Abstract
Aim: Type 2 Diabetes Mellitus (T2DM) is associated with both microvascular complications such as diabetic retinopathy (DR), and macrovascular complications like coronary artery disease (CAD). Genetic risk factors have a role in the development of these complications. In the present case-control study, we investigated genetic variations associated with DR and CAD in T2DM patients from the United Arab Emirates.
Methods: A total of 407 Emirati patients with T2DM were recruited. Categorization of the study population was performed based on the presence or absence of DR and CAD. Seventeen Single Nucleotide Polymorphisms (SNPs), were selected for association analyses through search of publicly available databases, namely GWAS catalog, infinome genome interpretation platform and GWAS Central database. A multivariate logistic regression test was performed to evaluate the association between the 17 SNPs and DR, CAD, or both. To account for multiple testing, significance was set at p < 0.00294 using the Bonferroni correction.
Results: The SNPs rs9362054 near the CEP162 gene and rs4462262 near the UBE2D1 gene were associated with DR (OR = 1.66, p = 0.001; OR = 1.37, p = 0.031; respectively), and rs12219125 near the PLXDC2 gene was associated (suggestive) with CAD (OR = 2.26, p = 0.034). Furthermore, rs9362054 near the CEP162 gene was significantly associated with both complications (OR = 2.27, p = 0.0021). The susceptibility genes for CAD (PLXDC2) and DR (UBE2D1) have a role in angiogenesis and neovascularization. Moreover, association between the ciliary gene CEP162 and DR was established in terms of retinal neural processing, confirming previous reports.
Conclusions: The present study reports associations of different genetic loci with DR and CAD. We report new associations between CAD and PLXDC2, and DR with UBE2D1 using data from T2DM Emirati patients.
Keywords: type 2 diabetes mellitus, diabetic retinopathy, coronary artery disease, single nucleotide polymorphism, United Arab Emirates, Arab population
Introduction
Diabetes is among the largest global health emergencies in the twenty-first century, as it creates a major human and financial burden worldwide (1). The International Diabetes Federation (IDF) has estimated that 415 million people have been diagnosed with diabetes in 2015, and the number is expected to rise to 642 million by 2040. The financial burden associated with diabetes worldwide has been estimated to be 5–20% of total health expenditure (1). Within the United Arab Emirates (UAE), the prevalence of diabetes in adults between the age of 20–79 years old, was found to be 19.3% in 2015, with diabetes associated costs per individual reaching 2155.9 USD (1). Diabetes prevalence in the UAE is among the top prevalence rates in the Middle East and North Africa (MENA) region, with comparable levels to Saudi Arabia (20.0%) and Kuwait (20.0%); while showing higher prevalence levels as compared to other countries in the MENA region such as Egypt (16.7%), Jordan (11.7%), and Oman (14.8%) as reported by IDF(1).According to the IDF, type 2 diabetes mellitus (T2DM) is the most common type of diabetes (1), accounting for more than 90% of patients diagnosed with diabetes (2). Contributing to the human and financial costs of T2DM is that it is a complex multifactorial disease resulting from a combination of genetic, environmental, and behavioral risk factors that manifest in multiorgan dysfunction (3).
T2DM can lead to a number of long-term serious complications and health problems. Consistently high blood glucose levels (BGL) can seriously damage the heart, blood vessels, vision, the nervous system, and kidney. Therefore, T2DM is commonly associated with microvascular as well as macrovascular complications including diabetic retinopathy (DR), and coronary artery disease (CAD), respectively.
CAD is defined as a complex disease resulting from an interplay between lifestyle, environmental and genetic factors (4). Globally, diabetes is recognized as a major cause of CAD (5), which in turn is ranked as number one cause of mortality worldwide (4–6). Adults with diabetes are reported to have a higher death rate by 2 to 4-folds from CAD, as compared to adults without diabetes (5). Identified common risk factors associated with CAD include hypertension, smoking, increased age, diabetes mellitus, male-gender, diet high in fat, elevated low-density lipoprotein (LDL) cholesterol, reduced plasma high-density lipoprotein (HDL) cholesterol, increased triglycerides, and increased plasma total cholesterol (4, 6).
A study by Al-Maskari et al. reported CAD prevalence levels of 14.4% among UAE residents with diabetes in Al-Ain city, with age (p = 0.04), diabetes duration (p = 0.002), and hypertension (p = 0.04) reported as major risk factors (7). CAD prevalence of 17.8% among diabetic population from Yemen was reported while a prevalence of 23.7% for CAD among diabetic individuals was reported in Iran (8). Reduction in mortality and morbidity by 30–40% has been reported by studies targeting such risk factors and illustrating the importance of preventive measures against diabetes and CAD (6). Yet, genetic predisposition is estimated to account for 40–60% of CAD susceptibility (6) as concluded from familial and twin studies (5). Such studies highlight the importance of investigating not only common risk factors, but also genetic risk factors associated with CAD in patients with diabetes in order to provide early prevention schemes to reduce the mortality rates caused by CAD among patients with diabetes.
Diabetic Retinopathy (DR) is another serious complication of T2DM and is the most common cause of blindness for adults in developed countries (9). Deterioration of vision implicated in DR is a gradual process starting from mild non-proliferative diabetic retinopathy (mild-NPDR), to moderate and severe non-proliferative diabetic retinopathy (NPDR), finally to proliferative diabetic retinopathy (PDR) (10). Prevalence of DR is related to a number of common risk factors including diabetes duration, poor glycemic control, hypertension and dyslipidemia (5, 9–11). In the UAE, the prevalence of DR with diabetes was found to be 19% in Al-Ain (12). Prevalence of DR among different diabetic populations was reviewed by Zabetian et al. (8), and reported as follows; Saudi Arabia (30.0%), Qatar (23.5%), and Oman (16.2%). In addition to common risk factors, genetic risk factors have been reported to play an important role in the development of DR, where their impact accounts for 25–50% of DR risk (11).
Several Genome Wide Association Studies (GWAS) have identified possible genes associated with DR and CAD (13–16), with different degrees of genetic associations. For instance, the GLUL gene in human endothelial cells is associated with CAD in the T2DM European population (17), while the GRB2 gene is associated with DR in T2DM Australian patients and upregulated in neovascularization and retinal stress (18). To date, GWAS has been conducted on different ethnic groups. However, no extensive studies from the Middle Eastern population have been conducted (19).
The aim of the present study is to investigate common genetic variants (17 single nucleotide polymorphisms, SNPs) that have been reported to increase risk of diabetic complications including diabetic retinopathy (DR), coronary artery disease (CAD), or a combination of these two (R+CAD) in a case-control study in an Arab population in the UAE. This can help in establishing a comprehensive prevention program in the future for the diabetic Emirati population by considering early detection for T2DM complications in patients with certain SNPs preventatively.
Methods
Subjects and Sample Collection
Study subjects were enrolled in during routine visits to the endocrinology and cardiology clinics at Sheikh Khalifa Medical Centre (SKMC) and Mafraq Hospital in Abu Dhabi city, in the period between July 2014 and May 2015. The study cohort consisted of 407 (234 females and 173 males), unrelated patients diagnosed with T2DM from the UAE. The Institutional Ethics Committees of SKMC and Mafraq Hospital both reviewed and approved the study (REC-04062014 and R292, respectively). All participants provided written consent in accordance with the Helsinki Declaration of ethical conduct in research.
A qualified physician confirmed the presence of diabetes associated complications, as outlined in the criteria by the World Health Organization (WHO) consultation group report (20). Subjects included in this study were UAE-born nationals, diagnosed with T2DM, can give consent and older than 18 years of age when enrolled in the study. Subjects who could not give consent, who were pregnant at the time of enrollment or who were diagnosed with other pathophysiology like cancer or psychosis, were excluded from this study.
Biochemistry tests were performed at the time of enrollment. Blood pressure was taken at two different time points, 48 h apart, and the average was taken. Diagnosis criteria of hypertension and dyslipidemia were previously defined in Jelinek et al. (21)
The major complications of T2DM considered in this study were coronary artery disease and retinopathy. Diagnosis of cardiovascular disease was obtained from the medical records using ICD-9 diagnostic codes 410–414, and verified by the consulting physician. Diagnosis of retinopathy was defined according to WHO criteria (22), by the presence of either white or red lesions (non-proliferative or proliferative retinopathy) or the presence of both in the retina.
Genotyping
The GWAS data were obtained from 490 individuals resident in the United Arab Emirates (UAE). Genotyping was performed on the Infinium Omni5ExomeHuman chip according to the manufacturer's protocols (Illumina Inc., San Diego, USA), and raw data was collated on the GenomeStudio v2010.3 (Illumina Inc., San Diego, USA). The microarray contained 4,641,218 SNPs. Quality control (QC) on the data was performed using the PLINK software (version 1.07) (23) to remove SNPs with a minor allele frequency (MAF) < 0.05, with >5% missing genotype rate, failing the Hardy-Weinberg equilibrium (HWE) test at the 0.000001 significance level and Mendelian error. Approximately 39% of SNPs passed these QC criteria. Samples that failed quality control were also excluded from the analysis. The average call rate for the remaining 490 samples was 98.99%. Out of the 490 genotyped samples available in the Emirates Family Registry (EFR) database (24), 411 genotyped samples included available data on diabetes-associated complications. As a final exclusion step, four individuals were reported as patients diagnosed with type 1 diabetes mellitus (T1DM) and were excluded; hence, resulting in the inclusion of 407 individuals in the study.
SNPs Selection
A search was performed using different resources, with the aim of selecting SNPs that have been reported with CAD or DR in individuals with T2DM. Further SNPs reported in association with CAD or retinopathy (R) in individuals without T2DM, were also selected. The following resources were utilized: GWAS catalog: https://www.ebi.ac.uk/gwas/home, infinome genome interpretation platform: https://www.infino.me/, and GWAS Central database: http://www.gwascentral.org/. Forty-three SNPs were initially retrieved from searching the aforementioned resources, reported in association either with CAD or R. Next, a search was performed on the database established using Emirates Family Registry (EFR)(24), which resulted in selecting 17 SNPs with significant and suggestive associations with either CAD or R (P<5 × 10−5). The 17 SNPs are reported in Table 2, as SNPs found in the Emirati cohort under study. The current study focused on DR and CAD as T2DM associated complications, given that DR was reported as the single most prevalent T2DM complication among Emiratis, followed by CAD prevalence, as reported by Jelinek et al. (21).
Table 2.
The study selected SNPs according to their previous reports, together with reported associated traits and ethnicities.
| SNP† ID | Gene(s) | Chr: BP | Associated population | Associated trait | References |
|---|---|---|---|---|---|
| rs7553035 | RD3 | 1: 211691706 | European | R‡ | (25) |
| rs10004839 | LOC105377441 | 4: 138154812 | European | R‡ | (25) |
| rs6472155 | LOC105375878; LOC105375879 | 8: 65730207 | European | R‡ | (25) |
| rs17194885 | SRC | 20: 36068389 | European | R‡ | (25) |
| rs2811893 | MYSM1 | 1: 59162148 | Taiwanese | DR | (14) |
| rs17376456 | KIAA0825; LOC105379087 | 5: 93557702 | Taiwanese | DR | (14) |
| rs12219125 | PLXDC2 | 10: 20593087 | Taiwanese | DR | (14) |
| rs4838605 | ARHGAP22 | 10: 49699957 | Taiwanese | DR | (14) |
| rs4462262 | LOC105378313; LOC105378314 | 10: 59189178 | Taiwanese | DR | (14) |
| rs2038823 | HS6ST3 | 13: 96951433 | Taiwanese | DR | (14) |
| rs9362054 | LINC01611 | 6: 85178268 | Japanese | DR | (13) |
| rs9543976 | UCHL3 | 13: 76136648 | Chinese | DR | (26) |
| rs646776 | CELSR2 | 1: 109818530 | European | CAD‡ | (15) |
| rs4977574 | CDKN2B-AS1 | 9: 22098574 | European | CAD‡ | (15) |
| rs8055236 | CDH13 | 16: 83212398 | European(British) | CAD‡ | (27) |
| rs10911021 | LOC105371642; ZNF648 | 1: 182081960 | European | CAD | (17) |
| rs7901695 | TCF7L2 | 10: 114754088 | African Americans | CAD | (16) |
BP, Base-pair position; CAD, Coronary artery disease; Chr., Chromosome number; DR, Diabetic retinopathy; R, retinopathy; SNP, Single Nucleotide Polymorphism. Individuals diagnosed with R or CAD but without T2DM.
Statistical Analysis
Statistical analyses for demographic, clinical and laboratory data were performed using Microsoft Excel and R (28) software was utilized in performing graphical normality tests of distribution. Continuous variables results were expressed as mean ± standard deviation, or as median and interquartile range (IQR) for highly-skewed distributions; while categorical variables were presented as counts and percentages. For comparisons between cases and controls, the Pearson chi-square test was utilized for categorical variables or Fisher's exact test when expected frequencies were < 5. For continuous variables statistical differences were assessed using two-tailed Student t-tests for normally-distributed data, or using the Wilcoxon rank-sum (Mann-Whitney) test for highly-skewed data. Results were considered of statistical significance when the p-value was < 0.05. Data quality control was implemented using the Hardy-Weinberg Equilibrium (HWE) test via Plink Software (23), where a P < 0.005 indicates significant deviations from HWE, reflecting issues such as population stratification (23, 29) or genotyping errors (29). Such SNPs were excluded from the analysis to avoid biased conclusions. For the purpose of assessing genetic risk factors that are associated with development of diabetic retinopathy (DR), or coronary artery disease (CAD), or both of these complications (DR+CAD), a multivariate logistic regression test in Plink Software (23) was performed to evaluate the association between a T2DM complication as a result (response or dependent variable) and the presence of a SNP (predictor or independent variable). Furthermore, adjusting for covariates was considered in generating measures of association and significance within the statistical modeling, where covariate adjustment is based on factors which showed a significant difference between cases and controls. Covariate adjustment was conducted for each group in the study cohort as follows; for the R vs. no R group, age and diabetes duration were considered as covariates; for the CAD vs. no. CAD group, gender, age, hypertension, dyslipidemia and smoking behavior were adjusted for as covariates; finally for the CAD+DR vs. no CAD and no DR. group, age, diabetes duration, hypertension, and dyslipidemia were considered as covariates. The Bonferroni correction was performed to account for multiple testing that is conducted in this study, where the α-value is adjusted from 0.05 to a new α-value = (0.05/N) (30), where N refers to the number of statistical test performed. In the current study, 17 SNPs were investigated in association with certain T2DM complications. Therefore, a new α-value = (0.05/17) = 0.002941 is set. Results from our multiple testing approach describe a SNP as “significantly” associated SNP when p < 0.002941. SNPs that show a direction of association (when a p < 0.05 yet did not reach significance of new α-value) are reported as SNPs that are “suggestively” associated with a complication. SNPs with suggestive association are worth investigating in the future with a larger cohort size.
Results
Characteristics of Study Subjects
Demographic data, clinical and laboratory data of participants are summarized in Table 1. A total sample of 407 T2DM Emirati patients were recruited. Categorization of study population into 3 groups was performed based on the presence of diabetes and the presence or absence of diabetic retinopathy (DR) and coronary artery disease (CAD). For category 1, patients diagnosed with DR had significantly higher age and a higher reported diabetes duration as compared to controls without DR. In category 2, T2DM patients with CAD had also significantly higher age with higher total cholesterol, LDL-cholesterol, hypertension, and presence dyslipidemia. Significantly more male patients presented with CAD and indicated longer smoking history. For category 3, patients with both conditions had similar results to category 2, with higher reported diabetes duration (Table 1).
Table 1.
Demographic, clinical, and laboratory data of the study subjects.
| Type of variables | Variable | DR‡ (n = 202) | No DR (n = 205) | P* | CAD (n = 160) | No CAD (n = 245) | P* | DR + CAD (n = 76) | No DR and No CAD (n = 121) | P* |
|---|---|---|---|---|---|---|---|---|---|---|
| Demographic | Gender: Female† | 124 (61%) | 110 (54%) | 0.11 | 74 (46%) | 159 (65%) | 0.00021 | 37 (49%) | 73 (60%) | 0.11 |
| Age (years) | 63 ± 11 | 60 ± 11 | 0.0034 | 66 ± 9.3 | 59 ± 11 | < 0.0001 | 67 ± 9.7 | 57 ± 12 | < 0.0001 | |
| Clinical variables | Diabetes duration (years)** | 14 (5.5, 20) | 7 (4, 15) | < 0.00001 | 9 (5, 20) | 12 (5, 20) | 0.070 | 13 (5, 20) | 9 (5, 16) | 0.022 |
| Clinical hypertension† | 169 (84%) | 167 (82%) | 0.71 | 152 (95%) | 183 (75%) | < 0.0001 | 74 (97%) | 89 (75%) | 0.0001 | |
| Dyslipidemia† | 193 (96%) | 184 (90%) | 0.052 | 158 (99%) | 218 (89%) | 0.0002 | 75 (99%) | 101 (84%) | 0.0005 | |
| Smoking† | 50 (25%) | 59 (29%) | 0.34 | 53 (33%) | 55 (23%) | 0.018 | 25 (33%) | 31 (26%) | 0.29 | |
| Diabetes family history† | 136 (68%) | 139 (68%) | 0.88 | 100 (63%) | 174 (71%) | 0.21 | 48 (64%) | 87 (72%) | 0.51 | |
| Diabetes complications family history† | 53 (26%) | 41 (20%) | 0.12 | 32 (20%) | 62 (25%) | 0.38 | 19 (25%) | 28 (23%) | 0.81 | |
| BMI (kg/m2) | 32 ± 6.1 | 32 ± 6.4 | 0.42 | 32 ± 6.0 | 32 ± 6.4 | 0.23 | 31 ± 5.7 | 32 ± 6.4 | 0.57 | |
| Waist circumference (cm) | 107 ± 12 | 106 ± 15 | 0.43 | 108 ± 14 | 105 ± 13 | 0.050 | 108 ± 12 | 104 ± 14 | 0.075 | |
| Laboratory variable Glycemic index | HbA1c (%) | 7.8 ± 1.5 | 7.7 ± 1.7 | 0.52 | 7.9 ± 1.6 | 7.7 ± 1.6 | 0.34 | 7.8 ± 1.5 | 7.5 ± 1.7 | 0.32 |
| Lipids profile (mg/dl) | Total-cholesterol | 148 ± 37 | 153 ± 44 | 0.21 | 142 ± 38 | 156 ± 41 | 0.0011 | 139 ± 32 | 159 ± 43 | 0.001 |
| Triglycerides** | 119 (90.3, 165) | 117 (88.6, 154) | 0.48 | 116 (90.8, 160) | 118 (87.7, 164) | 0.98 | 110 (90.3, 161) | 113 (81.5, 154) | 0.51 | |
| HDL-cholesterol | 47 ± 18 | 47 ± 21 | 0.83 | 45 ± 22 | 49 ± 18 | 0.052 | 45 ± 12 | 49 ± 15 | 0.073 | |
| LDL-cholesterol | 74 ± 30 | 81 ± 37 | 0.062 | 73 ± 33 | 80 ± 34 | 0.045 | 69.8 ± 28 | 85 ± 37 | 0.0062 |
P-value: for continuous variables, computed using two-tailed t-test for normally-distributed data, or using Mann-Whitney test for highly-skewed data**. For categorical variables, computed using the Pearson chi-square test or Fisher's exact test for expected frequencies < 5.
Categorical variables are presented as counts and percentages. All remaining continuous variables are expressed as mean ± standard deviation, or as median (IQR lower, upper) for highly-skewed distributions**.
‡BMI, Body Mass Index; CAD, Coronary Artery Disease; DR, Diabetic Retinopathy; HbA1c, glycosylated hemoglobin; HDL, high-density lipoprotein; LDL, low-density lipoprotein; n, number of individuals. Bold value indicates significance level and p < 0.05.
Association Between SNPs and T2DM Complications Under Study
The 17 SNPs within our database and associated with DR and CAD complications are listed in Table 2. The results of associations between SNPs and T2DM complications (DR and CAD) considering gender, age, diabetes duration, dyslipidemia, hypertension and smoking behavior as covariates are shown in Table 3, where covariate adjustment was performed for each group within the study cohort based on factors which showed a significant difference between cases and controls. Furthermore, ORs of confounding factors in Table 3 give an indication of the relative contribution of each factor to the others. Rs9362054 near the CEP162 gene, and rs4462262 near the UBE2D1 gene were associated with increased risks of DR (DR vs. no DR) with rs9362054 having an odds ratio of nearly 2 and highly significant. For CAD (CAD vs. no CAD) rs12219125 near the PLXDC2 gene showed suggestive association, with an increased risk of CAD with an odds ratio over 2. For the DR+CAD category (DR + CAD vs. no DR and no CAD), rs9362054 was significantly associated with the presence of both complications, while rs17376456 in the KIAA0825 gene showed suggestive association with both complications, with rs17376456 having an odds ratio below 1 and indicative of a possible protective function with both complications less likely to occur.rs9362054 again doubling the odds ratio for presence of the two complications.
Table 3.
Results of significant and suggestive associations between tested SNPs and T2DM complication: DR, CAD, or a combination of both.
| SNP† ID | Gene(s) | Chr: BP | A1/A2 | MAF Cases | MAF Controls | OR (CI 95%)‡ | P-value |
|---|---|---|---|---|---|---|---|
| DR vs. no DR | |||||||
| rs9362054 | LINC01611 | 6: 85178268 | A/G | 39% | 50% | 1.66 (1.23–2.24) | 0.001 |
| Age: 1.02 (1.00–1.04) | |||||||
| Diabetes duration: 1.05 (1.03–1.08) | |||||||
| Unadjusted: 1.62 (1.21–2.16) | 0.0011 | ||||||
| rs4462262 | LOC105378313; LOC105378314 | 10: 59189178 | A/G | 39% | 46% | 1.37 (1.03–1.81) | 0.031 |
| Age: 1.02 (1.00–1.70) | |||||||
| Diabetes duration: 1.05 (1.03–1.08) | |||||||
| Unadjusted: 1.31 (1.00–1.73) | 0.050 | ||||||
| CAD vs. no CAD | |||||||
| rs12219125 | PLXDC2 | 10: 20593087 | A/C | 3.8% | 6.3% | 2.26 (1.06–4.81) | 0.034 |
| Gender: 0.43 (0.24–0.75) | |||||||
| Age: 1.06 (1.03–1.08) | |||||||
| Hypertension: 3.74 (1.65–8.48) | |||||||
| Dyslipidemia: 5.20 (1.12–24.2) | |||||||
| Smoking: 0.85 (0.47–1.54) | |||||||
| Unadjusted: 1.79 (0.89–3.59) | 0.10 | ||||||
| DR + CAD vs. no DR and no CAD | |||||||
| rs17376456 | KIAA0825; LOC105379087 | 5: 93557702 | G/A | 14% | 9.9% | 0.429 (0.193–0.952) | 0.037 |
| Age: 1.08 (1.04–1.13) | |||||||
| Diabetes duration: 1.01 (0.97–1.05) | |||||||
| Hypertension: 5.60 (1.18–26.6) | |||||||
| Dyslipidemia: 4.56 (0.50–41.3) | |||||||
| Unadjusted: 0.607 (0.312–1.18) | 0.14 | ||||||
| rs9362054 | LINC01611 | 6: 85178268 | A/G | 36% | 50% | 2.27 (1.35–3.83) | 0.0021 |
| Age: 1.08 (1.04–1.13) | |||||||
| Diabetes duration: 1.02 (0.98–1.06) | |||||||
| Hypertension: 4.52 (0.95–21.5) | |||||||
| Dyslipidemia: 5.03 (0.55–45.7) | |||||||
| Unadjusted: 1.89 (1.21–2.93) | 0.0047 | ||||||
SNP, Single-nucleotide Polymorphism; Chr, chromosome number; A1, minor allele; A2, major allele; MAF, Minor Allele Frequency; OR, Odds Ratio; CI, Confidence Interval; DR, Diabetic Retinopathy; CAD, Coronary Artery Disease; LINC01611, long intergenic non-protein coding RNA 1611; LOC105378313, uncharacterized LOC105378313; LOC105378314, uncharacterized LOC105378314; PLXDC2, plexin domain containing 2; KIAA0825, protein-coding gene with uncharacterized protein KIAA0825; LOC105379087, uncharacterized LOC105379087. Adjusted OR values are illustrated, where adjustment was performed using factors that show a significant difference between cases and controls for each group within the study cohort.
A2, the major allele, was used as a reference allele in logistic regression tests; Bold and italic values indicates the significance level and p < 0.05.
Comparison Between Emirati Population and Reference Populations
We next compared the results of the identified SNPs based on results that were previously reported in different ethnic groups, as shown in Table 4. The SNP rs9362054 near CEP162 was previously reported by Awata et al. as a locus for DR in the Japanese population, with 1.40-fold increase in DR risk (13). Huang et al. reported significant association of the SNPs rs12219125 and rs17376456 with DR in the Taiwanese population with a 1.62- and 3.63-fold increases in DR risk, respectively (14). In the current Emirati patients study, rs12219125 is suggestively associated with CAD, with an increase of 2.26-fold.
Table 4.
Comparison between SNPs reported with T2DM complication in this study: DR, CAD, or both, with previous reports.
| SNP† ID | Chr. No. | Gene(s) | Ref. population p-value | Ref. population OR | Our reported p-value | Reported trait | Our reported trait | References |
|---|---|---|---|---|---|---|---|---|
| DR vs. no DR | ||||||||
| rs9362054 | 6 | LINC01611 | 1 × 10−6 | 1.40 | 0.001 | DR, T2DM, Japanese | DR, T2DM, Emirati | (13) |
| rs4462262 | 10 | LOC105378313; LOC105378314 | 9 × 10−8 | – | 0.031 | DR, T2DM,Taiwanese | DR, T2DM,Emirati | (14) |
| CAD vs. no CAD | ||||||||
| rs12219125 | 10 | PLXDC2 | 9 × 10−9 | 1.62 | 0.034 | DR, T2DM,Taiwanese | CAD, T2DM, Emirati | (14) |
| DR + CAD vs. no DR and no CAD | ||||||||
| rs17376456 | 5 | KIAA0825; LOC105379087 | 3 × 10−15 | 3.63 | 0.037 | DR, T2DM,Taiwanese | DR+CAD, T2DM, Emirati | (14) |
| rs9362054 | 6 | LINC01611 | 1 × 10−6 | 1.40 | 0.0021 | DR, T2DM, Japanese | DR+CAD, T2DM, Emirati | (13) |
CAD, Coronary artery disease; Chr.No, Chromosome number; DR, Diabetic retinopathy; KIAA0825, protein-coding gene with uncharacterized protein KIAA0825; LINC01611, long intergenic non-protein coding RNA 1611; LOC105378313,uncharacterized LOC105378313; LOC105378314,uncharacterized LOC105378314; OR, Odds ratio; PLXDC2,plexin domain containing 2; Ref, Reference; SNP, Single Nucleotide Polymorphism; T2DM, Type 2 diabetes mellitus.
Discussion
Diabetic retinopathy and coronary artery disease are clinically significant complications of T2DM that lead to increased morbidity and mortality. Genetic risk factors were found to play an important role in the susceptibility of these complications. Previous studies have reported a 25–50% genetic risk for DR (11) and 40–60% risk for CAD (6). Genetic risk factors may act solely or in combination with primary risk factors such as hypertension and level of cholesterol, leading to these diabetic complications.
In the present study, we investigated genetic risk factors associated with DR, CAD or a combination of both complications in a case-control study of T2DM Emirati patients. In the following discussion, we will also present the possible functions of the genetic loci associated with each category in an attempt to identify possible pathways that may be involved in the etiology of DR or CAD. Demographic, clinical, and laboratory data in the present study indicated a different distribution of risk factors that may contribute to the development of the tested traits. Although some studies report the “male gender” as a risk factor for T2DM as reported by Nordstrom et al. (31) yet it is attributed it to the effect of differences in visceral fat mass between males and females; indicative of visceral fat mass effect rather than gender. Nevertheless, studies from the UAE either do not report gender as a risk factor as reported recently by Sulaiman et al. (32) or report female predominance in terms of T2DM incidence among UAE residents (33). As reported by Ali et al. (33) restricted outdoor physical activities due to sociocultural norms as well as the lack of culturally-sensitive exercise facilities contribute to lower physical activity and higher predominance among female residents in the UAE.
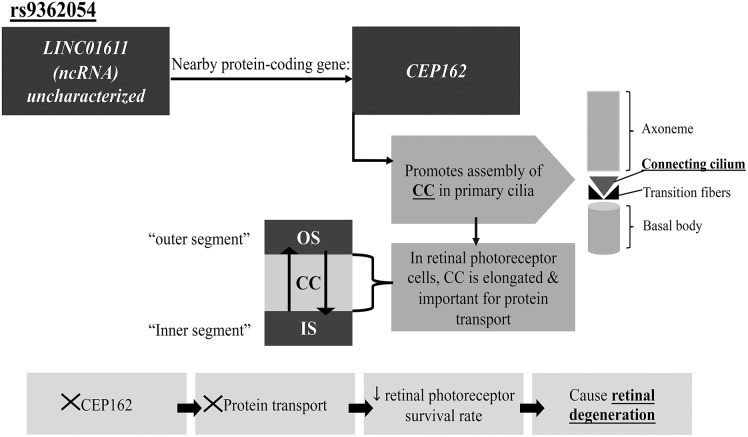
The top SNP, rs9362054, significantly associated with DR in the T2DM Emirati cohort, is located on the LINC01611 gene or alternatively called RP1-90L14.1, a long intergenic non-protein coding RNA 1611. Although the SNP, rs9362054 also shows a significant association with DR+CAD (in the DR+CAD vs. no DR and no CAD group), to our current knowledge its potential association with CAD has not yet been established in literature. Therefore, rs9362054 is discussed herein only in the context of DR. The SNP rs9362054 is an intronic variant, and was previously reported to be associated with DR in a Japanese population using the GWAS approach (13). The nearest protein-coding gene to LINC01611 (using UCSC database), is CEP162 (centrosomal protein 162), which is alternatively called QN1. A connection between the long non-coding RNA LINC01611 gene and CEP162 gene may be possible via cis-regulation exerted by cis-acting lncRNA, which controls expression of genes nearby the lncRNA transcription sites (34). CEP162 protein is utilized in promoting the assembly of the transition zone of the primary cilia found on the apical surface of the majority of mammalian cells in G0/G1 phase of the cell cycle (35). Primary cilia are important in cell signaling, thermo-, mechano- and chemosensation (30–32, 35, 36). Defects in cilia are found to be associated with several disorders, including retinal degeneration, liver, and kidney diseases, a group of disorders termed ciliopathies (36). Ciliary dysfunction affects retinal photoreceptors; where a transport process across the retinal connecting cilium is disrupted, leading to reduced survival of retinal photoreceptor cells (36, 37). CEP162 loss halts the ciliogenesis process, specifically the assembly of the connecting cilium stage (35), which in turn affects protein transport process and resulting in retinal degeneration (36) (Figure 1). However, the effect of dysfunctional neurons on retinal vasculature physiology and survival remains poorly understood (38). In transgenic rats (TGR) overexpressing a mutant cilia gene (38), encoding polycystin-2 protein, resulted in neuronal death and subsequent retinal degeneration. The ciliary protein polycystin-2 is located in the connecting cilium and its dysfunctional expression results in defective protein transport across the connecting cilium. The study further reported similar phenotypes to diabetic retinopathy including vasoregression, loss of endothelial capillary cells and pericytes (38). This association between a ciliary gene and DR was previously discussed in Awata et al. (13) and verifies our finding that a similar process is present in the Emirati patient group.
Figure 1.
Proposed pathway by which rs9362054 SNP can be associated with DR. CEP162, centrosomal protein 162; CC, Connecting cilium; IS, Inner segment; ncRNA, Non-coding RNA; OS, Outer segment. See text for further details.
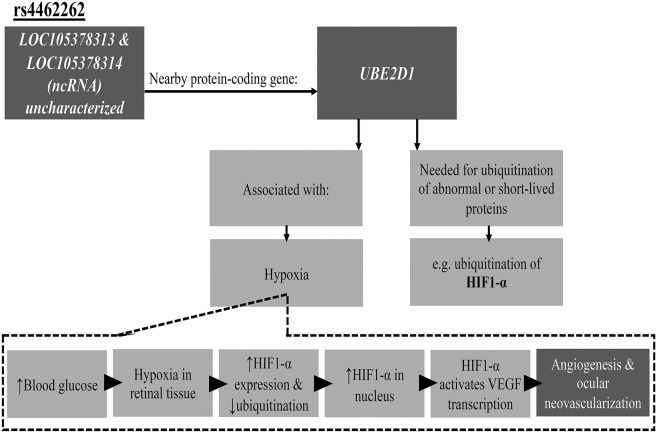
Another SNP suggestively associated with DR in our study is rs4462262. This is an intergenic SNP, which is located between the uncharacterized genes LOC105378313 and LOC105378314. This SNP was previously reported to be associated with DR in a Taiwanese population by a GWAS approach (14). A protein-coding gene, UBE2D1 (ubiquitin conjugating enzyme E2 D1), located near rs4462262 was previously investigated for function and possible involvement in the development of retinopathy. UBE2D1 is also known as UBCH5. UBE2D1 is one of the three classes of ubiquitination enzymes. It is a ubiquitin-conjugating enzyme (E2 class), which accepts ubiquitin from ubiquitin-activating enzymes (E1 class) and catalyzes the covalent attachment of ubiquitin to other proteins. Hence, contributing to the selective degradation of short-lived or abnormal proteins (39). UBE2D1 interacts with enzymes from E1 and E3 classes in the ubiquitination of hypoxia-inducible factor alpha subunit (HIF1-α). HIF1-α is an important regulator of oxygen homeostasis (40), and is associated with pathological conditions that are caused by hypoxia and retinal ischemia (41). The expression of HIF1-α is up-regulated by hypoxia during normal retinal development (41). In diabetes mellitus, elevated blood glucose levels negatively affect retinal capillaries, resulting in their incompetence on functional and anatomical levels that manifest as retinal hypoxia that can lead to proliferative retinopathy. Persistently-high glucose levels induce further damage to retinal capillaries and vessels that eventually result in hypoxia (41). Under hypoxic conditions, levels of HIF1-α protein are elevated and its ubiquitinated portion decreases, which results in HIF1-α accumulation in the nucleus (Figure 2). Consecutively, HIF1-α activates the transcription of different target genes, including the Vascular Endothelial Growth Factor (VEGF), which is involved in angiogenesis and ocular neovascularization (41, 42), a characteristic of diabetic retinopathy. The detailed hypothetical model of the association between ubiquitin-conjugating enzyme UBE2D1 and HIF1-α in the development of DR is shown in Figure 2.
Figure 2.
Proposed pathway illustrating association of rs4462262 SNP with DR. HIF1-α, hypoxia-inducible factor 1-alpha; ncRNA, Non-coding RNA; UBE2D1, ubiquitin conjugating enzyme E2 D1; VEGF, Vascular Endothelial Growth Factor. See text for further details.
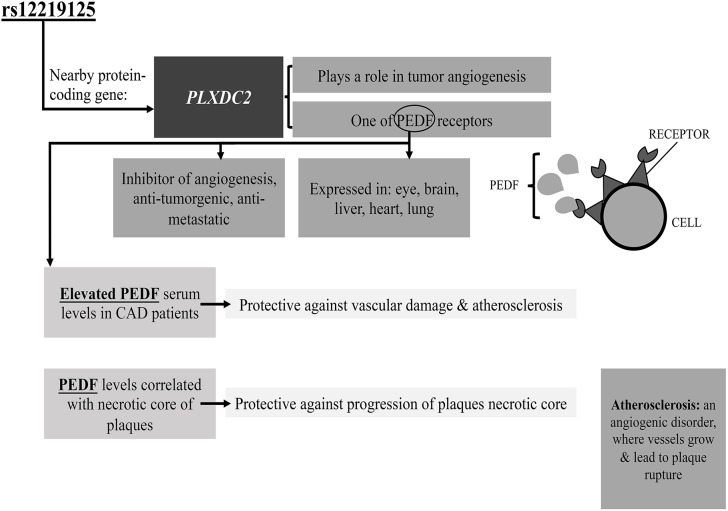
Finally, rs12219125 showed suggestive association with CAD in T2DM Emirati patients, and was previously reported to be significantly associated with DR in a Taiwanese population (14). This is the first study to investigate the association of the rs12219125 SNP with CAD in T2DM patients. This SNP is located in an intergenic region, and the nearest protein-coding gene is PLXDC2. The protein encoded by this gene, which is known as Plexin domain-containing protein 2, was suggested to play a role in tumor angiogenesis (43). Furthermore, a study by Cheng et al. identified the transmembrane protein PLXDC2 to be one of the cell-surface receptors for Pigment Epithelium Derived Factor (PEDF) (44). PEDF is expressed in different tissues including eye, brain, liver, heart, and lung, where it is an important inhibitor of angiogenesis, with an anti-tumorigenic, anti-thrombotic, and anti-metastatic function (44). It also has been utilized in the treatment of eye diseases including diabetic retinopathy, ischemic retinopathy and age-related macular degeneration, where reduced PEDF levels in the eye are associated with the susceptibility to these disorders (44). The hypothetical model for association between PLXDC2-PEDF and DR is shown in Figure 3. One previous study agreeing with our results, reported that elevated serum PEDF levels were observed in patients diagnosed with coronary artery disease (CAD) (45).These higher levels of PEDF may act as a protective mechanism in response to vascular damage and atherosclerosis (45). In addition to inhibition of angiogenesis, PEDF inhibits inflammation and cardiovascular remodeling and is an important target in the prevention of atherogenesis and necrotic core progression. Furthermore, PEDF serum level was found to be correlated with necrotic core progression during statin therapy, which suggests a protective response mechanism aimed at protecting against core progression (46).
Figure 3.
Proposed pathway by which rs12219125 SNP may be associated with CAD as a T2DM complication. PEDF, Pigment Epithelium Derived Factor; PLXDC2, plexin domain containing 2. See text for further details.
Studies investigating the association of the SNPs rs9362054, rs4462262, rs12219125, and rs17376456 are summarized in Table 4, including this study. In the Emirati population under study, rs12219125 is suggestively associated with CAD with increased risk of 2.26-fold, whereas in the Taiwanese population it was associated with a 1.62-fold increased risk of DR (14). Discrepancies in reported risk levels between our population and reference populations can be explained by the larger number of subjects in the Japanese and Taiwanese GWA studies in comparison to our study. Moreover, differences in confounding factors for adjustment of logistic regression models between our study cohort, the Japanese and the Taiwanese cohorts can further contribute to discrepancies, which is dependent on each cohort under study and is based on factors that show significant difference between cases and controls. For example, adjusting for hypertension state as a confounding factor was performed in the current study, while it was not considered as a confounder in the Japanese and Taiwanese GWA studies despite significant difference between cases and controls. One of the strengths of the present study is that patients were recruited from the two largest hospitals in Abu Dhabi, UAE; making our sample fairly representative of Emirati patients. Nonetheless, a limitation of this study would be the small sample size (n = 407). Another limitation inherent to the current study is that the logistic analyses performed did not include treatment approaches as most patients are diagnosed with multiple conditions and are undertaking several treatments which would affect the analyses stability if multiple drug classes were to be adjusted for. A future study with a larger sample size is important in confirming these findings in association with T2DM in the Emirati population A future study with a larger sample size is important in confirming these findings in association with T2DM in the Emirati population which can help in the future for early detection of complication for T2D patients with certain SNPs for a preventative manner.
In conclusion, our study findings contribute to the understanding of the genetic susceptibility of CAD and DR in patients with T2DM. We report associations of CAD with the PLXDC2 gene and DR with the UBE2D1 gene, both of these genes may contribute to DR and CAD as part of diabetes progression by playing a role in angiogenesis and neovascularization. Moreover, association between the ciliary gene CEP162 and DR was established in terms of neural processing in the retina, and confirms findings that were previously reported. Although our findings require further investigation using a larger sample size they present potential pathways that may contribute to the etiology of DR or CAD in patients with T2DM. Our study is the first in the Middle East region to report genetic risk factors associated with DR or CAD.
Ethics Statement
The Institutional Ethics Committees of Sheikh Khalifa Medical Center (SKMC) and Mafraq Hospital in Abu Dhabi city both reviewed and approved the study (REC-04062014 and R292, respectively). All participants provided written consent in accordance with the Helsinki Declaration of ethical conduct in research.
Author Contributions
SA wrote the manuscript and performed genetic association tests with support from HA. HA contributed to manuscript writing, analyses and study design. WO contributed to genetic association analyses and study design. SL, KK, AK, WA, and HJ have all contributed to the study design. HA supervised the project. All authors discussed the results and contributed to the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the contribution of participating patients whose cooperation made this study possible.
Glossary
Abbreviations
- UAE
United Arab Emirates
- T2DM
type 2 diabetes mellitus
- BGL
blood glucose levels
- DR
diabetic retinopathy
- CAD
coronary artery disease
- LDL
low-density lipoprotein
- HDL
high-density lipoprotein
- NPDR
non-proliferative diabetic retinopathy
- PDR
proliferative diabetic retinopathy
- GWAS
Genome Wide Association Studies
- SNPs
single nucleotide polymorphisms
- WHO
World Health Organization
- HWE
Hardy-Weinberg Equilibrium
- UBE2D1
ubiquitin conjugating enzyme E2 D1
- HIF1-α
hypoxia-inducible factor alpha subunit
- VEGF
Vascular Endothelial Growth Factor
- PLXDC2
Plexin domain-containing protein 2
- PEDF
Pigment Epithelium Derived Factor.
Footnotes
Funding. This study was supported by research incentive funds from Khalifa University Internal Research Fund Level 2 granted to HA.
References
- 1.IDF Diabetes Atlas - 7th Edition Int Diabetes Fed IDF-Diabetes Atlas 7th Ed 2015. Available online at: http://www.diabetesatlas.org/ (accessed August 15, 2016).
- 2.Widmaier EP, Raff H, Strang KT, Vander AJ. Vander's human physiology: the mechanisms of body function. In: Vander's Human Physiology: The Mechanisms of Body Function. 12th Ed. New York, NY: McGraw-Hill (2011). p. 581–583. [Google Scholar]
- 3.Chen L, Magliano DJ, Zimmet PZ. The worldwide epidemiology of type 2 diabetes mellitus—present and future perspectives. Nat Rev Endocrinol. (2011) 8:228–36. 10.1038/nrendo.2011.183 [DOI] [PubMed] [Google Scholar]
- 4.Wang Q. Molecular genetics of coronary artery disease. Curr Opin Cardiol. (2005) 20:182–8. 10.1097/01.hco.0000160373.77190.f1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murea M, Ma L, Freedman BI. Genetic and environmental factors associated with type 2 diabetes and diabetic vascular complications. Rev Diabet Stud RDS. (2012) 9:6–22. 10.1900/RDS.2012.9.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts R, Stewart AFR. The genetics of coronary artery disease: Curr Opin Cardiol. (2012) 27:221–7. 10.1097/HCO.0b013e3283515b4b [DOI] [PubMed] [Google Scholar]
- 7.Al-Maskari F, El-Sadig M, Norman JN. The prevalence of macrovascular complications among diabetic patients in the United Arab Emirates. Cardiovasc Diabetol. (2007) 6:24. 10.1186/1475-2840-6-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zabetian A, Keli HM, Echouffo-Tcheugui JB, Narayan KMV, Ali MK. Diabetes in the Middle East and North Africa. Diabetes Res Clin Pract. (2013) 101:106–22. 10.1016/j.diabres.2013.03.010 [DOI] [PubMed] [Google Scholar]
- 9.American Diabetes Association Microvascular complications and foot care. Diabet Care. (2016) 39:S72–80. 10.2337/dc16-S012 [DOI] [PubMed] [Google Scholar]
- 10.American Diabetes Association Diabetic retinopathy. Diabet Care. (2002) 25:S90–3. 10.2337/diacare.25.2007.S90 [DOI] [Google Scholar]
- 11.Simó-Servat O, Hernández C, Simó R. Genetics in diabetic retinopathy: current concepts and new insights. Curr Genomics. (2013) 14:289–99. 10.2174/13892029113149990008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Maskari F, El-Sadig M. Prevalence of diabetic retinopathy in the United Arab Emirates: a cross-sectional survey. BMC Ophthalmol. (2007) 7:11. 10.1186/1471-2415-7-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Awata T, Yamashita H, Kurihara S, Morita-Ohkubo T, Miyashita Y, Katayama S, et al. A genome-wide association study for diabetic retinopathy in a Japanese population: potential association with a long intergenic non-coding RNA. PloS ONE. (2014) 9:e111715. 10.1371/journal.pone.0111715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y-C, Lin J-M, Lin H-J, Chen C-C, Chen S-Y, Tsai C-H, et al. Genome-wide association study of diabetic retinopathy in a Taiwanese population. Ophthalmology. (2011) 118:642–8. 10.1016/j.ophtha.2010.07.020 [DOI] [PubMed] [Google Scholar]
- 15.Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet Lond Engl. (2011) 377:383–92. 10.1016/S0140-6736(10)61996-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. (2011) 7:e1001300. 10.1371/journal.pgen.1001300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi L, Qi Q, Prudente S, Mendonca C, Andreozzi F, di Pietro N, et al. Association between a genetic variant related to glutamic acid metabolism and coronary heart disease in individuals with type 2 diabetes. JAMA. (2013) 310:821–8. 10.1001/jama.2013.276305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burdon KP, Fogarty RD, Shen W, Abhary S, Kaidonis G, Appukuttan B, et al. Genome-wide association study for sight-threatening diabetic retinopathy reveals association with genetic variation near the GRB2 gene. Diabetologia. (2015) 58:2288–97. 10.1007/s00125-015-3697-2 [DOI] [PubMed] [Google Scholar]
- 19.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. (2016) 538:161–4. 10.1038/538161a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alberti KGMM, Zimmet PZ, WHO Consultation . Definition, diagnosis and classification of diabetes mellitus and its complications. part 1: diagnosis and classification of diabetes mellitus. Provisional report of a WHO Consultation. Diabet Med. (1998) 15:539–53. [DOI] [PubMed] [Google Scholar]
- 21.Jelinek HF, Osman WM, Khandoker AH, Khalaf K, Lee S, Almahmeed W, et al. Clinical profiles, comorbidities and complications of type 2 diabetes mellitus in patients from United Arab Emirates. BMJ Open Diabetes Res Care. (2017) 5:e000427. 10.1136/bmjdrc-2017-000427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarty CA, Taylor KI, McKay R, Keeffe JE. Diabetic retinopathy: effects of national guidelines on the referral, examination and treatment practices of ophthalmologists and optometrists. Clin Experiment Ophthalmol. (2001) 29:52–8. 10.1046/j.1442-9071.2001.d01-3.x [DOI] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. (2007) 81:559–75. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alsafar H, Jama-Alol KA, Hassoun AAK, Tay GK. The prevalence of type 2 diabetes mellitus in the United Arab Emirates: justification for the establishment of the Emirates family registry. Int J Diabetes Dev Ctries. (2012) 32:25–32. 10.1007/s13410-012-0062-6 [DOI] [Google Scholar]
- 25.Jensen RA, Sim X, Li X, Cotch MF, Ikram MK, Holliday EG, et al. Genome-wide association study of retinopathy in individuals without diabetes. PloS ONE. (2013) 8:e54232. 10.1371/journal.pone.0054232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheu WH-H, Kuo JZ, Lee I-T, Hung Y-J, Lee W-J, Tsai H-Y, et al. Genome-wide association study in a Chinese population with diabetic retinopathy. Hum Mol Genet. (2013) 22:3165–73. 10.1093/hmg/ddt161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. (2007) 447:661–78. 10.1038/nature05911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing (2009). Available online at: http://www.R-project.org (accessed March 5, 2019).
- 29.Namipashaki A, Razaghi-Moghadam Z, Ansari-Pour N. The essentiality of reporting hardy-weinberg equilibrium calculations in population-based genetic association studies. Cell J. (2015) 17:187–92. 10.22074/cellj.2016.3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bush WS, Moore JH. Chapter 11: genome-wide association studies. PLoS Comput Biol. (2012) 8:e1002822. 10.1371/journal.pcbi.1002822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordström A, Hadrévi J, Olsson T, Franks PW, Nordström P. Higher prevalence of type 2 diabetes in men than in women is associated with differences in visceral fat mass. J Clin Endocrinol Metab. (2016) 101:3740–6. 10.1210/jc.2016-1915 [DOI] [PubMed] [Google Scholar]
- 32.Sulaiman N, Mahmoud I, Hussein A, Elbadawi S, Abusnana S, Zimmet P, et al. Diabetes risk score in the United Arab Emirates: a screening tool for the early detection of type 2 diabetes mellitus. BMJ Open Diabetes Res Care. (2018) 6:e000489. 10.1136/bmjdrc-2017-000489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ali HI, Baynouna LM, Bernsen RM. Barriers and facilitators of weight management: perspectives of Arab women at risk for type 2 diabetes. Health Soc Care Commun. (2010) 18:219–28. 10.1111/j.1365-2524.2009.00896.x [DOI] [PubMed] [Google Scholar]
- 34.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. (2013) 15:7–21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- 35.Wang W-J, Tay HG, Soni R, Perumal GS, Goll MG, Macaluso FP, et al. CEP162 is an axoneme-recognition protein promoting ciliary transition zone assembly at the cilia base. Nat Cell Biol. (2013) 15:591–601. 10.1038/ncb2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. (2006) 7:125–48. 10.1146/annurev.genom.7.080505.115610 [DOI] [PubMed] [Google Scholar]
- 37.Wheway G, Parry DA, Johnson CA. The role of primary cilia in the development and disease of the retina. Organogenesis. (2014) 10:69–85. 10.4161/org.26710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Y, Wang Y, Stock O, Pfister F, Tanimoto N, Seeliger MW, et al. Vasoregression linked to neuronal damage in the rat with defect of polycystin-2. PLoS ONE. (2009) 4:e7328. 10.1371/journal.pone.0007328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Windheim M, Peggie M, Cohen P. Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem J. (2008) 409:723–29. 10.1042/BJ20071338 [DOI] [PubMed] [Google Scholar]
- 40.Gehrke SG, Riedel H-D, Herrmann T, Hadaschik B, Bents K, Veltkamp C, et al. UbcH5A, a member of human E2 ubiquitin-conjugating enzymes, is closely related to SFT, a stimulator of iron transport, and is up-regulated in hereditary hemochromatosis. Blood. (2003) 101:3288–93. 10.1182/blood-2002-07-2192 [DOI] [PubMed] [Google Scholar]
- 41.Semenza GL. HIF-1 and human disease: one highly involved factor. Genes Dev. (2000) 14:1983–91. 10.1101/gad.14.16.1983 [DOI] [PubMed] [Google Scholar]
- 42.Crawford TN, Alfaro DV, Kerrison JB, Jablon EP. Diabetic retinopathy and angiogenesis. Curr Diabetes Rev. (2009) 5:8–13. 10.2174/157339909787314149 [DOI] [PubMed] [Google Scholar]
- 43.Carson-Walter EB, Watkins DN, Nanda A, Vogelstein B, Kinzler KW, St Croix B. Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Res. (2001) 61:6649–55. [PubMed] [Google Scholar]
- 44.Cheng G, Zhong M, Kawaguchi R, Kassai M, Al-Ubaidi M, Deng J, et al. Identification of PLXDC1 and PLXDC2 as the transmembrane receptors for the multifunctional factor PEDF. eLife. (2014) 3:e05401. 10.7554/eLife.05401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang F, Ma X, Zhou M, Pan X, Ni J, Gao M, et al. Serum pigment epithelium-derived factor levels are independently correlated with the presence of coronary artery disease. Cardiovasc Diabetol. (2013) 12:56. 10.1186/1475-2840-12-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nozue T, Yamagishi S, Hirano T, Yamamoto S, Tohyama S, Fukui K, et al. Pigment epithelium-derived factor is associated with necrotic core progression during statin therapy: Coron Artery Dis. (2015) 26:107–13. 10.1097/MCA.0000000000000192 [DOI] [PubMed] [Google Scholar]