Abstract
UV light targets both membrane receptors and nuclear DNA, thus evoking signals triggering apoptosis. Although receptor-mediated apoptosis has been extensively investigated, the role of DNA damage in apoptosis is less clear. To analyze the importance of DNA damage induced by UV-C light in apoptosis, we compared nucleotide excision repair (NER)-deficient Chinese hamster ovary cells (lines 27-1 and 43-3B mutated for the repair genes ERCC3 and ERCC1, respectively) with the corresponding DNA repair-proficient fibroblasts (CHO-9 and ERCC1 complemented 43-3B cells). NER-deficient cells were hypersensitive as to the induction of apoptosis, indicating that apoptosis induced by UV-C light is due to unrepaired DNA base damage. Unrepaired lesions, however, do not activate the apoptotic pathway directly because apoptosis upon UV-C irradiation requires DNA replication and cell proliferation. It is also shown that in NER-deficient cells unrepaired lesions are converted into DNA double-strand breaks (DSBs) and chromosomal aberrations by a replication-dependent process that precedes apoptosis. We therefore propose that DSBs arising from replication of DNA containing nonrepaired lesions act as an ultimate trigger of UV-C–induced apoptosis. Induction of apoptosis by UV-C light was related to decline in the expression level of Bcl-2 and activation of caspases. Decline of Bcl-2 and subsequent apoptosis might also be caused, at least in part, by UV-C–induced blockage of transcription, which was more pronounced in NER-deficient than in wild-type cells. This is in line with experiments with actinomycin D, which provoked Bcl-2 decline and apoptosis. UV-C–induced apoptosis due to nonrepaired DNA lesions, replication-dependent formation of DSBs, and activation of the mitochondrial damage pathway is independent of functional p53 for which the cells are mutated.
INTRODUCTION
The main target of UV-C irradiation in living cells is nuclear DNA. The formation of DNA lesions such as pyrimidine dimers and TC(6-4) photoproducts inhibits DNA replication as well as transcription of RNA and causes chromosomal breakage, DNA recombination, mutations, and reproductive cell death (Friedberg et al., 1995). UV-C light also targets extranuclear macromolecules, inducing signal transduction pathways via the activation of membrane receptors such as insulin receptor, platelet-derived growth factor, fibroblast growth factor, and epidermal growth factor receptor (Canman and Kastan, 1996). Programmed cell death (apoptosis) induced by UV-C light in various cell types has been proposed to be a consequence of receptor activation (reviewed in Schwarz, 1998). This conclusion is based mainly on the finding that UV-C provokes clustering of CD95R, which leads to activation of the apoptotic caspase network (Rehemtulla et al., 1997; Aragane et al., 1998). Another line of evidence suggests activation of growth factor receptors to stimulate mitogen-activated protein kinase cascade, which activates transcription factors targeting genes that are involved in cell survival and apoptosis (Canman and Kastan, 1996). However, the model of receptor-mediated activation of apoptotic pathways in cells exposed to a DNA-damaging agent does not clearly consider nonrepaired DNA damage to be involved as a primary trigger of apoptosis. It also does not allocate DNA repair to play a role in defense against apoptosis.
A major defense mechanism against the deleterious effects of UV-C light is nucleotide excision repair (NER). NER-deficient cells are generally hypersensitive to the cytotoxic, mutagenic, and genotoxic effects of UV-C light (Wood and Burki, 1982; Westerveld et al., 1984; Stefanini et al., 1986; Weber et al., 1988; Weeda et al., 1990; reviewed in Friedberg et al., 1995). The hypersensitivity of repair-deficient cells to the cell-killing effect as measured by reproductive cell death (colony formation) could theoretically be due to apoptosis, necrosis, mitotic death, or irreversible cell cycle arrest (Orren et al., 1997). The contribution of nonrepaired DNA damage induced by various genotoxins to either one of these endpoints, notably apoptosis, still needs to be clarified. Also, it is not clear whether DNA damage-induced pathways interact, by cooperation or inhibition, with receptor-mediated signaling.
UV-C light activates the p53 tumor suppressor protein. This activation has been shown to control cell cycle arrest, blocking G1/S and G2/M transition (Agarwal et al., 1995; Guillouf et al., 1995). This is supposed to permit cells to repair DNA damage before passing through S phase or entering mitosis (Ponten et al., 1995; Poon et al., 1996). Furthermore, p53 has been shown to be directly involved in DNA repair (Ford and Hanawalt, 1997; Smith et al., 2000; Zhou et al., 2001) and in the regulation of the expression of DNA repair genes such as alkyltransferase (Rafferty et al., 1996; Grombacher et al., 1998). These functions of p53 are obviously protective, reducing genotoxicity of DNA-damaging agents. In contrast, p53 is a major player of apoptotic signaling (Reinke and Lozano, 1997), being involved in down-regulation of antiapoptotic Bcl-2 (Haldar et al., 1994; Miyashita et al., 1994) and up-regulation of proapoptotic Bax (Miyashita and Reed, 1995) and CD95R/FasR (Muller et al., 1998). It has been suggested that stalling of the transcription machinery by UV-C light-induced DNA lesions leads to activation of p53, thus initiating apoptosis (Ljungman et al., 1996, 1999). p53 induction by UV light has also been shown to occur in human epidermal cells where it causes apoptosis and sunburn (Brash et al., 1996). Apoptosis, however, can also be induced in a p53-independent manner upon exposure of cells to UV-C light and other kinds of DNA-damaging agents (Strasser et al., 1994), which was recently shown to be true also for p53-deficient mouse fibroblasts (Lackinger and Kaina, 2000). This indicates the existence of hitherto unknown p53-independent mechanisms involved in DNA damage-triggered apoptosis.
UV-C light blocks DNA replication, as all genotoxic agents do (Painter and Howard, 1982). Immediate replication blockage is due to inhibition of replication initiation and, at high-dose level, to DNA chain elongation (Kaufmann and Cleaver, 1981; Drissi and Lee, 1998; McGregor, 1999). UV-C induces DNA breaks and homologous as well as nonhomologous DNA recombination, which are detectable in the first post-treatment mitosis as chromatid-type aberrations. This process is strictly bound on replication of DNA containing nonrepaired lesions. It has been suggested that blockage of DNA replication is causally related to the formation of DNA double-strand breaks (DSBs) and chromosomal aberrations because of nuclease attack at stalled replication forks (Kaina, 1985, 1998). DSBs are also formed as a consequence of replication of DNA containing other kinds of damage, such as the alkylation lesion O6-methylguanine. This lesion has previously been shown to be majorly responsible for cell killing in alkyltransferase-deficient cells (reviewed in Kaina et al., 1997b) and to be ultimately involved in activating the apoptotic pathway (Tominaga et al., 1997; Kaina et al., 1997b; Meikrantz et al., 1998; Hickman and Samson, 1999; Ochs and Kaina, 2000). It has also been proposed that DSBs are the ultimate trigger of O6-methylguanine–induced apoptosis (Ochs and Kaina, 2000). In view of these findings, the question arises as to a possible involvement of DSBs in UV-C light-induced apoptosis. Herein, we show that apoptosis induced by UV-C light in fibroblasts requires cell cycle progression and DNA replication and is related to DNA strand break formation, which causes Bcl-2 decline and caspase activation in a p53-independent manner.
MATERIALS AND METHODS
Materials
All antibodies used for Western blotting were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). We used the antibodies anti-Bcl-2 (clone C-2), anti-Fas (clone M-20), and anti-ERK2 (C-14). The p53-promoter-mdm2-luciferase plasmid was provided by Dr. M. Oren (Rehovot, Israel). The general cell-permeable caspase inhibitor N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone (zVAD-FMK) was purchased from R & D Systems (Minneapolis, MN). Fluorescein isothiocyanate (FITC)-coupled zVAD-FMK is a product of Promega (Madison, WI). FITC-coupled anti-bromo-deoxyuridine (BrdU)-antibody is a product of BD Biosciences (San Jose, CA).
Cell Lines
In our experiments, we used the parental Chinese hamster ovary cell line CHO-9 and the UV-C–hypersensitive derivatives 43-3B and 27-1 (Wood and Burki, 1982). 43-3B cells are mutated in the ERCC1 gene (Westerveld et al., 1984), whereas 27-1 cells are mutated in the ERCC3 gene (Weeda et al., 1990). The corresponding gene products, ERCC1 (5′ endonuclease) and ERCC3 (helicase), are required for the incision step of the NER pathway. Therefore, cells in which these genes are inactivated are unable to perform NER. Stably complemented 43-3B cells by transfection with the cloned gene (herein designated as 43-3B/ERCC1 cells) were kindly provided by Dr. R. Wood (Pittsburgh, PA). Cells were cultured in DMEM/F-12 medium containing Glutamax (Invitrogen, Carlsbad, CA) and 5% fetal calf serum at 37°C in an atmosphere containing 7% CO2. Established mouse fibroblasts (the line BK4 generated in this laboratory) and primary mouse fibroblasts (Lackinger and Kaina, 2000) were cultured in DMEM containing 10% fetal calf serum. Transfected cell lines were selected by G418 (1.5 mg/ml).
UV-C Treatment
Before UV-C treatment, the culture medium was removed, cells were irradiated, and the medium was added again immediately after irradiation. The UV-C lamp used was calibrated routinely.
Survival Experiments
Four hundred cells were seeded into 6-cm dishes. Then 8–10 h later, the culture medium was removed and cells were irradiated with different doses of UV-C. One week later colonies were fixed with methanol and stained with 1.25% Giemsa, 0.125% crystal violet for counting. Survival was expressed in relation to the untreated control. Values are given as the mean of three independent experiments.
Determination and Quantification of Apoptosis
For quantification of apoptotic and necrotic cells, flow cytometric analyses with annexin V-FITC and propidium iodide-stained cells were performed. Treated and untreated cells were trypsinized, washed in ice-cold phosphate-buffered saline (PBS), and resuspended in 30 μl of cold annexin V binding buffer (10 mM HEPES pH 7.4, 0.14 M NaCl, 0.25 mM CaCl2 · 2H2O, 0.1% bovine serum albumin, wt/vol). After addition of 1.5 μl of annexin-V-FITC (BD PharMingen, San Diego, CA) cells were incubated for 15–30 min in the dark. At least 264 μl of binding buffer and 6 μl of propidium iodide (50 μg/ml) were added per sample. Samples were analyzed on a FACSort flow cytometer (BD Biosciences). For each sample, 10,000 cells were analyzed. The number of apoptotic and necrotic cells was calculated using a computer program (Cell Quest software; BD Biosciences). For semiquantitative determination of apoptosis, DNA laddering assays were performed as described (Ioannou and Chen, 1996). DNA of UV-C treated or untreated cells was isolated 48 h after irradiation.
Equal amounts of DNA of each sample were separated on a 1.5% agarose gel. To evaluate the number of cells with enhanced apoptotic caspase activity, treated and untreated cells were incubated in the dark in medium containing 10 μM FITC-VAD-FMK. Thereafter, cells were trypsinized and analyzed by flow cytometry. For simultaneous determination of DNA content and caspase activity, cells were trypsinized after incubation with FITC-VAD-FMK, washed with ice-cold PBS, and fixed in ice-cold 70% ethanol for a minimum of 1 h. Thereafter, cells were centrifuged, resuspended in 300 μl of PBS, and treated with RNase A for 30 min at room temperature. Finally, propidium iodide was added to a final concentration of 16 μg/ml and 10,000 cells were analyzed by flow cytometry.
BrdU–Enzyme-linked Immunosorbent Assay (ELISA)
For determination of DNA synthesis we used the Cell Proliferation ELISA BrdU assay (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's protocol. Briefly, cells cultured in 96-microwell plates were pulse labeled for 1 h with 10 μM BrdU. Thereafter, cells were fixed and DNA was denaturated for 30 min. After 1 h of incubation with a peroxidase-coupled anti-BrdU-antibody, cells were washed three times with PBS. Thereafter, peroxidase substrate (tetramethylbenzidine) was added for 30 min and measurements were performed on an ELISA reader (405 nm).
Neutral Single Cell Gel Electrophoreses (SCGE)
Treated and untreated cells were trypsinized and washed with PBS. Cell concentration was equilibrated to 1 × 106/ml. Cell suspension (10 μl) was resuspended in 120 μl of 0.5% low melting point agarose at 37°C, spotted onto a microscope slide, and covered with a coverslip. After keeping on ice for 5 min and removing the coverslip, slides were incubated in neutral lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Tris, 1% Na-laurylsarcosinate, pH 7.5) for 1 h. Thereafter, slides were placed into an electrophoreses chamber filled with electrophoreses buffer (90 mM Tris, 90 mM boric acid, 2 mM EDTA, pH 7.5). Thereafter, electrophoreses were performed, slides were washed in water, fixed in ethanol, and air dried. Analysis of 50 cells/sample was performed automatically after staining with 20 μg/ml ethidium bromide on a fluorescence microscope by using a charge-coupled device camera connected to a personal computer and analysis software (Fuji Imaging, Tokyo, Japan).
Chromosomal Preparations
Treated and untreated cells were incubated in medium in the presence of 50 ng/ml colcemid for 2 h. Thereafter, cells were trypsinized. Trypsinized cells were collected together with the medium, centrifuged, washed with PBS, and centrifuged again. Thereafter, the cell pellet was resuspended and incubated in 75 mM KCl for 7 min at room temperature and centrifuged. Afterward, the cell pellet was resuspended in 2 ml of 75 mM KCl, and 12 ml of ice-cold methanol/acetic acid (3:1) was added. After centrifugation, cells were resuspended in methanol/acetic acid and incubated at −20°C for a minimum of 30 min. Thereafter, cells were again centrifuged and resuspended in methanol/acetic acid. The cell pellet was resuspended in 1 ml of methanol/acetic acid (1:1), spotted onto an ice-cold wet microscope slide, and fixed by heating. After Giemsa staining, the preparations were analyzed under a microscope. At least 100 metaphases were scored per measure point.
BrdU-In Situ Staining
Immediately after irradiation of cells grown on a coverslip, BrdU was added into the culture medium up to a final concentration of 10 μM/ml. After a postincubation period of 36 h, cells were washed with cold PBS, fixed onto the coverslip, and DNA was denaturated for 30 min. Thereafter, cells were incubated with an FITC-coupled anti-BrdU antibody in the dark for 30 min. Then cells were washed three times for 5 min with PBS. For chromatin staining, cells were incubated for 5 min in a 10-μl solution (1 μg/ml). The percentage of BrdU-stained cells with apoptotic chromatin morphology was determined using a fluorescence microscope.
Transfection Experiments
Cells were stably transfected with Bcl-2 expression plasmid (Dimmeler et al. 1999) as previously described (Ochs and Kaina, 2000).
Reporter Gene Assay
Cells were transfected with a p53-driven mdm-2-promoter-luciferase plasmid by means of the calcium phosphate coprecipitation method as described above. Transfected cells were split into two culture dishes serving as a control and treatment dish, respectively. Ten hours thereafter cells were either treated with 15 J/m2 UV-C or left untreated. Twelve hours after treatment cells were trypsinized, washed in cold PBS, and resuspended in 0.25 M Tris pH 8.0. Thereafter, cells were frozen in liquid nitrogen and thawed quickly at 37°C for three times. Finally, the suspension was centrifuged for 10 min at 10,000 × g and protein concentration of the supernatant was determined. Protein (10 μg) of each probe in a total volume of 20 μl was used for luciferase activity measurement. Measurements were made using the Berthold luminometer Sirius.
Preparation of Cell Extracts
Trypsinized treated and untreated cells were washed with cold PBS, resuspended in sonification buffer (20 mM Tris-HCl pH 8.5, 1 mM EDTA, 5% glycerin, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride), and sonified. The resulting suspension was centrifuged with 20,000 × g for 15 min. Supernatants were collected and protein concentration was determined.
Western Blot Analysis
Protein (20–30 μg) of the probes was separated on a 7.5–12% SDS polyacrylamide gel. Thereafter, proteins were blotted onto a nitrocellulose transfer membrane (Protran; Schleicher & Schuell, Dassel, Germany) for 3 h or, in some experiments, overnight. Membranes were blocked for 2 h in 5% (wt/vol) milk powder in PBS containing 0.1% Tween 20 (PBT), incubated for 2 h with the primary antibody (1:3000–5000 dilution), washed three times with PBT, and incubated for 1 h with a horseradish peroxidase-coupled secondary antibody 1:3000 (Amersham Biosciences AB, Uppsala, Sweden). After final washing with PBT (3 times for 10 min each) blots were developed by using a chemiluminescence detection system (Amersham Biosciences AB).
Transcription Measurement
Treated and untreated cells were pulse labeled for 1 h with 0.5 μCi/ml [3H]uridine triphosphate. After trypsinization, cells were sucked onto glass microfiber filters, washed carefully with 10% trichloroacetic acid and three times with aqua dest., and finally with ethanol. Thereafter, filters were air dried and measured by scintillation counting.
RESULTS
NER-deficient Cells Are Hypersensitive to UV-C–induced Apoptosis
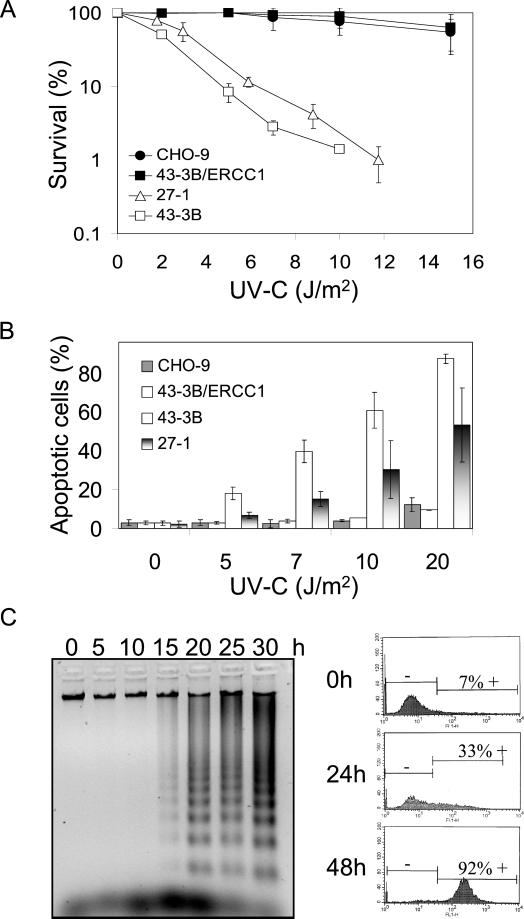
The NER-deficient CHO cell lines 27-1 and 43-3B mutated in ERCC3 and ERCC1 gene, respectively, are known to be hypersensitive to UV-C light. The killing response of these cells in comparison with the wild-type and ERCC1-complemented 43-3B cells as determined by reduction in colony formation is shown in Figure 1A. Annexin staining and flow cytometric measurement showed that these cells are also hypersensitive to the induction of apoptosis. Within the dose range in which survival of complemented and wild-type cells was not yet affected 27-1 and 43-3B cells showed a dose-dependent increase in apoptosis (Figure 1B). The frequency of necrosis (for quantification, see MATERIALS AND METHODS) was only slightly enhanced in the DNA repair-deficient cells (our unpublished data ).
Figure 1.
Cell-killing response and apoptosis in NER-deficient cells treated with UV-C light. (A) Survival of CHO-9, 43-3B/ERCC1, 27-1, and 43-3B cells as a function of UV-C dose, as determined by colony formation assay. (B) Frequency of apoptosis of CHO-9, 43-3B/ERCC1, 43-3B, and 27-1 cells as a function of dose of UV-C (measured 48 h after irradiation). (C) Apoptotic DNA laddering pattern in 27-1 cells (left) as a function of time after UV-C treatment (10 J/m2). Flow cytometric quantification of 27-1 cells with enhanced caspase activity 24 and 48 h after UV-C irradiation (10 J/m2) as determined by FITC-zVAD-FMK staining (right).
To confirm increase of the induction of apoptosis by UV-C light in the NER-deficient cells and to determine the time of initiation of apoptosis after UV-C treatment, DNA laddering assays were performed. As demonstrated in Figure 1C, apoptotic DNA degradation started ∼15 h after UV-C irradiation in the NER-deficient cells. We also measured overall caspase activation by flow cytometry of FITC-VAD-FMK–stained cells. Induction of caspases was first observed 15 h after UV-C treatment and further increased with the time of postincubation (Figure 1C, right).
UV-C–induced Apoptosis in NER-deficient Cells Requires Cell Cycle Progression
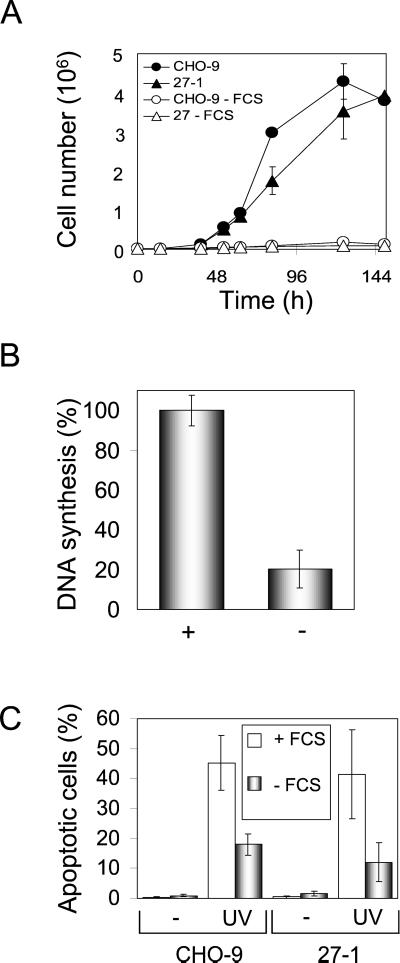
To address the question of whether cell cycle progression is necessary for UV-C–induced apoptosis, we comparatively investigated UV-C–induced apoptosis in serum-starved and nonstarved cells. Serum starvation of CHO-9 and 27-1 cells for 3 days strongly reduced cell proliferation (Figure 2A) as well as DNA synthesis (Figure 2B). Flow cytometric cell cycle analysis revealed accumulation of starved cells in G1 (>88% G1 fraction; our unpublished data). Starvation of CHO-9 and 27-1 cells followed by UV-C irradiation resulted in a significant reduction (by ∼50%) of apoptosis compared with proliferating cells (Figure 2C). In this experiment, we used an equitoxic dose (in terms of colony formation) of UV-C for irradiation of CHO-9 and 27-1 cells (40 and 10 J/m2, respectively), which yielded nearly identical rates of apoptosis in both cell types. Because with an equitoxic dose of UV light the same frequency of proliferating cells underwent apoptosis (Figure 2C), it is reasonable to conclude that apoptosis is a main cause of UV-C–induced cytotoxicity, both in NER-deficient 27-1 and wild-type cells. We should note that the frequency of necrosis was not significantly enhanced in 27-1 cells. Overall, the data obtained with the NER-deficient mutant clearly indicate that DNA damage is the main trigger of UV-C–induced apoptosis. The data also show that UV-C–induced apoptosis requires cell proliferation.
Figure 2.
UV-C–induced apoptosis in serum-starved cells. (A) Growth curves of CHO-9 and 27-1 cells cultured with or without fetal calf serum. (B) DNA synthesis of CHO cells cultured for 72 h with (+) or without (−) serum as determined by BrdU-ELISA. (C) Frequency of apoptosis in CHO-9 and 27-1 cells irradiated with 40 and 10 J/m2, respectively, as measured by annexin staining. Cells were cultured for 72 h with or without serum before irradiation and apoptosis was measured 48 h after irradiation.
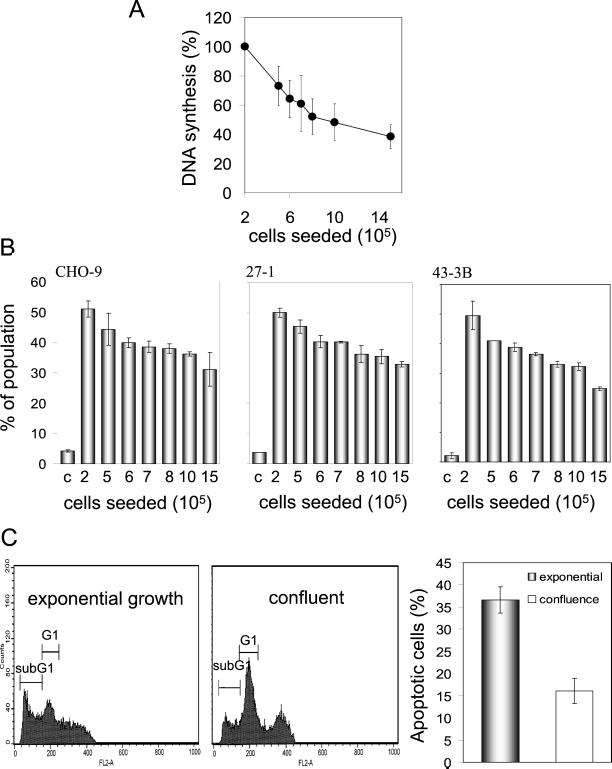
One could argue that serum starvation may alter the sensitivity of cells to UV-C light due to growth factor depletion. To reject this argument, we analyzed UV-C–induced apoptosis without changing the serum concentration as a function of cell density, which determines the confluence status of the adherent growing cells. As shown in Figure 3A, the DNA replication rate declined with increasing cell density. If cells seeded at different density were treated with UV-C light, the frequency of apoptosis induced in CHO-9, 27-1, and 43-3B cells was inversely related to cell density (Figure 3B). Obviously, the cells became more refractory to UV-C–induced apoptosis when they were not allowed to replicate extensively.
Figure 3.
Dependency of UV-C–induced apoptosis on proliferation. (A) DNA synthesis as a function of the number of cells seeded. (B) Frequency of apoptosis 48 h after UV-C irradiation (10 J/m2 for 27-1 and 43-3B cells; 40 J/m2 for CHO-9 cells) as a function of cell seeding number. (C) Flow cytometric quantification of UV-C (40 J/m2)-induced apoptotic sub-G1 cells in exponentially growing and confluent primary mouse fibroblasts as measured 48 h after irradiation.
To gain further support for the conclusion of proliferation dependence of the induction of apoptosis by UV-C light, we compared confluent and subconfluent primary mouse fibroblasts. These cells exhibit contact inhibition more severely than CHO cells. As shown in Figure 3C, contact-inhibited mouse fibroblasts displayed a very strong suppression of UV-C–induced apoptosis if they were treated in a confluent state. The effect was clearly more pronounced than in the CHO cell lines (38 vs. 55% reduction, respectively), which is likely due to nearly complete growth inhibition of mouse monolayers cultures.
UV-C Light Induces Proliferation-dependent DSBs in NER-deficient Cells
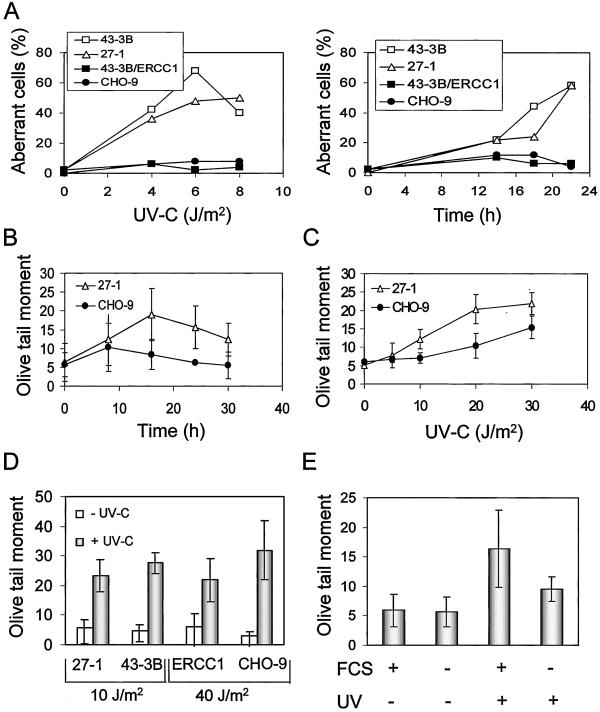
Previously, we proposed that DNA DSBs and/or chromosomal aberrations arising from them act as a distal trigger of genotoxin-induced apoptosis (Ochs and Kaina, 2000; Lips and Kaina, 2001). To see whether this could also be true for NER-deficient cells irradiated with UV-C, we analyzed both endpoints comparatively in CHO-9 and NER-deficient 27-1 cells as a function of dose and time. The data are shown in Figure 4A. Chromosomal aberrations were significantly induced by low doses of UV-C light in NER-deficient cells, whereas only a slight induction was observed within this dose range (0–10 J/m2) in CHO-9 and 43-3B/ERCC1 cells. Chromosomal aberrations were observed as early as 14 h after irradiation and increased in frequency with increasing postincubation time.
Figure 4.
UV-C–induced DNA breakage is proliferation dependent. (A) Frequency of UV-C–induced chromosomal aberrations in 43-3B, 27-1, 43-3B/ERCC1, and CHO-9 cells as a function of UV-C dose (left; 22 h post-treatment) and time (right; 8 J/m2 of UV-C). (B) UV-C–induced DSBs as a function of time. Cells were irradiated with a UV-C dose of 10 J/m2 and analyzed by means of neutral SCGE. (C) DSBs as a function of UV-C dose 20 h post-treatment, as determined by neutral SCGE. (D) UV-C light-induced DSBs as measured 20 h after UV-C irradiation in 27-1, 43-3B, 43-3B/ERCC1 (ERCC1), and CHO-9 cells, which were determined by neutral SCGE assay. (E) UV-C–induced DSBs 20 h after UV-C treatment (10 J/m2) in 27-1 cells cultivated in medium not containing (−) or containing (+) fetal calf serum, as determined by neutral SCGE.
To see whether NER-deficient cells are vulnerable to DSB formation, the frequency of DSBs was analyzed by means of neutral SCGE. DSB induction was observed already 8 h after UV-C irradiation in NER-deficient cells (Figure 4B). The induction was a function of dose; it increased notably in NER-deficient 27-1 cells with doses between 5 and 30 J/m2 (as measured 20 h after irradiation; Figure 4C). Increase of DSB frequency was also observed in 43-3B cells irradiated with 10 J/m2 (Figure 4D). In NER-proficient cells a similar frequency of DSBs was observed with 40 J/m2 (Figure 4D). To see whether the formation of DSBs by UV-C light depends on DNA synthesis and cell cycle progression, we compared serum-starved and nonstarved cells treated with UV-C light. The frequency of DSBs was clearly reduced in irradiated starved cells (Figure 4E).
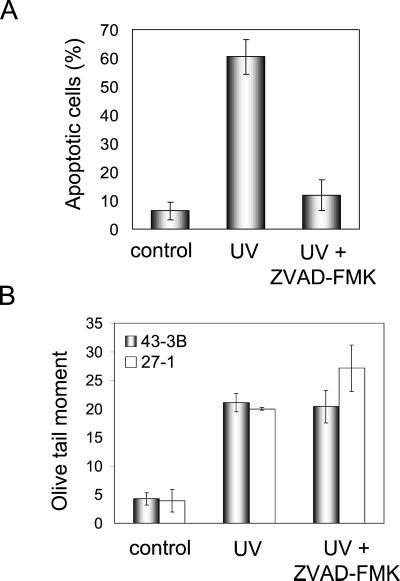
One could argue that DNA strand breaks observed by SCGE are merely a reflection of apoptotic internucleosomal DNA cleavage. Therefore, we compared the induction of DSBs in cells treated with UV-C alone and UV-C plus zVAD-FMK, which is a general caspase inhibitor. Post-treatment of UV-C–irradiated cells with zVAD-FMK reduced apoptosis frequency nearly to the level of untreated cells (Figure 5A). However, application of zVAD-FMK did not affect the level of UV-C–induced DSBs as determined by SCGE (Figure 5B). This indicates that DSBs measured by neutral SCGE were not resulting from apoptotic internucleosomal DNA fragmentation.
Figure 5.
Reduction of apoptosis but not DSBs after UV-C irradiation by zVAD-FMK. (A) Frequency of apoptosis in UV-C–irradiated (10 J/m2) 27-1 cells treated or not treated with the general caspase inhibitor zVAD-FMK (100 μM). Cells were analyzed 48 h after UV-C treatment by annexin staining and flow cytometry. (B) UV-C (10 J/m2) induced DSBs in 27-1 cells treated or not with the general caspase inhibitor zVAD-FMK (100 μM). Cells were harvested 20 h after irradiation and subjected to neutral SCGE.
Most UV-C–induced Apoptotic Cells Underwent S Phase after Irradiation
As revealed by time course experiments, DNA breaks and aberrations preceded the appearance of apoptotic cells. To elucidate whether apoptotic cells have passed through S phase after UV-C irradiation, we labeled UV-C–irradiated cells deficient in NER with BrdU immediately after irradiation and analyzed DNA synthesis and apoptosis (by means of apoptotic nuclear morphology) in the same individual cells by using anti-BrdU antibody as well as 4,6-diamidino-2-phenylindole (DAPI) staining in situ. As shown in Table 1, ∼70% of the apoptotic cells were clearly positive for BrdU staining, which is characteristic for replicating cells. This indicates that most cells have progressed through S phase before they were triggered to die by apoptosis.
Table 1.
Percentage of cells undergoing apoptosis after DNA replication Quantification was performed by simultaneous staining for DNA synthesis and apoptotic nuclear morphology after UV-C irradiation (36 h post-treatment; 10 J/m2) of 27-1 cells. BrdU-labelling had been performed for 36 h after UV-C irradiation.
| Expt. | Analyzed apoptotic cells | BrdU-positive apoptotic cells | % |
|---|---|---|---|
| 1 | 217 | 165 | 76, 0 |
| 2 | 122 | 77 | 63, 1 |
| 3 | 247 | 173 | 70, 0 |
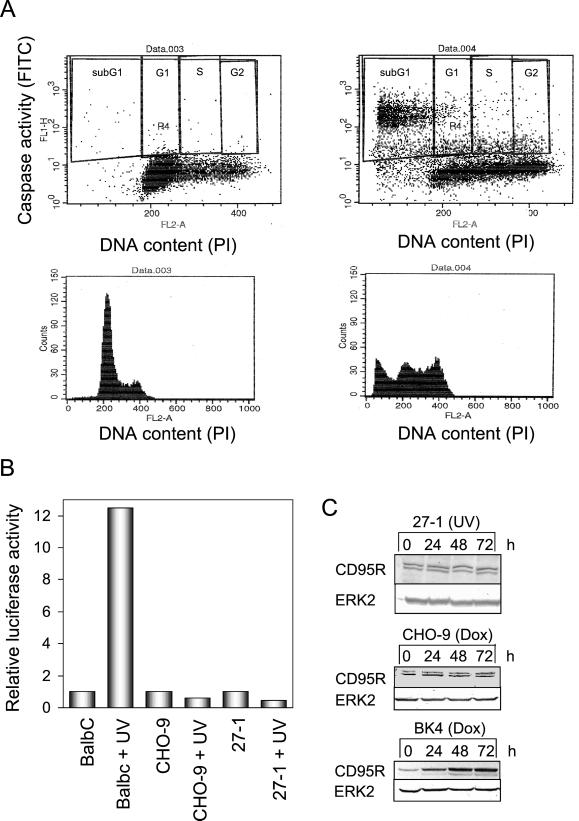
NER-deficient cells pass after UV-C irradiation through the cell cycle (otherwise chromosomal aberrations would not be detectable in the first post-treatment mitoses), but do not significantly proliferate later on (our unpublished data). Therefore, UV-C–induced apoptosis due to DNA damage is likely to be initiated in the postexposure cell cycle, after the cells have passed through mitosis. By simultaneous flow cytometric determination of DNA content and overall caspase activity in treated and untreated cells, we demonstrate that apoptotic caspase activation occurs in all cell cycle phases of UV-C—irradiated cells. Most caspase activity was detectable, however, in the apoptotic sub-G1 population and in cells out of G1 phase (Figure 6A). Because most of the apoptotic cells underwent S phase after irradiation (Table 1), it is reasonable to conclude that G1 phase cells exhibiting strongly enhanced caspase activity undergo apoptosis after the first post-treatment division, i.e., in the first postexposure cell cycle.
Figure 6.
Cell cycle-specific activation of caspases and p53 activity in UV-C–irradiated 27-1 cells. (A) Flow cytometric dual color staining for DNA content (PI) and caspase activity of untreated (top, left) and UV-C–treated (10 J/m2; top, right) 27-1 cells. The bottom panels show quantitative cell cycle distributions 36 h after treatment. (B) Determination of p53-regulated mdm2 promoter activity after transient transfection of the construct followed by UV-C irradiation (15 J/m2). Cells were harvested 12 h after treatment of BalbC, CHO-9, and 27-1 cells and analyzed for luciferase activity. (C) Expression of CD95R after UV-C treatment (10 J/m2) of 27-1 cells and of doxorubicin-treated (0.5 μg/ml) CHO-9 and BK4 cells, as shown by Western blot analysis.
UV-C–induced Apoptosis in NER-deficient Cells Does not Require Functional p53 and Fas Receptor (Fas-R) Up-Regulation
Because p53 is considered to be a key regulator of apoptosis upon DNA damage, we analyzed whether CHO-9 and 27-1 cells possess p53 transactivation activity. We used mdm2-promoter-luciferase construct, which is clearly inducible in p53-expressing BalbC cells upon UV-C irradiation. In CHO-9 and 27-1 cells, however, there was no induction of the p53-driven promoter (Figure 6B), supporting the view that p53 is mutated and therefore functionally inactive in CHO-9 cells. Obviously, p53 is not involved in UV-C–induced apoptosis in this cell type. We also measured the expression level of Fas-R (CD95R). As shown in Figure 6C, treatment of cells with UV-C did not provoke up-regulation of Fas-R. In a control experiment with doxorubicin, which was reported to induce Fas-R (Fulda et al., 2000), we also did not find induction, whereas in mouse fibroblasts (BK4 cells expressing p53 wild type) Fas-R was up-regulated (Figure 6C). These data indicate that CHO-9 cells and the corresponding NER-defective derivatives are unable to respond with Fas up-regulation to UV-C irradiation. Similar data were obtained with Fas ligand, which proved also not to be induced in CHO-9 cells upon UV-C treatment (our unpublished data).
Decline of Bcl-2 in NER-deficient Cells
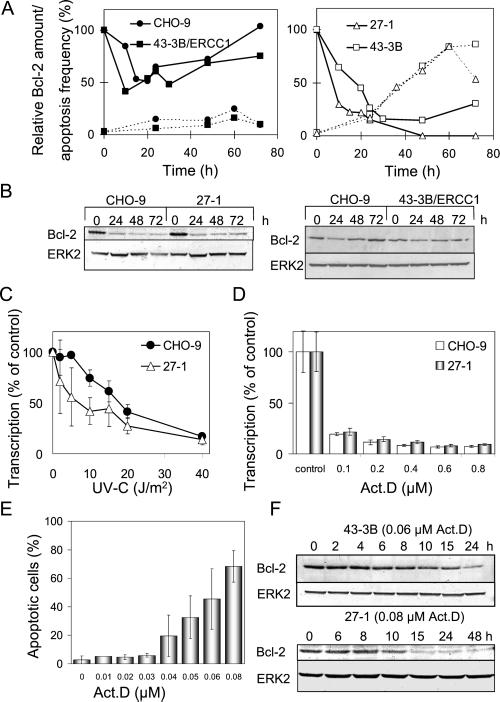
To see whether Bcl-2 level is related to UV-C–induced apoptosis, Bcl-2 was quantified as a function of time after UV-C exposure. As shown in Figure 7A, Bcl-2 decreased in the repair competent cells 10–20 h after irradiation and thereafter recovered, returning to control level (Figure 7A, left). In NER-deficient cells, however, a dramatic decrease of Bcl-2 was observed and recovery to the control level did not occur (Figure 7A, right). As revealed by the time course experiments, in both NER-deficient cell types decrease in Bcl-2 preceded the induction of apoptosis (Figure 7A, right). One could argue that NER-competent cells would respond in the same way as repair-deficient cells, albeit at higher dose level at which UV-C–induced DNA lesions cannot be repaired in time. This is indeed the case. As shown in Figure 7B, Bcl-2 declined in CHO-9 cells irradiated with 40 J/m2 to the same extent as in 27-1 cells irradiated with 10 J/m2. With this low dose, CHO-9 and ERCC-1–complemented 43-3B cells did not display significant Bcl-2 decline. Because NER-deficient cells showed decline of Bcl-2 at a lower UV-C dose level than repair-competent cells, the data indicate that Bcl-2 becomes down-regulated in response to nonrepaired UV-C–induced DNA damage.
Figure 7.
Decline of Bcl-2 in NER-deficient cells after UV-C irradiation. (A) Relative Bcl-2 amount (continuous line) and apoptosis frequency (dotted line) as a function of time after UV-C irradiation (10 J/m2) of CHO-9 and 43-3B/ERCC1 cells (left) and NER-deficient 27-1 and 43-3B cells (right). (B) Expression of Bcl-2 in CHO-9 and 27-1 cells treated with 40 and 10 J/m2, respectively (left), as well as expression of Bcl-2 in 10 J/m2 UV-C–treated CHO-9 and 43-3B/ERCC1 cells, as shown by Western blot analysis. (C) Transcriptional activity as a function of UV-C dose in CHO-9 and 27-1 cells 4 h after treatment. (D) Transcriptional activity as a function of concentration of actinomycin D in CHO-9 and 27-1 cells, as determined 4 h after treatment. (E) Frequency of apoptosis in 27-1 cells as a function of concentration of actinomycin D. (F) Expression of Bcl-2 in 43-3B and 27-1 cells after actinomycin D treatment, as shown by Western blot analysis. The ERK2 signal illustrates equal protein loading on the gel.
Trancription Blockage in NER-deficient Cells and Apoptosis
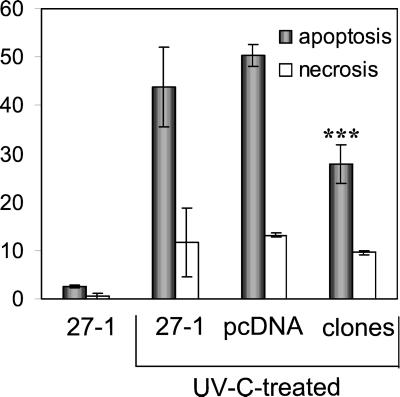
To elucidate the effect of UV-C irradiation on transcription, RNA synthesis was measured by uridine incorporation. Transcription was blocked significantly in the dose range of 2–20 J/m2 with 27-1 cells eliciting a clearly stronger response than the wild type (Figure 7C). This finding raised the question of whether increased transcription blockage due to nonrepaired DNA damage is related to Bcl-2 decline and the induction of apoptosis in NER-defective cells. If true one would suppose that inhibition of transcription by a treatment other than UV-C would cause the same effect. To prove this, we treated cells with the transcription inhibitor actinomycin D (Act D). The agent caused a clear suppression of transcription (Figure 7D) and, within the same concentration range, induction of apoptosis (Figure 7E). We next analyzed the effect of the RNA synthesis inhibitor on Bcl-2 expression, using a concentration of Act D that induced the same yield of apoptosis as 10 J/m2 of UV-C light (the Bcl-2 expression studies described above were performed by treating cells with 10 J/m2 UV-C). As shown in Figure 7F, Act D caused decrease in Bcl-2 with a time course similar to that observed after UV-C treatment of 27-1 and 43-3B cells. This supports the view that transcription blockage after exposure to UV-C light is involved in Bcl-2 decline and the induction of apoptosis in NER-deficient cells. To further prove the involvement of Bcl-2 in apoptosis, Bcl-2 was stably overexpressed by transfection in 27-1 cells (unpublished expression data), which were subjected for apoptosis measurements. Figure 8 shows the average response of Bcl-2 transfectants in comparison with 27-1 and vector-transfected 27-1 cells. Bcl-2 transfectants were significantly protected against the induction of apoptosis upon UV-C irradiation. They were not protected as to the level of UV-C–induced necrosis.
Figure 8.
Effect of Bcl-2 overexpression in 27-1 cells on UV-C–induced apoptosis. 27-1 cells were transfected with a myc-bcl-2 construct and seven Bcl-2-overexpressing clones were established. Clones, vector transfected cells, and untransfected cells were irradiated (10 J/m2 UV-C) and flow cytometric annexin measurements were performed 48 h later (∗∗∗, highly significant difference with p < 0.05). Ordinate: average yield (%) of apoptotic and necrotic cells.
DISCUSSION
Although it is known that UV-C light induces apoptosis in various cell types (reviewed in Schwarz, 1998), the mechanism still needs to be elucidated. UV-C light acts upon nuclear as well as cytoplasmic targets such as CD95/Fas (Rehemtulla et al., 1997; Aragane et al., 1998). Although several lines of evidence suggest that DNA damage and activation of the CD95/Fas receptor contribute independently to apoptosis (Schwarz, 1998), it is not known how important the DNA damage and the receptor triggered pathway for a given cell type are and how UV-C–induced DNA damage triggers the response. In this investigation, we demonstrate that hypersensitivity to UV-C light of NER mutants, which are deficient in the removal of UV-C–induced DNA damage (Weber et al., 1988; Hayashi et al., 1998), is due to massive induction of apoptosis (as determined by annexin staining, electrophoretic DNA laddering, and activation of caspases) but not to a significant extent to necrosis. The increased apoptotic response of NER-deficient cells indicates that nonrepaired UV-C–induced DNA damage acts as a primary trigger of apoptosis. The data confirm a recent report demonstrating repair of pyrimidine dimers by photolyase in mammalian cells to protect against apoptosis (Chigancas et al., 2000). One might argue that the apoptotic pathway is evoked specifically in repair-deficient cells. This however is not true. Treatment of DNA repair-proficient CHO-9 and ERCC1-complemented 43-3B cells with an equitoxic dose (as measured by loss of colony-forming ability) of UV-C resulted in a level of apoptosis comparable with that observed in the repair-deficient mutants. This indicates that reproductive cell death induced by UV-C light in normal repair-proficient CHO fibroblasts is also caused by apoptosis triggered by nonrepaired DNA damage.
The major cell-killing lesions induced by UV-C light are pyrimidine dimers and (6-4) photoproducts (Friedberg et al., 1995). If nonrepaired UV-C–induced DNA lesions trigger apoptosis, the question arises as to whether the primary DNA damage itself (i.e., the base damage) constitutes the signal activating the apoptotic pathway or whether other (secondary) lesions arising from them are involved. The primary base damage is very likely not sufficient in triggering apoptosis, at least in fibroblasts, because apoptosis is a rather late response that requires cell proliferation (as discussed below). A critical candidate of secondary lesions could be DSBs, which have previously been shown to be very efficient in inducing apoptosis (Lips and Kaina, 2001). UV-C light does not induce DSBs per se. However, DSBs can be formed during DNA replication due to nuclease attack at stalled replication forks, which has been proposed to be a critical event not only for UV-C but also for other genotoxic agents (Kaina et al., 1997a; Kaina, 1998). To examine whether unrepaired UV-C light-induced DNA lesions are converted into DSBs by means of DNA replication, we analyzed the induction of DSBs by neutral SCGE. We also analyzed chromosomal aberrations arising from them. It is shown that UV-C induces significantly more DSBs and chromosomal aberrations in NER-deficient cells compared with NER-proficient fibroblasts. This indicates that primary nonrepaired UV-C light–induced DNA lesions are indeed converted into DSBs. The induction of DSBs was dependent on DNA replication because confluent G1 phase-arrested and serum-starved cells suppressed in DNA replication exhibited a reduced rate of UV-C–induced DSBs. This supports the hypothesis that DSBs induced by UV-C light are generated in S phase during the replication of DNA, which contains lesions blocking replication.
It is of much interest that not only the frequency of DSBs but also the frequency of apoptosis was significantly reduced in UV-C light-treated cells that were nonreplicating due to serum starvation. This supports the hypothesis that DSBs are critically involved in UV-C light–induced apoptosis. Because serum starvation may cause side effects, we verified the finding by using cells of different confluence and proliferation status. These experiments revealed that UV-C–induced apoptosis is indeed a function of proliferation, because apoptosis in CHO cells was reduced with increasing confluence and reduced DNA replication capacity (as measured by BrdU incorporation) of the cell population. As demonstrated by simultaneous BrdU labeling and nuclear staining of apoptotic cells, at least 70% of apoptotic cells have passed through S phase upon UV-C treatment. Replication dependence of UV-C light-induced apoptosis was confirmed with primary mouse fibroblasts exhibiting strong contact inhibition. These cells become highly refractory to UV-C–induced apoptosis when they were kept in a confluent state upon irradiation. Overall, the data indicate that the induction of apoptosis by UV-C light requires DNA replication.
Apoptosis could be initiated in the postreplicative G2 phase or in the cell cycle thereafter. Because most UV-C–exposed cells undergo mitosis (in which chromosomal damage becomes visible) it is pertinent to conclude that apoptosis is initiated in the first postexposure cell cycle. This is supported by the fact that significant activation of caspases occurred in cells out of G1 phase 36 h after irradiation. Because the first postexposure mitotic cells exhibit massive chromosomal aberrations, it is tempting to speculate that loss of genetic material due to chromosomal deletions and translocations leading to a disturbed balance of gene expression could be critically involved in the initiation of apoptosis. Chromosomal aberrations as a trigger of apoptosis have been proposed before for another cellular system (Kaina et al., 1997a) and gain support by recent data from other investigations (De Santis et al., 2001).
To study in more detail the molecular mechanism of apoptosis due to UV-C–induced DNA damage, we proved the involvement of various pro- and anti-apoptotic proteins. One major player is p53, which is inducible by UV-C light leading either to cell cycle arrest or apoptosis, depending on the cellular system (Agarwal et al., 1995; Guillouf et al., 1995; Liebermann et al., 1995; Pellegata et al., 1996). CHO cells, however, do not express detectable p53 protein (our unpublished data). Also, luciferase assays with a p53-regulated mdm2-promoter construct showed lack of transactivating activity of p53 in UV-C–treated CHO cells (in contrast to p53-expressing mouse fibroblasts serving as a control). This demonstrates that p53 is functionally inactive in CHO-9 cells and the corresponding mutants we were working with. Inactivation of p53 in CHO cells has also been reported by another group (Lee et al., 1997). It indicates that p53 does not play a role in the induction of apoptosis in our cellular system. This is important to note because it has been shown that UV-C activates the CD95 receptor, which is p53 regulated (Muller et al., 1998). Furthermore, p53 has been shown to regulate the expression of proapoptotic Bax (Miyashita and Reed, 1995). We should note that we did not observe up-regulation of the CD95R and CD95L as well as Bax after UV-C irradiation in CHO cells undergoing apoptosis (this article; and our unpublished data).
There is increasing evidence that DNA damage-induced apoptosis is regulated via the mitochondrial apoptotic pathway, with Bcl-2 the most prominent protein involved. Bcl-2 is mainly localized in the outer mitochondrial membrane forming heterodimers with proapoptotic Bax, thus preventing homodimerization of Bax (reviewed in Antonsson and Martinou, 2000). Homodimerized Bax generates pores in the mitochondrial membrane, resulting in release of cytochrome c, apoptosis-inducing factor, and subsequent caspase-9 activation (Narita et al., 1998). Based on this model, down-regulation of Bcl-2 leads to induction of apoptosis, whereas up-regulation can prevent it. We show here that Bcl-2 is indeed down-regulated after UV-C irradiation in 27-1 and 43-3 cells. We also overexpressed Bcl-2 by transfection with an expression vector in NER-deficient 27-1 cells, thus protecting them from UV-C–induced apoptosis. In NER-proficient cells the decline in Bcl-2 level occurred only slightly and transiently and was observed only upon treatment with high doses of UV-C light. This is in contrast to NER-deficient 27-1 and 43-3B cells where Bcl-2 was not detectable anymore ∼72 h after irradiation. Bcl-2 decline was first observed ∼8 h after UV-C treatment and nearly paralleled the generation of DSBs. It therefore clearly preceded the induction of apoptosis. The results are in line with the view that Bcl-2 decline is a consequence of DSB formation upon UV-C treatment. The finding that ionizing irradiation as well as electroporation of PvuII, which both efficiently induce DSBs and apoptosis, are also able to induce down-regulation of Bcl-2 in p53-deficient cells (Lips and Kaina, 2001) supports this hypothesis.
It is well known that UV-C–induced DNA lesions block transcription, which has been suggested to act as a trigger of apoptosis (Ljungman and Zhang, 1996; Ljungman et al., 1999). To prove the involvement of transcription blockage in apoptosis in NER-deficient fibroblasts, we compared transcriptional activity of UV-C–treated 27-1 cells with the corresponding wild type. We observed a dramatic reduction of transcription in NER-deficient cells but not to the same extent in the wild type upon UV-C irradiation, which is obviously due to unrepaired DNA lesions. Therefore, transcription blockage must also be considered to be involved in the induction of apoptosis. This conclusion gains support from experiments reported herein in which transcription was inhibited by actinomycin D, which, at the same time, induced apoptosis. Actinomycin D also caused down-regulation of Bcl-2, indicating that transcriptional inhibition could be a second component activating the Bcl-2–dependent apoptotic pathway. We should note that we were unable to block UV-C–induced apoptosis completely in growth-arrested CHO cells. The residual amount of apoptotic cells observed after growth inhibition could thus be due to transcription blockage, which is not influenced by the state of proliferation of the cells.
The time sequence of events, as measured in NER-deficient cells, is summarized in Figure 9. It becomes obvious that the induction of DSBs and aberrations coincide with the Bcl-2 decline, which clearly precedes the appearance of apoptotic cells. The results therefore support a model suggesting that unrepaired UV-C–induced DNA lesions are converted into DSBs and aberrations by replication, leading to down-regulation of Bcl-2 and finally caspase activation. This model provides at the same time a simple explanation for the finding that UV-C–induced apoptosis in fibroblasts is, at least in part, replication dependent. Transcriptional inhibition may also come into play as a second (minor) component, causing activation of the mitochondrial apoptotic pathway upon the induction of DNA damage by UV-C light. This process appears to be independent of p53. It would be interesting to see whether the mechanism proposed is generally operating in fibroblasts and other cell types, notably p53-mutated tumor cells, and whether it competes with receptor-triggered apoptotic pathways.
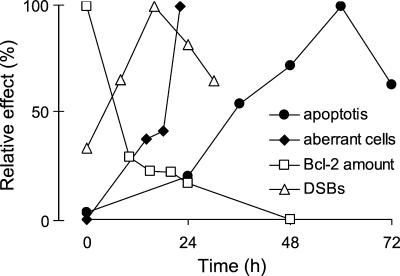
Figure 9.
Time course of effects induced by UV-C light in NER-deficient cells. Apoptosis, DSBs, chromosomal aberrations (percentage of aberrant cells) as well as Bcl-2 expression are shown at various time points after irradiation (10 J/m2) of 27-1 cells. Values are calculated as percentage relative to the maximum value.
ACKNOWLEDGMENTS
We gratefully acknowledge Dr. R. Wood for providing stably complemented 43-3B/ERRC1 cells, Dr. M. Oren for a gift of the mdm2-promotor reporter plasmid, and Dr. S. Dimmeler for Bcl-2 expression plasmid. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 519/B4 and Ka 724/8-4), by Stiftung Rheinland Pfalz for Innovation, and by a Ph.D. grant of the University of Mainz (to T.D.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–05-0225. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–05-0225.
REFERENCES
- Agarwal ML, Agarwal A, Taylor WR, Stark GR. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson B, Martinou JC. The Bcl-2 protein family. Exp Cell Res. 2000;256:50–57. doi: 10.1006/excr.2000.4839. [DOI] [PubMed] [Google Scholar]
- Aragane Y, Kulms D, Metze D, Wilkes G, Pöppelmann B, Luger T, Schwarz T. Ultraviolet light induces apoptosis via direct activation of CD95 (Fas/APO-1) independently of its ligand CD95L. J Cell Biol. 1998;140:171–182. doi: 10.1083/jcb.140.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brash DE, Ziegler A, Jonason AS, Simon JA, Kunala S, Leffell DJ. Sunlight and sunburn in human skin cancer: p53, apoptosis, and tumor promotion. J Invest Dermatol Symp Proc. 1996;2:136–142. [PubMed] [Google Scholar]
- Canman CE, Kastan MB. Signal transduction. Three paths to stress relief. Nature. 1996;384:213–214. doi: 10.1038/384213a0. [DOI] [PubMed] [Google Scholar]
- Chigancas V, Miyaji EN, Muotri AR, de Fatima Jacysyn J, Amarante-Mendes GP, Yasui A, Menck CF. Photorepair prevents ultraviolet-induced apoptosis in human cells expressing the marsupial photolyase gene. Cancer Res. 2000;60:2458–2463. [PubMed] [Google Scholar]
- De Santis LP, Garcia CL, Balajee AS, Calvo GTB, Bassi L. Transcription coupled repair deficiency results in increased chromosomal aberrations and apoptotic death in the UV61 cell line, the Chinese hamster homologue of Cockayne's syndrome B. Mutat Res. 2001;485:121–132. doi: 10.1016/s0921-8777(00)00065-3. [DOI] [PubMed] [Google Scholar]
- Dimmeler S, Breitschopf K, Haedeler J, Zeiher AM. Dephosphorylation targets Bcl-2 for ubiquitin-dependent degradation: a link between the apoptosome and the proteasome pathway. J Exp Med. 1999;189:1815–1822. doi: 10.1084/jem.189.11.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissi R, Lee SH. In vitro analysis of UV-damage-induced inhibition of replication. Biochem J. 1998;330:181–187. doi: 10.1042/bj3300181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford JM, Hanawalt PC. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. San Francisco: W H Freeman and Company; 1995. [Google Scholar]
- Fulda S, Strauss G, Meyer E, Debatin KM. Functional CD95 ligand and CD95 death-inducing signaling complex in activation-induced cell death and doxorubicin-induced apoptosis in leukemic T-cells. Blood. 2000;95:301–308. [PubMed] [Google Scholar]
- Grombacher T, Eichhorn U, Kaina B. p53 is involved in regulation of the DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) by DNA damaging agents. Oncogene. 1998;17:845–851. doi: 10.1038/sj.onc.1202000. [DOI] [PubMed] [Google Scholar]
- Guillouf C, Rosselli F, Krishnaraju K, Moustacchi E, Hoffman B, Liebermann DA. p53 involvement in control of G2 exit of the cell cycle: role in DNA damage-induced apoptosis. Oncogene. 1995;10:2263–2270. [PubMed] [Google Scholar]
- Haldar S, Negrini M, Monne M, Sabbioni S, Croce CM. Down-regulation of bcl-2 by p53 in breast cancer cells. Cancer Res. 1994;54:2095–2097. [PubMed] [Google Scholar]
- Hayashi T, Takao M, Tanaka K, Yasui A. ERCC1 mutations in UV-sensitive Chinese hamster ovary (CHO) cell lines. Mutat Res. 1998;407:269–276. doi: 10.1016/s0921-8777(98)00013-5. [DOI] [PubMed] [Google Scholar]
- Hickman MJ, Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc Natl Acad Sci. 1999;96:10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou YA, Chen FW. Quantitation of DNA fragmentation in apoptosis. Nucleic Acids Res. 1996;24:992–993. doi: 10.1093/nar/24.5.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaina B. The interrelationship between SCE induction, cell survival, mutagenesis, aberration formation and DNA synthesis inhibition in V79 cells treated with N-methyl-N-nitrosourea or N-methyl-N′-nitro-N-nitrosoguanidine. Mutat Res. 1985;142:49–54. doi: 10.1016/s0165-7992(85)80012-9. [DOI] [PubMed] [Google Scholar]
- Kaina B. Critical steps in alkylation-induced aberration formation. Mutat Res. 1998;404:119–124. doi: 10.1016/s0027-5107(98)00103-1. [DOI] [PubMed] [Google Scholar]
- Kaina B, Haas S, Kappes H. A general role for c-Fos in cellular protection against DNA-damaging carcinogens and cytostatic drugs. Cancer Res. 1997a;57:2721–2731. [PubMed] [Google Scholar]
- Kaina B, Ziouta A, Ochs K, Coquerelle T. Chromosomal instability, reproductive cell death and apoptosis induced by O6-methylguanine in Mex−, Mex+ and methylation-tolerant mismatch repair compromised cells: facts and models. Mutat Res. 1997b;381:227–241. doi: 10.1016/s0027-5107(97)00187-5. [DOI] [PubMed] [Google Scholar]
- Kaufmann WK, Cleaver JE. Mechanisms of inhibition of DNA replication by ultraviolet radiation in normal human and xeroderma pigmentosum fibroblasts. J Mol Biol. 1981;149:171–187. doi: 10.1016/0022-2836(81)90297-7. [DOI] [PubMed] [Google Scholar]
- Lackinger D, Kaina B. Primary mouse fibroblasts deficient for c-Fos, p53 or both proteins are hypersensitive to UV light and alkylating agent-induced chromosomal breakage and apoptosis. Mutat Res. 2000;457:113–123. doi: 10.1016/s0027-5107(00)00133-0. [DOI] [PubMed] [Google Scholar]
- Lee H, Larner JM, Hamlin JL. Cloning and characterization of Chinese hamster p53 cDNA. Gene. 1997;184:177–183. doi: 10.1016/s0378-1119(96)00592-6. [DOI] [PubMed] [Google Scholar]
- Liebermann DA, Hoffmann B, Steinmann RA. Molecular controls of growth arrest and apoptosis: p53-dependent and independent pathways. Oncogene. 1995;11:199–210. [PubMed] [Google Scholar]
- Lips J, Kaina B. DNA double-strand breaks trigger apoptosis in p53 deficient fibroblasts. Carcinogenesis. 2001;22:579–585. doi: 10.1093/carcin/22.4.579. [DOI] [PubMed] [Google Scholar]
- Ljungman M, Zhang F. Blockage of RNA polymerase as a possible trigger for UV light-induced apoptosis. Oncogene. 1996;13:823–831. [PubMed] [Google Scholar]
- Ljungman M, Zhang F, Chen F, Rainbow AJ, McKay BC. Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene. 1999;18:583–592. doi: 10.1038/sj.onc.1202356. [DOI] [PubMed] [Google Scholar]
- McGregor WG. DNA repair, DNA replication, and UV mutagenesis. J Investig Dermatol Symp Proc. 1999;4:1–5. doi: 10.1038/sj.jidsp.5640172. [DOI] [PubMed] [Google Scholar]
- Meikrantz W, Bergom MA, Memisoglu A, Samson L. O6-Alkylguanine DNA lesions trigger apoptosis. Carcinogenesis. 1998;19:369–372. doi: 10.1093/carcin/19.2.369. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Harigai M, Hanada M, Reed JC. Identification of a p53-dependent negative response element in the bcl-2 gene. Cancer Res. 1994;54:3131–3135. [PubMed] [Google Scholar]
- Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- Muller M, Wilder S, Bannasch, Israeli D, Lehlbach K, Li-Weber M, Friedman SL, Galle PR, Stremmel W, Oren M, Krammer PH. p53 activates the CD95 (Apo-1/Fas) gene in response to DNA damage by anticancer drugs. J Exp Med. 1998;188:2033–2045. doi: 10.1084/jem.188.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Shimizu S, Ito T, Chittenden T, Lutz RJ, Matsuda H, Tsujimoto Y. Bax interacts with the permeability transition pore to induce permeability transition and cytochrome c release in isolated mitochondria. Proc Natl Acad Sci USA. 1998;95:14681–14686. doi: 10.1073/pnas.95.25.14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs K, Kaina B. Apoptosis induced by DNA damage O6-methylguanine is Bcl-2 and caspase-9/-3 regulated and p53 and Fas/caspase-8 independent. Cancer Res. 2000;60:5815–5824. [PubMed] [Google Scholar]
- Orren KD, Petersen LN, Bohr VA. Persistent DNA damage inhibits S-phase and G2 progression, and results in apoptosis. Mol Biol Cell. 1997;8:1129–1142. doi: 10.1091/mbc.8.6.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter RB, Howard R. The HeLa DNA-synthesis test as a rapid screen for mutagenic carcinogens. Mutat Res. 1982;92:427–437. doi: 10.1016/0027-5107(82)90241-x. [DOI] [PubMed] [Google Scholar]
- Pellegata NS, Antoniono RJ, Redpath L, Stanbridge EJ. DNA damage and p53-mediated cell cycle arrest: a reevaluation. Proc Natl Acad Sci USA. 1996;93:15209–15214. doi: 10.1073/pnas.93.26.15209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponten F, Berne B, Ren ZP, Nister M, Ponten J. Ultraviolet light induces expression of p53 and p21 in human skin: effect of sunscreen and constitutive p21 expression in skin appendages. J Invest Dermatol. 1995;105:402–406. doi: 10.1111/1523-1747.ep12321071. [DOI] [PubMed] [Google Scholar]
- Poon RYC, Jiang W, Toyoshima H, Hunter T. Cyclin-dependent kinases are inactivated by a combination of p21 and Thr-14/Tyr-15 phosphorylation after UV-induced DNA damage. J Biol Chem. 1996;271:13283–13291. doi: 10.1074/jbc.271.22.13283. [DOI] [PubMed] [Google Scholar]
- Rafferty JA, Clarke AR, Sellappan D, Koref MS, Frayling IM, Margison GP. Induction of murine O-6-alkylguanine-DNA-alkyltransferase in response to ionizing radiation is p53 gene dose dependent. Oncogene. 1996;12:693–697. [PubMed] [Google Scholar]
- Rehemtulla A, Hamilton C, Chinnaiyan A, Dixit V. Ultraviolet radiation-induced apoptosis is mediated by activation of CD95 (Fas/APO-1) J Biol Chem. 1997;272:25783–25786. doi: 10.1074/jbc.272.41.25783. [DOI] [PubMed] [Google Scholar]
- Reinke V, Lozano G. Differential activation of p53 targets in cells treated with ultraviolet radiation that undergo both apoptosis and growth arrest. Radiat Res. 1997;148:115–122. [PubMed] [Google Scholar]
- Schwarz T. UV light affects cell membrane and cytoplasmic targets. J Photochem Photobiol. 1998;44:91–96. doi: 10.1016/S1011-1344(98)00126-2. [DOI] [PubMed] [Google Scholar]
- Smith ML, Ford JM, Hollander MC, Bortnick RA, Amundson SA, Seo YR, Deng CX, Hanawalt PC, Fornace AJ., Jr p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol Cell Biol. 2000;20:3705–3714. doi: 10.1128/mcb.20.10.3705-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanini M, Mondello C, Tessera L, Capuano Y, Guerra BR, Nuzzo F. Sensitivity to DNA-damaging agents and mutation induction by UV light in UV-sensitive CHO cells. Mutat Res. 1986;174:155–159. doi: 10.1016/0165-7992(86)90108-9. [DOI] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Jacks T, Cory S. DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell. 1994;79:329–339. doi: 10.1016/0092-8674(94)90201-1. [DOI] [PubMed] [Google Scholar]
- Tominaga Y, Tsuzuki T, Shiraishi T, Shiraishi A, Kawate H, Sekiguchi M. Alkylation-induced apoptosis in embryonic stem cells in which the gene for DNA repair, methyltransferase, had been disrupted by gene targeting. Carcinogenesis. 1997;18:889–896. doi: 10.1093/carcin/18.5.889. [DOI] [PubMed] [Google Scholar]
- Weber CA, Salazar EP, Stewart SA, Thompson LH. Molecular cloning and biological characterization of a human gene, ERCC2, that corrects the nucleotide excision repair defect in CHO UV5 cells. Mol Cell Biol. 1988;8:1137–1146. doi: 10.1128/mcb.8.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda G, van Ham RCA, Masurel R, Westerveld A, Odijk H, De Wit J, Bootsma D, van der Eb AJ, Hoeijmakers JHJ. Molecular cloning and biological characterization of the human excision repair gene ERCC-3. Mol Cell Biol. 1990;10:2570–2581. doi: 10.1128/mcb.10.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerveld A, Hoeijmakers JHJ, van Duin M, De Wit J, Odijk H, Pastink A, Wood D, Bootsma D. Molecular cloning of a human DNA repair gene. Nature. 1984;310:425–429. doi: 10.1038/310425a0. [DOI] [PubMed] [Google Scholar]
- Wood RD, Burki HJ. Repair capability and the cellular age response for killing and mutation induction after UV. Mutat Res. 1982;95:505–514. doi: 10.1016/0027-5107(82)90281-0. [DOI] [PubMed] [Google Scholar]
- Zhou J, Ahn J, Wilson SH, Prices C. A role for p53 in base excision repair. EMBO J. 2001;20:914–923. doi: 10.1093/emboj/20.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]