Abstract
Human beta-defensins (hBDs, −1, 2, 3) are a family of epithelial cell derived antimicrobial peptides (AMPs) that protect mucosal membranes from microbial challenges. In addition to their antimicrobial activities, they possess other functions; e.g., cell activation, proliferation, regulation of cytokine/chemokine production, migration, differentiation, angiogenesis, and wound healing processes. It has also become apparent that defensin levels change with the development of neoplasia. However, inconsistent observations published by various laboratories make it difficult to reach a consensus as to the direction of the dysregulation and role the hBDs may play in various cancers. This is particularly evident in studies focusing on oral squamous cell carcinoma (OSCC). By segregating each hBD by cancer type, interrogating methodologies, and scrutinizing the subject cohorts used in the studies, we have endeavored to identify the “take home message” for each one of the three hBDs. We discovered that (1) consensus-driven findings indicate that hBD-1 and−2 are down- while hBD-3 is up-regulated in OSCC; (2) hBD dysregulation is cancer-type specific; (3) the inhibition/activation effect an hBD has on cancer cell lines is related to the direction of the hBD dysregulation (up or down) in the cancer from which the cell lines derive. Therefore, studies addressing hBD dysregulation in various cancers are not generalizable and comparisons should be avoided. Systematic delineation of the fate and role of the hBDs in a specific cancer type may lead to innovative ways to use defensins as prospective biomarkers for diagnostic/prognostic purposes and/or in novel therapeutic modalities.
Keywords: hBD-1, hBD-2, hBD-3, cancer, migration, proliferation
Introduction
The discovery of human β-defensins (hBDs) in mucosa has led to recognition that they are integral in innate immune protection; shielding mucosal surfaces from microbial challenges. The three hBDs, hBD1–3, are cationic, beta-sheeted peptides varying in length from 33 to 47 amino acid residues that are primarily expressed by epithelial cells (1–4). Of the three defensins, hBD-3 is the most positively charged (+11), followed by hBD-2 (+6) and hBD-1 (+4) (4). In solution, hBD-1 and hBD-2 are monomers while hBD-3 exists as a dimer (4). All three proteins have broad-spectrum antimicrobial activity, with hBD-3 consistently being the most potent (4). In addition to their antimicrobial activity, the expression of hBDs in numerous tissues has been linked to their ability to cross-talk with the adaptive immune response by acting as chemokines (5–10). These, along with evidence that some also play a role in wound healing (11), have contributed to our understanding that the hBDs, function to protect us at mucosal surfaces. Interestingly, there is growing evidence that, in certain situations, hBDs can be involved in disease progression, primarily in neoplasia. Abiko et al. (12), were the first to demonstrate the differential expression of hBD-1 and−2 mRNAs in oral cancer cell lines and tumor samples. Since then, multiple studies have reported dysregulation of β-defensins in cancers from diverse anatomical locations within the human body; e.g., oral cavity, esophagus, skin, kidney, prostate, thyroid, liver, lung, colon, vulva, and cervix (Table 1). Human β-defensins can be up- or down-regulated depending on the specific cancer type and its anatomical location. What does this observation mean for hBD expression in the context of neoplasia? Do hBDs have a role to play in activating and/or inhibiting tumor progression? Do hBDs act in unison or does each have a separate and possibly divergent role? Are they only bystander molecules that do not affect the tumor to any appreciable degree? The literature currently is rife with contradictory findings that make it difficult to ascertain what role, if any, hBDs play in neoplasia. In this review, we attempt to organize these observations by scrutinizing the current literature. By segregating findings related to hBD dysregulation in different cancers based on the defensin type and the specific anatomical location of the different cancers, as well as delving into the methodologies used in the studies, we begin to unravel the discrepancies that have plagued this area of research over the last 15 years. We discovered that disparate published results were often due to: (1) studies using cell lines vs. human tissue samples; (2) results based on human tissue samples varied depending upon whether the comparisons were done with matched or unmatched control samples; (3) conclusions varied when using mRNA-based vs. protein-based studies and; (4) in protein-based studies varying methods to detect defensins [Western, Immunohistochemstry (IHC) or Immunofluorescence microscopy (IFM)] could contribute to disparate findings. As examples of contradictory results, some studies showed that hBDs can promote cancer cell migration/proliferation (30, 32, 36, 37) while others showed that defensins were inhibitory (15, 38–42). Interestingly, hBDs were reported to behave as tumor suppressors (15, 22, 39) as well as proto-oncogenes (36, 43, 44). While these results may, on-the-surface, appear to be inconsistent, if studies are segregated based on cell lines used, type of cancer being investigated, and the specific hBD studied, a clearer picture unfolds. In this review, we attempt to set the record straight, and, in summary propose a general model for the role and fate of hBDs in carcinogenesis.
Table 1.
Studies related to differential expression of hBDs (1, 2, and −3) in cancer.
| Cancer type/Location | Fate in cancer | Cell lines/Patient | mRNA/Protein | Method | Sample numbers | Significant? | References | |
|---|---|---|---|---|---|---|---|---|
| HBD-1 | Oral cavity (OSCC) | ↓ | Cell lines | mRNA | qRT-PCR | 16:15 | YES | (13) |
| ↓ | Patient (UM) | mRNA | qRT-PCR | 5:5 | ND | (14) | ||
| ↑ | Patient (UM) | Protein | IHC | 30:15 | YES | (15) | ||
| ↓ | Patient (UM) | Protein | IHC | 60:15 | YES | (16) | ||
| ↓ | Patient (UM) | mRNA | Gene-array | 355:131 | YES | (17, 18) | ||
| ↓ | Patient (M) | mRNA | Gene-array | 15 | YES | (19) | ||
| Skin(BCC) | ↓ | Patient (M+UM) | mRNA | RT-PCR | 22:27 | YES | (20) | |
| Skin (SCC) | ↓ | Patient (UM) | Protein | IHC | 23:10 | YES | (21) | |
| Renal Cell | ↓ | Patient (M) | Protein | IHC | 48 | ND | (22) | |
| Colon | ↓ | Patients (M) | Both | qRT-PCR/IHC | 40 | YES | (23) | |
| ↓ | Patients | mRNA | Gene-array | 769:200 | YES | (24) | ||
| Prostate | ↓ | Patient (M) | Protein | IHC | 100 | ND | (22) | |
| Lung | ↑ | Patients (UM) | Serum protein | RIA | 56:46 | YES | (25) | |
| ↑ | Patient (M) | mRNA | RT-PCR | 20 | ND | (26) | ||
| Liver | ↓ | Patient | mRNA | Gene-array | 733:656 | YES | (27) | |
| HBD-2 | Oral cavity (OSCC) | ↓ | Cell lines | mRNA | qRT-PCR | 16:15 | YES | (13) |
| ↓ | Cell lines | mRNA | qRT-PCR | 6:1 | YES | (28) | ||
| ↓ | Patient (UM) | mRNA | qRT-PCR | 5:5 | ND | (14) | ||
| ↓ | Patient (UM) | Protein | IHC | 60:15 | YES | (16) | ||
| Tonsil | ↓ | Patient (UM) | mRNA | RT-PCR | 8:8 | YES | (29) | |
| Esophagus (SCC) | ↑ | Patient (M) | Protein | IHC | 58 | YES | (30) | |
| ↑ | Cell lines | Protein | Western | 4:1 | ND | (30) | ||
| Skin (BCC) | ↑ | Patient (M+UM) | mRNA | RT-PCR | 22:27 | YES | (20) | |
| Skin (SCC) | ↑ | Patient (UM) | Protein | IHC | 23:09 | YES | (21) | |
| Colon | ↓ | Patient (M) | Both | qRT-PCR/IHC | 40 | YES | (23) | |
| Cervix (SCC) | ↑ | Patients (M) | mRNA | RT-PCR | 5 | ND | (31) | |
| ↑ | Patients(UM) | Protein | IHC | 60:10 | ND | (32) | ||
| Vulva | ↑ | Patients (M) | mRNA | RT–PCR | 5 | ND | (31) | |
| Lung | ↑ | Patients (UM) | Serum protein | RIA | 56:46 | YES | (25) | |
| ↑ | Patient (M) | mRNA | PCR | 20 | ND | (26) | ||
| HBD-3 | Oral cavity (OSCC) | – | Cell lines | mRNA | qRT-PCR | 16: 15 | ND | (13) |
| ↑ | Patient (UM) | mRNA | qRT-PCR | 5:5 | ND | (14) | ||
| ↓ | Patient (UM) | Protein | IHC | 60:15 | YES | (16) | ||
| ↑ | Patient (M) | mRNA | qRT-PCR | 4 | YES | (33) | ||
| ↑ | Patient (M) | Protein | IHC | 4 | ND | (33) | ||
| ↑ | Patient (M) | mRNA | qRT-PCR | 45 | YES | (34) | ||
| ↑ | Patient (M) | Protein | IHC | 45 | ND | (34) | ||
| ↑ | Patient (UM) | Protein | IHC | 29:23 | ND | (35) | ||
| Cervical (SCC) | ↑ | Patient (UM) | Protein | IHC | 37:22 | YES | (36) | |
| Skin (BCC) | – | Patient (M+UM) | mRNA | RT-PCR | 22:27 | ND | (20) | |
| Colon | ↓ | Patient (M) | Both | qRT-PCR/IHC | 40 | YES | (23) |
OSCC, Oral Squamous Cell carcinoma; BCC, Basal Cell carcinoma; SCC, Squamous Cell carcinoma ↓, Down regulated in cancer; ↑, upregulated in cancer; ND, Unchanged in cancer; UM, Unmatched samples; i.e. control samples from different subjects. M, Matched samples; control samples from same subjects, Both, both protein and mRNA; IHC, Immunohistochemistry; RIA, Radioimmunoassay; ND, Not determined/described.
Dysregulation of Human Beta-Defensins in Cancer:
Human Beta-Defensin 1
HBD-1, oddly enough was first described in the hemofiltrate of patients with end-stage kidney disease undergoing dialysis, suggesting renal epithelia as the possible source for hBD-1 (1). Indeed, by in situ hybridization, Valore et al. (45), showed that hBD-1 mRNA is present in the epithelial layers of the distal tubules of the loops of Henle, the collecting ducts of the kidney. HBD-1 was also shown to be expressed in human airway epithelia, gingival tissue, epithelia lining the female genitalia and respiratory epithelia of normal and cystic fibrosis (CF) lungs (45–48).
Since its discovery, research has primarily focused on antimicrobial properties of hBD-1 (49–51), although other functions have also been reported (51). These include, immunoregulation (52, 53), cellular differentiation (54, 55), and glucose metabolism (56). HBD-1 has also been shown to be differentially expressed in different types of cancers (Table 1) and has been proposed as a tumor suppressor, because it promotes cancer cell apoptosis (38, 39) and also inhibits migration and invasion of cancer cells (15).
In a large-scale gene expression profiling study of renal epithelial neoplasms, hBD-1 transcript was found to be significantly down-regulated in renal carcinoma (57). IHC staining of the hBD-1 peptide in clinical specimens of both renal cell carcinoma and prostate cancer tissue revealed that >80% of the samples were either completely devoid of hBD-1 expression, or showed marginal expression when compared to adjacent benign epithelium (22). Expression of hBD-1 mRNA and peptide were significantly reduced in basal cell carcinoma (BCC) of the skin (20) and in cutaneous squamous cell carcinoma (SCC), as compared to healthy skin and precursor lesions (21), and in colon cancer (23). Using publicly available gene-expression data sets of colorectal cancer patients from four different countries, Bonamy et al. (24) further corroborated that the DEFB1 gene was consistently and significantly downregulated in colon cancer compared to non-tumor colon specimens. Gene expression datasets also suggested that DEFB1 was significantly down-regulated in liver cancer (27).
On the other hand, hBD-1 peptide and mRNA were shown to be upregulated in surgically resected human lung squamous cell carcinoma (SCC) and adenocarcinoma (AC), when compared to normal tissue samples (26), and hBD-1 serum levels were found to be elevated in lung cancer patients (N = 56) when compared to healthy subjects (N = 46) (25). Therefore, based on the works cited, the production of hBD-1, in the context of cancer, appears to be cancer type and location dependent. This however, does not appear to be the case with oral cancer (oral squamous cell carcinoma; OSCC), as most of the reported studies showed a decrease in hBD-1 expression levels, while only one reported an increase (Table 1). In a study of five OSCC tumors, hBD-1 mRNA was significantly reduced when compared to healthy gingival tissue (14), and this was corroborated in OSCC cell lines by Joly et al. (13). IHC staining, followed by scoring by two independent pathologists, showed that OSCC tissue (N = 60) harbored reduced hBD-1 peptide expression when compared to tissue from 15 healthy subjects (16). Interestingly, Han et al. (15), using the same IHC technique, followed by scoring by three independent pathologists, reported just the opposite results; i.e., hBD-1 was induced in OSCC tissue biopsies (N = 30) compared to healthy control biopsies (N = 15). The authors conceded that they were unable to detect hBD-1 expression in all of the 15 normal subjects. This is quite unusual, as hBD-1 mRNA and peptide are known to be ubiquitously expressed in mucosal tissues (47, 58, 59). Dale et al. (58) and Kawsar et al. (60) using IHC and IFM, respectively, demonstrated that hBD-1 is expressed in the stratum granulosum and spinosum of normal oral epithelium. Moreover, Han et al. (15) showed that hBD-1 expression levels in OSCC were reduced when compared to oral leukoplakia, a dysplastic and premalignant disorder of the oral cavity; i.e., supporting the notion that hBD-1 is under-expressed in OSCC. It has also been shown by merging of multiple microarray datasets of differentially expressed genes (DEGs) that DEFB1 is down regulated in 355 OSCC tumor samples compared to 131 normal (healthy) controls (17, 18). Therefore, with the exception of the Han et al. (15) findings, the consensus appears to be that hBD-1 is under-expressed in OSCC. It is important to note that hBD-1 expression levels were not compared to matched controls in the studies cited above. Suhr et al. (19) however, compared hBD-1 transcript in OSCC tissue (15 subjects) with adjacent normal tissue and found hBD-1 transcript to be down regulated; further supporting the consensus that hBD-1 is reduced in OSCC.
Human Beta-Defensin 2
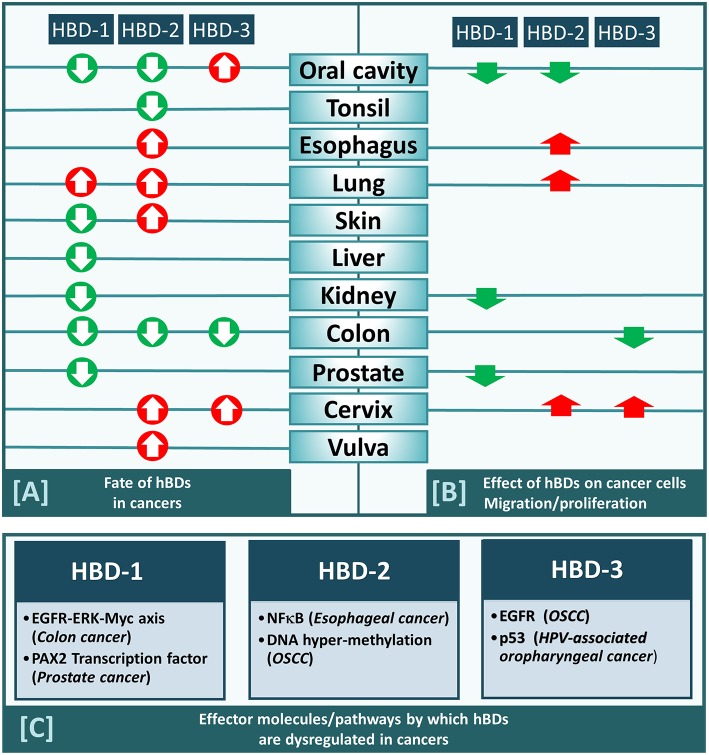
HBD-2 was discovered in human skin in 1997 (3), followed by descriptions of its expression throughout the epithelia of many organs, including the lung (61). Like hBD-1, the expression level of hBD-2 in cancer tissue appears to be dependent upon the type of cancer studied. While hBD-2 has been reported to be upregulated in several cancers, including esophageal (30), lung (25, 26), cervical (32), and skin cancer (BCC and SCC) (20, 21), it has been reported to be downregulated in colon cancer (23) (Table 1; Figure 1A).
Figure 1.
(A) HBD-1,-2, and−3 either increased (red arrow) or decreased (green arrow) in cancers from different anatomical locations within human body (as indicated). (B) HBD-1,-2, and−3 either promote (red arrow) or inhibit (green arrow) migration/proliferation of cancers cells derived from different anatomical locations (as indicated). (C) Effector molecules/pathways by which each of the three defensins (hBD-1,−2, and−3) are dysregulated in cancers.
In OSCC tissue, several investigators have reported hBD-2 (both mRNA and peptide) to be downregulated (Table 1) and its' mRNA is reduced in OSCC cell lines compared to normal oral cell lines (13). Its' mRNA was also shown to be reduced in tonsillar cancer (29). Consensus, therefore, supports the observation that hBD-2 is reduced in orally related cancers.
Lisovskiy et al. (31) reported that expression of hBD-2 mRNA is found in tumor cells of the cervix and vulva, but not in adjacent normal cervical and vulvar cells. HBD-2 mRNA absence from these normal cells is contradicted by Markeeva et al. (32) who reported that IHC analysis revealed hBD-2 expression in the adjacent normal cervical epithelium surrounding tumors. Interestingly, although hBD-2 was reported to be expressed primarily in the basal layer of normal cervical epithelial tissue, in carcinoma in situ (CIS), hBD-2 is expressed throughout all the tissue layers (32). These results are in stark contrast to immunofluorescence staining results by Jin et al. (44), showing that, in normal oral epithelium, hBD-2 was expressed mainly in the differentiated spinosum and granulosum layers (60), with markedly reduced expression in CIS.
Human Beta-Defensin 3
HBD-3 was first isolated from human psoriatic scales (2), but was also found in other epithelial tissues, including, tonsils, esophagus, trachea, colon and cervix (2, 23, 36, 62, 63). HBD-3 is particularly interesting in the context of mucosal dysplasia such as CIS and OSCC. It is primarily expressed in the basal keratinocyte region of the oral mucosa; i.e., in highly proliferating epithelial cells (60), has a role in normal wound healing (11), and is induced through activation of epidermal growth factor receptor (EGFR) (60), a process amplified in OSCC (34). Over the last decade, a number of investigators have interrogated the levels of hBD-3 expression in various stages of OSCC. An IHC study demonstrated higher levels of hBD-3 expression in OSCC compared to healthy and dysplastic oral tissues (35). Transcriptional studies demonstrated that hBD-3 mRNA expression is higher in OSCC lesions compared to oral tissue from healthy subjects (14). IHC analysis also showed that hBD-3 peptide was more prominent in oral tumor tissue compared to healthy oral mucosa in the same subjects (34), and that elevated expression of hBD-3 peptide localized exclusively to the cytoplasm of malignant epithelial cells (33). Collectively, these results (Table 1) suggest that hBD-3 is frequently overexpressed in OSCC, and this defensin may have a role in oncogenesis. In support of this hypothesis, Dasgupta et al. (43), reported that the oncogene E6 of HPV 16, etiologically associated with oropharyngeal cancers (64), increases hBD-3 mRNA and peptide expression in infected oral epithelial cells and that p53, an important tumor suppressor inhibited by E6 (65), blocks hBD-3 expression.
As mentioned earlier, Joly et al. (13), by comparing β-defensin mRNA levels in oral epithelial cell lines from 16 OSCC and 15 healthy subjects, found reduced levels of hBD-1 and−2 mRNA in OSCC cell lines compared to normal primary epithelial cells; however, they saw no differences in hBD-3 mRNA levels between the two cell types. While this may appear to contradict the patient-based results (14, 33–35), the Joly et al. (13) study was based on cell lines whose gene expression profiles may not always reflect a true representation of the disease process. As demonstrated by Ertel et al. (66) the signaling and metabolic pathways in transformed cell lines have distinctly different gene expression patterns than those associated with healthy and tumor tissue. In cervical cancer, similar contradictory results were reported in cell lines vs. tumor tissue samples; i.e., strong hBD-3 staining was observed in the cervical cancer tissues, while the peptide was not detectable in three cervical cancer cell lines (HeLa, CaSki, and SiHa) (36). Moreover, we find it perplexing that even though Joly et al. (13) reported no change in hBD-3 transcript expression between OSCC and normal cell lines, this is not reflected in the title of their study “Loss of human beta-defensin 1, 2, and 3 expression in oral squamous cell carcinoma.” Another perplexing result comes from an in situ hybridization study by Yoshimoto et al. (67). They reported that out of 20 OSCC samples, only 4 were positive for hBD-3. We find that quite unusual. While hBD-3 peptide may have varied between samples, all should have been hBD-3 transcript positive, suggesting that the in situ hybridization may not have been optimal in all the cases.
In an IHC study by Wang et al. (16) hBD-3 was shown to be under-expressed in OSCC when compared to healthy oral tissue. Examination of the details of this work reveals that: (1) the sample cohort was over-represented by OSCC subjects (60 OSCC vs. 15 healthy); (2) all 15 “healthy” samples were collected from surgical extractions of impacted third molars, a specific location of the oral cavity that may not be representative of the various sites from which the OSCC samples were taken and; (3) it is clear from (2) that the authors did not use paired samples to overcome inherent geographic differences in the oral cavity. On the other hand, observations reported for hBD-3 mRNA and peptide levels by Kestling et al. (34), and Shuyi et al. (33), were obtained from matched paired samples from the same subjects. Additionally, Jin et al. (44) demonstrated, by quantitative IFM, that hBD-3 is over-expressed in CIS compared to healthy tissue; hence, it is unlikely that hBD-3 would be overexpressed in CIS but not in OSCC. Finally, while unrelated to hBD-3 directly, it bears mentioning that Wang et al. (16), also reported that NOD1 and RIP2 were under-expressed in OSCC compared to healthy oral tissues; findings that are contradictory to more recent results showing that both proteins are over-expressed in OSCC (68). In summary, the majority of studies agree on hBD-3 overexpression in OSCC compared to healthy paired control tissue. Interestingly, hBD-3 overexpression has also been reported in cervical cancer (36) but not in colon cancer (23). Therefore, as we concluded for hBD-1 and−2, dysregulation of hBD-3 in cancer tissue appears to be associated with the type of cancer being studied and its location.
Mechanisms of Beta-Defensin Dyregulation in Cancers
The pathway by which a particular β-defensin is dysregulated in cancers has been studied in only a few cancer types and has been found to vary depending on the hBD and the cancer it is associated with (Figure 1C). For example, hBD-1 has been shown to be suppressed in colon cancer through the EGFR-ERK-MYC axis (24), while in prostate cancer it is suppressed by the PAX2 transcription factor (69). Mechanisms for dysregulation of hBD-2 and−3 are also cancer type dependent. For hBD-2, the nuclear factor kappa b (NF-κB) pathway was found to regulate it in esophageal cancer (30), while in OSCC it is regulated by DNA hypermethylation (28). For hBD-3 the EGFR pathway activates it in OSCC (33), while in HPV-associated oropharyngeal cancer it is regulated via the tumor suppressor p53 (43).
The Effect of Human Beta-Defensins on Cancer Cell Migration/Proliferation
Addition of exogenous synthetic hBD-1 peptide inhibits bladder cancer cell (TSU-Pr1) proliferation (38). Overexpression of hBD-1, by stable transfection of DEFB1 cDNA in renal cancer cells (SW156) resulted in caspase-3-mediated apoptosis (38). Induction of hBD-1 by ectopic expression of DEFB1 cDNA in prostate cancer cell lines PC3 and DU145 (39) also resulted in decreased cell growth and viability. HBD-1 also caused rapid induction of cytolysis and caspase-mediated apoptosis in these cell lines (39). Similarly, hBD-1 overexpression by stable transfection of DEFB1 has also been shown to inhibit migration of several oral cancer cell lines (HSC-3, UM1, SCC-9 and SCC-25) (15). RhoA and RhoC are likely to be associated with the regulatory effect of hBD-1 on the migration and invasion of OSCC cells (15). Thus, hBD-1 appears to have anti-cancer and/or tumor suppressive activities, and therefore its suppression in prostate, renal, bladder and/or oral cancer might contribute to cancer cell survival and tumor progression. Interestingly, hBD-1 is under-expressed in all these cancer types (e.g., renal, prostate and oral; Table 1; Figure 1A).
Increased expression of hBD-2 generated by gene transfection inhibits the proliferation and invasion of the OSCC cell line (SAS), possibly via G1/S arrest and pRB gene expression, indicating that hBD-2 may act as a tumor suppressor (28). Interestingly, as described above, hBD-2 has been reported to be under-expressed in OSCC (Table 1; Figure 1A). In contrast, it is overexpressed in esophageal, lung, and cervical cancer and it appears to promote the growth of cell lines from these cancers; i.e., esophageal (KYSE-150 cells, through NF-κB) (30), lung (A549 cells, through ATP-binding cassette transporter G2) (37), and cervical (M-HeLa) cancer cell lines (32). Thus, in cancers where hBD-2 under-expression has been reported, it inhibits migration and proliferation of cell lines emanating from those cancers, while in cancers where it is overexpressed it promotes respective cancer cell growth.
Over-expression of hBD-3 in HeLa cells (cervical cancer cell line), using lentiviral constructs, promoted cell proliferation by accelerating G1/S progression and enhanced cell migration and invasion (36), while addition of exogenous hBD-3 in colon cancer cell lines (SW480, SW620) inhibits their migration via downregulation of MTA2 (42). In vitro observations have also reported that exogenous hBD-3 promotes directed chemotaxis of myeloid cells via the chemotaxis receptor CCR2 (44). Moreover, in vivo observations show that in CCL2 (chemokine ligand for CCR2) deficient oral CIS lesions, hBD-3 over-expression acts to selectively recruit tumor-associated macrophages to CIS lesions (44). These results, collectively, suggest that in those cancers where hBD-3 over-expression has been reported, principally in oral, and cervical cancers, hBD-3 acts to exacerbate neoplasia, while in cancers where it is under-expressed, principally colon cancer, it is associated with inhibition of neoplasia.
Take Home Message
Based on the literature available, the apparent contradictory findings between labs regarding β-defensin expression levels in cancer tissue appear to be due to levels reported in specific cancer types (Figure 1A), where a given defensin is increased in one cancer but decreased in another. The mechanism by which hBDs are dysregulated in cancer also varies depending on which defensin and cancer type are being studied. Additionally, a specific beta defensin may promote or inhibit cancer cell proliferation/migration depending on the origin of the cancer cell, and the outcome may be associated with whether the defensin is increased or decreased in the tumor from which the cells are harvested (Figure 1B). Therefore, generalizations as to the role of β-defensins in cancer and neoplasia should be avoided, as the function of defensins may differ between cancers and cancer cells.
Concluding Remarks
We have surveyed the literature in an attempt to identify potential reasons behind contradictory findings related to differential regulation of β-defensins in different cancers. By mining the published information, interrogating methodologies and scrutinizing the subject cohorts used in the studies, we have endeavored to investigate the inconsistencies related to the fate of β-defensins in OSSC, and have concluded that hBD-1 and−2 are down while hBD-3 is upregulated in OSCC. Additionally, after evaluating each study based on the specific defensin and the anatomical location of the respective cancer, we conclude that generalizations as to the fate of the β-defensins in cancer should be avoided; they are defensin and cancer specific. We have previously stressed the promiscuous nature of human β-defensins (10), and are therefore not surprised that various cancers demonstrated differential expression of these molecules. These molecules are regulated differently (60, 70–72), they interact with receptors on cells differently and, while some function primarily as AMPs, others, in addition to antimicrobial activity, function in wound healing (11). Defining the role(s) AMPs play in cancer may lead to innovative ways to mine them as potential biomarkers for diagnostic/prognostic purposes and/or in novel therapeutic modalities.
Author Contributions
SG performed the literature search and drafted the manuscript. AW contributed to the discussions and writing of the article. TM provided critical revisions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. Supported by grants from the National Institutes of Health (P01DE019759) and Case-Coulter Transnational Research Partnership.
References
- 1.Bensch KW, Raida M, Mägert HJ, Schulz-Knappe P, Forssmann WG. hBD-1: a novel beta-defensin from human plasma. FEBS Lett. (1995) 368:331–5. 10.1016/0014-5793(95)00687-5 [DOI] [PubMed] [Google Scholar]
- 2.Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. (2001) 276:5707–13. 10.1074/jbc.M008557200 [DOI] [PubMed] [Google Scholar]
- 3.Harder J, Bartels J, Christophers E, Schroder JM. A peptide antibiotic from human skin. Nature. (1997) 387:861. 10.1038/43088 [DOI] [PubMed] [Google Scholar]
- 4.Schibli DJ, Hunter HN, Aseyev V, Starner TD, Wiencek JM, McCray PB, Jr, et al. The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus. J Biol Chem. (2002) 277:8279–89. 10.1074/jbc.M108830200 [DOI] [PubMed] [Google Scholar]
- 5.Diamond G, Ryan L. Beta-defensins: what are they really doing in the oral cavity. Oral Dis. (2011) 17:628–35. 10.1111/j.1601-0825.2011.01799.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greer A, Zenobia C, Darveau RP. Defensins and LL-37: a review of function in the gingival epithelium. Periodontol 2000. (2013) 63:67–79. 10.1111/prd.12028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarczak J, Kościuczuk EM, Lisowski P, Strzałkowska N, Józwik A, Horbanczuk J, et al. Defensins: natural component of human innate immunity. Hum Immunol. (2013) 74:1069–79. 10.1016/j.humimm.2013.05.008 [DOI] [PubMed] [Google Scholar]
- 8.Jin G, Weinberg A. Human antimicrobial peptides and cancer. Semin Cell Dev Biol. (2018) 88:156–62. 10.1016/j.semcdb.2018.04.006 [DOI] [PubMed] [Google Scholar]
- 9.Semple F, Dorin JR. β-Defensins: multifunctional modulators of infection, inflammation and more? J Innate Immun. (2012) 4:337–48. 10.1159/000336619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinberg A, Jin G, Sieg S, McCormick TS. The yin and yang of human Beta-defensins in health and disease. Front Immunol. (2012) 3:294. 10.3389/fimmu.2012.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangoni ML, McDermott AM, Zasloff M. Antimicrobial peptides and wound healing:biological and therapeutic considerations. Exp Dermatol. (2016) 25:167–73. 10.1111/exd.12929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abiko Y, Mitamura J, Nishimura M, Muramatsu T, Inoue T, Shimono M, et al. Pattern of expression of beta-defensins in oral squamous cell carcinoma. Cancer Lett. (1999) 143:37–43. 10.1016/S0304-3835(99)00171-8 [DOI] [PubMed] [Google Scholar]
- 13.Joly S, Compton LM, Pujol C, Kurago ZB, Guthmiller JM. Loss of human beta-defensin 1, 2, and 3 expression in oral squamous cell carcinoma. Oral Microbiol Immunol. (2009) 24:353–60. 10.1111/j.1399-302X.2009.00512.x [DOI] [PubMed] [Google Scholar]
- 14.Wenghoefer M, Pantelis A, Dommisch H, Reich R, Martini M, Allam JP, et al. Decreased gene expression of human beta-defensin-1 in the development of squamous cell carcinoma of the oral cavity. Int J Oral Maxillofac Surg. (2008) 37:660–63. 10.1016/j.ijom.2008.02.003 [DOI] [PubMed] [Google Scholar]
- 15.Han Q, Wang R, Sun C, Jin X, Liu D, Zhao X, et al. Human beta-defensin-1 suppresses tumor migration and invasion and is an independent predictor for survival of oral squamous cell carcinoma patients. PLoS ONE. (2014) 9:e91867. 10.1371/journal.pone.0091867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Jiang W, Duan N, Qian Y, Zhou Q, Ye P, et al. NOD1, RIP2 and Caspase12 are potentially novel biomarkers for oral squamous cell carcinoma development and progression. Int J Clin Exp Pathol. (2014) 7:1677–86. Available online at: http://www.ijcep.com/V7_No4.html [PMC free article] [PubMed] [Google Scholar]
- 17.Randhawa V, Acharya V. Integrated network analysis and logistic regression modeling identify stage-specific genes in Oral Squamous Cell Carcinoma. BMC Med Genomics. (2015) 8:39. 10.1186/s12920-015-0114-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randhawa V, Kumar Singh A, Acharya V. A systematic approach to prioritize drug targets using machine learning, a molecular descriptor-based classification model, and high-throughput screening of plant derived molecules: a case study in oral cancer. Mol Biosyst. (2015) 11:3362–77. 10.1039/C5MB00468C [DOI] [PubMed] [Google Scholar]
- 19.Suhr ML, Dysvik B, Bruland O, Warnakulasuriya S, Amaratunga AN, Jonassen I, et al. Gene expression profile of oral squamous cell carcinomas from Sri Lankan betel quid users. Oncol Rep. (2007) 18:1061–75. 10.3892/or.18.5.1061 [DOI] [PubMed] [Google Scholar]
- 20.Gambichler T, Skrygan M, Huyn J, Bechara FG, Sand M, Altmeyer P, et al. Pattern of mRNA expression of beta-defensins in basal cell carcinoma. BMC Cancer. (2006) 6:163. 10.1186/1471-2407-6-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scola N, Gambichler T, Saklaoui H, Bechara FG, Georgas D, Stücker M, et al. The expression of antimicrobial peptides is significantly altered in cutaneous squamous cell carcinoma and precursor lesions. Br J Dermatol. (2012) 167:591–7. 10.1111/j.1365-2133.2012.11110.x [DOI] [PubMed] [Google Scholar]
- 22.Donald CD, Sun CQ, Lim SD, Macoska J, Cohen C, Amin MB, et al. Cancer-specific loss of beta-defensin 1 in renal and prostatic carcinomas. Lab Invest. (2003) 83:501–5. 10.1097/01.LAB.0000063929.61760.F6 [DOI] [PubMed] [Google Scholar]
- 23.Semlali A, Al Amri A, Azzi A, Al Shahrani O, Arafah M, Kohailan M, et al. Expression and new exon mutations of the human Beta defensins and their association on colon cancer development. PLoS ONE. (2015) 10:e0126868. 10.1371/journal.pone.0126868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonamy C, Sechet E, Amiot A, Alam A, Mourez M, Fraisse L, et al. Expression of the human antimicrobial peptide β-defensin-1 is repressed by the EGFR-ERK-MYC axis in colonic epithelial cells. Sci Rep. (2018) 8:18043. 10.1038/s41598-018-36387-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arimura Y, Ashitani J, Yanagi S, Tokojima M, Abe K, Mukae H, et al. Elevated serum beta-defensins concentrations in patients with lung cancer. Anticancer Res. (2004) 24:4051–8. Available online at: http://ar.iiarjournals.org/content/24/6/4051.long [PubMed] [Google Scholar]
- 26.Shestakova T, Zhuravel E, Bolgova L, Alekseenko O, Soldatkina M, Pogrebnoy P. Expression of human beta-defensins-1, 2 and 4 mRNA in human lung tumor tissue: a pilot study. Exp Oncol. (2008) 30:153–6. Available online at: http://exp-oncology.com.ua/wp/wp-content/uploads/magazine/689.pdf?upload= [PubMed] [Google Scholar]
- 27.Ling YM, Chen JY, Guo L, Wang CY, Tan WT, Wen Q, et al. β-defensin 1 expression in HCV infected liver/liver cancer: an important role in protecting HCV progression and liver cancer development. Sci Rep. (2017) 7:13404. 10.1038/s41598-017-13332-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamino Y, Kurashige Y, Uehara O, Sato J, Nishimura M, Yoshida K, et al. HBD-2 is downregulated in oral carcinoma cells by DNA hypermethylation, and increased expression of hBD-2 by DNA demethylation and gene transfection inhibits cell proliferation and invasion. Oncol Rep. (2014) 32:462–8. 10.3892/or.2014.3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer JE, Harder J, Görögh T, Weise JB, Schubert S, Janssen D, et al. Human beta-defensin-2 in oral cancer with opportunistic Candida infection. Anticancer Res. (2004) 24:1025–30. Available online at: http://ar.iiarjournals.org/content/24/2B/1025.long [PubMed] [Google Scholar]
- 30.Shi N, Jin F, Zhang X, Clinton SK, Pan Z, Chen T. Overexpression of human β-defensin 2 promotes growth and invasion during esophageal carcinogenesis. Oncotarget. (2014) 5:11333–44. 10.18632/oncotarget.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lisovskiy IL, Soldatkina MA, Lytvyn DI, Markeeva NV, Turchak OV, Nespryadko SV, et al. Pattern of β-Defensin-2 (hBD-2) and EGFR mRNAs expression in cervical and vulval cancer cells. Exp Oncol. (2001) 23:248–52. Available online at: http://exp-oncology.com.ua/article/1039 [Google Scholar]
- 32.Markeeva N, Lysovskiy I, Zhuravel E, Soldatkina M, Lyzogubov V, Usenko V, et al. Involvement of human beta-defensin-2 in proliferation of transformed cells of human cervix. Exp Oncol. (2005) 27:308–13. Available online at: http://exp-oncology.com.ua/article/753 [PubMed] [Google Scholar]
- 33.Shuyi Y, Feng W, Jing T, Hongzhang H, Haiyan W, Pingping M, et al. Human beta-defensin-3 (hBD-3) upregulated by LPS via epidermal growth factor receptor (EGFR) signaling pathways to enhance lymphatic invasion of oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. (2011) 112:616–25. 10.1016/j.tripleo.2011.02.053 [DOI] [PubMed] [Google Scholar]
- 34.Kesting MR, Loeffelbein DJ, Hasler RJ, Wolff KD, Rittig A, Schulte M, et al. Expression profile of human beta-defensin 3 in oral squamous cell carcinoma. Cancer Invest. (2009) 27:575–81. 10.1080/07357900802620851 [DOI] [PubMed] [Google Scholar]
- 35.Hussaini HM, Lim PK, Rahman RA, Abraham T, Cheong CS, Zain RB, et al. Expression of human b-defensin-3 in oral squamous cell carcinoma using tissue microarray - A preliminary study. J Oral Pathol Med. (2006) 35:432–33. Available online at: https://works.bepress.com/rosnah_zain/69/ [Google Scholar]
- 36.Xu D, Zhang B, Liao C, Zhang W, Wang W, Chang Y, et al. Human beta-defensin 3 contributes to the carcinogenesis of cervical cancer via activation of NF-κB signaling. Oncotarget. (2016) 7:75902–13. 10.18632/oncotarget.12426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao C, Yue W, Tian H, Li L, Li S, Si L. Human beta-defensin 2 promotes the proliferation of lung cancer cells through ATP-binding cassette transporter G2. Int J Clin Exp Pathol. (2016) 9:5944–9. Available online at: http://www.ijcep.com/files/ijcep0022545.pdf [Google Scholar]
- 38.Sun CQ, Arnold R, Fernandez-Golarz C, Parrish AB, Almekinder T, He J, et al. Human beta-defensin-1, a potential chromosome 8p tumor suppressor: control of transcription and induction of apoptosis in renal cell carcinoma. Cancer Res. (2006) 66:8542–9. 10.1158/0008-5472.CAN-06-0294 [DOI] [PubMed] [Google Scholar]
- 39.Bullard RS, Gibson W, Bose SK, Belgrave JK, Eaddy AC, Wright CJ, et al. Functional analysis of the host defense peptide human beta defensin-1: new insight into its potential role in cancer. Mol Immunol. (2008) 45:839–48. 10.1016/j.molimm.2006.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhuravel OV, Gerashchenko OL, Khetsuriani MR, Soldatkina MA, Pogrebnoy PV. Expression of human beta-defensins-1-4 in thyroid cancer cells and new insight on biologic activity of hBD-2 in vitro. Exp Oncol. (2014) 36:174–8. Available online at: http://exp-oncology.com.ua/article/7089 [PubMed] [Google Scholar]
- 41.Gerashchenko O, Zhuravel E, Skachkova O, Khranovska N, Pushkarev V, Pogrebnoy P, et al. Involvement of human beta-defensin-2 in regulation of malignant potential of cultured human melanoma cells. Exp Oncol. (2014) 36:17–23. Available online at: http://exp-oncology.com.ua/article/6547 [PubMed] [Google Scholar]
- 42.Uraki S, Sugimoto K, Shiraki K, Tameda M, Inagaki Y, Ogura S, Kasai C, et al. Human β-defensin-3 inhibits migration of colon cancer cells via downregulation of metastasis-associated 1 family, member 2 expression. Int J Oncol. (2014) 45:1059–64. 10.3892/ijo.2014.2507 [DOI] [PubMed] [Google Scholar]
- 43.DasGupta T, Nweze EI, Yue H, Wang L, Jin J, Ghosh SK, et al. Human papillomavirus oncogenic E6 protein regulates human β-defensin 3 (hBD3) expression via the tumor suppressor protein p53. Oncotarget. (2016) 7:27430–44. 10.18632/oncotarget.8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin G, Kawsar HI, Hirsch SA, Zeng C, Jia X, Feng Z, et al. An antimicrobial peptide regulates tumor-associated macrophage trafficking via the chemokine receptor CCR2, a model for tumorigenesis. PLoS ONE. (2010) 5:e10993. 10.1371/journal.pone.0010993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valore EV, Park CH, Quayle AJ, Wiles KR, McCray PB, Jr, Ganz T. Human beta-defensin-1: an antimicrobial peptide of urogenital tissues. J Clin Invest. (1998) 101:1633–42. 10.1172/JCI1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCray PB, Jr, Bentley L. Human airway epithelia express a beta-defensin. Am J Respir Cell Mol Biol. (1997) 16:343–9. 10.1165/ajrcmb.16.3.9070620 [DOI] [PubMed] [Google Scholar]
- 47.Krisanaprakornkit S, Weinberg A, Perez CN, Dale BA. Expression of the peptide antibiotic human beta-defensin 1 in cultured gingival epithelial cells and gingival tissue. Infect Immun. (1998) 66:4222–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldman MJ, Anderson GM, Stolzenberg ED, Kari UP, Zasloff M, Wilson JM. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. (1997) 88:553–60. 10.1016/S0092-8674(00)81895-4 [DOI] [PubMed] [Google Scholar]
- 49.Fattorini L, Gennaro R, Zanetti M, Tan D, Brunori L, Giannoni F, et al. In vitro activity of protegrin-1 and beta-defensin-1, alone and in combination with isoniazid, against Mycobacterium tuberculosis. Peptides. (2004) 25:1075–7. 10.1016/j.peptides.2004.04.003 [DOI] [PubMed] [Google Scholar]
- 50.Pazgier M, Hoover DM, Yang D, Lu W, Lubkowski J. Human beta-defensins. Cell Mol Life Sci. (2006) 63:1294–313. 10.1007/s00018-005-5540-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prado-Montes de Oca E. Human beta-defensin 1: a restless warrior againstallergies, infections and cancer. Int J Biochem Cell Biol. (2010) 42:800–4. 10.1016/j.biocel.2010.01.021 [DOI] [PubMed] [Google Scholar]
- 52.Presicce P, Giannelli S, Taddeo A, Villa ML, Della Bella S. Human defensins activate monocyte-derived dendritic cells, promote the production of proinflammatory cytokines, and up-regulate the surface expression of CD91. J Leukoc Biol. (2009) 86:941–8. 10.1189/jlb.0708412 [DOI] [PubMed] [Google Scholar]
- 53.Yang D, Liu ZH, Tewary P, Chen Q, de la Rosa G, Oppenheim JJ. Defensin participation in innate and adaptive immunity. Curr Pharm Des. (2007) 13:3131–9. 10.2174/138161207782110453 [DOI] [PubMed] [Google Scholar]
- 54.Bick RJ, Poindexter BJ, Buja LM, Lawyer CH, Milner SM, Bhat S. Nuclear localization of hBD-1 in human keratinocytes. J Burns Wounds. (2007) 7:e3. [PMC free article] [PubMed] [Google Scholar]
- 55.Sayama K, Komatsuzawa H, Yamasaki K, Shirakata Y, Hanakawa Y, Ouhara K, et al. , New mechanisms of skin innate immunity: ASK1-mediated keratinocyte differentiation regulates the expression of beta-defensins LL37 and TLR2. Eur J Immunol. (2005) 35:1886–95. 10.1002/eji.200425827 [DOI] [PubMed] [Google Scholar]
- 56.Barnea M, Madar Z, Froy O. Glucose and insulin are needed for optimal defensin expression in human cell lines. Biochem Biophys Res Commun. (2008) 367:452–6. 10.1016/j.bbrc.2007.12.158 [DOI] [PubMed] [Google Scholar]
- 57.Young AN, Amin MB, Moreno CS, Lim SD, Cohen C, Petros JA, et al. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. Am J Pathol. (2001) 158:1639–51. 10.1016/S0002-9440(10)64120-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dale BA, Kimball JR, Krisanaprakornkit S, Roberts F, Robinovitch M, O'Neal R, et al. Localized antimicrobial peptide expression in human gingiva. J. Periodont. Res. (2001) 36:285–94. 10.1034/j.1600-0765.2001.360503.x [DOI] [PubMed] [Google Scholar]
- 59.Zhao C, Wang I, Lehrer RI. Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Lett. (1996) 396:319–22. 10.1016/0014-5793(96)01123-4 [DOI] [PubMed] [Google Scholar]
- 60.Kawsar HI, Weinberg A, Hirsch SA, Venizelos A, Howell S, Jiang B, et al. Overexpression of human beta-defensin-3 in oral dysplasia: potential role in macrophage trafficking. Oral Oncol. (2009) 45:50–56. 10.1016/j.oraloncology.2008.10.016 [DOI] [PubMed] [Google Scholar]
- 61.Bals R, Wang X, Wu Z, Freeman T, Bafna V, Zasloff M, et al. Human β-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J. Clin. Invest. (1998) 102:874–80. 10.1172/JCI2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.García JR, Jaumann F, Schulz S, Krause A, Rodríguez-Jiménez J, Forssmann U, et al. Identification of a novel, multifunctional β-defensin (human β-defensin 3) with specific antimicrobial activity. Its interaction with plasma membranes of Xenopus oocytes and the induction of macrophage chemoattraction. Cell Tissue Res. (2001) 306:257–264. 10.1007/s004410100433 [DOI] [PubMed] [Google Scholar]
- 63.Jia HP, Schutte BC, Schudy A, Linzmeier R, Guthmiller JM, Johnson GK, et al. Discovery of new human β-defensins using a genomics-based approach. Gene. (2001) 263:211–21. 10.1016/S0378-1119(00)00569-2 [DOI] [PubMed] [Google Scholar]
- 64.Syrjanen S. Human papillomaviruses in head and neck carcinomas. N Engl J Med. (2007) 356:1993–5. 10.1056/NEJMe078004 [DOI] [PubMed] [Google Scholar]
- 65.Lechner MS, Laimins LA. Inhibition of p53 DNA binding by human papillomavirus E6 proteins. J Virol. (1994) 68:4262–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ertel A, Verghese A, Byers SW, Ochs M, Tozeren A. Pathway-specific differences between tumor cell lines and normal and tumor tissue cells. Mol Cancer. (2006) 5:55. 10.1186/1476-4598-5-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshimoto T, Yamaai T, Mizukawa N, Sawaki K, Nakano M, Yamachika E, et al. Different expression patterns of beta-defensins in human squamous cell carcinomas. Anticancer Res. (2003) 23:4629–33. Available online at: https://europepmc.org/abstract/med/14981906 [PubMed] [Google Scholar]
- 68.Chan LP, Wang LF, Chiang FY, Lee KW, Kuo PL, Liang CH. IL-8 promotes HNSCC progression on CXCR1/2-meidated NOD1/RIP2 signaling pathway. Oncotarget. (2016) 7:61820–31. 10.18632/oncotarget.11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bose SK, Gibson W, Bullard RS, Donald CD. PAX2 oncogene negatively regulates the expression of the host defense peptide human beta defensin-1 in prostate cancer. Mol Immunol. (2009) 46:1140–8. 10.1016/j.molimm.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boughan PK, Argent RH, Body-Malapel M, Park JH, Ewings KE, Bowie AG, et al. Nucleotide-binding oligomerization domain-1 and epidermal growth factor receptor: critical regulators of beta-defensins during Helicobacter pylori infection. J Biol Chem. (2006) 281:11637–48. 10.1074/jbc.M510275200 [DOI] [PubMed] [Google Scholar]
- 71.Chung WO, Dale BA. Differential utilization of nuclear factor-kappaB signaling pathways for gingival epithelial cell responses to oral commensal and pathogenic bacteria. Oral Microbiol Immunol. (2008) 23:119–126. 10.1111/j.1399-302X.2007.00398.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sorensen OE, Thapa DR, Roupe KM, Valore EV, Sjobring U, Roberts AA, et al. Injury-induced innate immune response in human skin mediated by transactivation of the epidermal growth factor receptor. J Clin Invest. (2006) 116:1878–1885. 10.1172/JCI28422 [DOI] [PMC free article] [PubMed] [Google Scholar]