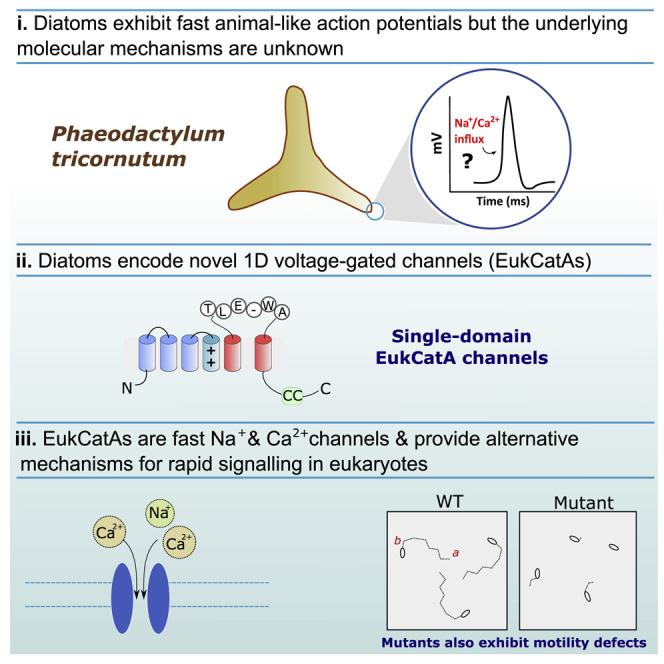
Summary
Rapid Na+/Ca2+-based action potentials govern essential cellular functions in eukaryotes, from the motile responses of unicellular protists, such as Paramecium [1, 2], to complex animal neuromuscular activity [3]. A key innovation underpinning this fundamental signaling process has been the evolution of four-domain voltage-gated Na+/Ca2+ channels (4D-Cavs/Navs). These channels are widely distributed across eukaryote diversity [4], albeit several eukaryotes, including land plants and fungi, have lost voltage-sensitive 4D-Cav/Navs [5, 6, 7]. Because these lineages appear to lack rapid Na+/Ca2+-based action potentials, 4D-Cav/Navs are generally considered necessary for fast Na+/Ca2+-based signaling [7]. However, the cellular mechanisms underpinning the membrane physiology of many eukaryotes remain unexamined. Eukaryotic phytoplankton critically influence our climate as major primary producers. Several taxa, including the globally abundant diatoms, exhibit membrane excitability [8, 9, 10]. We previously demonstrated that certain diatom genomes encode 4D-Cav/Navs [4] but also proteins of unknown function, resembling prokaryote single-domain, voltage-gated Na+ channels (BacNavs) [4]. Here, we show that single-domain channels are actually broadly distributed across major eukaryote phytoplankton lineages and represent three novel classes of single-domain channels, which we refer collectively to as EukCats. Functional characterization of diatom EukCatAs indicates that they are voltage-gated Na+- and Ca2+-permeable channels, with rapid kinetics resembling metazoan 4D-Cavs/Navs. In Phaeodactylum tricornutum, which lacks 4D-Cav/Navs, EukCatAs underpin voltage-activated Ca2+ signaling important for membrane excitability, and mutants exhibit impaired motility. EukCatAs therefore provide alternative mechanisms for rapid Na+/Ca2+ signaling in eukaryotes and may functionally replace 4D-Cavs/Navs in pennate diatoms. Marine phytoplankton thus possess unique signaling mechanisms that may be key to environmental sensing in the oceans.
Keywords: voltage-gated channel, diatoms, EukCats, signaling, BacNav, action potentials, ion selectivity, gliding motility, single-domain channel, calcium channel
Graphical Abstract
Highlights
-
•
Novel class of single-domain, voltage-gated channels (EukCatAs) identified in diatoms
-
•
EukCatAs are fast voltage-gated Na+- and Ca2+-permeable channels
-
•
EukCatAs underpin voltage-activated Ca2+ signaling and membrane excitability
-
•
EukCatAs may have functionally replaced 4D-Cav/Nav channels in pennate diatoms
Diatoms exhibit fast animal-like action potentials, but many species lack 4D-Cav/Nav channels that underpin membrane excitability in animals. Diatoms do encode novel 1D voltage-gated channels (EukCatAs). Helliwell, Chrachri et al. show that EukCatAs are fast Na+ and Ca2+ channels that provide alternative mechanisms for rapid signaling in eukaryotes.
Results and Discussion
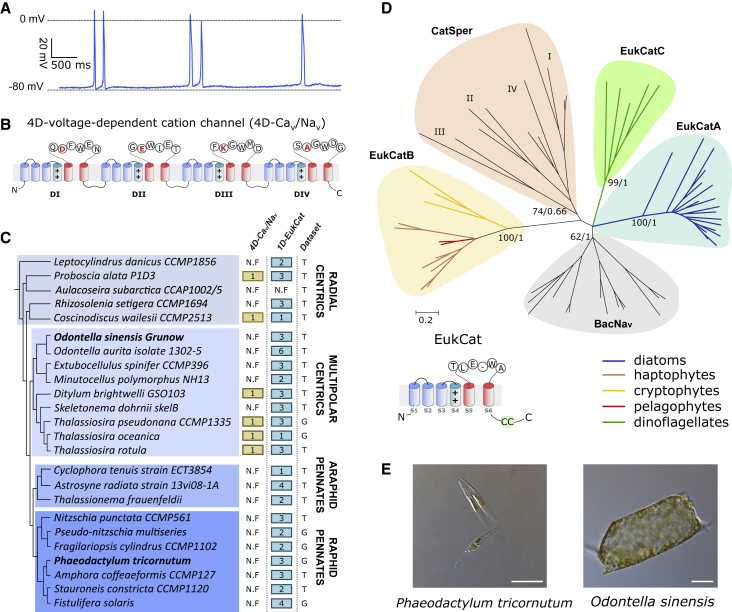
Diatoms are a diverse group of unicellular algae characterized by their ability to produce a silicified cell wall (frustule). They are abundant primary producers in marine and freshwater ecosystems, particularly in coastal waters [11]. Diatoms are typified by their ability to divide rapidly when they encounter favorable conditions, and sophisticated signaling mechanisms most likely contribute to their ecological success [12]. The centric diatom Odontella sinensis exhibits spontaneous action potentials resembling those produced by 4D-Cav/Navs [10, 13] (Figures 1A and 1B). However, the molecular basis and functional roles of action potentials in these non-motile phytoplankton cells are unknown. Our previous surveys of diatom genomes identified that the centric diatom Thalassiosira pseudonana, but not the model pennate Phaeodactylum tricornutum, encodes a 4D-Cav/Navs gene homolog [4]. In contrast, both genomes contain uncharacterized single-domain channels resembling prokaryote BacNav channels, first characterized from Bacillus halodurans (NaChBac) [14]. NaChBac yields voltage-gated, Na+ selective currents, with activation and inactivation kinetics typically 10–100 times slower than those of mammalian 4D-Navs [14], although representatives from marine bacteria (e.g., NavShep from Shewanella putrefaciens) are considerably faster [15]. We therefore reasoned that the single-domain channels identified in diatom genomes could contribute to membrane excitability if they are strongly voltage gated and exhibit appropriate activation and inactivation kinetics.
Figure 1.
Novel Classes of 1D Voltage-Gated Cation Channels (EukCats) Are Widely Distributed in Important Eukaryotic Phytoplankton Taxa
(A) Free running membrane potential (current clamp recording) demonstrating firing of spontaneous action potentials in the diatom O. sinensis [10].
(B) Schematic diagram of a four-domain, voltage-dependent cation channel (4D-Cav/Nav), showing SF with canonical “DEKA” locus of Na+ selective 4D-Nav1 s [4]. The voltage-sensing S4 domain is also highlighted (++).
(C) Distribution of 4D-Cav/Nav and novel eukaryote single-domain (1D) voltage-dependent cation channels (EukCat) in diatoms. The latter are ubiquitous in diatom genomes and transcriptomes, whereas 4D-Cav/Navs are restricted to certain centric diatoms. Data are derived from genome (G) and transcriptome (T) datasets, as indicated in the right column. N.F. denotes not found. Species from which genes were characterized in this study are highlighted in bold. Protein IDs for all species shown are in Data S1.
(D) Maximum likelihood phylogenetic tree of single-domain voltage-gated cation channels, including BacNavs, CatSpers, and novel eukaryote channels (EukCats). Three distinct classes (EukCatA-C) can be observed. ML bootstrap values (>70) and Bayesian posterior probabilities (>0.95) are indicated on selected nodes. Branch colors denote taxonomic group for EukCat channels. Details of sequences used to construct the phylogenetic tree are given in Data S2. Schematic diagram of a EukCat channel with SF of Phaeodactylum tricornutum protein (PtEUKCATA1; protein ID 43878) is also shown (lower left), indicating the position of transmembrane domains, selectivity filter, and coiled-coil domain (CC). By comparison, the OsEUKCATA1 (protein ID CAMPEP_0183296650) exhibits similar structural features to PtEUKCATA1 (50.7% amino acid identity) but has an SF motif of TLDAWAD.
(E) Differential interference contrast microscopy images of diatoms P. tricornutum (scale bar represents 10 μm) and Odontella sinensis (scale bar represents 50 μm) are shown.
To further examine the mechanisms underpinning membrane excitability in diatoms, we determined the broader distribution of 4D-Cav/Navs in available diatom genome and transcriptome databases. 4D-Cav/Navs were present in only 6/24 species surveyed (Figure 1C; Data S1), and these were confined to centrics, indicating that pennate diatoms have most likely lost 4D-Cav/Navs. Moreover, although two sequenced centric diatom genomes contain a 4D-Cav/Nav homolog (Figure 1C), a patchy distribution was seen in centric transcriptomes, indicating absence or poor expression. Notably, 4D-Cav/Navs are absent from the transcriptome of O. sinensis and several other mediophyte diatoms. Thus, alternative channels may underpin the fast Na+/Ca2+ action potentials of O. sinensis.
In contrast, single-domain channels were present in 6/6 of diatom genomes and 17/18 of transcriptomes examined, spanning all major classes (Figure 1C). We also identified similar channels in several other important eukaryote phytoplankton (including haptophytes, dinoflagellates, cryptophytes, and pelagophytes). Notably, these represent some of the most ecologically significant marine phytoplankton taxa [16] (Data S1). Phylogenetic analyses reveal that the single-domain channels group into three strongly supported clades (Figure 1D). Clade A includes diatoms; clade B representatives of the haptophytes, cryptophytes, and pelagophytes; and clade C dinoflagellates. These clades were phylogenetically distinct from BacNavs and CatSpers (a specialized family of weakly voltage-gated, single-domain channels present in mammalian sperm) [17, 18]. We have thus collectively termed these novel, single-domain eukaryote channels 1D-EukCats. Our findings highlight that single-domain, voltage-gated channels are far more prevalent in eukaryotes than previously recognized and thus warrant further attention.
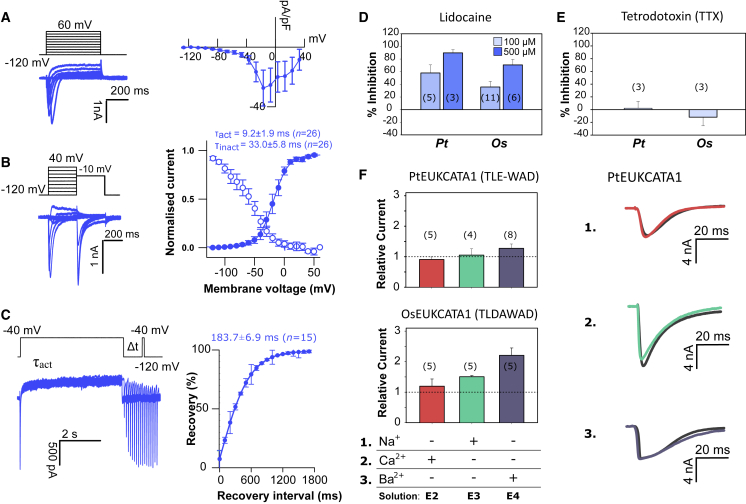
O. sinensis action potentials arise from rapid Na+- and Ca2+-based, depolarization-activated currents with activation and inactivation kinetics strongly resembling animal 4D-Nav/Cavs [10]. Furthermore, the anesthetic lidocaine significantly inhibited these currents, but not tetrodotoxin (TTX), the mammalian 4D-Nav1 blocker [3]. Diatom EukCatAs exhibit the typical organization of voltage-gated channel sub-units, with six predicted transmembrane segments (S1–S6), including the conserved arginine-rich S4 segment associated with voltage activation (Figures 1D and S1A) and a selectivity filter motif (SF) (Figure S1B) [14]. A coiled-coil domain is also present, which in BacNavs is involved in tetramerization and gating [19]. We generated codon-optimized constructs for heterologous expression in human HEK293 cells. We chose representative sequences from the genetically tractable diatom Phaeodactylum tricornutum [20] (PtEUKCATA1) and from O. sinensis (OsEUKCATA1) (Figures 1E, S1A, and S1B; Methods S1). These were expressed as C-terminal GFP fusions. To confirm PtEUKCATA1 and OsEUKCATA1 expression, only cells exhibiting GFP fluorescence were used for electrophysiological analysis (Figure S1C). Both constructs yielded robust depolarization-activated inward currents in HEK293 cells, with a voltage of half activation of −18.7 ± 0.4 mV (n = 14; PtEUKCATA1) and −23.8 ± 0.2 mV (n = 12; OsEUKCATA1; Figures 2A and S1D; Table S1). EukCatA channels also exhibited rapid kinetics (Figures 2B, 2C, and S1E): τactivation and τinactivation time constants (measured at −10 mV) were 9.2 ± 1.9 ms (PtEUKCATA1), 1.6 ± 0.1 ms (OsEUKCATA1), 33.0 ± 5.8 ms (PtEUKCATA1), and 52.6 ± 2.9 ms (OsEUKCATA1), respectively (Table S1). These are comparable to 4D-Cav/Navs and closely resemble those of the O. sinensis action potentials [10]. EukCatAs also exhibited sensitivity to lidocaine (Figure 2D), but not TTX (Figure 2E). These studies indicate that EukCatAs represent a novel class of voltage-gated channels in eukaryotes.
Figure 2.
Diatom EukCatAs Are Rapid-Depolarization-Activated Na+- and Ca2+-Permeable Channels
(A) Typical current traces for HEK293 cells expressing PtEUKCATA1 in response to membrane depolarization. Average peak current-voltage (I-V) curve (right), where current was normalized to cell capacitance. Peak current: −27.4 ± 12.0 pA/pF (error bars, SEM; n = 8).
(B) Steady-state inactivation: representative current traces (left) used to obtain steady-state inactivation curves (right) for PtEUKCATA1 (n = 17). Average normalized data were fitted using the Boltzmann equation (see STAR Methods) for both activation (solid circles) and steady-state inactivation (open circles) curves. Error bars: SEM (see Table S1 for time constants).
(C) Recovery from inactivation; superimposed currents obtained by a double-pulse protocol using a varying interval (Δt) between the two voltage pulses (left). Holding potential was −120 mV and the test pulse 40 mV for 100 ms, and the recovery pulse of −40 mV for 5 s was applied between 100 ms and 1,700 ms after the test pulse. The peak currents elicited by the recovery pulse were normalized in order to construct the recovery curve. A single exponential was fitted to the averaged normalized recovery curve yielding τ for recovery (first order exponential fit) of 650 ± 33.5 ms (error bars, SEM; n = 3).
(D and E) Impact of lidocaine (D) and tetrodotoxin TTX (10 μM; E) on diatom EukCatA channels: PtEUKCATA1 and OsEUKCATA1. Plots show mean % inhibition (relative to control current); error bars: SEM, n shown in parentheses. Inhibition of native HEK cell Na+ currents by TTX (10 μM) is also shown. TTX had no statistically significant impact on currents PtEUKCATA1 and OsEUKCATA1 (Student’s t test: p values: >0.5; n = 3).
(F) Mean peak currents generated following external cation substitution relative to the control external solution for PtEUKCATA1 (SF: TLE-WAD) and OsEUKCATA1 (SF: TLDAWAD; with representative traces for PtEUKCATA1, right; relative current was calculated by dividing the peak current in the test solution by the peak current in the control extracellular solution E1; Table S2). Replacement of extracellular Na+ with NMDG did not affect EukCatA currents. Similarly, removal of Ca2+ had no effect. Replacement of Ca2+ with Ba2+ (in the absence of Na+) slightly enhanced the EukCatA currents. Error bars: SEM; nl shown in parentheses. Extracellular and intracellular (pipette) solutions are given in Table S2.
See also Figure S1, Methods S1, and Tables S1 and S2.
Na+ selectivity of BacNav channels is mediated by a conserved SF. Modification of the B. halodurans NaChBac SF from TLESWAS to TLDDWAD caused this highly selective Na+ channel to become Ca2+ permeable [21, 22]. The SFs of diatom EukCatAs (PtEUKCATA1, TLE-WAD; OsEUKCATA1, TLDAWAD) differ from the Na+-selective BacNavs (Figure S1B). To determine Na+ and Ca2+ dependency of EukCatA currents, we performed several cation replacements in the external solution. Replacement of Na+ with N-methyl-D-glucamine (NMDG), leaving only Ca2+ as the major extracellular cation, did not affect the voltage-dependent inward current for PtEUKCATA1 or OsEUKCATA1 (Figure 2F). Moreover, no reduction in maximal current was seen for either EukCatA channel following removal of extracellular Ca2+, leaving Na+ as the major external cation (Figure 2F). This demonstrates that EukCatAs are permeable to both Na+ and Ca2+. In summary, EukCatAs expressed in heterologous systems exhibit similar voltage dependency, kinetics, pharmacology, and selectivity to O. sinensis action potentials (Table S1) and thus offer alternative mechanisms to 4D-Cav/Navs for rapid Na+/Ca2+ membrane excitability in diatoms.
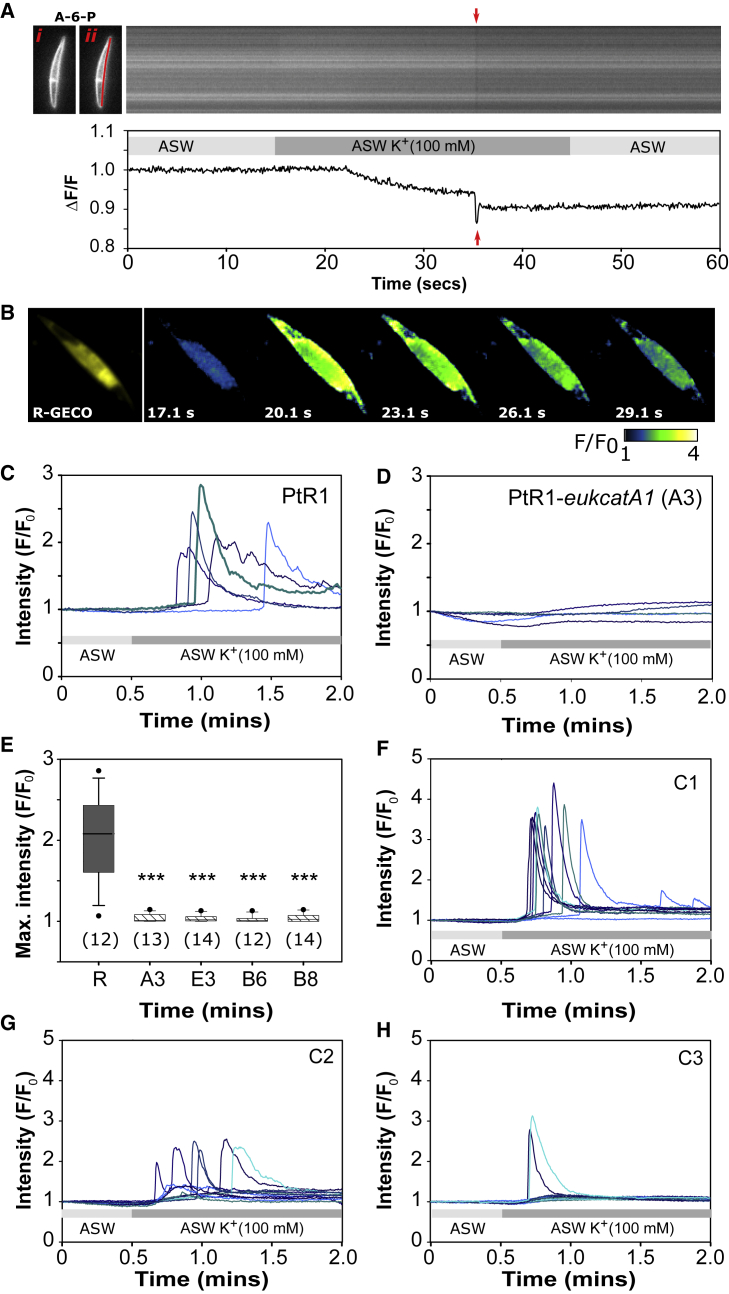
Techniques for genetic manipulation are currently unavailable in O. sinensis and other large diatoms amenable to electrophysiological approaches. We therefore used the model pennate P. tricornutum to test the role of EukCatAs in diatom excitability. This species encodes three EukCatA isoforms, but 4D-Cav/Navs are absent [4]. We examined membrane potential in wild-type P. tricornutum cells using the voltage-sensitive dye Annine-6-Plus (A-6-P) [23, 24] (Figure 3A). Perfusion of P. tricornutum with elevated K+ (100 mM), which depolarizes the plasma membrane in other diatoms [25], resulted in a gradual depolarization in the majority of cells (84%; n = 37 cells; Figure 3A). In addition, we detected very rapid depolarization events resembling action potentials in 23% of cells (n = 35 cells; 12.5 fps; 15 replicate experiments). This indicates that P. tricornutum cells exhibit membrane excitability, although it is likely that limitations in image acquisition speed and/or sensitivity resulted in under-sampling of action potentials using voltage imaging [26, 27].
Figure 3.
PtEUKCATA1 Is Required for Depolarization-Activated Ca2+ Signaling
(A) Left: (i) shows an epifluorescence microscopy image of a Phaeodactylum tricornutum cell stained with the voltage-sensing dye Annine-6-Plus (A-6-P) [23, 24]. The kymograph shows changes in A-6-P fluorescence along the highlighted section of the plasma membrane (ii) following exposure to ASW + 100 mM K+ (after 15 s for 30 s; 10 frames/s). Transient decreases in fluorescence (red arrows), indicative of rapid depolarization of membrane potential, can be observed following the much slower initial depolarization in response 100 mM K+. ΔF/F over time (s) is plotted below. Calibration of A-6-P in rat INS-1 cells using a similar imaging configuration indicated a decrease in fluorescence of 10% for membrane depolarization of 100 mV [23].
(B) Left: an epifluorescence microscopy image of a P. tricornutum (PtR1) cell expressing cytoplasmic-localized R-GECO Ca2+ reporter. Right: pseudocolored time-lapse images following membrane depolarization by perfusion with ASW containing elevated K+ (100 mM). Time (s) after treatment is indicated. Pseudocolor represents the change in fluorescence (F/F0), indicating a rise in cytosolic Ca2+.
(C) Representative cytosolic Ca2+ elevations due to membrane depolarization. Change in fluorescence intensity of R-GECO in 5 representative PtR1 cells exposed to 100 mM K+ is shown. The experiment was carried out on three independent subcultures of the PtR1 line, with similar results.
(D) The effect of membrane depolarization on PtR1-eukcatA1 knockout mutant (line A3). 100 mM K+ did not induce cytosolic Ca2+ elevations in the PteukcatA1 mutant; 5 representative cells are shown. The experiment was carried out on three independent subcultures of the mutant line A3, with similar results.
(E) Mean maximal change in fluorescence of R-GECO in response to depolarization (100 mM K+) for PtR1 and four independent PtR1-eukcat-A1 mutant lines (A3, E3, B6, and B8). Error bars: SEM; number of cells examined per line (n) over 3 independent experiments; p values (Student’s t test): ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(F–H) Complementation of depolarization-activated Ca2+ signaling phenotype in PtR1-eukcatA1 mutant A3 transformed with WT PtEUKCATA1 gene. Representative fluorescence traces of cells of three independent complemented lines are shown: C1 (F), C2 (G), and C3 (H) exposed to ASW with elevated K+ (100 mM) after 30 s for 90 s. The experiment was carried out on three independent occasions with similar results.
See also Figures S3 and S4.
We therefore developed tools for Ca2+ imaging in single P. tricornutum cells because EukCatAs are Ca2+ permeable and [Ca2+]cyt elevations are routinely used as measurements of neuronal membrane excitability [26]. We generated a transgenic strain of P. tricornutum stably expressing the intensiometric fluorescent Ca2+ indicator R-GECO (line PtR1; Figure 3B). To validate the ability of PtR1 cells to reproducibly report [Ca2+]cyt, we treated cells with a hypo-osmotic shock, which induces robust [Ca2+]cyt elevations in aequorin expressing P. tricornutum [12]. Treatment with 90% artificial seawater (ASW; diluted with deionized water) resulted in a single large transient increase in R-GECO fluorescence (average maximum intensity 2.53 ± 0.844; n = 12; SEM), which was dependent on external Ca2+ (Figure S2) [28]. We next examined the response of PtR1 cells to membrane depolarization. Compared to hypo-osmotic shock, treatment with 100 mM K+ resulted in smaller (average maximum intensity 2.05 ± 0.1; n = 12; SEM) and more sustained Ca2+ elevations (94% of cells; n = 54) that typically initiated after a short delay (average delay to maximal fluorescence: 33.4 ± 4.77 s; Figures 3B and 3C). The observed variability in the timing of this response is most likely due to differences in resting potential and/or the threshold for depolarization between cells. Direct imaging of membrane potential in PtR1 cells indicated that action potentials, when observed, directly preceded the [Ca2+]cyt elevation in each case (n = 4 cells exhibiting action potentials; Figure S3A). Given that PtEUKCATA1 is Na+ and Ca2+ permeable, we examined whether action potentials could occur without external Ca2+. Treatment of wild-type (WT) cells with ASW containing 100 mM K+, but no Ca2+ (+200 μM EGTA), led to multiple action potentials (45% of n = 23 cells; over 6 replicate experiments; Figures S3B–S3D). This indicates that Na+ conductance is sufficient to generate action potentials in P. tricornutum, as has also been reported for O. sinensis when Ca2+ is removed [29]. Moreover, multiple action potentials were more frequently observed in the absence of Ca2+ (7/10 cells exhibited multiple action potentials, compared to 1/8 cells in the presence of Ca2+). In contrast, K+-mediated Ca2+ elevations were not observed in PtR1 cells in the absence of Ca2+ (Figure S3E), indicating that external Ca2+ is necessary for depolarization-activated Ca2+ signaling in P. tricornutum. Together, these results suggest that a Ca2+-dependent negative feedback mechanism may regulate P. tricornutum membrane excitability. Thus, despite lacking 4D-Cav/Nav channels, plasma membrane depolarization in P. tricornutum leads to rapid action-potential-like depolarization events and [Ca2+]cyt elevations.
In addition to PtEUKCATA1, the P. tricornutum genome contains two further EukCatA isoforms that share 49.7% and 50.0% amino acid sequence identity (JGI protein IDs 54164 and 43828, respectively; Figure S1). Transcriptome data indicate that PtEUKCATA1 is expressed in P. tricornutum cells in standard liquid culture [30], and we confirmed its expression using RT-PCR. To examine the role of PtEUKCATA1 in generating depolarization-activated Ca2+ elevations, we employed CRISPR-Cas9 gene editing to generate bi-allelic knockout mutants in the PtR1 line (Figures S4A and S4B; STAR Methods). PtR1-eukcatA1 mutants exhibited only a modest reduction in specific growth rate in liquid culture (Figure S4C). However, four mutants were unable to generate depolarization-activated Ca2+ elevations (Figures 3D and 3E). In contrast, the PtR1-eukcatA1 mutants showed no defect in their response to hypo-osmotic stress when compared to WT PtR1 cells (Figures S4D–S4F). This indicates that the Ca2+ signaling phenotype in PtR1-eukcatA1 mutants is due to specific defects in depolarization-activated Ca2+ signaling, rather than broader defects in cellular Ca2+ signaling. The results are consistent with a role for voltage-dependent EukCatA channels in generation of action potentials and initiating Ca2+ elevations. Because two of the PtR1-eukcatA1 mutants (A3 and E3) did not retain the Cas9 gene (Figure S4A), we reintroduced the WT PtEUKCATA1 gene into mutant A3 under its native promoter and confirmed expression of the WT PtEUKCATA1 transcript in these lines (Figure S4G). This resulted in partial or complete complementation of the defective phenotype (n = 3; Figures 3F–3H). These data provide conclusive evidence that PtEUKCATA1 is required for depolarization-activated Ca2+ signaling in P. tricornutum.
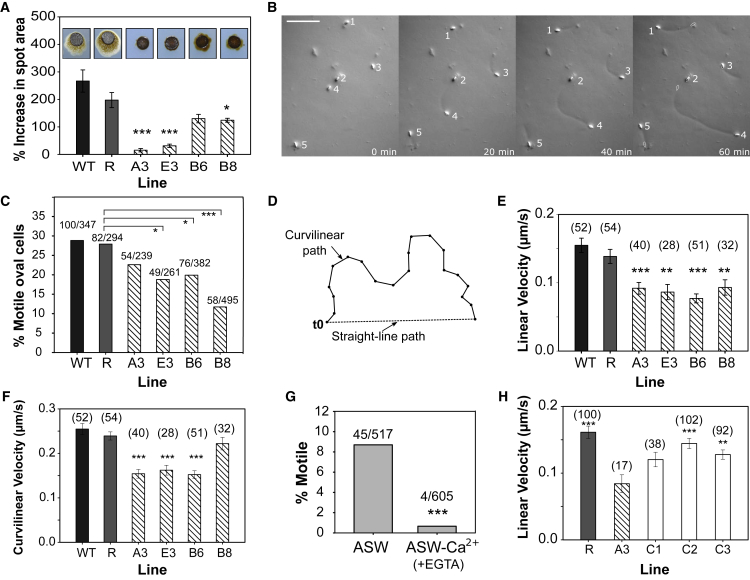
In addition, we noticed a potential defect in cell motility in cells grown on agar plates. P. tricornutum transitions from its planktonic fusiform morphotype to a motile oval morphotype on solid media [31]. The oval cells possess a partially silicified cell wall with a raphe [32], which enables the cell to glide across a solid surface. Evidence suggests that diatom gliding motility is Ca2+ dependent [33]. When uniform numbers of P. tricornutum cells (1 × 104) were spotted onto agar plates, “halos” of motile cells were visible in WT and PtR1 lines after 27 days, but these were consistently reduced in diameter in the PtR1-eukcatA1 mutants (Figure 4A). To investigate whether a defect in motility could account for this observation, we used video microscopy to track motility of individual oval cells (Figure 4B). The PtR1-eukcatA1 mutants exhibited significant defects in the proportion of motile cells (Figure 4C) and in gliding velocity (μm/s) compared to PtR1 and WT (Figures 4D–4F). Video microscopy also confirmed that gliding is Ca2+ dependent in P. tricornutum, as the cells exhibited little or no motility in ASW without Ca2+ (+200 μM EGTA; Figure 4G). Furthermore, a significant restoration of linear velocity was observed for two of the three complemented lines (C2 and C3) compared to mutant A3 (Figure 4H). Together, these results provide strong evidence that PtEUKCATA1 is involved in Ca2+-dependent locomotion in P. tricornutum.
Figure 4.
Gliding Motility, but Not Growth, Is Impaired in PtR1-eukcatA1 Mutants
(A) Cumulative movement of P. tricornutum cells on solid agar. P. tricornutum cells on agar plates adopt the oval morphotype and exhibit gliding motility (demonstrated by a diffuse “halo” around colonies spotted onto an agar plate—inset). % increase in spot area is shown after 27 days. The area of the halo is reduced in PtR1-eukcatA1 mutants compared to PtR1. Error bars indicate SEM; n = 4; p values (Student’s t test): ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. No statistical difference was evident between WT and PtR1 (R).
(B) Time-lapse video microscopy of gliding oval P. tricornutum cells. Oval morphotypes cells (numbered) were placed onto a thin agarose layer and covered with ASW. Gliding cells leave tracks in the agarose, indicating their path. The starting position of the cells is indicated in the final image (white oval). In the images shown, cells either exhibit constant forward motility (cell 4), multiple reversals of direction (cell 5), or no motility (cell 2). Scale bar represents 50 μm.
(C) Percentage of motile cells for WT, PtR1, and four PtR1-eukcatA1 mutant lines over 10-min experiments. p values (Fisher’s exact test): ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(D) Description of metrics used to quantify gliding motility in oval P. tricornutum cells. Linear velocity indicates the displacement from the starting position. Curvilinear velocity is the total distance traveled.
(E) Live-cell imaging of oval cells shows that mean linear velocity is significantly reduced in PtR1-eukcatA1 mutants compared to the PtR1 line. n, total number of cells quantified across 4 independent experiments; error bars indicate SEM; p values (Student’s t test): ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(F) Comparison of curvilinear velocity of the same lines. Error bars indicate SEM; p values (Student’s t test): ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(G) Gliding locomotion in P. tricornutum cells is inhibited in the absence of Ca2+. Cells were prewashed with either ASW or ASW-Ca2+ + 200 μM EGTA prior to motility assay (∗Fisher’s exact test: ∗∗∗p < 0.001).
(H) Mean linear velocity of PtR1-eukcatA1 mutant A3 compared to PtR1 (R) and three independent complemented lines: C1; C2; and C3. Error bars indicate SEM; p values (Student’s t test): ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p ≤ 0.001.
See also Figure S4.
Our work demonstrates that EukCatAs are important in voltage-regulated Ca2+ signaling and in the Ca2+-dependent gliding motility. Diatom gliding is vital for vertical migration in sediments [34] and migration toward nutrients [35]. Inhibition of Ca2+ influx leads to a reduced gliding speed in Navicula perminuta, although it does not impair the photophobic response [33]. Ca2+ influx through EukCatAs may therefore be important for processes associated with propulsion, e.g., the secretion of mucopolysaccharide or regulation of cytoskeleton dynamics [36]. However, it is also clear that EukCatAs most likely play wider roles in diatom biology. Gliding is only observed in the oval morphotype in P. tricornutum, although PtEUKCATA1 clearly contributes to Ca2+ signaling in response to membrane depolarization in fusiform cells (Figure 3B), which are non-motile. Furthermore, gliding motility is only found in pennate diatoms, whereas EukCatAs appear to be present in all diatoms. Fast Ca2+-dependent signaling processes have been identified in non-motile diatoms that could be mediated by EukCatAs. For example, the centric diatom Pleurosira laevis rapidly re-distributes its chloroplasts following mechanostimulation that is proposed to be mediated by a Ca2+ influx following plasma membrane depolarization [37]. Depolarization-regulated channels are prevalent in plant and animal lineages and critically underpin many cellular processes, including solute transport [38], pathogen detection [39, 40], and cellular wounding responses [41]. The ubiquitous presence of EukCatAs within diatoms suggests that they may also contribute to diverse signaling processes.
Notably, our findings demonstrate that EukCatAs provide alternative mechanisms for fast Na+/Ca2+-based electrical excitability in eukaryotes. Unlike EukCatAs, 4D-Cav/Navs are absent in many diatom taxa and appear to have been lost entirely in pennate species. However, although the model pennate P. tricornutum lacks 4D-Cav/Nav channels, it still exhibits very rapid depolarization events (Figure 3A). This suggests that EukCatAs can functionally replace 4D-Cav/Navs and may have contributed to their loss or functional diversification in the pennate diatoms. This functional redundancy means that, in contrast to land plants, the loss of 4D-Cav/Navs has not resulted in the loss of rapid Na+/Ca2+-based excitability in the pennate diatoms. Excitability is observed in certain specialized plant cells, such as the Venus flytrap sensory hair cell that displays somewhat slower action potentials (τ1/2 of ∼0.3 s), with the depolarizing phase most likely carried through rapid R-type Cl− channels [42]. Further work is required to establish the nature of the repolarizing outward conductance of diatom action potentials, which is typically carried by voltage-dependent K+ outward rectifier channels in excitable animal cells.
Outside of the diatom lineages, we identified two other classes of EukCats, found in haptophytes, cryptophytes, and pelagophytes (EukCatBs) and dinoflagellates (EukCatCs). The relationships between the EukCat clades were not well resolved, and they may not share common ancestry. However, it is interesting that the EukCats are found only in photosynthetic lineages with red-alga-derived plastids that dominate marine phytoplankton communities [43]. The broad distribution of EukCats among major phytoplankton lineages suggests that 1D voltage-gated cation channels are likely to play important and previously unforeseen roles in eukaryote biology. Gaining further insight of the wider properties of the different EukCat classes is therefore vital to determine their functional roles and better understand the evolution of environmental sensing mechanisms in the oceans more broadly.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli Top10 | Thermo Fisher Scientific | Cat. No. C404003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tetrodotoxin (citrate) | Tocris Bioscience | Cat. No. 1078 |
| Nifedipine | Sigma-Aldrich | Cat. No. N7634 |
| Spermidine | Sigma-Aldrich | Cat. No. S2626 |
| Tungsten M-10 Microcarriers | BioRad | Cat. No. 1652266 |
| Nourseothricin | Jena BioScience | Cat. No. AB-102L |
| Poly-L-lysine | Sigma Aldrich | CAS 25988-63-0 |
| Zeocin | InvivoGen | Cat. No. ant-zn-1 |
| Antibiotic Antimycotic (from GIBCO) | LIFETECH | A5955-100ML |
| Opti Mem (from GIBCO) | LIFETECH | 31985062 |
| Fetal Bovine Serum (from GIBCO) | LIFETECH | 10270-098 |
| Critical Commercial Assays | ||
| Phire Plant Direct PCR Kit | Thermo Fisher Scientific | Cat. No. F130WH |
| Q5 Site-Directed Mutagenesis Kit | (New England BioLabs, Hitchin, UK) | E0554S |
| SuperScript III Reverse Transcriptase | Thermo-Fisher Scientific | 18080093 |
| ISOLATE II RNA Mini Kit | Bioline | BIO-52071 |
| Experimental Models: Cell Lines | ||
| HEK293 Cells | Public Health England Culture Collection or The American Type Culture Collection (ATCC) | ECACC 85120602 |
| Experimental Models: Organisms/Strains | ||
| Phaeodactylum tricornutum (strain CCAP1055/1) | Culture Collection of Algae & Protozoa | https://www.ccap.ac.uk/ |
| Oligonucleotides | ||
| Single guide A RNA oligo 1 (ccPt43878A_F) | Eurofins | TCGAGATGATGACATTGGAATGGG |
| Single guide RNA A oligo 2 (ccPt43878A_R) | Eurofins | AAACCCCATTCCAATGTCATCATC |
| Single guide RNA B oligo 1 (ccPt43878B_F) | Eurofins | TCGAGGAGGAATACTACTGGGCCT |
| Single guide RNA B oligo 1 (ccPt43878B_F) | Eurofins | AAACCCCATTCCAATGTCATCATC |
| Cas9_F | Eurofins | CTTCGACCTTGCGGAAGATG |
| Cas9_R | Eurofins | CCGGACGAGAGCTTTAAGGA |
| PtEUKCATA1_F | Eurofins | TTTTGGTGCTTATTCTCTACGTC |
| PtEUKCATA1_R | Eurofins | TATGCGTTCTTGGGTCTCCT |
| Recombinant DNA | ||
| pcDNA3.1-C-eGFP | Genscript | N/A |
| NaChBac pTracer CMV2 | Addgene [14] | 60835 |
| pCLS16604_pNAT | Fayza Daboussi (French National Institute for Agricultural Research) [44] | N/A |
| pKSdiaCas9_sgRNA | Addgene [20] | 74923 |
| pKSdiaCas9_sgRNA ccPt43878A | This study | N/A |
| pKSdiaCas9_sgRNA ccPt43878B | This study | N/A |
| Pphat1 | https://www.ncbi.nlm.nih.gov/nuccore/AF219942 | AF219942 |
| pcDNA3.1-C-Egfp_PtEUKCATA1 | This study | N/A |
| pcDNA3.1-C-Egfp_OsEUKCATA1 | This study | N/A |
| Software and Algorithms | ||
| Geneious | Biomatters Limited | https://www.geneious.com/ |
| Clampex 10.2 acquisition software | Molecular Devices, Sunnyvale, California | N/A |
| pClamp software | Molecular Devices, Sunnyvale, California | N/A |
| NIS-ELEMENTS v.3.1 software | Nikon Instruments Europe B.V. | https://www.nikoninstruments.com/en_GB/Products/Software/NIS-Elements-Advanced-Research/NIS-Elements-Viewer |
| ImageJ | [45] | N/A |
| MtrackJ | [46] | https://imagescience.org/meijering/software/mtrackj/ |
| MEGA7 | [47] | https://www.megasoftware.net/ |
| BEAST v1.8 | [48] | http://beast.community/ |
| SigmaPlot (version 11) | http://www.sigmaplot.co.uk/ | |
| PHYTOCRISPEX | [49] | https://www.phytocrispex.biologie.ens.fr/CRISP-Ex/ |
| The Broad Institute sgRNA design program | https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design | |
| Other | ||
| 35 mm glass-bottomed dishes | IBL Baustoff + Labor GmbH | D35-10-0-N |
| PDS-1000/He Particle Delivery System | Bio-Rad, Hercules, CA, USA | N/A |
| 1350 PSI Rupture Disks | BioRad | 1652330 |
| He Macrocarriers | BioRad | 165-2257 |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Colin Brownlee (cbr@mba.ac.uk).
Experimental Model and Subject Details
Strains and culturing
Algal strains
Phaeodactylum tricornutum strain CCAP1055/1 was obtained from the Culture Collection of Algae and Protozoa (SAMS limited, Scottish Marine Institute (Oban, UK)) (Key Resources Table). P. tricornutum cells were maintained in either filtered seawater (FSW) or artificial seawater, ASW (450 mM NaCl, 30 mM MgCl2, 16 mM MgSO4, 8 mM KCl, 10 mM CaCl2, 2 mM NaHCO3, and 97μM H3BO3), supplemented with f/2 nutrients [50], with 100cμM Na2SiO3.5H2O, but not vitamins). Cultures were grown routinely in FSW, but were acclimated to ASW for 1-2 weeks prior to Ca2+ imaging experiments that required defined media. Cultures were maintained at 18°C under 50-80 μmol m−2 s−1 light on a 16τ8 h light:dark cycle.
Cell lines
HEK293 cells (ATCC CRL-1573) were grown in a humidified incubator at 37°C in 5% CO2 and 95% O2. Growth medium consisted of high glucose DMEM–Dulbecco’s Modified Eagle Medium with Antibiotic Antimycotic (GIBCO), and 10% FBS (GIBCO). Cells were passaged every 3 – 4 days at 1:6 or 1:12 dilutions (cell/mm2).
Method Details
Bioinformatics analysis
Sequence similarity searches were carried out to survey a broad range of eukaryote genomes and transcriptomes for single and four-domain voltage-gated ion channels (Data S1; Figure 1). Query sequences from Bacillus halodurans C-125 NaChBac (protein id: BAB05220.1) and BacNav-like sequences previously identified in diatom (P. tricornutum protein id: 43878) [4, 51], in addition to the 4D-Cav/Nav sequence of T. pseudonana (protein id: 22071) were used. Transcriptome databases surveyed were obtained from the Marine Microbial Eukaryote Sequencing Project (MMETSP, https://www.imicrobe.us/#/projects/104) [52]. The MMETSP databases provide transcriptome sequencing resources for ecologically significant marine microbial eukaryotes spanning the breadth of the eukaryotic tree of life (with representatives from all the major eukaryote super groups including the Archaeplastida, Alveolates, Stramenopiles, Rhizaria, Opisthokonts, Amoebozoa and Excavata). Eukaryote genomes from a range of sources were also used. The genomes of E. huxleyi [51], T. pseudonana [53], P. tricornutum [54], Pavlovales sp. CCMP2436, Aureococcus anaphagefferens clone 1984, Volvox carteri f. nagariensis EVE [55], Coccomyxa subelipsoida C-169 [56], Ostrecocccus lucimarinus CCMP2514, Ostrecocccus tauri OTH95 [57], Micromonas sp. RCC299 [58], Porphyra umbilicalis [59], Pelagophyceae sp. CCMP2097, Pseudo-Nitzschia multiseries CLN-47, Guillardia theta CCMP2712, and Bigelowiella natans CCMP2755 were obtained from Joint Genome Institute http://genome.jgi.doe.gov/. Further searches were performed at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Thalassiosira oceanica), the Cyanidioschyzon merolae genome project (http://merolae.biol.s.u-tokyo.ac.jp/) [60], the Cyanophora Genome project (http://cyanophora.rutgers.edu/cyanophora/home.php) [61], Symbiodinium microadriaticum portal (http://reefgenomics.org/blast/) [62], DictyBase (http://dictybase.org/; Dictyostelium discoideum), EnsemblProtists (http://protists.ensembl.org/index.html; Trypanosoma brucei, Naegleria gruberi and Acanthamoeba castellanii) or the Ectocarpus siliculosus genome portal (http://bioinformatics.psb.ugent.be/orcae/) [63].
Databases were searched using BLASTP and TBLASTN with an E-value cut off score of 1E−10. Each hit was inspected manually for relevant protein domains using Interpro [64], looking specifically for voltage-sensing domain (IPR005821), ion transport domain (IPR027359) and EF hands (IPR011992). The presence of a minimum of three pore domains was used as a threshold for candidate 4D-Cav/Navs in order to distinguish them from other voltage-gated cation channels. Protein hits of all ids reported in this study are given in Data S1.
Phylogenetic analyses of EukCat, Catsper and BacNav sequences were performed using multiple sequence alignments generated with MUSCLE via the Molecular Evolutionary Genetics Analysis (MEGA7) software [47]. After manual refinement, GBLOCKS0.91B was employed to remove poorly aligned residues, using the least stringent parameters [65], resulting in an alignment of 172 amino acid residues. Maximum likelihood trees were generated using MEGA7 with 100 bootstraps. Model analysis was performed in MEGA7 to determine an appropriate substitution model (WAG+G+I). Bayesian posterior probabilities were additionally calculated using BEAST v1.8.4 [48] running for 10000000 generations.
Synthesis of heterologous expression plasmids for HEK293 cells
Amino acid sequences of proteins used for heterologous expression are described in Methods S1. Coding sequences for OsEUKCATA1 were obtained from MMETSP transcriptomic datasets: O. sinensis (protein id: CAMPEP_0183296650; transcriptome database id: MMETSP0160). To confirm these sequences we amplified the open reading frame (ORF) from cDNA made from liquid cultures of O. sinensis (using the primers: Osinensis_F1: ATGAAGGACGAGAACAGCATCCC, Osinensis_R1: AGAATCAGTCTGGTTTTGTTGAAGATGCAC). The coding sequence for PtEUKCATA1 (protein id: 43878) was predicted by the JGI genome project for P. tricornutum. To confirm correct prediction of intron/exon boundaries for this gene model, the predicted ORF of PtEUKCATA1 was amplified from cDNA made from a liquid culture of P. tricornutum CCAP1055/1 (using the primers: Pt43878_F: GCCATCCGATGATGCAAGGAATCGTGGAG and Pt43878_R: AAACATTCTCGGGGACTTCTC). cDNA was synthesized using SuperScript III reverse transcriptase from RNA extracted using ISOLATE II RNA Mini Kit (Bioline) following the manufacturer’s instructions. Codon-optimized versions of the transcripts were then synthesized (GenScript, Piscataway, NJ) for characterization in human expression systems, and sub-cloned into pcDNA3.1-C-eGFP using HindIII and BamHI. A 6 bp Kozak sequence (GCCACC) was included upstream of the ATG, and the stop codon removed.
Transfection of HEK293 cells
HEK293 cells were plated for transfection onto glass-bottom (35mm) Petri-dishes coated with poly-L-lysine (ibidi GmbH, Germany) to help with cell adhesion. Transfections of HEK293 were performed with 4 μL Lipofectamine 2000 (Invitrogen) and 1-2.5 μg plasmid DNA per 35 mm2, each prepared separately with Opti-MEM (GIBCO). The lipofectamine and DNA were mixed and allowed to rest for 5 min. before 200 μL of the mixture was added to each plate. After 12-30 h of incubation, cells were rinsed and maintained with fresh growth media and kept in the incubator at 37°C with 5% CO2/95% O2 until used for electrophysiological experiments. Expression of the transgene was confirmed by fluorescence microscopy.
HEK293 whole cell patch-clamp electrophysiology
Electrophysiological recordings were carried out at room temperature with an Axopatch 200B or Multiclamp 700B amplifier (Molecular Devices, Sunnyvale, California) through a PC computer equipped with a Digidata 1332 analog-to-digital converter in conjunction with pClamp 9.2 or pClamp10.1 software (Molecular Devices, Sunnyvale, California). Patch electrodes were pulled from filamented borosilicate glass (1.5 mm OD, 0.86mm ID) using a P-97 puller (Sutter Instruments, Novato, CA, USA) to resistances of 2-5 MΩ. For analysis of OsEUKCATA1), unpolished electrode tips were coated with beeswax to minimize pipette capacitance. Voltage errors incurred from the liquid junction potentials (LJPs) and series resistance (recorded from the amplifier) were corrected by subtraction post hoc. These corrected voltages were used to plot IV curves and in all subsequent investigations. The amplitudes of the currents were measured from the baseline to the peak value and were normalized for cell capacitance as whole-cell current densities (pA/pF). Activation curves were derived by plotting normalized sodium conductance (GNa) as a function of test potential and fitted with the Boltzmann equation:
| (Equation 1) |
where I is the peak current at the given test potential Vm, Vrev is the reversal potential, Gmax is the maximal slope conductance, V0.5 is the half-point of the relationship, and k is the slope factor. Voltage-dependent inactivation was similarly determined with a Boltzmann equation:
| (Equation 2) |
where V0.5 isis the voltage at which 50% of the current is inactivated (the midpoint of the inactivation curve), and k is the slope factor. Statistical analyses were performed with Sigma Plot 11.0 (Systat Software, Inc., Chicago, IL). Data are shown as the mean ± SEM (n, number of experiments).
Native phytoplankton cell patch-clamp recording and analysis
O. sinensis single electrode voltage clamp recordings were obtained as previously described [10, 13]. Briefly, cells were plated into coverslip dishes in ASW consisting of; 450 mM NaCl, 30 mM MgCl2, 16 mM MgSO4, 8 mM KCl, 10 mM CaCl2, 2 mM NaHCO3 pH 8.0. Cells were impaled through the girdle band with a sharp microelectrode filled with 1 M KCl (resistance 10 MΩ) mounted on the headstage of an Axoclamp 900A amplifier (Molecular Devices), controlled with a Sutter MP285 motorized micromanipulator (Sutter Instruments, Petaluma, CA). Current and voltage signals were pre-amplified 5- 10 × before being acquired using a Digidata 1200 with Clampex 10.2 acquisition software (Molecular Devices, Sunnyvale, CA). The gain for the switch clamp was between 5-10 and switching frequency > 13 kHz for the data presented.
Generation of P. tricornutum constructs
We employed the CRISPR-Cas9 vector developed by Nymark et al., (2016) for editing the PtEUKCATA1 gene (protein id: 43878) in P. tricornutum [20]. We designed two sgRNAs targeted to generate a short deletion, which has proven efficient for high-throughput screening for bi-allelic mutants via PCR in diatoms [66]. A library of candidate sgRNAs was generated using the PHYTOCRISPEX [49] web tool with default parameters (NGG PAM, and CRISPR start from ‘G’). The Broad Institute sgRNA design program (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) was used subsequently to obtain ‘on-target’ efficiency scores. Two 20 bp guide RNAs (ccPt43878A: GATGATGACATTGGAATGGG and ccPt43878B: GGAGGAATACTACTGGGCCT) that passed the PHYTOCRISPEX OFF-target criteria were chosen based on their ON-target scores (0.45 and 0.51, respectively) and position within the gene. Target sgRNAs were predicted to disrupt the region encoding the pore domain of the protein in order to maximize disruption of channel function. Complementary oligos containing flanking overhang sequences corresponding to the pKS diaCas9_sgRNA plasmid (Addgene: 74923) were then synthesized. One μg of complementary oligos for ccPt43878A (ccPt43878A_F: TCGAGATGATGACATTGGAATGGG and ccPt43878A_R: AAACCCCATTCCAATGTCATCATC) and ccPt43878B (ccPt43878B_F: TCGAGGAGGAATACTACTGGGCCT and ccPt43878B_R: AAACCCCATTCCAATGTCATCATC) were annealed in a reaction mix containing 1 × T4 Ligase Buffer (NEB) in a total volume of 50cμl, incubated for 10 min at 85°C and allowed to cool to room temperature. The resulting annealed oligos were ligated into pKS diaCas9_sgRNA using a molar vector to insert ratio of 1:20, and a T4 DNA ligase (Fermentas). Plasmids were verified via Sanger sequencing.
For the PtEUKCATA1 complementation construct, we amplified the PtEUKCATA1 gene from 747 bp upstream of the ATG up to, but not including, the stop codon (TGA) using primers: PtEUKCATA1_comp_F-AACCAATGCATTGGCTGCAGGTCGACTAGGGCCACAGGTA and PtEUKCATA1_comp_R-AAACATTCTCGGGGACTTCTC. The forward primer includes a flanking PstI site, for downstream cloning into a derivative of the pPha-T1 vector (accession AF219942): pPha-T1-Venus vector, using PstI and StuI sites. To make the pPha-T1-Venus vector we synthesized a codon-optimized Venus sequence (accession AJN91098.1) incorporating an EcoR1 and StuI site upstream of the ATG, and a 3′ BamH1 site (GenScript, Piscataway, NJ). The codon optimized Venus construct was then sub-cloned into the pPha-T1 (accession AF219942) vector using EcoR1 and BamH1 sites.
To generate the PtR-GECO1 construct we synthesized (GenScript, Piscataway, NJ) the 1251 bp coding sequence (accession AEO16866.1), which was sub-cloned into the P. tricornutum shuttle vector pPha-T1 (accession AF219942) conferring resistance to the antibiotic zeocin [67] via EcoR1 and BamH1. The construct was transformed into WT P. tricornutum strain CCAP1055/1, using zeocin for selection (see ‘Biolistic Transformation of P. tricornutum’).
Biolistic transformation of P. tricornutum
P. tricornutum cells were grown in liquid culture for 5 days. Cell density was adjusted to 1 × 109 cells/mL and 100 μL spread (in a 3 cm diameter circle) in the center of an f/2 1% agar plate (made up with 50% diluted seawater without Si and vitamins), and left for 24 h in standard growth conditions prior to transformation. The plated cells were transformed via biolistic particle bombardment using the PDS-1000/He Particle Delivery System (Bio-Rad, Hercules, CA, USA). To prepare the DNA-loaded microparticles, 60 mg tungsten (0.6 μm in diameter) particles were washed 3 times in 100% ethanol and twice with sterile deionised water before being re-suspended in 1 mL sterile deionised water and distributed into 50 μL aliquots. A single aliquot of washed tungsten was then coated with 1.5 μg of the relevant plasmid DNA using 2.5 M CaCl2 and 20 mM spermidine (BioUltra, Sigma-Aldrich, cat no. 85558) with continuous vortexing. Coated particles were washed once in 100% ethanol and then re-suspended in 60 μL of 100% ethanol that was subsequently distributed between three macrocarrier disks for particle bombardment. The P. tricornutum agar plates were positioned on the second shelf of the PDS-1000/He Particle Delivery chamber, ∼7.5 cm from the stopping screen. Helium supply with minimum pressure of 1600 psi (300 psi above the burst pressure of the 1350 psi rupture disk) and a vacuum of 23 inch Hg was used to fire the DNA-coated microparticles toward cells. Following particle bombardment cells were incubated for 24 h under standard culturing conditions before being transferred to selection plates (1% agar f/2 -Si and vitamins in 50% diluted seawater) with 300 μg mL−1 nourseothricin65 or 75 μg mL−1zeocin depending on the plasmid. After three weeks colonies were re-streaked onto fresh selection plates.
To generate PtEUKCATA1 mutant lines we co-transformed PtR1 cells with pNAT66 and pKSdiaCas9_sgRNA constructs, using biolistic transformation selecting for nourseothricin resistant colonies. Putative PteukcatA1 mutants were screened via PCR using the Phire Plant Direct PCR Kit (ThermoFisher Scientific) with primers designed to amplify Cas9 (Cas9_F: CTTCGACCTTGCGGAAGATG and Cas9_R: CCGGACGAGAGCTTTAAGGA) and then subsequently with primers flanking the target region for deletion (PtEUKCATA1_F: TTTTGGTGCTTATTCTCTACGTC and PtEUKCATA1_R: TATGCGTTCTTGGGTCTCCT) to identify bi-allelic polymorphisms in this region. The mutant line A3 was subsequently cotransformed with the PtEUKCATA1 complementation construct and a selection plasmid conferring resistance to Blasticidin [68].
Epifluorescence imaging in P. tricornutum
P. tricornutum cells grown in liquid culture (ASW supplemented with f/2 nutrients [50], with 100cμM Na2SiO3.5H2O, but not vitamins) for 72 h were placed in a 35 mm glass-bottomed dish (In Vitro Scientific, Sunnyvale, CA, USA) coated with 0.01% poly-L-lysine (Sigma-Aldrich, St Louis, MO, USA). Cells adhered to the bottom of the dish were imaged at 20°C using epifluorescence microscopy using a Nikon Eclipse Ti microscope with a 40 ×, 1.30 NA oil immersion objective and detection with a Photometrics Evolve EM-CCD camera (Photometrics, Tucson, AZ, USA). Excitation of R-GECO (PtR1) cells was performed using a pE2 excitation system (CoolLED, Andover, UK) with 530-555 nm excitation and 575-630 nm emission filters. Images were captured using NIS-ELEMENTS v.3.1 software (Nikon, Japan) with a 300 ms camera exposure (frame rate of 3.33 frames s−1). For membrane potential imaging cells were stained for 5 min with voltage-sensitive dye Annine-6-Plus (A-6-P; final concentration 0.8 μg/mL), centrifuged for 2 min at 10,000 rpm and resuspended in ASW without A-6-P to minimize background A-6-P fluorescence. Excitation of A-6-P cells was performed in the same manner as for the calcium imaging, using an excitation wavelength of 475-495 nm and emission wavelength of 575-615 nm. Images were recorded at 12.5 frames s−1 with 2 × 2 binning.
For simultaneous determination of membrane potential and [Ca2+]cyt, we loaded PtR1 cells expressing R-GECO with A-6-P. Although there is some overlap in emission spectra of A-6-P and R-GECO, we were able to spatially distinguish between A-6-P localized to the membrane and R-GECO fluorescence in the cytosol. A-6-P fluorescence contributes to background fluorescence in the cytosol, but as A-6-P has a much lower dynamic range than R-GECO, this did not interfere with the detection of [Ca2+]cyt transients. Moreover, membrane depolarization results in a decrease in A-6-P fluorescence, whereas an increase in [Ca2+]cyt causes an increase in R-GECO fluorescence, allowing us to confirm that there was minimal interference between these fluorophores. An excitation wavelength of 475-495 nm and emission wavelength of 575-615 nm were used, with defined regions of interest within the cell.
During imaging, cells were continuously perfused with ASW (3 mL min−1). Depolarization treatments were delivered by switching the perfusion from ASW to ASW with elevated K+ (100 mM) (358 mM NaCl, 30 mM MgCl2, 16 mM MgSO4, 100 mM KCl, 10 mM CaCl2, 2 mM NaHCO3, and 97μM H3BO3 supplemented with f/2 nutrients [50], with 100cμM Na2SiO3.5H2O, but not vitamins) after 30 s for 90 s. For the hypoosmotic shock experiments multiple exposures to ASW diluted with deionised water were carried out. Cells exposed to hypoosmotic shock in the absence of Ca2+ were perfused with at least 50 mL Ca2+ free media (+200 μM EGTA) in order to minimize carryover of residual Ca2+ from the ASW growth medium.
Images were processed using NIS-ELEMENTS v.3.1 software. The mean fluorescence intensity within a region of interest over time was measured for each cell. Change in fluorescence intensity of R-GECO was calculated by normalizing each trace by the initial value (F/F0) (calcium imaging). For membrane potential imaging, ΔF/F was calculated by dividing the time varying fluorescence by the baseline fluorescence.
P. tricornutum motility assays
Solid plate assays
P. tricornutum cells grown in liquid culture (sub-cultured two times previously at 3 day intervals, to ensure maximal and uniform physiological health) under standard growth conditions for 48 h were diluted to uniform cell density (5 × 105 cells/mL). Twenty μL of cells were then spotted on f/2 1% square agar plates, with each plate containing a spot for each of the six genotypic lines examined. To account for population variability the response was examined over cultures derived from four independent clones per cell line with six replicates per clone, and a total of 24 spots assayed for each genotypic line. Plates were positioned at alternating right angles toward the light source to minimize positional effects. After 27 days photographs were taken and ImageJ [45] was used to quantify original spot area, spot spread and calculate percentage increase in area ((Total area-original spot area/original spot area) × 100).
Video Microscopy
Prior to the assay cells were passaged 3 times on solid agar (at 5 day intervals) to ensure maximal transition to oval morphotype and uniform good physiological health between lines. On the third transition, 5 day old cells were scraped from the plate and resuspended into 5 μL f/2 medium before being spread on glass-bottomed dishes with 0.5 mL f/2 1% agar, and covered with liquid f/2 media. Care was taken to ensure that cells were only removed from controlled growth conditions just prior to the motility assay. Cells were then viewed by differential interference contrast microscopy using a Leica DMi8 microscope equipped with a 20 × objective and an environmental chamber to control temperature (18°C). The proportion of oval cells (compared to fusiform and triradiate morphotypes) in the populations used for the assay was calculated and did not vary significantly between lines (accounting for 94%, 95%, 93%, 92%, 99% and 99% of cells for WT, PtR1, mutant A3, E3, B6, and B8 respectively (Figures 4E and 4F). For each analysis, 10 min videos were recorded at four frames/minute, with samples illuminated during image acquisition (camera exposure 200 ms). ImageJ [45] was used with the ImageJ plugin MTRACKJ [46] to track oval cell motility over the time-course of the experiment. Only tracks of oval cells that were not in contact with other cells at the beginning of the video and did not cross paths with other cells were quantified to avoid physical and biological interactions that might influence cell speed and/or path. Linear and curvilinear velocity were determined for each cell and the overall percentage of motile cells was calculated for each line totaling four independent videos for each line. Motility assays both carried out on plates and via video microscopy were done on well-established (at least 12 months old, sub-cultured weekly) transgenic lines (PtR1 and mutant lines A3, E3, B6, and B8) to ensure a stable and robust phenotype.
Quantification and Statistical Analysis
Statistical Analyses
Quantification of data are presented as mean ± standard error of the mean (SEM) with the precise number (n) indicated in the figure legends and where relevant the main text. Statistical analyses were performed using a Student’s t test or a Fisher’s exact test in SigmaPlot. Statistical differences are represented as p∗, < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001.
Data and Software Availability
The accession number for sequences/plasmids in GenBank or Fasta format from this study are given in the Key Resources Table.
Acknowledgments
We thank Dr. Hopes and Professor Mock (UEA, UK) for guidance on CRISPR-Cas9 strategy. We are also grateful to Dr. Bártulos and Professor Kroth for sharing the blasticidin resistance plasmid. We acknowledge support from the European Research Council grant ERC-ADG-670390 (C.B.) and NSF grants 0949744 and 1638838 (A.T.).
Author Contributions
Conceptualization, C.B., G.L.W., K.E.H., A.C., A.R.T., and F.V.; Methodology, K.E.H., A.C., J.A.K., G.L.W., and A.R.T.; Investigation, K.E.H., A.C., J.A.K., A.R.T., C.B., and G.L.W.; Writing, K.E.H., C.B., G.L.W., A.C., A.R.T., and J.A.K.; Funding Acquisition, C.B., G.L.W., and A.R.T.
Declaration of Interests
The authors declare no competing interests.
Published: April 18, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2019.03.041.
Contributor Information
Glen L. Wheeler, Email: glw@mba.ac.uk.
Colin Brownlee, Email: cbr@mba.ac.uk.
Supplemental Information
A. Presence of EukCat channels in eukaryotic phytoplankton taxa. The presence of 4D-Cav/Nav in diatom taxa is also shown. Single-domain channels resembling cation channels of sperm (CatSper) are also indicated. Data source: for transcriptome data, the dataset identifier (MMET ID) of each species is listed. Genome indicates a genome database was searched (see STAR Methods for details of relevant genome databases). Results reported in previous studies are indicated as supplemental references. N.F, not found. B. Protein Ids of EukCat hits identified in this study, and/or source for relevant primary reference. IDs of diatom 4D-Cav/Nav are also given.
The corresponding nucleotide sequences that were codon optimized for expression in HEK293 cells are also given.
References
- 1.Fujiu K., Nakayama Y., Yanagisawa A., Sokabe M., Yoshimura K. Chlamydomonas CAV2 encodes a voltage- dependent calcium channel required for the flagellar waveform conversion. Curr. Biol. 2009;19:133–139. doi: 10.1016/j.cub.2008.11.068. [DOI] [PubMed] [Google Scholar]
- 2.Lodh S., Yano J., Valentine M.S., Van Houten J.L. Voltage-gated calcium channels of Paramecium cilia. J. Exp. Biol. 2016;219:3028–3038. doi: 10.1242/jeb.141234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hille B. Third Edition. Sinauer; 2001. Ion Channels of Excitable Membranes. [Google Scholar]
- 4.Verret F., Wheeler G., Taylor A.R., Farnham G., Brownlee C. Calcium channels in photosynthetic eukaryotes: implications for evolution of calcium-based signalling. New Phytol. 2010;187:23–43. doi: 10.1111/j.1469-8137.2010.03271.x. [DOI] [PubMed] [Google Scholar]
- 5.Liebeskind B.J., Hillis D.M., Zakon H.H. Phylogeny unites animal sodium leak channels with fungal calcium channels in an ancient, voltage-insensitive clade. Mol. Biol. Evol. 2012;29:3613–3616. doi: 10.1093/molbev/mss182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pozdnyakov I., Matantseva O., Skarlato S. Diversity and evolution of four-domain voltage-gated cation channels of eukaryotes and their ancestral functional determinants. Sci. Rep. 2018;8:3539. doi: 10.1038/s41598-018-21897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edel K.H., Marchadier E., Brownlee C., Kudla J., Hetherington A.M. The evolution of calcium-based signalling in plants. Curr. Biol. 2017;27:R667–R679. doi: 10.1016/j.cub.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Oami K., Naitoh Y., Sibaoka T. Modification of voltage-sensitive inactivation of Na+ current by external Ca2+ in the marine dinoflagellate Noctiluca miliaris. J. Comp. Physiol. A. 1995;176:635–640. [Google Scholar]
- 9.Taylor A.R., Brownlee C. A novel Cl- inward-rectifying current in the plasma membrane of the calcifying marine phytoplankton Coccolithus pelagicus. Plant Physiol. 2003;131:1391–1400. doi: 10.1104/pp.011791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor A.R. A fast Na+/Ca2+-based action potential in a marine diatom. PLoS ONE. 2009;4:e4966. doi: 10.1371/journal.pone.0004966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malviya S., Scalco E., Audic S., Vincent F., Veluchamy A., Poulain J., Wincker P., Iudicone D., de Vargas C., Bittner L. Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. USA. 2016;113:E1516–E1525. doi: 10.1073/pnas.1509523113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falciatore A., d’Alcalà M.R., Croot P., Bowler C. Perception of environmental signals by a marine diatom. Science. 2000;288:2363–2366. doi: 10.1126/science.288.5475.2363. [DOI] [PubMed] [Google Scholar]
- 13.Kitchen S.A., Bourdelais A.J., Taylor A.R. Interaction of a dinoflagellate neurotoxin with voltage-activated ion channels in a marine diatom. PeerJ. 2018;6:e4533. doi: 10.7717/peerj.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren D., Navarro B., Xu H., Yue L., Shi Q., Clapham D.E. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- 15.Irie K., Kitagawa K., Nagura H., Imai T., Shimomura T., Fujiyoshi Y. Comparative study of the gating motif and C-type inactivation in prokaryotic voltage-gated sodium channels. J. Biol. Chem. 2010;285:3685–3694. doi: 10.1074/jbc.M109.057455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falkowski P.G., Katz M.E., Knoll A.H., Quigg A., Raven J.A., Schofield O., Taylor F.J.R. The evolution of modern eukaryotic phytoplankton. Science. 2004;305:354–360. doi: 10.1126/science.1095964. [DOI] [PubMed] [Google Scholar]
- 17.Ren D., Navarro B., Perez G., Jackson A.C., Hsu S., Shi Q., Tilly J.L., Clapham D.E. A sperm ion channel required for sperm motility and male fertility. Nature. 2001;413:603–609. doi: 10.1038/35098027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai X., Clapham D.E. Evolutionary genomics reveals lineage-specific gene loss and rapid evolution of a sperm-specific ion channel complex: CatSpers and CatSperbeta. PLoS ONE. 2008;3:e3569. doi: 10.1371/journal.pone.0003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arrigoni C., Rohaim A., Shaya D., Findeisen F., Stein R.A., Nurva S.R., Mishra S., Mchaourab H.S., Minor D.L., Jr. Unfolding of a temperature-sensitive domain controls voltage-gated channel activation. Cell. 2016;164:922–936. doi: 10.1016/j.cell.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nymark M., Sharma A.K., Sparstad T., Bones A.M., Winge P. A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci. Rep. 2016;6:24951. doi: 10.1038/srep24951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yue L., Navarro B., Ren D., Ramos A., Clapham D.E. The cation selectivity filter of the bacterial sodium channel, NaChBac. J. Gen. Physiol. 2002;120:845–853. doi: 10.1085/jgp.20028699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koishi R., Xu H., Ren D., Navarro B., Spiller B.W., Shi Q., Clapham D.E. A superfamily of voltage-gated sodium channels in bacteria. J. Biol. Chem. 2004;279:9532–9538. doi: 10.1074/jbc.M313100200. [DOI] [PubMed] [Google Scholar]
- 23.Collingridge P., Brownlee C., Wheeler G.L. Compartmentalized calcium signaling in cilia regulates intraflagellar transport. Curr. Biol. 2013;23:2311–2318. doi: 10.1016/j.cub.2013.09.059. [DOI] [PubMed] [Google Scholar]
- 24.Fromherz P., Hübener G., Kuhn B., Hinner M.J. ANNINE-6plus, a voltage-sensitive dye with good solubility, strong membrane binding and high sensitivity. Eur. Biophys. J. 2008;37:509–514. doi: 10.1007/s00249-007-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gradmann D., Boyd C.M. Electrophysiology of the marine diatom Coscinodiscus wailesii II. Potassium currents. J. Exp. Bot. 1999;50:453–459. [Google Scholar]
- 26.Kulkarni R.U., Miller E.W. Voltage imaging: pitfalls and potential. Biochemistry. 2017;56:5171–5177. doi: 10.1021/acs.biochem.7b00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storace D., Sepehri Rad M., Kang B., Cohen L.B., Hughes T., Baker B.J. Toward better genetically encoded sensors of membrane potential. Trends Neurosci. 2016;39:277–289. doi: 10.1016/j.tins.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vardi A., Formiggini F., Casotti R., De Martino A., Ribalet F., Miralto A., Bowler C. A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biol. 2006;4:e60. doi: 10.1371/journal.pbio.0040060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor A.R., Chrachri A., Wheeler G., Goddard H., Brownlee C. A voltage-gated H+ channel underlying pH homeostasis in calcifying coccolithophores. PLoS Biol. 2011;9:e1001085. doi: 10.1371/journal.pbio.1001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maheswari U., Montsant A., Goll J., Krishnasamy S., Rajyashri K.R., Patell V.M., Bowler C. The diatom EST database. Nucleic Acids Res. 2005;33:D344–D347. doi: 10.1093/nar/gki121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewin J.C., Lewin R.A., Philpott D.E. Observations on Phaeodactylum tricornutum. J. Gen. Microbiol. 1958;18:418–426. doi: 10.1099/00221287-18-2-418. [DOI] [PubMed] [Google Scholar]
- 32.De Martino A., Bartual A., Willis A., Meichenin A., Villazán B., Maheswari U., Bowler C. Physiological and molecular evidence that environmental changes elicit morphological interconversion in the model diatom Phaeodactylum tricornutum. Protist. 2011;162:462–481. doi: 10.1016/j.protis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 33.McLachlan D.H., Underwood G.J.C., Taylor A.R., Brownlee C. Calcium release from intracellular stores is necessary for the photophobic response in the benthic diatom Navicula perminuta (Bacillariophyceae)(1) J. Phycol. 2012;48:675–681. doi: 10.1111/j.1529-8817.2012.01158.x. [DOI] [PubMed] [Google Scholar]
- 34.Consalvey M., Paterson D.M., Underwood G.J.C. The ups and downs of life in a benthic biofilm: migration of benthic diatoms. Diatom Res. 2004;19:181–202. [Google Scholar]
- 35.Bondoc K.G.V., Heuschele J., Gillard J., Vyverman W., Pohnert G. Selective silicate-directed motility in diatoms. Nat. Commun. 2016;7:10540. doi: 10.1038/ncomms10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poulsen N.C., Spector I., Spurck T.P., Schultz T.F., Wetherbee R. Diatom gliding is the result of an actin-myosin motility system. Cell Motil. Cytoskeleton. 1999;44:23–33. doi: 10.1002/(SICI)1097-0169(199909)44:1<23::AID-CM2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 37.Makita N., Shihira-Ishikawa I. Chloroplast assemblage by mechanical stimulation and its intercellular transmission in diatom cells. Protoplasma. 1997;197:86–95. [Google Scholar]
- 38.Lichtner F.T., Spanswick R.M. Electrogenic sucrose transport in developing soybean cotyledons. Plant Physiol. 1981;67:869–874. doi: 10.1104/pp.67.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kikuyama M., Kuchitsu K., Shibuya N. Membrane depolarization induced by N-acetylchitooligosaccharide elicitor in suspension-cultured rice cells. Plant Cell Physiol. 1997;38:902–909. [Google Scholar]
- 40.Pavlovkin J., Novacky A., Ullrich-Eberius C.I. Membrane potential changes during bacteria-induced hypersensitive reaction. Physiol. Mol. Plant Pathol. 1986;28:125–135. [Google Scholar]
- 41.Lew R.R., Dearnaley J.D.W. Extracellular nucleotide effects on the electrical properties of growing Arabidopsisthaliana root hairs. Plant Sci. 2000;153:1–6. [Google Scholar]
- 42.Hedrich R., Neher E. Venus flytrap: how an excitable, carnivorous plant works. Trends Plant Sci. 2018;23:220–234. doi: 10.1016/j.tplants.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Dorrell R.G., Smith A.G. Do red and green make brown?: perspectives on plastid acquisitions within chromalveolates. Eukaryot. Cell. 2011;10:856–868. doi: 10.1128/EC.00326-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daboussi F., Leduc S., Maréchal A., Dubois G., Guyot V., Perez-Michaut C., Amato A., Falciatore A., Juillerat A., Beurdeley M. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat. Commun. 2014;5:3831. doi: 10.1038/ncomms4831. [DOI] [PubMed] [Google Scholar]
- 45.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meijering E., Dzyubachyk O., Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- 47.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rastogi A., Murik O., Bowler C., Tirichine L. PhytoCRISP-Ex: a web-based and stand-alone application to find specific target sequences for CRISPR/CAS editing. BMC Bioinformatics. 2016;17:261. doi: 10.1186/s12859-016-1143-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guillard R.R.L., Ryther J.H. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- 51.Read B.A., Kegel J., Klute M.J., Kuo A., Lefebvre S.C., Maumus F., Mayer C., Miller J., Monier A., Salamov A., Emiliania huxleyi Annotation Consortium Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature. 2013;499:209–213. doi: 10.1038/nature12221. [DOI] [PubMed] [Google Scholar]
- 52.Keeling P.J., Burki F., Wilcox H.M., Allam B., Allen E.E., Amaral-Zettler L.A., Armbrust E.V., Archibald J.M., Bharti A.K., Bell C.J. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Armbrust E.V., Berges J.A., Bowler C., Green B.R., Martinez D., Putnam N.H., Zhou S., Allen A.E., Apt K.E., Bechner M. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 54.Bowler C., Allen A.E., Badger J.H., Grimwood J., Jabbari K., Kuo A., Maheswari U., Martens C., Maumus F., Otillar R.P. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 55.Prochnik S.E., Umen J., Nedelcu A.M., Hallmann A., Miller S.M., Nishii I., Ferris P., Kuo A., Mitros T., Fritz-Laylin L.K. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blanc G., Agarkova I., Grimwood J., Kuo A., Brueggeman A., Dunigan D.D., Gurnon J., Ladunga I., Lindquist E., Lucas S. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012;13:R39. doi: 10.1186/gb-2012-13-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Derelle E., Ferraz C., Rombauts S., Rouzé P., Worden A.Z., Robbens S., Partensky F., Degroeve S., Echeynié S., Cooke R. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Worden A.Z., Lee J.H., Mock T., Rouzé P., Simmons M.P., Aerts A.L., Allen A.E., Cuvelier M.L., Derelle E., Everett M.V. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 59.Brawley S.H., Blouin N.A., Ficko-Blean E., Wheeler G.L., Lohr M., Goodson H.V., Jenkins J.W., Blaby-Haas C.E., Helliwell K.E., Chan C.X. Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta) Proc. Natl. Acad. Sci. USA. 2017;114:E6361–E6370. doi: 10.1073/pnas.1703088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsuzaki M., Misumi O., Shin-I T., Maruyama S., Takahara M., Miyagishima S.-Y., Mori T., Nishida K., Yagisawa F., Nishida K. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 61.Price D.C., Chan C.X., Yoon H.S., Yang E.C., Qiu H., Weber A.P., Schwacke R., Gross J., Blouin N.A., Lane C. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science. 2012;335:843–847. doi: 10.1126/science.1213561. [DOI] [PubMed] [Google Scholar]
- 62.Aranda M., Li Y., Liew Y.J., Baumgarten S., Simakov O., Wilson M.C., Piel J., Ashoor H., Bougouffa S., Bajic V.B. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 2016;6:39734. doi: 10.1038/srep39734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cock J.M., Sterck L., Rouzé P., Scornet D., Allen A.E., Amoutzias G., Anthouard V., Artiguenave F., Aury J.-M., Badger J.H. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- 64.Apweiler R., Attwood T.K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D., InterPro Consortium InterPro--an integrated documentation resource for protein families, domains and functional sites. Bioinformatics. 2000;16:1145–1150. doi: 10.1093/bioinformatics/16.12.1145. [DOI] [PubMed] [Google Scholar]
- 65.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 66.Hopes A., Nekrasov V., Kamoun S., Mock T. Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana. Plant Methods. 2016;12:49. doi: 10.1186/s13007-016-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaslavskaia L.A., Lippmeier J.C., Kroth P.G., Grossman A.R., Apt K.E. Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J. Phycol. 2000;36:379–386. [Google Scholar]
- 68.Buck J.M., Río Bártulos C., Gruber A., Kroth P.G. Blasticidin-S deaminase, a new selection marker for genetic transformation of the diatom Phaeodactylum tricornutum. PeerJ. 2018;6:e5884. doi: 10.7717/peerj.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Presence of EukCat channels in eukaryotic phytoplankton taxa. The presence of 4D-Cav/Nav in diatom taxa is also shown. Single-domain channels resembling cation channels of sperm (CatSper) are also indicated. Data source: for transcriptome data, the dataset identifier (MMET ID) of each species is listed. Genome indicates a genome database was searched (see STAR Methods for details of relevant genome databases). Results reported in previous studies are indicated as supplemental references. N.F, not found. B. Protein Ids of EukCat hits identified in this study, and/or source for relevant primary reference. IDs of diatom 4D-Cav/Nav are also given.
The corresponding nucleotide sequences that were codon optimized for expression in HEK293 cells are also given.
Data Availability Statement
The accession number for sequences/plasmids in GenBank or Fasta format from this study are given in the Key Resources Table.