P. putida lysine metabolism can produce multiple commodity chemicals, conferring great biotechnological value. Despite much research, the connection of lysine catabolism to central metabolism in P. putida remained undefined. Here, we used random barcode transposon sequencing to fill the gaps of lysine metabolism in P. putida. We describe a route of 2-oxoadipate (2OA) catabolism, which utilizes DUF1338-containing protein P. putida 5260 (PP_5260) in bacteria. Despite its prevalence in many domains of life, DUF1338-containing proteins have had no known biochemical function. We demonstrate that PP_5260 is a metalloenzyme which catalyzes an unusual route of decarboxylation of 2OA to d-2-hydroxyglutarate (d-2HG). Our screen also identified a recently described novel glutarate metabolic pathway. We validate previous results and expand the understanding of glutarate hydroxylase CsiD by showing that can it use either 2OA or 2KG as a cosubstrate. Our work demonstrated that biological novelty can be rapidly identified using unbiased experimental genetics and that RB-TnSeq can be used to rapidly validate previous results.
KEYWORDS: biochemistry, biotechnology, genomics, metabolism, transposons
ABSTRACT
Despite intensive study for 50 years, the biochemical and genetic links between lysine metabolism and central metabolism in Pseudomonas putida remain unresolved. To establish these biochemical links, we leveraged random barcode transposon sequencing (RB-TnSeq), a genome-wide assay measuring the fitness of thousands of genes in parallel, to identify multiple novel enzymes in both l- and d-lysine metabolism. We first describe three pathway enzymes that catabolize l-2-aminoadipate (l-2AA) to 2-ketoglutarate (2KG), connecting d-lysine to the TCA cycle. One of these enzymes, P. putida 5260 (PP_5260), contains a DUF1338 domain, representing a family with no previously described biological function. Our work also identified the recently described coenzyme A (CoA)-independent route of l-lysine degradation that results in metabolization to succinate. We expanded on previous findings by demonstrating that glutarate hydroxylase CsiD is promiscuous in its 2-oxoacid selectivity. Proteomics of selected pathway enzymes revealed that expression of catabolic genes is highly sensitive to the presence of particular pathway metabolites, implying intensive local and global regulation. This work demonstrated the utility of RB-TnSeq for discovering novel metabolic pathways in even well-studied bacteria, as well as its utility a powerful tool for validating previous research.
INTRODUCTION
Pseudomonas putida is a ubiquitous saprophytic soil bacterium and is a model organism for bioremediation (1). Interest in utilizing P. putida KT2440 as a chassis organism for metabolic engineering has recently surged due to the existence of well-established genetic tools and to its robust metabolism of aromatic compounds that resemble lignin hydrolysis products (2–4). As lignin valorization remains essential for the economic feasibility of cellulosic bioproducts, a nuanced and predictable understanding of P. putida metabolism is highly desirable (5).
Although its aromatic metabolism has garnered much attention, the lysine metabolism of P. putida has also been rigorously studied for over 50 years (6). An understanding of lysine metabolism has had biotechnological value, as it has been used to produce glutarate and 5-aminovalerate (5AVA) as well as valerolactam in P. putida and in the other bacteria (7–10). However, our current understanding of lysine catabolism remains incomplete. In particular, the connection between d-lysine metabolism and central metabolism in P. putida is unclear and has not been fully characterized.
P. putida employs bifurcating pathways to catabolize lysine, separately metabolizing the l-isomers and d-isomers (see Fig. S1a in the supplemental material) (11). The l-lysine degradation pathway proceeds to glutarate, which can then be degraded either to acetyl-coenzyme A (acetyl-CoA) via a glutaryl-CoA intermediate or to succinate without a CoA-bound intermediate (Fig. S1a) (9). Characterization of the final steps of d-lysine catabolism remains more elusive. The initial steps of d-lysine catabolism are well described, but the genetic basis stops at 2-aminoadipate (2AA) (12). Furthermore, 13C labeling experiments performed by Revelles et al. demonstrated a putative metabolic connection between the d- and l-lysine pathways at 2AA (11). The subsequent steps to central carbon metabolism have never been fully validated. (6, 11–13). Given the importance of lysine metabolism and the recent availability of high-throughput genetic tools, we sought to identify the steps in d-lysine metabolism that have remained missing despite 50 years of research.
Metabolic pathways of lysine catabolism in P. putida KT2440. (A) The l-lysine metabolic pathway is shown in blue, while the known steps of d-lysine metabolism are shown in red. (B) Proposed route of 2AA metabolism in P. putida. Download FIG S1, TIF file, 3.9 MB (4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
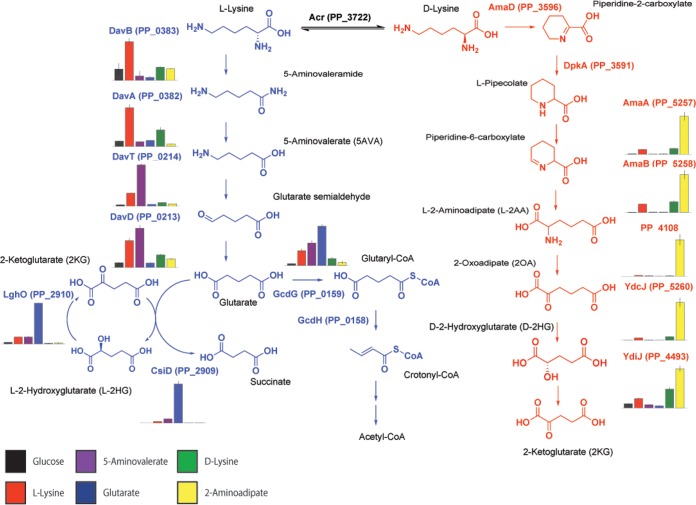
Random barcode transposon sequencing (RB-TnSeq) is a genome-wide approach that measures the importance of each gene to growth (or fitness) in a massively parallel assay (14). RB-TnSeq can identify phenotypes for thousands of previously uncharacterized genes (14, 15), including the levulinic acid degradation pathway in P. putida KT2440 (16). In this study, we applied RB-TnSeq to uncover multiple novel genes implicated in l- and d-lysine metabolism in P. putida. We first describe a three-enzyme route connecting l-2AA to 2-ketoglutarate (2KG) (Fig. S1B). Within this pathway, d-lysine metabolism connects to central metabolism through a 2-hydroxyglutarate (2HG) intermediate, which is directly produced from 2-oxoadipate (2OA) in a reaction catalyzed by a DUF1338-containing protein. The function of this protein family, widely distributed across many domains of life, was previously unknown. Subsequently, we further characterize the glutarate hydroxylase CsiD by demonstrating its 2-oxoacid promiscuity during the hydroxylation of glutarate. Finally, we show that the expression levels of all of the newly discovered enzymes change significantly in response to specific metabolites within the two catabolic pathways.
RESULTS
Identification of lysine catabolism genes via RB-TnSeq.
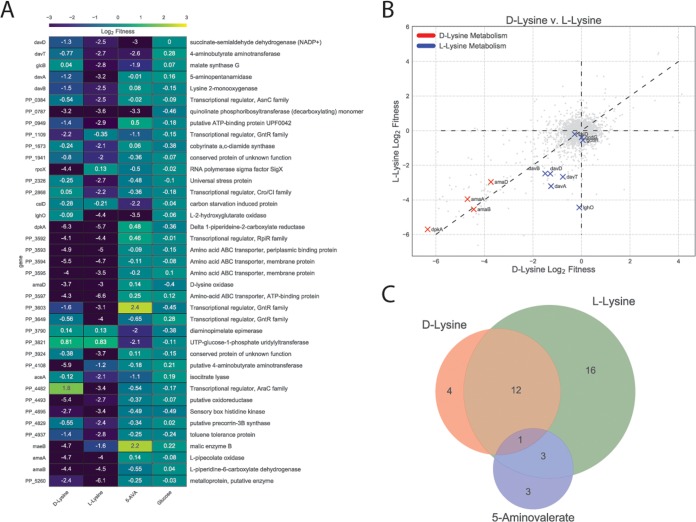
To identify mutants defective in lysine catabolism in P. putida KT2440, an RB-TnSeq library representing this bacterium (16) was grown on minimal medium supplemented with either d-lysine or l-lysine as the sole carbon source. To evaluate whether d-lysine metabolism was required for the metabolism of other downstream metabolites of l-lysine, the library was also grown on 5AVA. As a control, we also grew the library on glucose. Fitness was calculated as the log2 ratio of the strain and gene abundances at the end of selective growth to the initial abundance (14). Fitness profiling revealed 39 genes with significant fitness values below −2 for 5AVA, d-lysine, or l-lysine and no lower than −0.5 for glucose (Fig. 1A; see also Table S1 in the supplemental material). Within this set, 10 of the 12 known lysine degradation genes were identified, with the exceptions corresponding to the genes encoding two enzymes in the CoA-dependent route of glutarate degradation (i.e., genes gcdH and gcdG), which both had significant fitness values (t below −4) but with fitness values greater than −2. Instead, we identified the recently characterized genes involved in the CoA-independent pathway (namely, csiD and lghO) (9).
FIG 1.
Results of RB-TnSeq screen. (A) Genes that showed less than −2 log2 fitness on d-lysine, l-lysine, or 5AVA but showed a fitness defect of no less than −0.5 log2 when grown on glucose. (B) Plot of genome-wide fitness values of libraries grown on either l-lysine or d-lysine. Genes encoding enzymes known to be involved in d-lysine metabolism are shown in red, while those known to be involved in l-lysine metabolism are shown in blue. (C) Venn diagram of genes with significant fitness defects when grown on d-lysine, l-lysine, or 5AVA.
Quantitative results of RB-TnSeq screen. Download Table S1, CSV file, 0.00 MB (4KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The fitness data corroborated previous work showing that a functional d-lysine pathway is required for l-lysine catabolism (6, 11). None of the known l-lysine catabolic genes showed fitness defects for growth on d-lysine, but transposon insertions in all previously identified d-lysine genes showed negative fitness scores under conditions of growth on l-lysine (Fig. 1B). No known d-lysine catabolic enzymes showed fitness defects when grown on 5AVA, suggesting that the d-lysine dependence of l-lysine catabolism may occur only for early catabolic steps (Fig. 1C).
In addition to catabolic enzymes, lysine transporters and multiple transcriptional regulators were identified (Fig. 1A). The putative lysine amino acid ABC transporter system (P. putida 3593 [PP_3593], PP_3394, and PP_3395) showed significant fitness defects under conditions of growth with either isomer of lysine. Some of the transcriptional regulators were located near known catabolic or transport enzymes (PP_0384, PP_3592, and PP_3603), while others were not clustered with any obviously related genes (PP_1109, PP_2868, PP_3649, and PP_4482). Two known global regulators were identified in our screen: cbrA (PP_4695), encoding a histidine kinase sensor that showed fitness defects on both lysine isomers, and the alternative sigma factor rpoX (PP_2088) which had fitness defects only when grown on d-lysine.
Additionally, there were 15 genes which, when disrupted, displayed fitness advantages (values) greater than 2 on 5AVA, d-lysine, or l-lysine and a fitness value of less than 0.5 when grown on glucose. This positive fitness value indicates that these mutations confer a competitive advantage compared to other strains when grown on these carbon sources. Most striking among these genes were the sigma factor rpoS and the LPS export system (PP_1778/9) genes; when disrupted, both displayed fitness benefits on all three nonglucose carbon sources (see Fig. S2 in the supplemental material).
Results of RB-TnSeq screen. Data represent genes that showed greater than 2 log2 fitness on d-lysine, l-lysine, or 5AVA but a fitness defect of no less than 0.5 log2 when grown on glucose. Download FIG S2, TIF file, 3.0 MB (3.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Only one gene (PP_0787, encoding a quinolinate phosphoribosyltransferase) showed fitness defects on all three nonglucose carbon sources (Fig. 1C). However, disruption of PP_0787 also showed a significant fitness defect under conditions of growth on levulinic acid, suggesting that it is unlikely to be uniquely important to lysine metabolism (16). Only 3 genes shared fitness defects between 5AVA and l-lysine (davT, davD, and lghO), and all three have been previously implicated in 5AVA metabolism (Fig. 1C) (9).
PP_4108 is a l-2AA aminotransferase.
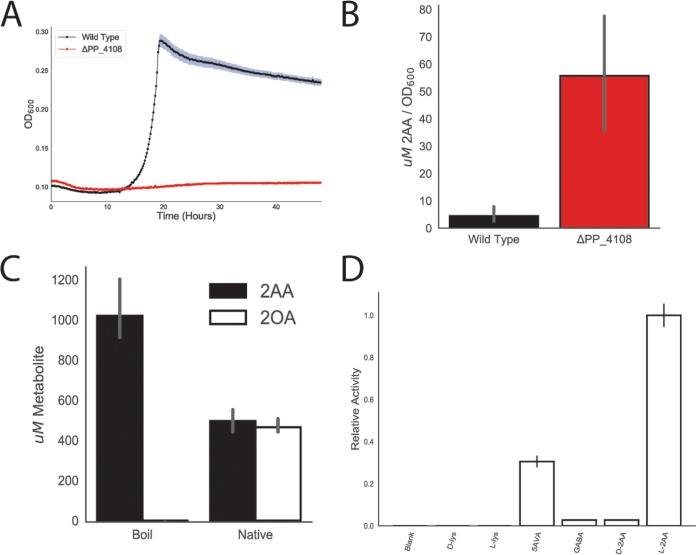
In humans and other animals, l-lysine degradation proceeds through a 2AA intermediate, which a transaminase converts to 2OA (9, 11, 17). Yet no such transaminase has been identified in P. putida. We identified a candidate aminotransferase, PP_4108, for which gene inactivation showed a significant growth defect on d-lysine (fitness value of −5.9) and a relatively minor growth defect on l-lysine (−1.2). To corroborate our RB-TnSeq fitness data, we constructed a deletion mutant of PP_4108 that failed to grow in a plate reader assay on 10 mM dl-2AA (Fig. 2A). The mutant showed a severe growth defect on 10 mM d-lysine and an increased lag time when grown on 10 mM l-lysine (Fig. S3).
FIG 2.
Identification of PP_4108 as an l-2AA aminotransferase. (A) Growth of wild-type KT2440 and the PP_4108 mutant on 2AA as a sole carbon source. The shaded area represents the 95% confidence interval (CI); n = 3. (B) In vivo accumulation of 2AA in wild-type KT2440 and a PP_4108 mutant after 12 h of growth on minimal medium supplemented with 10 mM glucose and 10 mM d-lysine. Bars represent 1og10 transformed spectral counts; error bars show 95% CI; n = 3. (C) In vitro transamination reactions of PP_4108 with 2KG as an amino acceptor. Bars represent metabolite concentrations (in micromoles) of either 2OA (black) or 2AA (white) in either boiled or native protein reactions. Error bars show 95% CI; n = 3. (D) In vitro transaminations of PP_4108 incubated with different possible amino donors and 2KG as the acceptor. Bars represent relative activity levels of enzyme standardized to l-2AA after 16 h of incubation. Error bars show standard deviations; n = 2.
Growth of PP_4108 mutants on lysine. (A) Growth of wild-type KT2440 and the PP_4108 mutant on d-lysine as a sole carbon source. The shaded area represents 95% CI; n = 3. (B) Growth of wild-type KT2440 and the PP_4108 mutant on l-lysine as a sole carbon source. The shaded area represents 95% CI; n = 3. (C) Time course of PP_4108 colorimetric glutamate formation. Error bars represent standard deviations; n = 2. Download FIG S3, TIF file, 2.2 MB (2.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To further validate this hypothesis, the ΔPP_4108 strain was subjected to metabolomics analysis to monitor the accumulation of its expected substrate, 2AA, under conditions of growth on glucose and d-lysine. After 12 h of growth on minimal media supplemented with 10 mM (each) glucose and d-lysine, the PP_4108 deletion strain showed a 6.3-fold increase (P = 0.00016) in the normalized concentration of intracellular 2AA compared to the wild-type (WT) strain (Fig. 2B). Next, PP_4108 was expressed and purified from Escherichiacoli for biochemical characterization. After incubation of the purified enzyme with dl-2AA, 2KG, and pyridoxal phosphate (PLP) for 16 h, the reaction mixture was analyzed by the use of the liquid chromatography-time of flight (LC-TOF) method. The expected product, 2OA, was detected in the enzymatic reaction but not in a boiled enzyme control, confirming PP_4108 to be a transaminase that converts 2AA to 2OA (Fig. 2C). As many transaminases have broad substrate specificity (18), we also probed the substrate range of PP_4108 using a colorimetric assay for glutamate, a stoichiometric product of the transamination reaction (Fig. 2D). The enzyme was most active on l-2AA and showed only 2.8% relative activity (P = 0.0057) on its enantiomer, d-2AA. This specificity for the l-2AA isomer may explain why only 50% of the Dl-2AA was transformed in the previous experiment (Fig. 2C). No activity was observed on either lysine isomer; however, the enzyme showed slight activity toward 4-aminobutyrate/γ-aminobutyrate (GABA) (2.8% relative activity, P = 0.0057) and moderate activity on 5AVA (30.5% relative activity, P = 0.0139). Over smaller time scales, PP_4108 had no activity on any substrate except l-2AA (Fig. S3c). These results suggest that P. putida KT2440 metabolizes d-lysine to l-2AA, which is then converted to 2OA by the transaminase PP_4108.
PP_5260 is a novel DUF1338 family enzyme that catalyzes the conversion of 2OA to 2HG.
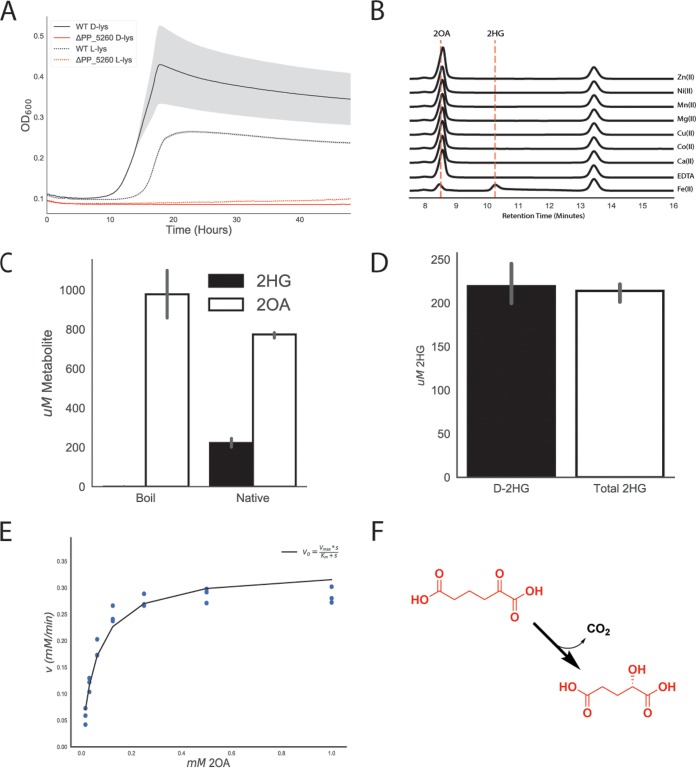
Early work proposed that 2OA is converted to 2KG via a 2HG intermediate (13, 19), while later results suggested a direct conversion of 2OA to glutarate (11). Either route likely requires decarboxylation of 2OA, so we initially searched for decarboxylases within our data set. Our fitness data on either lysine isomer revealed no obvious decarboxylases or enzymes likely to contain a thiamine pyrophosphate (TPP) cofactor, which is commonly employed by decarboxylases. However, a gene near other d-lysine catabolic genes in the P. putida genome, PP_5260, showed a significant fitness defect. A ΔPP_5260 strain was unable to grow on either isomer of lysine, verifying its importance in lysine degradation (Fig. 3A).
FIG 3.
Identification of ydcJ (PP_5260) as a 2OA decarboxylase/hydroxylase. (A) Growth of the wild-type strain (black) and the PP_5260 mutant (red) on d-lysine (line) or l-lysine (dashed line) as a sole carbon source. The shaded area represents 95% CI; n = 3. (B) HPLC traces representing results of in vitro reactions run with apo PP_5260 with exogenous metals added at 50 μM. Retention times for 2OA and 2HG are shown by vertical dashed lines. Metal or EDTA control is indicated to the right of traces. (C) In vitro assay of 2OA conversion to 2HG by purified PP_5260 protein analyzed via the LC-TOF method. 2OG in white, 2HG in black. (D) In vitro assay of purified PP_5260 protein with 2OA as the substrate. The black bar represents the concentration of d-2HG measured by enzyme coupled assay. The white bar represents the total 2HG concentration as measured by the LC-TOF method. Error bars represent 95% CI; n = 3. (E) Initial velocity of reaction catalyzed by PP_5260 as a function of 2OA concentration. Blue dots represent individual measurements, while the black fit line represents a Michaelis-Menten fit. (F) Chemical reaction catalyzed by PP_5260; 2OA is decarboxylated to d-2HG.
PP_5260 belongs to the DUF1338 protein family (http://pfam.xfam.org/family/PF07063). Although several unpublished crystal structures of DUF1338 domain-containing proteins have been deposited into the Protein Data Bank, characterizations of their biological function remain elusive. However, these structures, combined with protein sequence alignments, suggest that a putative metal binding site is conserved throughout the DUF1338 family. As we had hypothesized PP_5260 serves as the missing decarboxylase in d-lysine metabolism, we purified the enzyme for biochemical analysis. Enzymatic activity on 2OA was probed and analyzed via the LC-TOF method. After incubation of 2OA with PP_5260, we observed an ∼92% (P = 0.00034) reduction in the abundance of 2OA, whereas no 2OA was consumed in a boiled enzyme control or in an enzyme treated with EDTA, confirming it to be a metalloenzyme (Fig. S4a). We initially believed that the product would be either glutarate or glutarate semialdehyde; however, neither of these was detected in the reaction. Early biochemical work suggested that 2HG is a potential intermediate in pipecolate metabolism (19), and when the enzymatic product was compared to a racemic 2HG standard, they were found to share the same mass, retention time, and mass-to-charge ratio (Fig. S4b), as well as identical isotopic distributions of [M-H] peaks in the mass spectra (Fig. S4c).
Characterization of PP_5260. (A) In vitro reactions of boiled or native PP_5260 incubated with 2OA with 50 μM EDTA (white) or without EDTA (black). Bars represent 1og10 transformed spectral counts; errors bars represent 95% CI; n = 3 (B) LC-TOF analysis of a 2HG standard, and products of PP_5260 incubated with 2OA, and boiled control. (C) Mass spectra of 2HG standard and product of PP_5260 in vitro reaction. (D) Standard curves of d-2HG and l-2HG determined using a d-2HG-specific enzymatic detection assay. Shaded areas represent 95% CI; n = 3. Download FIG S4, TIF file, 3.8 MB (3.9MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To identify the metal cofactor, the enzyme was dialyzed against EDTA to remove metals, and individual divalent metals were added back. Only the addition of Fe(II) restored enzymatic activity, as measured by high-performance liquid chromatography (HPLC) (Fig. 3B). Subsequent reactions quenched after 5 min showed 200 μM 2HG formed and 800 μM 2OA remaining, demonstrating 1:1 2OA-to-2HG reaction stoichiometry (Fig. 3C). Whether the product of the PP_5260 reaction is l-2HG or is d-2HG is critical to understanding the eventual fate of d-lysine, as lghO is specific for l-2HG (Fig. S1a). An enzyme coupled assay specific for the detection of d-2HG was used to assess the stereochemistry of the PP_5260 product. Standard curves of d-2HG and l-2HG showed that the assay was responsive only to d-2HG (Fig. S4d). The concentrations of in vitro PP_5260 reaction mixtures were then measured by both the LC-TOF method and the enzyme coupled assay, revealing that all of the 2HG present represented the d-isomer (Fig. 3D).
Kinetic parameters of PP_5260 were determined using an enzyme-coupled assay to spectrophotometrically measure CO2 evolution via NADH oxidation (20). PP_5260 displayed a Vmax of 0.33 mM/min (± 0.08 mM), a Km of 0.06 mM (± 0.03 mM), and a Kcat of 330 m−1 using 2OA as the substrate. Taken together, these results reveal that PP_5260 is a novel Fe(II)-dependent decarboxylase that converts 2OA to d-2HG (Fig. 3F), a chemical reaction not previously observed in nature.
DUF1338 proteins represent a widely distributed enzyme family with a putative conserved role in amino acid catabolism.
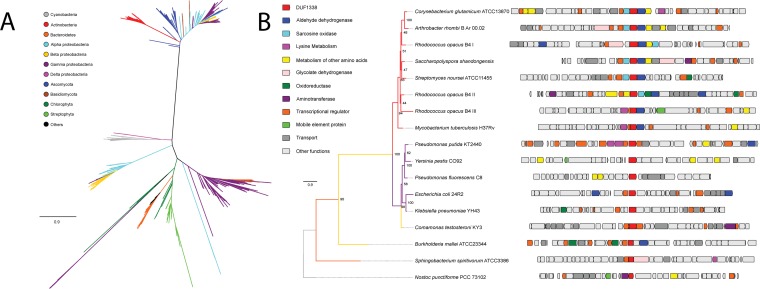
After functional characterization of PP_5260, we use phylogenomics to propagate the annotation and to further explore the biological role of DUF1338 proteins found in other organisms. We found that DUF1338 proteins are widely distributed across the tree of life, with homologs of PP_5260 found in plants and green algae (21), fungi, and bacteria, though they were not found in animals or archaea (Fig. 4A). Homologs are widely distributed among bacteria, with the Firmicutes being a notable exception. PP_5260 homologs within the plant group Streptophyta, as well as the bacterial groups Actinobacteria, Cyanobacteria, and Bacteroidetes, formed monophyletic clades, while homologs from other taxonomic groups were not monophyletic (Fig. 4A). DUF1338 homologs are found in bacteria important to biotechnology (Corynebacterium glutamicum), the environment (Nostoc punctiforme), and medicine (Yersinia pestis, Mycobacterium tuberculosis, Burkholderia pseudomallei).
FIG 4.
Phylogenomics of the DUF1338 enzyme family. (A) Phylogenetic relationships among DUF1338 homologs and their distribution among major phyla. Branches in the tree are colored by phylum. DUF1338 is found in most bacterial phyla as well as in plants and fungi. Nonmonophyletic clades suggest pervasive horizontal gene transfer events in the family. (B) Phylogenomics of selected DUF1338 homologs in bacteria. The phylogeny at the left shows the phylogenetic relationships between selected homologs; the branches have been colored according to their adscription to a given phylum, and the support values are shown at the nodes. The boxes in the right represent the gene neighborhood for each homolog. The genes have been colored to represent their annotated functions.
Publicly available fitness data show that both Pseudomonas fluorescens FW300-N2C3 and Sinorhizobium meliloti PP_5260 homologs have l-lysine-specific defects when interrupted (15). Genomic contexts within other bacteria suggest that many DUF1338-containing enzymes may be involved in the metabolism of lysine or other amino acids (Fig. 4B). Within the Actinobacteria, DUF1338 homologs are often found adjacent to sarcosine oxidases, aldehyde dehydrogenases, and transaminases, implying an additional catabolic amino acid function. In both the oleaginous bacterium Rhodococcus opacus B4 and M. tuberculosis, DUF1338 homologs are found next to predicted l-lysine aminotransferases, suggesting that an ancestral homolog functioned in lysine catabolism. Interestingly, the R.opacus B4 genome has three DUF1338 homologs, only one of which corresponds to genes predicted to be specific to lysine catabolism. The other two gene neighborhoods are similar in their functional content, mainly differing by encoding an oxidoreductase or glycolate dehydrogenase, either of which may perform the same biochemical function. In Alphaproteobacteria, Betaproteobacteria, and Cyanobacteria, the presence of aldehyde dehydrogenases, oxidoreductases, glycolate dehydrogenases, and aminotransferases implies a metabolic function similar to that of PP_5260.
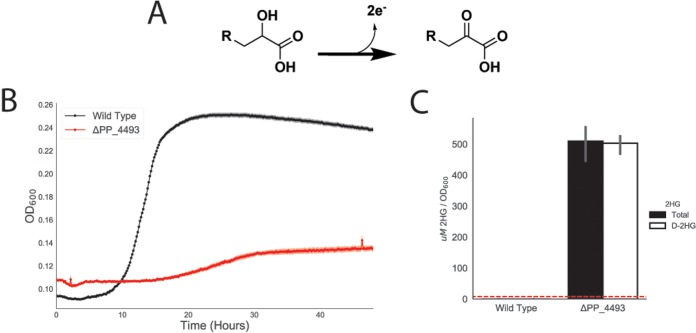
PP_4493 putatively oxidizes d-2HG to 2KG and connects d-lysine to central metabolism.
In the CoA-independent route of glutarate metabolism, LghO oxidizes l-2HG to 2KG; however, this enzyme is highly specific to the l-2HG isomer and showed no fitness defect on d-lysine in our RB-TnSeq data (Fig. S1a). A putative flavin adenine dinucleotide (FAD)-dependent and 4Fe-4S cluster-containing glycolate dehydrogenase, PP_4493, did show fitness defects on both d-lysine and l-lysine (fitness scores of −5.4 and −2.7, respectively) (Fig. 1A). Glycolate dehydrogenases are members of a larger family of enzymes that oxidize the alcohol group of an alpha-hydroxyacid to the corresponding alpha-ketoacid (Fig. 5A). Therefore, we hypothesized that PP_4493 could potentially oxidize a similar 2-hydroxyacid, 2HG, to the corresponding alpha-ketoacid, 2KG. Moreover, many PP_5260 homologs were located next to or near putatively annotated glycolate dehydrogenases in other bacteria, underscoring their potential metabolic link (Fig. 4B). To confirm these hypotheses, we again constructed a deletion strain, P. putida ΔPP_4493, which could not grow on d-lysine as a sole carbon source (Fig. 5B) and which showed attenuated growth on l-lysine (Fig. S5). Furthermore, when grown on 10 mM glucose and 10 mM d-lysine, the mutant accumulated ∼500 μM 2HG (normalized to optical density [OD]), whereas wild-type P. putida did not accumulate any detectable 2HG (Fig. 5C). Subsequent analysis of accumulated 2HG via the use of a d-2HG-specific detection kit revealed that this accumulated 2HG was indeed d-2HG (Fig. 5C). These data and the conserved function and genomic context of glycolate dehydrogenases strongly suggest tha PP_4493 catalyzes the last step of l-2AA metabolism, oxidizing d-2HG to 2KG (Fig. S1b).
FIG 5.
Identification of PP_4493 as a putative d-2HG dehydrogenase. (A) General chemical reaction of a dehydrogenase converting a 2-hydroxyacid to a 2-ketoacid. (B) Growth of P. putida KT2440 and the PP_4108 mutant on d-lysine as a sole carbon source. The shaded area represents 95% CI; n = 3. (C) In vivo accumulation of 2HG in wild-type KT2440 and a PP_4108 mutant after 12 h of growth on minimal medium supplemented with 10 mM glucose and 10 mM d-lysine. The white bar represents the concentration of d-2HG measured by enzyme coupled assay. The black bar represents the total 2HG concentration as measured by the LC-TOF method. Red line represents the limit of detection of the enzyme coupled assay for d-2HG. Bars represent 1og10 transformed spectral counts; error bars show 95% CI; n = 3.
Growth of the PP_4493 mutant on l-lysine and of wild-type KT2440 and the PP_4493 mutant on l-lysine as a sole carbon source. The shaded area represents 95% CI; n = 3. Download FIG S5, TIF file, 3.5 MB (3.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
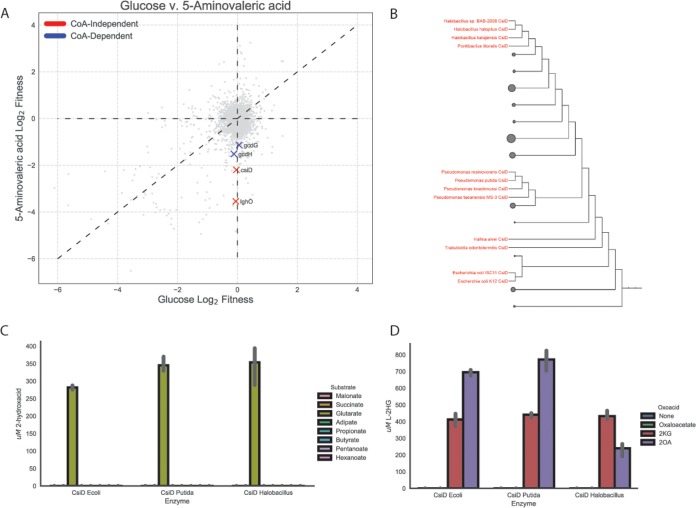
CsiD is highly specific for glutarate hydroxylation but promiscuous in 2-oxoacid selectivity.
During the initial preparation of the manuscript, Zhang et al. discovered a novel pathway of glutarate metabolism in P. putida (9). They described a cyclic reaction cascade wherein a novel 2KG-dependent nonheme Fe(II) oxygenase, PP_2909 (CsiD), hydroxylates glutarate to form 2HG and succinate using 2KG as a cosubstrate. PP_2910 (LghO), a putative l-2HG oxidase, then subsequently converts l-2HG to 2KG, regenerating the 2KG consumed in the initial reaction. These reactions result in the net incorporation of succinate into central metabolism (Fig. S1). Our fitness results of the library grown on 5AVA also identified both csiD and lghO, in addition to the two enzymes from the CoA-dependent glutarate pathway, glutaryl-CoA ligase (gcdG) and glutaryl-CoA dehydrogenase (gcdH), mutants of which showed mild fitness defects when grown on 5AVA (Fig. 6A). We also purified csiD and confirmed that it hydroxylated glutarate in a 2KG-dependent manner (Fig. S6a). HPLC analysis demonstrated that as glutarate was consumed, equimolar quantities of succinate and l-2HG were produced (Fig. S6b). Additionally, a csiD deletion mutant showed an increased lag time when grown on either l-lysine or 5AVA. By deleting the glutaryl-CoA ligase gcdG and disrupting the CoA-dependent glutarate pathway, we completely prevented growth on 5AVA or l-lysine (Fig. S6c). These results are in agreement with those reported by Zhang et al. (9).
FIG 6.
Role of CsiD in P. putida lysine metabolism. (A) Plot of genome-wide fitness values of libraries grown on either 5AVA or glucose. CoA-dependent glutarate degradation genes are shown in red, while those known to be involved succinate producing metabolism are shown in blue. (B) Phylogenetic tree of bacterial CsiD homologs. Homologs used in in vitro assays are highlighted in red. (C) In vitro reactions of CsiD with different substrates using 2KG as a 2-oxoacid. Bars show the peak area of 2-hydroxyacid; error bars show 95% CI; n = 3. (D) In vitro reactions of CsiD homologs with different 2-oxoacids. Bars represent spectral counts of l-2HG. Error bars show 95% CI; n = 3.
Characterization of PP_2909 (CsiD) in P. putida. (A) In vitro PP_2909 reactions. Bars show concentrations (in millimoles) of succinate (blue) or 2HG (green) formed by boiled enzyme control, no-2KG control, or native enzyme with 2KG added. Errors bars show 95% CI; n = 3. (B) Time course of PP_2909 in vitro reaction. The plot shows 2HG, 2KG, succinate, and glutarate levels over time. The shaded region shows 95% CI; n = 3. (C) Growth curves of wild-type KT2440 or PP_0158, PP_2909, and PP_2910 single mutants or PP_2909/PP_0158 double mutants grown on either l-lysine (left column) or 5AVA (right column). The shaded region shows 95% CI; n = 3. Download FIG S6, TIF file, 5.9 MB (6.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Because nonheme Fe(II) oxidases can be promiscuous with respect to the 2-oxoacid cosubstrate (21, 22), we evaluated the 2-oxoacid specificity of CsiD. First, we evaluated the hydroxyl acceptor substrate specificity of CsiD family proteins by purifying two additional homologs from E. coli and a halophilic bacterium, Halobacillus sp. BAB-2008 (Fig. 6B). We probed the activity of the homologs against a panel of 3 to 6 carbon fatty acids and diacids in the presence of 2KG and found that only glutarate served as a hydroxylation substrate (Fig. 6C). These results are consistent with the work recently reported by Zhang et al. (9) and further suggest that the specificity of CsiD homologs is conserved across phyla. Although extremely specific for the hydroxylation substrate, all three CsiD homologs could utilize both 2OA and 2KG, but not oxaloacetate, as a cosubstrate for l-2HG formation (Fig. 6D). The coproduct of the reaction using 2OA as a 2-oxoacid would be glutarate rather than succinate. This result is particularly interesting as it provides a possible mechanism for the previously observed metabolic link between d-lysine catabolism and l-lysine catabolism. Growth defects observed in a ΔPP_2909 ΔPP_0158 double mutant grown on d-lysine also support this hypothesis (Fig. S7a).
Growth of PP_2909/PP_0158 and PP_2088 mutants on d-lysine. (A) Growth of wild-type KT2440 and the PP_2909/PP_0158 mutant on d-lysine as a sole carbon source. The shaded area represents 95% CI; n = 3. (A) Growth of wild-type KT2440 and the PP_2088 mutant on d-lysine as a sole carbon source. The shaded area represents 95% CI; n = 3. Download FIG S7, TIF file, 2.1 MB (2.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Expression of lysine metabolic proteins is responsive to pathway metabolites.
Multiple studies have demonstrated that the expression of lysine catabolic genes is upregulated in the presence of pathway metabolites (9, 12, 23). To investigate the regulation of the newly discovered lysine catabolic enzymes from this study, wild-type P. putida KT2440 was grown in minimal media on glucose or a single lysine metabolite (e.g., d-lysine, l-lysine, 5AVA, 2AA, or glutarate) as a sole carbon source until the cultures reached an optical density at 600 nm (OD600) of 1.0. We then quantified the relative abundances of d-lysine and l-lysine catabolic proteins via targeted proteomics (Fig. 7). All pairwise statistical comparisons of different carbon sources for each protein can be found in Table S2. All five d-lysine pathway proteins examined (AmaA [PP_5257], AmaB [PP_5258], PP_4108, YdcJ [PP_5260], and YdiJ [PP_4493[) were upregulated when grown on l-lysine, d-lysine, or 2AA compared to the glucose control. Neither 5AVA nor glutarate significantly induced expression of any of the d-lysine proteins analyzed. Of all the targeted proteins, the three identified in this study that directly degraded 2AA were most strongly induced by 2AA. Somewhat surprisingly, we also found that the two enzymes involved in 2AA formation, AmaA and AmaB, were also more highly expressed in the presence of 2AA, suggesting the possible involvement of a global regulator. An interesting finding from our initial screen was that sigma factor RpoX (PP_2088) was required for fitness on d-lysine (Fig. 1A). Deletion mutants of rpoX were severely attenuated in their ability to grown on d-lysine as a sole carbon source (Fig. S7b). Further work will be necessary to examine the complex regulatory network that controls d-lysine metabolism.
FIG 7.
Expression of lysine degradation pathways in response to different lysine metabolites. Data represent the relative abundances of selected lysine degradation enzymes expressed in wild-type KT2440 in response to different carbon sources. Bars show spectral counts of proteins after 36 h of growth on 10 mM glucose (black), 5AVA (purple), d-lysine (green), l-lysine (red), glutarate (blue), or 2AA (yellow). Error bars show 95% CI; n = 3.
Statistical analysis of proteomics. Download Table S2, CSV file, 0.05 MB (48.9KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The initial two enzymes from l-lysine metabolism, DavA and DavB, were most highly expressed in the presence of l-lysine but also were also significantly expressed in the presence of d-lysine. As previously observed, DavT and DavD were most strongly upregulated on 5AVA, moderately upregulated on l-lysine, and upregulated to a lesser extent on d-lysine. The induction levels of LhgO and CsiD were highest when grown on glutarate, although these proteins were also moderately upregulated by 5AVA and l-lysine. By comparison, PP_0159 (GcdG) expression in the presence of glutarate was stimulated to a lesser extent than LhgO and CsiD expression; in addition, GcdG was slightly upregulated on 5AVA and l-lysine.
DISCUSSION
Despite intensive study, complete biochemical and genetic understanding of d-lysine catabolism in P. putida has remained elusive. A 2OA degradation pathway has been extensively characterized in mammals, because of its implications in human disease (24). In the mammalian pathway, l-lysine is metabolized to 2OA and eventually converted to acetyl-CoA via a glutaryl-CoA intermediate (24). However, this pathway has not been observed in bacteria. Previous work suggested that 2OA is converted either via decarboxylation to glutarate or through several enzymatic steps to 2HG (11, 19, 25), and yet none of those studies conclusively demonstrated a genetic and biochemical basis for those hypotheses. In this work, we demonstrated plausible biochemical routes to account for both of these previously hypothesized pathways.
The first route, catalyzed by the DUF1338-containing metalloenzyme PP_5260, involves the direct conversion of 2OA to d-2HG. The formation of the d-2HG isomer by PP_5260 maintains stereochemical separation from the l-2HG formed by l-lysine degradation, thus requiring the dehydrogenase PP_4493 gene rather than the l-2HG-specific oxidase lghO gene. This transformation seemingly involves two separate reactions: a decarboxylation and a hydroxylation. Hydroxymandelate synthase has been shown to catalyze a similar enzymatic reaction, via an intramolecular oxidative decarboxylation, similarly to 2KG dependent Fe(II) oxidases (26). PP_5260 is also a Fe(II)-dependent decarboxylase, and the two share similar Kcat values for their given substrates (330 m−1 for PP_5260 and 270 m−1 for hydroxymandelate synthase) (27). Though PP_5260 and hydroxymandelate synthase share little sequence homology, this enzyme may give us insight into the molecular mechanism of DUF1338 enzymes. We have given PP_5260 the tentative title of 2-hydroxyglutarate synthase (hglS) and will use that name until further mechanistic studies (under way in our group) are completed and a more accurate enzyme name can be assigned.
In bacteria, homologs of PP_5260 appear widely distributed with their genomic contexts, suggesting functions both within and beyond lysine metabolism. Genomic contexts in other bacteria, particularly Actinobacteria, suggest that these homologs may be involved in other amino acid catabolic pathways. Unfortunately, there is scant evidence for homologous function in model organisms. For example, although DUF1338 proteins are present in other Ascomycota, there is no homolog in Saccharomyces cerevisiae. Interestingly, the E. coli homolog of PP_5260 is located next to a potential glucan biosynthesis gene: glucan biosynthesis protein D (28). Another DUF1338-containing protein from rice has been characterized and was implicated in starch granule formation (29). These results suggest that DUF1338 proteins could play a role in sugar metabolism.
Recently, Zhang et al. thoroughly characterized a CoA-independent glutarate catabolism route ending at succinate and involving the Fe(II)-dependent oxygenase CsiD (9). Our RB-TnSeq screening convergently uncovered this pathway, and our biochemical and physiological results fully corroborate their findings. While both works showed that multiple CsiD homologs from divergent bacteria are highly specific to glutarate as a hydroxyl acceptor, all three homologs that we tested showed promiscuous activity toward 2-oxoacid cosubstrates. The ability of the P. putida CsiD to utilize 2OA as a cosubstrate is particularly interesting as it may directly connect l-lysine metabolism and d-lysine metabolism. The promiscuity of CsiD may explain reports of glutarate formation from d-lysine in earlier studies (11). Further studies involving labeled substrates may help elucidate the potential link between the two pathways. While CsiD plays a clear role in l-lysine metabolism in P. putida, its role in other organisms remains a mystery. In E. coli, RpoS controls the expression of CsiD, but rpoS mutants showed fitness benefits on all three lysine metabolites tested in our RB-TnSeq data (30). Recent work has shown that E. coli also uses CsiD to metabolize lysine, suggesting a possible conserved role for this pathway across bacteria (31).
Work presented here and previous reports have shown the expression levels of both lysine catabolism pathways are highly responsive to their respective metabolites. While this metabolism appears highly coordinated, the genes themselves are dispersed across the genome, with both PP_4018, and PP_4493 found at locations that were outside operons and relatively distant from other lysine catabolic genes. At least two global regulators appeared to be important for lysine metabolism based on our Rb-TnSeq data, namely, cbrA (PP_4695) and rpoX (PP_2088). The two-component system CbrAB has been implicated in catabolite repression and C/N balance in Pseudomonasaeruginosa, with mutants unable to grown on multiple amino acids (32). Further work in P. putida KT2440 showed that the CbrAB system behaved similarly to that in P. aeruginosa (33). It would be unsurprising if this regulator were found to control the expression of various genes within lysine catabolism; more work into uncovering the regulon is warranted. RpoX, on the other hand, has been implicated in osmotic tolerance in P. aeruginosa (34, 35). This is interesting, as lysine metabolism, and, specifically, pipecolate metabolism, has been found to be associated with osmotic tolerance across multiple bacteria (36). As an rpoX deletion mutant was unable to grow on d-lysine, these results suggest that d-lysine metabolism of P. putida may be involved in adaptation to saline conditions or to other osmotically stressful environments.
Interesting issues remain as to why P. putida maintains separate metabolic pathways for d- and l-lysine and why l-lysine metabolism seems dependent on the presence of an intact d-lysine metabolism. Previously work proposed that the d-lysine pathway may provide a way of resolving the C/N imbalance that may occur when l-lysine is metabolized. However, we believe that this is unlikely, as both lysine degradation pathways contain one deamination reaction and one transamination reaction (11). Our fitness results indicate that d-lysine metabolism is dispensable for growth on 5AVA. This would suggest that only the initial two steps of l-lysine metabolism, the oxidation of lysine to 5-aminopentanamide by DavB and its subsequent deamination to 5AVA by DavA, are dependent on d-lysine catabolism. We propose that the adjacent AsnC family regulator PP_0384 likely responds to l-lysine, given that many proteins within this family respond to amino acids, including lysine (37, 38), and that expression of these two enzymes is most responsive to l-lysine. To our knowledge, there has been no rigorous characterization of the regulation of the davAB operon or of the biochemical activities of these two enzymes in vitro. Future studies to uncover the mechanistic regulation at the transcriptional and posttranslational levels in these two steps may uncover the necessity of d-lysine dependency of the l-lysine catabolic pathway. Overall, our work highlights the utility of global fitness profiling for discovering novel, complex metabolic pathways in even well-characterized bacteria.
MATERIALS AND METHODS
Media, chemicals, and culture conditions.
Bacterial cultures were routinely grown in Luria-Bertani (LB) Miller medium (BD Biosciences, USA). E. coli was grown at 37°C, while P. putida was grown at 30°C unless otherwise noted. When indicated, P. putida was grown on modified MOPS (morpholinepropanesulfonic acid) minimal medium (39). Cultures were supplemented with kanamycin (Sigma-Aldrich, USA) (50 mg/liter), gentamicin (Fisher Scientific, USA) (30 mg/liter), or carbenicillin (Sigma-Aldrich, USA) (100 mg/liter) when indicated. d-2AA was purchased from TaKaRa Bioscience (USA). All other compounds were purchased through Sigma-Aldrich.
Strains and plasmids.
All bacterial strains and plasmids used in this work are listed in Table 1. All strains and plasmids created in this work are publicly available through the Joint BioEnergy Institute (JBEI) registry (https://public-registry.jbei.org/folders/391). All plasmids were designed using Device Editor and Vector Editor software, while all primers used for the construction of plasmids were designed using j5 software (40–42). Synthetic DNA coding for the Halobacillus sp. BAB-2008 csiD homolog was purchased from Integrated DNA Technologies (IDT, Coralville, IA). Plasmids were assembled via Gibson assembly using standard protocols (43) or via Golden Gate assembly using standard protocols (44). Plasmids were isolated routinely using a Qiaprep Spin Miniprep kit (Qiagen, USA), and all primers were purchased from Integrated DNA Technologies (IDT, Coralville, IA).
TABLE 1.
Strains and plasmids used in this studya
| Strains or plasmid | JBEI part ID | Source or reference |
Genotype |
|---|---|---|---|
| Strains | |||
| E. coli DH10B | Thermo Fisher | ||
| E. coli S17 | ATCC 47055 | ||
| E. coli BL21(DE3) | Novagen | ||
| P. putida KT2440 | ATCC 47054 | ||
| P. putida ΔPP_0158 | JPUB_010967 | This work | |
| P. putida ΔPP_2088 | JPUB_013224 | This work | |
| P. putida ΔPP_2909 | JPUB_010968 | This work | |
| P. putida ΔPP_2910 | JPUB_010969 | This work | |
| P. putida ΔPP_0158 ΔPP_2909 | JPUB_010970 | This work | |
| P. putida ΔPP_4108 | JPUB_010971 | This work | |
| P. putida ΔPP_4493 | JPUB_010972 | This work | |
| P. putida ΔPP_5260 | JPUB_010973 | This work | |
| Plasmids | |||
| pMQ30 | 45 | Gm, SacB | |
| pET28 | Novagen | Kan | |
| pET21b | Novagen | Amp | |
| pMQ30-PP_0158 | JPUB_010989 | This work | Gm, SacB |
| pMQ30-PP_2088 | JPUB_013222 | This work | Gm, SacB |
| pMQ30-PP_2909 | JPUB_010991 | This work | Gm, SacB |
| pMQ30-PP_2910 | JPUB_010995 | This work | Gm, SacB |
| pMQ30-PP_4108 | JPUB_010981 | This work | Gm, SacB |
| pMQ30-PP_4493 | JPUB_010979 | This work | Gm, SacB |
| pMQ30-PP_5260 | JPUB_010977 | This work | Gm, SacB |
| pET28-CsiD_Halo | JPUB_010987 | This work | Kan |
| pET28-CsiD_Ecoli | JPUB_010993 | This work | Kan |
| pET28-CsiD_Pput | JPUB_010975 | This work | Kan |
| pET21b-PP_4108 | JPUB_010983 | This work | Amp |
| pET21b-PP_5260 | JPUB_010985 | This work | Amp |
ID, identifier; Amp, ampicillin; Gm, gentamicin; Kan, kanamycin.
Random barcode TnSeq experiments.
P. putida RB-TnSeq library JBEI-1 was created by diluting a 1-ml aliquot of the previously described P. putida RB-TnSeq library (16) in 500 ml of LB media supplemented with kanamycin; the library was then grown to an OD600 of 0.5 and frozen as 1-ml aliquots after adding glycerol to reach a final concentration of 20% (vol/vol). Libraries were stored at −80°C until use. A 1-ml aliquot of P. putida RB-TnSeq library JBEI-1 was thawed on ice and diluted in 25 ml of LB supplemented with kanamycin. The culture was grown until it reached an OD600 of 0.5, at which point three 1-ml aliquots were removed, pelleted, decanted, and then stored at −80°C for use as a time zero control. The library was then washed once in MOPS minimal medium without any carbon source and then diluted 1:50 into 10 ml MOPS minimal medium supplemented with 10 mM glucose or 5AVA or d-lysine or l-lysine. Cells were grown in 50-ml culture tubes for 48 h at 30°C with shaking at 200 rpm. After growth, 2-ml aliquots from the culture tubes were pelleted, decanted, and frozen at −80°C for barcode sequencing (BarSeq). We performed DNA barcode sequencing as previously described (14, 16). The fitness of a strain is defined here as the normalized log2 ratio of barcode reads in the experimental sample to barcode reads in the time zero sample. The fitness of a gene is defined here as the weighted average of the strain fitness for insertions in the central 10% to 90% of the gene. The gene fitness values are normalized such that the typical gene has a fitness of zero. The primary statistic t value represents the form of fitness divided by the estimated variance across different mutants of the same gene. Statistic t values of >|4| were considered significant. All experiments described here passed testing using the quality metrics described previously unless noted otherwise. All fitness data in this work is publicly available at http://fit.genomics.lbl.gov.
Construction of deletion mutants.
Deletion mutants in P. putida were constructed by homologous recombination and sacB counterselection using allelic exchange vector pMQ30 (45). Briefly, homology fragments ranging in size from 1 kbp to 2 kbp upstream and downstream of the target gene, including the start and stop codons, respectively, were cloned into pMQ30. An exception to these design parameters was plasmid pMQ-PP_5260, which maintained an additional 21 nucleotides (nt) at the 5′ end of the gene in addition to the stop codon. Plasmids were then transformed via electroporation into E. coli S17 and then mated into P. putida via conjugation. Transconjugants were selected for on LB agar plates supplemented with 30 mg/ml gentamicin and 30 mg/ml chloramphenicol. Transconjugants were then grown overnight on LB media also supplemented with 30 mg/ml gentamicin and 30 mg/ml chloramphenicol and were then plated on LB agar with no NaCl that was supplemented with 10% (wt/vol) sucrose. Putative deletions were restreaked on LB agar with no NaCl supplemented with 10% (wt/vol) sucrose and then were screened via PCR with primers flanking the target gene to confirm gene deletion.
Plate-based growth assays.
Growth studies of bacterial strains were conducted using microplate reader kinetic assays. Overnight cultures were inoculated into 10 ml of LB medium from single colonies and were grown at 30°C. These cultures were then washed twice with MOPS minimal media without any added carbon and diluted 1:100 into 500 μl of MOPS medium with a 10 mM concentration of a carbon source in 48-well plates (Falcon; catalog no. 353072). Plates were sealed with a gas-permeable microplate adhesive film (VWR, USA), and then optical density was monitored for 48 h using a BioTek Synergy 4 plate reader (BioTek, USA) at 30°C with fast continuous shaking. Optical density was measured at 600 nm, and all OD600 measurements are reported without path length corrections.
Expression and purification of proteins.
A 5-ml overnight culture of E. coli BL21(DE3) containing the expression plasmid was used to inoculate a 500-ml culture of LB. Cells were grown at 37°C to an OD600 of 0.6 and were then induced with isopropyl-β-d-1-thiogalactopyranoside to reach a final concentration of 1 mM. The temperature was lowered to 30°C, and cells were allowed to express for 18 h before being harvested via centrifugation. Cell pellets were stored at −80°C until purification. For purification, cell pellets were resuspended in lysis buffer (50 mM sodium phosphate, 300 mM sodium chloride, 10 mM imidazole, 8% glycerol, pH 7.5) and sonicated to lyse cells. Insoluble content was pelleted via centrifugation (30 min at 40,000 × g). The supernatant was applied to a fritted column containing nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen, USA) which had been preequilibrated with several column volumes of lysis buffer. The resin was washed with lysis buffer containing 50 mM imidazole, and then the protein was eluted using a stepwise gradient of lysis buffer containing increasing imidazole concentrations (100 mM, 200 mM, and 400 mM). Fractions were collected and analyzed via SDS-PAGE. Purified protein was dialyzed overnight at 4˚C against 50 mM HEPES (pH 7.5)–5% glycerol.
CsiD in vitro assays.
The activity of purified CsiD homologs was analyzed in 100-μl reaction mixtures containing 50 mM HEPES (pH 7), 5 mM glutarate, 5 mM 2KG, 25 μM FeSO4, 0.1 mM ascorbate, and 0.5 mM dithiothreitol. For negative-control reactions, each of the respective reaction components was omitted. To initiate reactions, CsiD was added to reach a final concentration of 7 μM. For the no-enzyme control, CsiD was denatured at 98°C for 10 min prior to addition to the reaction mix. Reactions were allowed to proceed at 22°C for 3 h. Products were analyzed via LC-TOF method 3 after quenching was performed via the addition of acetonitrile and methanol for a final ACN/H2O/MeOH ratio of 6:3:1 To analyze products from the substrate range as well as the 2-oxoacid specificity experiments, reactions were measured via LC-TOF method 1.
Transamination assays.
To determine product formation via PP_4108, assays were conducted using 50 mM HEPES (pH 7.5) with 5 mM 2KG, 0.1 mM PLP, 5 mM substrate, and 10 μM purified enzyme or boiled enzyme control in 100-μl volumes. Reaction mixtures were incubated at 30°C for 16 h and quenched via the addition of ACN and MeOH for a final ACN/H2O/MeOH ratio of 6:3:1 for LC-TOF method 3. To determine substrate specificity, reactions were set up at the 75-μl scale and carried out at 30°C for up to 16 h before freezing. For analysis, reaction mixtures were diluted 15-fold in water and assessed by a colorimetric assay for glutamate (Sigma MAK004) in 96-well format via the use of a Spectramax M4 plate reader (Molecular Devices, USA).
PP_5260 in vitro assays.
The activity of PP_5260 was initially assessed using 50 mM HEPES with 5 mM 2OA as the substrate and 10 μM purified enzyme or boiled enzyme control. Reaction mixtures were incubated for 16 h at 30°C. To test the necessity of metal cofactors, EDTA was added to reach a final concentration of 50 μM. Reactions were quenched via the addition of ACN and methanol MeOH for a final ACN/H2O/MeOH ratio of 6:3:1 for LC-TOF analysis using method 3 or via the addition of an equal volume of ice-cold methanol for HPLC analysis and LC-TOF using method 2.
To determine the metal cofactor, after purification over Ni-NTA resin, the protein was concentrated and dialyzed overnight against 50 mM HEPES–100 mM NaCl (pH 7.5). To generate apo-enzyme, the protein was then dialyzed four times at a protein/dialysis buffer ratio of 1:300 against the same buffer containing 5 mM EDTA in order to remove any bound metal. The enzyme was dialyzed once more against buffer without EDTA overnight in order to remove any remaining chelating reagent. The apo-enzyme was then assayed in the presence of 50 μM concentrations of a variety of potential metal cofactors using 50 mM HEPES with 10 mM 2OA as the substrate and 10 μM purified enzyme. Reaction mixtures were incubated for 30 min at 30°C, and activity was assessed via HPLC analysis.
Determination of enzyme stoichiometry was performed in 50 mM HEPES–50 μM FeCl2 with 1 mM 2OA as the substrate and 0.1 μM purified enzyme or boiled enzyme control. Reaction mixtures were incubated for 5 min at 30°C and then quenched with an equal volume of ice-cold methanol and quantified using LC-TOF method 2.
Enzyme coupled decarboxylation assays were carried out as previously described (20). Reaction mixtures contained 100 mM Tris-HCl (pH 7), 10 mM MgCl2, 0.4 mM NADH, 50 μM FeCl2, 4 mM phosphoenol pyruvate (PEP), 100 U/ml pig heart malate dehydrogenase (Roche), 2 U/ml microbial PEP carboxylase (Sigma), and 10 mM 2OA. Reactions were initiated by the addition of purified PP_5260 or boiled enzyme controls, and absorbance at 340 nm was measured using a Spectramax M4 plate reader (Molecular Devices, USA). Michaelis-Menten behavior was formulated as previously described (46). Least-squares minimization was used to derive Km and Kcat values. Determination of d-2HG concentration was performed with a d-2-hydroxyglutarate (d-2HG) assay kit (Sigma MAK320).
Additional experimental methods. Download Text S1, DOCX file, 0.01 MB (15.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ACKNOWLEDGMENTS
This article is dedicated to the memory of Eugene Madsen.
We thank Morgan Price, John Hangasky, Jamie Meadows, Robert Haushalter, Bo Pang, Nick Weathersby, Mary Thompson, and Catharine Adams for their helpful discussions in preparing the manuscript. We thank the SMART program of the University of California, Berkeley (UC Berkeley), for providing support for R.N.K. to conduct summer research.
This work was funded in part by the DOE Joint BioEnergy Institute (https://www.jbei.org) supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research; and protein purification and homology modeling components were funded by the Agile BioFoundry (http://agilebiofoundry.org) supported by the U.S. Department of Energy, Energy Efficiency and Renewable Energy, Bioenergy Technologies Office, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy.
Our views and opinions expressed here do not necessarily state or reflect those of the United States Government or any agency thereof. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, expressed or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights.
The Joint Program in Bioengineering is a joint venture between the University of California, Berkeley, and the University of California, San Francisco.
We were responsible for performing the study as follows: conceptualization, M.G.T. and J.M.B.-H.; methodology, M.G.T., J.M.B.-H., J.F.B., P.C.-M., S.C.C., N.C.H., C.B.E., E.E.K.B., C.J.P., and A.M.D.; investigation, M.G.T., J.M.B.-H., W.A.S., R.N.K., J.F.B., V.T.B., P.C.-M., J.W.G., C.J.P., N.C.H., F.F.T., J.H.P., W.S., and E.E.K.B.; writing—original draft—M.G.T.; writing—review and editing—all of us; resources and supervision, P.D.A., A.P.A., A.M.D., and J.D.K.
J.D.K. has financial interests in Amyris, Lygos, Constructive Biology, Demetrix, Napigen, and Maple Bio.
Footnotes
Citation Thompson MG, Blake-Hedges JM, Cruz-Morales P, Barajas JF, Curran SC, Eiben CB, Harris NC, Benites VT, Gin JW, Sharpless WA, Twigg FF, Skyrud W, Krishna RN, Henrique Pereira J, Baidoo EEK, Petzold CJ, Adams PD, Arkin AP, Deutschbauer AM, Keasling JD. 2019. Massively parallel fitness profiling reveals multiple novel enzymes in Pseudomonas putida lysine metabolism. mBio 10:e02577-18. https://doi.org/10.1128/mBio.02577-18.
REFERENCES
- 1.Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martins dos Santos VAP, Fouts DE, Gill SR, Pop M, Holmes M, Brinkac L, Beanan M, DeBoy RT, Daugherty S, Kolonay J, Madupu R, Nelson W, White O, Peterson J, Khouri H, Hance I, Lee PC, Holtzapple E, Scanlan D, Tran K, Moazzez A, Utterback T, Rizzo M, Lee K, Kosack D, Moestl D, Wedler H, Lauber J, Stjepandic D, Hoheisel J, Straetz M, Heim S, Kiewitz C, Eisen J, Timmis KN, Dusterhoft A, Tummler B, Fraser CM. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 2.Nikel PI, Chavarría M, Danchin A, de Lorenzo V. 2016. From dirt to industrial applications: Pseudomonas putida as a synthetic biology chassis for hosting harsh biochemical reactions. Curr Opin Chem Biol 34:20–29. doi: 10.1016/j.cbpa.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 3.Loeschcke A, Thies S. 2015. Pseudomonas putida-a versatile host for the production of natural products. Appl Microbiol Biotechnol 99:6197–6214. doi: 10.1007/s00253-015-6745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiménez JI, Miñambres B, García JL, Díaz E. 2002. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol 4:824–841. doi: 10.1046/j.1462-2920.2002.00370.x. [DOI] [PubMed] [Google Scholar]
- 5.Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, Langan P, Naskar AK, Saddler JN, Tschaplinski TJ, Tuskan GA, Wyman CE. 2014. Lignin valorization: improving lignin processing in the biorefinery. Science 344:1246843. doi: 10.1126/science.1246843. [DOI] [PubMed] [Google Scholar]
- 6.Chang YF, Adams E. 1971. Induction of separate catabolic pathways for L- and D-lysine in Pseudomonas putida. Biochem Biophys Res Commun 45:570–577. doi: 10.1016/0006-291X(71)90455-4. [DOI] [PubMed] [Google Scholar]
- 7.Chae TU, Ko Y-S, Hwang K-S, Lee SY. 2017. Metabolic engineering of Escherichia coli for the production of four-, five- and six-carbon lactams. Metab Eng 41:82–91. doi: 10.1016/j.ymben.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Barajas JF, Burdu M, Wang G, Baidoo EE, Keasling JD. 2017. Application of an acyl-CoA ligase from Streptomyces aizunensis for lactam biosynthesis. ACS Synth Biol 6:884–890. doi: 10.1021/acssynbio.6b00372. [DOI] [PubMed] [Google Scholar]
- 9.Zhang M, Gao C, Guo X, Guo S, Kang Z, Xiao D, Yan J, Tao F, Zhang W, Dong W, Liu P, Yang C, Ma C, Xu P. 2018. Increased glutarate production by blocking the glutaryl-CoA dehydrogenation pathway and a catabolic pathway involving L-2-hydroxyglutarate. Nat Commun 9:2114. doi: 10.1038/s41467-018-04513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HT, Khang TU, Baritugo K-A, Hyun SM, Kang KH, Jung SH, Song BK, Park K, Oh M-K, Kim GB, Kim HU, Lee SY, Park SJ, Joo JC. 2019. Metabolic engineering of Corynebacterium glutamicum for the production of glutaric acid, a C5 dicarboxylic acid platform chemical. Metab Eng 51:99–109. doi: 10.1016/j.ymben.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Revelles O, Espinosa-Urgel M, Fuhrer T, Sauer U, Ramos JL. 2005. Multiple and interconnected pathways for L-lysine catabolism in Pseudomonas putida KT2440. J Bacteriol 187:7500–7510. doi: 10.1128/JB.187.21.7500-7510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Revelles O, Wittich R-M, Ramos JL. 2007. Identification of the initial steps in D-lysine catabolism in Pseudomonas putida. J Bacteriol 189:2787–2792. doi: 10.1128/JB.01538-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perfetti R, Campbell RJ, Titus J, Hartline RA. 1972. Catabolism of pipecolate to glutamate in Pseudomonas putida. J Biol Chem 247:4089–4095. [PubMed] [Google Scholar]
- 14.Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, Hoover CA, Blow MJ, Bristow J, Butland G, Arkin AP, Deutschbauer A. 2015. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6:e00306. doi: 10.1128/mBio.00306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, Kuehl JV, Melnyk RA, Lamson JS, Suh Y, Carlson HK, Esquivel Z, Sadeeshkumar H, Chakraborty R, Zane GM, Rubin BE, Wall JD, Visel A, Bristow J, Blow MJ, Arkin AP, Deutschbauer AM. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557:503–509. doi: 10.1038/s41586-018-0124-0. [DOI] [PubMed] [Google Scholar]
- 16.Rand JM, Pisithkul T, Clark RL, Thiede JM, Mehrer CR, Agnew DE, Campbell CE, Markley AL, Price MN, Ray J, Wetmore KM, Suh Y, Arkin AP, Deutschbauer AM, Amador-Noguez D, Pfleger BF. 2017. A metabolic pathway for catabolizing levulinic acid in bacteria. Nat Microbiol 2:1624–1634. doi: 10.1038/s41564-017-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuno E, Tsujimoto M, Nakamura M, Kido R. 1993. 2-Aminoadipate-2-oxoglutarate aminotransferase isoenzymes in human liver: a plausible physiological role in lysine and tryptophan metabolism. Enzyme Protein 47:136–148. doi: 10.1159/000468669. [DOI] [PubMed] [Google Scholar]
- 18.Wilding M, Peat TS, Newman J, Scott C. 2016. A β-alanine catabolism pathway containing a highly promiscuous ω-transaminase in the 12-aminododecanate-degrading Pseudomonas sp. strain AAC. Appl Environ Microbiol 82:3846–3856. doi: 10.1128/AEM.00665-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopchick JJ, Hartline RA. 1979. alpha-Hydroxyglutarate as an intermediate in the catabolism of alpha-aminoadipate by Pseudomonas putida. J Biol Chem 254:3259–3263. [PubMed] [Google Scholar]
- 20.Witkowski A, Joshi AK, Smith S. 2002. Mechanism of the β-ketoacyl synthase reaction catalyzed by the animal fatty acid synthase. Biochemistry 41:10877–10887. doi: 10.1021/bi0259047. [DOI] [PubMed] [Google Scholar]
- 21.Martinez S, Hausinger RP. 2015. Catalytic mechanisms of Fe(II)- and 2-oxoglutarate-dependent oxygenases. J Biol Chem 290:20702–20711. doi: 10.1074/jbc.R115.648691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez S, Hausinger RP. 2016. Biochemical and spectroscopic characterization of the non-heme Fe(II)- and 2-oxoglutarate-dependent ethylene-forming enzyme from Pseudomonas syringae pv. phaseolicola PK2. Biochemistry 55:5989–5999. doi: 10.1021/acs.biochem.6b00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Revelles O, Espinosa-Urgel M, Molin S, Ramos JL. 2004. The davDT operon of Pseudomonas putida, involved in lysine catabolism, is induced in response to the pathway intermediate delta-aminovaleric acid. J Bacteriol 186:3439–3446. doi: 10.1128/JB.186.11.3439-3446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danhauser K, Sauer SW, Haack TB, Wieland T, Staufner C, Graf E, Zschocke J, Strom TM, Traub T, Okun JG, Meitinger T, Hoffmann GF, Prokisch H, Kölker S. 2012. DHTKD1 mutations cause 2-aminoadipic and 2-oxoadipic aciduria. Am J Hum Genet 91:1082–1087. doi: 10.1016/j.ajhg.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reitz M, Rodwell V. 1969. a-Hydroxyglutarate oxidoreductase of Pseudomonas putida. J Bacteriol 100:708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wójcik A, Broclawik E, Siegbahn PEM, Borowski T. 2012. Mechanism of benzylic hydroxylation by 4-hydroxymandelate synthase. A computational study. Biochemistry 51:9570–9580. doi: 10.1021/bi3010957. [DOI] [PubMed] [Google Scholar]
- 27.Di Giuro CML, Konstantinovics C, Rinner U, Nowikow C, Leitner E, Straganz GD. 2013. Chiral hydroxylation at the mononuclear nonheme Fe(II) center of 4-(S) hydroxymandelate synthase–a structure-activity relationship analysis. PLoS One 8:e68932. doi: 10.1371/journal.pone.0068932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lequette Y, Odberg-Ferragut C, Bohin J-P, Lacroix J-M. 2004. Identification of mdoD, an mdoG paralog which encodes a twin-arginine-dependent periplasmic protein that controls osmoregulated periplasmic glucan backbone structures. J Bacteriol 186:3695–3702. doi: 10.1128/JB.186.12.3695-3702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Ren Y, Lu B, Yang C, Feng Z, Liu Z, Chen J, Ma W, Wang Y, Yu X, Wang Y, Zhang W, Wang Y, Liu S, Wu F, Zhang X, Guo X, Bao Y, Jiang L, Wan J. 2016. FLOURY ENDOSPERM7 encodes a regulator of starch synthesis and amyloplast development essential for peripheral endosperm development in rice. J Exp Bot 67:633–647. doi: 10.1093/jxb/erv469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vijayakumar SRV, Kirchhof MG, Patten CL, Schellhorn HE. 2004. RpoS-regulated genes of Escherichia coli identified by random lacZ fusion mutagenesis. J Bacteriol 186:8499–8507. doi: 10.1128/JB.186.24.8499-8507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knorr S, Sinn M, Galetskiy D, Williams RM, Wang C, Müller N, Mayans O, Schleheck D, Hartig JS. 2018. Widespread bacterial lysine degradation proceeding via glutarate and L-2-hydroxyglutarate. Nat Commun 9:5071. doi: 10.1038/s41467-018-07563-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishijyo T, Haas D, Itoh Y. 2001. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol Microbiol 40:917–931. doi: 10.1046/j.1365-2958.2001.02435.x. [DOI] [PubMed] [Google Scholar]
- 33.Valentini M, Garcia-Maurino SM, Perez-Martinez I, Santero E, Canosa I, Lapouge K. 2014. Hierarchical management of carbon sources is regulated similarly by the CbrA/B systems in Pseudomonas aeruginosa and Pseudomonas putida. Microbiology 160:2243–2252. doi: 10.1099/mic.0.078873-0. [DOI] [PubMed] [Google Scholar]
- 34.Bouffartigues E, Gicquel G, Bazire A, Bains M, Maillot O, Vieillard J, Feuilloley MGJ, Orange N, Hancock REW, Dufour A, Chevalier S. 2012. Transcription of the oprF gene of Pseudomonas aeruginosa is dependent mainly on the SigX sigma factor and is sucrose induced. J Bacteriol 194:4301–4311. doi: 10.1128/JB.00509-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blanka A, Schulz S, Eckweiler D, Franke R, Bielecka A, Nicolai T, Casilag F, Düvel J, Abraham W-R, Kaever V, Häussler S. 2014. Identification of the alternative sigma factor SigX regulon and its implications for Pseudomonas aeruginosa pathogenicity. J Bacteriol 196:345–356. doi: 10.1128/JB.01034-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neshich IAP, Kiyota E, Arruda P. 2013. Genome-wide analysis of lysine catabolism in bacteria reveals new connections with osmotic stress resistance. ISME J 7:2400–2410. doi: 10.1038/ismej.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brinkman AB, Bell SD, Lebbink RJ, de Vos WM, van der Oost J. 2002. The Sulfolobus solfataricus Lrp-like protein LysM regulates lysine biosynthesis in response to lysine availability. J Biol Chem 277:29537–29549. doi: 10.1074/jbc.M203528200. [DOI] [PubMed] [Google Scholar]
- 38.Thaw P, Sedelnikova SE, Muranova T, Wiese S, Ayora S, Alonso JC, Brinkman AB, Akerboom J, van der Oost J, Rafferty JB. 2006. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res 34:1439–1449. doi: 10.1093/nar/gkl009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaBauve AE, Wargo MJ. 2012. Growth and laboratory maintenance of Pseudomonas aeruginosa. Curr Protoc Microbiol Chapter 6:Unit 6E.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ham TS, Dmytriv Z, Plahar H, Chen J, Hillson NJ, Keasling JD. 2012. Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. Nucleic Acids Res 40:e141. doi: 10.1093/nar/gks531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, Densmore D, Ham TS, Keasling JD, Hillson NJ. 2012. DeviceEditor visual biological CAD canvas. J Biol Eng 6:1. doi: 10.1186/1754-1611-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hillson NJ, Rosengarten RD, Keasling JD. 2012. j5 DNA assembly design automation software. ACS Synth Biol 1:14–21. doi: 10.1021/sb2000116. [DOI] [PubMed] [Google Scholar]
- 43.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 44.Engler C, Kandzia R, Marillonnet S. 2008. A one pot, one step, precision cloning method with high throughput capability. PLoS One 3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shanks RMQ, Kadouri DE, MacEachran DP, O’Toole GA. 2009. New yeast recombineering tools for bacteria. Plasmid 62:88–97. doi: 10.1016/j.plasmid.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cornish-Bowden A. 2012. Fundamentals of enzyme kinetics, 4th ed John Wiley & Sons, Incorporated, Hoboken, NJ. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metabolic pathways of lysine catabolism in P. putida KT2440. (A) The l-lysine metabolic pathway is shown in blue, while the known steps of d-lysine metabolism are shown in red. (B) Proposed route of 2AA metabolism in P. putida. Download FIG S1, TIF file, 3.9 MB (4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Quantitative results of RB-TnSeq screen. Download Table S1, CSV file, 0.00 MB (4KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Results of RB-TnSeq screen. Data represent genes that showed greater than 2 log2 fitness on d-lysine, l-lysine, or 5AVA but a fitness defect of no less than 0.5 log2 when grown on glucose. Download FIG S2, TIF file, 3.0 MB (3.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Growth of PP_4108 mutants on lysine. (A) Growth of wild-type KT2440 and the PP_4108 mutant on d-lysine as a sole carbon source. The shaded area represents 95% CI; n = 3. (B) Growth of wild-type KT2440 and the PP_4108 mutant on l-lysine as a sole carbon source. The shaded area represents 95% CI; n = 3. (C) Time course of PP_4108 colorimetric glutamate formation. Error bars represent standard deviations; n = 2. Download FIG S3, TIF file, 2.2 MB (2.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Characterization of PP_5260. (A) In vitro reactions of boiled or native PP_5260 incubated with 2OA with 50 μM EDTA (white) or without EDTA (black). Bars represent 1og10 transformed spectral counts; errors bars represent 95% CI; n = 3 (B) LC-TOF analysis of a 2HG standard, and products of PP_5260 incubated with 2OA, and boiled control. (C) Mass spectra of 2HG standard and product of PP_5260 in vitro reaction. (D) Standard curves of d-2HG and l-2HG determined using a d-2HG-specific enzymatic detection assay. Shaded areas represent 95% CI; n = 3. Download FIG S4, TIF file, 3.8 MB (3.9MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Growth of the PP_4493 mutant on l-lysine and of wild-type KT2440 and the PP_4493 mutant on l-lysine as a sole carbon source. The shaded area represents 95% CI; n = 3. Download FIG S5, TIF file, 3.5 MB (3.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Characterization of PP_2909 (CsiD) in P. putida. (A) In vitro PP_2909 reactions. Bars show concentrations (in millimoles) of succinate (blue) or 2HG (green) formed by boiled enzyme control, no-2KG control, or native enzyme with 2KG added. Errors bars show 95% CI; n = 3. (B) Time course of PP_2909 in vitro reaction. The plot shows 2HG, 2KG, succinate, and glutarate levels over time. The shaded region shows 95% CI; n = 3. (C) Growth curves of wild-type KT2440 or PP_0158, PP_2909, and PP_2910 single mutants or PP_2909/PP_0158 double mutants grown on either l-lysine (left column) or 5AVA (right column). The shaded region shows 95% CI; n = 3. Download FIG S6, TIF file, 5.9 MB (6.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Growth of PP_2909/PP_0158 and PP_2088 mutants on d-lysine. (A) Growth of wild-type KT2440 and the PP_2909/PP_0158 mutant on d-lysine as a sole carbon source. The shaded area represents 95% CI; n = 3. (A) Growth of wild-type KT2440 and the PP_2088 mutant on d-lysine as a sole carbon source. The shaded area represents 95% CI; n = 3. Download FIG S7, TIF file, 2.1 MB (2.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Statistical analysis of proteomics. Download Table S2, CSV file, 0.05 MB (48.9KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Additional experimental methods. Download Text S1, DOCX file, 0.01 MB (15.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.