SUMMARY
Nucleoporins (Nups) are involved in neural development and alterations in Nup genes are linked to human neurological diseases. However, physiological functions of specific Nups and the underlying mechanisms involved in these processes remain elusive. Here we show that tissue-specific depletion of the nucleoporin Seh1 causes dramatic myelination defects in the Central Nervous System (CNS). While proliferation is not altered in Seh1-deficient Oligodendrocyte Progenitor Cells (OPCs), they fail to differentiate into mature oligodendrocytes, which impairs myelin production and remyelination after demyelinating injury. Genome-wide analyses show that Seh1 regulates a core myelinogenic regulatory network and establishes an accessible chromatin landscape. Mechanistically, Seh1 regulates OPCs differentiation by assembling Olig2 and Brd7 into a transcription complex at nuclear periphery. Together, our results reveal that Seh1 is required for oligodendrocyte differentiation and myelination by promoting assembly of an Olig2-dependent transcription complex and define a nucleoporin as key player in the CNS.
Graphical Abstract
In Brief
Liu et al. demonstrate that the nucleoporin Seh1 functions in oligodendrocyte differentiation, myelination, and post-injury remyelination by assembling a Seh1/Olig2/Brd7 transcription complex at nuclear periphery.
INTRODUCTION
In the CNS, oligodendrocytes form myelin sheath to wrap axons and promote rapid propagation of action potentials and proper CNS functions (Bercury and Macklin, 2015). Defective remyelination by oligodendrocytes disrupts salutatory nerve conduction and cognitive or motor functions, which contributes to neurological disorders such as multiple sclerosis and leukodystrophies (Trapp and Nave, 2008). OPCs are abundant in demyelinated regions of patients with multiple sclerosis, however they fail to differentiate into mature oligodendrocytes (Chang et al., 2002). This differentiation process is accompanied by large-scale nuclear reorganization and alterations in global gene expression (Yu et al., 2013). These processes rely on the precise control of networks of transcriptional factors, chromatin modifiers, and signaling pathways.
As components of Nuclear Pore Complexes (NPCs), Nups have been shown to play prominent roles in cell-type-specific transcriptional regulation (Ibarra and Hetzer, 2015). It has long been observed, by electron microscopy, that decondensed chromatin surrounded the NPCs (Blobel, 1985). More recent findings have shown that active transcription occurs at the nuclear pore complex and that Nups have a role in this process (Pascual-Garcia and Capelson, 2014). Nups can be classified as core scaffold, central channel, peripheral and trans-membrane Nups (Wente and Rout, 2010). Peripheral and trans-membrane Nups, such as Nup153, Nup98 and Nup210 have been found to directly regulate gene expression by interacting with transcription factors such as sox2, mef2c, etc.(Pascual-Garcia et al., 2017; Raices et al., 2017; Toda et al., 2017). However, it remains unclear whether core scaffold Nups are involved in developmental gene regulation.
Nups have been linked to neuronal development. These findings were mainly based on analyzes of homozygous embryonic development or knockdown approaches (Lupu et al., 2008; Toda et al., 2017). Mutations on Nup genes are linked to developmental defects and neurological diseases (Basel-Vanagaite et al., 2006). However, it remains unknown the relationship of these mutations with Nup functions in the CNS. Here we show that Seh1, a core scaffold nucleoporin, plays a critical role in oligodendrocyte lineage progression and CNS myelination. Seh1 depletion in oligodendrocyte-lineage dramatically impairs the OPC differentiation and myelin production during mouse development and remyelination after demyelination injury. Using a combination of chromatin immunoprecipitation, genome-wide profiling and global chromatin accessibility analysis, we show that Seh1 affects the transcriptional landscape of OPCs. Mechanistic studies revealed that Seh1 interacts with Olig2 and promote the assembly of the Olig2/Brd7 transcriptional complex. Our findings demonstrate a previously unknown role for the nucleoporin Seh1 in the differentiation of OPCs and provide insights into how oligodendrocyte master regulators orchestrate with nuclear structural proteins at the nuclear periphery to regulate the transcriptional program and cell fate.
RESULTS
Seh1 Is Required for Myelination in the CNS
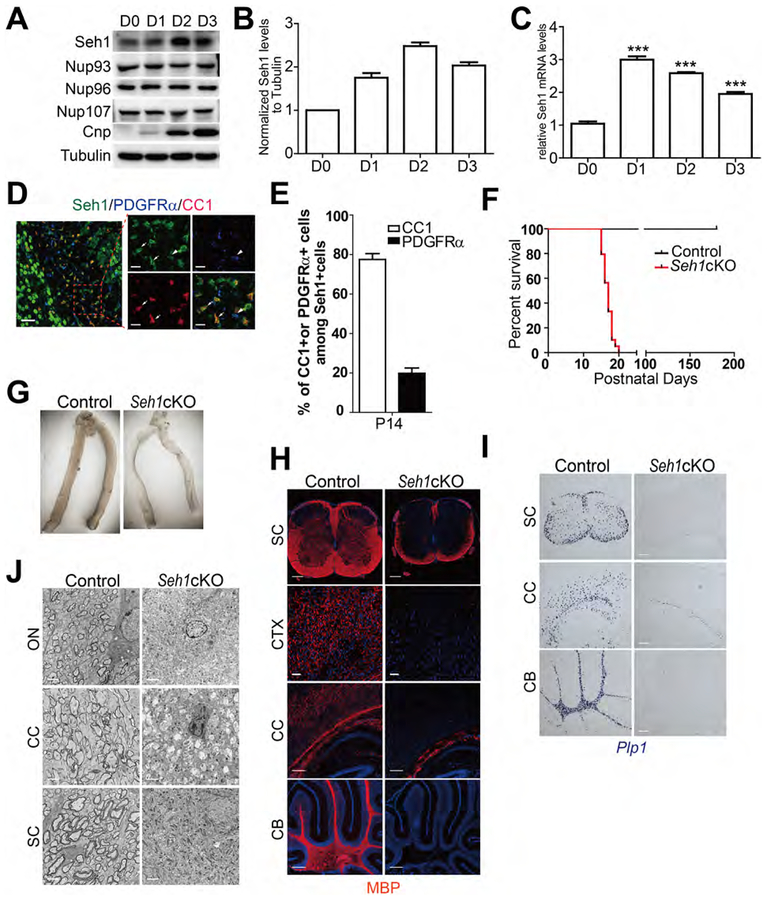
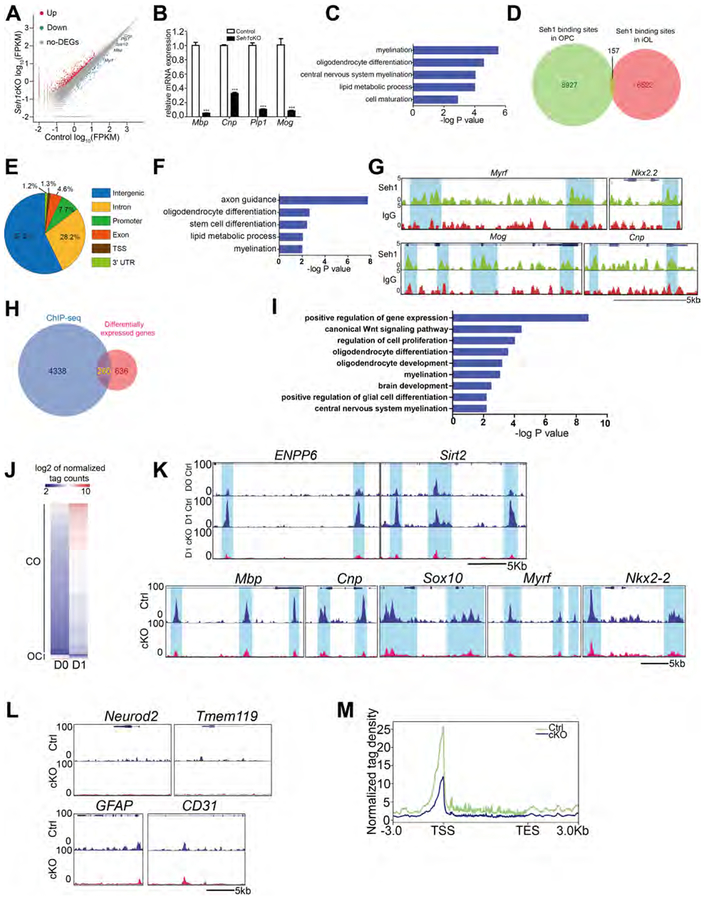
Nucleoporins are linked to neuronal development. While Nups have been reported to regulate neural progenitor cells maintenance and differentiation (Toda et al., 2017), the roles of Nups in oligodendrocytes development have not been studied. We first analyzed Nup protein levels in OPCs and mature oligodendrocyte. During rat OPCs in vitro differentiation, Seh1, a core scaffold nucleoporin, was upregulated, which was similar to the oligodendrocyte differentiation marker CNP. In contrast, the protein levels of other scaffold nucleoporins: Nup93, Nup96 and Nup107 were constant during the differentiation process (Figure 1A–1C). High Seh1 levels were also detected in CC1+ mature oligodendrocyte, but its expression levels were low in PDGFRα+ OPCs in the P14 corpus callosum (Figure 1D). The proportion of CC1+ among Seh1+ cells in the corpus callosum at P14 is 77.5%, suggesting that Seh1 is largely confined to differentiating cells in the oligodendrocyte lineage (Figure 1E). These data suggest that the Seh1 is selectively upregulated upon oligodendrocyte differentiation.
Figure 1. Seh1 Is Required for Myelination in the CNS.
(A) Immunoblot of indicated proteins in cultured rat OPCs and differentiating oligodendrocytes after T3 treatment.
(B) Histograms show Seh1 fold changes measured by densitometry after normalization with Tubulin.
(C) qRT-PCR analysis of Seh1 in cultured rat OPCs and differentiating oligodendrocytes after T3 treatment (n= 3 experiments from 6 rats. D1 versus D0, ***p<0.0001; D2 versus D0, ***p=<0.0001; D3 versus D0, ***p=0.0006).
(D) Co-immunostaining of Seh1, PDGFRα and CC1 in the corpus callosum of wild-type mice at P14. Boxed image is shown on the right. Arrow indicates Seh1+/CC1+ cells; arrowheads indicate Seh1+/PDGFRα+ cells. Scale bars=50 μm in left and 15 μm in right panel.
(E) Quantification of CC1+ or PDGFRα+ cells among Seh1+ cells in the corpus callosum at P14 (n=3 mice).
(F) Survival curves of control and Seh1cKO mice (n=50 for control and n=38 for mutant mice).
(G) Representative images of optic nerve from control and Seh1cKO mice at P14.
(H) Immunostaining of MBP in the spinal cord (SC), cortex (CTX), corpus callosum (CC) and cerebellum (CB) of control and Seh1cKO at P14. Nuclei were stained with DAPI. Scale bars=200 μm.
(I) In situ hybridization of Plp1 in the spinal cord (SC), corpus callosum (CC) and cerebellum (CB) of control and Seh1cKO at P14. Scale bars=200 μm.
(J) Electron micrograph analysis of optic nerve (ON), corpus callosum (CC) and spinal cord (SC) of control and Seh1cKO at P14. Scale bars=2 μm.
Data are represented as mean ± SEM. ***p<0.001, two-tailed unpaired Student’s t test.
See also Figure S1.
To investigate the physiological function of Seh1, we flanked exon 6 and 7 of the Seh1 gene by loxP sequences to generate Seh1loxP/loxP mice. Cre-mediated recombination caused premature termination of Seh1 (Figure S1A). Seh1−/− mice was not viable after crossing with EIIa-Cre transgenic mice, which indicates that Seh1 is essential for embryonic development (Figure S1B). Instead, we generated whole body-inducible Seh1 mutants by breeding Seh1loxP/loxP mice with CMV-CreERT mice, which carry a CMV tamoxifen-inducible Cre (Hayashi and McMahon, 2002). Tamoxifen was administered in CMV-CreERT: Seh1loxP/locP mutants (Seh1 CMV-iKO) and Seh1loxP/loxP pups from P3 to P5 and tissue was harvested at P12 (Figure S1C). Comparing to control mice, Seh1 was substantially reduced and loss of Seh1 led to translucent optic nerve, a well-characterized CNS white matter tract, which indicates a sign of severe deficiency in myelin formation (Figures S1D and S1E). Immunostaining followed by fluorescence microscopy also showed that expression of myelin proteins MBP was decreased in mutant mice (Figure S1F). These data prompted us to further investigate the function of Seh1 in oligodendrocyte development. We then selectively ablated Seh1 in oligodendrocyte lineage cells by breeding mice with the floxed Seh1 allele with an oligodendrocyte lineage expressing Olig1-Cre line that commences in primitive OPCs prior to PDGFRα+ OPCs (Xin et al., 2005). Seh1 expression was substantially reduced in the corpus callosum of the Seh1 conditional knockout mice (Seh1loxP/loxP; Olig1-Cre+/−, Seh1cKO) at P14 (Figure S1G). Seh1cKO mice were born at Mendelian frequencies and appeared to be normal at birth but developed tremor, ataxia, and seizures from postnatal week 2, and died during the third postnatal week (Figure 1F). The optic nerve from Seh1cKO mice was translucent compared to the control, which is consistent with the Seh1 CMV-iKO phenotype (Figure 1G). Indeed, levels of myelin genes such as MBP and Plp are essentially decreased in the spinal cord, cortex, corpus callosum and cerebellum of mutant mice at P14 compared to control mice (Figures 1H, 1I and S1H). To confirm the defective myelination phenotypes, myelin sheath assembly was examined by electron microscopy. In contrast to the large number of myelinated axons that were observed in control mice at P14, myelinated axons were hardly detectable in the optic nerve, corpus callosum or spinal cord of mutant mice (Figure 1J). Together, these data suggest that Seh1 is important for myelinogenesis in the CNS.
Seh1 Is Required for OPC Differentiation in a Cell Autonomous Manner
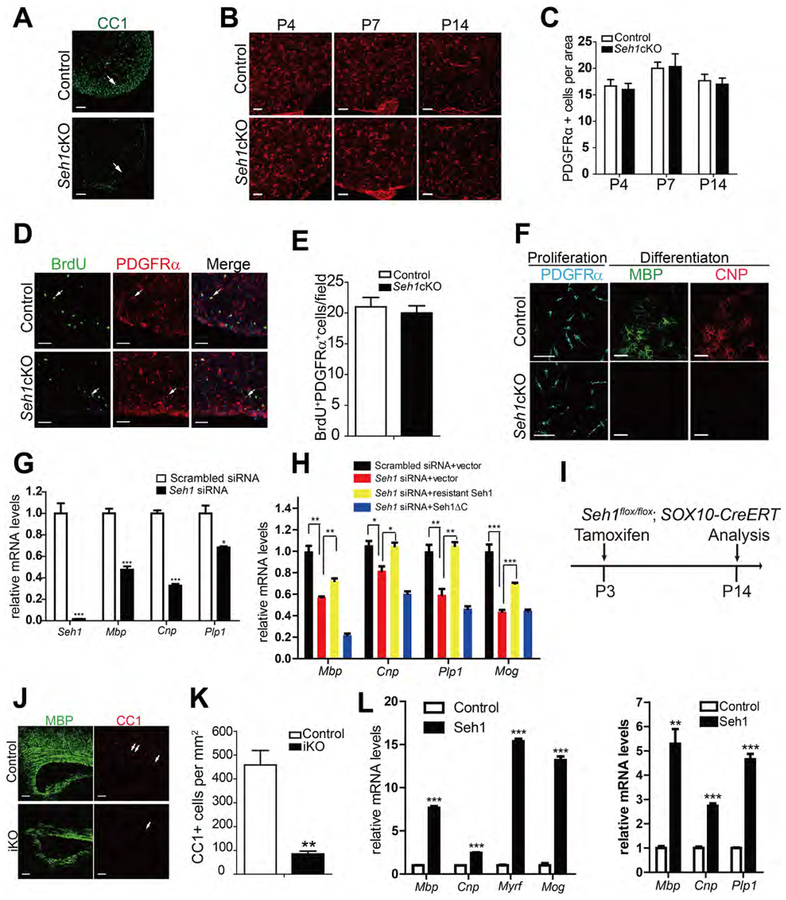
The hypomyelinating phenotype of Seh1 mutant mice suggest a role for Seh1 in the maturation of OPCs into myelin-forming oligodendrocyte cells. To test this hypothesis, we analyzed the number of OPCs and mature oligodendrocytes in control and Seh1cKO mice. In contrast with the markedly reduction of mature oligodendrocytes (CC1+), the OPC marker PDGFRα was detected in the spinal cord and brain of Seh1cKO at P4, P7 and P14 (Figures 2A–2C and S2A–2C). Similar OPC proliferation rates (BrdU+ proliferative OPCs) were observed between control and mutant mice (Figures 2D and 2E). Additionally, the NeuN, glial fibrillary acidic protein (GFAP), and Iba1 staining, which recognize mature neuron, astrocytes, and microglia respectively, were comparable between control and mutant mice (Figure S2D). These data suggest that ablation of Seh1 prevents OPC differentiation. To investigate whether defects in the differentiation capacity of OPCs are a cell-autonomous effect, we purified OPCs from control and Seh1cKO neonatal cortex using immunopanning. After triiodothyronine (T3) induction, control OPCs readily differentiated into MBP+, CNP+ OLs. In contrast, no MBP+ oligodendrocytes were detected in mutant OPCs (Figure 2F). Reduction of Seh1 by siRNA-mediated knockdown in rat OPCs also caused significant inhibition in the expression of myelination-related genes (Figure 2G). In addition, reintroduction of siRNA-resistant Seh1 could rescue the defects in myelin gene expression (Figure 2H and S2G). We further examined the effect of inactivation of Seh1 on OPC differentiation during early postnatal development in a time-controlled manner. At postnatal 3, Sox10-expressing cells were mainly OPCs (Figure S2E and S2F). We therefore bred Seh1loxP/loxP mice with Sox10-CreERT mice and induced recombination of Seh1 in OPCs at P3 by tamoxifen administration (Figure 2I) (McKenzie et al., 2014). A marked reduction in myelination and mature oligodendrocytes was observed in Sox10-CreERT: Seh1loxPlloxP mutants (Seh1-iKO) mice compared with control littermates at P14 (Figures 2J and 2K). To determine whether Seh1 is sufficient to promote OPC differentiation, rat OPCs were transfected with control or Seh1-expressing plasmids. We found that overexpression of Seh1 significantly induced the expression of myelin genes in OPCs under both differentiation- and proliferation-promoting conditions (Figure 2L), indicating that Seh1 can alter the timing of differentiation. Together, these observations demonstrate that Seh1 is both essential and sufficient for promoting OPC differentiation and myelination.
Figure 2. Seh1 Is Required for the OPC Differentiation in a Cell-Autonomous Manner.
(A) Immunostaining of CC1 in the spinal cord of control and Seh1cKO at P14. Arrows indicate the labeled cells. Scale bars=100 μm.
(B) Immunostaing of PDGFRα in the spinal cord of control and Seh1cKO at P4, P7, P14. Scale bars=50 μm.
(C) Quantification of PDGFRα+ OPCs per area in the spinal cord from P4, P7, P14 control and Seh1cKO mice (P4, n=3 for control and n=3 for mutant mice, p=0.7096; P7, n=3 for control and n=3 for mutant mice, p=0.9066; P14, n=3 for control and n=3 for mutant mice, , p=0.7096).
(D) Immunostaining of BrdU and PDGFRα in the spinal cord from P14 control and Seh1cKO mice. Arrows indicate the double-labeled cells. Scale bars=50 μm.
(E) Quantification of BrdU+l PDGFRα+ cells per field (0.25 mm2) in the spinal cord of control and Seh1cKO (n= 3 for control and n=3 for mutant mice, p=0.6291).
(F) Immunolabeling of PDGFRα, MBP, and CNP in control and Seh1cKO OPCs under proliferative condition (PDGFRα) or 4 day in differentiation medium (MBP, CNP). Scale bars=50 μm.
(G) qRT-PCR validation of knockdown efficiency of Seh1 in primary rat OPCs and qRT-PCR analysis of myelination-associated genes in primary rat OPCs under differentiation conditions following treatments with scrambled or Seh1 siRNAs (n= 3 independent experiments from 3 rats. Seh1, ***p=0.0005; Mbp, ***p=0.0006; Cnp, ***p<0.0001; Plp1, *p=0.0126).
(H) qRT-PCR analysis of myelination-associated genes in primary rat OPCs under differentiation conditions following co-transfection with the indicated plasmids and siRNAs (n= 3 independent experiments from 8 rats. **p=0.0011, **p=0.0038, *p=0.036, *p=0.011, **p=0.0072, **p=0.0021, ***p=0.0010, ***p<0.001). Seh1ΔC= Seh1 (1–258 aa).
(I) Diagram showing tamoxifen administration to induce Seh1 deletion.
(J) Immunostaining of MBP and CC1 on the corpus callosum of control and Seh1iKO mice at P14. Arrows indicate CC1+ cells. Scale bars=100 μm.
(K) Quantification of CC1+ oligodendrocyte cell numbers in the corpus callosum of tamoxifen-treated control and Seh1iKO mice at P14. (n= 3 for control and n=3 for Seh1iKO mice. **p=0.0038)
(L) qRT-PCR analysis of myelination-associated genes in primary rat OPCs under differentiation (left panel) or proliferation (right panel) conditions following Seh1 expression (n= 3 independent experiments from 3 rats (left). n= 3 independent experiments from 3 rats (right).. Left: Mbp, ***p<0.0001; Cnp, ***p<0.0001; Myrf, ***p<0.0001; Mog, ***p=0.0002; Right, Mbp, **p=0.0021; Cnp, ***p=0.0001; Plp1, ***p<0.0001).
Data are represented as mean ± SEM; *p<0.05, **p<0.01, ***p<0.001, two-tailed unpaired Student’s t test.
See also Figure S2.
Seh1 Is Critical for Adult Myelinogenesis and OL Remyelination after Demyelination
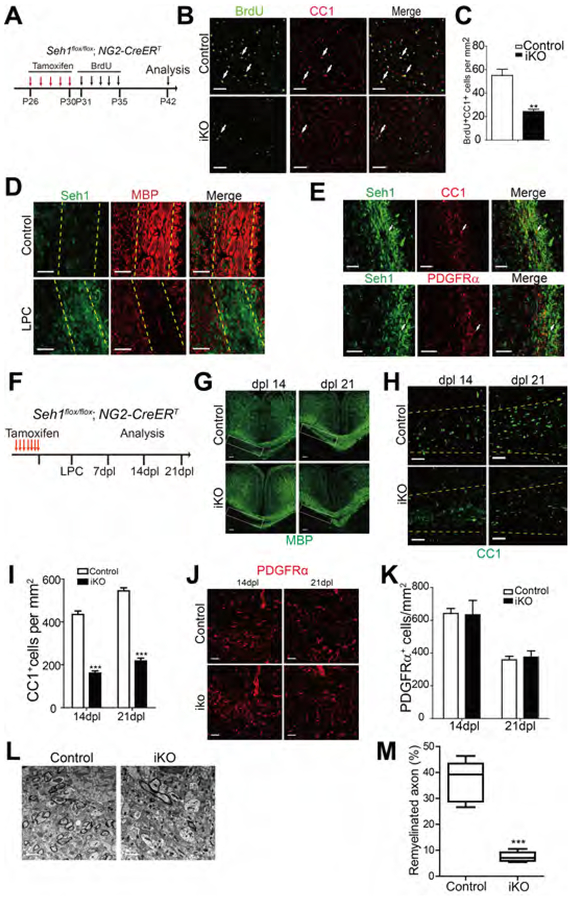
Many OPCs persist in the adult mouse CNS and continue to divide and differentiate into myelinating oligodendrocytes throughout life (Richardson et al., 2011; Rivers et al., 2008). Since Seh1 is essential for OPCs differentiation during development, we further assessed the function of Seh1 in the adult myelinogenesis by crossing Seh1loxP/loxP mice with NG2-CreERT transgenic mice, which express tamoxifen-inducible Cre in OPCs (Zhu et al., 2008). A 5 day tamoxifen injection was performed in Seh1 OPC-iKO (NG2-CreERT: Seh1loxP/loxP) and heterozygous controls mice from P26 to P30. BrdU was injected for the following 5 days (P31 to P35) to label dividing adult OPCs (Figure 3A). This genetic approach allowed us to perform long-term lineage-tracing analysis since mature CC1+ oligodendrocytes would not be labeled by BrdU. At P42, the number of BrdU+ CC1+ double positive cells in Seh1 OPC-iKO was greatly reduced compared to control, indicating that acute inactivation of Seh1 in the adult OPCs impaired adult OPC differentiation (Figures 3B and 3C). We then hypothesized that Seh1 might have a role in the remyelination process following lysolecithin (LPC)-induced demyelination. LPC injection induces rapid myelin breakdown followed by remyelination (Franklin, 2002). Seh1 level was significantly up-regulated following demyelination and it was expressed in the oligodendrocyte lineage (Figures 3D and 3E). To determine the potential role of Seh1 in remyelination, we examined the re-appearance of the myelin protein and OLs in the lesion during remyelination. Seven tamoxifen injections were administered prior to LPC in control and Seh1 OPC-iKO mice (Figure 3F). At 14 and 21 days post-lesion, there was significantly less MBP protein in the corpus callosum of Seh1 OPC-iKO mice than in control mice (Figure 3G). Consistently, the number of CC1+ differentiating oligodendrocytes was markedly reduced (Figures 3H and 3I). In contrast, depletion of Seh1 did not impair the recruitment of PDGFRα+ OPCs and the number was comparable between control and Seh1 OPC-iKO (Figures 3J and 3K). Ultrastructural analysis by electron microscopy further indicated that far fewer myelinated axons were detected in the lesions of Seh1 OPC-iKO mice than in control (Figure 3L and 3M). Similar results showing decrease in remyelination and OL regeneration were also obtained in the spinal cord of Seh1 OPC-iKO mice (Figure S3). Altogether, these observations suggest that Seh1 is critical for adult myelinogenesis and remyelination after white matter injury.
Figure 3. Seh1 Is Critical for Adult Myelinogenesis and OL Remyelination after Demyelination.
(A) Diagram showing tamoxifen administration from P26 to P30 and BrdU labeling from P31 to P35 in the inducible mouse models.
(B) Immunostaining of BrdU and CC1 on the corpus callosum of control and Seh1 OPC-iKO mice at P42 after tamoxifen and BrdU injection. Arrows indicate BrdU+/CC1+ cells. Scale bars=50 μm.
(C) Quantification of BrdU+/CC1+ cells per mm2 in the corpus callosum of tamoxifen-treated control and Seh1 OPC-iKO mice at P42 (n= 3 for control and n=3 for Seh1-iKO mice. **p=0.0054).
(D) Immunostaining of Seh1 and MBP at Dpl 7 in the corpus callosum from non-lesion control and mice with LPC lesion. Yellow dashed lines indicate corpus callosum. Scale bars=50 μm.
(E) Immunostaining of Seh1, CC1, and PDGFRα at Dpl 14 in the LPC lesions of the corpus callosum. Arrows indicate Seh1+/CC1+ (top) and Seh1+/PDGFRα+ (bottom) cells. Scale bars=50 μm.
(F) Diagram showing tamoxifen administration and LPC injection schedule.
(G, H and J) Immunostaining of MBP (G), CC1 (H) and PDGFRα (J) in the corpus callosum lesions of control and Seh1 OPC-iKO mice at Dpl 14 and 21. Scale bars =100 μm in (G) and 50 μm in (H) and (J).
(I and K) Quantification of CC1+ oligodendrocytes (I) and PDGFRα+ OPCs (K) in LPC-induced lesions at Dpl 14 and 21 (I: n= 3 for control and n=3 for Seh1 OPC-iKO mice at 14dpl, ***p=0.0001; n= 3 for control and n=3 for Seh1 OPC-iKO mice at 21 dpl, ***p<0.0001; K: n= 3 for control and n=3 for Seh1 OPC-iKO mice at 14dpl, p=0.9330; n= 3 for control and n=3 for Seh1 OPC-iKO mice at 21 dpl, p=0.7247).
(L) Electron microscopy of LPC lesions from control and Seh1 OPC-iKO corpus callosum at Dpl 14. Scale bars=2 μm
(M) Quantification of remyelinated axons in LPC-induced lesion of control and Seh1 OPC-iKO corpus callosum at Dpl 14. (n= 3 for control and n=3 for Seh1 OPC-iKO mice at 14dpl, ***p<0.0001)
Data are represented as mean ± SEM; **p<0.01, ***p<0.001, two-tailed unpaired Student’s t test.
See also Figure S3.
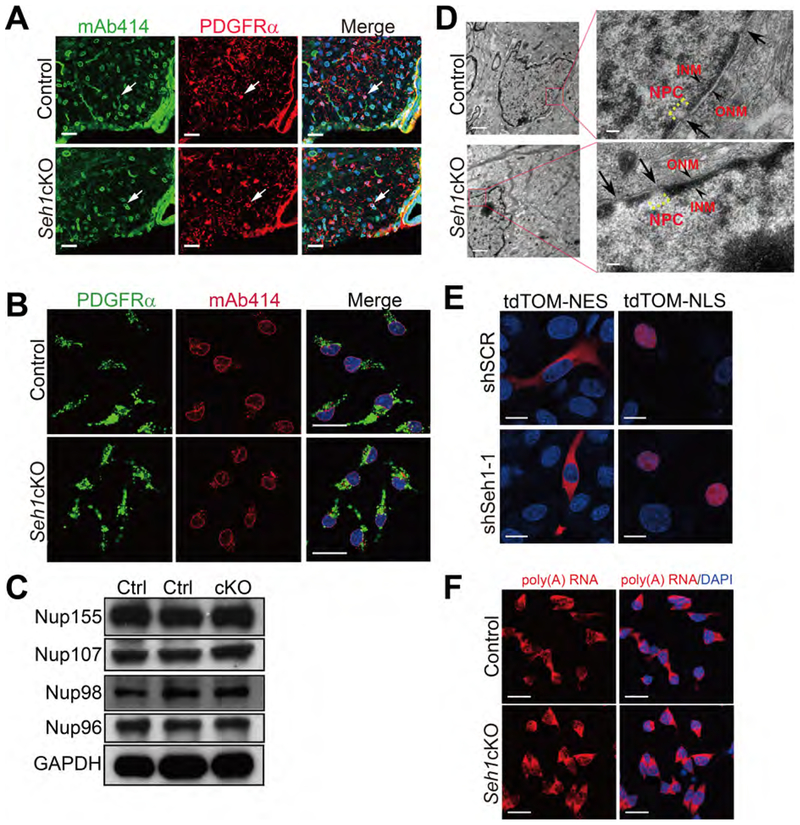
Ablation of Seh1 Does not Cause Major Defects in NPC Assembly or Function
Since Seh1 is a component of the Nup107–160 complex, which is important for NPC structure (Walther et al., 2003), we asked whether the observed phenotype was a result of defects in nuclear transport. As shown in Figure 4A and 4B, an antibody that detects a subset of FG-Nups (mAb414) showed comparable staining intensity and localization between control and mutant PDGFRα+ oligodendrocytes (Figures 4A and 4B). Furthermore, the levels of several nucleoporins were similar in control and mutant spinal cord protein lysate, indicating that other nucleoporins were present at relatively normal ratios in the absence of Seh1 (Figure 4C). Electron microscopy also showed normal nuclear envelope and NPC structure in Seh1-deficient optic nerve (Figure 4D). As NPCs are critical for maintaining proper nucleocytoplasmic transport, we tested the integrity of the nuclear permeability barrier. The cellular localization of tdTomato containing a Nuclear Localization Signal (NLS) or Nuclear Export Signal (NES) sequence was examined in a Seh1-knockdown oligodendrocyte cell line, Oli-neu cells (Pereira et al., 2011) (Figure S4A). We found no significant difference in the localization of tdTomato signals upon Seh1 knockdown (Figure 4E). In addition, the nuclear export inhibitor, LMB (Kudo et al., 1998) is able to block the export of tdTomato-NES (Figure S4B). Intracellular distribution of poly(A)+ RNA was also assessed by oligo-dT in situ hybridization and it showed no significant difference between control and mutant OPCs or Seh1-knockdown Oli-neu cells (Figure 4F and S4C). Instead, ectopic expression of the influenza virus NS1 protein, a well-known blocker of cellular mRNA nuclear export (Satterly et al., 2007; Zhang et al., 2012), induced inhibition of mRNA nuclear export and decreased the overall poly(A) RNA levels in Oli-neu cells (Figure S4D). However, we cannot exclude the possibility that transport of specific proteins or RNAs, or the general nuclear transport rates are altered upon Seh1 depletion. Overall, these data suggest that the phenotypes observed in oligodendrocytes differentiation are unlikely to be the consequence of major alternations in nucleocytoplasmic transport.
Figure 4. Ablation of Seh1 Does not Cause Major Defects in NPC Assembly or Function.
(A) Immunostaining of mAb414 and PDGFRα in spinal cord of P14 control and Seh1cKO mice. Arrows indicate mAb414+/PDGFRα+ cells. Nuclei were stained with DAPI. Scale bars=25 μm.
(B) Immunostaining of mAb414 and PDGFRα in primary control and Seh1cKO OPCs. Scale bars=25 μm.
(C) Immunoblot was performed to detect the indicated proteins in the spinal cord lysate of control and Seh1cKO at P14. Ctrl: control; cKO: Seh1cKO.
(D) Electron microscopy images of oligodendrocytes from control and Seh1cKO optic nerve. Boxed image is shown in the right. Arrows indicate NPCs. Arrowheads indicate ONM and INM. ONM/INM, outer/inner nuclear membrane. Scale bars=1 μm in left panel and 0.1 μm in right panel.
(E) Immunofluorescence of tdTomato-NES and tdTomato-NLS signals in Oli-neu cells transduced with scrambled or Seh1 shRNAs. NES, nuclear export signal; NLS, nuclear localization signal. Scale bars=15 μm.
(F) Oligo-dT In situ hybridization followed by fluorescence microscopy was performed in control and Seh1cKO OPCs. Scale bars=25 μm.
See also Figure S4.
It has been shown that Seh1 is a component of the GATOR2 complex and positively regulate mTOR signaling (Bar-Peled et al., 2013). We thus tested the phosphorylation status of the mTOR substrate S6K and S6, and also assessed total S6 or S6K levels. We found that lack of Seh1 in spinal cord or isolated OPCs did not significantly affect S6K and S6 phosphorylation or their total levels (Figures S4E and S4F). Thus, mTOR signaling may not be involved in Seh1-mediated regulation of oligodendrocyte differentiation. We then performed systematic phosphoproteomic analysis of primary OPCs from control or Seh1cKO mice. In total, 3,112 phosphopeptides were quantified and no significant changes in phosphorylation levels were observed between control and mutant samples, especially given that the proportions of altered phosphopeptides between two biological replicates were similar to those between control and mutant samples (3.2%=101/3112 between wild-type1/wild-type2 and 2.8%=87/3112 mutant1/mutant2; 3.6%=111/3112 between wild-type1/mutant1 and 3.5%=110/3112 between wild-type2/mutant2) (Figure S4G). We further analyzed the specific phosphosite intensities of phosphorylated myelination regulators and found that depletion of Seh1 did not cause significant changes in their phosphorylation levels (Figure S4H). Thus the observed phenotypes upon depletion of Seh1 are unlikely to be attributed to significant alterations in signaling.
Seh1 Regulates a Core Network of Genes that Drive OPCs Differentiation
Nups have recently been shown to play an important role in regulation of gene expression (Toda et al., 2017). Thus, we tested whether Seh1 might regulate the expression of genes important for oligodendrocyte differentiation and myelination. We carried out transcriptome profiling and analyzed global gene expression patterns in OPCs and immature oligodendrocytes (iOLs) from control or Seh1cKO mice. We found that 145 mRNAs were upregulated and 57 were downregulated genes in the mutant OPCs, and 659 mRNAs were upregulated and 237 were downregulated in the mutant iOLs compared with control (fold change>2, FDR<0.001) (Figure S5A and 5A). As expected, the downregulated mRNAs in iOLs encode a cohort of myelin structural proteins and critical myelination regulators, including Mbp, Cnp, Plp1, Myrf and Sox10 (Emery et al., 2009; Stolt et al., 2002), consistent with the deficient myelin formation phenotype (Figure 5A). Similarly, quantitative real-time PCR (qRT-PCR) analysis confirmed a marked reduction in myelination-associated genes, such as Mbp, Cnp, Plp1 and Mog (Figure 5B). Gene Ontology (GO) analysis showed that the downregulated genes in iOLs are enriched for those that function in myelination and oligodendrocyte differentiation (Figure 5C). By contrast, we did not see the similar enrichment in OPCs. Together, these data suggest that Seh1 regulates a core network of genes that control OPC differentiation and myelination.
Figure 5. Seh1 Regulates a Core Network of Genes that Drive OPCs Differentiation.
(A) Differentially expressed transcripts (highlighted in color; fold change >2, false discovery rate<0.001) between control and Seh1cKO iOLs.
(B) qRT-PCR analysis of myelination-associated genes in control and Seh1cKO iOLs. (n= 3 independent experiments from 6 mice per group. Mbp, ***p<0.0001, Cnp, ***p<0.0001, Plp1, “p<0.0001, Mog, “p=0.0005).
(C) The gene ontology (GO) analysis of the significantly downregulated genes between control and Seh1cKO iOLs.
(D) Venn diagram showing the global Seh1-binding sites in OPCs and iOLs.
(E) Fractions of Seh1 ChIP-seq peaks in different regions of the genome.
(F) GO of Seh1 targeted genes.
(G) ChIP-seq showing Seh1 enrichment at selected gene loci (Myrf, Nkx2.2, Mog, Cnp).
(H) Venn diagram showing the overlap between Seh1-bound genes and differentially expressed genes in control and Seh1cKO iOLs.
(I) GO functional categories analysis of Seh1 directly targeted genes.
(J) Heatmap of CO/OC loci. Loci of open chromatin were arranged into groups depending on changing to more open (CO) or more close (OC). Units are in log2 of normalized tag count
(K) Top panel: representative ATAC-seq tracks of myelination-associated genes in control OPCs (D0), iOLs (D1) and Seh1cKO iOLs (D1). Bottom panel: fractions of ATAC-seq peaks which become more open (CO) or more closed (OC) in different regions of the genome of control and Seh1cKO iOLs.
(L) Representative ATAC-seq tracks of neuronal, microglial, astrocytic and endothelial genes in control and Seh1cKO iOLs.
(M) Distribution of ATAC-seq reads around Seh1-directly upregulated genes in control and Seh1cKO iOLs.
Data are represented as mean ± SEM; ***p<0.001, two-tailed unpaired Student’s t test.
See also Figure S5.
We next asked whether Seh1 regulates the transcriptional program of oligodendrocytes via direct association with the genome. Chromatin immunoprecipitation and sequencing (ChlP-seq) was performed to assess Seh1 genome-wide occupancy in OPCs and iOLs. We identified 9,084 Seh1-binding sites within OPCs data sets and 6,679 binding sites within the iOLs data sets. Strikingly, only 157 binding sites were occupied by Seh1 at both stages, indicating a dynamic change of Seh1-targeting sites at each stage (Figure 5D). In iOLs, the majority of Seh1-binding peaks were present in intronic (28.2%) and intergenic (57.0%) regions, and only 7.7% of Seh1-binding sites located at promoter regions (Figure 5E). GO analysis showed a significant enrichment for genes involved in axon guidance and oligodendrocyte differentiation, suggesting potential roles for Seh1 in the regulation of the myelinogenic network (Figure 5F). Among Seh1-targeted genes were those encoding factors involved in myelination regulators (Myrf and Nkx2.2) and myelin components (Mog and Cnp; Figure 5G). We also exogenously expressed HA-tagged Seh1 and validated a subset of genes by ChIP-qPCR (Figure S5B). To identify potential Seh1 direct target genes, the Seh1-bound genes were intersected with those misregulated genes following Seh1 deletion. We found that 260 genes were potential targets for Seh1 regulation in iOLs (Figure 5H). Among them, 197 genes were upregulated and 63 genes were downregulated after Seh1 depletion (Figure S5C). In the case of OPCs, we found that 68 genes were candidates for Seh1-mediated regulation. Among them, 53 genes were upregulated and 15 genes were downregulated (Figure S5C). GO analysis of potential Seh1 targets in iOLs were overrepresented by those involved in oligodendrocyte differentiation and oligodendrocyte development (Figure 5I).
Seh1 Establishes an Accessible Chromatin Landscape to Promote Transcriptional Programs
To identify the global chromatin accessibility dynamics during the transition of OPC differentiation, OPCs isolated from cortex of control and Seh1cKO neonatal mice were cultured in differentiation medium for 1 day to induce OPC differentiation. We then harvested cells at day 0 and 1, and performed an assay for transposase-accessible chromatin using sequencing (ATAC-seq) (Buenrostro et al., 2013). Peaks of open chromatin were identified. By comparing the peak intensity at each locus between OPCs and iOLs from control mice, the dataset reveals two peak groups: becoming more open (closed to open, CO) or becoming more closed (open to closed, OC) during this differentiation. We observed 4,158 CO peaks and 122 OC peaks (Figure 5J). CO peaks outnumber OC peaks in the initial phase of differentiation, suggesting that the chromatin became more permissive during this process. Among these peaks, we identified early differentiation marker genes such as Enpp6 and Sirt2, which acquired open chromatin (Figure 5K). GO analysis showed that the CO loci correspond to genes related to cell differentiation and axon guidance (Figure S5D). Depletion of Seh1 caused altered chromatin accessibility in 1,372 genes in iOLs. Notably, 30% of them (407 of 1,372 genes) were Seh1-bound genes. In addition, among Seh1-bound genes, 8.9% (407 of 4,598 genes) showed differential chromatin accessibility upon depletion of Seh1, which is significantly higher than the proportion among Seh1-unbound genes (4.7%, 965 of 20,350 genes, p value<0.00001, 2-sample z-test for 2 population proportions), suggesting a specific role of Seh1 in chromatin remodeling. Ablation of Seh1 dramatically reduced chromatin accessibility of myelin structural genes and myelination regulators, such as Mbp, Cnp, Sox10, Myrf and Nkx2–2, which suggests that Seh1 is required for maintenance of an open chromatin structure of these genes (Figure 5K). In contrast, the signals due to chromatin accessibility from neuron-enriched genes (Neurod2), astrocyte genes (GFAP), microglia genes (Tmem119) and endothelial genes (CD31) were unchanged in Seh1 mutant iOLs (Figure 5L). Importantly, for those genes bound by Seh1 and down-regulated upon Seh1 depletion, chromatin accessibility around transcription start site (TSS) was markedly decreased in mutant iOLs (Figure 5M). These data suggest that Seh1 is required for establishment of accessible chromatin landscape to promote oligodendrocyte transcriptional progression.
Seh1 Interacts with Olig2 at the Nuclear Periphery
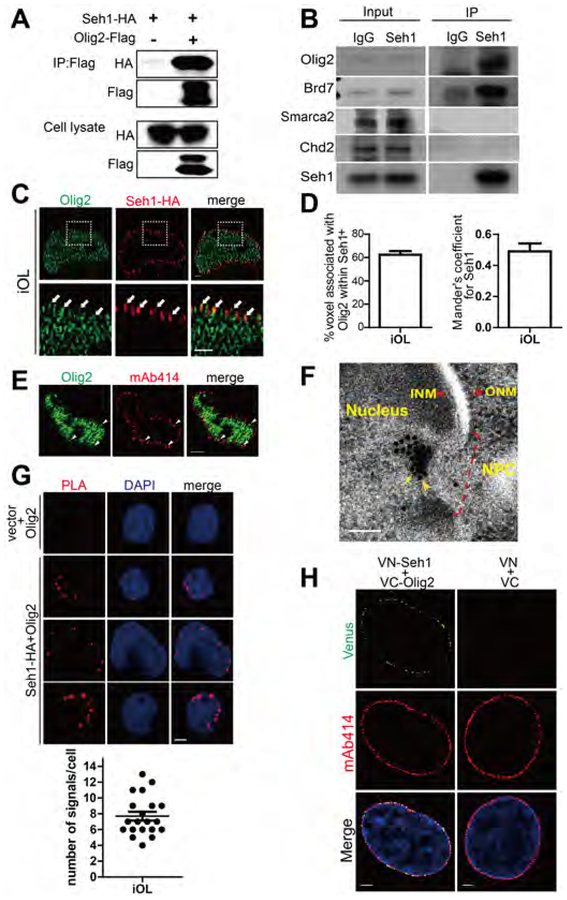
Nups have been shown to regulate cell-type-specific gene expression by interacting with master transcription factors (Raices et al., 2017; Toda et al., 2017). We therefore tested whether Seh1 functionally associates with key factors involved in oligodendrocytes lineage progression. Co-immunoprecipitation of ectopically expressed Seh1 and Olig2 in 293T cells or pull-down of endogenous Olig2 using a Seh1 antibody in rat iOLs revealed that Seh1 binds Olig2, a key fate-determining transcription factor, but not other myelination regulators including Sox10, Nkx2–2, and Myrf (Figures 6A and S6A). Seh1 also bound Olig1, but with much lower affinity compared to Olig2 (Figures S6A) (Lu et al., 2002; Zhou and Anderson, 2002). Super-resolution imaging further showed that Seh1 co-localizes with Olig2 at the nuclear periphery (Figure 6C, 6D, S6B and S6C). In spinal cord, Olig2 could be found at the mAb414 positive foci, indicating its nuclear pore distribution (Figure 6E). During OPC differentiation, Seh1 was mainly found at the nuclear periphery (Figure S6D). In addition, we examined the subcellular localization of Seh1 and Olig2 in rat differentiating iOLs by immunogold EM. Both proteins were detected close to the nuclear pores and in close proximity to each other, indicating a potential association between Seh1 and Olig2 at the nuclear periphery (Figure 6F). We have also utilized the proximity ligation assay (PLA) to test the interaction between Seh1 and Olig2. Consistent with the immunostaining results, PLA signals were observed at the nuclear periphery in iOLs (Figure 6G), suggesting a strong association between Seh1 and Olig2. To further confirm our observations, we employed a biomolecular fluorescence complementation (BiFC) assay that visualizes the direct interaction of proteins (Kodama and Hu, 2010). Again, we found that Seh1 interacted with Olig2 at the nuclear periphery (Figure 6H). Taken together, these data demonstrate that Seh1 interacts with Olig2 at the nuclear periphery.
Figure 6. Seh1 Interacts with Olig2 at the Nuclear Periphery.
(A and B) Co-immunoprecipitation of Seh1-HA with Olig2-Flag from transiently transfected 293T cells (A) or endogenous Olig2 with Seh1 in rat iOLs (B).
(C) Super-resolution images of Seh1 and Olig2 at the nuclear periphery in rat differentiating iOLs. The areas within the boxes are magnified and are shown in the bottom panels. Arrowheads indicate associated signals. Scale bars=1 μm.
(D) Left panel: Quantification of associated voxels with Olig2 among Seh1-positive voxels; Right panel: Quantitative analysis of association of Seh1 and Olig2 signals by Mander’s coefficient analyzing the ratio of Seh1-positive voxel with Olig2 signals.
(E) Immunostaining of Olig2 and mAb414 in spinal cord of P2 control mice. Arrowheads indicate Olig2+/mAb414+ cells. Scale bars=2 μm.
(F) Immunoelectron micrographs of transfected Seh1-HA and Olig2-Flag in rat differentiating iOLs. Light yellow arrows indicate Seh1 and Dark yellow arrowheads indicate Olig2. Small particles (10nm) label Seh1 and big particles (15nm) label Olig2. NPC, INM and ONM were highlighted. NPC=nuclear pore complex; ONM/INM, outer/inner nuclear membrane. Scale bars=100 μm.
(G) Top panel: PLA signal detection for Seh1 and Olig2 in iOLs following expression of Seh1-HA. Scale bars=2 μm; Bottom panel: quantification of PLA signal numbers.
(H) BiFC analysis of Seh1-Olig2 interactions. Transiently expression of indicated combination of split Venus plasmids in iOLs. Scale bars=2.5 μm.
See also Figure S6.
Olig2 is phosphorylated at serine 147 during motor neuron production and dephosphorylated at the onset of OPC genesis (Li et al., 2011). We next tested whether the phosphorylation status of Olig2 affected its interaction with Seh1. Unphosphorylated Olig2 (S147A) and constitutive phosphorylated Olig2 (S147E) were used in immunoprecipitation assay. As shown in Figure S6E, Seh1 bound Olig2 regardless of the phosphorylation status of serine 147 (Figure S6E). We have also tested whether Seh1 is phosphorylated during OPC differentiation. Seh1 was immunoprecipitated and detected by western blot with phospho-serine/threonine/tyrosine antibody. Seh1 was upregulated upon OPC differentiation, but phosphorylated Seh1 was not detected in OPCs or iOLs (Figure S6F).
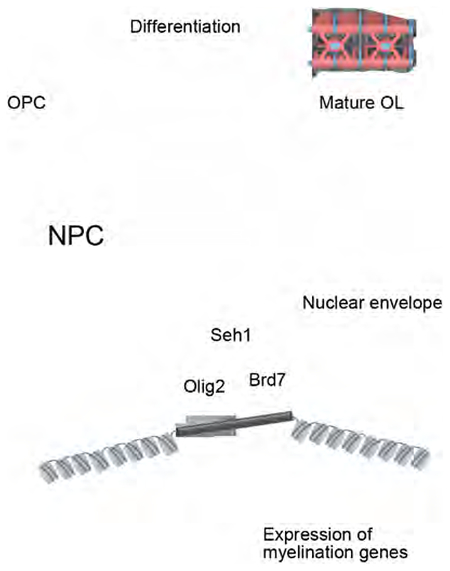
Seh1 Promotes Myelination-related Gene Transcription by Assembling an Olig2/Brd7 Transcription Complex
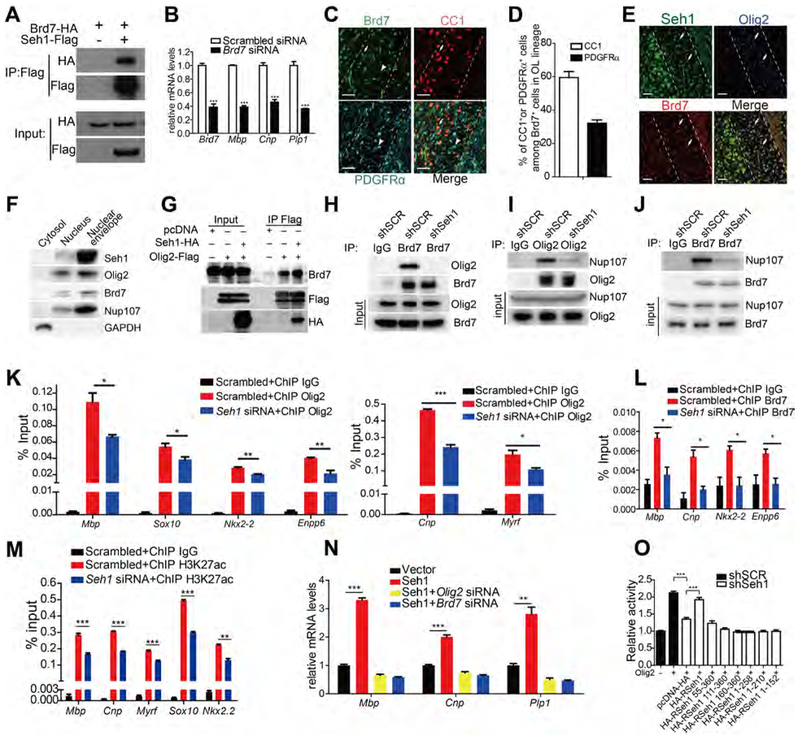
NPCs have been reported to act as a platform for transcription regulation (Pascual-Garcia and Capelson, 2014). Since Seh1 is required for establishment of accessible chromatin landscape to promote oligodendrocyte transcriptional progression and does not possess any reported transcriptional modulation activity on its own, we reasoned that Seh1 may act as a scaffold to assemble a transcription complex at the nuclear periphery. To test our hypothesis, we performed immunoprecipitation using Seh1 antibody or IgG in differentiated rat OPCs followed by mass-spectrometry. The Seh1-binding candidates are listed in Figure S6G. Nup85 and Nup160, which are known Seh1-binding proteins, were identified as Seh1-binding partners, thus validating our mass-spectrometry results (Wente and Rout, 2010). Notably, chromatin remodelers, such as Brd7, Chd2 and SMARCA2, were among the Seh1-binding candidates and we found that Seh1 bound Olig2 and Brd7, but not other chromatin remodelers (Figures 6B and 7A). As a component of the switch/sucrose nonfermentable (SWIlSNF) chromatin remodeling complex, Brd7 has been shown to act as a transcription activator in cell differentiation and is found at early stages in the germinal zone of developing spinal cord (Fu et al., 2009; Kaeser et al., 2008). We then knocked down Brd7 in rat OPCs and observed a significant decrease in expression of oligodendrocyte differentiation-associated genes Mbp, Cnp and Plp1 upon T3-induced differentiation (Figure 7B). BI-7273 is a potent and selective inhibitor of Brd7 and BI-7273 treatment of rat OPCs caused a similar defect (Figure S7A) (Martin et al., 2016). Furthermore, Brd7 is detected in the OL lineage and is mainly expressed in CC1+ oligodendrocytes (Figures 7C and 7D). Immunostaining also shows that Brd7 co-localizes with Seh1 and Olig2 at the mouse corpus callosum (Figure 7E). We also assessed the subcellular localization of Olig2 and Brd7 by nuclear envelope fractionation. Indeed, Olig2 and Brd7 co-purified with Seh1, which is consistent with the immunostaining studies (Figure 7F). Taken together, these results suggest that Seh1 may function together with Olig2 and Brd7.
Figure 7. Seh1 Promotes Myelination-related Gene Transcription by Assembling an Olig2/Brd7 Transcription Complex.
(A) Co-immunoprecipitation of Brd7-HA with Seh1-Flag from transiently transfected 293T cells.
(B) qRT-PCR validation of knockdown efficiency of Brd7 in primary rat OPCs and qRT-PCR analyses of myelination-associated genes in primary rat OPCs under differentiation conditions following treatments with scrambled or Brd7 siRNAs. (n= 3 independent experiments from 3 rats. Brd7, ***p=0.0004, Mbp, ***p<0.0001, Cnp, ***p=0.0005, Plp1, ***p=0.0004).
(C) Co-immunostaining of Brd7, CC1 and PDGFRα in the corpus callosum of wild-type mice at P14. Arrow indicates Brd7+/CC1+ cells; arrowheads indicate Brd7+/PDGFRα+ cells. Scale bars=50 μm.
(D) Quantification of CC1+ or PDGFRα+ cells among Brd7+ cells in the corpus callosum at P14 (n=3 mice).
(E) Immunostaining of Seh1, Olig2 and Brd7 in corpus callosum of wild-type mice at P14. Arrows indicate Seh1+/Olig2+/Brd7+ cells. Scale bars=50 μm.
(F) Immunoblot was performed with the indicated antibodies following nuclear envelope extraction of iOLs. GAPDH is a positive control for cytoplasmic extraction; Nup107 is a positive control for nuclear envelope fraction.
(G) Co-immunoprecipitation of Brd7 with Olig2-Flag from transiently transfected 293T cells with the indicated plasmids.
(H) Co-immunoprecipitation of Olig2 with Brd7 from Oli-neu cells transfected with the indicated shRNAs. SCR, scrambled.
(I) Co-immunoprecipitation of Nup107 with Olig2 from Oli-neu cells transfected with the indicated shRNAs.
(J) Co-immunoprecipitation of Nup107 with Brd7 from Oli-neu cells transfected with indicated shRNAs.
(K) Enrichment of Olig2 at promoter regions of oligodendrocyte genes in Seh1 knockdown iOLs compared to control cells. (n= 3 experiments from 6 rats. Mbp, *p=0.019, Sox10, *p=0.033, Nkx2–2, **p=0.0014, Enpp6, **p=0.0069, Cnp, ***p=0.0001, Myrf, *p=0.0264).
(L) Enrichment of Brd7 at promoter regions of oligodendrocyte genes in Seh1 knockdown iOLs compared to control cells. SCR, scrambled. (n= 3 experiments from 6 rats. Mbp, *p=0.0143, Cnp, *p=0.01, Nkx2–2, *p =0.0176, Enppβ, *p =0.0128).
(M) Enrichment of H3K27ac at promoter regions of oligodendrocyte genes in Seh1 knockdown iOLs compared to control cells. (n= 3 experiments from 6 rats. Mbp, ***p=0.0006, Cnp, ***p<0.0001, Myrf, “p=0.0007, Sox10, “*p<0.0001, Nkx2.2, **p=0.0012).
(N) qRT-PCR analysis of myelination-associated genes in primary rat OPCs under differentiation conditions following co-transfection with indicated plasmids and siRNAs (n= 3 experiments from 6 rats. Mbp, ***p<0.0001, Cnp, ***p=0.0002, Plp1, **p=0.0018).
(O) Luciferase activity of Cnp promoter-driven reporters in control (shSCR, scrambled) or Seh1 knockdown (shSeh1) 293T cells transfected with Olig2 and with the indicated plasmids expressing different regions of Seh1 (n= 3 experiments. shSCR versus shSeh1+pcDNA, ***p<0.0001, shSeh1+pcDNA versus shSeh1+HA-RSeh1, ***p<0.0001).
Data are represented as mean ± SEM; *p<0.05, **p<0.01, ***p<0.001, two-tailed unpaired Student’s t test. See also Figure S7.
To further study the formation of the Seh1/Olig2/Brd7 complex, we performed coimmunoprecipitation assay and found that the association between Olig2 and Brd7 was increased by Seh1 overexpression and impaired by knockdown of Seh1, indicating that Seh1 likely mediates the assembly of the Olig2/Brd7 complex (Figures 7G and 7H). Furthermore, the association between Olig2 and nucleoporins or the association between Brd7 and nucleoporins was impaired by knockdown of Seh1 (Figure 7I and 7J), suggesting that Seh1 is required for the interaction between these factors and nucleoporins. In contrast, knockdown of Olig2 did not affect the association between Brd7 and Seh1 or the association between Brd7 and nucleoporins (Figure S7B–S7D). Similarly, the interaction between Olig2 and Seh1 or the interaction between Olig2 and nucleoporins was not affected by knockdown of Brd7 (Figure S7E and S7F). We also show, by immunofluorescence microscopy, that only intact Seh1, and not its truncated mutants, was able to localize at the nuclear periphery (Figure S7G and S7H). Meanwhile, a truncated mutant could not rescue the defects in myelin gene expression caused by knockdown of Seh1 (Figure 2H). Through a series of truncations followed by co-IP, we also found that only intact Seh1 was able to bind Olig2 and Brd7 (Figures S7I and S7J). These data indicate that Seh1 recruits Olig2 and Brd7 to form a multi-protein regulatory complex at the nuclear periphery.
We next tested whether Seh1 played a role in Olig2/Brd7 complex-mediated transcription of myelination-associated genes and chromatin modification. Indeed, the enrichment of Olig2 and Brd7 in the promoter regions of myelination-related genes was impaired by knockdown of Seh1 in iOLs (Figure 7K and 7L). Furthermore, the enrichment of H3K27 acetylation, which induces transcription activation, was also significantly decreased in these regions (Figure 7M). We also performed additional ChIP-qPCR assay using antibody against Nup93, which strictly localizes at the nuclear pore complex (Brown et al., 2008). As shown in Figure S7K, we found that Seh1-bound genes were associated with Nup93, indicating that the function of Seh1 in the transcriptional regulation of these genes likely takes place at the nuclear periphery. Overexpression of Seh1 in rat OPCs promoted myelin genes expression and this induction was abolished by knockdown of Olig2 or Brd7 (Figure 7N and S7L). These data indicate that the regulatory role of Seh1 depends on Olig2 and Brd7. We have also found that knockdown of Seh1 significantly impaired Olig2-mediated Cnp promoter reporter activity. Expression of shRNA-resistant Seh1 effectively rescued the impaired Olig2 transcription activity but expression of the truncated shRNA-resistant Seh1, which did not bind Olig2 or Brd7, failed to do so (Figure 7O). In contrast, the reporter activity of a non-oligodendroglial related gene (IFN-β) was not affected by Seh1 (Figure S7M). Together, these results suggest that Seh1 acts as a scaffold protein to assemble the Olig2-Seh1-Brd7 transcription complex to promote myelination-related gene expression and oligodendrocyte differentiation.
DISCUSSION
Seh1 Has a Physiological Function in the Central Nervous System
Connections between Nups and neurological diseases have recently emerged. Aladin gene variants are linked to the Triple A syndrome, an autosomal recessive disorder characterized by abnormal autonomic nervous system development (Kallabi et al., 2016). A Nup62 homozygous variant is also associated with autosomal recessive infantile bilateral striatal necrosis (Basel-Vanagaite et al., 2006). A syndromic form of autosomal recessive congenital microcephaly has been recently mapped to chromosome 18p11.22-q12.3 and Seh1 lies in this region (Hassan et al., 2008). Whereas no exonic sequence variants was identified, the possibility of a functional variant in the regulatory regions needs further investigation. Nup133, Nup153 and Nup210 have been implicated in the regulation of neural differentiation (D’Angelo et al., 2012; Lupu et al., 2008; Toda et al., 2017). However, evidence from tissue-specific depletion in mouse models is still missing. Here we used the Cre-loxP system to specifically deplete Seh1 expression in the oligodendrocyte lineage. The mutant mice exhibited generalized tremors and severe demyelinating phenotypes. By inducible depletion of Seh1 in adult OPCs, we showed that Seh1 is important for adult oligodendrocyte production and myelinogenesis. Furthermore, ablation of Seh1 in OL lineage impaired remyelination after LPC-induced demyelination in the adult mammalian CNS. Recently, depletion of the nucleoporins Nup210, Nup96 and Sec13 in mice was found to affect the immune system including CD4+ T cell homeostasis and MHC expression (Borlido et al., 2018; Faria et al., 2006; Moreira et al., 2015). Thus, it will be interesting to test if these nucleoporins have physiological functions in other tissues.
Seh1 Functions as a Scaffold for the Assembly of Transcription Complexes
The NPC has been linked to genome compartmentalization and acts as a platform for gene regulation (Cabal et al., 2006; Menon et al., 2005). Our data indicate that during oligodendrocyte development, Seh1 interacts with Olig2 at the nuclear periphery and recruits the chromatin remodeler Brd7 to assemble a transcription complex. Using BiFC and PLA assay, we observed that these two proteins interacted at the nuclear periphery. Seh1 truncations, which were not localized at the nuclear periphery, did not bind Olig2 or Brd7, suggesting that the nuclear pore localization is necessary for assembly of this transcription complex. This interaction could be structurally important to activate the cell-type-specific gene transcription, since truncated Seh1 could not rescue the defects in myelin gene expression caused by knockdown of Seh1. The nuclear architecture composed of Seh1, Olig2, and Brd7 could provide a transcription-friendly microenvironment to the genome and maintain a high concentration of transcription factors and co-factors for efficient transcription.
An Accessible Chromatin Landscape Is Established during Oligodendrocyte Differentiation
It has been known that genome reorganizations occurs during neuronal differentiation. Our results from ATAC-seq revealed dynamic changes in chromatin states. Notably, the CO peaks outnumber OC peaks, indicating that the chromatin becomes more opened during the initial stages of OPC differentiation. The result also indicates certain pioneer factors mainly displace nucleosome in closed chromatin and initiate the opening up of chromatin at the early phases of differentiation. It will be interesting to identify the transcription factors which drive these chromatin dynamics. In addition, we showed that loss of Seh1 resulted in closed chromatin conformation of myelin-related genes, suggesting that Seh1 is essential for increasing chromatin accessibility near lineage-specific regulatory genes to promote the OPC differentiation programs.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Liang Zhang (lzhangxmu@xmu.edu.cn).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mice were maintained in the core animal facility at Xiamen University, and all experimental procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee at Xiamen University. Mice were housed in a vivarium with 12 h light/dark cycle with free access to water and food. Both male and female mice were used. The ages of mice used were from P4 to P84 as stated in text and figures. Seh1loxP/loxP mice were generated using homologous recombination in ES cells. To generate Seh1−/− mice, Seh1loxP/loxP mice were crossed with EIIA-Cre mice (Lakso et al., 1996). Seh1loxP/loxP mice were crossed with Olig1-cre+/− mice to generate Seh1cKO (Seh1loxP/loxP; Olig1-Cre+/−) and heterozygous (Seh1loxP/+; Olig1-Cre+/−) mice (Xin et al., 2005). We used heterozygous mice as control since they developed and behaved the same as WT. A similar mating strategy was used for generating Seh1 inducible mice: CMV-CreERT, NG2-CreERT and Sox10-CreERT mice were crossed with Seh1loxP/loxP mice (Hayashi and McMahon, 2002; McKenzie et al., 2014; Zhu et al., 2011). Mice were maintained on the mixed C57BL/6;129Sv background.
Cells and cell cultures
Mouse OPCs were obtained from cortices of mouse brains as described (Dugas et al., 2006). Briefly, dissociated P4–P7 mouse cortices cells were immunopanned on anti-GalC, and then placed on anti-O4 antibody coated plates to harvest OPCs. Purified OPCs were trypsinized and plated onto poly-D-lysine-coated plates. Cells were grown in DMEM/F-12 (GIBCO) with addition of 2% B-27 (GIBCO), 1% N2 (GIBCO), 20 ng/ml PDGF-AA (Peprotech, 100–13A), 10 ng/ml CNTF (Peprotech, 450–13), 20 ng/ml bFGF (Sino Biological, 10014-HNAE), and 1 ng/ml NT3 (Peprotech, 450–03). For OPCs differentiation, triiodothyronine (T3) (60 nM) was added to the media and PDGF-AA, FGF and NT3 were removed. Rat OPCs were obtained from P2–P4 rat brains as mouse OPCs with slight modifications. Briefly, dissociated rat cortices cells were immunopanned on anti-GalC, and then placed on anti-A2B5 antibody coated plates to harvest OPCs. Oli-neu cells were maintained in DMEM/F-12 supplemented with 2% B-27, 1% N2, 5% FBS (GIBCO), 1% horse serum (GIBCO), 7.2 mM Glucose and 1× penicillin-streptomycin. 293T (from the American Type Culture Collection) cells were cultured in DMEM supplemented with 10% FBS and 1× penicillin-streptomycin. Transfections of 293T cells were performed using Polyetherimide (Sigma-Aldrich).
Approximately 2.0 × 10^6 OPCs or Oli-neu were resuspended in 100 μL Basic Nucleofector™ Kit for Primary Mammalian Glial Cells (Lonza, O-017). Additionally, 5μg of the appropriate plasmid or 5 μl of 20 μM siRNA (siSeh1: GGCTCAGTTTGATAATCAT; siOlig2. GCCAGAACCCGATGATCTT; siBrd7: GCCAAGATTACCCGTATGT, Ribobio) was added to the solution. The solution was then placed in a cuvette provided in the Nucleofector Kit and electroporated using a Lonza Nucleofector 2b device (LONZA) according to the manufacturer’s protocol and program O-017. Next, cells were resuspended in medium and incubated for 30 min at 37°C, 5% CO2, before changing to new media to remove dead cells.
METHOD DETAILS
Tissue and Immunohistochemistry
Mice at defined ages were anesthetized before sacrifice and then perfused with ice-cold phosphate-buffered saline (PBS) followed by 2% paraformaldehyde (PFA). The brain and spinal cord were dissected, fixed in 2% paraformaldehyde overnight, dehydrated in 25% sucrose at 4°C, embedded in OCT (Leica, 14020108926) and processed for cryo-sections at 12 μm. For immunohistochemistry, cryo-sections were permeabilized and blocked in blocking buffer (0.4% Triton X-100 and 3% normal BSA in PBS) for 1h at room temperature (RT) and overlaid with primary antibodies overnight at 4°C. After washing with PBS, sections were incubated with secondary antibodies conjugated to Cy2, Cy3 or Cy5 (Jackson ImmunoResearch Laboratories, 1:1000) for 1h at RT, stained with DAPI for 10min, washed three times in PBS and then mounted in Mounting Medium. For Immunocytochemistry, cells were fixed with PFA for 30min at RT, permeabilized with pre-cold buffer (0.5% Triton X-100 in PBS) at 4°C, and blocked in blocking buffer (5% BSA in PBS). Coverslips were covered with diluted primary antibodies overnight at 4°C. Second ary antibodies were incubated for 1h at RT and nuclei were stained with DAPI for 10 min. Immunofluorescence images were obtained with a confocal laser microscope (LEICA, SP8).
For RNA in situ hybridization, cryosections of brain and spinal cord tissues were incubated with digoxigenin (DIG)-labeled antisence riboprobes for murine Plp1/Dm-20 as described previously (Ma et al., 1997). The probe was synthesized using T3 RNA polymerase (Promega, P208C) and labeled with DIG RNA label mix (Roche, 11277073910). An anti-DIG antibody conjugated with alkaline phosphatase (Roche, 11093274910) was used to probe sections, which were stained with 5-bromo-4-cloro-3-indolyl phosphate (BCIP) (Solarbio, B8090) lnitro blue tetrazolium (NBT) (Solarbio, N8140) chromogenic substrates.
Microscopy
Fluorescence microscopy was performed using a Leica SP8 and Zeiss LSM 780/880 microscope. Images were obtained with 20× and 63× objectives, and colocalization were confirmed by 3-D reconstruction of Z-series. For super-resolution imaging, a Leica SP8 Hyvolution (63× objective lens) and a Zeiss LSM 880 Airy scan (63× objective lens) were used. Cells were stained with primary antibodies overnight at 4°C in a humid box, washed 3 times, incubated with secondary antibodies for 1 hr at RT, and stained with DAPI. Cell-culture slides were mounted with Prolong Gold (Invitrogen). Nuclei were imaged with a Zeiss LSM 880 airyscan (63× object lens) and a Leica SP8 Hyvolution (63× object lens). After 3-D reconstruction, colocalization between two proteins obtained from Zeiss LSM 880 airyscan was quantified using automatic thresholding tools or a surface-colocalization tool with distance map in Imaris (Bitplane). The images from Leica Hyvolution were deconvolved using Huygens Essential (Scientific Volume Imaging (SVI)).
Electron microscopy
Tissue processing was performed as described previously (Yu et al., 2013). Briefly, mice were deeply anesthetized, perfused with pre-cold sodium cacodylate buffer. Optic nerve and spinal cord were dissected immediately and fixed with 2.5% glutaraldehyde overnight at 4°C. Next, samples were treated with 1% osmium tetroxide, dehydrated, and embedded into PolyBed resin. 70-nm ultrathin sections were then stained with lead citrate for electron microscopy imaging using the JEM-2100HC microscope (JEOL, Akishima, Tokyo, JAPAN).
For immunogold assay, rat OPC culture and transfection conditions were performed as described above. Cells were fixed in 4% PFA. After dehydration and permeation, LP White Resin (London Resin Company) was used for polymerization for 48 h at −20°C. 80-nm ultrathin sections were double-labeled with rabbit Flag and mouse HA antibodies. Bound antibodies were detected with goat anti-rabbit IgG conjugated with 15 nm gold and goat anti-mouse IgG conjugated with 10 nm gold (Electron Microscopy Sciences)
Lysolecithin-induced demyelinating injury
Lysolecithin (LPC)-induced demyelination was carried out in the corpus callosum (CC) and spinal cord (SC) of 8-week-old Seh1loxP/loxP; NG2-creERT mice and Seh1loxP/+; NG2-creERT control mice as described previously (He et al., 2016). Briefly, for CC injury, after exposing the skull, 1.5 μl of 1% (W/V) LPC (Sigma) in 0.9% NaCl was stereotactic injected into CC. The coordinates were: 1mm backward to bregma, 1 mm lateral to bregma, and 1.5mm deep relative to the skull surface. Brain tissue carrying the lesions were harvested at different time points. For SC injury, after exposing the spinal vertebrae at the level of T9–T12, meningeal tissue in the intervertebral space was cleared. 0.5μl of 1% (W/V) LPC (Sigma) in 0.9% NaCl was stereotactically injected into the ventrolateral white matter. SC tissues carrying the lesions were collected at different time points.
Tamoxifen Administration
Tamoxifen (sigma) was dissolved in corn oil at a concentration of 20 mg/ml and stored at −20°C. Tamoxifen was administered to neonatal mi ce through i.p. injection at a dosage of 40 μg/g (gram, body weight). Single injection was administered daily for three consecutive days from P3 to P5. Young adult mice received daily tamoxifen i.p. injection at a dosage of 5 mg per 40 g body weight for five consecutive days from P26 to P30. For LPC assay, tamoxifen was administered for 8- to 10-week-old mice through gavage at a dosage of 200 μg/g, single time daily for seven consecutive days.
BrdU Assay
For proliferation assay, mice were pulsed with 100 μg/g (gram, body weight) of BrdU at 2 h before sacrifice. For differentiation assay, mice were pulsed with 250 μg/g of BrdU twice daily at 12 h intervals for five consecutive days from P31 to P35.
Plasmid Preparation
Mouse Seh1 (gene ID: 72124), rat Seh1 (gene ID: 100911798), human Seh1 (gene ID: 81929), truncated Seh1, Olig2 (gene ID: 250913) and Brd7 (gene ID: 29117) were constructed using either pcDNA3.3, pLV or EGFP-C1 vectors. Mutations in Seh1 were generated by a PCR-based site-directed mutagenesis method using Primestar polymerase (Takara). The 21-nucleotide sequence for shRNA against human Seh1 is 5′-ATAGTGGCTCAGTTCGATAAT -3′, shRNA against mouse Seh1 is 5′-CGATGTGTCTTTCGACTTCCA -3′, shRNA against mouse Olig2 is 5′-GCCACGTCTTCCACCAAG -3′, shRNA against mouse Brd7 is 5′-GCCAAGATTACCCGTATGTTA -3′. Packaging of lentiviruses were obtained by inserting corresponding cDNAs into pLKO.1 or pLKO.3g vectors. Lentiviruses were generated by transfecting 293T cells with the lentiviral vector and packaging plasmids.
Proximity Ligation Assay
The procedures followed the Duolink PLA Fluorescence protocol from Sigma-Aldrich. Cells were fixed with 2% paraformaldehyde in PBS for 20 min at RT, and then permeabilized with 0.5% Triton X-100 for 5 min at 4°C, and blocked for 1 hr at 37°C with a blocking solution (Sigma). Cells were washed with PBS for 5 min. The primary antibodies were diluted in the Duolink Antibody Diluent, added to the samples, which were then incubated in a humidity chamber overnight at 4°C. Subsequently, the PLUS and MINUS PLA probe were diluted in Duolink Antibody Diluent, were then added to the cells and incubated in a pre-heated humidity chamber for 1 hr at 37 °C. Next, cells were washed with Wash Buffer A (Sigma) and incubated with the ligation solution (Sigma) for 30 min at 37 °C. The ligation solution was removed and the slides were washed with Wash Buffer A twice. Then, cells were incubated with the amplification solution (Sigma) for 100 min at 37°C in the dark. Cells were washed with 1× Wash Buffer B twice and 0.1× Wash Buffer B once at RT. Finally, cells were incubated with DAPI and mounted with a coverslip using a Duolink mounting medium.
Biomolecular Fluorescence Complementation Assay
The cDNA sequences of mouse Seh1 and Olig2 cDNA were subcloned into pcDNA-VN-155 and pcDNA-VC155 plasmid vectors, respectively (Shyu et al., 2006). The pcDNA-VN155 has an I152L mutation to reduce the self-assembly background (Kodama and Hu, 2010). The concentration for pcDNA-VN-155 and pcDNA-VC155 plasmids for transfection was titrated until BiFC signals became negligible, and the same molar concentrations were applied for other plasmids. 5 μg of plasmid was transfected into 2 × 106 OPCs using Amaxa Nucleofector (Lonza, O-017) according to the manufacturer′s protocol. After 48 h transfection, cells were induced by T3 for 24 h, and then fixed with 2% PFA/PBS. Nuclei were stained with DAPI for imaging.
Nuclear Envelope Isolation
NEs were isolated according to the manufacturer’s protocol (Inventbiotech). Briefly, 1.5*10^7 iOLs were resuspended in 500μl buffer A (with 1 mM PMSF) and incubated on ice for 10 min. The tube was vortexed vigorously for 20 seconds. Then, the cell suspension was transferred to the filter cartridge and centrifuged at 14,000 rpm for 30 seconds. The supernatant was cytosol. The pellet was washed with PBS (with 1 mM PMSF) once. Next, 300 μl buffer B (with 1 mM PMSF) was added to resuspend the pellet by vigorously vortexing for 10 seconds. After incubating on ice for 5 min, the suspension was vigorously vortexed for 10 seconds. The suspension was incubated again for 5 minutes on ice and vigorously vortexed for 10 seconds. We then took 30 μl of the supernatant, which constituted the nuclear fraction. After centrifugation at 8,000 rpm for 5 min, the supernatant was transferred to a fresh 2.0 ml microcentrifuge tube. 720 μl cold PBS (with 1 mM PMSF) was added to the tube, which was inverted 10 times. Centrifugation was then performed at 14,000 rpm for 15 min. The pellet constituted the nuclear envelope fraction. NEs were finally resuspended in 50 μl PBS (with 0.5% Triton X-100 and 1 mM PMSF). Protein concentration was quantified with BCA protein assay kit (Sangon) and equal amounts of proteins were subjected to SDS-PAGE.
RNA extraction and qRT-PCR
Total RNA was extracted using Eastep®Super Total RNA Extraction Kit (Promega) and reverse transcribed into cDNA with the GoScript™ Reverse Transcription System (Promega). qRT-PCR was performed using the Bio-Rad Real-Time PCR System. Primers used for mouse gene sequences were: Plp-f, TGCTCGGCTGTACCTGTGTACATT, Plp-r, TACATTCTGGCATCAGCGCAGAGA; Mbp-f, TCACAGAAGAGACCCTCACA; Mbp-r, GCCGTAGTGGGTAGTTCTTG;Cnp1-f TCCACGAGTGCAAGACGCTATTCA, Cnp1-r, TGTAAGCATCAGCG GACACCATCT; Gapdh-f, TGCCTATGATGACATCAAGAA, Gapdh-r, GGAGTGGGTGTCGCTGTTG; Mag-f, TGGTGTGTGGCTGAGAACCAGTAT, Mag-r TGCACAGTGTGACTCCAGAAGGAT; Mog-f, AGATGGCCTGTTTGTGGAG, Mog-r, TTCATCCCCAACTGCCC; for rat gene sequences were: Gapdh-f, TCCAGTATGACTCTACCCACG, Gapdh-r, CACGACATACTCAGCACCAG; Mbp-f, TTGACTCCATCGGGCGCTTCTTTA, Mbp-r, TTCATCTTGGGTCCTCTGCGACTT. Cnp-f, CTACTTTGGCAAGAGACCTCC, Cnp-r, AGAGATGGACAGTTTGAAGGC; Plp-f, TCTTTGGCGACTACAAGACCACCA, Plp-r, CCAATGACA CACCCGCTCCAA; Seh1-f, CCCAAGCACATGGGTCTGAT, Seh1-r, GTCATCACTTCCCACAGCGA; Brd7-f, TCTGACCTTCCCAATTTTAGC, Brd7-r, ATCTTCAGGTGACGGCATG.
Luciferase Reporter Assay
HEK 293T Cells were transfected with pGL3-Cnp-Luc reporter constructs (He et al., 2016) or IFN-β reporter constructs (Lienenklaus et al., 2009), β-galactosidase (β-gal) expression vectors and other indicated plasmids. At 48 h post-transfection, cells were harvested and luciferase activities were measured in triplicate and normalized for transfection efficiency with β-gal activity.
Phosphoproteomic analysis
OPCs cells were seeded at 1×106 cells/well in 6-well plates. Cells were treated with T3 for 0 or 24 h and collected in biological duplicates. Cells were washed with ice-cold PBS for three times. 200 μl lysis buffer was added into one well. The lysed cells were transferred to 1.5 ml EP tubes, which was subjected to 95 °C heat for 5 min and sonicated. The protein concentration was measured with the Pierce 660 nm protein assay reagent (Thermo). 50 μg proteins were used for digestion. Trypsin (Sigma) was added at a ratio of 1:100 (trypsin:protein). The tubes were kept at 37 °C for 12–16 h. After digestion, phosphopeptides were enriched using TiO2. Briefly, peptide solutions were added with an equal volume of 4% TFA/2 mM KH2PO4isopropanol (ISO). After removing the pellet by centrifugation at 12,000 rpm for 5 min, the supernatants were used for enrichment. 2 mg TiO2 was added into one digestion solution. The tubes were incubated at 40 °C shaking (2,000 rpm) for 5 m in. Beads were collected by centrifugation and the supernatant was discarded. Beads were then washed with 500 μl of 2%TFAl50%ISO for three times. Phosphopeptides were eluted with 60 μl of 20% NH3. H2Ol32%ACN, followed by concentration using an evaporative concentrator. Phosphopeptides were then desalted using SDB-RPS StageTips. Phosphopeptides were dissolved in 0.1% formic acid and analyzed by Oribtrap Fusion Lumos Tribrid mass spectrometry (Thermo Fisher). The gradient time was 60 min. A DDA scheme was used with an MS1 full scan solution of 120,000 FWHM (at mlz 200) followed by as many subsequent MS2 scans on selected precursors as possible within a 3 second maximum cycle time. MSlMS was performed in the Orbitrap using HCD at a resolution of 30,000 FWHM. Spectral. raw files were analyzed using Maxquant software (1.6.0.16) (Cox and Mann, 2008). The search database was swissprot human sequences including common contaminants. The fixed modification was Carbamidomethyl(C), and variable modifications were Oxidation(M), Acetyl(protein N-term) and Phospho(STY). LFQ quantification and match between runs were enabled. The search results were added to the Perseus platform (Tyanova et al., 2016). Phosphopeptides with localization probability >0.75 were kept for further analysis. Volcano plots were generated with default setting.
RNA-seq and data analysis
RNA from control and Seh1cKO spinal cord or OPCs were prepared using Illumina RNA-Seq Preparation Kit and subjected to 50-bp single-end sequencing with a BGISEQ-500 sequencer as previously described (Chen et al., 2017). At least 20 million clean reads of sequencing depth were obtained for each sample. RNA-seq raw data were initially filtered to obtain clean data after quality control. Clean data were aligned to the mouse genome (mm10) by HISAT2 (Langmead and Salzberg, 2012). Raw counts for each gene were calculated by Htseq (Anders et al., 2015). StringTie was used to estimate the expression level of detected genes (Pertea et al., 2015). DEGs were defined as genes with FDR less than 0.001 and fold change larger than 2.
ChIP-seq and ChIP-qPCR
ChIP-seq was performed as previously described (Yu et al., 2013) with slight modifications. Briefly, approximately 1 x10^7 cells were cross-linked with 1% formaldehyde for 10 min at room temperature and quenched with 125 mM glycine for 5 min. Sonicated chromatin was used for immunoprecipitation by incubation with antibodies (4 μg) overnight at 4°C. Immunoprecipitated complexes were collected using protein 40μl A/G plus agarose beads (Millipore). Subsequently, beads were washed sequentially with low-salt buffer, high-salt buffer, LiCl buffer, twice with TE, and then eluted in 500 μl of elution buffer (1% SDS, 0.1M NaHCO3). The eluates were heated at 42°C for 2h and treated with protein ase K followed by incubation at 65°C for 10 h to reverse the cross-linking. Next, e luates were treated with RNaseA for 30 min before DNA was extracted and purified. The ChIP libraries were prepared using KAPA HyperPrep Kits (Roche, 07962347001) and then run on the Illumina sequencer Hiseq-Xten PE150. Primers used in ChIP-qPCR are: Rat Mbp-f. GCTTCTCTCCCTCTTCAGCA, Mbp-r. TGGTTTCTCACGACGTG; Cnp-f. GGCTAGGGCTGCGTACATTA, Cnp-r. GAGTCTGAAGCCAGCCTCTC; Myrf-t AACCTCATTGTGAGG CACCC, Myrf-r. TCCTCCCTCCTTCTCAGTG G; Sox10-f: CCTAATTCACTGGGCTCTGC, Sox10-r: AAGGCCTCAGGGTGGATAGT; Nkx2-2-f: AGGACCTTGCAGAGACG, Nkx2-2-r: GGCGTCTTACAGGTCCA.
ChIP-Seq Analysis
The Primary Analysis of ChIP-Seq Data sets were performed by using Illumina’s Genome Analysis pipeline. The sequencing reads were aligned to the human genome UCSC build hg18 by using Bowtie2 alignment programs in two ways: only uniquely aligned reads were kept or both uniquely aligned reads and the sequencing reads that align to repetitive regions were kept for downstream analysis (if a read aligns to multiple genome locations, only one location is arbitrarily chosen). The multiple reads were collapsed in order to reduce the PCR biases. The aligned reads were used for peak finding with HOMER (Heinz et al., 2010; Wang et al., 2011) (http://biowhat.ucsd.edu/homer). The identification of ChIP-seq peaks was performed using HOMER following protocols as described previously (Wang et al., 2011). Peaks were identified by searching locations of high read density using a 200-bp sliding window. Regions of maximal density exceeding a given threshold were called as peaks, and we required that adjacent peaks to be at least 500 bp away to avoid redundant detection. Only one tag from each unique position was considered to avoid clonal artifacts from sequencing. The threshold for the number of tags that determined a valid peak was selected at a false discovery rate of 0.001 determined by peak finding using randomized tag positions in a genome with an effective size of 2×109 bp. We also required peaks to have at least 4-fold more tags (normalized to total count) than input control samples. In addition, we considered 4-fold more tags relative to the local background region (10 kb) to avoid identifying regions with genomic duplications or nonlocalized binding. Annotated positions for promoters, exons, introns and other features were based on RefSeq transcripts and repeat annotations from the University of California, Santa Cruz. Peaks from separate experiments were considered equivalent/co-bound if their peak centers were located within 200 bp of each other. Read density heat maps were created by first using HOMER to generate read densities and then visualized using Java TreeView (http://jtreeview.sourceforge.net).
ATAC-seq
In brief, 50,000 mouse OPCs were washed once with cold PBS and resuspended in 50 μL lysis buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.2% (v/v) IGEPAL CA-630) to isolate nuclei. The suspension of nuclei was then centrifuged for 10 min at 500 g at 4°C, followed by the addition of 50 μL transposition reaction mix (10 μL 5 × TTBL, 5 μL TTE Mix V50 and 35 μL nuclease-free H2O) of TruePrep™ DNA Library Prep Kit V2 for Illumina’s instructions (TD501, Vazyme). Samples were then PCR amplified and incubated at 37°C for 30 min. DNA was isolated using VAHTSTM DNA Clean Beads (Vazyme #N411). ATAC-seq libraries were first subjected to 15 cycles of pre-amplification. Libraries were purified with a VAHTSTM DNA Clean Beads. Library concentration was measured using a Qubit™ dsDNA HS Assay Kit (Invitrogen, Q32854) according to the manufacturer’s instructions. Finally, the ATAC library was sequenced on an illumina Hiseq-Xten PE150 by Novogene to generate 2× 150-bp paired-end reads.
ATAC-Seq Analysis
FASTQ files of ATAC-seq data were aligned to the mouse mm10 reference using Bowtie2 with the following options: --very-sensitive (-D 20 -R 3 -N 0 -L 20 -i S,1,0.50). Low quality mapped reads were removed using samtools (view −q 35) and only unique reads mapping to a single genomic location and strand were kept. Resulting SAM files were converted to BAM format using SAM tools. Peak calling was performed using MACS2 with parameters call peak -f BAMPE -n NAME -g <int> --keep-dup all. Peak sets called in individual replicates were combined and individual peaks merged if overlapping within 300 bp to form a union peak set. (https://informatics.fas.harvard.edu/atac-seq-guidelines.html#alignments)
QUANTIFICATION AND STATISTICAL ANALYSIS
All data presented are expressed as arithmetic mean ± SEM, and the exact values of n (sample size) are provided in the Result section and figure legends. All statistical analyses were performed using GraphPad Prism 5. Null hypotheses were rejected at p values equal to or higher than 0.05. Statistical significance was determined using two-tailed unpaired Student’s t tests. Quantifications were performed from at least three experimental groups in a blinded fashion. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those generally employed in the field. No randomization was used to collect all the data; they were quantified blindly.
DATA AND SOFTWARE AVAILABILITY
The accession number for all the sequencing data reported in this paper is at NCBI GEO: GSE119816
Supplementary Material
Highlights.
Nucleoporin Seh1 is required for OPC differentiation
Seh1 is essential for CNS myelination and remyelination
Seh1 forms a complex with Olig2 at nuclear periphery
Seh1 recruits Olig2 and Brd7 to promote oligodendrocyte development
ACKNOWLEDGMENTS
We thank Liping Xie, Luming Yao, Caiming Wu, Qingfeng Liu, Changchuan Xie, Liqiong Yuan for technical support; Guang Li for Venus plasmid; Xin Chen for antibodies; William D Richardson and Huiliang Li for providing SOX10-CreERT mice, Chong Liu for providing NG2-CreERT mice, Q Richard Lu for providing Olig1-Cre mice. This study was funded by grants from the National Natural Science Foundation of China 31872642 to L.Z., 81472725 to W.M., the Fundamental Research Funds for the Central Universities 20720160072 to L.Z. and W.M., and NIH 1R01 GM113874–01 to B.M.A.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, and Sabatini DM (2013). A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Muncher L, Straussberg R, Pasmanik-Chor M, Yahav M, Rainshtein L, Walsh CA, Magal N, Taub E, Drasinover V, et al. (2006). Mutated nup62 causes autosomal recessive infantile bilateral striatal necrosis. Annals of neurology 60, 214–222. [DOI] [PubMed] [Google Scholar]
- Bercury KK, and Macklin WB (2015). Dynamics and mechanisms of CNS myelination. Developmental cell 32, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G (1985). Gene gating: a hypothesis. Proceedings of the National Academy of Sciences of the United States of America 82, 8527–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlido J, Sakuma S, Raices M, Carrette F, Tinoco R, Bradley LM, and D’Angelo MA (2018). Nuclear pore complex-mediated modulation of TCR signaling is required for naive CD4(+) T cell homeostasis. Nature immunology 19, 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, and Silver PA (2008). Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes & development 22, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. (2006). SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441, 770–773. [DOI] [PubMed] [Google Scholar]
- Chang A, Tourtellotte WW, Rudick R, and Trapp BD (2002). Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. The New England journal of medicine 346, 165–173. [DOI] [PubMed] [Google Scholar]
- Chen K, Liu J, Liu S, Xia M, Zhang X, Han D, Jiang Y, Wang C, and Cao X (2017). Methyltransferase SETD2-Mediated Methylation of STAT1 Is Critical for Interferon Antiviral Activity. Cell 170, 492–506 e414. [DOI] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, and Hetzer MW (2012). A change in nuclear pore complex composition regulates cell differentiation. Developmental cell 22, 446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Tai YC, Speed TP, Ngai J, and Barres BA (2006). Functional genomic analysis of oligodendrocyte differentiation. The Journal of neuroscience : the official journal of the Society for Neuroscience 26, 10967–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, and Barres BA (2009). Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell 138, 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria AM, Levay A, Wang Y, Kamphorst AO, Rosa ML, Nussenzveig DR, Balkan W, Chook YM, Levy DE, and Fontoura BM (2006). The nucleoporin Nup96 is required for proper expression of interferon-regulated proteins and functions. Immunity 24, 295–304. [DOI] [PubMed] [Google Scholar]
- Franklin RJ (2002). Why does remyelination fail in multiple sclerosis? Nature reviews Neuroscience 3, 705–714. [DOI] [PubMed] [Google Scholar]
- Fu H, Cai J, Clevers H, Fast E, Gray S, Greenberg R, Jain MK, Ma Q, Qiu M, Rowitch DH, et al. (2009). A genome-wide screen for spatially restricted expression patterns identifies transcription factors that regulate glial development. The Journal of neuroscience : the official journal of the Society for Neuroscience 29, 11399–11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MJ, Chishti MS, Jamal SM, Tariq M, and Ahmad W (2008). A syndromic form of autosomal recessive congenital microcephaly (Jawad syndrome) maps to chromosome 18p11.22-q11.2. Human genetics 123, 77–82. [DOI] [PubMed] [Google Scholar]
- Hayashi S, and McMahon AP (2002). Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Developmental biology 244, 305–318. [DOI] [PubMed] [Google Scholar]
- He D, Marie C, Zhao C, Kim B, Wang J, Deng Y, Clavairoly A, Frah M, Wang H, He X, et al. (2016). Chd7 cooperates with Sox10 and regulates the onset of CNS myelination and remyelination. Nature neuroscience 19, 678–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A, and Hetzer MW (2015). Nuclear pore proteins and the control of genome functions. Genes & development 29, 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser MD, Aslanian A, Dong MQ, Yates JR 3rd, and Emerson BM (2008). BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. The Journal of biological chemistry 283, 32254–32263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallabi F, Belghuith N, Aloulou H, Kammoun T, Ghorbel S, Hajji M, Gallas S, Chemli J, Chabchoub I, Azzouz H, et al. (2016). Clinical and Genetic Characterization of 26 Tunisian Patients with Allgrove Syndrome. Archives of medical research 47, 105–110. [DOI] [PubMed] [Google Scholar]
- Kodama Y, and Hu CD (2010). An improved bimolecular fluorescence complementation assay with a high signal-to-noise ratio. BioTechniques 49, 793–805. [DOI] [PubMed] [Google Scholar]
- Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, and Yoshida M (1998). Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Experimental cell research 242, 540–547. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, and Westphal H (1996). Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proceedings of the National Academy of Sciences of the United States of America 93, 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, de Faria JP, Andrew P, Nitarska J, and Richardson WD (2011). Phosphorylation regulates OLIG2 cofactor choice and the motor neuron-oligodendrocyte fate switch. Neuron 69, 918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienenklaus S, Cornitescu M, Zietara N, Lyszkiewicz M, Gekara N, Jablonska J, Edenhofer F, Rajewsky K, Bruder D, Hafner M, et al. (2009). Novel reporter mouse reveals constitutive and inflammatory expression of IFN-beta in vivo. J Immunol 183, 3229–3236. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, and Rowitch DH (2002). Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109, 75–86. [DOI] [PubMed] [Google Scholar]
- Lupu F, Alves A, Anderson K, Doye V, and Lacy E (2008). Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Developmental cell 14, 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Sommer L, Cserjesi P, and Anderson DJ (1997). Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. The Journal of neuroscience : the official journal of the Society for Neuroscience 17, 3644–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LJ, Koegl M, Bader G, Cockcroft XL, Fedorov O, Fiegen D, Gerstberger T, Hofmann MH, Hohmann AF, Kessler D, et al. (2016). Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor. Journal of medicinal chemistry 59, 4462–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie IA, Ohayon D, Li H, de Faria JP, Emery B, Tohyama K, and Richardson WD (2014). Motor skill learning requires active central myelination. Science 346, 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, and Santangelo GM (2005). Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proceedings of the National Academy of Sciences of the United States of America 102, 5749–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira TG, Zhang L, Shaulov L, Harel A, Kuss SK, Williams J, Shelton J, Somatilaka B, Seemann J, Yang J, et al. (2015). Sec13 Regulates Expression of Specific Immune Factors Involved in Inflammation In Vivo. Scientific reports 5, 17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Garcia P, and Capelson M (2014). Nuclear pores as versatile platforms for gene regulation. Current opinion in genetics & development 25, 110–117. [DOI] [PubMed] [Google Scholar]
- Pascual-Garcia P, Debo B, Aleman JR, Talamas JA, Lan Y, Nguyen NH, Won KJ, and Capelson M (2017). Metazoan Nuclear Pores Provide a Scaffold for Poised Genes and Mediate Induced Enhancer-Promoter Contacts. Molecular cell 66, 63–76 e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira GB, Dobretsova A, Hamdan H, and Wight PA (2011). Expression of myelin genes: comparative analysis of Oli-neu and N20.1 oligodendroglial cell lines. Journal of neuroscience research 89, 1070–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, and Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature biotechnology 33, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raices M, Bukata L, Sakuma S, Borlido J, Hernandez LS, Hart DO, and D’Angelo MA (2017). Nuclear Pores Regulate Muscle Development and Maintenance by Assembling a Localized Mef2C Complex. Developmental cell 41, 540–554 e547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson WD, Young KM, Tripathi RB, and McKenzie I (2011). NG2-glia as multipotent neural stem cells: fact or fantasy? Neuron 70, 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, and Richardson WD (2008). PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nature neuroscience 11, 1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, and Fontoura BM (2007). Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proceedings of the National Academy of Sciences of the United States of America 104, 1853–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu YJ, Liu H, Deng X, and Hu CD (2006). Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques 40, 61–66. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, and Wegner M (2002). Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes & development 16, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Hsu JY, Linker SB, Hu L, Schafer ST, Mertens J, Jacinto FV, Hetzer MW, and Gage FH (2017). Nup153 Interacts with Sox2 to Enable Bimodal Gene Regulation and Maintenance of Neural Progenitor Cells. Cell stem cell 21, 618–634 e617. [DOI] [PubMed] [Google Scholar]
- Trapp BD, and Nave KA (2008). Multiple sclerosis: an immune or neurodegenerative disorder? Annual review of neuroscience 31, 247–269. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, and Cox J (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, et al. (2003). The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell 113, 195–206. [DOI] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. (2011). Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, and Rout MP (2010). The nuclear pore complex and nuclear transport. Cold Spring Harbor perspectives in biology 2, a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, and Lu QR (2005). Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. The Journal of neuroscience : the official journal of the Society for Neuroscience 25, 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, et al. (2013). Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 152, 248–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Das P, Schmolke M, Manicassamy B, Wang Y, Deng X, Cai L, Tu BP, Forst CV, Roth MG, et al. (2012). Inhibition of pyrimidine synthesis reverses viral virulence factor-mediated block of mRNA nuclear export. The Journal of cell biology 196, 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, and Anderson DJ (2002). The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109, 61–73. [DOI] [PubMed] [Google Scholar]
- Zhu X, Bergles DE, and Nishiyama A (2008). NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development 135, 145–157. [DOI] [PubMed] [Google Scholar]
- Zhu X, Hill RA, Dietrich D, Komitova M, Suzuki R, and Nishiyama A (2011). Age-dependent fate and lineage restriction of single NG2 cells. Development 138, 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for all the sequencing data reported in this paper is at NCBI GEO: GSE119816