Abstract
Blast disease caused by fungal pathogen Pyricularia oryzae is a major threat to rice productivity worldwide. The rice-blast pathogen can infect both leaves and panicle neck nodes. Nearly, 118 genes for resistance to leaf blast have been identified and 25 of these have been molecularly characterized. A great majority of these genes encode nucleotide-binding site–leucine-rich repeat (NBS–LRR) proteins and are organized into clusters as allelic or tightly linked genes. Compared to ever expanding list of leaf-blast-resistance genes, a few major genes mediating protection to neck blast have been identified. A great majority of the genetic studies conducted with the genotypes differing in the degree of susceptibility/resistance to neck blast have suggested quantitative inheritance for the trait. Several reports on co-localization of gene/QTLs for leaf- and neck-blast resistance in rice genome have suggested the existence of common genes for resistance to both phases of the disease albeit inconsistencies in the genomic positions leaf- and neck-blast-resistance genes in some instances have presented the contrasting scenario. There is a strong evidence to suggest that developmentally regulated expression of many blast-resistance genes is a key determinant deciding their effectiveness against leaf or neck blast. Testing of currently characterized leaf-blast-resistance genes for their reaction to neck blast is required to expand the existing repertoire resistance genes against neck blast. Current developments in the understanding of molecular basis of host–pathogen interactions in rice-blast pathosystem offer novel possibilities for achieving durable resistance to blast through exploitation of natural or genetically engineered loss-of-function alleles of host susceptibility genes.
Keywords: Pyricularia oryzae, Oryza sativa, Leaf blast, Neck blast, QTLs, Mapping
Introduction
Rice (Oryza sativa L.) is one of the most important food crops, feeding more than 50% of the world’s population. Rice is affected by several biotic and abiotic stresses amongst which blast disease caused by the fungal pathogen Pyricularia oryzae Cavara (Telomorph, Magnaporthe oryzae) is a major biotic stress that threatens rice production worldwide (Ashkani et al. 2015; Wang et al. 2015). The disease causes yield losses ranging from 10 to 30% each year which translates into a loss of about 157 million tonnes of rice worldwide (Talbot 2003). The disease has two commonly recognized phases: leaf blast and neck blast. Leaf blast occurs most often during the plant’s vegetative stage causing characteristic spindle shaped lesions on leaf blade and necrotic lesions at leaf collar. The neck blast (a near synonym of panicle blast), which is the most destructive form of the disease, occurs during the reproductive stage and is characterized by fungal infection at the panicle base and plant nodes. The fungal infection at the panicle base restricts the flow of photosynthates to the developing grains resulting in chaffy grains or empty panicles. The yield reduction inflicted by neck-blast infection is twice as severe as leaf blast with losses approaching up to 70% of the anticipated yield under epidemic conditions (Puri et al. 2009). Although wide range of effective fungicides are available to control the disease, their use is not favoured by the farmers due to cost, and associated environmental and health issues. The development and deployment of rice varieties fortified with a high level of resistance to both leaf and panicle blast is one of the most economical ways to manage the disease. Identification of effective sources of resistance genes and their precise mapping in rice genome is one of the basic requirements for effective manipulations of these genes in resistance breeding programmes. In the following sections, we have presented a detailed update on identification, mapping and molecular characterization of leaf, and panicle blast-resistance genes in rice.
Genetics and mapping of genes governing resistance to leaf-blast disease
The discovery and utilization of disease-resistance genes for breeding broad-spectrum-resistant genotypes is the most preferred strategy to manage the blast disease. Genetic investigations on blast resistance started in the early 1920s when Sasaki (1922) for the first time observed that rice varieties differed in their response to different isolates of rice-blast fungus P. oryzae. Extensive genetic studies that followed led to the identification of first leaf-blast-resistance gene Pi-a from japonica rice variety, Aichi Asahi (Kiyosawa 1967). First attempt to map a blast-resistance gene was made by Yu et al. (1991), who mapped two blast-resistance genes, Pi-2(t) and Pi-4(t) using Restriction Length Fragment Polymorphism (RFLP) analysis of a set of near isogenic lines. Since the development of first genetic map of rice based on RFLP markers in 1988 (McCouch et al. 1988), a great variety of genetic markers, including simple sequence repeats (SSRs), single-nucleotide polymorphisms (SNPs), and small insertions/deletions (InDels), amplified fragment length polymorphisms (AFLPs), random amplified polymorphic DNAs (RAPDs), cleaved amplified polymorphic sequences (CAPS), and RFLPs have been used for genetic mapping in rice culminating in development of high density linkage maps of rice (Inoue et al. 1994; Chen et al. 1997; Harushima et al. 1998; Monna et al. 1994; Mackill et al. 1996; Cho et al. 2000; McCouch et al. 2002). In the current decade, the accessibility of complete genome sequences of rice subspecies indica and japonica (http://rgp.dna.affrc.go.jp; http://www.genomics.org.cn) has enabled the rice researchers to exploit DNA polymorphisms such as SNPs and InDels for the fine scale mapping of the targeted genes (Lei et al. 2013; Wang et al. 2016b; Liu et al. 2013; Hu et al. 2018). These developments have helped mapping a number of blast-resistance genes that have been precisely localized on the rice chromosomes. In rice, nearly, 118 genes for resistance to leaf-blast disease have been identified through genetic analyses of rice varieties belonging to both japonica and indica subspecies and 25 of them have been cloned and characterized until date (Tables 1, 2). Of the total identified leaf-blast-resistance genes, a great majority is located on chromosomes 6, 11, and 12 which harbour 21, 27, and 27 leaf-blast-resistance genes, respectively. Chromosomes 3 and 7 contain minimum number of blast-resistance genes with only one gene in each. Classical and molecular genetic analyses have revealed that a great majority of blast-resistance genes are distributed into clusters consisting of allelic or tightly linked genes. At least three major clusters of blast-resistance genes have been detected in rice on chromosomes 6, 11, and 12 (Zhou et al. 2006; Zhai et al. 2011; Yang et al. 2009). For example, ten blast-resistance genes, Pi8, Pi9, Pi26(t), Pi27(t), Pi40, Piz, Pi-2, Piz-t, Pigm(t), and Pi59(t), have been mapped at the Piz locus on chromosome 6 (Koide et al. 2013) and at least 9 resistance genes have mapped to R-gene cluster on the telomeric end of rice chromosome 11 and seven of them viz., Pi-k, Pi-ks, Pi-kp, Pi-km, Pi-kh, Pi-kl, and Pi-1 have been shown to be the alleles of Pi-k locus (Zhai et al. 2011; Hua et al. 2012; Singh et al. 2015). Similarly, at least nine blast-resistance specificities, Pi12, Pita, Pita2, Pi39, Pi42, Pi24, Pi20, PiGD3, and Pi157 have been mapped to R-gene cluster at the centeromeric region of rice chromosome 12 (Bryan et al. 2000; Zhuang et al. 2002; Liu et al. 2004, 2007b; Li et al. 2008; Kumar et al. 2010). A chromosomal map depicting distribution of leaf-blast-resistance genes across different rice chromosomes is shown in Fig. 1.
Table 1.
List of blast-resistance genes mapped through molecular markers
| Sr. no. | Gene | Donor | Genomic position (Mb) | Chromosome | Linked marker | References |
|---|---|---|---|---|---|---|
| 1 | Pit | K-59, Tjahaja, K-59 | 2.27 | 1 | RFLP, SNP | Kaji et al. (1997) and Hayashi et al. (2006) |
| 2 | Pi27(t) | Q14 | 5.55 | 1 | SSR | Zhu et al. (2004) |
| 3 | Pi-h2(t) | HR4 | 7.90 | 1 | SSR | Xiao et al. (2015) |
| 4 | Pi-tp(t) | Tetep | 25.13 | 1 | SSR | Barman et al. (2004) |
| 5 | Pi35(t) | Hokkai 188 | 32.1 | 1 | SSR | Nguyen et al. (2006) |
| 6 | Pi64 | Yangmaogu | 32.31 | 1 | SSR, Indel | Ma et al. (2015) |
| 7 | Pi 37(t) | St. No. 1 | 33.1 | 1 | SSR | Chen et al. (2005) |
| 8 | Pi-sh | Akihikari | 33.3 | 1 | SSR | Fukuta (2004) |
| 9 | Pir2-3(t) | IR64 | – | 2 | SSR | Dwinita et al. (2008) |
| 10 | Pirf2-1(t) | O. rufipogan | – | 2 | SSR | Dwinita et al. (2008) |
| 11 | Pi-Da(t) | Dacca 6 | 2.21 | 2 | SSR | Shi et al. (2012) |
| 12 | Pig(t) | Guangchangzhan | 34.34 | 2 | SSR | Zhou et al. (2004) |
| 13 | Pi-25(t) | IR64 | 34.36 | 2 | QTL | Sallaud et al. (2003) |
| 14 | Pi-tq5 | Teqing | 34.61 | 2 | RFLP | Tabien et al. (2000) |
| 15 | Pi-14(t) | Maowangu | 34.94 | 2 | RFLP, Isozyme | Pan et al. (1998) and Zhou et al. (2004) |
| 16 | Pi-16(t) | AUS373 | 34.94 | 2 | RFLP, Isozyme | Pan et al. (1999) and Zhou et al. (2004) |
| 17 | Pi-d1(t) | Digu | 34.94 | 2 | SSR, RFLP | Chen et al. (2004) |
| 18 | Pi-y2(t) | Yanxian No. 1 | 35.03 | 2 | SSR | Lei et al. (2005) |
| 19 | Pi-y1(t) | Yanxian No. 1 | 35.03 | 2 | SSR | Lei et al. (2005) |
| 20 | Pi-b | Tohoku, Koshiihikari | 35.10 | 2 | SNP | Hayashi et al. (2006) |
| 21 | Pi66(t) | AS20-1 | 26.78 | 3 | SSR | Liang et al. (2016) |
| 22 | Pikur 1 | Kuroka | – | 4 | Isozyme | Fukuoka et al. (2009) |
| 23 | pi-21 | Owarihatamochi | 19.81 | 4 | RFLP, SSR | Fukuoka and Okuno (2001) |
| 24 | Pias(t) | Asominori | 31.26 | 4 | SSR, CAPS | Endo et al. (2012) |
| 25 | Pi-45(t) | Moroberekan | 31.49 | 4 | SSR | Kim et al. (2011) |
| 26 | Pikahei-1(t) | Kahei | 31.67 | 4 | SSR, SNP | Xu et al. (2008a) |
| 27 | Pi-39(t) | Chubu 111 | 32.68 | 4 | SSR | Terashima et al. (2008) |
| 28 | Pi26(t) | IR64 | 2.78 | 5 | RFLP,RAPD, | Sallaud et al. (2003) |
| 29 | Pi23 | Suweon 365 | 10.75 | 5 | RFLP, SSR | Ahn et al. (1997) |
| 30 | Pi-10(t) | Tongil | 14.52 | 5 | RAPD | Naqvi et al. (1995) |
| 31 | Pi22 | Suweon 365 | 4.89 | 6 | RFLP | Ahn et al. (1997) |
| 32 | Pi27(t) | IR64 | 6.92 | 6 | RFLP | Sallaud et al. (2003) |
| 33 | Pi-40(t) | IR65482 | 9.86 | 6 | STS, SSR | Jeung et al. (2007) |
| 34 | Pi2-1 | Tianjingyeshengdao | 10.08 | 6 | SSR, SFP | Wang et al. (2012) |
| 35 | Pi2-2 | Jefferson | 10.20 | 6 | SSR | Jiang et al. (2012) |
| 36 | Pigm(t) | Gumei 4 | 10.36 | 6 | CAPS, InDel | Deng et al. (2006) |
| 37 | Pi-9(t) | IR31917 | 10.38 | 6 | STS | Qu et al. (2006) |
| 38 | Pi51(t) | D69 | 10.38 | 6 | InDel, SSR | Xiao et al. (2012) |
| 39 | Pi2 | 5173, C101A51 | 10.39 | 6 | SSR, STS, RFLP | Jiang and Wang (2002) and Zhou et al. (2006) |
| 40 | Piz | Fukinishiki | 10.39 | 6 | STS | Zhou et al. (2006) |
| 41 | Piz-t | Toride No. 1 | 10.39 | 6 | STS | Zhou et al. (2006) |
| 42 | Pi50(t) | EBZ | 10.41 | 6 | SSR, CAPS | Zhu et al. (2012) |
| 43 | Pi59(t) | Hoaru | 10.82 | 6 | SSR | Koide et al. (2013) |
| 44 | Pi26(t) | Gumei 2 | 11.06 | 6 | RFLP, SSR | Wu et al. (2005) |
| 45 | Pi8 | Kasalath | 11.36 | 6 | Isozyme markers, RFLP | Pan et al. (1996a) and Takehisa et al. (2009) |
| 46 | Pi-25(t) | Gumei 2 | 12.33 | 6 | RFLP, RGA, SSR | Wu et al. (2005) |
| 47 | Pid3 | Digu | 13.05 | 6 | STS | Shang et al. (2009) |
| 48 | Pi-13 | Kasalath | 15.83 | 6 | SSR | Ebitani et al. (2011) |
| 49 | Pi-dt(2) | Digu | 17.16 | 6 | SSR, RGA | Chen et al. (2004) |
| 50 | Pid2 | Digu | 17.16 | 6 | CAPS | Chen et al. (2006) |
| 51 | Pi-tq1 | Teqing | 29.02 | 6 | RFLP | Tabien et al. (2000) |
| 52 | Pi-17(t) | DJ 123 | 22.25 | 7 | Isozyme marker | Pan et al. (1996b) |
| 53 | Pi-11(t) | Zhai-Ye-Quing | 13.93 | 8 | RFLP, RAPD | Causse et al.(1994) |
| 54 | Pi-33 | IR64, Bala | 7.56 | 8 | SSR, RFLP | Berruyer et al. (2003) |
| 55 | Pi-29(t) | IR64 | 13.93 | 8 | RFLP, RAPD, Isozyme | Sallaud et al. (2003) |
| 56 | PiGD-1(t) | Sanhuangzhan 2 | 16.37 | 8 | SSR, RFLP, RGA | Liu et al. (2004) |
| 57 | Pi-36(t) | Q61 | 2.87 | 8 | SSR, CRG | Liu et al. (2005) |
| 58 | pi55(t) | Yuejingsimiao 2 | 25.58 | 8 | SSR, STS | Xiu-Ying et al. (2012) |
| 59 | Pi-5(t) | RIL249 | 9.77 | 9 | AFLP, RFLP, CAPS | Jeon et al. (2003) |
| 60 | Pi15 | GA25 | 9.61 | 9 | SSR, CRG | Lin et al. (2007b) |
| 61 | Pi56(t) | SHZ-2 | 9.77 | 9 | SSR, CRG, SNP | Liu et al. (2013) |
| 62 | Pihk2 | Heikezijing | 10.17 | 9 | SSR, ILP, InDel | He et al. (2017) |
| 63 | PiGD-2(t) | Sanhuangzhan 2 | – | 10 | SSR, RFLP, RGA | Liu et al. (2004) |
| 64 | Pi28(t) | Azucena | 21.04 | 10 | RFLP, RAPD | Sallaud et al. (2003) |
| 65 | Pi-30(t) | IR64 | 4.41 | 11 | RFLP, RAPD, Isozyme | Sallaud et al. (2003) |
| 66 | Pi-a | Aichi Asahi | 6.49 | 11 | SSR, Indel | Zeng et al. (2011) |
| 67 | Pi60(t) | 93-11 | 6.62 | 11 | SSR, InDel | Lei et al. (2013) |
| 68 | Pi-CO39(t) | Co39 | 6.66 | 11 | SSR, RFLP | Chauhan et al. (2002) |
| 69 | Pi-7(t) | Moroberekan | 18.64 | 11 | RFLP | Wang et al. (1994) |
| 70 | Pi-34 | Chubu-32 | 19.96 | 11 | SSR | Zenbayashi et al. (2007) |
| 71 | Pi-38 | Tadukan | 22.48 | 11 | SSR, AFLP | Gowda et al. (2006) |
| 72 | Pik-h | IRBLkh-K3 | 24.99 | 11 | SNP | Xu et al. (2008b) |
| 73 | Pi54 | Tetep | 25.26 | 11 | SSR | Sharma et al. (2005) |
| 74 | Pik-s | Shin 2 | 27.31 | 11 | SSR | Fjellstrom et al. (2004) |
| 75 | Pi-jnw1 | Jiangnanwan | 27.36 | 11 | SSR, InDel | Wang et al. (2016b) |
| 76 | Pi-hk1 | Heikezijing | 27.66 | 11 | SSR | Wu et al. (2013) |
| 77 | Pi43(t) | Zhe733 | 27.67 | 11 | SSR | Lee et al. (2009a) |
| 78 | Pi-47 | Xiangzi 3150 | 27.67 | 11 | SSR | Huang et al. (2011) |
| 79 | Pik-l | Liziangxintuanheigu | 27.69 | 11 | SSR, STS, CAPS | Singh et al. (2015) |
| 80 | Pi46(t) | H4 | 27.74 | 11 | SSR, InDel | Xiao et al. (2011) |
| 81 | Pi-1(t) | Apura, C101LAC | 28.00 | 11 | STS, RFLP, SSR, CAPS | Parco (1995), Yu et al. (1996), Fuentes et al. (2008) and Hua et al. (2012) |
| 82 | Pik-m | Tohoku IL4, Tsuyuake | 28.00 | 11 | RFLP, SSR | Kaji and Ogawa (1996) and Li et al. (2007) |
| 83 | Pik-e | Xiangzao 143 | 28.00 | 11 | SSR, InDel | Chen et al. (2015) |
| 84 | Pi-k | Kusabue, Kanto 51 | 28.01 | 11 | RFLP, InDel, SNP | Hayasaka et al. (1996) and Hayashi et al. (2006) |
| 85 | Pik-p | K60 | 28.05 | 11 | SSR, CAPS | Wang et al. (2009) |
| 86 | Pi-h1(t) | HR4 | 28.11 | 11 | SSR, InDel | Xiao et al. (2015) |
| 87 | Pi65(t) | Gangyu 129 | 28.22 | 11 | SNP, InDel | Zheng et al. (2016) |
| 88 | Pi49 | Mowanggu | 28.80 | 11 | SSR | Sun et al. (2013) |
| 89 | Pi-44(t) | Moroberekan | 28.93 | 11 | RFLP, STS, AFLP | Chen et al. (1999) |
| 90 | Pi18 | Suweon365 | 28.93 | 11 | RFLP | Ahn et al. (2000) |
| 91 | Pi-lm2 | Lemont | 28.93 | 11 | RFLP | Tabien et al. (2000) |
| 92 | Pi-6(t) | Apura | 7.73 | 12 | RFLP | Yu et al. (1996) |
| 93 | Pi12 | Hong Jiao Zhan | 7.73 | 12 | RFLP | Zhuang et al. (1998) |
| 94 | Pi62(t) | Yashiromochi | 7.73 | 12 | RAPD, RFLP | Wu et al. (1996) |
| 95 | Pi12 | Hong Jiao Zhan | 7.73 | 12 | RFLP | Zhuang et al. (1998) |
| 96 | Pi62(t) | Yashiromochi | 7.73 | 12 | RAPD, RFLP | Wu et al. (1996) |
| 97 | Pi-tq6 | Teqing | 7.73 | 12 | RFLP | Tabien et al. (2000) |
| 98 | Pitb | Zixuan | 9.37 | 12 | SSR, InDel | Sun et al. (2016) |
| 99 | Pita3(t) | IRBLta2-Re | 9.89 | 12 | SSR | Chen et al. (2014) |
| 100 | Pi61(t) | 93-11 | 9.98 | 12 | InDel, SSR | Lei et al. (2013) |
| 101 | Pi58(t) | Haoru | 10.42 | 12 | SSR | Koide et al. (2013) |
| 102 | Pita | Yashiromochi | 10.60 | 12 | RFLP, RAPD, SNP | Rybka et al. (1997) and Hayashi et al. (2006) |
| 103 | Pita-2 | Yashiromochi, Pi No. 4 | 10.60 | 12 | RFLP, RAPD, SNP | Rybka et al. (1997) and Hayashi et al. (2006) |
| 104 | Pi-24(t) | Zhong 156 | 10.60 | 12 | RFLP, RAPD, RGA | Zhuang et al. (2002) |
| 105 | Pi-39 | Q-15 | 10.61 | 12 | SSR | Liu et al. (2007b) |
| 106 | Pi-42(t) | DHR9 | 10.62 | 12 | RAPD, SSR, STS | Kumar et al. (2010) |
| 107 | Pi-19(t) | IRBL19-A | 10.73 | 12 | SSR | Koide et al. (2011) |
| 108 | Pi57(t) | IL-E1454 | 10.80 | 12 | SSR, STS | Dong et al. (2017) |
| 109 | Pi-31(t) | IR64 | 11.93 | 12 | RFLP, RAPD, | Sallaud et al. (2003) |
| 110 | Pi-48 | Xiangzi 3150 | 11.95 | 12 | SSR | Huang et al. (2011) |
| 111 | Pi51(t) | Tianjingyeshengdao | 11.95 | 12 | SSR, SFP | Wang et al. (2012) |
| 112 | Pi67 | Tetep | 12.09 | 12 | SSR | Joshi et al. (2019) |
| 113 | Pi157 | Moroberekan | 12.37 | 12 | RFLP | Naqvi and Chattoo (1996) |
| 114 | Pi-20(t) | IR64 | 12.95 | 12 | SSR | Li et al. (2008) |
| 115 | Pih3(t) | HR4 | 12.95 | 12 | SSR | Xiao et al. (2015) |
| 116 | PiGD-3(t) | Sanhuangzhan 2 | 14.45 | 12 | SSR, RFLP, RGA | Liu et al. (2004) |
| 117 | Pi-41 | 93-11 | 16.74 | 12 | SSR, STS | Yang et al. (2009) |
| 118 | Pi-32(t) | IR64 | 21.24 | 12 | RFLP, RAPD | Sallaud et al. (2003) |
Table 2.
List of leaf-blast-resistance genes cloned and characterized from rice
| Sr no. | Gene designation | Chromosome | Cloning strategy | Protein type | References |
|---|---|---|---|---|---|
| 1 | Pi37 | 1 | MB | NBS–LRR | Lin et al. (2007a) |
| 2 | Pit | 1 | MB | CC–NBS–LRR | Hayashi and Yoshida. (2009) |
| 3 | Pi-sh | 1 | Mutant screening | CC–NBS–LRR | Takahashi et al. (2010) |
| 4 | Pi64 | 1 | MB | CC–NBS–LRR | Ma et al. (2015) |
| 5 | Pi-b | 2 | MB | NBS–LRR | Wang et al. (1999) |
| 6 | pi21 | 4 | MB | Proline-rich heavy-metal–binding protein | Fukuoka et al. (2009) |
| 7 | Pi63 | 4 | MB | CC–NBS–LRR | Xu et al. (2014) |
| 8 | Pid-2 | 6 | MB | Lectin receptor | Chen et al. (2006) |
| 9 | Pi9 | 6 | MB | NBS–LRR | Qu et al. (2006) |
| 10 | Pi-2 | 6 | MB | NBS–LRR | Zhou et al. (2006) |
| 11 | Piz-t | 6 | MB | NBS–LRR | Zhou et al. (2006) |
| 12 | Pid3 | 6 | In silico analysis | NBS–LRR | Shang et al. (2009) |
| 13 | Pigm | 6 | MB | NBS–LRR | Deng et al. (2009) |
| 14 | Pi25 | 6 | MB | CC–NBS–LRR | Chen et al. (2011) |
| 15 | Pi36 | 8 | MB | CC–NBS–LRR | Liu et al. (2007a) |
| 16 | Pi5 | 9 | MB | CC–NBS–LRR | Lee et al. (2009b) |
| 17 | Pi-54/Pi-kh | 11 | MB | NBS–LRR | Sharma et al. (2005) |
| 18 | Pik-m | 11 | MB | NBS–LRR | Ashikawa et al. (2008) |
| 19 | Pi-k | 11 | MB | CC–NBS–LRR | Zhai et al. (2011) |
| 20 | Pik-p | 11 | MB | CC–NBS–LRR | Yuan et al. (2011) |
| 21 | Pik-e | 11 | MB | CC–NBS–LRR | Chen et al. (2015) |
| 22 | Pi-a | 11 | MB and mutant screening | CC–NBS–LRR | Okuyama et al. (2011) |
| 23 | Pi1 | 11 | MB | CC–NBS–LRR | Hua et al. (2012) |
| 24 | Pita | 12 | MB | NBS–LRR | Bryan et al. (2000) |
| 25 | Pitr | 12 | MB | Putative E3 ligase | Zhao et al. (2018) |
MB map based cloning
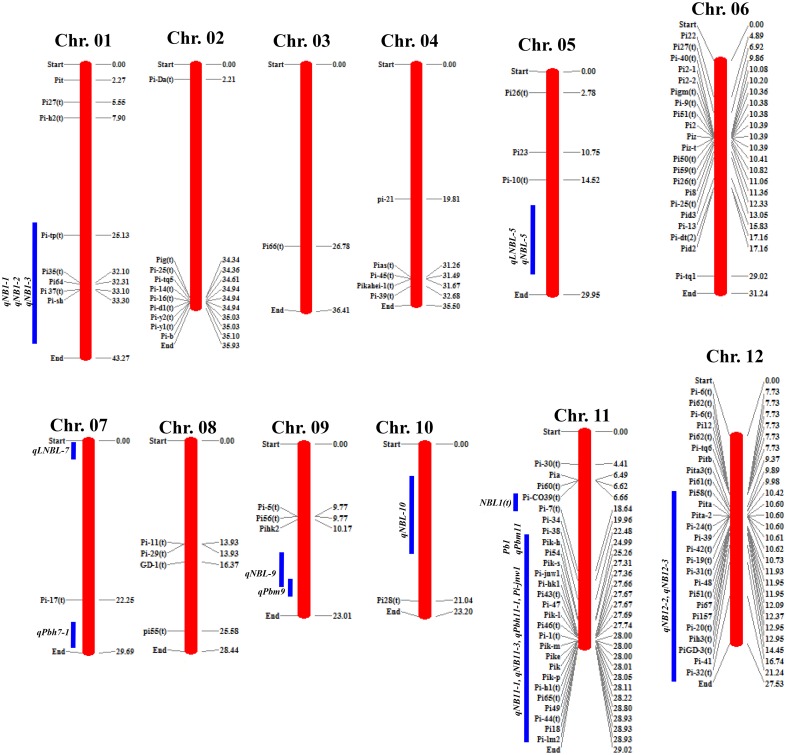
Fig. 1.
Chromosomal locations of leaf-blast R genes and quantitative trait loci (QTLs) for neck-blast resistance in rice. The chromosomal locations for R genes and QTLs were deduced by projecting the sequences of closely linked/flanking markers on the genome sequence of cv. Nipponbare released by International Rice Genome Sequencing Project (http://rapdb.dna.affrc.go.jp). The physical position of each gene/QTL in million base pair (Mb) units is shown on right side of each chromosome. The blue bars indicate the estimated location of QTLs for neck-blast resistance as deduced from their flanking markers
Amongst the molecularly characterized major leaf-blast R genes, 22, namely, Pi37, Pit, Pi-sh, Pi64, Pi-b, Pi63, Pi9, Pi-2, Piz-t, Pid3, Pigm, Pi25, Pi36, Pi5, Pi-54, Pik-m, Pi-k, Pik-p, Pik-e, Pi-a, Pi1, and Pita, belong to the largest class of plant R genes that encode proteins with nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains (Lin et al. 2007a; Hayashi and Yoshida 2009; Takahashi et al. 2010; Ma et al. 2015; Wang et al. 1999; Xu et al. 2014; Qu et al. 2006; Zhou et al. 2006; Shang et al. 2009; Deng et al. 2009; Chen et al. 2011; Liu et al. 2007a; Lee et al. 2009b; Sharma et al. 2005; Ashikawa et al. 2008; Zhai et al. 2011; Yuan et al. 2011; Chen et al. 2015; Okuyama et al. 2011; Hua et al. 2012; Bryan et al. 2000), whereas one, Pid2, encodes serine–threonine–kinase membrane spanning protein (Chen et al. 2006). pi21, the only field blast-resistance gene cloned until date, encodes a protein with heavy-metal binding and proline-rich domains (Fukuoka et al. 2009) and Pitr, an atypical resistance gene, encodes a putative E3 ligase with four Armadillo repeats (Zhao et al. 2018).
Molecular mapping and cloning of blast-resistance genes have provided several opportunities to augment the conventional disease-resistance breeding by enabling efficient selection of resistance genes in breeding programmes (Singh et al. 2012), pyramiding of resistance genes for achieving broad spectrum and durable resistance (Hittalmani et al. 2000), cataloguing of gene bank collections for resistance genes (Vasudevan et al. 2014; Yadav et al. 2017), and unraveling allelic diversity for resistance genes in germplasm collections through sequencing-based allele mining (Vasudevan et al. 2015; Lv et al. 2017). Genetic mapping and molecular cloning of different blast-resistance genes have provided an array of gene-linked, gene based, or functional markers for the efficient selection of resistance genes in breeding programmes (Jia et al. 2002; Shang et al. 2009; Hayashi et al. 2010). Structural comparisons of the cloned members of multi-allelic resistance loci have also provided information on the DNA regions within these genes responsible for their distinct-resistance specificities and offered the possibility of using these as the basis for identification of different resistance alleles in breeding programmes (Zhou et al. 2006; Zhai et al. 2011; Hua et al. 2012).
Marker-assisted backcross breeding has been successfully used to transfer single or a combination of various leaf-blast-resistance genes such as Pi1, Piz-5, and Pita into rice variety Co39 (Hittalmani et al. 2000), Pi2, Pi9, Pi1, Pi54, Pita, Pi-b, and Pi5 genes into varieties such as Pusa Basmati 1 (Khanna et al. 2015), Pi2, and Pi54 into Pusa Basmati 1121, Pusa Basmati 6, and parental lines of Basmati hybrids (Singh et al. 2012; Ellur et al. 2016). The detailed information on the applications of molecular markers in blast-resistance breeding has been recently reviewed by Ashkani et al. (2015) and Srivastva et al. (2017).
Genetics and mapping of genes governing resistance to neck-blast disease
Of the two known phases of rice blast, the neck blast is economically more significant causing yield losses up to 70–100% under epidemic conditions (Puri et al. 2009). Although tremendous success has been achieved in identification and characterization of genes governing resistance against leaf blast, genetic studies on neck-blast resistance have lagged far behind. Though there are around 118 R genes identified for leaf-blast resistance, very few genes mediating resistance to neck blast have been identified and precisely mapped in rice genome (Table 3). The reports on leaf-blast-resistant cultivars being susceptible to neck blast and vice versa (Sirithunya et al. 2002; Zhuang et al. 2002; Puri et al. 2009; Ishihara et al. 2014), imply that there may be inherent variation in the mechanisms of resistance to leaf and neck blast.
Table 3.
List of neck-blast-resistance genes and QTLs identified from rice
| Sr no. | Gene/QTL | Donor | Chrm. No. | Genomic position (Mb)a | Linked marker | Phenotypic variation explained (%) | References |
|---|---|---|---|---|---|---|---|
| 1 |
qNB1-1 qNB1-2 qNB1-3 |
Jao Hom Nin | 1 |
23.97–40.16 23.97–40.16 26.81–40.16 |
SSR |
16.57 16.75 9.02 |
Noenplab et al. (2006) |
| 2 | Pi64 | Yangmaogu | 1 | 32.31–34.34 | SSR, CAPS | –b | Ma et al. (2015) |
| 3 | qNBL-5 | IR64 | 5 | 19.60–27.81 | RFLP, RAPD, SSR | 11.10 | Hittalmani et al. (2003) |
| 4 | qLNBL-5 | Akhanaphou | 5 | 19.72–23.84 | SSR | 13.54–26.23 | Aglawe et al. (2017) |
| 5 | Pi25(t) | Gumei 2 | 6 | 13.05–13.06 | RFLP, RGA | –b | Zhuang et al. (2002) |
| 6 | qLNBL-7 | Akhanaphou | 7 | 0.46–2.67 | SSR | 11.14–31.04 | Aglawe et al. (2017) |
| 7 | qPbh7-1 | Heikezijing | 7 | 25.65–29.56 | SSR | 17.74 | Fang et al. (2016) |
| 8 | qNBL-9 | IR64 | 9 | 15.54–20.48 | RFLP, RAPD, SSR | 8.50 | Hittalmani et al. (2003) |
| 9 | qPbm9 | Miyazakimochi | 9 | 19.74–21.00 | SSR, SNP | 5.70 | Ishihara et al. (2014) |
| 10 | qNBL-10 | IR64 | 10 | 5.48–16.65 | RFLP, RAPD, SSR | 24.10 | Hittalmani et al. (2003) |
| 11 | NBL1(t) | IR64 | 11 | 7.90–10.13 | RFLP, RAPD, SSR | –b | Hittalmani et al. (2003) |
| 12 | qPbm11 | Miyazakimochi | 11 | 22.48–24.68 | SSR, SNP | 30.80 | Ishihara et al. (2014) |
| 13 | qNB11-1 qNB11-3 | Jao Hom Nin | 11 |
22.48–28.80 24.23–28.80 |
SSR |
34.58 22.12 |
Noenplab et al. (2006) |
| 14 | Pb-1 | Modan | 11 | 22.88–22.92 |
RFLP SNP, Indel |
–b | Fujii et al. (2000) and Hayashi et al. (2010) |
| 15 | qPbh11-1 | Heikezijing | 11 | 24.23–28.96 | SSR | 14.19–34.00 | Fang et al. (2016) |
| 16 | Pi-jnw1 | Jiangnanwan | 11 | 24.68–28.93 | SSR, Indel | 39.92–53.68 | Wang et al. (2016b) |
| 17 | qNB12-2 | KDML105 | 12 | 10.08–21.48 | SSR | 6.03 | Noenplab et al. (2006) |
| 18 | qNB12-3 | Jao Hom Nin | 12 | 10.08–21.48 | SSR | 17.51 | Noenplab et al. (2006) |
aThe chromosomal locations were assigned by projecting the sequences of flanking markers on the genome sequence of cv. Nipponbare released by International Rice Genome Sequencing Project (http://rapdb.dna.affrc.go.jp)
bMajor genes exhibiting Mendelian inheritance
Sirithunya et al. (2002) have identified quantitative trait loci (QTLs) associated with leaf- and neck-blast resistance from a resistance donor CT9993-5-10-M (CT). Two QTLs for broad-spectrum leaf resistance have been located on chromosomes 7 and 9, whereas two neck-blast QTLs mapped to chromosomes 5 and 6. The inconsistencies in map locations of leaf- and neck-blast-resistance QTLs have suggested the presence of different genetic mechanisms for leaf- and neck-blast resistance. Alternately, there are several instances where the gene/QTLs for panicle blast resistance have been mapped on the same genomic region, where major leaf-blast-resistance genes have been located in the rice genome, suggesting the involvement of common genes for resistance to both leaf and neck blast (Noenplab et al. 2006).
A gene Pb1 for adult-stage panicle blast resistance has been identified from an indica cultivar ‘Modan’ (Fujii et al. 2000). The gene has been localized on the long arm of chromosome 11 and is known to encode a coiled coil–nucleotide-binding site–leucine-rich repeat (CC–NBS–LRR) protein (Hayashi et al. 2010). The gene has been introduced into several elite varieties commercially cultivated in Japan and not shown any signs of breakdown of resistance for almost 30 years (Hayashi et al. 2010). Pb1 is known to exhibit low level of expression at the early vegetative stages (two-and six-leaf stages) and is gradually up-regulated in later developmental stages, reaching its peak expression levels at full heading stage. Interestingly, the Pb1 transformants of a susceptible cultivar Nipponbare that constitutively over express the gene have also shown strong resistance to leaf blast at early vegetative stages (Hayashi et al. 2010). These findings clearly suggest that the expression pattern of a blast-resistance gene is a major determinant in deciding whether the gene will mediate protection to leaf or neck blast or both phases of the disease. The genes that are primarily expressed at early vegetative phase are most likely to mediate protection to leaf blast, those expressed during the flag leaf and heading stages shall display neck and panicle blast resistance, while those displaying constitutive expression throughout the plant are expected to provide protection against both the phases of the disease.
Zhuang et al. (2002) have identified a blast-resistance gene Pi25(t) from a durably resistant semi-dwarf indica variety ‘Gumei-2’. The gene provides resistance to both leaf and neck blast and is located in the genetic interval known to harbour well-known leaf-blast-resistance genes Pi-2(t) and Pi-9(t) on chromosome 6. Hittalmani et al. (2003) have reported that neck-blast resistance in a famous indica variety IR64 is controlled by both major and minor genes. A major gene NBL1(t) for neck-blast resistance has been associated with marker Nbp44 on chromosome 11. Besides this, three other quantitative trait loci (QTLs) qNBL-10, qNBL-9, and qNBL-5 explaining 8.5–24.10% of the phenotypic variations for neck-blast resistance have been located on chromosomes 10, 9, and 5, respectively (Table 3).
Noenplab et al. (2006) have reported the co-localization of genetic factors responsible for leaf- and neck-blast resistance in a Thai rice variety Jao Hom Nin (JHN). A total of 14 QTLs, representing seven each for leaf- and neck-blast resistance, have been mapped on three chromosomes, namely, 1, 11, and 12. The six QTLs, three each against leaf (qLB1-1, qLB1-2, and qLB1-3), and neck-blast resistance (qNB1-1, qNB1-2, and qNB1-3) on chromosome 1 are located near the marker RM212 close to a major blast-resistance gene Pi37. Similarly, four QTLs representing two each for leaf (qLB11-1 and qLB11-3) and neck-blast resistance (qNB11-1 and qNB11-3) are present on chromosome 11 in the vicinity of three major blast-resistance genes, Pi7(t), Pi1, and Pilm2. For the QTLs detected on chromosome 12, the genomic locations have shown an overlap with the location of major leaf-blast-resistance genes such as Pita and Pi20(t).
Ishihara et al. (2014) have identified two QTLs for panicle blast resistance from japonica cultivar Miyazakimochi: a major QTL, qPbm11, on chromosome 11 with a contribution of 30.8% and a minor QTL, qPbm9, on chromosome 9 contributing 5.7% to phenotypic variance for panicle blast resistance. The phenotypic analysis of BC2F7 lines introgressed with these QTLs has indicated that the level of panicle blast resistance conferred by qPbm11 is very similar to the level of resistance in donor Miyazakimochi, whereas qPbm9 makes small contribution to overall resistance. The genomic position of major QTL qPbm11 coincides with that of panicle blast-resistance locus, Pb1, previously identified from indica cultivar Modan. However, the absence of Pb1 encoded transcripts in the panicles of Miyazakimochi as revealed through reverse transcriptase PCR has suggested that the qPbm11 is different from Pb1.
Ma et al. (2015) have identified a rice-blast-resistance gene Pi64 which confers resistance to both leaf and neck blast from a broad-spectrum-resistant japonica landrace Yangmaogu (YMG). The gene is located on chromosome 1 and encodes a CC–NBS–LRR protein. The expression studies have suggested that Pi64 is constitutively expressed at all development stages and in all tissues examined. The observed constitutive expression pattern has been suggested to as the key factor responsible for its effectiveness against both leaf and neck blast.
Fang et al. (2016) have identified two QTLs for panicle blast resistance from a landrace genotype Heikezijing. One of these QTLs, qPbh-11-1, located on long arm of chromosome 11 contributes 14.19–34.00% to phenotypic variance and occupies different genomic position compared to two panicle blast-resistance loci Pb1 and qPbm11 previously identified from the same chromosome. qPbh-11-1 is located in a genomic region harbouring Pi-k gene cluster from where a major leaf-blast-resistance gene Pi-hk1(t) has also earlier been identified in donor genotype Heikezijing (Wu et al. 2013). Pi-jnw1 gene conferring resistance to panicle and leaf blast has been identified from a japonica landrace Jiangnanwan (Wang et al. 2016b). Pi-jnw1 is located between markers RM27273 and RM27381 on chromosome 11 and explains 39.92–53.68% of phenotypic variation for panicle blast resistance and 10.91–23.60% of variation for the leaf-blast resistance. The gene has been fine mapped to a 282 kb region on chromosome 11 from where three leaf-blast-resistance genes Pi-k, Pi34, and Pi-hk1 have previously been identified in different rice genotypes (Hayashi et al. 2006; Zenbayashi et al. 2007; Wu et al. 2013). Two novel QTLs qLNBL-5 and qLNBL-7 conferring resistance to leaf as well as neck blast have been identified from an Indian landrace Akhanaphou, having a high level of field resistance to blast (Aglawe et al. 2017). Both of these QTLs confer resistance to leaf as well as neck blast contributing 26% and 25% to phenotypic variance for the resistance. The genomic position of qLNBL-5 located on chromosome 5 coincides with an earlier reported meta-QTL for leaf blast on the chromosome 5 (Ballini et al. 2008), whereas the qLNBL-7 identified from chromosome 7 is positioned within the genetic interval RM7161-RM21328 previously known to harbour one more meta-QTL for leaf blast. These observations reinforce the inferences of several earlier studies that suggested common genes for resistance to both leaf and neck blast (Noenplab et al. 2006; Ishihara et al. 2014; Fang et al. 2016).
Conclusions and future guidelines for blast-resistance breeding
Recent advancements in rice genomics, especially the accessibility of complete genome sequences of rice subspecies japonica and indica and concomitant availability of an array of sequence-based molecular markers, have greatly facilitated detailed genetic analysis of blast resistance in rice. These developments have culminated in identification of nearly 118 blast-resistance genes, 25 of which have been cloned and molecularly characterized. Genetic mapping and molecular cloning of different blast-resistance genes have provided a gamut of tightly linked or functional gene-derived markers for use in marker-assisted breeding. Compared to the success achieved in identification and characterization of resistance genes against leaf blast, a few major genes mediating protection to neck blast have been identified; of the some 118 major blast-resistance genes identified from rice, only three, Pb1, Pi25, and Pi64, are known to provide resistance to panicle and neck blast. Most of the genetic studies on neck and panicle blast resistance have been conducted with the genotypes differing in degree of susceptibility/resistance to disease and consequently have resulted in the identification of QTLs providing quantitative resistance to neck and panicle blast. However, there are several instances, where the gene(s)/QTLs governing neck-blast and leaf-blast resistance have been mapped to same genomic region in rice suggesting the possibility of involvement of common genes for resistance to both leaf and neck blast. There are also evidences that gene for gene resistance mediating protection to leaf blast also operates in the roots and other parts of rice plant (Sesma and Osbourn 2004). These studies have indicated the possibility that the leaf-blast-resistance genes that are constitutively expressed in rice genome may mediate resistance to neck blast as well. Precise studies involving the characterization of neck-blast resistance of rice isogenic lines introgressed with major leaf-blast-resistance genes against a diverse spectrum of pathogen races are needed to ascertain their role in mediating protection to neck blast. These studies will also clarify whether the shifts in race composition of pathogen and environmental conditions during crop season are the sole factors that predispose certain leaf-blast resistant varieties to neck blast and vice versa or the developmentally regulated expression of the resistance genes has some role to play in the outcome.
The clustering of leaf-blast-resistance genes into complex loci and overlapping in the genomic position of leaf- and neck-blast-resistance genes have potential implications in resistance breeding. The genomic region harbouring these gene clusters can be targeted for introgression into susceptible varieties to achieve broad-spectrum resistance, as has been demonstrated for the Pita resistance gene complex located near the centromeric region of rice chromosome 12. The Pita gene complex derived from a broad-spectrum-resistant genotype Tetep has provided a stable resistance to blast disease in range of indica and japonica rice varieties since the 1960s (Moldenhauer et al.1990; Zhao et al. 2018).
Durable resistance to blast has been a priority area in rice breeding. The efforts to achieve durable resistance using major race-specific genes are often frustrated due to evolution of new races of the pathogen that negate the effect of resistance genes. Marker-assisted pyramiding of race-specific resistance genes is widely practiced to increase the durability of resistance. However, most of the gene pyramiding projects rely only on gene number per se as selection criterion to ensure durability, while the attempts to assess the magnitude of fitness penalty that these genes impose on the pathogen populations have rarely been conducted beforehand to guide the selection of effective gene combinations for pyramiding. The resistance gene combinations that impose substantial fitness costs on pathogen variants evolving matching virulences against such genes are most likely to provide durable resistance, e.g., a strong virulence dissociation against major blast-resistance genes Pi1 and Pi2 has been reported in the rice-blast populations in different parts of the world (Mekwatanakarn et al. 2000; Rathour et al. 2006). It is, therefore, imperative that the choice of resistance gene combinations for pyramiding should be based on the sound knowledge of their effect on pathogen fitness to ensure durability.
Recent studies on molecular basis of host–pathogen interactions in rice have identified plant disease-susceptibility genes (S) that promote disease in the host following activation by pathogen effectors. These genes promote disease either by promoting pathogen growth and development or acting as negative regulators of basal defense response. The recessive loss-of-function alleles of rice susceptibility genes Pi21, Bsr-d1, and Bsr-k1 have been shown to mediate non-race-specific resistance to blast (Fukuoka et al. 2009; Li et al. 2017; Zhou et al. 2018). Consistent with these findings, the CRISPR/Cas9-targeted knockouts of ERF transcription factor gene OsERF922 and Bsr-d1, both of which promote susceptibility to blast, have demonstrated enhanced resistance to rice P. oryzae (Wang et al. 2016a; Li et al. 2017). These case studies have suggested the possibility of using spontaneous or genetically engineered loss-of-function mutant alleles of S genes for achieving durable non-race-specific resistance to blast.
Of the over 120 rice-blast-resistance gene identified until date, only few (less than 4%) have been sourced from the wild species. As many beneficial alleles must have been left behind in wild species during evolution and domestication of rice, these untapped genetic resources should be explored to identify new genes for resistance to rice blast. The efforts should also be made to unravel new allelic variants of already characterized resistance genes from unexplored landraces and wild species through sequencing-based allele mining. While the mining of coding region is expected to provide new alleles possessing varied resistance specificities, the prompter mining will uncover variants differing in expression patterns. The novel allelic variants with differing resistance specificities and expression patterns can be deployed in the field either as gene pyramids through transgenic approach or as multilines to achieve broad-spectrum resistance to blast. Furthermore, detailed understanding of polymorphic sites controlling resistance specificities of different alleles will facilitate intragenic allele pyramiding for developing chimeric alleles having broader recognition spectrum compared to parental alleles, as has been demonstrated for the alleles of powdery mildew-resistance gene Pm3 in wheat (Brunner et al. 2010).
Acknowledgements
The senior author gratefully acknowledges the Inspire Fellowship received from the Department of Science and Technology, Ministry of Science and Technology, Government of India for her Ph.D programme.
Author contributions
SK formulated the initial draft and RR corrected and wrote the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest to declare.
References
- Aglawe SB, Umakanth B, Rama Devi SJS, Vishalakshi B, Bhadana VP, Sharma SK, Sharma PK, Kumar S, Prasad MS, Madhav MS. Identification of novel QTLs conferring field resistance for rice leaf and neck blast from an unique landrace of India. Gene Rep. 2017;7:35–42. [Google Scholar]
- Ahn SN, Kim YK, Hong HC, Han SS, Choi HC, McCouch SR, Moon HP. Mapping of genes conferring resistance to Korean isolates of rice blast fungus using DNA markers. Korean J Breed. 1997;29(4):416–423. [Google Scholar]
- Ahn SN, Kim YK, Hong HC, Han SS, Kwon SJ, Choi HC, Moon HP, McCouch SR. Molecular mapping of a new gene for resistance to rice blast (Pyricularia grisea Sacc.) Euphytica. 2000;116:17–22. [Google Scholar]
- Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu J, Matsumoto T, Ono K, Yano M. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pik-m specific rice blast resistance. Genetics. 2008;180:2267–2276. doi: 10.1534/genetics.108.095034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkani S, Rafil MY, Shabanimofrad M, Miah G, Sahebi M, Azizi P, Tanweer FA, Akhtar MS, Nasehi A. Molecular breeding strategy and challenges towards the improvement of blast disease resistance in rice crop. Front Plant Sci. 2015;6:1–14. doi: 10.3389/fpls.2015.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballini E, Morel JB, Droc G, Price A, Courtois B, Notteghem JL, Tharreau D. A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol Plant Microbe Interact. 2008;21(7):859–868. doi: 10.1094/MPMI-21-7-0859. [DOI] [PubMed] [Google Scholar]
- Barman SR, Gowda M, Venu RC, Chattoo BB. Identification of a major blast resistance gene in the rice cultivar Tetep. Plant Breed. 2004;123:300–302. [Google Scholar]
- Berruyer R, Adreit H, Milazzo J, Gaillard S, Berger A, Dioh W, Lebrun MH, Tharreau D. Identification and fine mapping of Pi33 the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor Appl Genet. 2003;107:1139–1147. doi: 10.1007/s00122-003-1349-2. [DOI] [PubMed] [Google Scholar]
- Brunner S, Hurni S, Streckeisen P, Mayr G, Albrecht M, Yahiaoui N, Keller B. Intragenic allele pyramiding combines different specificities of wheat Pm3 resistance alleles. Plant J. 2010;6:433–445. doi: 10.1111/j.1365-313X.2010.04342.x. [DOI] [PubMed] [Google Scholar]
- Bryan GT, Wu KS, Farrall L, Jia Y, Hershey HP, McAdams SA, Valent B. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell. 2000;12:2033–2045. doi: 10.1105/tpc.12.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu K, Xiao J, Yu ZH, Ronald PC, Harrington SE, Second G, McCouch SR, Tanksley SD. Saturated molecular map of the rice genome based on an inter-specific backcross population. Genetics. 1994;138:1251–1274. doi: 10.1093/genetics/138.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan R, Farman M, Zhang HB, Leong S. Genetic and physical mapping of a rice blast resistance locus Pi-CO39(t) that corresponds to the avirulence gene AVR1-CO39 of Magnaporthe grisea. Mol Genet Genom. 2002;267:603–612. doi: 10.1007/s00438-002-0691-4. [DOI] [PubMed] [Google Scholar]
- Chen X, Temnykh S, Xu Y, Gho YG, McCouch SR. Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.) Theor Appl Genet. 1997;95:553–567. [Google Scholar]
- Chen DH, Vina MD, Inukai T, Mackill DJ, Ronald PC, Nelson RJ. Molecular mapping of the blast resistance gene Pi44(t) in a line derived from a durably resistant rice cultivar. Theor Appl Genet. 1999;98:1046–1053. [Google Scholar]
- Chen XW, Li SG, Xu JC, Zhai WX, Ling ZZ, Ma BT, Wang YP, Wang WM, Cao G, Ma YQ, Shang JJ, Zhao XF, Zhou KD, Zhu LH. Identification of two blast resistance genes in a rice variety Digu. J Phytopathol. 2004;152:77–85. [Google Scholar]
- Chen S, Wang L, Que ZQ, Pan R, Pan Q. Genetic and physical mapping of Pi37(t), a new gene conferring resistance to rice blast in the famous cultivar St. No. 1. Theor Appl Genet. 2005;111:1563–1570. doi: 10.1007/s00122-005-0086-0. [DOI] [PubMed] [Google Scholar]
- Chen XW, Shang JJ, Chen DX, Lei CL, Zou Y, Zhai WX, Liu GZ, Xu JC, Ling ZZ, Cao G, Ma BT, Wang YP, Zhao XF, Li SG, Zhu LH. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006;46:794–804. doi: 10.1111/j.1365-313X.2006.02739.x. [DOI] [PubMed] [Google Scholar]
- Chen J, Shi Y, Liu W, Chai R, Fu Y, Zhuang J, Wu J. A Pid3 allele from rice cultivar Gumei 2 confers resistance to Magnaporthe oryzae. J Genet Genom. 2011;38:209–216. doi: 10.1016/j.jgg.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Chen S, Su J, Han J, Wang W, Wang C, Yang J, Zeng L, Wang X, Zhu X, Yang C. Resistance spectrum assay and fine mapping of the blast resistance gene from a rice experimental line, IRBLta2-Re. Euphytica. 2014;195:209–216. [Google Scholar]
- Chen J, Peng P, Tian J, He Y, Zhang L, Liu Z, Yin D, Zhang Z. Pike, a rice blast resistance allele consist of two adjacent NBS-LRR genes, was identified as a novel allele at the Pik locus. Mol Breed. 2015;35:117. [Google Scholar]
- Cho YG, Ishii T, Emnykh S, Chen X, Lipovich L, McCouch SR, Park WO, Ayres N, Cartinhour S. Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.) Theor Appl Genet. 2000;100:697–712. [Google Scholar]
- Deng Y, Zhu X, Shen Y, He Z. Genetic characterization and fine mapping of the blast resistance locus Pigm(t) tightly linked to Pi2 and Pi9 in a broad-spectrum resistant Chinese variety. Theor Appl Genet. 2006;113:705–713. doi: 10.1007/s00122-006-0338-7. [DOI] [PubMed] [Google Scholar]
- Deng Y, Zhu X, Xu J, Chen H, He Z. Map-based cloning and breeding application of a broad-spectrum resistance gene Pigm to rice blast. In: Wang GL, Valent B, editors. Advances in genetics, genomics and control of rice blast disease. Dordrecht: Springer; 2009. pp. 161–171. [Google Scholar]
- Dong L, Liu S, Xu P, Deng W, Li X, Tharreau D, Li J, Zhou J, Wang Q, Tao D, Yang Q. Fine mapping of Pi57(t) conferring broad spectrum resistance against Magnaporthe oryzae in introgression line IL-E1454 derived from Oryza longistaminata. PLoS One. 2017;12(10):e0186201. doi: 10.1371/journal.pone.0186201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwinita WU, Sugiono M, Hajrial A, Asep S, Ida H. Blast resistance genes in wild rice Oryza rufipogon and rice cultivar IR64. Indones J Agric Sci. 2008;1:71–76. [Google Scholar]
- Ebitani T, Hayashi N, Omoteno M, Ozaki H, Yano M, Morikawa M, Fukuta Y. Characterization of Pil3, a blast resistance gene that maps to chromosome 6 in indica rice (Oryza sativa L. variety, Kasalath) Breed Sci. 2011;61:251–259. [Google Scholar]
- Ellur RK, Khanna A, Yadav A, Pathania S, Rajashekara H, Singh VK, Krishnan SG, Bhowmick PK, Nagarajan M, Vinod KK, Prakash G. Improvement of Basmati rice varieties for resistance to blast and bacterial blight diseases using marker assisted backcross breeding. Plant Sci. 2016;242:330–341. doi: 10.1016/j.plantsci.2015.08.020. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamaguchi M, Kaji R, Nakagomi K, Yokogami M, Nakamura T, Ishikawa G, Yonimaru J, Nishio T. Close linkage of a blast resistance gene, Pias(t), with a bacterial leaf blight resistance gene, Xa1-as(t), in a rice cultivar ‘Asominori’. Breed Sci. 2012;62:334. doi: 10.1270/jsbbs.62.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang N, Wang R, He W, Yin C, Guan C, Chen H, Huang J, Wang J, Bao Y, Zhang H. QTL mapping of panicle blast resistance in japonica landrace Heikezijing and its application in rice breeding. Mol Breed. 2016;36:171–179. [Google Scholar]
- Fjellstrom R, Conaway-Bormans CA, McClung AM, Marchetti MA, Shank AR, Park WD. Development of DNA markers suitable for marker assisted selection of three genes conferring resistance to multiple pathotypes. Crop Sci. 2004;44:1790–1798. [Google Scholar]
- Fuentes LJ, Correa-Victoria FJ, Escobar F, Prado G, Aricapa G, Duque MC, Tohme J. Identification of microsatellite markers linked to the blast resistance gene Pi-1(t) in rice. Euphytica. 2008;160:295–304. [Google Scholar]
- Fujii K, Hayano-Saito Y, Saito K, Sugiura N, Hayashi N, Tsuji T, Izawa T, Iwasaki M. Identification of a RFLP marker tightly linked to the panicle blast resistance gene Pb1 in rice. Breed Sci. 2000;50(3):183–188. [Google Scholar]
- Fukuoka S, Okuno K. QTL analysis and mapping of pi21 a recessive gene for field resistance to rice blast in Japanese upland rice. Theor Appl Genet. 2001;103:85–190. [Google Scholar]
- Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K, Yano M. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009;325:998–1001. doi: 10.1126/science.1175550. [DOI] [PubMed] [Google Scholar]
- Fukuta Y (2004) The Pish blast resistance gene of rice (Oryza sativa L.) is located on the long arm of chromosome 1. Japan International Research Center for Agricultural Sciences (JIRCAS), Annual Report, pp 46–47
- Gowda M, Roy-Barman S, Chattoo BB. Molecular mapping of a novel blast resistance gene Pi38 in rice using SSLP and AFLP markers. Plant Breed. 2006;125:596–599. [Google Scholar]
- Harushima Y, et al. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka H, Miyao A, Yano M, Matsunaga K, Sasaki T. RFLP mapping of a rice blast resistance gene Pik. Breed Sci. 1996;46:68. [Google Scholar]
- Hayashi K, Yoshida H. Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J. 2009;57:413–425. doi: 10.1111/j.1365-313X.2008.03694.x. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshida H, Ashikawa I. Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes. Theor Appl Genet. 2006;113:251–260. doi: 10.1007/s00122-006-0290-6. [DOI] [PubMed] [Google Scholar]
- Hayashi N, Inoue H, Kato T, Funao T, Shirota M, Shimizu T, Kanamori H, Yamane H, Hayano-Saito Y, Matsumoto T, Yano M, Takatsuji H. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010;64:498–510. doi: 10.1111/j.1365-313X.2010.04348.x. [DOI] [PubMed] [Google Scholar]
- He WW, Fang NY, Wang RS, Wu YY, Zen GY, Guan CH, Chen H, Huang J, Wang JF, Bao YM, Zhang HS. Fine mapping of a new race-specific blast resistance gene, Pi-hk2, in japonica Heikezijing from Taihu region of China. Phytopathology. 2017;107:84–91. doi: 10.1094/PHYTO-03-16-0151-R. [DOI] [PubMed] [Google Scholar]
- Hittalmani S, Parco A, Mew TV, Zeigler RS, Huang N. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor Appl Genet. 2000;100(7):1121–1128. [Google Scholar]
- Hittalmani S, Srinivasachary BP, Shashidhar HE, et al. Identifying major genes and QTLs for field resistance to neck blast in rice. In: Khush GS, et al., editors. Advances in rice genetics. Los Banos: Rice Genetics Collection; 2003. pp. 248–250. [Google Scholar]
- Hu J, Chang X, Zou L, Tang W, Wu W. Identification and fine mapping of Bph33, a new brown planthopper resistance gene in rice (Oryza sativa L.) Rice. 2018;11:55. doi: 10.1186/s12284-018-0249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua L, Wu J, Chen C, Wu W, He X, Lin F, Wang L, Ashikawa I, Matsumoto T, Wang L, Pan Q. The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor Appl Genet. 2012;125(5):1047–1055. doi: 10.1007/s00122-012-1894-7. [DOI] [PubMed] [Google Scholar]
- Huang H, et al. Molecular mapping of the new blast resistance genes Pi47 and Pi48 in the durably resistant local rice cultivar Xiangzi 3150. Phytopathology. 2011;101:620–626. doi: 10.1094/PHYTO-08-10-0209. [DOI] [PubMed] [Google Scholar]
- Inoue T, Zhong HS, Miyao A, Ashikawa I, Monna L, Fukuoka S, Miyadera N, Nagamura Y, Kurata K, Sasaki T, Minobe Y. Sequence tagged sites (STSs) as standard landmarks in the rice genome. Theor Appl Genet. 1994;89:728–734. doi: 10.1007/BF00223712. [DOI] [PubMed] [Google Scholar]
- Ishihara T, Saito YH, Oide S, Ebana K, La NT, Hayashi K, Ashizawa T, Suzuki F, Koizumi S. Quantitative trait locus analysis of resistance to panicle blast in the rice cultivar Miyazakimochi. Rice. 2014;7:2. doi: 10.1186/s12284-014-0002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Chen D, Yi GH, Wang GL, Ronald PC. Genetic and physical mapping of Pi5(t) a locus associated with broad-spectrum resistance to rice blast. Mol Genet Genom. 2003;269:280–289. doi: 10.1007/s00438-003-0834-2. [DOI] [PubMed] [Google Scholar]
- Jeung JU, Kim BR, Cho YC, Han SS, Moon HP, Lee YT, Jena KK. A novel gene Pi40(t) linked to the DNA markers derived from NBS-LRR motifs confers broad spectrum blast resistance in rice. Theor Appl Genet. 2007;115:1163–1177. doi: 10.1007/s00122-007-0642-x. [DOI] [PubMed] [Google Scholar]
- Jia Y, Wang Z, Singh P. Development of dominant rice blast Pi-ta resistance gene markers. Crop Sci. 2002;42:2145–2149. [Google Scholar]
- Jiang J, Wang S. Identification of a 118 kb DNA fragment containing the locus of blast resistance gene Pi-2(t) in rice. Mol Genet Genom. 2002;268:249–252. doi: 10.1007/s00438-002-0742-x. [DOI] [PubMed] [Google Scholar]
- Jiang N, et al. Molecular mapping of the Pi2/9 allelic gene Pi2-2 conferring broad-spectrum resistance to Magnaporthe oryzae in the rice cultivar Jefferson. Rice. 2012;5:1–7. doi: 10.1186/1939-8433-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Dhatwalia S, Kaachra A, Sharma KD, Rathour R. Genetic and physical mapping of a new rice blast resistance specificity Pi-67 from a broad spectrum resistant genotype Tetep. Euphytica. 2019;215:9. doi: 10.1007/s10681-018-2332-y. [DOI] [Google Scholar]
- Kaji R, Ogawa T. RFLP mapping of a blast resistance gene Pi-km in rice. Breed Sci. 1996;46(Suppl. 1):70. [Google Scholar]
- Kaji R, Ogawa T, Nishimura M. RFLP mapping of a blast resistance gene, Pit, in rice. Breed Sci. 1997;47(Suppl. 1):37. [Google Scholar]
- Khanna A, Sharma V, Ellur RK, Shikari AB, Krishnan SG, Singh UD, Prakash G, Sharma TR, Rathour R, Variar M, Prashanthi SK. Development and evaluation of near-isogenic lines for major blast resistance gene(s) in Basmati rice. Theor Appl Genet. 2015;128(7):1243–1259. doi: 10.1007/s00122-015-2502-4. [DOI] [PubMed] [Google Scholar]
- Kim DM, Ju HG, Yang P, Han SS, Roh JH, Ahn SN. Mapping and race specific reaction of the resistance gene Pi45(t) in rice. Korean J Breed Sci. 2011;43:42–49. [Google Scholar]
- Kiyosawa S (1967) Genetic studies on host-pathogen relationship in the rice blast disease. In: Proceedings of symposium of rice diseases and their control by growing resistant varieties and other measures. Tokyo, pp 137–153
- Koide Y, Telebanco-Yanoria MJ, Fukuta Y, Kobayashi N. Detection of novel blast resistance genes Pi58(t) and Pi59(t) in a Myanmar rice landrace based on a standard differential system. Mol Breed. 2013;32:241–252. [Google Scholar]
- KoideY Telebanco-Yanoria MJ, Pena FD, Fukuta Y, Kobayashi N. Characterization of rice blast isolates by the differential system and their application for mapping a resistance gene, Pi19 (t) J Phytopathol. 2011;159:85–93. [Google Scholar]
- Kumar P, Pathania S, Katoch P, Sharma TR, Plaha P, Rathour R. Genetic and physical mapping of blast resistance gene Pi-42(t) on the short arm of rice chromosome 12. Mol Breed. 2010;25:217–228. [Google Scholar]
- Lee S, Wamishe Y, Jia Y, Liu G, Jia MH. Identification of two major resistance genes against race 1E–1k Magnoporthe oryzae in the indica rice cultivar Zhe733. Mol Breed. 2009;24:127–134. [Google Scholar]
- Lee SK, Song MY, Seo YS, Kim HK, Ko S, Cao PJ, Suh JP, Yi G, Roh JH, Lee S, An G, Hahn TR, Wang GL, Ronald P, Jeon JS. Rice Pi5 mediated resistance to Magnaporthe oryzae requires the presence of two Nucleotide-Binding-Leucine-Rich-Repeat genes. Genetics. 2009;181:1627–1638. doi: 10.1534/genetics.108.099226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei CL, Huang DY, Li W, Wang LJ, Liu ZL, Wang XT, Shi K, Cheng ZJ, Zhang X, Ling ZZ, Wan JM. Molecular mapping of a blast resistance gene in an indica rice cultivar Yanxian No. 1. Rice Genet Newsl. 2005;22:76–77. [Google Scholar]
- Lei C, Hao K, Yang Y, Ma J, Wang S, Wang J, Cheng Z, Zhao S, Zhang X, Guo X, Wang C, Wan J. Identification and fine mapping of two blast resistance genes in rice cultivar 93-11. Crop J. 2013;1:2–14. [Google Scholar]
- Li LY, Wang L, Jing JX, Li ZQ, Lin F, Huang LF, Pan QH. The Pik-m gene, conferring stable resistance to isolates of Magnaporthe oryzae was finely mapped in a crossover cold region on rice chromosome 11. Mol Breed. 2007;20:179–188. [Google Scholar]
- Li W, Lei C, Cheng Z, Jia Y, Huang D, Wang J, Wang J, Zhang X, Su N, Guo X, Zhai H, Wan J. Identification of SSR markers for a broad-spectrum blast resistance gene Pi20(t) for marker assisted breeding. Mol Breed. 2008;22:141–149. [Google Scholar]
- Li W, et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell. 2017;114:116–126. doi: 10.1016/j.cell.2017.06.008. [DOI] [PubMed] [Google Scholar]
- Liang Z, Wang L, Pan Q. A new recessive gene conferring resistance against rice blast. Rice. 2016;9:47. doi: 10.1186/s12284-016-0120-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Chen S, Que Z, Wang L, Liu X. The blast resistance gene Pi37 encodes an NBS-LRR protein and is a member of a resistance gene cluster on rice chromosome 1. Genetics. 2007;177:1871–1880. doi: 10.1534/genetics.107.080648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LY, Jing JX, Li ZQ, Lin F, Huang LF, Pan QH. A high-resolution map of the rice blast resistance gene Pi15 constructed by sequence-ready markers. Plant Breed. 2007;126:287–290. [Google Scholar]
- Liu B, Zhang S, Zhu X, Yang Q, Wu S, Mei M, Mauleon R, Leach J, Mew T, Leung H. Candidate defense genes as predictors of quantitative blast resistance in rice. Mol Plant Microbe Interact. 2004;17:1146–1152. doi: 10.1094/MPMI.2004.17.10.1146. [DOI] [PubMed] [Google Scholar]
- Liu XQ, Wang L, Chen S, Lin F, Pan QH. Genetic and physical mapping of Pi36(t) a novel rice blast resistance gene located on rice chromosome 8. Mol Genet Genom. 2005;274:394–401. doi: 10.1007/s00438-005-0032-5. [DOI] [PubMed] [Google Scholar]
- Liu X, Lin F, Wang L, Pan Q. The in silico map-based cloning of Pi36, a rice coiled-coil-nucleotide-binding site-leucine-rich repeat gene that confers race-specific resistance to the blast fungus. Genetics. 2007;176:2541–2549. doi: 10.1534/genetics.107.075465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang Q, Lin F, Hua L, Wang C, Wang L, Pan Q. Identification and fine mapping of Pi39(t) a major gene conferring the broad-spectrum resistance to Magnaporthe oryzae. Mol Genet Genom. 2007;278:403–410. doi: 10.1007/s00438-007-0258-5. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu B, Zhu X, Yang J, Bordeos A, Wang G, Leach JE, Leung H. Fine-mapping and molecular marker development for Pi56(t), a NBS-LRR gene conferring broad-spectrum resistance to Magnaporthe oryzae in rice. Theor Appl Genet. 2013;126(4):985–998. doi: 10.1007/s00122-012-2031-3. [DOI] [PubMed] [Google Scholar]
- Lv Q, Huang Z, Xu X, Tang L, Liu H, Wang C, Zhou Z, Xin Y, Xing J, Peng Z, Li X, Zheng T, Zhu L. Allelic variation of the rice blast resistance gene Pid3 in cultivated rice worldwide. Sci Rep. 2017;7:10362. doi: 10.1038/s41598-017-10617-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, et al. Pi64, encoding a novel CC-NBS-LRR protein, confers resistance to leaf and neck blast in rice. Mol Plant Microbe Interact. 2015;28(5):558–568. doi: 10.1094/MPMI-11-14-0367-R. [DOI] [PubMed] [Google Scholar]
- Mackill DJ, Zhang Z, Redona ED, Colowit PM. Level of polymorphism and genetic mapping of AFLP markers in rice. Genome. 1996;39:969–977. doi: 10.1139/g96-121. [DOI] [PubMed] [Google Scholar]
- McCouch SR, Kochert G, Yu ZH, Wang ZY, Khush GS, Coffman WR, Tanksley SD. Molecular mapping of rice chromosomes. Theor Appl Genet. 1988;76(6):815–829. doi: 10.1007/BF00273666. [DOI] [PubMed] [Google Scholar]
- McCouch SR, et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.) DNA Res. 2002;9(6):199–207. doi: 10.1093/dnares/9.6.199. [DOI] [PubMed] [Google Scholar]
- Mekwatanakarn P, Kositratana W, Levy M, Zeigler RS. Pathotype and avirulence gene diversity in Thailand as determined by rice lines near-isogenic for major resistance genes. Plant Dis. 2000;84:60–70. doi: 10.1094/PDIS.2000.84.1.60. [DOI] [PubMed] [Google Scholar]
- Moldenhauer KAK, Lee FN, Norman RJ, Helms RS, Wells BR, Dilday RH, Rohman PC, Marchetti MA. Registration of “Katy” rice. Crop Sci. 1990;30:747–748. [Google Scholar]
- Monna L, Miyao A, Inoue T, Fukuoka S, Yamazaki M, Zhong HS, Sasaki T, Minobe Y. Determination of RAPD markers in rice and their conversion into sequence tagged sites (STSs) and STS-specific primers. DNA Res. 1994;1:139–148. doi: 10.1093/dnares/1.3.139. [DOI] [PubMed] [Google Scholar]
- Naqvi NI, Chattoo BB. Molecular genetic analysis and sequence characterized amplified region-assisted selection of blast resistance in rice. Rice Genet. 1996;3:570–576. doi: 10.1139/g96-004. [DOI] [PubMed] [Google Scholar]
- Naqvi NI, Bonman JM, Mackill DJ, Nelson RJ, Chattoo BB. Identification of RAPD markers linked to a major blast resistance gene in rice. Mol Breed. 1995;1:341–348. [Google Scholar]
- Nguyen TTT, Koizumi S, La TN, Zenbayashi KS, AshizawaT Yasuda N, Imazaki I, Miyasaka A. Pi35(t) a new gene conferring partial resistance to leaf blast in the rice cultivar Hokkai 188. Theor Appl Genet. 2006;113:697–704. doi: 10.1007/s00122-006-0337-8. [DOI] [PubMed] [Google Scholar]
- Noenplab A, Vanavichit A, Toojinda T, Sirithunyad P, Tragoonrung S, Sriprakhon S, Vongsaprom C. QTL mapping for leaf and neck blast resistance in Khao Dawk Mali 105 and Jao Hom Nin recombinant inbred lines. Sci Asia. 2006;32:133–142. [Google Scholar]
- Okuyama Y, Kanzaki H, Abe A, Yoshida K, Tamiru M, Saitoh H, Fujibe T, Matsumura H, Shenton M, Galam DC, Undan J, Ito A, Sone T, Terauchi R. A multifaceted genomics approach allows the isolation of the rice Pia blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 2011;66:467–479. doi: 10.1111/j.1365-313X.2011.04502.x. [DOI] [PubMed] [Google Scholar]
- Pan Q, Wang L, Ikehashi H, Tanisaka T. Identification of a new blast resistance gene in the indica rice cultivar Kasalath using Japanese differential cultivars and isozyme markers. Phytopathology. 1996;86:1071–1075. [Google Scholar]
- Pan QH, Tanisaka T, Ikehashi H. Studies on the genetics and breeding of blast resistance in rice VI. Gene analysis for the blast resistance of two Yunnan native cultivars GA20 and GA25. Breed Sci. 1996;46(Suppl. 2):70. [Google Scholar]
- Pan QH, Wang L, Ikehashi H, Yamagata H, Tanisaka T. Identification of two new genes conferring resistance to rice blast in the Chinese native cultivar ‘Maowangu’. Plant Breed. 1998;117:27–31. [Google Scholar]
- Pan QH, Wang L, Tanisaka T. A new blast resistance gene identified in the Indian native rice cultivar Aus373 through allelism and linkage tests. Plant Pathol. 1999;48(2):288–293. [Google Scholar]
- Parco A. STS markers for the blast resistance gene Pi-1(t) Rice Genome. 1995;4:9. [Google Scholar]
- Puri KD, Shreshta SM, Chhetri GBK, Joshi KD. Leaf and neck blast resistance reaction in Tropical rice lines under greenhouse condition. Euphytica. 2009;165(3):523–532. [Google Scholar]
- Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L, Han B, Wang GL. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics. 2006;172:1901–1914. doi: 10.1534/genetics.105.044891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathour R, Singh BM, Plaha P. Virulence structure of the Magnaporthe grisea rice population from the Northwestern Himalayas. Phytoparasitica. 2006;34(3):281–291. [Google Scholar]
- Rybka K, Miyamoto M, Ando I, Saito A, Kawasaki S. High resolution mapping of the indica-derived rice blast resistance genes II. Pi-ta2 and Pi-ta and a consideration of their origin. Mol Plant Microbe Interact. 1997;10:517–524. [Google Scholar]
- Sallaud C, Lorieux M, Roumen E, Tharreau D, Berruyer R, Svestasrani P, Garsmeur O, Ghesquiere A, Notteghem JL. Identification of five new blast resistance genes in the highly blast resistant rice variety IR64 using a QTL mapping strategy. Theor Appl Genet. 2003;106:794–803. doi: 10.1007/s00122-002-1088-9. [DOI] [PubMed] [Google Scholar]
- Sasaki R. Existence of strains in rice blast fungus. Int J Plant Prot Tokoyo. 1922;9:631–644. [Google Scholar]
- Sesma A, Osbourn AE. The rice leaf blast pathogen undergoes developmental processes typical of root-infecting fungi. Nature. 2004;431:582–586. doi: 10.1038/nature02880. [DOI] [PubMed] [Google Scholar]
- Shang J, Tao Y, Chen X, Zou Y, Lei C, Wang J, Li X, Zhao X, Zhang M, Lu Z, Xu J, Cheng Z, Wan J, Zhu L. Identification of a new rice blast resistance gene, Pid3, by genome-wide comparison of paired nucleotide-binding site-leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes. Genetics. 2009;182:1303–1311. doi: 10.1534/genetics.109.102871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma TR, Madhav MS, Singh BK, Shanker P, Jana TK, Dalal V, Pandit A, Singh A, Gaikwad K, Upreti HC, Singh NK. High-resolution mapping, cloning and molecular characterization of the Pi-kh gene of rice, which confers resistance to Magnaporthe grisea. Mol Genet Genom. 2005;274:569–578. doi: 10.1007/s00438-005-0035-2. [DOI] [PubMed] [Google Scholar]
- Shi BH, Zhang JH, Zheng YM, Liu YQ, Vera Cruz CM, Zheng TQ, Zhao MF. Identification of a new resistance gene Pi-Da(t) from Dacca6 against rice blast fungus Magnaporthe oryzae in Jin23B background. Mol Breed. 2012;30:1089–1096. [Google Scholar]
- Singh VK, et al. Incorporation of blast resistance into “PRR78”, an elite Basmati rice restorer line, through marker assisted backcross breeding. Field Crops Res. 2012;128:8–16. [Google Scholar]
- Singh WH, Kapila RK, Sharma TR, Rathour R. Genetic and physical mapping of a new allele of Pik locus from japonica rice ‘Liziangxintuanheigu’. Euphytica. 2015;205:889–901. [Google Scholar]
- Sirithunya P, Tragoonrung S, Vanavichit A, Pa-In N, Vongsaprom C, Toojinda T. Quantitative Trait loci associated with leaf and neck blast resistance in recombinant inbred line population of rice (Oryza sativa) DNA Res. 2002;9:79–88. doi: 10.1093/dnares/9.3.79. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Shamim M, Kumar M, Mishra A, Pandey P, Kumar D, Yadav P, Siddiqui MH, Singh KN. Current status of conventional and molecular interventions for blast resistance in rice. Rice Sci. 2017;24(6):299–321. [Google Scholar]
- Sun P, Liu J, Wang Y, Jiang N, Wang S, Dai Y, Gao J, Li Z, Pan S, Wang D, Li W, Liu X, Xiao Y, Liu E, Wang GL, Dai L. Molecular mapping of the blast resistance gene Pi49 in the durably resistant rice cultivar Mowanggu. Euphytica. 2013;192:45–54. [Google Scholar]
- Sun H, Wu L, Guo L, Song F, Zheng Z, Cheng Z. Genetic analysis and molecular mapping of Pitb with broad-spectrum resistance to Magnaporthe oryzae in rice. Euphytica. 2016;209:547–554. [Google Scholar]
- Tabien RE, Li Z, Paterson AH, Marchetti MA, Stansel JW, Pinson SRM. Mapping of four rice blast resistance genes from Lemont and Teqing and evaluation of their combinatorial effect for field resistance. Theor Appl Genet. 2000;101:1215–1225. [Google Scholar]
- Takahashi A, Hayashi N, Miyao A, Hirochika H. Unique features of the rice blast resistance Pish locus revealed by large scale retrotransposon-tagging. BMC Plant Biol. 2010;10:175. doi: 10.1186/1471-2229-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehisa H, Yasuda M, Fukuta Y, Kobayashi N, Hayashi N, Nakashita H, Abe T, Sato T. Genetic analysis of resistance genes in an Indica-type rice (Oryza sativa L.), Kasalath, using DNA markers. Breed Sci. 2009;59:253–260. [Google Scholar]
- Talbot NJ. On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu Rev Microbiol. 2003;57:177–202. doi: 10.1146/annurev.micro.57.030502.090957. [DOI] [PubMed] [Google Scholar]
- Terashima T, Fukuoka S, Saka N, Kudo S. Mapping of a blast field resistance gene Pi39(t) of elite rice strain Chubu 111. Plant Breed. 2008;127:485–489. [Google Scholar]
- Vasudevan K, Vera Cruz CM, Gruissem W, Bhullar NK. Large scale germplasm screening for identification of novel rice blast resistance sources. Front Plant Sci. 2014;5:505. doi: 10.3389/fpls.2014.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan K, Gruissem W, Bhullar NK. Identification of novel alleles of the rice blast resistance gene Pi54. Sci Rep. 2015;5:15678. doi: 10.1038/srep15678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Mackill DJ, Bonman JM, McCouch SR, Champoux MC, Nelson RJ. RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics. 1994;136:1421–1434. doi: 10.1093/genetics/136.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZX, Yano M, Yamanouchi U, Iwamoto M, Monna L, Hayasaka H, Katayose Y, Sasaki T. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 1999;19:55–64. doi: 10.1046/j.1365-313x.1999.00498.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Xu X, Lin F, Pan Q. Characterization of rice blast resistance genes in the Pik cluster and fine mapping of the Pik-p locus. Phytopathology. 2009;99:900–905. doi: 10.1094/PHYTO-99-8-0900. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. Molecular mapping of the blast resistance genes Pi2-1 and Pi51(t) in the durably resistant rice Tianjingyeshengdao. Phytopathology. 2012;102:779–786. doi: 10.1094/PHYTO-03-12-0042-R. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhao JM, Zhang LX, Wang P, Wang SW, Wang H, Wang XX, Liu ZH, Zheng WJ. Analysis of the diversity and function of the alleles of the rice blast resistance genes Piz-t, Pita and Pik in 24 rice cultivars. J Integr Agric. 2015 doi: 10.1016/s2095-3119(15)61207-2. [DOI] [Google Scholar]
- Wang F, Wang C, Liu P, Lei C, Hao W, Gao Y, Liu YG, Zhao K. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One. 2016;11:e0154027. doi: 10.1371/journal.pone.0154027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Fang W, Guan C, He W, Bao Y, Zhang H. Characterization and fine mapping of a blast resistant gene Pi-jnw1 from the japonica rice landrace Jiangnanwan. PLoS One. 2016;11(12):e0169417. doi: 10.1371/journal.pone.0169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K. S., Martinez C., Lentini Z., Tohme J., Chumley F. G., Scolnik P. A., Valent B. Rice Genetics Collection. 2008. Cloning a blast resistance gene by chromosome walking; pp. 669–674. [Google Scholar]
- Wu JL, Fan YY, Li DB, Zheng KL, Leung H, Zhuang JY. Genetic control of rice blast resistance in the durably resistant cultivar Gumei 2 against multiple isolates. Theor Appl Genet. 2005;111:50–56. doi: 10.1007/s00122-005-1971-2. [DOI] [PubMed] [Google Scholar]
- Wu YY, Bao YM, Xie LJ, Su YY, Chu RZ, He WW, Huang J, Wang JF, Zhang HS. Fine mapping and identification of blast resistance gene Pi-hk1 in a broad-spectrum resistant japonica rice landrace. Phytopathology. 2013;103(11):1162–1168. doi: 10.1094/PHYTO-02-13-0044-R. [DOI] [PubMed] [Google Scholar]
- Xiao W, Yang Q, Wang H, Guo T, Liu Y, Zhu X, Chen Z. Identification and fine mapping of a resistance gene to Magnaporthe oryzae in a space-induced rice mutant. Mol Breed. 2011;28:303–312. [Google Scholar]
- Xiao W, Yang Q, Wang H, Duan J, Guo T, Liu Y, Zhu X, Chen Z. Identification and fine mapping of a major R gene to Magnaporthe oryzae in a broad-spectrum resistant germplasm in rice. Mol Breed. 2012;30:1715–1726. [Google Scholar]
- Xiao W, Yang Q, Sun D, Wang H, Guo T, Liu Y, Zhu X, Chen Z. Identification of three major R genes responsible for broad-spectrum blast resistance in an indica rice accession. Mol Breed. 2015;35:49. [Google Scholar]
- Xiu-Ying H, Liu X, Wang L, Wang L, Lin F, Cheng Y, Pan Q. Identification of the novel recessive gene pi55(t) conferring resistance to Magnaporthe oryzae. Sci China Life Sci. 2012;55(2):141–149. doi: 10.1007/s11427-012-4282-2. [DOI] [PubMed] [Google Scholar]
- Xu X, Chen H, Fujimura T, Kawasaki S. Fine mapping of a strong QTL of field resistance against rice blast Pikahei-1(t) from upland rice Kahei utilizing a novel resistance evaluation system in the greenhouse. Theor Appl Genet. 2008;117:997–1008. doi: 10.1007/s00122-008-0839-7. [DOI] [PubMed] [Google Scholar]
- Xu X, Hayashi N, Wang CT, Kato H, Fujimura T, Kawasaki S. Efficient authentic fine mapping of the rice blast resistance gene Pik-h in the Pik cluster, using new Pik-h differentiating isolates. Mol Breed. 2008;22:289–299. [Google Scholar]
- Xu X, Hayashi N, Wang CT, Fukuoka S, Kawasaki S, Takasuji H, Jiang CJ. Rice blast resistance gene Pikahei-1(t), a member of a resistance gene cluster on chromosome 4, encodes a nucleotide-binding site and leucine-rich repeat protein. Mol Breed. 2014;34:691–700. [Google Scholar]
- Yadav MK, Aravindan S, Ngangkham U, Subudhi HN, Bag MK, Adak T, Munda S, Samantaray S, Jena M. Use of molecular markers in identification and characterization of resistance to rice blast in India. PLoS One. 2017;12(4):e0176236. doi: 10.1371/journal.pone.0176236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Lin F, Wang L, Pan Q. Identification and mapping of Pi41 a major gene conferring resistance to rice blast in the Oryza sativa subspecies indica reference cultivar 93-11. Theor Appl Genet. 2009;118:1027–1034. doi: 10.1007/s00122-008-0959-0. [DOI] [PubMed] [Google Scholar]
- Yu ZH, Mackill DJ, Bonman JM, Tanskley SD. Tagging of blast resistance in rice via linkage to RFLP markers. Theor Appl Genet. 1991;81:471–476. doi: 10.1007/BF00219436. [DOI] [PubMed] [Google Scholar]
- Yu ZH, Mackill DJ, Bonman JM, McCouch SR, Guiderdoni E, Notteghem JL, Tanksley SD. Molecular mapping of genes for resistance to rice blast (Pyricularia grisea Sacc.) Theor Appl Genet. 1996;93:859–863. doi: 10.1007/BF00224086. [DOI] [PubMed] [Google Scholar]
- Yuan B, Zhai C, Wang W, Zeng X, Xu X, Hu H, Lin F, Wang L, Pan Q. The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor Appl Genet. 2011;122:1017–1028. doi: 10.1007/s00122-010-1506-3. [DOI] [PubMed] [Google Scholar]
- Zenbayashi K, Fukuoka S, Katagiri S, Fujisawa M, Matsumoto T, Ashizawa T, Koizumi S. Genetic and physical mapping of the partial resistance gene Pi34 to blast in rice. Phytopathology. 2007;97:598–602. doi: 10.1094/PHYTO-97-5-0598. [DOI] [PubMed] [Google Scholar]
- Zeng XS, Yang XF, Zhao ZH, Lin F, Wang L, Pan Q. Characterization and fine mapping of the rice blast resistance gene Pia. Sci China Life Sci. 2011;54:372–378. doi: 10.1007/s11427-011-4154-1. [DOI] [PubMed] [Google Scholar]
- Zhai C, Lin F, Dong Z, He X, Yuan B, Zeng X, Wang L, Pan Q. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 2011;189:321–334. doi: 10.1111/j.1469-8137.2010.03462.x. [DOI] [PubMed] [Google Scholar]
- Zhao H, Wang X, Jia Y, Minkenberg B, Wheatley M, Fan J, Jia MH, Famoso A, Edwards D, Wamishe Y, Valent B, Wang GL, Yang Y. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat Commun. 2018 doi: 10.1038/s41467-018-04369-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Wang Y, Wang L, Ma Z, Zhao J, Wang P, Zhang L, Liu Z, Lu X. Genetic mapping and molecular marker development for Pi65(t), a novel broad-spectrum resistance gene to rice blast using next-generation sequencing. Theor Appl Genet. 2016;129:1035–1044. doi: 10.1007/s00122-016-2681-7. [DOI] [PubMed] [Google Scholar]
- Zhou JH, Wang JL, Xu JC, Lei CL, Ling ZZ. Identification and mapping of a rice blast resistance gene Pi-g(t) in the cultivar Guangchangzhan. Plant Pathol. 2004;53(2):191–196. [Google Scholar]
- Zhou B, Qu S, Liu G, Dolan M, Sakai H, Lu G, Bellizzi M, Wang GL. The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol Plant Microbe Interact. 2006;19:1216–1228. doi: 10.1094/MPMI-19-1216. [DOI] [PubMed] [Google Scholar]
- Zhou X, et al. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc Natl Acad Sci USA. 2018;115(12):3174–3179. doi: 10.1073/pnas.1705927115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Wang L, Pan Q. Identification and characterization of a new blast resistance gene located on rice chromosome 1 through linkage and differential analyses. Phytopathology. 2004;94:515–519. doi: 10.1094/PHYTO.2004.94.5.515. [DOI] [PubMed] [Google Scholar]
- Zhu X, Chen S, Yang J, Zhou S, Zeng L, Han J, Jing S, Ling W, Pan Q. The identification of Pi50(t), a new member of the rice blast resistance Pi2/Pi9 multigene family. Theor Appl Genet. 2012;124:1295–1304. doi: 10.1007/s00122-012-1787-9. [DOI] [PubMed] [Google Scholar]
- Zhuang JY, Lu J, Qian HR, Lin HX, Zheng KL (1998) Tagging of blast resistance gene(s) to DNA markers and marker-assisted selection (MAS) in rice improvement. In: IAEA-TECDOC:1010. International Atomic Energy Agency (IAEA)
- Zhuang JY, Ma WB, Wu JL, Chai RY, Lu J, Fan YY, Jin MZ, Leung H, Zheng KL. Mapping of leaf and neck blast resistance genes with resistance gene analog, RAPD and RFLP in rice. Euphytica. 2002;128(3):363–370. [Google Scholar]