV3101 and transformed into Arabidopsis Col‐0 using the floral dip method (Clough & Bent, 1998).
Genotyping the CLE5 CRISPR alleles was performed by using the primers CLE5CR_FW and CLE5CR_RV (Supporting Information Table S1) in a PCR reaction to amplify a 727 base pair (bp) product. Digesting the PCR product with HphI yielded 384 bp and 343 bp bands from wild‐type tissue, whereas the product from mutant tissue remained undigested. Genotyping the CLE6 CRISPR alleles was performed by using the primers CLE6CR_FW and CLE6CR_RV (Supporting Information Table S1) in a PCR reaction to amplify an 882 bp product. Digesting the PCR product with MslI yielded 545 bp and 337 bp bands from wild‐type tissue, whereas the product from mutant tissue remained undigested. T2 mutant plants lacking the Cas9 cassette were identified by PCR using primers sgRNA_FW and sgRNA_RV specific to the Cas9 sequence.
2.7. Phenotypic analysis
Leaf morphometrics experiments were conducted using LeafAnalyser software as described (Weight, Parnham, & Waites, 2008), using 50 landmarks per leaf sample and >50 leaf samples per genotype. Principal component analyses were also conducted using LeafAnalyser by calculating eigenvectors (principal components) and eigenvalues (variances) from the covariance matrix generated by LeafAnalyser of each landmark from each leaf (see Weight et al., 2008; Supporting Information Appendix S2). Histological analysis was performed as described (Ha et al., 2003). Scanning electron microscopy was performed as described (Fiume, Pires, Kim, & Fletcher, 2010) and samples visualized on a Hitachi S4700 scanning electron microscope.
3. RESULTS
3.1. CLE5 and CLE6 have distinct yet overlapping expression patterns
To investigate CLE5 and CLE6 transcription patterns in detail throughout the Arabidopsis life cycle, we performed promoter:GUS and in situ hybridization analysis of the two genes in aerial tissues. In wild‐type Col‐0 plants pCLE6:GUS specific promoter activity was detected at the base of young rosette leaves (Figure 1a) and at the base of the floral organs (Figure 1b). Similarly, specific pCLE5:GUS activity was detected at the base of young rosette leaves (Figure 1c), at the base of the cotyledons in mature embryos (Figure 1d), and at the base of the cauline leaves (Figure 1e). pCLE6:GUS promoter activity was also found at the base of the cotyledons. Previously reported pCLE6:GUS expression included the base of the cauline leaves at the primary branching point on the inflorescence stem and pCLE5:GUS expression in floral organs (Jun, Fiume, et al., 2010). Overall, activity of the CLE5 and CLE6 promoters was restricted to the most proximal region of lateral organ primordia, adjacent to the boundary with the shoot meristem.
Figure 1.
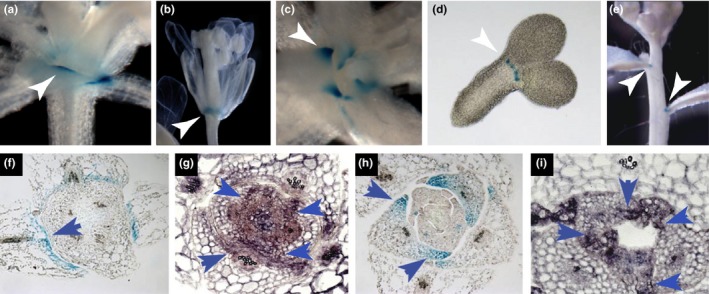
CLE5 and CLE6 expression in wild‐type Arabidopsis plants. (a–b) pCLE6:GUS promoter activity at the base of wild‐type Col‐0 (a) rosette leaves and (b) floral organs. (c–e) pCLE5:GUS promoter activity at the base of Col‐0 (c) rosette leaves, (d) embryo cotyledons, and (e) cauline leaves. (f–g) CLE6 mRNA expression in transverse sections of leaves from 7‐day‐old wild‐type plants. (h–i) CLE5 mRNA expression in transverse sections of leaves from 7‐day‐old wild‐type plants. Arrowheads indicate gene expression at the base of the organ
Transverse sections through wild‐type vegetative shoot apices revealed further region‐specific expression of CLE5 and CLE6. pCLE6:GUS promoter activity in the developing rosette leaves was confined to the adaxial domain (Figure 1f), and this expression pattern was confirmed using RNA in situ hybridization (Figure 1g). In contrast, CLE5 expression occurred in both the adaxial and abaxial domains (Figure 1h,i). Furthermore, CLE5 and CLE6 displayed reciprocal expression patterns along the medio‐lateral axis of developing rosette leaf primordia. CLE6 was transcribed predominantly within the medial domain above the midvein (Figure 1a,f,g) whereas CLE5 transcription was restricted to the lateral domain at the very edges of the primordia (Figure 1c,h,i). Thus, although the promoter activity patterns of CLE5 and CLE6 overlap extensively, the two genes display distinct expression patterns along the adaxial‐abaxial and medial‐lateral polarity axes within the developing leaf primordia.
3.2. Known leaf patterning transcriptional factors regulate CLE5 and CLE6 expression
The CLE5 and CLE6 expression patterns at the base of developing lateral organs were very similar to those reported for the BOP1 and BOP2 leaf patterning genes (Ha et al., 2004; Hepworth et al., 2005; Norberg et al., 2005), as well as for their downstream target AS2 (Byrne et al., 2000; Iwakawa et al., 2007). Given the overlap in expression patterns between these genes, we examined whether CLE5 and/or CLE6 transcription was regulated by BOP or AS2 gene activity in developing rosette leaf primordia using RT‐qPCR. Compared to wild‐type 10‐day‐old Col‐0 seedlings, CLE5 and CLE6 expression was reduced by 25%–50% in bop1‐4 bop2‐11 (b1b2) null mutant seedlings (Figure 2a), indicating that BOP1 and BOP2 are positive regulators of CLE5 and CLE6 transcription. Conversely, in as2‐1 seedlings, CLE5 and CLE6 expression levels were elevated compared to wild‐type (Figure 2a), indicating that AS2 negatively regulates CLE5 and CLE6 transcription. Taken together, these results indicate that CLE5 and CLE6 function downstream of BOP1/2 and AS2 transcriptional regulation as direct or indirect targets of these key leaf patterning factors.
Figure 2.
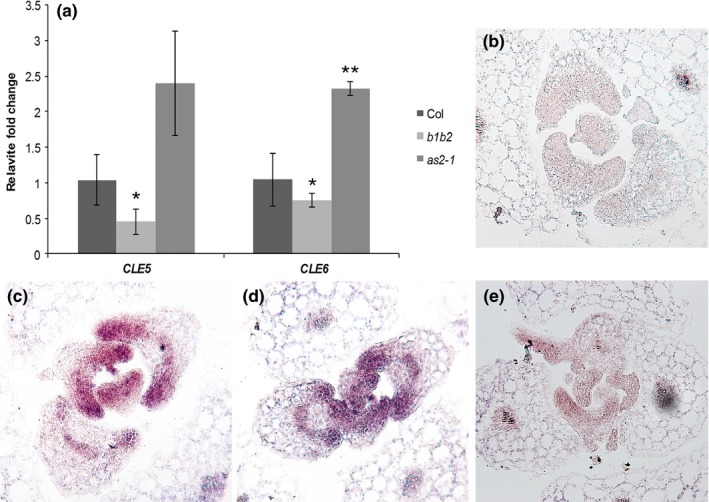
CLE5 and CLE6 expression in leaf patterning mutants. (a) Relative fold change in CLE5 and CLE6 transcript levels in 10‐day‐old Col‐0, bop1‐4 bop2‐11 (b1b2), and as2‐1 seedlings. Expression values (mean ± SD) were normalized to MON1 and asterisks indicate a significant difference from the wild‐type mean (*p < 0.05; **p < 0.01) using Student's t test. (b) Transverse section of a Col‐0 seedling hybridized with a CLE6 sense probe. (c) Transverse section of a Col‐0 seedling hybridized with a CLE6 antisense probe. (d) Transverse section of a b1b2 seedling hybridized with a CLE6 antisense probe. (e) Transverse section of an as2‐1 seedling hybridized with a CLE6 antisense probe
The BOP1/2 and AS2 proteins could regulate CLE5 and CLE6 transcription by affecting their mRNA levels, their expression domains within developing leaves, or both. To determine which, we performed in situ hybridization experiments using 10‐day‐old Col‐0, b1b2, and as2‐1 seedling tissues. Compared to the sense probe, which showed no specific CLE6 expression in wild‐type Col‐0 leaves (Figure 2b), the antisense probe detected strong CLE6 expression across the adaxial domain of young Col‐0 leaf primordia and the marginal region (tips) of older primordia (Figure 2c). This expression pattern was unchanged in b1b2 leaf primordia (Figure 2d), indicating that BOP1 and BOP2 induce CLE6 mRNA expression levels without altering its expression domain. Similarly, we detected no difference in the CLE6 expression domain between developing Col‐0 and as2‐1 leaves (Figure 2e), showing that the elevation in CLE6 mRNA levels in as2 leaves does not result from an enlarged expression domain. These data indicate that neither BOP1/2 nor AS2 affect the CLE6 expression pattern but that a combination of activators and repressors is required to restrict CLE5 and CLE6 transcription to the appropriate levels in developing rosette leaves.
Next, we investigated whether the regulation of the two CLE genes by BOP1 was direct or indirect by conducting a time course of CLE5 and CLE6 transcription in p35S:BOP1‐GR bop1‐1 transgenic plants. In this system, application of the hormone dexamethasone (Dex) translocates the ectopically produced BOP1 transcriptional regulatory protein into the nucleus, rescuing the dominant negative bop1‐1 ectopic leaf outgrowth phenotype (Jun, Ha, et al., 2010) that phenocopies the b1b2 null mutant phenotype (Ha et al., 2004). We first examined AS2 transcription over the 4‐hr time course as a positive control. Consistent with previous data (Jun, Ha, et al., 2010), we found that AS2 transcription was induced by BOP1‐GR after 30 min of 10 μM Dex application and continued to be induced over the full 4 hr of Dex treatment (Figure 3a). CLE6 transcription was significantly upregulated by BOP1 after 1 hr of Dex treatment, and its expression levels continued to increase to >2 fold after 4 hr (Figure 3a). Thus BOP1 is sufficient to induce CLE6 transcription, although only to moderate levels. The rapid activation of CLE6 transcription after 1 hr of Dex application suggests that CLE6 could be an immediate target of BOP1 induction. In contrast, CLE5 expression was slightly downregulated after 30 min of Dex treatment and remained steady thereafter (Figure 3a), indicating that BOP1 is insufficient to induce CLE5 transcription on its own.
Figure 3.
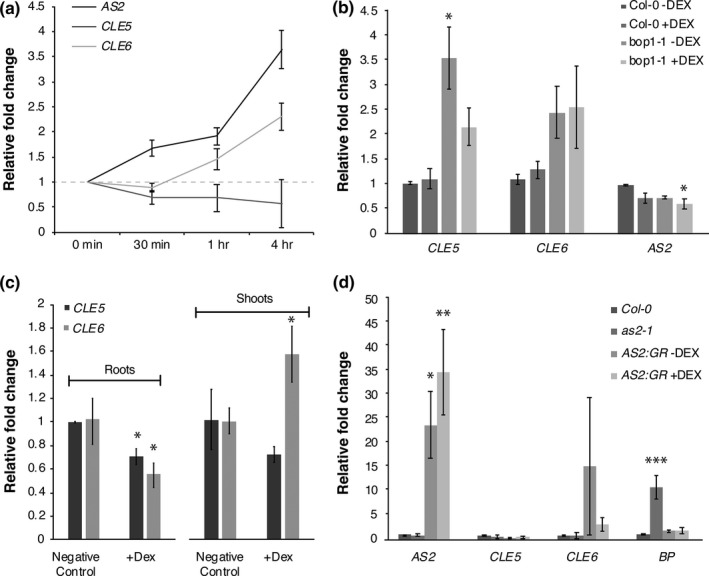
CLE5 and CLE6 expression in response to BOP1 or AS2 induction. (a) Time course of AS2,CLE5 and CLE6 transcript levels in p35S:BOP1‐GR bop1‐1 seedlings treated with Dex for 0 to 4 hr. (b) Relative fold change in AS2, CLE5 and CLE6 transcript levels in Col‐0 and bop1‐1 seedlings treated with Dex for 4 hr. (c) Relative fold change in CLE5 and CLE6 transcript levels in root or shoot tissues from p35S:BOP1‐GR bop1‐1 seedlings treated with Dex for 4 hr. (d) Relative fold change in AS2, CLE5, CLE6 and BP transcript levels in shoot tissues from as2‐1 and p35S:AS2‐GR as2‐1 seedlings treated with Dex for 2 or 4 hr. Expression values (mean ± SD) were normalized to TUB2 or MON1 and asterisks indicate a significant difference from the wild‐type mean (*p < 0.05; **p < 0.01; ***p < 0.001)
As a control for the Dex treatment itself we applied 10 μM Dex for 4 hr to Col‐0 and bop1‐1 plants and quantified AS2, CLE5, and CLE6 mRNA levels. We found that Dex application to wild‐type plants had no effect on AS2, CLE5, or CLE6 transcript abundance (Figure 3b), although we observed considerable variability in the transcript levels within each genotype because only a very small number of cells within the total leaf tissue assayed express CLE5 or CLE6. Dex application to bop1‐1 plants lacking the p35S:BOP1‐GR transgene also did not affect AS2 or CLE6 mRNA levels; however, CLE5 mRNA levels were reduced by approximately 40% compared to mock‐treated bop1‐1 plants. Thus the slight reduction in CLE5 mRNA levels during the time course can be attributed to a repressive effect of Dex application to bop1‐1 plants rather than of BOP1‐GR activity.
Both CLE5 and CLE6 are expressed in root tissues as well as in shoot tissues (Jun, Fiume, et al., 2010), so we determined whether the effect of BOP1 on CLE5 and/or CLE6 expression was limited to one of the two tissue types. We treated p35S:BOP1‐GR bop1‐1 plants with 10 μM Dex for 4 hr, isolated shoot and root tissues, and measured CLE5 and CLE6 mRNA levels using RT‐qPCR. We found that although CLE5 expression levels were unaltered in shoot versus root tissues, BOP1 induction of CLE6 expression occurred in the shoot tissues but not in the root tissues (Figure 3c). Thus, the regulation of CLE6 transcription by BOP1 is restricted to above‐ground shoot tissues that contain the leaf primordia.
To determine if CLE6 induction by BOP1 was due to direct transcriptional regulation, we tested whether BOP1 protein directly associated with CLE6 regulatory sequences. We performed chromatin immunoprecipitation (ChIP‐qPCR) assays with Dex‐treated p35S:BOP1‐GR bop1‐1 seedlings using primer sets spanning 3.2 kilobases (kb) upstream of the CLE5 coding region, the intergenic region between CLE5 and CLE6, and 1 kb downstream of the CLE6 coding region. No BOP1 binding to these CLE5 and CLE6 regulatory regions was detected (Supporting Information Figure S1), although BOP1 binding was detected as expected to regulatory sites within the AS2 promoter (Jun, Ha, et al., 2010). Therefore the regulation of CLE6 by BOP1 appears to occur indirectly through an intermediary factor. We also attempted to rescue the bop1‐4 bop2‐11 ectopic blade outgrowth phenotype (Ha et al., 2007) by directing CLE6 expression within the BOP domain under the control of 6.0 kb of BOP1 promoter sequence. This promoter region is sufficient to drive BOP1 transcription in its native domain (Jun, Ha, et al., 2010). However, none of the pBOP1:CLE6 bop1‐4 bop2‐11 lines exhibited rescue of the ectopic blade outgrowth phenotype, indicating that CLE6 expression in the BOP1 domain alone is not sufficient to restore wild‐type petiole identity. Thus it is likely that rescue of the bop phenotype requires CLE6 expression beyond the BOP expression domain and/or other factors in addition to CLE6.
The observation that BOP1 was not a direct regulator of CLE5 or CLE6 transcription suggested that role would fall to a downstream component of the BOP1 regulatory pathway. Our time course showed that AS2 induction by BOP1‐GR occurred prior to CLE6 induction, so we tested whether the AS2 transcription factor might directly regulate CLE5 and/or CLE6 transcription. We analyzed CLE5 and CLE6 mRNA levels in shoots of 11‐day‐old p35S:AS2‐GR as2‐1 seedlings after 2 and 4 hr of Dex induction using RT‐qPCR. At both time points we observed a slight decrease in CLE5 mRNA levels compared to mock‐treated seedlings (Figure 3d); however, this decline was not statistically significant. We also detected no significant change in CLE6 mRNA levels at either time point (Figure 3d). These data show that AS2 alone is not sufficient to affect CLE5 and/or CLE6 transcription, either because the CLE genes are not direct AS2 regulatory targets or because the amount of its partner protein AS1 becomes rate‐limiting when AS2 is over‐expressed.
Other well‐characterized players in CLE gene regulation are members of the WOX family of transcription factors. Among these, WOX1 and PRESSED FLOWER (PRS), also known as WOX3, have described roles in early leaf development (Nakata et al., 2012), and act in blade outgrowth and leaf margin formation. To assess whether these WOX transcription factors regulated CLE5 and CLE6 transcription, we measured CLE5 and CLE6 mRNA levels in the shoots of 10‐day‐old wox1‐1 and prs‐1 null mutant seedlings. Expression of CLE5 and CLE6 was reduced by about 50% in both prs and wox1 seedlings compared to wild‐type Col‐0 (Figure 4a), indicating that WOX1 and PRS are each positive regulators of CLE5 and CLE6 transcription. In situ hybridization showed no alteration in the CLE6 expression domain in 10‐day‐old prs wox1 seedlings compared to Col‐0 (Figure 4c,d), indicating that PRS and WOX1 do not affect the CLE6 spatial domain. We also noted that WOX1 transcripts were absent from prs‐1 seedlings and PRS transcripts were absent from wox‐1 seedlings (Figure 4a). Thus PRS and WOX1 are required for one another's expression in developing leaf primordia.
Figure 4.
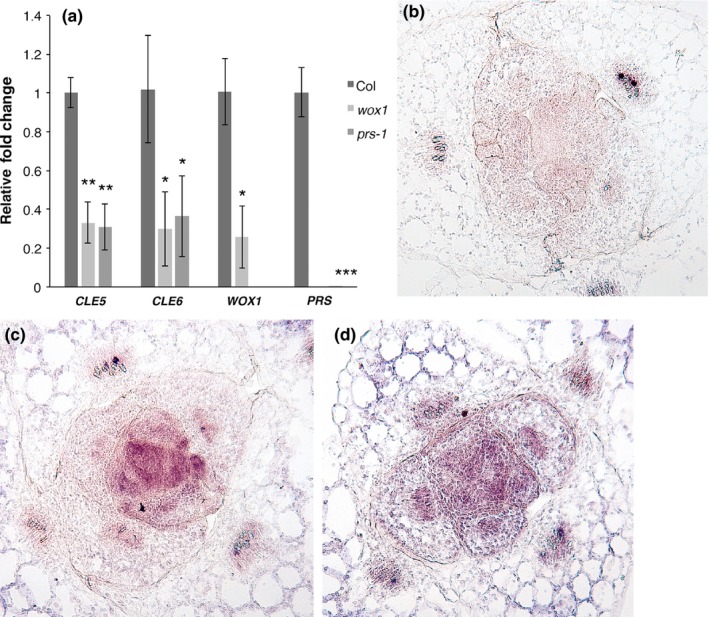
CLE5 and CLE6 expression in prs and wox1 mutants. (a) CLE5 and CLE6 transcript levels in 10‐day‐old Col‐0 and prs‐1 and wox1‐1 seedlings. Expression values (mean ± SD) were normalized to MON1 and asterisks indicate a significant difference from the wild‐type mean (*p < 0.05; **p < 0.01; ***p < 0.001). (b) Transverse section of a Col‐0 seedling hybridized with a CLE6 sense probe. (c) Transverse section of a Col‐0 seedling hybridized with a CLE6 antisense probe. (d) Transverse section of a prs‐1 wox1‐1 seedling hybridized with a CLE6 antisense probe
3.3. CLE5 and CLE6 transcription is regulated by plant hormones
Because hormones have long been implicated in regulation of leaf development and architecture, we sought to define the effect of various hormones on CLE5 and CLE6 expression in developing Arabidopsis rosette leaves. First, we treated 11‐day‐old wild‐type seedlings with 30 μM gibberellin (GA4), 2 μM brassinolide (BL), or 10 μM indole‐3‐acetic acid (IAA), a naturally occurring auxin, for 4 hr and Col‐0 quantified CLE5 and CLE6 transcription levels using RT‐qPCR. We found that the mRNA levels of both genes were slightly elevated in after application of both GA4 and BL (Figure 5a), indicating that the two CLE genes respond to hormones that regulate leaf formation. Interestingly, IAA treatment led to a much higher upregulation of CLE5 and CLE6 expression (Figure 5a), with CLE6 transcription showing a ~24‐fold induction. Thus both the CLE5 and especially the CLE6 gene appear to be auxin responsive.
Figure 5.
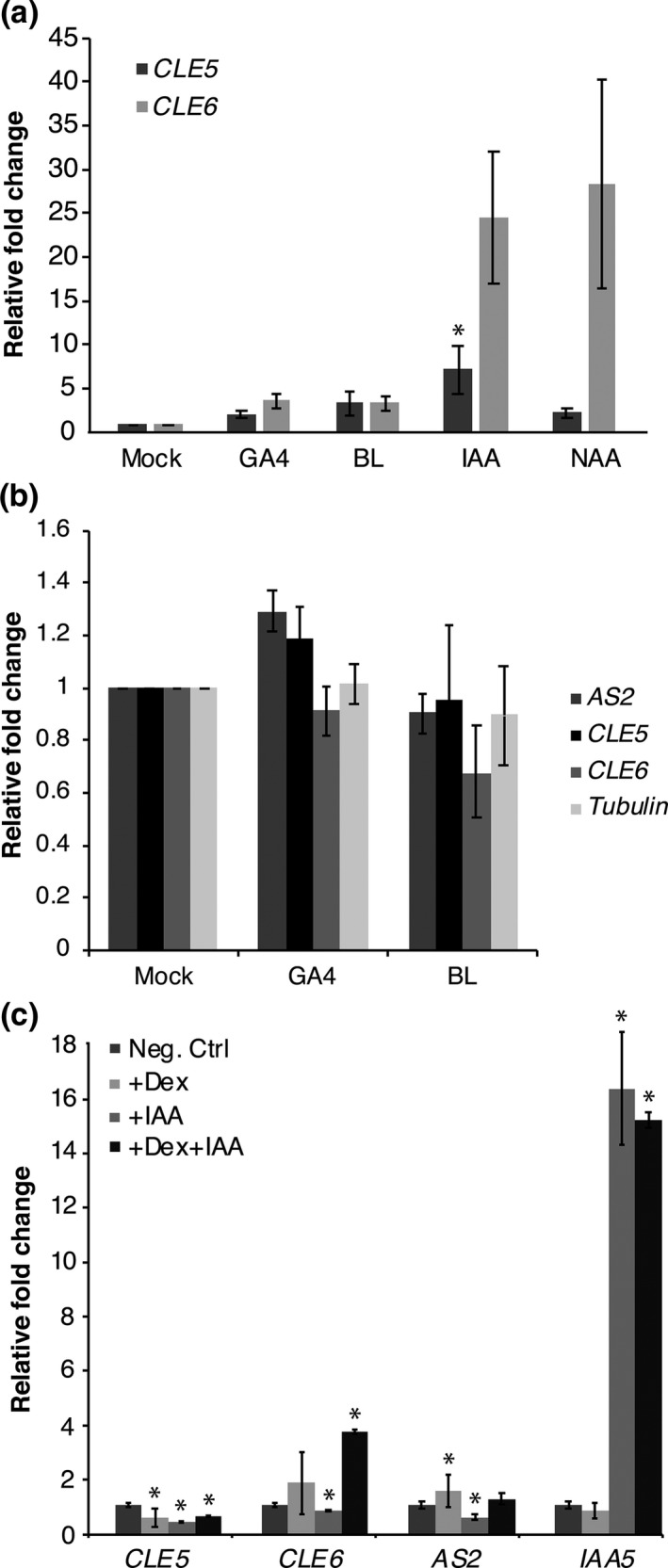
CLE5 and CLE6 expression in response to BOP1 induction in the absence or presence of hormones. (a) Relative fold change in CLE5 and CLE6 transcript levels in 11‐day‐old Col‐0 seedlings treated with gibberellin (GA 4), brassinolide (BL), or auxin (IAA or NAA) for 4 hr. (b) Relative fold change in AS2, CLE5 and CLE6 transcript levels 11‐day‐old p35S:BOP1‐GR bop1‐1 seedlings treated with Dex plus either GA 4 or BL for 4 hr. (c) Relative fold change in CLE5, CLE6, AS2, and IAA5 transcript levels of 11‐day old p35S:BOP1‐GR bop1‐1 seedlings treated with Dex and/or IAA for 4 hr. Expression values (mean ± SD) were normalized to MON1 and asterisks indicate a significant difference from the wild‐type mean at p < 0.05
Next we examined whether phytohormones played a role in BOP1‐mediated regulation of CLE5 and CLE6 expression. We treated 11‐day‐old p35S:BOP1‐GR bop1‐1 seedlings with 10 μM Dex alone or together with 20 μM GA4, 2 μM BL, or 10 μM IAA for 4 hr and quantified CLE5 and CLE6 transcription levels using RT‐qPCR. No significant change in CLE5 or CLE6 mRNA expression levels was observed following application of both +Dex and +GA, or of both +Dex and +BL (Figure 5b). This indicates that neither GA nor BL affects the BOP1‐mediated regulation of CLE5 or CLE6 transcription, nor vice versa. In contrast, the application of both +Dex and +IAA resulted in differential effects on CLE5 and CLE6 expression. For CLE5, BOP1‐GR induction by 4 hr of Dex treatment led to a moderate reduction in CLE5 transcript levels, as did 4 hr application of 10 μM IAA (Figure 5c). Simultaneous treatment with both Dex and IAA for 4 hr did not further reduce CLE5 mRNA levels, indicating that the two treatments did not have cumulative effects. Our data indicate that in a bop1‐1 background CLE5 is repressed by Dex application, as shown earlier (Figure 3b), as well as by exogenous auxin. In the case of CLE6, BOP1‐GR induction by Dex treatment alone led to no significant change in CLE6 transcript levels, whereas IAA treatment resulted in a slight reduction in CLE6 mRNA levels (Figure 5c). CLE6 transcript levels were much more mildly affected by IAA treatment in bop1‐1 plants (Figure 5c) than in Col plants (Figure 5a). Upon simultaneous treatment with both Dex and IAA for 4 hr, CLE6 transcription was elevated beyond the levels detected upon BOP1‐GR induction alone (Figure 5c). These results suggest that CLE6 transcript levels are independently regulated by BOP1 and auxin. Thus the CLE5 and CLE6 genes show differential regulation by BOP1 and phytohormones in developing leaves.
3.4. CLE5 and CLE6 have mild effects on overall leaf shape
In order to determine the function of CLE5 and CLE6 during Arabidopsis development we generated null alleles of the two genes using the CRISPR/Cas9 genome engineering technology (Feng et al., 2013; Nekrasov, Staskawicz, Weigel, Jones, & Kamoun, 2013). Transformation of wild‐type Col‐0 plants with a single guide RNA (sgRNA) construct targeted to the CLE5 or CLE6 coding sequences yielded multiple independent transformants. Independent T1 plants were self‐fertilized and homozygous individuals were identified in the T2 or T3 generations by restriction enzyme digestion and sequencing.
Two independent CLE5 and two independent CLE6 homozygous lines were chosen for further study (Figure 6a). One cle5 allele consisted of an insertion of an “A” nucleotide at position +215 downstream of the translation start site, and was designated cle5‐1. A second allele contained a deletion of a five nucleotides starting at position +210 and was designated cle5‐2. The cle5‐1 mutation introduces a frameshift that alters the amino acid sequence of the CLE domain beginning at the third residue, while the cle5‐2 mutation deletes the first three residues of the CLE domain and also introduces a frameshift. The first cle6 allele consisted of an insertion of an “A” nucleotide at position +106 downstream of the translation start site, and was designated cle6‐1. A second allele contained a deletion of four nucleotides starting at position +102 and was designated cle6‐2. Each of these mutations generates a frameshift in the CLE6 coding sequence upstream of the CLE domain. Due to the nature of the mutations none of these cle5 or cle6 alleles produces a functional CLE polypeptide, and thus they represent loss‐of‐function alleles.
Figure 6.
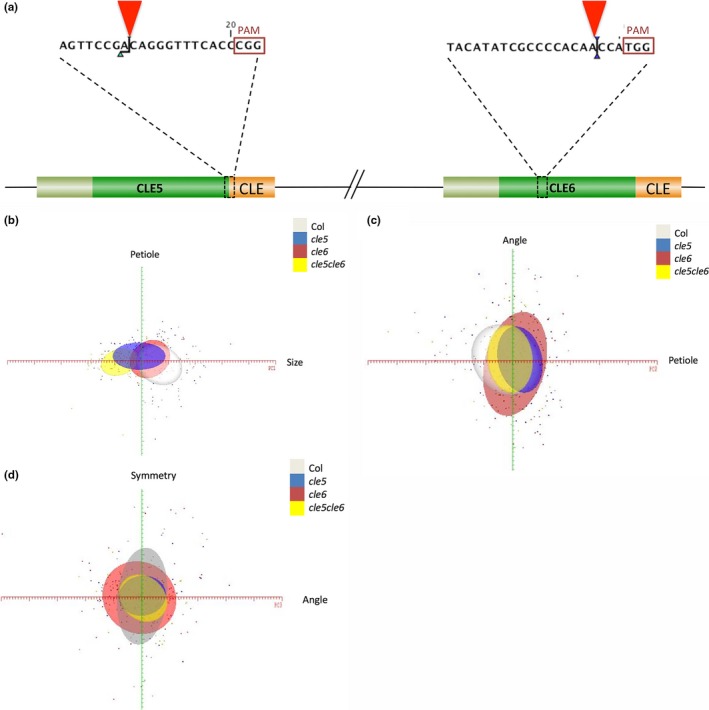
CLE5 and CLE6 loss‐of‐function allele generation and role in leaf formation. (a) Locations of the cle5 and cle6 CRISPR‐Cas9 induced mutations (red arrowheads) upstream of the PAM site (red box) within the sgRNA for the CLE5 and CLE6 coding sequences. The coding sequences of the signal peptides are represented in olive, the variable domains in green, and the CLE domains in orange. (b–d) Two‐dimensional PC maps generated using ≥5 standard deviations from the mean leaf along the X‐axis PC (red) and ≥2.5 standard deviations along the Y‐axis PC (green). Each standard deviation is represented by a major tick on the axis, and each leaf measured is represented by a single colored point. (b) Variation along PC1 and PC2 for Col‐0 (white oval), cle5 (blue oval), cle6 (red oval), and cle5 cle6 (yellow oval) leaves. (c) Variation along PC2 and PC3 for each genotype. (d) Variation along PC3 and PC4 for each genotype
Because CLE5 and CLE6 loci are located approximately 1.7 kb apart in a head‐to‐tail arrangement on chromosome 2, their proximity makes it improbable to produce double mutants by conventional genetic crosses. Therefore we took advantage of genome engineering to target both CLE5 and CLE6 for simultaneous mutation by transforming wild‐type Col‐0 plants with a construct that contains both of the sgRNAs used in the previous experiments, the one targeted to the CLE5 coding sequence and the one targeted to the CLE6 coding sequence. Three doubly homozygous mutants were obtained, one of which contained an insertion of an “A” nucleotide at position +215 downstream of the CLE5 translation start site as well as an insertion of an “A” nucleotide at position +106 downstream of the CLE6 translation start site. Because each mutation generates a frameshift in the respective CLE5 or CLE6 coding sequence upstream of the CLE domain, no functional polypeptides are generated and this cle5‐3 cle6‐3 double mutant represents a knockout of both genes.
We next performed a large‐scale morphological analysis of Col‐0, cle5‐1, cle6‐1, and cle5‐3 cle6‐3 plants from germination through the vegetative phase of development. We observed no significant differences in leaf initiation rate, total rosette leaf number, mature rosette leaf size, rosette diameter, or flowering time under different photoperiods between Col‐0 and cle5, cle6, or cle5 cle6 plants. We also analyzed the size, number, composition, and morphology of wild‐type and mutant rosette leaf cells using scanning electron microscopy (SEM) and histological analysis but detected no differences between the genotypes. WOX1 and PRS promote leaf margin cell file formation (Nakata et al., 2012), with prs wox1 rosette leaves having fewer and p35S:PRS leaves having more margin cell files than wild‐type rosette leaves (Supporting Information Figure S2). Because wox1‐1 and prs‐1 plants display reduced CLE5 and CLE6 expression, we used SEM to assess whether mutations in CLE5 and/or CLE6 caused leaf margin cell file defects. However, we found no reproducible differences in margin cell file number, morphology or fate between Col‐0, cle5, cle6, and cle5 cle6 rosette leaves (Supporting Information Figure S2). Thus CLE5 and CLE6 separately or together have no macroscopic effects on vegetative development under normal growth conditions.
Finally, we conducted a leaf morphometric study to identify any subtle phenotypes attributable to the loss of CLE5 or CLE6 activity. The LeafAnalyser image processing program (Weight et al., 2008) was used to quantify the shape of Col‐0, cle5‐1, cle6‐1, and cle5‐3 cle6‐3 first through fourth rosette leaves by performing a principal component (PC) analysis of distinct aspects of overall rosette leaf shape among the different genotypes. LeafAnalyser parsed the major sources of variation in leaf shape into four principal components—leaf size, width, petiole angle, and tip‐to‐base asymmetry—that together account for 95% of the total Arabidopsis leaf shape variation. Using the LeafAnalyser software we measured each of the four PCs for at least 50 leaves per genotype, and then generated two‐dimensional PC maps by plotting the values for each genotype for two of the PCs (leaf size versus width, width versus petiole angle, etc.). Each oval represented one standard deviation from the mean leaf for one genotype, such that the extent of overlap between ovals showed the relative similarity in phenotype.
This morphological analysis revealed modest effects of CLE5 and CLE6 on leaf shape (Figure 6b–d). The first component, which accounted for almost 66% of the variation, was overall leaf size and was likely due to leaf age, independent of genotype. The second component was variation in leaf width that accounted for 12% of the variability and was most apparent in the cle5 and cle5 cle6 genotypes (Figure 6b). The third component was leaf curvature due to petiole angle and accounted for almost 10% of the total leaf shape variability. This variance was most evident in cle6 leaves (Figure 6c). The fourth component, leaf symmetry, accounted for 7% of the variability and was detected in both single and double mutants (Figure 6d). While these effects are subtle, they do distinguish possible distinct roles for CLE5 and CLE6 in regulating leaf shape during development.
4. DISCUSSION
Vegetative development is a highly coordinated series of events that starts with a primordium initiated from the shoot apical meristem. This primordium undergoes pattern formation along three polarized axes, the establishment of the basic cell types of the petiole, lamina, and marginal structures, and a maturation process that involves cell differentiation and expansion to achieve the mature leaf shape (Bar & Ori, 2014). Long‐range hormonal signals such as auxin, GA, and BR are well‐characterized players in leaf development; however, their relative ubiquity and omnipresence due to diffusion and active transport likely limit their ability to regulate the highly coordinated and precise events required in organ development alone. While small peptides are known integral signals in plant root and vasculature cell development, we pose a model of CLE signaling that plays a role in regulating shoot organ formation in plants.
Analyses of CLE gene expression reveal CLE5 and CLE6 promoter activity in the aerial tissues of wild‐type Arabidopsis seedlings was uniquely confined to the area around the shoot apex, the base of the lateral organs, and the leaf hydathodes (Jun, Fiume, et al., 2010). We investigated the expression of CLE5 and CLE6 at higher resolution in aerial tissues of wild‐type Arabidopsis plants using promoter:GUS and in situ hybridization (Figure 1). Interestingly, the two closely related genes displayed distinct expression patterns. CLE6 was expressed exclusively within the adaxial domain of developing rosette leaf primordia, while CLE5 transcription was restricted to the lateral regions of the primordia but was detected in both the adaxial and abaxial domains. Thus, the upper, outer periphery of the developing leaves express both CLE5 and CLE6, whereas the underside of the leaf margins expresses only CLE5 and the midvein region expresses only CLE6. Our data show subtle but distinct differences between the CLE5 and CLE6 expression patterns despite the high degree of similarity between the two transcription units. The CLE5 and CLE6 genes encode precursor proteins with 77% identity and 84% similarity (Sharma, Ramirez, & Fletcher, 2003), and produce predicted mature CLE peptides with identical amino acid sequences (Cock & McCormick, 2001). The two genes, along with CLE4 and CLE7, lie within a 16.5 kb region on chromosome 2. Within this gene cluster, the CLE5, CLE6, and CLE7 coding regions are more similar to one another than to other members of the family, suggesting they may have arisen by local gene duplication events (Cock & McCormick, 2001). Nonetheless, analysis of 1.7 kb of upstream promoter sequence, corresponding to the distance between the CLE5 stop codon and the CLE6 start codon, between these three genes shows only 49.6% similarity between the CLE5 and CLE6 promoters. This is comparable to the 45.2% similarity between the CLE5 and CLE7 promoters, despite CLE7 expression being restricted to the root (Jun, Fiume, et al., 2010). Therefore the divergence of the upstream regulatory regions between the CLE5 and CLE6 genes may account for the differences in their expression patterns. Still, determining how highly similar peptides are differentially regulated in space and time is crucial to understanding the roles of such signaling molecules in the larger context of plant development.
Our study reveals that CLE5 and CLE6 are downstream targets of two transcriptional regulators that affect leaf patterning, BOP1/2 and AS2. Both CLE5 and CLE6 are downregulated in bop1 bop2 seedlings, showing that BOP1 and BOP2 are positive regulators of CLE5 and CLE6 transcription (Figure 2). Conversely, an increase in CLE5/6 transcript levels in as2‐1 seedlings indicates that AS2 negatively regulates CLE5 and CLE6 transcription. AS2 transcripts are present throughout the adaxial leaf domain (Byrne et al., 2000; Iwakawa et al., 2007), whereas BOP1 and BOP2 expression is confined to the most proximal end of the adaxial domain (Ha et al., 2004; Norberg et al., 2005). This suggested that BOP1/2 might induce CLE5/6 transcription at the base of the petiole and AS2 might repress it in more distal positions along the petiole and in the blade. However, in situ hybridization experiments indicated that the absence of neither BOP1/2 nor AS2 altered the CLE6 expression domain (Figure 2), indicating that BOP1/2 and AS2 affect CLE5 and CLE6 transcript levels within their native domain rather than establishing the boundaries of their spatial expression domains.
Additional experiments showed that CLE5 and CLE6 are differentially regulated by BOP1/2 and AS2. CLE6 transcription was upregulated in shoot tissues within 1 hr in response to BOP1‐GR translocation (Figure 3), suggesting that CLE6, like AS2, might be a direct target of BOP1 activation. However, using ChIP‐qPCR we detected no BOP1 binding to the CLE6 (or the CLE5) regulatory region (Supporting Information Figure S1). Thus either the regulation of CLE6 transcription by the BOP proteins is indirect or additional rate‐limiting factors are required for BOP1 binding to the CLE6 locus. Our data also show that the inductive effect of BOP1 on CLE6 transcription overcomes the repressive effect of AS2 we observed in the as2‐1 background (Figure 2). This oppositional effect of AS2 and BOP1/2 on CLE6 expression indicates that a combination of activators and repressors likely controls CLE gene transcription in developing leaves.
In contrast to CLE6, CLE5 expression was not upregulated upon BOP1‐GR translocation (Figure 3), which indicates that BOP1 is sufficient to induce CLE6 but not CLE5 transcription in leaves. Based on control experiments (Figure 3), the slight reduction observed in CLE5 transcript levels is more likely to be a result of Dex application itself, as has been noted elsewhere (Kang, Fang, & Singh, 1999), than of targeted transcriptional regulation by BOP1. Finally, we detected no significant affect on either CLE5 or CLE6 transcription in the AS2‐GR system (Figure 3). One possible explanation is that over‐expression of AS2 alters the expression or activity of its partner AS1. Alternatively, the amount of endogenous AS1 may be a rate‐limiting factor for the activity of the complex, rendering the excess AS2 protein unable to affect target gene transcription.
WOX transcription factors are known key components of CLE signaling pathways, and WOX transcription factor genes have been linked to the promotion of leaf blade outgrowth in several plant species. These include LAM1 in Nicotiana sylvestris (Lin et al., 2013), STF in Medicago truncatula (Tadege et al., 2011), narrow sheath1 and 2 in maize (Nardmann, Ji, Werr, & Scanlon, 2004), and narrow leaf2 and 3 in rice (Ishiwata et al., 2013). In Arabidopsis, the closely related WOX1 and PRS (aka WOX3) genes have described roles in leaf development (Nakata et al., 2012), during which their expression is induced by auxin (Caggiano et al., 2017). prs wox1 leaves display reduced blade outgrowth and fewer margin cell files (Nakata et al., 2012) while PRS over‐expressor lines show an increase in leaf margin cell file number (Supporting Information Figure S2), indicating that PRS and WOX1 promote blade outgrowth and margin cell file formation. Our expression analysis reveals that both WOX1 and PRS are positive regulators of CLE5 and CLE6 transcription (Figure 4). This is consistent with a role for a WOX‐CLE signaling module in leaf development and provides further evidence that multiple factors regulate CLE gene expression and organize leaf development. Based on the subtle leaf shape defects in cle5 cle6 seedlings we propose that CLE5 and CLE6 may act downstream of PRS and WOX1 in regulating lamina outgrowth. In contrast, no obvious leaf margin cell file phenotype was observed in cle5 or cle6 single or double mutant plants (Supporting Information Figure S2), either because CLE5 and CLE6 are not required for margin cell development or because their loss is compensated by other CLE genes.
In addition to transcriptional regulators, several long‐range hormones have well‐established roles in plant organ development. GA acts subsequent to initial patterning events to regulate the rate of cell proliferation and expansion during leaf outgrowth (Achard et al., 2009). In addition, auxin, ABA and BL all play roles in regulating the transition from leaf growth by cell division to growth by cell expansion (Kalve et al., 2014). GA is known to promote CLE6 transcription in the root stele, and ectopic expression of CLE6 has been shown to partially compensate for GA‐deficiency during vegetative growth as well (Bidadi et al., 2014). Our work shows that CLE5 and CLE6 are indeed responsive to GA in shoot tissues, as well as to BL and IAA (Figure 5). CLE5 and CLE6 expression is slightly induced by GA (Figure 5), which promotes leaf differentiation, and thus the CLE genes may act downstream of GA (and/or other hormones) during the later stages of leaf differentiation to achieve the mature leaf morphology.
The transcription of both CLE5 and CLE6 is also auxin responsive, with CLE6 responding more strongly to IAA application than CLE5 (Figure 5). Interestingly, simultaneous BOP1‐GR induction and auxin application resulted in differential effects on CLE5 and CLE6 transcription. Whereas CLE5 mRNA expression levels were slightly reduced, CLE6 transcription levels were elevated in the presence of both BOP1 and auxin (Figure 5). These data reveal that CLE5 and CLE6 undergo differential regulation by BOP1 and phytohormones, again indicating that a combination of activators and repressors are required to control CLE gene expression in developing leaves.
Although gross morphological phenotypes were not visible in cle5/6 single or double mutant plants, more sensitive morphometric analysis showed that the two genes affect the final shape of the rosette leaves. Specifically, variation in leaf petiole width (PC2) accounted for 12% of the variability in leaf shape and was most apparent in the cle5 and cle5 cle6 genotypes (Figure 6). This subtle effect on petiole width is reminiscent of the blade‐on‐petiole phenotype of bop leaves, where formation of ectopic blade tissue along the petiole results in a wider than normal leaf base (Ha et al., 2003, 2007). Thus, the regulation of CLE5 and CLE6 expression by the BOP proteins appears to be important for fine‐tuning leaf formation by limiting the lateral growth of the petiole. Collectively, our data are consistent with a scenario in which BOP1/2, AS2, PRS/WOX1, and phytohormones act combinatorially to modulate CLE5 and CLE6 transcription at the leaf base to levels appropriate to produce the final leaf shape, although additional experiments will be necessary to fully clarify the relationships between these various factors. A role for CLE5 and CLE6 in regulating the activity of differentiating cells during the later stages of leaf development contrasts with that of most other CLE genes functionally characterized to date, which predominantly affect undifferentiated, meristematic cells (Fletcher, Brand, Running, Simon, & Meyerowitz, 1999; Gutierrez‐Alanis et al., 2017; Hirakawa et al., 2008; Stahl et al., 2009; Whitford et al., 2008).
The fact that two genes that encode identical CLE peptides are expressed in overlapping but not identical patterns and are differentially regulated by upstream hormones and transcription factors may be an evolutionary artifact or may have functional significance. To date we have not observed a unique function for either CLE5 or CLE6 in organ development. However, overlapping domains of genes expressing secreted peptides may contribute to dose‐dependent signaling events that functional analysis at the level of single cells may be required to uncover. Such multiple independent yet cross‐talking regulatory mechanisms may provide the range of distinct signaling events required for the highly coordinated development of an organ with three polar axes and multiple specific cell types.
ACCESSION NUMBERS
AS2 (At1g65620), BOP1 (At3g57130), BOP2 (At2g41370), CLE5 (At2g31083), CLE6 (At2g31085), MON1 (At2g28390), PRS (At2g28610), TUB2 (At5g62690), WOX1 (At3g18010).
AUTHOR CONTRIBUTIONS
PD, EG, JHJ and JCF designed the research; PD, EG, TD and JHJ performed the research; EG contributed new tools; PD, EG, JHJ, TD and JCF analyzed the data; and PD and JCF wrote the paper with input from the other authors.
Supporting information
ACKNOWLEDGMENTS
We thank Dr. Jian‐Kang Zhu and Dr. Brian Staskawicz for providing CRISPR/Cas9 vectors. This work was supported by the US National Science Foundation (IOS‐1257429 to JCF) and the US Department of Agriculture (CRIS 2030‐21000‐041‐00D to JCF).
DiGennaro P, Grienenberger E, Dao TQ, Jun JH, Fletcher JC. Peptide signaling molecules CLE5 and CLE6 affect Arabidopsis leaf shape downstream of leaf patterning transcription factors and auxin. Plant Direct. 2018;2:1–14. 10.1002/pld3.103
REFERENCES
- Achard, P. , Gusti, A. , Cheminant, S. , Alioua, M. , Dhondt, S. , Coppens, F. , … Genschik, P. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Current Biology, 19, 1188–1193. 10.1016/j.cub.2009.05.059 [DOI] [PubMed] [Google Scholar]
- Bar, M. , & Ori, N. (2014). Leaf development and morphogenesis. Development, 141, 4219–4230. 10.1242/dev.106195 [DOI] [PubMed] [Google Scholar]
- Bidadi, H. , Matsuoka, K. , Sage‐Ono, K. , Fukushima, J. , Pitaksaringkarn, W. , Asahina, M. , … Satoh, S. (2014). CLE6 expression recovers gibberellin deficiency to promote shoot growth in Arabidopsis. Plant Journal, 78, 241–252. 10.1111/tpj.12475 [DOI] [PubMed] [Google Scholar]
- Brand, U. , Fletcher, J. C. , Hobe, M. , Meyerowitz, E. M. , & Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science, 289, 617–619. 10.1126/science.289.5479.617 [DOI] [PubMed] [Google Scholar]
- Brand, U. , Grunewald, M. , Hobe, M. , & Simon, R. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiology, 129, 565–575. 10.1104/pp.001867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, M. E. , Barley, R. , Curtis, M. , Arroyo, J. M. , Dunham, M. , Hudson, A. , & Martienssen, R. A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis . Nature, 408, 967–971. 10.1038/35050091 [DOI] [PubMed] [Google Scholar]
- Caggiano, M. P. , Yu, X. , Bhatia, N. , Larsson, A. , Ram, H. , Ohno, C. K. , … Heisler, M. (2017). Cell type boundaries organize plant development. eLife, 6, e27421 10.7554/eLife.27421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S. J. , & Bent, A. F. (1998). Floral dip: A simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant Journal, 16, 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cock, J. M. , & McCormick, S. (2001). A large family of genes that share homology with CLAVATA3 . Plant Physiology, 126, 939–942. 10.1104/pp.126.3.939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski, T. , Stitt, M. , Altmann, T. , Udvardi, M. K. , & Scheible, W.‐R. (2005). Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology, 139, 5–17. 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells, J. P. , Provost, C. M. , Mishra, L. , & Turner, S. (2013). WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organization. Development, 140, 2224–2234. 10.1242/dev.091314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Z. , Zhang, B. , Ding, W. , Liu, X. , Yang, D. L. , Wei, P. , … Zhu, J. K. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Research, 23, 1229–1232. 10.1038/cr.2013.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiume, E. , Pires, H. R. , Kim, J. S. , & Fletcher, J. C. (2010). Analyzing floral meristem development In Hennig L., & Kohler C. (Eds.), Methods in Molecular Biology (pp. 130–152). Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Fletcher, J. C. , Brand, U. , Running, M. P. , Simon, R. , & Meyerowitz, E. M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science, 283, 1911–1914. 10.1126/science.283.5409.1911 [DOI] [PubMed] [Google Scholar]
- Grienenberger, E. , & Fletcher, J. C. (2015). Peptide signaling molecules in plant development. Current Opinion in Plant Biology, 23, 8–14. 10.1016/j.pbi.2014.09.013 [DOI] [PubMed] [Google Scholar]
- Gutierrez‐Alanis, D. , Yong‐Villalobos, L. , Jimenez‐Sandoval, P. , Alatorre‐Cobos, F. , Oropeza‐Aburto, A. , Mora‐Macias, J. , … Herrera‐Estrella, L. (2017). Phosphate starvation‐dependent iron metabolism induces CLE14 expression to trigger root meristem differentiation through CLV2/PEPR2 signaling. Developmental Cell, 41, 555–570. 10.1016/j.devcel.2017.05.009 [DOI] [PubMed] [Google Scholar]
- Ha, C. M. , Jun, J. H. , Nam, H. G. , & Fletcher, J. C. (2004). BLADE‐ON‐PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana . Plant and Cell Physiology, 45, 1361–1370. 10.1093/pcp/pch201 [DOI] [PubMed] [Google Scholar]
- Ha, C. M. , Jun, J. H. , Nam, H. G. , & Fletcher, J. C. (2007). BLADE‐ON‐PETIOLE1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial‐abaxial polarity genes. Plant Cell, 19, 1809–1825. 10.1105/tpc.107.051938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, C. M. , Kim, G.‐T. , Kim, B. C. , Jun, J. H. , Soh, M. S. , Ueno, Y. , … Nam, H. G. (2003). The BLADE‐ON‐PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis . Development, 130, 161–172. 10.1242/dev.00196 [DOI] [PubMed] [Google Scholar]
- Haecker, A. , Grob‐Hardt, R. , Geiges, B. , Sarkar, A. , Breuninger, H. , Herrmann, M. , & Laux, T. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana . Development, 131, 657–668. 10.1242/dev.00963 [DOI] [PubMed] [Google Scholar]
- Hepworth, S. R. , Zhang, Y. , McKim, S. , Li, X. , & Haughn, G. (2005). BLADE‐ON‐PETIOLE‐dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell, 17, 1434–1448. 10.1105/tpc.104.030536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa, Y. , Kondo, Y. , & Fukuda, H. (2010). TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis . Plant Cell, 22, 2618–2629. 10.1105/tpc.110.076083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa, Y. , Shinohara, H. , Kondo, Y. , Inoue, A. , Nakanomyo, I. , Ogawa, M. , … Fukuda, H. (2008). Non‐cell‐autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proceedings of the National Academy of Sciences of the United States of America, 105, 15208–15213. 10.1073/pnas.0808444105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwata, A. , Ozawa, M. , Nagasaki, H. , Kato, M. , Noda, Y. , Yamaguchi, T. , & Hirano, H. Y. (2013). Two WUSCHEL‐related homeobox genes, narrow leaf2 and narrow leaf3, control leaf width in rice. Plant and Cell Physiology, 54, 779–792. 10.1093/pcp/pct032 [DOI] [PubMed] [Google Scholar]
- Iwakawa, H. , Iwasaki, M. , Kojima, S. , Ueno, Y. , Soma, T. , Tanaka, H. , … Machida, C. (2007). Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant Journal, 51, 173–184. 10.1111/j.1365-313X.2007.03132.x [DOI] [PubMed] [Google Scholar]
- Iwakawa, H. , Ueno, Y. , Semiarti, E. , Onouchi, H. , Kojima, S. , Tsukaya, H. , … Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant and Cell Physiology, 43, 467–478. 10.1093/pcp/pcf077 [DOI] [PubMed] [Google Scholar]
- Jun, J. H. , Fiume, E. , Roeder, A. H. K. , Meng, L. , Sharma, V. K. , Osmont, K. S. , … Fletcher, J. C. (2010). Comprehensive analysis of CLE polypeptide signaling gene expression and over‐expression activity in Arabidopsis . Plant Physiology, 154, 1721–1736. 10.1104/pp.110.163683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun, J. H. , Ha, C. M. , & Fletcher, J. C. (2010). BLADE‐ON‐PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell, 22, 62–76. 10.1105/tpc.109.070763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalve, S. , De Vos, D. , & Beemster, G. T. S. (2014). Leaf development: A cellular perspective. Frontiers in Plant Science, 5, e362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H. G. , Fang, Y. , & Singh, K. B. (1999). A glucocorticoid‐inducible transcription system causes severe growth defects in Arabidopsis and induces defense‐related genes. Plant Journal, 20, 127–133. 10.1046/j.1365-313X.1999.00575.x [DOI] [PubMed] [Google Scholar]
- Laux, T. , Mayer, K. F. X. , Berger, J. , & Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development, 122, 87–96. [DOI] [PubMed] [Google Scholar]
- Leibfried, A. , To, J. P. C. , Busch, W. , Stehling, S. , Kehle, A. , Demar, M. , … Lohmann, J. U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin‐inducible response regulators. Nature, 438, 1172–1175. 10.1038/nature04270 [DOI] [PubMed] [Google Scholar]
- Lin, H. , Niu, L. , McHale, N. A. , Ohme‐Takagi, M. , Mysore, K. S. , & Tadege, M. (2013). Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Proceedings of the National Academy of Sciences of the United States of America, 110, 366–371. 10.1073/pnas.1215376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W. , Shuai, B. , & Springer, P. S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES‐domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and adaxial‐abaxial patterning. Plant Cell, 15, 2241–2252. 10.1105/tpc.014969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K. J. , & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods, 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Machida, C. , Nakagawa, A. , Kojima, S. , Takahashi, H. , & Machida, Y. (2015). The complex of ASYMMETRIC LEAVES (AS) proteins plays a central role in antagonistic interactions of genes for leaf polarity specification in Arabidopsis . WIREs Developmental Biology, 4, 655–671. 10.1002/wdev.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi, Y. (2014). Posttranslationally modified small‐peptide signals in plants. Annual Review of Plant Biology, 65, 385–413. 10.1146/annurev-arplant-050312-120122 [DOI] [PubMed] [Google Scholar]
- Miyawaki, K. , Tabata, R. , & Sawa, S. (2013). Evolutionarily conserved CLE peptide signaling in plant development, symbiosis, and parasitism. Current Opinion in Plant Biology, 16, 598–606. 10.1016/j.pbi.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Nakata, M. , Matsumoto, N. , Tsugeki, R. , Rikirsch, E. , Laux, T. , & Okada, K. (2012). Roles of the middle domain‐specific WUSCHEL‐RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell, 24, 519–535. 10.1105/tpc.111.092858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardmann, J. , Ji, J. , Werr, W. , & Scanlon, M. J. (2004). The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development, 131, 2827–2839. 10.1242/dev.01164 [DOI] [PubMed] [Google Scholar]
- Nekrasov, V. , Staskawicz, B. , Weigel, D. , Jones, J. D. G. , & Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA‐guided endonuclease. Nature Biotechnology, 31, 691–693. 10.1038/nbt.2655 [DOI] [PubMed] [Google Scholar]
- Norberg, M. , Holmlund, M. , & Nilsson, O. (2005). The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development, 132, 2203–2213. 10.1242/dev.01815 [DOI] [PubMed] [Google Scholar]
- Pallakies, H. , & Simon, R. (2014). The CLE40 and CRN/CLV2 signaling pathways antagonistically control root meristem growth in Arabidopsis. Molecular Plant, 7, 1619–1636. 10.1093/mp/ssu094 [DOI] [PubMed] [Google Scholar]
- Rice, P. , Longden, I. , & Bleasby, A. (2000). EMBOSS: The European Molecular Biology Open Software Suite. Trends in Genetics, 16, 276–277. 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- Sarkar, A. K. , Luijten, M. , Miyashima, S. , Lenhard, M. , Hashimoto, T. , Nakajima, K. , … Laux, T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature, 446, 811–814. 10.1038/nature05703 [DOI] [PubMed] [Google Scholar]
- Schoof, H. , Lenhard, M. , Haecker, A. , Mayer, K. F. X. , Jurgens, G. , & Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell, 100, 635–644. 10.1016/S0092-8674(00)80700-X [DOI] [PubMed] [Google Scholar]
- Sharma, V. K. , Ramirez, J. , & Fletcher, J. C. (2003). The Arabidopsis CLV3‐like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Molecular Biology, 51, 415–425. 10.1023/A:1022038932376 [DOI] [PubMed] [Google Scholar]
- Stahl, Y. , Wink, R. H. , Ingram, G. C. , & Simon, R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Current Biology, 19, 909–914. 10.1016/j.cub.2009.03.060 [DOI] [PubMed] [Google Scholar]
- Strabala, T. J. , O'Donnell, P. J. , Smit, A.‐M. , Ampomah‐Dwamena, C. , Martin, E. J. , Netzler, N. , … Hudson, K. R. (2006). Gain‐of‐function phenotypes for many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiology, 140, 1331–1344. 10.1104/pp.105.075515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege, M. , Lin, H. , Bedair, M. , Berbel, A. , Wen, J. , Rojas, C. M. , … Mysore, K. S. (2011). STENOFOLIA regulates blade outgrowth and leaf vascular patterning in Medicago truncatula and Nicotiana sylvestris . Plant Cell, 23, 2125–2142. 10.1105/tpc.111.085340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya, H. (2013). Leaf development. The Arabidopsis Book, 11, e0163 10.1199/tab.0163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Zhang, G. , & Wu, M. (2016). CLE peptide signaling and crosstalk with phytohormones and environmental stimuli. Frontiers in Plant Science, 6, e1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weight, C. , Parnham, D. , & Waites, R. (2008). LeafAnalyzer: A computational method for rapid and large‐scale analyses of leaf shape variation. Plant Journal, 53, 578–586. [DOI] [PubMed] [Google Scholar]
- Whitford, R. , Fernandez, A. , De Groodt, R. , Ortega, E. , & Hilson, P. (2008). Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proceedings of the National Academy of Sciences of the United States of America, 105, 18625–18630. 10.1073/pnas.0809395105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Xu, Y. , Dong, A. , Sun, Y. , Pi, L. , Xu, Y. , & Huang, H. (2003). Novel as1 and as2 defects in leaf adaxial‐abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying adaxial identity. Development, 130, 4097–4107. 10.1242/dev.00622 [DOI] [PubMed] [Google Scholar]
- Yadav, R. K. , Perales, M. , Gruel, J. , Girke, T. , Jonsson, H. , & Reddy, G. V. (2011). WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes & Development, 25, 2025–2030. 10.1101/gad.17258511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, Y. L. , Ishida, T. , Yoshimura, M. , Imamura, Y. , Shimaoka, C. , & Sawa, S. (2017). A collection of mutants for CLE‐peptide‐encoding genes in Arabidopsis generated by CRISPR/Cas9‐mediated gene targeting. Plant and Cell Physiology, 58, 1848–1856. 10.1093/pcp/pcx139 [DOI] [PubMed] [Google Scholar]
- Yamaguchi, N. , Winter, C. M. , Wu, M.‐F. , Kwon, C. S. , William, D. A. , & Wagner, D. (2014). Chromatin immunoprecipitation from Arabidopsis tissues. The Arabidopsis Book, 12, e0170 10.1199/tab.0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


