Abstract
A full-length cDNA of phyA gene of Aspergillus niger, encoding phytase enzyme, was cloned and expressed in E. coli BL21 cells and assayed for its activity. The phyA cDNA consisted of 1404 bp, which encoded 467 amino acid residues. The phytase activity of purified phytase was 826.33 U/mL. The phyA gene under the control of endosperm-specific promoters was transformed into an Indian maize inbred line, UMI29, using particle bombardment-mediated transformation method to generate transgenic maize plants over-expressing phytase in seeds. PCR and GUS analyses demonstrated the presence of transgenes in T0 transgenic plants and their stable inheritance in the T1 progenies. Three transgenic events expressing detectable level of A. niger phytase were characterized by western blot analysis. Phytase activity of 463.158 U/kg of seed was observed in one of the events, JB-UMI29-Z17/2. The phytase activity of transgenic maize seeds was 5.5- to 7-fold higher than the wild-type UMI29 seeds and, consequently, the seeds had 0.6- to 5-fold higher inorganic phosphorus content.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1731-7) contains supplementary material, which is available to authorized users.
Keywords: A. niger, PHYA, Maize, Phytase, Endosperm-specific expression
Introduction
Maize grains are produced primarily to be used as feed for poultry and livestock in several countries including India. Though maize grains are rich in mineral nutrients such as phosphorous, calcium, zinc, iron and magnesium, the bioavailability of these minerals are limited due to complexation of the divalent mineral nutrients with phytic acid (myo-inositol 1,2,3,4,5,6-hexakisphosphate; Ins P6) to form an insoluble complex called phytate. Phosphorous is one of the most important minerals in animal nutrition and the second most abundant element in animals’ body after calcium. Phosphorous plays a key metabolic role and has more physiological function than any other mineral (Adedokun and Adeola 2013). Phytic acid, the storage form of phosphorus, is often considered anti-nutrient as it binds minerals, making them less available to monogastric animals. This problem is overcome by supplementing maize grains with phytase enzyme in animal feed (Simell et al. 1989). However, supplementation of the phytase enzyme in animal feed becomes expensive. An alternative to the supplementation strategy is to develop crop varieties with reduced levels of phytic acid by exploring naturally available variability through conventional plant breeding strategies (Cichy and Raboy 2008) or engineering crops to express phytase in their seeds (Raboy et al. 2000). Genes encoding phytase have been cloned from diverse sources and characterized, which include fungal phytase from Aspergillus (Ullah 1988), bacterial phytase from Escherichia coli (Greiner et al. 1993) and a mammalian phytase (Craxton et al. 1997).
Phytase genes cloned from various species of Aspergillus were over-expressed in crop species such as tobacco, soybean, alfalfa, rice and canola (Pen et al. 1993; Verwoerd et al. 1995; Ullah et al. 1999, 2002; Denbow et al. 1998; Brinch-Pedersen et al. 2000; Zhang et al. 2000; Lucca et al. 2001; Ponstein et al. 2002; Hong et al. 2004). Poultry-feeding studies showed that the plant-delivered phytase can substitute a proportional measure of inorganic phosphate (Pen et al. 1993; Denbow et al. 1998; Zhang et al. 2000). The present study is an attempt to express A. niger phytase in an Indian tropical maize inbred (UMI 29) line with a view to improving its nutritional quality through increasing the bioavailability of mineral nutrients.
Materials and methods
Microbial strains
The phytase (phyA) cDNA was isolated from A. niger 563 strain obtained from the National Chemical Laboratory (NCL), Pune, India. E.coli DH5α was utilized for the maintenance and manipulation of plasmids.
Phytase gene isolation from A. niger
Total RNA was extracted from A. niger, following the TRIzol method as per manufacturer’s instructions (MBI Fermentas, Germany). A full-length cDNA sequence of phyA was amplified by polymerase chain reaction with an RT-PCR kit (RevertAiD™H Minus First Strand cDNA Synthesis Kit; MBI Fermentas, Germany) using oligonucleotide primers (PHYF—5′-ATGGGCGTCTCTGCTGTTCTACTTC-3′ and PHYR—5′-CTAAGCAAAACACTCCGCCCAATC-3′). Amplification was performed in a thermal cycler using a temperature profile of pre-incubation at 94 °C for 5 min, followed by 35 cycles of melting at 94 °C for 1 min, annealing at 54 °C for 1 min and synthesis at 72 °C for 1 min and finally an extension of 72 °C for 10 min. The PCR product was purified and cloned into vector, pTZ57R/T (MBI Fermentas, Germany), and the vector was named as pTA-phyA. Plasmid DNA sequencing was done and the identity of the cloned phyA gene was confirmed through NCBI BLAST search. Phytase gene sequence was aligned with seven homologous known genes from other microorganisms by clustalW (Thompson et al. 1997; Larkin et al. 2007). The neighbour joining tree (NJ) was constructed based on the p-distance in software MEGA 5 (Tamura et al. 2007).
Expression of phyA gene in E. coli
Phytase gene was amplified from the pTA-phyA plasmid using the primers ECOF2 (5′-TCCGAATTCCTGGCAGTCCCCGCCTCGAGA-3′) and HINR2 (5′-CGCAAGCTTAGCTAAGCAAAACACTCCGCC-3′) with sequences for EcoRI and HindIII restriction sites at the 5′ end. Amplification was performed in a thermal cycler using a temperature profile of pre-incubation at 94 °C for 5 min, leading to 35 cycles of melting at 94 °C for 1 min, annealing at 54 °C for 1 min and synthesis at 72 °C for 1 min followed by an extension of 72 °C for 10 min. The EcoRI and HindIII enzymes were used to digest the amplified product as well as E. coli expression vector, pET-28a(+) (Novagen, Madison, WI). Then, vector pET-28a(+)-phyA was created by cloning the coding sequence of phyA into the expression vector, pET-28a(+), and confirmed by restriction digestion and sequencing. Finally, pET-28a(+)-phyA was transformed into BL21 (E. coli) competent cells, in which the expression of the phytase protein was induced at 30 °C by 1 mM isopropyl-β-d-thiogalacto pyranoside (IPTG) at different induction times (1, 2, 3, 4, 5, 6 and 7 h). The protein was analysed on SDS-PAGE (12%) with the pET-28a(+) vector as control. The resultant phytase protein was purified using BugBuster® His·Bind Purification Kit (Novagen, Germany). The recombinant protein was assayed for phytase activity and monitored for the release of organic phosphorus from phytic acid as described by Chen et al. (2004). One unit of phytase is defined as the enzyme required to release 1 µmol of inorganic phosphorus per minute from sodium phytate at pH 5.5 and 37 °C.
Genetic transformation of phyA in maize
Plant expression vector
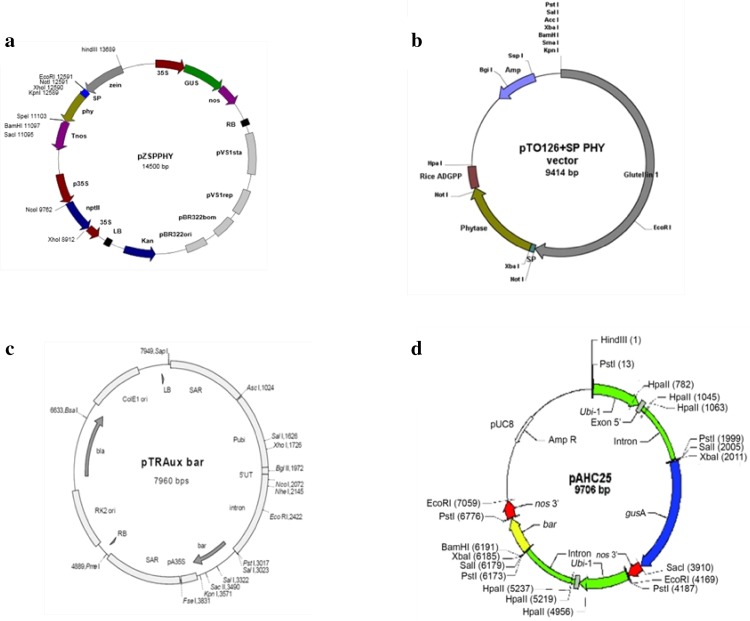
The phytase constructs used for maize transformation consisted of the 1404 bp phyA gene fused with a 90 bp α-amylase signal peptide to facilitate secretion of the phytase protein into the intracellular space. Two constructs were used in the study, viz., pZSPPHY (phyA gene under the control of maize α-zein promoter and nos terminator) and pTOSPPHY (phyA gene under the control rice glutelin promoter and ADPGP terminator) (Fig. 1a, b). The phosphinothricin resistance gene (bar) was used as the plant selectable marker. Maize zein promoter was isolated earlier in our laboratory (Geetha 2011) and its endosperm-specific expression pattern in the rice model system was demonstrated in our laboratory (Joshi et al. 2015). The pZSPPHY construct harbours gusA besides phyA gene (Fig. 1a) and was co-bombarded with pTRAUX harbouring bar gene (Fig. 1c). The pTOSPPHY construct (Fig. 1b) was co-bombarded with pAHC25 harbouring bar and gusA genes (Fig. 1d).
Fig. 1.
Physical map of vectors used in the study. a pZSPPHY, b pTOSPPHY, c pTRAUX, d pAHC25
Plant materials and genetic transformation
Maize plants of UMI 29 inbred line, developed by Tamil Nadu Agricultural University, Coimbatore, India, grown under well-managed garden land (vertisol) conditions were used as a source of immature embryos. Immature ears were harvested 8–10 days after self-pollination and sterilized using 70% ethanol for 1 min, followed by 2.5% sodium hypochlorite for 7 min. The sterilized ears were rinsed four times with sterilized distilled water under aseptic conditions. Immature embryos of 1.5 mm length were excised from kernels aseptically and pre-cultured for 3–4 days on N6 callus induction medium (Chu et al. 1975; containing N6 macro- and micronutrients, N6 vitamins, 20 g/L sucrose, 0.3 g/L casein hydrolysate, 1 mg/L 2,4-D and 0.4% (w/v) Gelrite. The pre-cultured immature maize embryos were given 4 h of osmoticum treatment on osmoticum medium (N6 callus induction medium supplemented with 36.4 g/L mannitol and 36.4 g/L sorbitol) in dark and bombarded at 6 cm microcarrier flying distance at a rupture pressure of 1100 psi twice with DNA-coated gold particles (0.9 µm diameter) using the PDS 1000/He device (Biorad, USA) with 4 h interval, as previously described (Joshi et al. 2016).
The bombarded embryos were transferred onto fresh N6 callus induction medium for resting and incubated in dark for 16 h. After three rounds of selection on selection medium (N6 callus induction medium supplemented with 3 mg/L phosphinothricin; Sigma-Aldrich), the transformed calli were transferred onto regeneration medium (MS medium; Murashige and Skoog 1962) containing MS macro- and micronutrients, MS vitamins, 30 g/L sucrose, 1 mg/L kinetin and 1 mg/L BAP, 3 mg/L phosphinothricin and 3.8 g/L Phytagel) and incubated under 16 h/8 h of light and dark at 25 ± 2 °C in a plant growth chamber. The shoots which were 3–5 cm tall with primary roots were transferred to culture bottles containing regeneration medium and incubated at 25 ± 2 °C with a photoperiod of 16 h/8 h light and dark in a plant growth chamber for elongation of shoots and induction of secondary roots. The plantlets with two to three well-developed leaves were hardened and transferred to a transgenic greenhouse (Joshi et al. 2016).
Molecular and biochemical analyses
PCR analysis
PCR analysis was done to check the presence of phyA and gusA genes in putative transgenic (T0) maize plants using total genomic DNA isolated from leaves following the SDS method [Tris–HCl, pH 8.0 (0.1 M); EDTA, pH 8.0 (0.02 M); NaCl (0.1 M); SDS (1%); Salgado et al. 2006]. PCR amplification of phyA, gusA gene and phyA-ADPGP terminator junction sequence was carried out using sequence-specific primers, PHYF (ACATCGAAGCCAATTTCACC), PHYR (CATGGGTGAACAGGTCACAG); GUS1F (CAACGAACTGAACTGGCAGA), GUS1R (TTTTTGTCACGCGCTATCAG) and INPF2 (GAAGATAGCGAATTGGCCGATGAC), ADGPR (GTGCCTTGAACTGCTTTTATTCTT), respectively. DNA amplifications were performed in a thermal cycler using the following temperature profile: initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 1 min, annealing at 59 °C (phyA) or 58 °C (gusA) or 62 °C (phytase and ADPGP terminator) for 1 min and extension at 72 °C for 1 min. An additional complete extension cycle was performed at 72 °C for 10 min.
High inorganic phosphate content (HIP) assay
The inorganic phosphate (Pi) content in the maize endosperm tissue was determined using single seed HIP assay (Raboy et al. 2000). Ten seeds were randomly selected from each of the pZSPPHY transformed events (JB-UMI29-Z17, JB-UMI29-Z30) and pTOSPPHY transformed event (JB-UMI29-G22) and six seeds from pTOSPPHY transformed event (JB-UMI29-G20). The endosperm portion was removed from the single seed (T1) using a sterile scalpel blade. Ten milligrams of endosperm tissue was weighed, pounded and extracted overnight in 100 µl of HCl (0.4 M) at 4 °C and 10 μl of extracts were assayed for Pi following the method described by Chen et al. (1956), modified for microtitre plates. To each microtitre plate, 10 μl of extract, 90 μl deionized water and 100 μl of colorimetric reagent [consisting of a 1:1:1:2 mixture of 10% (w/v) ascorbic acid, 6 N H2SO4, 2.5% (w/v) ammonium molybdate and distilled, deionized water] were added. Besides, in each microtitre plate, five wells were prepared to contain KH2PO4 standard. Following colour development at ambient temperature, the results were quantified using a microtitre plate spectrophotometer (660 nm). Each experiment was replicated thrice along with wild-type UMI29 as negative and lpa2 low phytate mutant as a positive control.
Western blot analysis
Total protein from the endosperm tissues (T1) of transgenic maize events, JB-UMI29-Z17/2, JB-UMI29-Z17/3, JB-UMI29-Z30/6, JB-UMI29-G20/4, JB-UMI29-G20/5, JB-UMI29-G22/1 and non-transformed UMI29 was isolated as described by Karaman et al. (2012). An aliquot of 100 µl of protein extraction buffer [200 mM Tris–HCl (pH 8.0), 100 mM NaCl, 400 mM sucrose and 10 mM EDTA, 14 mM 2-BME, 0.05% Tween 20] was added to 10 mg of powdered endosperm tissue and incubated on vortex at room temperature for 1 h. Later, the suspension was kept at 4 °C for 16 h and centrifuged at 13,000 rpm for 15 min and the supernatant collected in a fresh tube. Equal quantity of total protein was loaded in the SDS-PAGE for analysis. After separation on SDS-PAGE, the protein samples were transferred onto a nitrocellulose membrane and western blotting was carried out (Burnette 1981) using primary antibody (polyclonal antibody raised against purified phytase in rabbit) at a dilution of 1:500 and secondary antibody (goat anti-rabbit IgG antibody, alkaline phosphatase conjugate) at a dilution of 1:20,000. Purified phytase (obtained in this study) and commercial wheat phytase (Sigma) were used as controls.
Phytase assay
Phytase assay was carried out in the endosperm tissue (T1) of transgenic maize events, JB-UMI29-Z17/2, JB-UMI29-Z17/3, JB-UMI29-Z30/6, JB-UMI29-G20/4, JB-UMI29-G20/5, JB-UMI29-G22/1 and non-transformed UMI29 as described by Chen et al. (2008). An aliquot of 100 µl of extraction buffer (50 mM sodium acetate, 1 mM CaCl2; pH 5.5) was added to 10 mg of powdered endosperm tissue and incubated under a shaker at room temperature for 1 h. The tubes were then centrifuged at 3000g for 10 min. The supernatant was transferred into fresh tubes and mixed with 900 µl of 5 mM phytic acid and incubated for 30 min at 37 °C. The reaction was stopped by adding 1 mL of 15% (v/v) aqueous trichloroacetic acid (TCA). In controls, TCA was added to the supernatant first, followed by phytic acid substrate and incubated under conditions mentioned above. The released Pi was quantified colorimetrically using fresh colour reagent [0.6 M H2SO4 + 2% ascorbic acid + 0.5% ammonium molybdate]. Purified phytase and wheat phytase (Sigma) were used as controls. Standard solutions of potassium phosphate were used as reference. Phytase activity (U/mL) was calculated using the formula: (µ moles of Pi released × dilution factor)/(duration of assay in min × volume of sample in mL).
Transgene inheritance
The segregation of transgene was studied in the T1 generation of JB-UMI29-G20 and JB-UMI29-G22 events transformed with the pTOSPPHY construct. Since a limited number of seeds were available, ten seeds in each of the above lines were planted. PCR analysis was done to check the presence of phyA and gusA genes in the T1 generation plants using total genomic DNA isolated from leaves.
Statistical analysis
The data were analysed using AgRes Statistical Software, Version 3.01 (Pascal International Software Solutions). ANOVA was conducted on the data transformed by arcsine or square root transformation of the percentage or count data, followed by least significant difference (LSD) test to select the best treatment. Other statistical analysis was performed in worksheet format using data analysis tool pack feature available in MS Office Excel 2007 software.
Results
Isolation and sequence analysis of the phyA gene
A full-length cDNA of the phytase gene was isolated and designated as phyA (GenBank accession no. JQ241266). This cDNA contained 1404 bp ORF encoding 467 amino acid proteins with a calculated molecular weight of 52 kDa. The predicted PhyA protein shared 100% identity with A. awamori phytase (ABA29207) and 99% with A. niger (XP0014017130, XM001401676 and AAG40885). The amino acid sequence of the PhyA contained the consensus motifs RHGXRXP and HD which are conserved among histidine acid phosphatases (Fig. S1a). Phylogenetic analysis showed that the phyA formed two clusters. Cluster dendrogram results revealed that the cloned phyA from A. niger formed a separate cluster with the phytase sequences of other Aspergillus species (Fig. S1b).
PhyA expression in E. coli
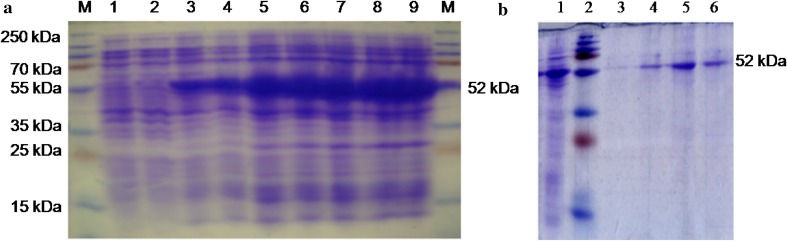
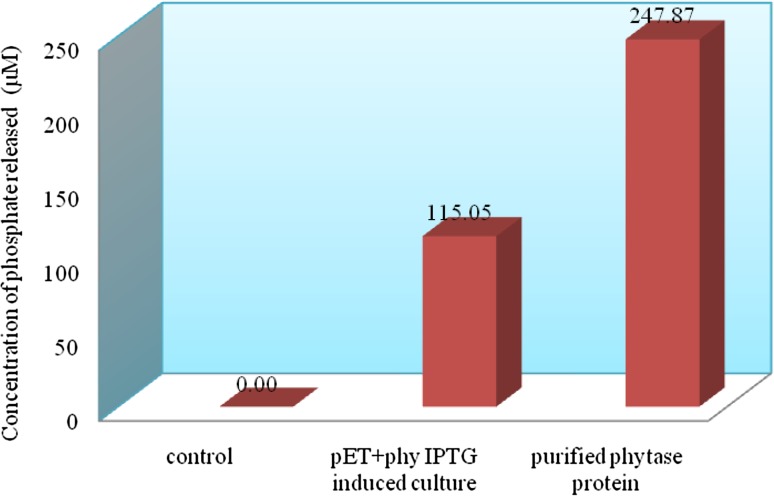
The recombinant E. coli containing pET28a(+)-phyA expressed a protein of the expected 52 kDa upon induction by IPTG (Fig. 2a). The expression of the 52 kDa protein increased with induction time. This protein was absent in non-induced E. coli cells as well as in E. coli harbouring pET28a(+) alone. A higher level of expression of the target protein was observed after 4 to 7 h of induction (Fig. 2a). The phytase protein was purified from the 100 mL culture (6 h after induction with 50 mM IPTG). The purified recombinant enzyme, on SDS-PAGE, showed a single band of 52 kDa (Fig. 2b). In phytase assay, the concentration of liberated inorganic phosphate was calculated based on a standard curve generated with 9 mM potassium monobasic phosphate standard, and based on the liberated Pi (Fig. 3), the phytase activity was calculated as 826.33 U/mL for purified phytase and 383.5 U/mL for crude extract of E. Coli (pET-phyA).
Fig. 2.
Expression of phyA in E. coli BL21 cells. a SDS-PAGE analysis of phytase protein from crude lysate; M protein marker, 1 control-pET28a(+), 2 pET28a(+)-phyA (Uninduced), 3–9 phytase expression after IPTG induction of 1, 2, 3, 4, 5, 6 and 7 h, respectively. b SDS-PAGE analysis of purified phytase protein; 1 pET28a(+)-phyA (uninduced crude lysate), 2 prestained protein marker, 3–6 purified phytase protein
Fig. 3.
Determination of liberated inorganic phosphorus
Generation and screening of transgenic maize plants
The embryogenic calli obtained from immature embryos after bombardment were sub-cultured thrice on selection medium. Phosphinothricin-resistant embryogenic calli on selection media grew well, whereas non-transformed calli turned brown, watery and later dried (Fig. 4). Totally, 36 events were regenerated with the pZSPPHY and 32 events with the pTOSPPHY construct (Supplementary Table 1).
Fig. 4.
Biolistic-mediated genetic transformation of maize inbred UMI29. a Immature embryos on N6 medium containing 2,4-D (1.0 mg/L) and AgNO3 (10 mg/L), b immature embryos kept on osmoticum medium containing mannitol (36.4 g/L) and sorbitol (36.4 g/L) 4 h prior to the bombardment, c immature embryo-derived calli after the first round of selection on N6 selection medium (PPT 3 mg/mL), d immature embryo-derived calli after the first round of selection on N6 selection medium (PPT 3 mg/mL), e immature embryo-derived calli after the second round of selection on regeneration medium (PPT 3 mg/mL), f regeneration of shoots on MS medium containing BAP (10 mg/L) and PPT (3 mg/L), g plantlets on half strength MS basal medium containing PPT (3 mg/L), h putative transgenic plants in transgenic greenhouse
PCR analysis of the generated transformants
PCR analysis of the 36 putative pZSPPHY transformants using gene-specific primers resulted in an amplification of 878 bp (gusA) and 190 bp (phyA) in 13 events with a transformation efficiency of 1.60% (Supplementary Table 1; Figs. S2, S3). Among the 32 events generated using pTOSPPHY construct, 8 events were positive for gusA (878 bp), phyA + ADPGP (917 bp) and phyA (190 bp) gene sequences with a transformation efficiency of 1.26% (Supplementary Table 1; Figs. S4, S5).
Inorganic phosphate level in transformed events
In HIP assay, seeds from transgenic plants had higher Pi levels compared to non-transformed UMI29 control. Among the different transgenic events screened, JB-UMI29-Z17 was found to have higher Pi level which was on par with the lpa2 mutant (Table 1) and the level was fivefold higher compared to the non-transformed UMI29 control.
Table 1.
HIP assay in transgenic maize seeds expressing A. niger phytase
| Event no. | Construct | Seed no. | Amount of inorganic phosphate (mg/g)ǂ |
|---|---|---|---|
| JB-UMI29-17Z | pZSPPHY | 2 | 2.54 ± 0.05a |
| 3 | 2.35 ± 0.06b | ||
| 7 | 1.89 ± 0.05c | ||
| JB-UMI29-30Z | pZSPPHY | 6 | 0.90 ± 0.02e |
| 8 | 0.79 ± 0.01e | ||
| JB-UMI29-20G | pTOSPPHY | 4 | 0.97 ± 0.01e |
| 5 | 1.34 ± 0.01d | ||
| JB-UMI29-22G | pTOSPPHY | 1 | 0.98 ± 0.02e |
| UMI29 control | 0.51 ± 0.08f | ||
| lpa2 | 2.73 ± 0.27a |
ǂEach treatment was replicated three times and the values presented are mean ± SE, followed by alphabets to imply significant difference (p ≤ 0.05) after grouping of treatment means after ANOVA by LSD and values with the same letter are not significantly different
Expression of phyA gene in transgenic maize (T1) seeds
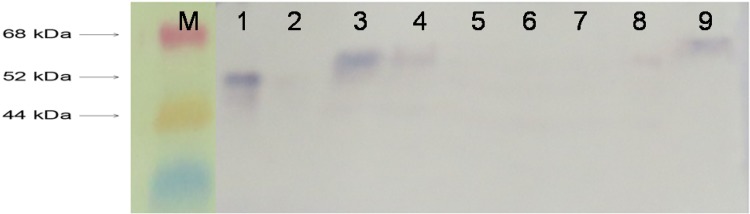
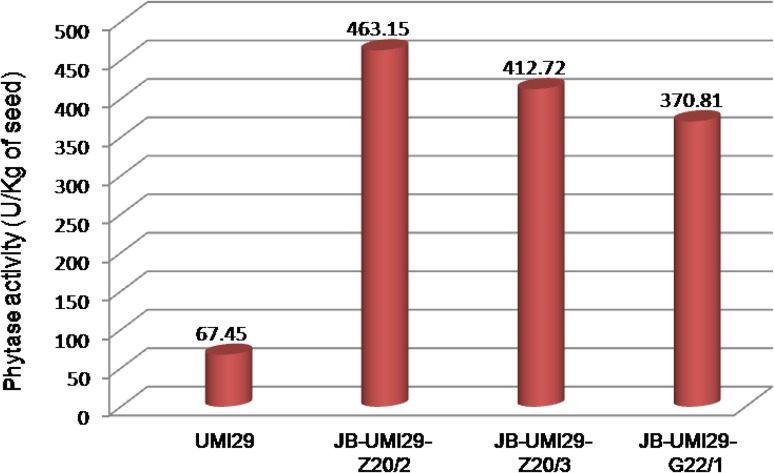
In western blot analysis, total protein extract from seed endosperm showed a strong signal in JB-UMI29-Z17/2 and JB-UMI29-Z17/3 and a weak signal in JB-UMI29-G22/1. The size of phytase protein produced in transgenic maize plant was slightly higher compared to the purified phytase and was detected at ~ 62 kDa. No signal was observed in the non-transformed UMI29 control (Fig. 5). The phytase activity in T1 seeds of three transgenic events, JB-UMI29-Z17/2, JB-UMI29-Z17/3 and JB-UMI29-G22/1, was 463.15, 412.72 and 370.81 U/kg of seed, respectively. The increase in phytase activity was about 5.5- to 7-fold in JB-UMI29-G22/1, JB-UMI29-Z17/3 and JB-UMI29-Z17/2 compared to wild-type UMI29 seeds (67.45 U/kg of seed; Fig. 6).
Fig. 5.
Western blot analysis of transgenic maize seeds. M protein marker, 1 purified phytase, 2 non-transformed UMI29, 3 JB-UMI29-Z17/2, 4 JB-UMI29-Z17/3, 5 JB-UMI29-Z30/6, 6 JB-UMI29-G20/4, 7 JB-UMI29-G20/5, 8 JB-UMI29-G22/1, 9 wheat phytase
Fig. 6.
Phytase activity in transgenic maize seeds
Transgene inheritance
PCR and GUS analyses showed that the T1 plants of JB-UMI29-G20 and JB-UMI29-G22 events segregated for the transgene, phyA and gusA genes (Supplementary Table 2).
Discussion
Maize kernel, despite containing a large quantity of carbohydrates, proteins, vitamins and fats, has nutritional limitation as food or feed due to non-availability of phosphorus present in the phytate complex. Further, when maize is consumed as food or feed, the phytic acid complex also chelates calcium, iron and zinc and reduces the bioavailability of these minerals as well. To overcome these limitations, phytase enzyme produced by recombinant microbes is generally added to the maize-containing feed of monogastric animals such as pig and poultry. An alternative strategy to overcome the non-availability of essential minerals is to engineer plants for enhanced phytase activity in their seeds (Dionisio et al. 2011). The present study is an attempt to genetically engineer a local tropical maize inbred line for endosperm-specific expression of phytase with a view to improving the bioavailability of P and other micronutrients in monogastric animals.
In maize, the expression of two endogenous phytases, ZmPHYTI and ZmPHYTII, has been reported in the developing maize seedlings and roots (Maugenest et al. 1999) and no phytase activity has been detected in the mature endosperm of maize (Dionisio et al. 2011). Among the phytases isolated from different organisms, A. niger phytase, phyA, was found to have the major advantage of being active in two pH ranges, 2.5 and 5.0 (Holm et al. 2002). In plants, the expressed A. niger phytase is active at pH 5.0, thereby reducing the phytate complex in the seed and in turn increasing the availability of Pi during seed development. The A. niger phytase expressed in the seeds is also active in the gut pH 2.5–3.0 of monogastric animals, and thereby digesting the residual phytate in the whole seed.
In this study, a 1.4 kbp-long cDNA of phyA gene was isolated from A. niger and the cDNA encodes an approximately 52 kDa protein having 467 amino acids. Bacterial phytases are smaller in size (40–55 kDa) than the fungal phytases, because of glycosylation differences (Choi et al. 2001; Rodriguez et al. 2000). Sequence evaluation of phyA gene displayed the consensus motifs RHGXRXP and HD that are conserved among histidine acid phosphatases. These motifs count on a crucial element within the phosphorylation (Ullah et al. 1991; Van Etten et al. 1991; Kostrewa et al. 1997; Oh et al. 2001). The RHGXRXP motif is located at the dynamic sites of phytase. Furthermore, phyA contains a remote C-terminal His-Asp motif (HD motif) that is also likely to take part in the catalysis. Based on the above evidences, it is suggested that the cloned phyA belongs to a member of the phytase sub-family of histidine acid phosphatases (Mitchell et al. 1997).
Aspergillus niger phyA gene has been cloned and over-expressed in several microbial hosts, including S. cerevisiae (Han et al. 1999), P. pastoris (Han and Lei 1999), A. niger (Van Dijck 1999;) and E. coli (Phillippy and Mullaney 1997). As a first step towards demonstrating the potential of the cloned phytase gene, the cDNA was expressed and purified in E. coli system. The phytase activity of purified phytase was 826.33 U/mL. Comparable results were reported by earlier worker in the methylotrophic yeast, Pichia pastoris, by way of the heterologous expression of Debaryomyces castellii CBS 2923 phytase and the maximum production level obtained was 476 U/mL (Ragon et al. 2008). Xiong et al. (2005) and Bei et al. (2001) reported 865 U/mL and 165 U/mL of enzymatic activity, respectively, with A. niger phytase in P. pastoris.
Transgenic expression of phytase in maize seeds is a simple and less expensive strategy to enhance the bioavailability of phosphorus in feed compared to the supplementation of microbial phytase. In cereals, 90% of the seed phytic acid is in the aleurone, and the rest of the 10% in the scutellum; this is in contrast to maize, 90% is observed in scutellum and 10% in aleurone (O’Dell et al. 1972). We, therefore, used endosperm-specific promoters such as rice glutelin, a popular endosperm-specific promoter and maize α-zein promoter for driving endosperm-specific expression. Earlier, we have demonstrated that the α-zein promoter isolated from maize could drive endosperm-specific expression in transgenic rice (Joshi et al. 2015). For genetic transformation studies, we used a tropical inbred UMI29, which was shown to be amenable to genetic transformation in our laboratory (Joshi et al. 2014, 2016).
Thirty-six transgenic plants were obtained using the pZSPPHY construct and 32 plants using the pTOSPPHY construct. PCR and stable GUS expression studies showed the presence of transgenes in some of the transformants. To conclude whether the ectopically expressed phytase can effectively release Pi from the phytate complex, the Pi content of the transgenic seeds was estimated in JB-UMI29-Z17, JB-UMI29-Z30, JB-UMI29-G20 and JB-UMI29-G22 events. The Pi level in the transgenic seeds ranged from 0.79 to 2.54 mg/g of seed endosperm, while in control seeds the Pi level was 0.51 mg/g. The transgenic seeds showed a 0.6- to 5-fold increase in Pi level compared to the wild type. Among the different events screened, the event JB-UMI29-Z17/2 had a maximum Pi content of 2.54 mg/g of seed endosperm. This shows that the expression of A. niger phyA gene in endosperm released Pi and in turn enhanced the content of Pi. Similarly, the expression of A. niger phyA gene using an endosperm-specific EGH5 promoter in maize seeds increased the Pi level compared to the control (Shen et al. 2008). Earlier reports on soybean, Arabidopsis and maize over-expressing phytase additionally confirmed its effectiveness (Chiera et al. 2004; Coello et al. 2001; Chen et al. 2008).
Western blot analysis of the total protein of transgenic maize seed endosperm showed a detectable level of phytase protein expression in seeds of three events, viz., JB-UMI29-Z17/2, JB-UMI29-Z17/3 and JB-UMI29-G22/1. The plant-expressed A. niger phytase was higher in molecular weight compared to the E. coli-expressed protein (52 kDa). The increase in molecular weight of maize-expressed A. niger phytase may be due to glycosylation. Molecular weight shift of ectopically expressed phytases has also been observed in other expression systems such as tobacco (Verwoerd et al. 1995), wheat (Brinch-Pedersen et al. 2000) and maize (Chen et al. 2008), and the plant-produced A. phytase molecular size ranged from 60 to 71 kDa. It was also reported that though there was a difference in glycosylation pattern in the plant-expressed A. niger phyA and E. coli appA, they had the same catalytic property as that of the native enzyme (Coello et al. 2001; Ullah et al. 2002). Pen et al. (1993) studied the impact of plant-expressed phytase on broiler diet and observed that the inclusion of the transgenic tobacco seeds expressing phyA in animal feeds improved the phosphorus availability and broiler growth rate. A similar observation by Nyannor and Adeola (2008) suggests that E. coli phytase expressed in corn is effective in phosphorous usage and might minimize the need for supplemental phosphorous in broiler diets.
The phytase activity was assayed in the maize seeds that showed detectable amount of phytase protein in western blot analysis. A maximum phytase activity of 463.15 U/kg of seed was observed in event JB-UMI29-Z17/2 transgenic seed, and the phytase activity of event JB-UMI29-Z17/3 and event JB-UMI29-G22/1 was 412.72 and 370.81 U/kg of seed, respectively. The expression of A. niger phytase protein in transgenic maize seeds increased the phytase activity by 5.5- to 7-fold compared to non-transformed UMI29 seeds (67.45 U/kg of seed). Previous studies to express A. niger phytase protein in maize seeds resulted in a maximum increase of phytase activity by 2200–2900 U/kg of seed (Chen et al. 2008; Drakakaki et al. 2005) and least increase in phytase activity by 20.67 U/kg of seed (Shen et al. 2008).
Evaluation of T1 progenies confirmed stable integration of transgene and its inheritance in the subsequent generation. The average germination percentage of the T1 seeds was 90% and seeds were able to produce normal plants. This shows that the endosperm-specific expression has no negative effect on seed germination, growth and development of seedlings.
Conclusion
A full-length cDNA (phyA), encoding a phytase protein, was isolated from A. niger and heterologously expressed in E. coli and the phytase activity of the purified protein was estimated as 826.33 U/mL. Subsequently, the phyA gene was expressed in a tropical maize inbred line UMI29 in a seed-specific manner using maize α-zein or rice glutelin promoter. The T1 progeny evaluation showed stable integration and expression of transgenes. Western blot analysis confirmed the expression of A. niger phytase enzyme in transgenic maize seeds. Among different events generated, the event, JB-UMI29-Z17/2 had higher level of phytase activity (463.15 U/kg of seed) with fivefold increase in Pi level (2.54 mg/g) compared to untransformed control maize seeds (0.51 mg/g). The study demonstrated that it is possible to achieve a desirable level of phytase enzyme activity in an Indian transgenic tropical maize inbred line. This event could be a potential donor for transfer of phyA gene into other elite maize inbred lines through backcross breeding.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge the Department of Biotechnology, Government of India, New Delhi, for funding this research.
Abbreviations
- bp
Base pair
- cDNA
Complementary deoxyribonucleic acid
- lpa
Low phytic acid
- Pi
Inorganic phosphorus
- phyA
Phytase A gene
- U
Unit
- µM
Micromolar
- UMI
University maize inbred
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Adedokun SA, Adeola O. Calcium and phosphorus digestibility: metabolic limits. J Appl Poultry Res. 2013;22:600–608. doi: 10.3382/japr.2013-00740. [DOI] [Google Scholar]
- Bei JL, Chen Z, Yang L, Liao XZ, Wang Jiang ZY. Over-expression of artificial synthetic gene of Aspergillus niger NRRL3135 phytase in Pichia pastoris. Sheng Wu Gong Cheng Xue Bao. 2001;17:254–258. [PubMed] [Google Scholar]
- Brinch-Pedersen H, Olesen A, Rasmussen SK, Holm PB. Generation of transgenic wheat (Triticum aestivum L.) for constitutive accumulation of an Aspergillus phytase. Mol Breed. 2000;6:195–206. doi: 10.1023/A:1009690730620. [DOI] [Google Scholar]
- Burnette WN. Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate—polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Chen PS, Torribara TY, Warner H. Micro determination of phosphorous. Anal Chem. 1956;28:1756–1758. doi: 10.1021/ac60119a033. [DOI] [Google Scholar]
- Chen CC, Wu PH, Huang CT, Cheng KJ. A Pichia pastoris fermentation strategy for enhancing the heterologous expression of an Escherichia coli phytase. Enzyme Microbial Technol. 2004;35:315–320. doi: 10.1016/j.enzmictec.2004.05.007. [DOI] [Google Scholar]
- Chen R, Xue G, Che P, Yao B, Yang W, Ma Q, Fan Y, Zhao Z, Tarczynski MC, Shi J. Transgenic maize plants expressing a fungal phytase gene. Transgenic Res. 2008;17:633–643. doi: 10.1007/s11248-007-9138-3. [DOI] [PubMed] [Google Scholar]
- Chiera JM, Finer JJ, Grabau EA. Ectopic expression of a soybean phytase in developing seeds of Glycine max to improve phosphorus availability. Plant Mol Biol. 2004;56:895–904. doi: 10.1007/s11103-004-5293-6. [DOI] [PubMed] [Google Scholar]
- Choi YM, Suh HJ, Kim JM. Purification and properties of extracellular phytase from Bacillus sp. KHU-10. J Protein Chem. 2001;20:287–292. doi: 10.1023/A:1010945416862. [DOI] [PubMed] [Google Scholar]
- Chu CC, Wang CC, Sun CS, Hsu C, Yin KC, Chu CY, Bi FY. Establishment of an efficient medium for anther culture of rice through comparative experiments on the nitrogen source. Sci Sin. 1975;18:659–668. [Google Scholar]
- Cichy KA, Raboy V. Evaluation and development of low phytate crops. In: Krishnan H, editor. Modification of seed composition to promote health and nutrition, agronomy monograph 51. San Antonio: American Society of Agronomy and Crop Science Society of America; 2008. pp. 177–200. [Google Scholar]
- Coello P, Maughan JP, Mendoza A, Philip R, Bollinger DW, Veum TL, Vodkin LO, Polacco JC. Generation of low phytic acid Arabidopsis seeds expressing an E. coli phytase during embryo development. Seed Sci Res. 2001;11:285–291. [Google Scholar]
- Craxton A, Caffrey JJ, Burkhart W, Safrany ST, Shears SB. Molecular cloning and expression of a rat hepatic multiple inositol polyphosphate phosphatase. Biochem J. 1997;328:75–81. doi: 10.1042/bj3280075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denbow DM, Grabau EA, Lacy GH, Kornegay ET, Russell DR, Umbeck PF. Soybeans transformed with a fungal phytase gene improve phosphorus availability for broilers. Poult Sci. 1998;77:878–881. doi: 10.1093/ps/77.6.878. [DOI] [PubMed] [Google Scholar]
- Dionisio G, Madsen CK, Holm PB, Welinder KG, Jorgensen M, Stoger E, Arcalis E, Brinch-Pedersen H. Cloning and characterization of purple acid phosphatase phytases from wheat, barley, maize, and rice. Plant Physiol. 2011;156:1087–1100. doi: 10.1104/pp.110.164756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G, Marcel S, Glahn RP, Lund EK, Pariagh S, Fischer R, Christou P, Stoger E. Endosperm-specific co-expression of recombinant soybean ferritin and Aspergillus phytase in maize results in significant increases in the levels of bioavailable iron. Plant Mol Biol. 2005;59:869–880. doi: 10.1007/s11103-005-1537-3. [DOI] [PubMed] [Google Scholar]
- Geetha S (2011) Development of nutrient rich maize cultivar meant for poultry feed through transgenic means. Dissertation, Tamil Nadu Agricultural University, Coimbatore, India
- Greiner R, Konietzny U, Jany KD. Purification and characterization of two phytases from escherichia coli. Arch Biochem Biophys. 1993;303:107–113. doi: 10.1006/abbi.1993.1261. [DOI] [PubMed] [Google Scholar]
- Han YM, Lei XG. Role of glycosylation in the functional expression of an Aspergillus niger phytase (phyA) in Pichia pastoris. Arch Biochem Biophys. 1999;364:83–90. doi: 10.1006/abbi.1999.1115. [DOI] [PubMed] [Google Scholar]
- Han YM, Wilson DB, Lei XG. Expression of Aspergillus niger phytase gene (phyA) in Saccharomyces cerevisiae. Appl Environ Microbiol. 1999;65:1915–1918. doi: 10.1128/aem.65.5.1915-1918.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm PB, Kristiansen KN, Pedersen HB. Transgenic approaches in commonly consumed cereals to improve iron and zinc content and bioavailability. J Nutr. 2002;132:514–516. doi: 10.1093/jn/132.3.514S. [DOI] [PubMed] [Google Scholar]
- Hong CY, Cheng KJ, Tseng TH, Wang CS, Liu LF, Yu SM. Production of two highly active bacterial phytases with broad pH optima in germinated transgenic rice seeds. Transgenic Res. 2004;13:29–39. doi: 10.1023/B:TRAG.0000017158.96765.67. [DOI] [PubMed] [Google Scholar]
- Joshi JB, Yathish KR, Joel AJ, Kumar KK, Kokiladevi E, Arul L, Gnanam R, Balasubramanian P, Sudhakar D. A high-throughput regeneration protocol for recalcitrant tropical Indian maize (Zea mays L) inbreds. Maydica. 2014;59:211–216. [Google Scholar]
- Joshi JB, Geetha S, Singh Birla, Kumar KK, Kokiladevi E, Arul L, Balasubramanian P, Sudhakar D. A maize α-zein promoter drives an endosperm-specific expression of transgene in rice. Physiol Mol Biol Plants. 2015;21:35–42. doi: 10.1007/s12298-014-0268-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi JB, Nallathambi G, Kumar KK, Kokiladevi E, Arul L, Balasubramanian P, Sudhakar D. An efficient recovery of transgenic plants from a tropical Indian maize inbred line. J Microbiol Biotechnol Food Sci. 2016;5:335–340. doi: 10.15414/jmbfs.2016.5.4.335-340. [DOI] [Google Scholar]
- Karaman S, Cunnick J, Wang K. Expression of the cholera toxin B subunit (CT-B) in maize seeds and a combined mucosal treatment against cholera and traveler’s diarrhoea. Plant Cell Rep. 2012;31:527–537. doi: 10.1007/s00299-011-1146-3. [DOI] [PubMed] [Google Scholar]
- Kostrewa D, Leitch FG, D’Arcy A, Broger C, Mitchell D, Loon APGM. Crystal structure of phytase from Aspergillus ficuum at 2.5 Å resolution. Nat Struct Biol. 1997;4:185–190. doi: 10.1038/nsb0397-185. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lucca P, Hurrell R, Potrykus I. Genetic engineering approaches to improve the bioavailability and the level of iron in rice grains. Theor Appl Genet. 2001;102:392–397. doi: 10.1007/s001220051659. [DOI] [Google Scholar]
- Maugenest S, Martinez I, Godin B, Perez P, Lescure AM. Structure of two maize phytase genes and their spatio-temporal expression during seedling development. Plant Mol Biol. 1999;39:503–514. doi: 10.1023/A:1006131506193. [DOI] [PubMed] [Google Scholar]
- Mitchell DB, Vogel K, Weimann BJ, Pasamontes L, van Loon AP. The phytase sub-family of histidine acid phosphatases: isolation of two genes for two novel phytases from the fungi Aspergillus terrus and Myceliophthora thermophila. Microbiol-SGM. 1997;143:245–252. doi: 10.1099/00221287-143-1-245. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plantarum. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Nyannor EKD, Adeola O. Corn expressing an Escherichia coli-derived phytase gene: comparative evaluation study in broiler chicks. Poult Sci. 2008;87:2015–2022. doi: 10.3382/ps.2007-00501. [DOI] [PubMed] [Google Scholar]
- O’Dell B, de Boland AR, Koirtyohann SR. Distribution of phytate and nutritionally important elements among the morphological components of cereal grains. J Agric Food Chem. 1972;20:718–721. doi: 10.1021/jf60181a021. [DOI] [Google Scholar]
- Oh BC, Chang BS, Park KH, Ha NC, Kim HK, Oh BH, Oh TK. Calcium-dependent catalytic activity of a novel phytase from Bacillus amyloliquefaciens DS11. Biochemistry. 2001;40:9669–9676. doi: 10.1021/bi010589u. [DOI] [PubMed] [Google Scholar]
- Pen J, Verwoerd TC, Vanparidon PA, Beudeker RF, Vandenelzen PJM, Geerse K, Vanderklis JD, Versteegh HAJ, Vanooyen AJJ, Hoekema A. Phytase containing transgenic seeds as a novel feed additive for improved phosphorus utilization. Biotechnology. 1993;11:811–814. [Google Scholar]
- Phillippy BQ, Mullaney EJ. Expression of an Aspergillus niger phytase (phyA) in E. coli. J Agric Food Chem. 1997;45:3337–3342. doi: 10.1021/jf970276z. [DOI] [Google Scholar]
- Ponstein AS, Bade JB, Verwoerd TC, Molendijk L, Storms J, Beudeker RF, Pen J. Stable expression of phytase (phyA) in canola (Brassica napus) seeds towards a commercial product. Mol Breed. 2002;10:31–44. doi: 10.1023/A:1020326219687. [DOI] [Google Scholar]
- Raboy V, Gerbasi PF, Young KA, Stoneberg SD, Pickett SG, Bauman AT, Murthy PPN, Sheridan WF, Ertl DS. Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiol. 2000;124:355–368. doi: 10.1104/pp.124.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragon M, Roux VN, Chemardin P, Moulin G, Boze HH. Molecular gene cloning and overexpression of the phytase from Debaryomyces castellii CBS 2923. Protein Express Purif. 2008;58:275–283. doi: 10.1016/j.pep.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Rodriguez E, Mullaney EJ, Lei XG. Expression of Aspergillus fumigates phytase gene in Pichia pastoris and characterization of the recombinant enzyme. Biochem Biophys Res Commun. 2000;268:373–378. doi: 10.1006/bbrc.2000.2121. [DOI] [PubMed] [Google Scholar]
- Salgado KC, Vieira MC, VonPinho E, Guimaraes CT, VonPinho R, Sousa LV. Genetic purity certificate in seeds of hybrid maize using molecular markers. Revista Brasileira de Sementes. 2006;28:1. doi: 10.1590/S0101-31222006000100024. [DOI] [Google Scholar]
- Shen Y, Wang H, Pan G. Improving inorganic phosphorous content in maize seeds by introduction of phytase gene. Biotechnology. 2008;7:323–327. doi: 10.3923/biotech.2008.323.327. [DOI] [Google Scholar]
- Simell M, Turunen M, Piironen J, Vaara T (1989) Feed and food applications of phytase. Lecture at 3rd Meet. Industrial Applications of Enzymes, Barcelona, Spain
- Tamura K, Dudley J, Nei M, Kurmar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah AHJ. Production, rapid purification and catalytic characterization of extracellular phytase from Aspergillus ficuum. Prep Biochem. 1988;18:443–458. doi: 10.1080/00327488808062543. [DOI] [PubMed] [Google Scholar]
- Ullah AHJ, Cummins BJ, Dischinger HC., Jr Cyclohexanedione modification of arginine at the active site of Aspergillus ficuum phytase. Biochem Biophys Res Commun. 1991;178:45–53. doi: 10.1016/0006-291X(91)91777-A. [DOI] [PubMed] [Google Scholar]
- Ullah AHJ, Sethumadhavan K, Mullaney EJ, Ziegelhoffer T, Phillips SA. Characterization of recombinant fungal phytase (phyA) expressed in tobacco leaves. Biochem Biophys Res Comm. 1999;264:201–206. doi: 10.1006/bbrc.1999.1501. [DOI] [PubMed] [Google Scholar]
- Ullah AHJ, Sethumadhavan K, Mullaney EJ, Ziegelhoffer T, Austin-Phillips S. Cloned and expressed fungal phyA gene in alfalfa produces a stable phytase. Biochem Biophys Res Commun. 2002;290:1343–1348. doi: 10.1006/bbrc.2002.6361. [DOI] [PubMed] [Google Scholar]
- Van Dijck PWM. Chymosin and phytase: made by genetic engineering. J Biotechnol. 1999;67:77–80. doi: 10.1016/S0168-1656(98)00124-2. [DOI] [Google Scholar]
- Van Etten RL, Davidson R, Stevis PE, MacArthur H, Moore DL. Covalent structure, disulfide bonding, and identification of reactive surface and active site residues of human prostatic acid phosphatase. J Biol Chem. 1991;266:2313–2319. [PubMed] [Google Scholar]
- Verwoerd TC, van Paridon PA, van Ooyen AJ, van Lent JW, Hoekema A, Pen J. Stable accumulation of Aspergillus niger phytase in transgenic tobacco leaves. Plant Physiol. 1995;109:1199–1205. doi: 10.1104/pp.109.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong AS, Yao QH, Peng RH, Han PL, Cheng ZM, Li Y. High level expression of a recombinant acid phytase gene in Pichia pastoris. J Appl Microbiol. 2005;98:418–428. doi: 10.1111/j.1365-2672.2004.02476.x. [DOI] [PubMed] [Google Scholar]
- Zhang ZB, Kornegay ET, Radcliffe JS, Denbow DM, Veit HP, Larsen CT. Comparison of genetically engineered microbial and plant phytases for young broilers. Poult Sci. 2000;79:709–717. doi: 10.1093/ps/79.5.709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.