Summary
2-oxoglutarate:ferredoxin oxidoreductase (OGOR) is a thiamine pyrophosphate (TPP) and [4Fe-4S] cluster-dependent enzyme from the reductive tricarboxylic acid (rTCA) cycle that fixes CO2 to succinyl-CoA, forming 2-oxoglutarate and CoA. Here we report an OGOR from the rTCA cycle of Magnetococcus marinus MC-1, along with all three potential ferredoxin (Fd) redox partners. We demonstrate MmOGOR operates bidirectionally (both CO2-fixing and 2-oxoglutarate oxidizing), and that only one Fd (MmFd1) supports efficient catalysis. Our 1.94-Å and 2.80-Å resolution crystal structures of native and substrate-bound forms of MmOGOR reveal the determinants of substrate specificity and CoA-binding in an OGOR, and illuminate the [4Fe-4S] cluster environment, portraying the electronic conduit allowing MmFd1 to be wired to the bound-TPP. Structural and biochemical data further identify Glu45α as a mobile residue that impacts catalytic bias toward CO2-fixation although it makes no direct contact with TPP-bound intermediates, indicating that reaction directionality can be tuned by second layer interactions. (149 of 150 words limit)
Keywords: CO2 fixation, reductive tricarboxylic acid (rTCA) cycle, 2-oxoglutarate:ferredoxin oxidoreductase, ferredoxin, thiamine pyrophosphate, iron-sulfur clusters, electron-transfer, catalytic bias, Magnetococcus marinus MC-1
Introduction
CO2 reduction is a thermodynamically and kinetically challenging process,1 but nature has developed ways of transforming CO2 into different organic molecules. In the biosphere, the Calvin cycle of photosynthesis converts CO2 and ribulose-1,5-biphosphate into 3-phosphoglycerate with ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo),2 an enzyme with cross-reactivity towards oxygen and a reaction rate of a few turnovers per second.2 Five other biological pathways found in archaea and anaerobic bacteria, also assimilate CO2 into biomass,3 including the reductive tricarboxylic acid (rTCA) cycle (Figure S1), which is energetically more efficient than the Calvin cycle.3–4 To overcome thermodynamically disfavored CO2 reduction, two key steps of CO2 fixation in the rTCA cycle are catalyzed by members of the 2-oxoacid:ferredoxin oxidoreductase (OFOR) superfamily (Figure 1B and S2): pyruvate:ferredoxin oxidoreductase (PFOR) and 2-oxoglutarate:ferredoxin oxidoreductase (OGOR), use the reducing power of ferredoxin (Fd, a low potential electron carrier), to catalyze the reduction of CO2 with acetyl-CoA and succinyl-CoA, to form pyruvate and 2-oxoglutarate, respectively (Figure 1A,B). As a pyruvate synthase, PFOR also participates in the dicarboxylate/4-hydroxybutyrate cycle, and links the Wood-Ljungdahl pathway to the incomplete rTCA cycle in methanogens and acetogens.3, 5 A molecular view of how OFORs achieve reversibility has yet to be revealed, in part due to the paucity of data on enzymes that run natively in the reductive direction. Here we report the first ‘rTCA OFOR’, revealing molecular details essential to the OFOR family, which also appear critical for the reduction of CO2.
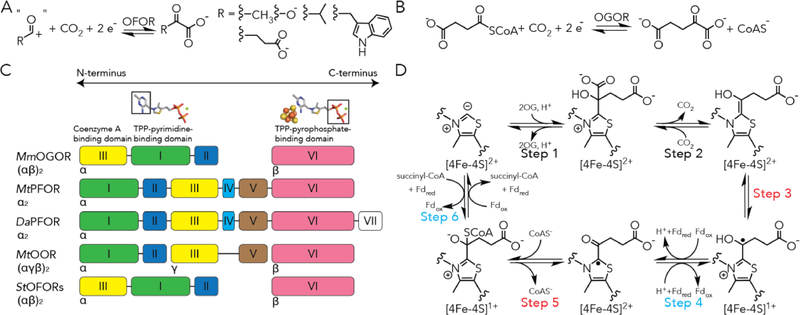
Figure 1. 2-oxoacid:ferredoxin oxidoreductases (OFORs) and the domain arrangements of structurally characterized OFORs.
(A) General scheme of OFOR reactions. (B) 2-oxoglutarate:ferredoxin oxidoreductase (OGOR) reversibly reduce carbon dioxide and succinyl-CoA into 2-oxoglutarate. (C) The domain arrangements of structurally characterized OFORs. Domain III is a coenzyme A binding domain. Domain I binds the pyrimidine moiety of thiamine pyrophosphate (TPP). Domain VI binds the pyrophosphate of TPP and a [4Fe-4S] cluster. Domain V, which is absent in MmOGOR, adapts a ferredoxin fold that binds two [4Fe-4S] clusters in PFOR and OOR. (D) The proposed reaction mechanism of carbon fixation and 2-oxoglutarate oxidation by OGOR based on studies from MtPFOR.14,19.20 Steps involving Fd-based electron transfer are labeled with teal, and those involving internal electron transfer are labeled with red.
Divided by substrate specificity, OFORs can be categorized into 2-oxoglutarate:ferredoxin oxidoreductase (OGOR),6 pyruvate:ferredoxin oxidoreductase (PFOR),7 2-oxoisovalerate:ferredoxin oxidoreductase (VOR),8 indopyruvate:ferredoxin oxidoreductase (IOR),9–10 and oxalate oxidoreductase (OOR)11–14 (Figure S2). Notably, OFORs adapt different oligomeric states (Figure 1C) and are mostly coenzyme A (CoA)-dependent except for OOR,15 which is specific for cleaving oxalate into CO2 without CoA.11, 14 Although PFOR and OGOR from the rTCA cycle engage in CO2 fixation, on many occasions, OFORs catalyze the reversal reaction to oxidize an 2-oxo-acid and provide low potential electrons for downstream reactions, including sulfate reduction,16 dinitrogen reduction,17 and aromatic compound reduction.18 Yet, the basis of why OFORs display preferences for either CO2-reduction or 2-oxoacid oxidation in any specific pathway is still unknown.
Our current understanding on the OFOR superfamily is predominantly based on OFORs engaged in oxidative chemistry, such as the PFOR from Moorella thermoacetica (MtPFOR),19–21 which reversibly oxidizes pyruvate to generate CO2 and electrons that are localized on the Fd redox partner, which are then used for all reductive reactions of physiology. MtPFOR contains one thiamine pyrophosphate (TPP) (like all OFORs) and three [4Fe-4S] clusters per catalytic unit (like many OFORs). For pyruvate oxidation, MtPFOR utilizes TPP to activate pyruvate. The lactyl-TPP intermediate formed undergoes decarboxylation and transfers an electron to one of the three [4Fe-4S] clusters to form a radical intermediate that is tractable via electron paramagnetic resonance (EPR) spectroscopy.21 The TPP-radical species then decays to transfer the second electron to another of the three [4Fe-4S] clusters.20 However, the radical decay process is gated when CoA is absent, and the electron transfer is accelerated by 105 -fold in the presence of CoA.19–20 Electronic repulsion between the thiolate of CoA and the radical intermediate shifts the equilibrium toward electron transfer and product formation.14, 19 Acetyl-CoA is released in the last step, and electrons housed on the internal [4Fe-4S] clusters are delivered to an external Fd (or Fds), which are then used in other pathways.
In this way, MtPFOR provides a great mechanistic framework to probe CO2 reduction by OFORs presuming microscopic reversibility (Figure 1D). However, OGOR enzymes found in the rTCA largely are composed of enzymes that differ from other OFORs in that they lack the ferredoxin domain (domain V) that harbors two [4Fe-4S] clusters (Figure 1C).15 In other words, bioinformatics predicts that an OGOR is an OFOR with only one [4Fe-4S] cluster, which should be insufficient for the two-electron process. We have turned to investigating the structural and biochemical properties of a native rTCA OGOR, as well as its physiologically relevant Fd, in order to understand how an OFOR can be biased toward CO2 reduction, and how electron transfer chemistry may operate in support of the rTCA cycle.
Prior to our work, structures of OFORs have been limited to three different types, two PFORs (MtPFOR19 and the cognate enzyme from Desulfovibiro africanus (DaPFOR)22–24), an oxaloacetate oxidoreductase (OOR) from M. thermoacetica (MtOOR),12–13 and two OFORs from Sulfolobus tokodaii of cryptic function (StOFORs).25 Even with these structures in hand, many open questions about OFOR chemistry remain, including the issues of electron transfer described above. For example, how OFORs differentiate 2-oxoacids and/or their acyl-CoAs to proceed in either CO2-reduction or 2-oxoacid-oxidaton is still unknown. This question is particularly critical to the exploitation of the rTCA in any kind of bioenergy application, where both PFOR and OGOR (Figure S1) are required: if the wrong substrate is channeled by either enzyme down the oxidative direction, it is easy to imagine that the pathway may short-circuit. The structures that are available report on enzymes that bind small substrates, 2-oxoacids with six non-H atoms, instead of larger substrates, i.e. 2-oxoglutarate, so we do not yet understand how larger substrates are accommodated. And even though StOFOR is reactive toward 2-oxoglutarate,25 they are not substrate specific, so identification of a physiological substrate and specific function has been difficult; mutational studies shift reactivity in ways that the structures cannot account for. Consequently, a structure of a substrate-specific OGOR with a full description of structure-function relationships will greatly expand our understanding of OFOR chemistry. Additionally, except for MtPFOR, which is able to catalyze both reactions at different growth conditions, all the current structurally characterized OFORs catalyze 2-oxoacid oxidation in vivo. Therefore, characterization of a substrate-specific OGOR, which at the physiological conditions reduces CO2 will help us understand the traits of OFORs that favor CO2 reduction vs 2-oxoacid oxidation.
Here, we report the first crystal structural of an OGOR from the rTCA cycle, OGOR from Magnetococcus marinus MC-1 (MmOGOR), a magnetotactic mesophile that has an established rTCA pathway used for carbon uptake.26–27 We present not only a series of structures, but corresponding spectroscopic and biochemical characterizations, including the identification of the cognate physiological redox partner of MmOGOR that is essential for the native CO2 fixation activity.
Results
MmOGOR is a substrate-specific OFOR with one [4Fe-4S] cluster per catalytic unit
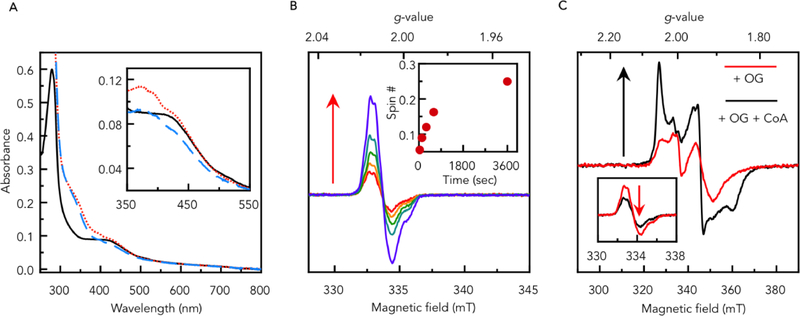
Recombinant MmOGOR (Mmc1_1749, 1750) was expressed in E. coli and purified as a holo-protein.27 The iron and sulfur quantification assays show 3.6 ± 0.2 iron and 3.4 ± 0.1 inorganic sulfur per αβ heterodimer, which indicate MmOGOR binds one [4Fe-4S] cluster per catalytic unit, in agreement with protein sequence analyses for predicted OGORs in the OFOR superfamily. As-purified MmOGOR was EPR silent (data not shown) and displayed a broad UV-vis absorption band around 400 nm (Figure 2A, black solid line), suggesting the [4Fe-4S] cluster of MmOGOR was purified in the oxidized state ([4Fe-4S]2+).
Figure 2. Spectroscopic properties of MmOGOR.
(A) UV-vis spectra of 5 μM as-purified MmOGOR (black solid line), first incubated with 5 mM 2-oxoglutarate (OG) for 15 min (red dotted line), and then supplied with 0.1 mM coenzyme A (CoA) for additional 5 min (blue dashed line). The spectra between 350 nm and 550 nm are shown in the inset with the background absorbance of OG and CoA subtracted. (B) EPR spectra of 70 μM MmOGOR incubated with 20 mM OG at room temperature for 30 sec, 2 min, 5 min, 10 min and 60 min measured at 52K. Spin quantification of each time point is shown in the inset. Experimental conditions: microwave frequency, 9.386 GHz; microwave power, 10 μW; modulation amplitude, 0.2 mT. (C) EPR spectra of 70 μM MmOGOR incubated with 20 mM OG at room temperature for 30 sec (red) and the same sample supplied with 1 mM CoA for additional 10 sec (black) measured at 15K. Experimental conditions: microwave frequency, 9.384 GHz; microwave power, 4 mW; modulation amplitude, 0.5 mT. The same samples measured at the same condition as panel (B) are shown in the inset.
Recombinant MmOGOR is highly active when compared to other OFORs (Table S2). As-purified MmOGOR exhibited a kcat of 1820 min−1 (Vmax = 19.5 μmol min−1 mg−1, Figure S3A) and an apparent KM of 4.4 mM for 2-oxoglutarate oxidation at pH 8.5 (Table 1). The activity was favored at alkaline conditions with the maximum activity observed between pH 8.5 and 10.5 (Figure S3B). The high KM indicates MmOGOR is a CO2-fixing 2-oxoglutarate synthase in vivo, which is in agreement with two CO2-reducing OGORs from Hydrogenobacter thermophilus involved in the rTCA cycle exhibiting KM values of 1.4 and 2.9 mM towards 2-oxoglutarate.28 In contrast, a 2-oxoglutarate-oxidizing OGOR from Thauera aromatica has a KM of 110 μM.18
Table 1.
Kinetic properties for MmOGOR and its molecular variants
| Direction | Substrates | Enzyme | kcat (min−1) | KM (μM) | kcat/KM (s−1 M−1) | Buffer pH |
|---|---|---|---|---|---|---|
| 2-oxoglutarate oxidation | 2-oxoglutarate | WT | 1820 | 4400 | 6.9 × 103 | 8.5 |
| WT | 650 | 970 | 1.1 × 104 | 7.0 | ||
| WT | 1334 | 4900 | 4.5 × 103 | 7.5 | ||
| Y436αF | 1068 | 1800 | 9.9 × 103 | 7.5 | ||
| E45αQ | 407 | 640 | 1.1 × 104 | 7.5 | ||
| CoA | WT | 1480 | 6.8 | 3.6 × 106 | 8.5 | |
| WT | 674 | 10 | 1.1 × 106 | 7.0 | ||
| MmFd1 | WT | 735 | 1.8 | 6.8 × 106 | 7.0 | |
| CO2 reduction | Succinyl-CoA | WT | 26.8 | 32 | 1.4 × 104 | 7.0 |
| MmFd1 | WT | 30.6 | 1.3 | 3.9 × 105 | 7.0 | |
Incubating MmOGOR with 2-oxoglutarate alone, we found the characteristic absorption of the TPP-based radical around 370 nm in the UV-vis spectrum (Figure 2A, red dotted line).29 The TPP-based radical was monitored by X-band electron paramagnetic resonance (EPR) spectroscopy at 52K. After incubating with 2-oxoglutarate at RT for 30 sec, OGOR generated ~0.06 spin per αβ heterodimer (g = 2.01), and the spin number increased to 0.25 spin after incubated at RT for 1 hr (Figure 2B, with EPR simulations shown as Figure S4). Features of reduced [4Fe-4S] clusters can be detected by EPR at 15 K (S = 1/2 for [4Fe-4S]+, g = 2.04, 1.94, and 1.89, Figure 2C, red dotted line and Figure S4), and there was ~0.02 spin per αβ heterodimer after incubating for 30 sec (or longer). Although the ratio of the TPP radical to [4Fe-4S]+ is non-stoichiometric, the parallel changes suggest the formation of radical and the reduction of the [4Fe-4S]2+ cluster is a concomitant process in the OGOR reaction (Step 3, Figure 1D). Indeed, this transformation has never been captured previously via spectroscopy for (αβ)2 type OFORs, but only inferred.30–32 The lower spin number of [4Fe-4S]+ compared to that of the radical over the course of 1 hr could be attributed to the latent instability of the cluster, perhaps via an intrinsic hydrogenase activity of the reduced [4Fe-4S] cluster(s) in OFORs33–34 in the absence of their physiological redox partners.
Upon CoA addition into the mixture of OGOR and 2-oxoglutarate, the UV-vis absorption features around 370nm decreased (Figure2A, blue dashed line), suggesting the TPP-based radical was turned-over (Step 5, Figure 1D). EPR at 52K indicates the spin number for the TPP-based radical dropped from 0.06 to 0.03 per αβ heterodimer (Figure 2C inset) by supplying CoA to MmOGOR pre-incubated with 2-oxoglutarate for 30 sec. Conversely, the signal for [4Fe-4S]+ (g = 2.05, 1.94, 1.86) increased from 0.02 to 0.07 per αβ heterodimer. These results confirm that CoA promotes the electron transfer from the TPP-based radical to the [4Fe-4S] cluster (Step 5, Figure 1D).20 Thus, 2-oxoglutarate oxidation catalyzed by MmOGOR appears to follow the mechanism that has been proposed for MtPFOR, which is the first demonstration of mechanistic conservation across the OFOR superfamily (Figure 1D).
In terms of substrate specificity, in steady-state assays MmOGOR demonstrated high substrate specificity towards 2-oxoglutarate with negligible activity with other 2-oxoacids, including pyruvate-like substrates (glyoxylate, pyruvate, 2-oxobutyrate, 2-oxoisovalerate, 3-methyl-2-oxovalerate, phenylpyruvate) and 2-oxoglurate-like substrates (oxaloacetate and 2-oxoadipate) (Table S3).
MmOGOR requires a cognate electron donor Fd for CO2 reduction
Fds with [4Fe-4S] clusters are the hypothesized or known redox partners for OFORs.5, 28, 35–36 In the M. marinus MC-1 strain genome,27 we identified three genes that encode [4Fe-4S] Fds (Figure S5B–D and Table S1). MmFd1 (Mmc1_0249) is located in the gene cluster that comprises genes related to arginine biosynthesis (argC) and regulation of cell metabolism under nutritional stress such as amino-acids deprivation (spoT/relA),37–39 which suggests MmFd1 could play a key role in the autotrophic growth of the cell where CO2 is the only carbon source. MmFd2 (Mmc1_1207) and MmFd3 (Mmc1_1191) are in the nif-gene clusters known to be involved in the FeMo cofactor biosynthesis and nitrogen fixation.40–42 M. marinus MC-1 is able to fix nitrogen under both heterotrophic and autotrophic growth conditions,27 but it is not clear if and how these Fds from the nif-gene clusters participate in the CO2 fixation pathway.
Sequence analysis suggests all three Mm Fds contain two [4Fe-4S] clusters. MmFd1 and MmFd2 belong to a well-characterized AlvinFd type,43 whereas MmFd3 belong to a type exemplified by the nif-specific FdIII (Figure S6).41 For further characterization, we recombinantly expressed and purified all the three Fds. Electrochemical measurement showed both MmFd1 and MmFd3 exhibited two distinct reduction potentials for each of their clusters where MmFd1 has much lower reduction potentials (MmFd1: Em,1= −635 mV, Em,2= −485 mV, vs Standard Hydrogen Electrode (SHE); MmFd3: Em,1= −380 mV, Em,2= −233 mV, vs SHE). In contrast the two clusters of MmFd2 exhibited the same reduction potential (Em,1=Em,2=−520 mV, vs SHE) (Figure S7), as has been demonstrated by other two-cluster Fds.44–45
To kinetically implicate a natively functional Fd, we compared the rates of MmOGOR in both 2-oxoglutarate oxidation and CO2 reduction directions, using the three MmFds as the electron acceptors and donors, respectively. In the 2-oxoglutarate oxidation direction, we equate the rates of MmOGOR to the rates of Fd reduction which were measured directly through the absorbance change at 400 nm caused by the reduction of [4Fe-4S] cluster of Fds (Figure S8) and indirectly through a coupled assay that re-oxidizes Fds using metronidazole (which loses its characteristic absorption at 320 nm when reduced) (Figure S8 and S9A).46 In both assays, MmOGOR demonstrated the highest rate when MmFd1 served as the electron acceptor (Figure S8 and S9). On the other hand, the rates of CO2 reduction can be monitored by the generation of 2-oxoglutarate using glutamate dehydrogenase (GDH) and NADPH (Figure S10). GDH reduces 2-oxoglutarate by NADPH, which loses its characteristic absorption at 340 nm when oxidized. MmFd1 could support a CO2 reduction activity of 27.2 ± 0.9 min−1, higher than that of MmFd2 (6.3 ± 1.1min−1) and MmFd3 (8.1± 0.4 min−1). The kinetic differences between the Fds suggest MmFd1 is a better redox partner to MmOGOR and that CO2 reduction requires a cognate electron donor. Hence, we further obtained kinetic parameters of MmOGOR in 2-oxoglutarate oxidation and CO2 reduction at pH 7.0 using MmFd1 as the redox partner. (Table 1, Figure S4E–F). Because the upstream reaction in the CO2 reduction assay also generates CO2 (Figure S9B), we did not measure the KM towards CO2.28
Even though the catalytic efficiency (kcat/KM) of MmOGOR measured in our in vitro assays is higher in the 2-oxoglutarate oxidation direction as compared to the CO2 reduction direction (Table 1), there are many factors indicating MmOGOR favors CO2 fixation in vivo. First, the concentration for CO2 in the assay condition is maintained at ~2.2 mM through chemical equilibrium with 20 mM bicarbonate, whereas the cellular CO2 concentration can be higher under autotrophic conditions. For example, the cellular concentration of CO2 could reach up to 33 mM in the case of M. thermoacetica growing under 100% CO2 atmosphere.5 The higher CO2 concentration in the cellular environment is expected to favor CO2 reduction. Second, MmOGOR exhibits a low KM toward succinyl-CoA (32 μM) versus a high KM for 2-oxoglutarate (970 μM). The difference in KM indicates CO2 reduction can reach its Vmax with a low substrate concentration. Lastly, a succinyl-CoA synthethase (SucCD) is identified upstream of MmOGOR in the genome of M. marinus MC-1 (Figure S3A and Table S1),26–27 which suggests OGOR-catalyzed CO2 reduction as part of the rTCA cycle (Figure S1) is a highly-concerted process, with succinyl-CoA synthethase delivering succinyl-CoA to MmOGOR to ensure supply of co-substrate for CO2 fixation.
Structure of MmOGOR reveals a common domain arrangement and a tailored active site
MmOGOR was crystallized anaerobically, and its structure was determined to 1.94-Å resolution (Table S4 and S5). MmOGOR is a dimer of dimers with an overall oligomeric structure of (αβ)2 (Figure 3A). Each αβ heterodimer forms a catalytic unit composed of one TPP molecule, one [4Fe-4S] cluster and four domains. Interestingly, the order of the domains in the primary sequence varies among OGORs (See Figure 1C). In MmOGOR, domain III (residues 2–197) of α comes first, followed by domain I (residues 198–460), and then domain II (residues 461–573), whereas the β subunit contains domain VI. Despite these differences in primary structure, the arrangement of the domains is the same in all structurally characterized family members (Figure 3A and S11). As observed previously, domain I, II, and VI form the dimer interface and domain III is distal to this interface. Notably, the common ferredoxin domain (V) that binds two [4Fe-4S] clusters is absent in MmOGOR (Figure S11). The roles that the various domains play is conserved; domain I binds the pyrimidine moiety of TPP; domain VI binds the pyrophosphate moiety of TPP and the only [4Fe-4S] cluster in MmOGOR, which is equivalent to the proximal [4Fe-4S] cluster in PFOR and OOR (Figure S12). The rearrangement of domains is achieved simply by rewiring the flexible regions in both ends of each domain. A similar domain allocation was observed in StOFOR1 (29% identity for chain α; 39% identity for chain β) and StOFOR2 (29% identity for chain α; 38% identity for chain β). Although StOFORs are active toward 2-oxoglutarate, both StOFORs lack substrate specificity, and no 2-oxoglutarate or succinyl-CoA bound structure is available.
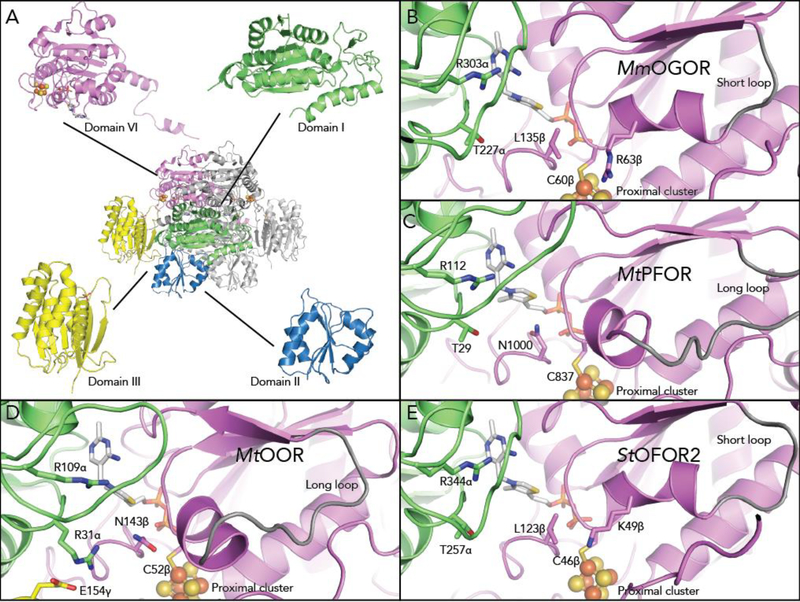
Figure 3. The structure of MmOGOR and the comparison of active sites between OFORs.
(A) The crystal structure and domain arrangement of MmOGOR. MmOGOR is a dimer of heterodimers. Each heterodimer forms a catalytic unit that is comprised of chain α (domain III-I-II from N to C terminus), chain β (domain VI), one TPP molecule bound to domain I and VI, and one [4Fe-4S] cluster anchored by domain VI. (B) The active site of MmOGOR contains two positively charged residues, Arg303 and Arg63β, for binding of succinyl-CoA or 2-oxoglutarate, a dicarboxylic acid. Thr227α is a part of a conserved YPTIP motif in which Thr facilitates binding of carboxylic acid substrate moiety. The active site is larger due to the presence of a short loop (gray) that follows conserved residue Cys60β and restricts the position of a helix on the outer edge of the active site, increasing the space for substrate binding. (C) The active site of MtPFOR (PDB ID: 6CIN19) uses Arg112, Thr29 and Asn1000 to bind pyruvate. The single positively charged residue suites the singly negatively-charged pyruvate. MtPFOR contains a longer loop (gray, extended outside the frame) that positions a helix into the active site, effectively shrinking the active site such that it accommodates a smaller substrate. (D) The active site of MtOOR (PDB ID: 5C4I12) contains two positively charged residues, Arg109α and Arg31α, for binding oxalate, a dicarboxylic acid. Similar to MtPFOR, OOR contains a longer loop, and thus smaller active site pocket. (E) The active site of StOFOR2 (PDB ID: 5B4625) contains two positively charged resides and a large active site pocket, but the origin of its lack of specificity is unclear
The space in the active site of MmOGOR is greater compared to that of OOR and PFOR (Figure 3B–D), which allows larger substrates, both 2-oxoglutarate and a succinyl group, to enter the binding pocket. The differences in binding pockets sizes result from differences in the helix-loop motif following Cys60β, an absolutely conserved residue that binds the proximal [4Fe-4S] cluster in all OFORs. In MmOGOR, the motif is composed of 10 residues before it links to the beta sheet of the Rossmann-like domain VI, whereas the same motif is 6 and 12 residues longer in MtOOR and MtPFOR. The extension in MtOOR or MtPFOR forms a longer loop, which pushes the helix part of the helix-loop motif toward to the active site, effectively shrinking the space available for substrate binding (Figure 3C,D). The shorter loop in MmOGOR pulls the helix back from the active site afforded room for larger substrates to bind (Figure 3B). StOFOR2 also has a short loop (Figure 3E), perhaps explaining its promiscuous behavior with larger substrates.
Arg303α and Thr227α, which are conserved 2-oxoacid binding residues in all structurally characterized CoA-dependent OFOR,15 are also found in MmOGOR (Figure 3B). MmOGOR has a second Arg in the active site, Arg63β from the helix-loop motif, which is 10.7 Å from C2 of TPP. The distance is reasonable to bind C5 of 2-oxoglutatate or the succinyl group of succinyl-CoA, and will later be shown to bind both. The active site design is akin to OOR, which utilizes two positively charged residues to bind oxalate, a dicarboxylic-acid, but the position of the second positively charged residue (Arg31α in OOR) is not the same (Figure 3B,D). In StOFORs, a lysine (Lys48β) was also proposed to play a similar role (Figure 3E).25
The environment of the [4Fe-4S] cluster is different for enzymes that lack a Fd domain
The [4Fe-4S] cluster in MmOGOR locates 10.6 Å away from the TPP active site (from the edge of the cluster to the C2 position of TPP), which is also the position equivalent to the proximal cluster in PFOR and OOR. The proximity between cofactors indicates the [4Fe-4S] cluster is capable of facile electron-transfer and thereby playing an essential role in the two distinct one-electron processes of OFOR reactions (Figure 1D).
The environment of the cluster in MmOGOR is more similar to StOFOR1 than it is to OFORs that contain a domain V (Figure 4). Hydrophobic residues surround the [4Fe-4S] cluster in MmOGOR (Figure 4A). Ile46α from domain III is positioned next to the [4Fe-4S] cluster, and the closest residues in the cluster-binding motif of MmOGOR are bulky and hydrophobic residues, which restrict the solvent accessibility of the cluster. Sequence alignment of reported (αβ)2 type OFORs reveals that those bulky and hydrophobic residues from domain III and [4Fe-4S] cluster binding motif are well conserved (Figure S13). Although not commented on previously, the [4Fe-4S] cluster in StOFOR is similarly hydrophobic (Figure 4B). In contrast, in MtPFOR and MtOOR, the residues surrounding the proximal cluster are all small or hydrophilic residues (Figure 4C and 4D). A previous study on StOFOR1, an (αβ)2 type OFOR, showed a low reduction potential of −545 mV (vs SHE),32 which likely arises from the limited solvent accessibility and lack of positively charged residues near the cluster, in comparison to the Mt and Da enzymes. Since the cluster environments are similar in both MmOGOR and StOFOR1 (Figure 4B); it is expected that the reduction potential of the MmOGOR cluster would also be similar. (Efforts to measure the redox potential of the MmOGOR cluster by protein film electrochemistry were unsuccessful.)
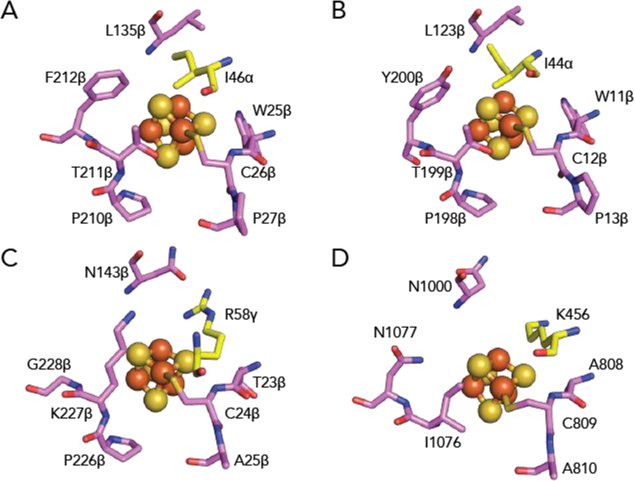
Figure 4. The environment of the proximal [4Fe-4S] cluster in structurally characterized OFORs.
(A) In MmOGOR, the proximal cluster is surrounded by hydrophobic residues. The hydrophobic interactions include Leu135β and two loops formed by cluster-ligating motifs, 25βWCP27β and 209βCPTF212β. Additionally, Ile46α, which is a positively charged residue in PFOR and OOR, is contributed by domain III. (B) In StOFOR1 (PDB ID: 5B4825), the proximal cluster is also surrounded by hydrophobic residues. The hydrophobic interactions include Leu123β, two neighboring loops that are part of cluster-ligating motifs, 11βWCP13β, and 197βCPTY200β, and Ile44α from domain III. (C) In MtOOR (PDB ID: 5C4I12), a conserved positively-charged arginine from domain III, Arg58γ, resides next to the proximal cluster. Other residues surrounding the proximal cluster are smaller in comparison to those of MmOGOR and StOFORs. (D) In MtPFOR (PDB ID: 6CIN19), a conserved positively-charged lysine from domain III, Lys456, is within hydrogen-bonding distance with the proximal cluster. Other residues surrounding the proximal cluster are smaller in comparison to those of MmOGOR and StOFORs. Domain coloring as in Figure 1A.
To examine the importance of these hydrophobic residues in MmOGOR, Ile46α, a residue not responsible for [4Fe-4S] binding (Figure 4A and S13A), was mutated to an alanine. The Ile46αAla variant was fully active compared to the wild type when benzyl viologen (BV) was used as the electron acceptor in the standard assay (2033±206 min−1, vs 1330±34 min−1 for the wild type), indicating the enzyme overall fold was not disrupted through the mutation. However, the activity diminished when methyl viologen (MV) was used as the electron acceptor (8.6±0.4 min−1, vs 1250±25 min−1 for the wild type). The difference in activity with different redox mediators (MV, −444 mV; BV, −360 mV, vs SHE) suggests the reduction potential of the [4Fe-4S] cluster in the Ile46αAla variant was higher than that of the wild type, further implicating Ile46α in modulating the reduction potential of the [4Fe-4S] cluster in MmOGOR.
Co-crystal structures show substrate-binding determinants and reveal domain movement
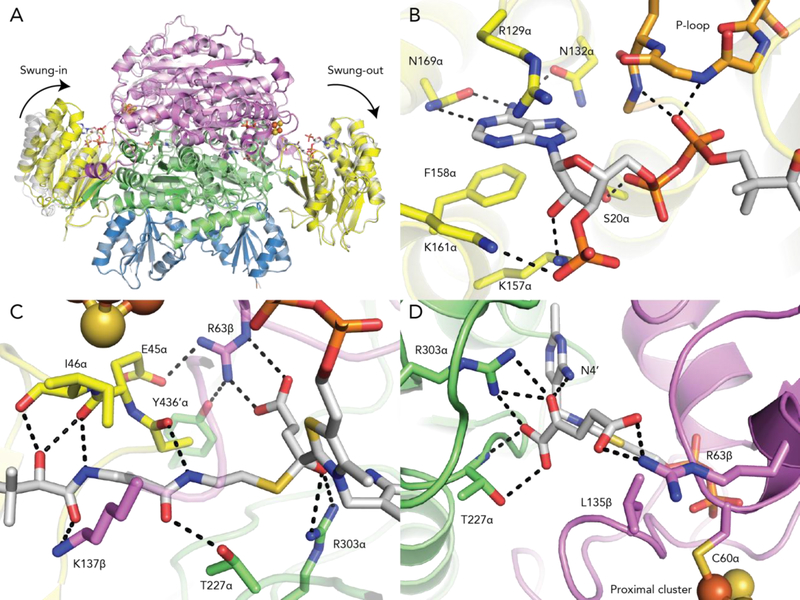
To demonstrate the molecular basis of substrate recognition, the structure of MmOGOR co-crystallized with 2-oxoglutarate and CoA was solved to 2.80-Å resolution (Table S4 and S5). The overall structure of MmOGOR co-crystallized with substrates is similar to the native structure without substrate (Figure 5A). However, we were fortunate to catch MmOGOR in two different states: in one αβ heterodimer, succinyl-CoA is bound and domain III adopts a ‘swung-in’ conformation, whereas in the other heterodimer, 2-oxoglutarate and CoA are bound and domain III is ‘swung-out’ (Figure 5A, S14). Both catalytic units reveal key components for substrate/product binding.
Figure 5. Structure of MmOGOR with 2-oxoglutarate and succinyl-CoA bound.
(A) Two conformations of domain III are observed. In the absence of substrate or product, domain III is swung-out (native structure in grey). In the co-crystal structure (colored), domain III is swung-in when succinyl-CoA is bound and swung-out in the presence of 2-oxoglutarate and CoA. (B) Domain III binds the adenosine end of succinyl-CoA similarly to what was observed in MtPFOR. The P-loop, 12αGEGGEG17α, and Ser13α contact the pyrophosphate. 129αRx2N132α and 158αFx10N169α recognize the adenine ring and Lys157α and Lys161α are positioned to make charge-charge interactions with the 3′-phospho group of succinyl-CoA. (C) The other end of succinyl-CoA extends into the active site formed by domain I and VI. The side chain of Lys137β and the backbone carbonyl atoms of Ala44α, Glu45α and Ile46α form hydrogen bonds with the pantothenate moiety of CoA. Thr227α also contact the extended CoA moiety. Residues Arg303α and Arg63β contact the succinyl group. (D) 2-oxoglutarate is bound to the MmOGOR active site by the same two positively-charged residues, Arg303α and Arg63β, which also contact the succinyl moiety of succinyl-CoA. Thr227α, which is a part of previously identified YPITP motif, also contributes to substrate binding. Leu135β provide hydrophobic contact with 2-oxoglutarate. Domain coloring as in Figure 1A.
In the αβ heterodimer that has succinyl-CoA bound, domain III has moved in toward the active site such that the succinyl-CoA molecule can reach from domain III, where the 3’-phosphoadenosyl end of the CoA is anchored, across and into the active site housed in domains I and VI. A similar domain III movement is observed when CoA binds to MtPFOR.19 All three motifs that were previously discovered to bind the 3’-phosphoadenosyl pyrophosphate moiety in MtPFOR are found in domain III of MmOGOR, 12αGEGGEG17α, 129αRx2N132α and 158αFx10N169α (Figure 5B). 12αGEGGEG17α, also known as the P-loop (Phosphate-binding loop) motif, binds phosphate through the electronic dipole of the subsequent alpha helix and via hydrogen bonds from backbone amides. Asn132α and Asn169α form hydrogen bonds with the adenine base, whereas Arg129α and Phe158α form cation-pi and pi-pi interaction with the adenine. In addition to previously identified motifs, Ser20α of MmOGOR forms a hydrogen bond with the CoA pyrophosphate, and the side chains of Lys157α and Lys161α on domain III bind 3 ´-phospho group of CoA (Figure 5B). The latter interactions are akin to that observed in MtPFOR through Arg1016 on domain VI (Figure S14A and S14B). The finding that these two OFORs utilize residues from different domains to bind the 3 ´-phospho group of CoA reveals a certain degree of modularity in CoA substrate binding.
The pantothenate of CoA also makes a number of interactions as it stretches from domain III into the active site, including with Thr227α (Figure 5C) as also observed in MtPFOR.19 The pantothenate also forms a hydrogen bond with Lys137β from domain VI (Figure 5C). In StOFOR1, chemical modification of the residue corresponding to Lys137β was shown to inactivate the enzyme by 58%.47 The structure shown here suggests the chemical modification likely inactivates the enzyme by hindering CoA from accessing active site. In both MtPFOR and MmOGOR, domain III movement also results in the repositioning of a few residues from domain III toward the active site where backbone atoms from these residues form hydrogen bonds with the pantothenate of CoA (Figure 5C). Unique to MmOGOR, a side chain from one of these residues, Glu45α, reaches into the active site and directly contacts an important active site residue. Glu45α orients Arg63β such that it now contacts Tyr435α′, in addition to contacting the succinyl group of succinyl-CoA. This second-layer hydrogen-bonding network creates a binding pocket that can incorporate a succinyl group but is not spatially suitable for a 2-oxoglutarate molecule. C2 of TPP binds the CS1 atom of succinyl-CoA, and Arg303α and N4´ of TPP form hydrogen bonds with the OS1 atom of succinyl-CoA (Figure 5C).
In the αβ heterodimer that displays a swung-out domain III, CoA is bound to domain III but the CoA does not extend into the active site nor does it make all of the contacts observed for succinyl-CoA bound to MmOGOR or for CoA bound to MtPFOR. In particular, the interactions with P-loop residues and with Asn132α, Ser20α and Lys157α are preserved, but several of the other residues responsible for anchoring CoA to domain III in MmOGOR (Arg129α, Asn169α, Phe158α and Lys161α) have not undergone the conformational rearrangements necessary to contact the CoA (Figure S14C). It appears that the binding of CoA such that it extends into the active site requires the swinging-in of domain III in addition to local residue arrangements; the same is true for MtPFOR. Thus, this snapshot of CoA-bound to MmOGOR would seem to represent a lower-affinity product-bound state where CoA is ready for release or a lower-affinity substrate-bound state that is pre-catalytic, whereas the succinyl-CoA bound structure, which shows the β-cystamine moiety stretching into the active site, would seem to represent a substrate-state that is ready for catalysis or represents a product-state that just completed catalysis.
In the CoA-bound MmOGOR structure, an intact 2-oxoglutarate is also found in the active site (Figure 5D and S14D). The oxygen atoms on C1 and C2 of 2-oxoglutarate form hydrogen bonds with TPP, Arg303α and Thr227α, which are similar interactions to those made by substrate in MtPFOR. The C5 carboxylic group of 2-oxoglutarate also forms hydrogen bond with Arg63β. Although 2-oxoglutarate and the succinyl moiety of succinyl-CoA both interact with Arg303α and Arg63β, the whole succinyl moiety fits between these two Arg residues whereas the larger 2-oxoglutarate extends down into the extended CoA binding site where it interacts with Thr227α (Figure 5C,D). It is the 2-oxoglutarate carboxylate that will be lost as CO2 that extends into this CoA binding site, suggesting that decarboxylation occurs before the cysteamine end of CoA enters the active site.
Based on aforementioned interactions between MmOGOR, and 2-oxoglutarate and succinyl-CoA (Figure 5D), we prepared OGOR variants of residues that directly interact with 2-oxoglutarate (Thr227αAla, Arg303αAla, Arg63βAla and Arg63βLeu) and residues that form second coordination sphere interactions with Arg63β (Glu45αGln and Tyr436αPhe), and measured their 2-oxoglutarate oxidation activities. The first group of variants that directly interact with 2-oxoglutarate displayed no detectable activity in assay conditions, likely the mutations were too strongly disruptive to substrate binding. Nevertheless, both second coordination-sphere variants show lower KM for 2-oxoglutarate than WT MmOGOR (Table 1, Figure S4C) while kcat was diminished, the magnitude of the change was less than that for KM. It is possible to rationalize the lower KM values by considering the effect on Arg63β when Glu45α and Tyr436α are not available for interactions; without the other residues Arg63β would be effectively strengthened in its ability to bind the carboxylate group of the 2-oxoglutarate-derived intermediates along the reaction pathway (See Figure 1D), thus lowering KM values. Loss of the negatively charged side chain in Glu45αGln-MmOGOR would be expected to have a larger effect on Arg63β than loss of a hydrogen bond interaction in Tyr436αPhe-MmOGOR, and the data support this prediction (Table 1). We would also expect that the loss of a negative charge near the TPP in Glu45αGln-MmOGOR to decrease kcat by removing a potential facilitating factor for TPP radical oxidation, and that decrease is observed (Table 1). In contrast, the Tyr436αPhe substitution would not be expected to effect TPP radical oxidation rates as much due to a more minimal electrostatic change in the active site, and kcat is in fact largely unchanged (Table 1). Overall, both substitutions lead to an increased kcat/KM, suggesting that second coordination sphere interactions are important to dictating whether an OGOR functions as a CO2-reducing or an 2-oxogluarate-oxidizing enzyme, where the KM alone may be sufficient to tune bias. Most interestingly, the 2-oxoglutarate-oxidizing OGOR from T. aromatica contains a glutamine instead of a glutamate on the corresponding site of Glu45α,18 whereas the CO2-fixing OGOR from H. thermophilus contains an aspartate on the corresponding site of Glu45α.28 Both observations strengthen our proposal that the second layer interaction steers the directionality of an OGOR. To our knowledge, this is the first direct evidence showing the influence of second layer interactions in the OFOR superfamily.
Discussion
MmOGOR, an (αβ)2 type OFOR from the rTCA cycle, provides a simple OFOR model system for understanding biological CO2 fixation with only four domains instead of the more typical six domains and only one [4Fe-4S] cluster to reduce to drive CO2 reduction instead of three. In this study, we sought to identify the physiological electron donor Fd for MmOGOR and to probe enzymatic activity in both directions, utilizing spectroscopic, biochemical and structural approaches. Additionally, we have investigated substrate specificity and the role of conformational change in catalysis by obtaining structures of MmOGOR with substrates and products bound.
MmOGOR is the first structurally characterized OFOR to operate physiologically in the CO2-reducing direction. The redox active [4Fe-4S]2+/1+ cluster of MmOGOR, which delivers electrons between Fd and the active site, is expected to have a low reduction potential based on its hydrophobic environment, which is far more similar to the iron-sulfur cluster environment in StOFOR than it is to the OFORs that have multiple clusters. Interestingly, the location of the ferredoxin-like domain V in PFORs and OOR is empty in MmOGOR (and StOFOR), suggesting that Fd binds to the same location as domain V. Modeling indicates protein-protein interactions are likely to be a key factor in affording specificity with multiple surfaces of the Fd and of the OFOR making contact in order to bring the clusters in close proximity for electron transfer (Figure S15). Thus, surface complementary and reduction potential of the Fd must be considered in assessing the molecular basis of electron donor specificity. In support of this idea, we find considerable differences in the ability of the three MmFds to achieve CO2 fixation. We also find that these three Fds have substantially different reduction potentials, ranging from −233mV to −635 mV. For MmFd3, the reduction potentials (−235 mV, −380 mV) are likely too high to serve as an electron donor for MmOGOR. Although the reduction potentials of MmFd2 ( −520 mV) match the expected potential for MmOGOR (~ −545 mV), this Fd is not a good electron donor or acceptor for MmOGOR, which implies a structural mismatch. MmFd1 has appropriate reduction potentials ( −635 mV, −435 mV) and displays the highest catalytic numbers in both reactions directions, allowing us to identify MmFd1 as the physiological redox partner of MmOGOR. Based on the difference in kinetics for these three Fds, we predict MmFd1 plays an essential role in cell growth and CO2 fixation by being the cognate redox partner for MmOGOR.
Focusing in on the details in the MmOGOR-MmFd1 model (Figure S15), a C-terminal extension found in MmFd1 (Figure S6) is near the domain III of MmOGOR, and the Cluster II of MmFd1 is 12.5 Å away from the only [4Fe-4S] cluster in MmOGOR, forming the entry point of electron transfer (Figure S6 and S15). The 12.5-Å distance is within reasonable range for biological electron transfer.48 It is also worth noting that it is Cluster II, the one with higher potential (−485 mV), not Cluster I, the one with the lower potential (−635 mV) that was modeled to interact with the cluster in MmOGOR (Figure S7 and S15). This model agrees with a previous finding on another CO2-fixing OGOR from H. thermophilus, whose Fd donor is a monocluster [4Fe-4S] Fd exhibiting a reduction potential of −485 mV.49
The active site MmOGOR is larger than the active sites of MtOOR or MtPFOR, consistent with their substrate preferences. As described above, the shortening of the helix-loop motif following Cys60β effectively pulls the helix of the motif away from the active site, allowing for the binding of the larger succinyl-CoA or 2-oxoglutarate. Revisiting the sequence analysis previous published,15 we find that shorter loop lengths correlate to larger substrates as a general trend in the OFOR family – the helix-loop motifs of OGOR, VOR and IOR are all shorter than that of OOR and PFOR (Figure S16), and the substrates of OGOR, VOR and IOR (2-oxoglutarate, 2-oxoisovalerate and indolepyruvate) are all larger than that of OOR or PFOR (oxalate and pyruvate). This may well be an important observation for future strategies where OFORs might be manipulated in biofuels or bioenergy applications.
Beside active site size, charge distribution is another critical factor determining the substrate specificity of an OFOR. Previous structural studies on OOR established that two positively charged residues are utilized to attract oxalate, a doubly negatively charged molecule at physiological pH.12, 15 2-oxoglutarate, which is the only other doubly negatively charged molecule under physiological pH in the enzyme family, and succinyl-CoA, which is the only negatively charged acyl substrate in this enzyme family, are, not surprisingly, bound to another positively charged residue, Arg63β, beside the conserved Arg302α. Again, prior sequence analyses of OFORs15 further show that residues equivalent to Arg63β can be found in all OGORs, but not other members in this enzyme family (Figure S16). Taken together, these structural data help inform engineering efforts for fixing CO2 on to different substrates.
The 2.80-Å resolution structure of OGOR with substrates bound, together with the spectroscopic analyses, supports a common mechanism of CoA binding and CoA-gated electron transfer. Domain III undergoes the same conformational change in MmOGOR that was observed in MtPFOR upon CoA binding,19 and in both cases, CoA binding promotes an electron transfer event. Although many of the interactions between succinyl-CoA and MmOGOR depicted here are also seen in the CoA-bound MtPFOR structure, there are some subtle differences that appear to tune reactivity. In particular, domain III motion upon CoA binding in MmOGOR repositions Glu45α toward the active site altering the hydrogen-bonding and electrostatic environment of the substrate-binding residue Arg63β, and creating a less positively-charged environment around the TPP. This change increases the 2-oxoglutarate KM and also increases kcat; the latter presumably due to the promotion of TPP-intermediate oxidation. The detrimental affect of Glu45α on the KM for the negatively charged substrate 2-oxoglutarate, however, dominates the catalytic efficiency, resulting in a WT MmOGOR that is a worse 2-oxoglutarate oxidase than Glu44αGln-MmOGOR.
The rearrangement of Glu45α that accompanies domain III movement is interesting in that it borrows pages out of the MtOOR playbook. MtOOR does not utilize CoA, but domain III ‘swinging’ is still involved in catalysis, opening and closing the active site for substrate binding and product release. And of relevance for this discussion, domain III movement in MtOOR alternatively positions domain III residue Glu154γ into and out of the active site, which in turns alters the position of the side chain of substrate-binding residue Arg31α (Figure 3D).13 Thus, both MtOOR and MmOGOR have mobile Glu residues on domain III that can interact with substrate-binding Arg residues during catalysis, although the locations of these Arg and Glu residues are not the same. Additionally, MtOOR undergoes a second conformational change following substrate binding, a negatively charged Asp residue on the so-called ‘switch loop’ flips into the active site varying the electrostatics and putatively driving TPP radical oxidation through a charge repulsion mechanism.13 MtPFOR does not have a ‘switch loop’ or a Glu on domain III that interacts with an active site Arg. Instead, it has been proposed that the negatively charged thiolate of the CoA itself can drive TPP radical oxidation.19 In MmOGOR, we have a hybrid of MtPFOR and MtOOR; CoA-dependence with the addition of a mobile Glu.
It is also interesting to consider the CO2 reduction direction in MmOGOR. Here the presence of Glu44α nearby might lower the affinity of the active site for the negatively charged succinyl moiety of succinyl-CoA, but CoA itself makes so many interactions with the enzyme that the impact of Glu44α would likely be small, and once the succinyl moiety has reacted with the TPP, CoA should depart, removing the negatively charged CoA thiolate and repositioning the negatively charged Glu44α. The net result would be an increase in the positive charge near the TPP-bound intermediate, prompting electron flow to the TPP, affording CO2 reduction.
In summary, these data (in addition to previously available structural and biochemical data12–13, 15, 19) reveal how changes in the length of a loop, or positioning of a charged residue, or installation of second layer interactions can dramatically impact substrate preference or even reaction direction. We find similar mechanistic strategies co-opted by different family members and merged with other strategies to afford altered reactivities. The modularity of OFORs is emphasized by the replacement of a Fd domain V with a binding site for an external Fd protein. Importantly, however, one size doesn’t fit all and we find that only one MmFd appears able to afford relevant rates of CO2 reduction. As we begin to unravel the determinants of catalytic bias, sequence alignments take on new meaning and identification of the physiological reductant takes on new importance. We also see that one structure of one enzyme is not enough as domain and residue movements are key to the tuning the chemistry of the OFOR superfamily.
Experimental procedures
For full details please refer to Supplemental Experimental procedures.
Supplementary Material
Acknowledgements
B.L. and S.J.E. are supported by Department of Energy Office of Science (BES DE-SC0012598). C.L.D. is a Howard Hughes Medical Investigator. The crystallographic study was supported in part by the National Institutes of Health (NIH) R35 GM126982 (C.L.D.). CLD is a Senior Fellow of the Bio-inspired Solar Energy Program, Canadian Institute for Advanced Research (CIFAR). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the NIH (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Software used in the project was installed and configured by SBGrid.
Footnotes
Declaration of Interests
The authors declare no conflict of interest.
References
- 1.Appel AM; Bercaw JE; Bocarsly AB; Dobbek H; DuBois DL; Dupuis M; Ferry JG; Fujita E; Hille R; Kenis PJ, et al. (2013) Frontiers, opportunities, and challenges in biochemical and chemical catalysis of CO2 fixation. Chem. Rev 113, 6621–6658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spreitzer RJ; Salvucci ME (2002) Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu. Rev. Plant. Biol 53, 449–475 [DOI] [PubMed] [Google Scholar]
- 3.Fuchs G (2011) Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu. Rev. Microbiol 65, 631–658 [DOI] [PubMed] [Google Scholar]
- 4.Evans MC; Buchanan BB; Arnon DI (1966) A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc. Natl. Acad. Sci. USA 55, 928–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furdui C; Ragsdale SW (2000) The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J. Biol. Chem 275, 28494–28499 [DOI] [PubMed] [Google Scholar]
- 6.Mai X; Adams M (1996) Characterization of a fourth type of 2-keto acid-oxidizing enzyme from a hyperthermophilic archaeon: 2-ketoglutarate ferredoxin oxidoreductase from Thermococcus litoralis. J. Bacteriol 178, 5890–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ragsdale SW (2003) Pyruvate ferredoxin oxidoreductase and its radical intermediate. Chem. Rev 103, 2333–2346 [DOI] [PubMed] [Google Scholar]
- 8.Heider J; Mai X; Adams MW (1996) Characterization of 2-ketoisovalerate ferredoxin oxidoreductase, a new and reversible coenzyme A-dependent enzyme involved in peptide fermentation by hyperthermophilic archaea. J. Bacteriol 178, 780–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mai X; Adams MW (1994) Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. A new enzyme involved in peptide fermentation. J. Biol. Chem 269, 16726–16732 [PubMed] [Google Scholar]
- 10.Tersteegen A; Linder D; Thauer RK; Hedderich R (1997) Structures and functions of four anabolic 2-oxoacid oxidoreductases in Methanobacterium thermoautotrophicum. Eur. J. Biochem 244, 862–868 [DOI] [PubMed] [Google Scholar]
- 11.Pierce E; Becker DF; Ragsdale SW (2010) Identification and characterization of oxalate oxidoreductase, a novel thiamine pyrophosphate-dependent 2-oxoacid oxidoreductase that enables anaerobic growth on oxalate. J. Biol. Chem 285, 40515–40524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson MI; Brignole EJ; Pierce E; Can M; Ragsdale SW; Drennan CL (2015) The structure of an oxalate oxidoreductase provides insight into microbial 2-oxoacid metabolism. Biochemistry 54, 4112–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson MI; Chen PY-T; Johnson AC; Pierce E; Can M; Ragsdale SW; Drennan CL (2016) One-carbon chemistry of oxalate oxidoreductase captured by X-ray crystallography. Proc. Natl. Acad. Sci. USA 113, 320–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pierce E; Mansoorabadi SO; Can M; Reed GH; Ragsdale SW (2017) Properties of intermediates in the catalytic cycle of oxalate oxidoreductase and its suicide inactivation by pyruvate. Biochemistry 56, 2824–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson MI; Chen PY-T; Drennan CL (2016) A structural phylogeny for understanding 2-oxoacid oxidoreductase function. Curr. Opin. Struct. Biol 41, 54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pieulle L; Guigliarelli B; Asso M; Dole F; Bernadac A; Hatchikian EC (1995) Isolation and characterization of the pyruvate-ferredoxin oxidoreductase from the sulfate-reducing bacterium Desulfovibrio africanus. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol 1250, 49–59 [DOI] [PubMed] [Google Scholar]
- 17.Wahl RC; Orme-Johnson W (1987) Clostridial pyruvate oxidoreductase and the pyruvate-oxidizing enzyme specific to nitrogen fixation in Klebsiella pneumoniae are similar enzymes. J. Biol. Chem 262, 10489–10496 [PubMed] [Google Scholar]
- 18.Dorner E; Boll M (2002) Properties of 2-oxoglutarate:ferredoxin oxidoreductase from Thauera aromatica and its role in enzymatic reduction of the aromatic ring. J. Bacteriol 184, 3975–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen PY-T; Aman H; Can M; Ragsdale SW; Drennan CL (2018) Binding site for coenzyme A revealed in the structure of pyruvate:ferredoxin oxidoreductase from Moorella thermoacetica. Proc. Natl. Acad. Sci. USA 115, 3846–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furdui C; Ragsdale SW (2002) The roles of coenzyme A in the pyruvate: ferredoxin oxidoreductase reaction mechanism: rate enhancement of electron transfer from a radical intermediate to an iron-sulfur cluster. Biochemistry 41, 9921–9937 [DOI] [PubMed] [Google Scholar]
- 21.Menon S; Ragsdale SW (1997) Mechanism of the Clostridium thermoaceticum pyruvate:ferredoxin oxidoreductase: evidence for the common catalytic intermediacy of the hydroxyethylthiamine pyropyrosphate radical. Biochemistry 36, 8484–8494 [DOI] [PubMed] [Google Scholar]
- 22.Chabriere E; Charon MH; Volbeda A; Pieulle L; Hatchikian EC; Fontecilla-Camps JC (1999) Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. Nat. Struct. Biol 6, 182–190 [DOI] [PubMed] [Google Scholar]
- 23.Chabriere E; Vernede X; Guigliarelli B; Charon MH; Hatchikian EC; Fontecilla-Camps JC (2001) Crystal structure of the free radical intermediate of pyruvate:ferredoxin oxidoreductase. Science 294, 2559–2563 [DOI] [PubMed] [Google Scholar]
- 24.Cavazza C; Contreras-Martel C; Pieulle L; Chabriere E; Hatchikian EC; Fontecilla-Camps JC (2006) Flexibility of thiamine diphosphate revealed by kinetic crystallographic studies of the reaction of pyruvate-ferredoxin oxidoreductase with pyruvate. Structure 14, 217–224 [DOI] [PubMed] [Google Scholar]
- 25.Yan Z; Maruyama A; Arakawa T; Fushinobu S; Wakagi T (2016) Crystal structures of archaeal 2-oxoacid: ferredoxin oxidoreductases from Sulfolobus tokodaii. Sci. Rep 6, 33061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams TJ; Zhang CL; Scott JH; Bazylinski DA (2006) Evidence for autotrophy via the reverse tricarboxylic acid cycle in the marine magnetotactic coccus strain MC-1. Appl. Environ. Microbiol 72, 1322–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bazylinski DA; Williams TJ; Lefevre CT; Berg RJ; Zhang CL; Bowser SS; Dean AJ; Beveridge TJ (2013) Magnetococcus marinus gen. nov., sp. nov., a marine, magnetotactic bacterium that represents a novel lineage (Magnetococcaceae fam. nov., Magnetococcales ord. nov.) at the base of the Alphaproteobacteria. Int. J. Syst. Evol. Microbiol 63, 801–808 [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto M; Ikeda T; Arai H; Ishii M; Igarashi Y (2010) Carboxylation reaction catalyzed by 2-oxoglutarate:ferredoxin oxidoreductases from Hydrogenobacter thermophilus. Extremophiles 14, 79–85 [DOI] [PubMed] [Google Scholar]
- 29.Kerscher L; Oesterhelt D (1981) The catalytic mechanism of 2-oxoacid:ferredoxin oxidoreductases from Halobacterium halobium. One-electron transfer at two distinct steps of the catalytic cycle. Eur. J. Biochem 116, 595–600 [DOI] [PubMed] [Google Scholar]
- 30.Cammack R; Kerscher L; Oesterhelt D (1980) A stable free radical intermediate in the reaction of 2-oxoacid: ferredoxin oxidoreductases of Halobacterium halobium. FEBS Lett. 118, 271–273 [Google Scholar]
- 31.Zhang Q; Iwasaki T; Wakagi T; Oshima T (1996) 2-oxoacid:ferredoxin oxidoreductase from the thermoacidophilic archaeon, Sulfolobus sp. strain 7. J. Biochem 120, 587–599 [DOI] [PubMed] [Google Scholar]
- 32.Yan Z; Fushinobu S; Wakagi T (2014) Four Cys residues in heterodimeric 2-oxoacid: ferredoxin oxidoreductase are required for CoA-dependent oxidative decarboxylation but not for a nonoxidative decarboxylation. Biochim. Biophys. Acta, Proteins Proteomics 1844, 736–743 [DOI] [PubMed] [Google Scholar]
- 33.Menon S; Ragsdale SW (1996) Unleashing hydrogenase activity in carbon monoxide dehydrogenase/acetyl-CoA synthase and pyruvate:ferredoxin oxidoreductase. Biochemistry 35, 15814–15821 [DOI] [PubMed] [Google Scholar]
- 34.Reed GH; Ragsdale SW; Mansoorabadi SO (2012) Radical reactions of thiamin pyrophosphate in 2-oxoacid oxidoreductases. Biochim. Biophys. Acta, Proteins Proteomics 1824, 1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gehring U; Arnon DI (1972) Purification and properties of α-ketoglutarate synthase from a photosynthetic bacterium. J. Biol. Chem 247, 6963–6969 [PubMed] [Google Scholar]
- 36.Yoon KS; Bobst C; Hemann CF; Hille R; Tabita FR (2001) Spectroscopic and functional properties of novel 2[4Fe-4S] cluster-containing ferredoxins from the green sulfur bacterium Chlorobium tepidum. J. Biol. Chem 276, 44027–44036 [DOI] [PubMed] [Google Scholar]
- 37.Chun JY; Lee EJ; Lee HS; Cheon CI; Min KH; Lee MS (1998) Molecular cloning and analysis of the argC gene from Corynebacterium glutamicum. Biochem. Mol. Biol. Int 46, 437–447 [DOI] [PubMed] [Google Scholar]
- 38.Traxler MF; Summers SM; Nguyen HT; Zacharia VM; Hightower GA; Smith JT; Conway T (2008) The global, ppGpp‐mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol 68, 1128–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hauryliuk V; Atkinson GC; Murakami KS; Tenson T; Gerdes K (2015) Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol 13, 298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jimenez-Vicente E; Navarro-Rodriguez M; Poza-Carrion C; Rubio LM (2014) Role of Azotobacter vinelandii FdxN in FeMo-co biosynthesis. FEBS Lett. 588, 512–516 [DOI] [PubMed] [Google Scholar]
- 41.Jouanneau Y; Meyer C; Gaillard J; Forest E; Gagnon J (1993) Purification and characterization of a novel dimeric ferredoxin (FdIII) from Rhodobacter capsulatus. J. Biol. Chem 268, 10636–10644 [PubMed] [Google Scholar]
- 42.Konig S; Gros O; Heiden SE; Hinzke T; Thurmer A; Poehlein A; Meyer S; Vatin M; Mbeguie AMD; Tocny J, et al. (2016) Nitrogen fixation in a chemoautotrophic lucinid symbiosis. Nat. Microbiol 2, 16193. [DOI] [PubMed] [Google Scholar]
- 43.Huber JG; Gaillard J; Moulis JM (1995) NMR of Chromatium vinosum ferredoxin: evidence for structural inequivalence and impeded electron transfer between the two [4Fe-4S] clusters. Biochemistry 34, 194–205 [DOI] [PubMed] [Google Scholar]
- 44.Kummerle R; Gaillard J; Kyritsis P; Moulis JM (2001) Intramolecular electron transfer in [4Fe-4S] proteins: estimates of the reorganization energy and electronic coupling in Chromatium vinosum ferredoxin. J. Biol. Inorg. Chem 6, 446–451 [DOI] [PubMed] [Google Scholar]
- 45.Prince RC; Adams MW (1987) Oxidation-reduction properties of the two Fe4S4 clusters in Clostridium pasteurianum ferredoxin. J. Biol. Chem 262, 5125–5128 [PubMed] [Google Scholar]
- 46.Chen J-S; Kay Blanchard D (1979) A simple hydrogenase-linked assay for ferredoxin and flavodoxin. Anal. Biochem 93, 216–222 [PubMed] [Google Scholar]
- 47.Luo J; Fukuda E; Takase H; Fushinobu S; Shoun H; Wakagi T (2009) Identification of the lysine residue responsible for coenzyme A binding in the heterodimeric 2-oxoacid: ferredoxin oxidoreductase from Sulfolobus tokodaii, a thermoacidophilic archaeon, using 4-fluoro-7-nitrobenzofurazan as an affinity label. Biochim. Biophys. Acta, Proteins Proteomics 1794, 335–340 [DOI] [PubMed] [Google Scholar]
- 48.Moser CC; Keske JM; Warncke K; Farid RS; Dutton PL (1992) Nature of biological electron transfer. Nature 355, 796–802 [DOI] [PubMed] [Google Scholar]
- 49.Li B; Elliott SJ (2016) The catalytic bias of 2-oxoacid: ferredoxin oxidoreductase in CO2: evolution and reduction through a ferredoxin-mediated electrocatalytic assay. Electrochim. Acta 199, 349–356 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.