Abstract
In order to characterize unauthorized genetically modified petunia, an integrated strategy has been applied here on several suspected petunia samples from the European market. More precisely, DNA fragments of interest were produced by DNA walking anchored on key targets, earlier detected by real-time PCR screening analysis, to be subsequently sequenced using the MinION platform from Oxford Nanopore Technologies. This way, the presence of genetically modified petunia was demonstrated via the characterization of their transgene flanking regions as well as unnatural associations of elements from their transgenic cassette.
Subject terms: Molecular engineering in plants, Next-generation sequencing
Introduction
Recently, genetically modified (GM) petunia plants, imported from firms located in Germany and the Netherlands, have been found on the Finnish market by the Finnish Food Safety Authority1. Afterwards, GM petunia plants have also been detected in other EU countries. Regarding the environment as well as the food and feed chain, GM petunia plants have been assessed by experts as representing a negligible risk. However, the presence of GM petunia plants on the European (EU) market is considered as illegal because no authorization has been delivered by the EU competent authorities regarding their cultivation and commercialization. Consequently, all GM petunia plants identified on the EU market have to be discarded and destroyed. However, given that no registration of commercial names for petunia varieties is currently mandatory, the potential identification of GM petunia events based on commercial names is complicated by the fact that a single petunia variety may have several different commercial names and vice versa1–5.
To screen the potential presence of GM organisms (GMO) in the food and feed chain, the enforcement laboratories are usually using real-time PCR methods targeting transgenic elements commonly found in GMO. Subsequently, GM events are identified using the corresponding real-time PCR event-specific method6. In the context of GM petunia, the same screening strategy can be employed in targeting transgenic elements previously used in the generation of GM petunia, such as p35S (Cauliflower mosaic virus (CaMV) 35S promoter), t35S (CaMV 35S terminator), pNOS (Agrobacterium tumefaciens nopaline synthase promoter), tNOS (Agrobacterium tumefaciens nopaline synthase terminator), nptII (neomycin phosphotransferase II gene) and tOCS (Agrobacterium tumefaciens octopine synthase terminator). In addition, GM petunia plants harboring the maize A1 gene (dihydroflavonol 4-reductase), allowing to modify the flower pigmentation, have previously been reported. Therefore, a screening marker targeting this maize A1 gene can also be interesting to be used3–5,7–16. Although it is necessary for the identification as well as to trace the origin, no real-time PCR construct-specific method and event-specific method is available for GM petunia. Moreover, the development of real-time PCR construct-specific and event-specific methods requires first of all an access to their genome or at least to crucial sequences such as their transgene flanking regions and unnatural associations of elements from their transgenic cassette(s). Moreover, given that the GM petunia plants may have different origins of transformation, more than one method could be necessary to develop.
With the aim to get the sequences of interests corresponding to transgene flanking regions and unnatural associations of transgenic elements, we have applied on petunia samples an integrated strategy previously developed to detect and demonstrate the presence of unauthorized GMO in the food and feed chain17–24. This strategy consists of two main successive steps. First, the presence of GMO is detected through a real-time PCR screening targeting key transgenic elements potentially commonly found in GMO, especially in GM petunia. Second, a PCR-based DNA walking method anchored on earlier detected key transgenic elements is applied bi-directionally to obtain information about the sequences flanking the known transgenic elements via the sequencing of the resulting PCR products using a long-length Next Generation Sequencing (NGS) technology, which was in this study the MinION platform from Oxford Nanopore Technologies. In order to detect and trace GM petunia plants, the proposed integrated strategy has thus been in this study applied on several petunia plants commercialized on the EU market.
Results and Discussion
Several petunia plants commercialized on the EU market were collected in Hungary and the Czech Republic (Fig. 1). First, the petunia samples were screened for the potential presence of GM petunia by real-time PCR screening using markers targeting elements that are either commonly found in GMO (p35S, pNOS, tNOS, Cry1Ab/Ac and t35S) or only present in EU unauthorized GMO (t35S pCAMBIA). The screening marker targeting the maize A1 gene, was also integrated in the analysis given its reported presence in GM petunia plants. An additional category of screening markers specific to the Cauliflower Mosaic Virus (CaMV) (CAP and CRT2) was used to discard false positive signals coming from transgenic sequences derived from CaMV, such as p35S (Table 1)4,5,17–24. As expected, the wild-type (WT) petunia plants (samples n°19–20) were negative for all tested screening markers. All other samples (n°1–18 and n°21–23) presented a similar pattern of real-time PCR amplification. These latter presented a positive signal for the p35S, pNOS, t35S and A1 screening markers and a negative signal for the tNOS, Cry1Ab/Ac, t35S pCAMBIA, CAP and CRT2 screening markers (Table 2).
Figure 1.
List of petunia samples. For each sample, the commercial name, origin and distributor are indicated in that order below the corresponding picture.
Table 1.
Oligonucleotides used for the real-time PCR screening, DNA walking and PCR assays.
| Assays | Methods | Oligonucleotide names | Oligonucleotide sequences | References |
|---|---|---|---|---|
| Real-time PCR screening | p35S | p35S-F | AAAGCAAGTGGATTGATGTGATA | 27 |
| p35S-R | GGGTCTTGCGAAGGATAGTG | 27 | ||
| Real-time PCR screening | pNOS | pNOS-F | CGTTTTACGTTTGGAACTGACA | 28 |
| pNOS-R | CTCATTAAACTCCAGAAACCCG | 28 | ||
| Real-time PCR screening | tNOS | tNOS-F | GATTAGAGTCCCGCAATTATACATTTAA | 27 |
| tNOS-R | TTATCCTAGKTTGCGCGCTATATTT | 27 | ||
| Real-time PCR screening | t35S pCAMBIA | t35S pCAMBIA-F | CGGGGGATCTGGATTTTAGTA | 17 |
| t35S pCAMBIA-R | AGGGTTCCTATAGGGTTTCGCTC | 17 | ||
| Real-time PCR screening | Cry1Ab/Ac | Cry1Ab/Ac-F | ACCGGTTACACTCCCATCGA | 29 |
| Cry1Ab/Ac-R | CAGCACCTGGCACGAACTC | 29 | ||
| Real-time PCR screening | t35S | t35S-F | In-house | Unpublished data |
| t35S-R | In-house | Unpublished data | ||
| Real-time PCR screening | CAP_CaMV | CAP-F | In-house | Unpublished data |
| CAP-R | In-house | Unpublished data | ||
| Real-time PCR screening | CRT2_CaMV | CRT2-F | CCAGAAGAACATTGGGTCAATGC | 30 |
| CRT2-R | CTATGTCTTTGCAGACTTTGCTGAT | 30 | ||
| Real-time PCR screening | A1 | qPhA1_F4 | CGACTTCTGCCGTCGCG | 5 |
| qPhA1_R4 | GATGATGGTGACCAGGTCCAG | 5 | ||
| DNA Walking | p35S-F | p35S-F a | GGGTCTTGCGAAGGATAGTG | 27 |
| p35S-F b | TGTGCGTCATCCCTTACGTCAGT | 19 | ||
| p35S-F c | TATCACATCAATCCACTTGCTTT | 27 | ||
| DNA Walking | p35S-R | p35S-R a | AAAGCAAGTGGATTGATGTGATA | 27 |
| p35S-R b | ACTGACGTAAGGGATGACGCACA | 19 | ||
| p35S-R c | CACTATCCTTCGCAAGACCC | 27 | ||
| PCR confirmation assay | GM petunia transgene flanking region | Junction-F | CATTTCGCCCTCATGAAAATGAT | This study |
| Junction-R | GGTCGCCGCATACACTATTC | This study |
Table 2.
Results of the real-time PCR screening analysis, using the p35S, pNOS, tNOS, t35S pCAMBIA, Cry1Ab/Ac, t35S, A1, CAP and CRT markers, and the PCR confirmation assay, targeting the GM petunia transgene flanking region (junction), applied on the petunia samples.
| Samples | Real-time PCR screening analysis | PCR confirmation assay | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| p35S | pNOS | tNOS | t35S pCAMBIA | Cry1Ab/Ac | t35S | CAP | CRT2 | A1 | Junction | |
| 1_GM | + | + | − | − | − | + | − | − | + | + |
| 2_GM | + | + | − | − | − | + | − | − | + | + |
| 3_GM | + | + | − | − | − | + | − | − | + | + |
| 4_GM | + | + | − | − | − | + | − | − | + | + |
| 5_GM | + | + | − | − | − | + | − | − | + | + |
| 6_GM | + | + | − | − | − | + | − | − | + | + |
| 7_GM | + | + | − | − | − | + | − | − | + | + |
| 8_GM | + | + | − | − | − | + | − | − | + | + |
| 9_GM | + | + | − | − | − | + | − | − | + | + |
| 10_GM | + | + | − | − | − | + | − | − | + | + |
| 11_GM | + | + | − | − | − | + | − | − | + | + |
| 12_GM | + | + | − | − | − | + | − | − | + | + |
| 13_GM | + | + | − | − | − | + | − | − | + | + |
| 14_GM | + | + | − | − | − | + | − | − | + | + |
| 15_GM | + | + | − | − | − | + | − | − | + | + |
| 16_GM | + | + | − | − | − | + | − | − | + | + |
| 17_GM | + | + | − | − | − | + | − | − | + | + |
| 18_GM | + | + | − | − | − | + | − | − | + | + |
| 21_GM | + | + | − | − | − | + | − | − | + | + |
| 22_GM | + | + | − | − | − | + | − | − | + | + |
| 23_GM | + | + | − | − | − | + | − | − | + | + |
| 19_WT | − | − | − | − | − | − | − | − | − | − |
| 20_WT | − | − | − | − | − | − | − | − | − | − |
Positive and negative signals are respectively indicated by + and −. The samples are indicated as genetically modified (GM) or wild-type (WT).
In a second step, the samples suspected to be GM petunia (n°1–18 and n°21–23) were analyzed with the available bidirectional DNA walking method anchored on the p35S transgenic element that presented a positive signal during the previous real-time PCR screening step19,21,24. All the generated PCR products presented a size range going approximately from 100 to 1500 bp. The obtained PCR products of each petunia sample were pooled and barcoded to be then sequenced using the MinION platform from Oxford Nanopore Technologies (Supplementary File 1).
Among the generated DNA fragments from each tested petunia sample, the ones related to the DNA walking p35S-R method were all characterized by a sequence successively corresponding to the p35S element [GenBank: MF521566.1; AY615305.1] followed by an uncharacterized vector sequence [GenBank: MF521566.1] and the maize A1 gene [GenBank: MF521566.1; NM_001158995.2]. This association of sequences was previously mentioned as being used to produce GM petunia presenting a novel flower pigmentation7. Regarding the sequences of the DNA fragments produced with the DNA walking p35S-F method, the p35S element [GenBank: KY964325.1] was followed by an uncharacterized vector sequence [GenBank: KY964325.1] and the beta-lactamase (bla) gene [GenBank: KY964325.1; KT405500.1], conferring a resistance to ampicillin (Fig. 2, Supplementary Files 2–4). The identification of these unnatural associations of elements allows confirming that these petunia plants were actually GM. These results suggest also that there is the possibility that the same transgenic cassette may have been used in all the GM petunia tested in this study. Moreover, these sequences could be useful to develop a construct-specific method. For most of the petunia samples (n°1–3, n°5, n°7–9, n°11–14, n°16–18 and n°21–23), longest sequences were generated by the DNA walking p35S-F method, allowing to reach the transgene flanking region between the bla element from the transgenic cassette [GenBank: KY964325.1; KT405500.1] and the petunia host genome [GenBank: AY136628.2; CP023763.1; HG975447.1] (Fig. 2, Supplementary Files 3, 5).
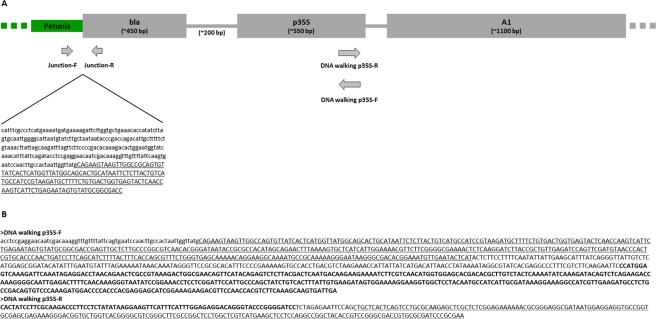
Figure 2.
Schematic representation of the identified transgenic cassette of the tested GM petunia plants, based on the sequence information generated in this study. (A) The petunia part is represented in green while the transgenic cassette is indicated in grey. The approximate size of the transgenic elements is indicated in base-pairs (bp). The beta-lactamase gene (bla); the Cauliflower Mosaic Virus (CaMV) 35S promoter (p35S); the maize A1 gene (A1). The starting positions and walking directions of the applied DNA walking methods anchored on the p35S element are indicated by arrows (Table 1). The positions of primers (Junction-F and Junction-R) used to confirm the observed transgene flanking region with the MinION platform are indicated by arrows (Table 1). The consensus sequences of the transgene flanking region confirmed by PCR amplification and Sanger sequencing for the petunia samples n°1–18, 21–23 (see Supplementary File 5) is indicated. (B) The consensus sequences produced by the MinION platform with the DNA walking p35S-F (see Supplementary Files 3–4) and the DNA walking p35S-R (see Supplementary File 2) methods applied on all GM petunia samples are indicated. In all these sequences, the petunia part is indicated in lower case while the transgenic cassette, containing bla (underlined), uncharacterized vector part, p35S (in bold) and the maize A1 gene (dotted underlined), is indicated in upper case.
As the identified transgene flanking region was highly similar between all these samples (n°1–3, n°5, n°7–9, n°11–14, n°16–18 and n°21–23), the same origin of transformation was suspected (Supplementary File 6). The confirmation of this transgene flanking region was obtained by PCR amplifications followed by Sanger sequencing using two primers, one designed on the petunia sequence and one designed on the bla gene (Fig. 2, Table 1, Supplementary File 6). Due to the sequence similarity between the p35S and bla elements for all tested GM petunia samples (Supplementary File 4), this analysis was also applied on GM petunia samples presenting shorter sequences characterizing only the transgenic cassette (n°4, n°6, n°10 and n°15) (Fig. 2, Supplementary File 3). All these GM petunia samples presented a PCR amplicon with the expected size (402 bp) and with the same transgene flanking region (Table 2, Fig. 2, Supplementary File 5). The identical insertion site of these samples suggests a similar origin of GM event transformation.
In order to collect additional information related to the petunia sequence interrupted by the inserted transgenic cassette, further analysis were performed. This petunia sequence presented a similarity (77%) with a transposon (GenBank: RVW93168.1) (Fig. 2; Supplementary File 7). The insertion of the transgenic cassette expressing the maize A1 gene on a transposable element, well-known to be able to influence gene expression, could explain the flower color variations observed among the tested GM petunia plants25. However, further analysis are needed to confirm this hypothesis.
Regarding the transgenic cassette of all tested GM petunia samples, the GM petunia described in Meyer et al., 1987 represents a possible origin7. This hypothesis is based on the similarity of the unnatural association of elements (bla, p35S and A1 gene) (Fig. 2, Supplementary Files 2–4) as well as to the presence and absence of transgenic elements determined using the real-time PCR screening (positive for p35S, pNOS and t35S and negative for tNOS) (Table 2). However, the analysis of the GM petunia material published by Meyer et al., 1987 and its comparison to the GM petunia plants tested in this study are needed to confirm this hypothesis7.
At the technical level, the used MinION platform from the Oxford Nanopore technologies presents the advantage to be able to deal with heterogenic DNA libraries, to provide data in real-time of long read-lengths and to be easily available in-house due to the small size and portability of the device and the attractive price. Moreover, this high-throughput technology allows to sequence several samples simultaneously. In contrast to the Sanger sequencing platform, the use of a NGS platform, such as the MinION device, for the sequencing of DNA libraries generated through the proposed PCR-based DNA walking methods allows also to reduce the laborious aspect of the experimental procedure. More precisely, as each DNA walking PCR product presents in general several amplicons, an individual purification of each of these amplicons from an electrophoresis gel is required for their subsequent sequencing using the Sanger technology while the entire PCR product can directly be sequenced using the MinION device. However, according to previous studies, this NGS technology presents a relatively high error rate of generated raw data (~10–20%)21,24,26. In the present study, despite the high similarity between the sequences of the transgene flanking region generated by the MinION and Sanger platforms, the error rate was varying approximately between 1 to 22% (Supplementary File 6). Nevertheless, in contrast to a single mutation point, the detection of transgenic cassettes and transgene flanking regions from GMO as well as the determination of the transformation origin were still possible with such a kind of error rate. These generated sequences could also be used to develop real-time PCR construct-specific and event-specific methods in order to be used in GMO routine analysis by the enforcement laboratories for the survey of GM petunia plants on the market. For such purpose, the use of sequences confirmed by the Sanger sequencing technology, presenting a lower error rate than the MinION platform, is strongly advised for the oligonucleotide design of these real-time PCR construct-specific and event-specific methods24,26.
Conclusion
The proposed integrated strategy, combining a PCR-based DNA walking, anchored on earlier detected key transgenic elements, with the MinION sequencing platform from Oxford Nanopore Technologies, was successfully applied on several suspected GM petunia samples circulating on the EU market. Their presence was demonstrated by the characterization of their transgene flanking regions as well as unnatural associations of elements from their transgenic cassette. Moreover, the high similarity between the transgene flanking region from all tested GM petunia samples strongly suggests their common origin of transformation although these GM petunia samples had different commercial names. Given that the tested GM petunia plants were also presenting some flower color variations among themselves (Fig. 1), any petunia plants could be relevant for such investigation5. In addition, with the aim to increase the characterization of sequences of interest, additional DNA walking methods anchored on other transgenic elements observed in GM petunia, such as bar, pNOS and t35S, could be useful to develop.
Regarding the choice of the NGS platform, the MinION device allowed in-house generation of a long-read sequencing data for several samples simultaneously in a short-time frame (maximum 48 hours), which is highly interesting because such time-frame is compatible with the workflow of enforcement laboratories. However, the bioinformatics analysis still required a certain level of expertise necessary to be developed in enforcement laboratories. Moreover, given its relative high-error rate, the re-sequencing of fragments of interest using for instance the Sanger technology is advised for further applications, such as the development of real-time PCR construct-specific and event-specific methods.
Materials and Methods
Plant materials, DNA extraction, DNA concentration and DNA purity
Twenty-three petunia samples were collected from the Hungarian (samples n°1–20) and Czech Republic (samples n°21–23) markets (Fig. 1). Based on the real-time PCR assay (Table 2), the petunia plants corresponding to the samples n°1–18 and n°21–23 were suspected to be GM while the petunia plants corresponding to the samples n°19 and n°20 were wild-type (WT). The commercial names of the GM petunia samples were “Petunia hybrida Potunia Plus Papaya”, “Petunia Potunia Plus Papaya”, “Petunia Bingo Orange”, “Petunia Hells Fruit Punch”, “Petunia hybrida MyLove Orange”, “Petunia hybrida Go!Tunia Orange”, “Petunia hybrida Viva Orange”, “Petunia Orange Coloured flower”, “Petunia Top-Tunia Salmon Orange”, “Petunia Orange coloured flower (with dark vein in the middle)”, “Petunia atkinsiana Pegasus Orange”, “Petunia atkinsiana Pegasus Orange Morn”, “Petunia Surfinia (Harmónia trio mix)”, “Petunia Yellow and orange coloured flower”, “Petunia Papaya”, “Petunia Pegasus Table Orange” and “Solomon” (Fig. 1). These samples were sampled by the Hungarian and Czech Republic competent authorities in the GMO control context. Leaves from each sample were grounded individually to obtain a fine homogenous powder. The subsequent DNA extraction step was performed using the DNA Wizard Clean up system for genomic DNA kit (Promega) for the samples n°1–20 and a CTAB-based procedure (ISO 21571) for the samples n°21–23 (International Standard ISO 21571, 2005). DNA concentration was measured by spectrophotometry using the Nanodrop® 2000 (ThermoFisher) and DNA purity was evaluated using the A260/A280 and A260/A230 ratios.
Real-time PCR assay
For the real-time PCR screening, 25 ng of DNA from each sample were tested using the p35S, pNOS, tNOS, t35S pCAMBIA, Cry1Ab/Ac, t35S, A1, CAP and CRT2 markers (Table 1). The real-time PCR mix contained 1X SYBR®Green PCR Mastermix (Diagenode) and 250 nM of each primer in a total volume of 25 µl. The real-time PCR program consisted of initial DNA polymerase activation for 10 min at 95 °C followed by 40 amplification cycles of 15 sec at 95 °C (denaturing step) and 1 min at 60 °C, for the p35S, pNOS, tNOS, t35S pCAMBIA, Cry1Ab/Ac, t35S, CAP and CRT2 markers, or 65 °C, for the A1 marker, (annealing-extension step). Melting curve analyses were performed by gradually increasing the temperature from 60 to 95 °C in 20 min (±0.6°/20 sec). All runs were performed on an ABI 7300 real-time PCR system (Life Technologies). For each assay, a “No Template Control” (NTC) was included.
DNA walking
To isolate unknown sequences flanking the known sequences, a PCR-based DNA walking was performed as previously described17–19,21,23,24. Briefly, a target-specific primer (a) and one kind of the degenerated random tagging primer (DRT) mixes (A-D) were first used to amplify the sequences of interest. In a second and third semi-nested PCR, target-specific primers (b and c) were sequentially combined with universal tagging primers, UAP-N1 and UAP-N2 respectively, in order to increase the yield of the sequences of interest and to decrease the background. For each petunia sample (n°1–18 and n°21–23) presenting earlier a positive signal in real-time PCR for the p35S marker, 100 ng of DNA were tested using the bidirectional DNA walking method anchored on p35S. The final PCR products were visualized by electrophoresis using the Tapestation 4200 device with the associated High Sensitivity D5000 Screen Tape and reagents (Agilent) (Fig. 2, Supplementary File 1). The resulting PCR products were sequenced using the MinION platform from Oxford Nanopore technologies.
MinION sequencing and data analysis of PCR products generated by DNA walking
For each GM petunia sample (n°1–18 and n°21–23), all PCR products generated by the bidirectional DNA walking method anchored on p35S were pooled to be purified using the AGENCOURT® AMPURE® XP Kit (Beckman Coulter Life Sciences) according to the manufacturer’s instructions. The concentration of each purified pool was measured using the Qubit dsDNA HS Assay Kits by the Qubit 3.0 Fluorometer (Life Technologies) according to the manufacturer’s instructions. For each purified pool, a sequencing library was prepared from 1 µg of DNA using the 1D amplicon by ligation sequencing kit (SQK-LSK108; Oxford Nanopore Technologies, Oxford, UK) and the 1D Native barcoding kit (EXP-NBD103; Oxford Nanopore Technologies, Oxford, UK) according to the manufacturer’s instructions. More precisely, each purified pool was initially end-repaired and dA-tailed to be subsequently ligated individually to a unique barcode. All these barcoded samples were then pooled to be ligated to adapters in order to constitute the sequencing library. As only 12 different barcodes were available, two sequencing libraries were prepared, one for the samples n°1–11 and one for the samples n°12–18 and n°21–23 (Supplementary File 8). Finally, each sequencing library was loaded into an individual MinION flow cell of the MinION sequencing device (Oxford Nanopore Technologies, Oxford, UK) to be sequenced for 48 hours. The basecalling of the generated raw data (fast5 format) was performed using Albacore (version 2.0.1). The demultiplexing of the different petunia samples, individually barcoded with a unique barcode during the library preparation, was automatically carried out during this step. The individual fastq files were extracted from the base-called fastq5 files with poretools (version 0.6.0). The raw data are available on NCBI (SRA file accession number: PRJNA523486).
The generated sequences were analyzed via a rapid workflow using CLC Genomics Workbench (Qiagen). The sequences were mapped to available petunia genomes [https://solgenomics.net/organism/Petunia_inflata/genome; https://solgenomics.net/organism/Petunia_axillaris/] at default parameters to find the transgene flanking regions. The sequences presenting a partial similarity to petunia genomes were blasted to the NCBI database to characterize the part corresponding to the transgenic cassettes. In addition, the sequence presenting no correspondence to the petunia genomes were blasted to the NCBI database, allowing to characterize the transgenic cassettes. Additional sequence analyses (alignment and creation of consensus sequences) were performed using CLC Genomics Workbench (Qiagen) (Fig. 2, Supplementary Files 2–7).
Verification of transgene flanking regions by PCR amplification and Sanger sequencing
The identified transgene flanking region from the GM petunia samples n°1–3, n°5, n°7–9, n°11–14, n°16–18 and n°21–23 observed with the MinION sequencing platform were verified by PCR. For this PCR confirmation assay, a forward primer (Junction-F) and a reverse primer (Junction-R) were designed using the software Primer3 to anneal respectively to the sequences from the petunia host and the transgenic cassette in order to amplify by PCR a DNA fragment of 402 bp (Fig. 2, Table 1, Supplementary File 6). In addition to these petunia samples, this PCR confirmation assay was applied on the petunia samples n°4, n°6, n°10, n°15 and n°19–20. An NTC also was included.
For all PCR assays, a standard 25 µl reaction volume was applied containing 1X Green DreamTaq PCR Master Mix (ThermoFisher Scientific), 250 nM of each primer (Eurogentec) and 5 µl of DNA (5 ng/µl). The PCR program consisted of a single cycle of 1 min at 95 °C (initial denaturation) followed by 35 amplification cycles of 30 sec at 95 °C (denaturation), 30 sec at 50 °C (annealing) and 1 min at 72 °C (extension) and finishing by a single cycle of 5 min at 72 °C (final extension). The run was performed on a Swift MaxPro Thermal Cycler (Esco). The PCR products were visualized by electrophoresis using the Tapestation 4200 device with the associated High Sensitivity D1000 Screen Tape and reagents (Agilent) (Supplementary File 5). The sequencing of the PCR products, earlier purified using USB ExoSAP-IT PCR Product Cleanup (Affymetrix) according to the manufacturer instructions, was performed on a Genetic Sequencer 3130XL using the Big Dye Terminator Kit v3.1 (Applied Biosystems). The resulting sequences were analyzed using CLC Genomics Workbench (Qiagen) (Fig. 2, Supplementary Files 5,6).
The sequences of the transgene flanking regions observed with the MinION (Fig. 2, Supplementary File 3) and Sanger platforms (Fig. 2, Supplementary File 5) were compared by alignment using CLC Genomics Workbench (Qiagen) and Nucleotide BLAST NCBI (Supplementary File 6).
Supplementary information
Acknowledgements
The authors would like to thank Stefan Hoffman for his technical assistance as well as Raf Winand and Kevin Vanneste for their Bioinformatics support. The Sanger sequencing was performed at the Transversal activities in Applied Genomics Service at Sciensano.
Author Contributions
M.A.F. performed the experiment, analyzed data and drafted the manuscript. D.V.G. participated to the real-time PCR screening analysis. G.U. and J.O. provided the petunia samples and the associated pictures indicated in Fig. 1. G.U., J.O., S.D.K., A.S., N.P. and N.H.C.R. helped to design the study, participated in the interpretation of the results and drafted the manuscript. All authors have read and approved the final manuscript.
Data Availability
All data generated or analyzed during this study are included in this published article [and its Supplementary Information Files] or is available from the corresponding author.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-43463-5.
References
- 1.EVIRA Current issues: Evira removes genetically modified orange petunias from sale, https://www.evira.fi/en/plants/current-issues/2017/evira-removes-genetically-modified-orange-petunias-from-sale/ (2017).
- 2.COGEM Cogem advice CGM/170522-04: Unauthorised GM garden petunia varieties with orange flowers, https://www.cogem.net/index.cfm/en/publications/publication/unauthorised-gm-garden-petunia-varieties-with-orange-flowers (2017).
- 3.Bashandy H, Teeri TH. Genetically engineered orange petunias on the market. Planta. 2017;246:277–280. doi: 10.1007/s00425-017-2722-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Working Group of the Regulatory Committee for Directive 2001/18/EC. Working Group of the Regulatory Committee for Directive 2001/18/EC on 26 january 2018 in Brussels (2018).
- 5.Haselmair-Gosch C, et al. Great Cause—Small Effect: Undeclared Genetically Engineered Orange Petunias Harbor an Inefficient Dihydroflavonol 4-Reductase. Front. Plant Sci. 2018;9:149. doi: 10.3389/fpls.2018.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broeders, S., Papazova, N., Van den Bulcke, M. & Roosens, N. Development of a molecular platform for GMO detection in food and feed on the basis of “combinatory qPCR” technology. In: Hernandez-Rodriguez, P. & Gome, A. P. R. (Eds), Polymerase Chain Reaction, vol. 1 (pp. 363–404). Rijeka, Croatia (2012).
- 7.Meyer P, Heidman I, Forkmann G, Saedler H. A new petunia flower colour generated by transformation of a mutant with a maize gene. Nature. 1987;330:662678. doi: 10.1038/330677a0. [DOI] [PubMed] [Google Scholar]
- 8.Van der Krol AR, et al. An antisense chalcone synthase gene in transgenic plants inhibits flower pigmentation. Nature. 1988;333:866–869. doi: 10.1038/333866a0. [DOI] [Google Scholar]
- 9.Van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2:291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linn F, Heidmann I, Saedler H, Meyer P. Epigenetic changes in the expression of the maize A1 gene in Petunia hybrida: role of numbers of integrated gene copies and state of methylation. Mol Gen Genet. 1990;222(2-3):329–336. doi: 10.1007/BF00633837. [DOI] [PubMed] [Google Scholar]
- 11.Napoli C, Lemieux C, Jorgensen R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. The Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brugliera F, et al. Isolation and characterization of a cDNA clone corresponding to the Rt locus of Petunia hybrida. Plant J. 1994;5(1):81–92. doi: 10.1046/j.1365-313X.1994.5010081.x. [DOI] [PubMed] [Google Scholar]
- 13.Elomaa P, et al. Transgene inactivation in Petunia hybrida is influenced by the properties of the foreign gene. Mol Gen Genet. 1995;248(6):649–656. doi: 10.1007/BF02191704. [DOI] [PubMed] [Google Scholar]
- 14.Shimada Y, et al. Expression of chimeric P450 genes encoding flavonoid-3′, 5′-hydroxylase in transgenic tobacco and petunia plants. FEBS Lett. 1999;461(3):241–245. doi: 10.1016/S0014-5793(99)01425-8. [DOI] [PubMed] [Google Scholar]
- 15.Tsuda S, et al. Flower color modification of Petunia hybrida commercial varieties by metabolic engineering. Plant. Biotechnology. 2004;21(5):377–386. [Google Scholar]
- 16.Gargul JM, Mibus H, Serek M. Manipulation of MKS1 gene expression affects Kalanchoë blossfeldiana and Petunia hybrida phenotypes. Plant Biotechnol J. 2015;13:51–61. doi: 10.1111/pbi.12234. [DOI] [PubMed] [Google Scholar]
- 17.Fraiture MA, et al. An innovative and integrated approach based on DNA walking to identify unauthorized GMOs. Food Chemistry. 2014;147:60–69. doi: 10.1016/j.foodchem.2013.09.112. [DOI] [PubMed] [Google Scholar]
- 18.Fraiture MA, et al. Validation of a sensitive DNA walking strategy to characterise unauthorized GMOs using model food matrices mimicking common rice products. Food Chemistry. 2015;173:1259–1265. doi: 10.1016/j.foodchem.2014.09.148. [DOI] [PubMed] [Google Scholar]
- 19.Fraiture MA, et al. Integrated DNA walking system to characterize a broad spectrum of GMOs in food/feedmatrices. BMC Biotechnology. 2015;15:76. doi: 10.1186/s12896-015-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraiture MA, et al. Biotech Rice: Current developments and future detection challenges in food and feed chain. Trends in Food and Technology. 2016;52:66–79. doi: 10.1016/j.tifs.2016.03.011. [DOI] [Google Scholar]
- 21.Fraiture MA, et al. An integrated strategy combining DNA walking and NGS to detect GMO. Food Chemistry. 2017;232:351–358. doi: 10.1016/j.foodchem.2017.03.067. [DOI] [PubMed] [Google Scholar]
- 22.Fraiture MA, Herman P, De Loose M, Debode F, Roosens NH. How can we better detect unauthorized GMO in the food and feed chain. Trends in Biotechnology. 2017;35:508–517. doi: 10.1016/j.tibtech.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Fraiture MA, Vandamme J, Herman P, Roosens NH. Development and validation of an integrated DNA walking strategy to detect GMO expressing Cry genes. BMC Biotechnology. 2018;18:40. doi: 10.1186/s12896-018-0446-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraiture MA, et al. Nanopore sequencing technology: a new route for the fast detection of unauthorized GMO. Scientific reports. 2018;8:7903. doi: 10.1038/s41598-018-26259-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz-Sommer Z, et al. Influence of transposable elements on the structure and function of the A1 gene of Zea mays. EMBO J. 1987;6:287–294. doi: 10.1002/j.1460-2075.1987.tb04752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weirather JL, et al. Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Research. 2017;6:100. doi: 10.12688/f1000research.10571.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbau-Piednoir E, et al. SYBR®Green qPCR screening methods for the presence of 35S promoter and NOS terminator elements in food and feed products. European Food Research and Technology. 2010;230:383–93. doi: 10.1007/s00217-009-1170-5. [DOI] [Google Scholar]
- 28.Broeders S, et al. New SYBR®Green methods targeting promoter sequences used for screening of several GM events pending for authorisation in Europe. European Food Research and Technology. 2013;236(3):537–547. doi: 10.1007/s00217-013-1910-4. [DOI] [Google Scholar]
- 29.Barbau-Piednoir E, et al. Four new SYBR Green qPCR screening methods for the detection of Roundup Ready, LibertyLink, and CryIAb traits in genetically modified products. European Food Research and Technology. 2012;234:13–23. doi: 10.1007/s00217-011-1605-7. [DOI] [Google Scholar]
- 30.Barbau-Piednoir E, et al. Inter-laboratory Testing of GMO Detection by Combinatory SYBR®Green PCR Screening (CoSYPS) Food Analytical Methods. 2014;7(8):1719–1728. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its Supplementary Information Files] or is available from the corresponding author.