Abstract
The Nopp140 gene of Drosophila maps within 79A5 of chromosome 3. Alternative splicing yields two variants. DmNopp140 (654 residues) is the sequence homolog of vertebrate Nopp140. Its carboxy terminus is 64% identical to that of the prototypical rat Nopp140. DmNopp140-RGG (688 residues) is identical to DmNopp140 throughout its first 551 residues, but its carboxy terminus contains a glycine/arginine-rich domain that is often found in RNA-binding proteins such as vertebrate nucleolin. Both Drosophila variants localize to nucleoli in Drosophila Schneider II cells and Xenopus oocytes, specifically within the dense fibrillar components. In HeLa cells, DmNopp140-RGG localizes to intact nucleoli, whereas DmNopp140 partitions HeLa nucleoli into phase-light and phase-dark regions. The phase-light regions contain DmNopp140 and endogenous fibrillarin, whereas the phase-dark regions contain endogenous nucleolin. When coexpressed, both Drosophila variants colocalize to HeLa cell nucleoli. Both variants fail to localize to endogenous Cajal bodies in Xenopus oocyte nuclei and in HeLa cell nuclei. Endogenous HeLa coilin, however, accumulates around the periphery of phase-light regions in cells expressing DmNopp140. The carboxy truncation (DmNopp140ΔRGG) also fails to localize to Cajal bodies, but it forms similar phase-light regions that peripherally accumulate endogenous coilin. Conversely, we see no unusual accumulation of coilin in cells expressing DmNopp140-RGG.
INTRODUCTION
Our traditional understanding of nucleolar function has been the multistage biosynthesis of ribosomes (reviewed by Busch and Smetana, 1970; Hadjiolov, 1985). rRNA transcription occurs on the boundaries between the fibrillar centers (FCs) and the dense fibrillar components (DFCs) (Dundr and Raska, 1993; Hozák et al., 1994; Shaw and Jordan, 1995). Site-specific ribose methylation (reviewed by Smith and Steitz, 1997; Weinstein and Steitz, 1999) and pseudouridine conversion (reviewed by Bachellerie and Cavaillé, 1997) are two posttranscriptional modifications of the pre-rRNA that occur within the DFCs. Cleavage of the pre-rRNA, also within the DFCs, yields mature 18S, 5.8S, and 28S rRNAs (Kass et al., 1990; Mougey et al., 1993; Peculis and Steitz, 1993). Approximately 80 ribosomal proteins assemble with these rRNAs and the extranucleolar-expressed 5S rRNA to form the small (18S rRNA) and large (5S, 5.8S, and 28S) ribosomal subunits. Immature subunits appear in the peripheral granular components of nucleoli before their further maturation and export to the cytoplasm. Besides ribosome biosynthesis, however, novel functions have been ascribed to nucleoli. They include nuclear import and export, gene silencing, assembly of signal recognition particles, modifications to small nonnucleolar RNAs, cell cycle regulation, and aging (reviewed by Pederson, 1998; Garcia and Pillus, 1999; Johnson et al., 1999; Pederson and Politz, 2000; Visintin and Amon, 2000). Since the early 1970s (Orrick et al., 1973), several nonribosomal nucleolar proteins have been described in yeasts and metazoans (reviewed by Olson, 1990; Olson et al., 2000). How these various proteins participate in the diversified tasks of the nucleolus remains the focus of intense investigation.
Nucleolin, fibrillarin, B23, and Nopp140 are the most extensively studied nucleolar proteins in vertebrates. Nucleolin (110 kDa/pI 5.5; its homologs are Nsr1 in Saccharomyces cerevisiae and gar2 in Schizosaccharomyces pombe) is modular in composition (Lapeyre et al., 1987). Its amino terminal third contains alternating acidic and basic domains, its central domain contains four consensus RNA-binding domains (RBDs; Burd and Dreyfuss, 1994), and its carboxy terminus is rich in glycine and dimethylarginine residues that form several Arg-Gly-Gly (RGG) motifs. Such RGG motifs are common to a variety of RNA-binding proteins (Burd and Dreyfuss, 1994). Nucleolin interacts with nascent pre-rRNA (Herrera and Olson, 1986; Ghisolfi-Nieto et al., 1996) to facilitate early site-specific cleavages and perhaps other processing events (Ginisty et al., 1998; reviewed by Ginisty et al., 1999). Fibrillarin (34 kDa/pI 8.5; its homolog is Nop1 in yeast) is intimately associated with box C/D small nuclear ribonucleoprotein particles (snoRNP) for either cleavage or site-specific methylation of the pre-rRNA. In fact, the fibrillarin homolog in the hyperthermophile Methanococcus jannaschii may be the methyltransferase itself (Wang et al., 2000). Like nucleolin, fibrillarin contains RGG motifs, but within its amino terminus instead of its carboxy terminus. B23 (38 kDa/pI 5.1) is a putative ribosome assembly factor (reviewed by Olson et al., 2000). It contains two acidic regions, but no RBDs or RGG motifs. B23 nevertheless binds nucleic acids (Dumbar et al., 1989; Wang et al., 1994) and displays nuclease activity (Herrera et al., 1995; Savkur and Olson, 1998) as well as chaperone functions (Szebeni and Olson, 1999). Interestingly, B23 may also participate in centrosome duplication in early to mid-G1 (Zatsepina et al., 1999; Okuda et al., 2000).
Meier and Blobel (1990, 1992) described the prototypical nucleolar phosphoprotein of 140 kDa (Nopp140) in rat. More recently, vertebrate homologs have been identified in Xenopus (xNopp180; Cairns and McStay, 1995) and human (p130; Pai et al., 1995; Chen et al., 1999). Other vertebrate proteins similar and perhaps identical to Nopp140 have been described (Pfeifle and Anderer, 1984; Pfeifle et al., 1986; Vandelaer and Thiry, 1998; Isaac et al., 2000). Nopp140 contains a large central region consisting of several (10–18) alternating acidic and basic regions. The acidic regions contain exclusively aspartic acid, glutamic acid, and serine. The serines are phosphorylated in vivo by casein kinase type II (CKII) enzymes (Meier, 1996; Li et al., 1997), and the resulting phosphoserine residues lend to the acidic property of the region. The interspersed basic regions are rich in lysine, alanine, and proline. A conserved carboxy terminus follows the central acidic and basic domain (Meier, 1996). A putative protein kinase C phosphorylation site is conserved within the carboxy domain, suggesting that Nopp140 is a terminal substrate in signal transduction phosphorylation cascades (Meier, 1996). Like B23, Nopp140 contains no consensus RBDs or RGG motifs. Unlike B23, however, Nopp140 does not appear to be an RNA-binding protein. Srp40 (41 kDa) in S. cerevisiae is the immunological and structural homolog of mammalian Nopp140 (Meier, 1996). Srp40 consists of two relatively long acidic regions that alternate with two short basic regions. The carboxy terminal region of Srp40 is 59% identical to the prototypical terminus in rat Nopp140. Deletion of the SRP40 gene causes minor growth impairment, whereas overproduction of Srp40 causes severe growth impairment (Meier, 1996).
The precise functions of Nopp140 remain uncertain, and our best understanding regarding its function derives from its associations with other nuclear and nucleolar proteins. First, Nopp140 localizes to nucleolar DFCs (Meier and Blobel, 1992). Reports indicate that Nopp140 interacts with the largest subunit of RNA polymerase I (RPA194) (Chen et al., 1999). It may also interact with C/EBPβ and TFIIB to activate the alpha-1-acid glycoprotein gene (agp) in mammalian liver (Miau et al., 1997). Based upon these observations, Nopp140 may function in transcription regulation of rRNA genes within nucleoli and certain nonribosomal genes presumably outside the nucleolus.
Alternatively, Nopp140 may function in pre-rRNA processing and ribosome biogenesis within the DFCs. Nopp140 associates with both classes of mammalian snoRNP particles (box H/ACA and box C/D snoRNPs) as determined by coimmunoprecipitations (Yang et al., 2000). For example, Nopp140 associates in stoichiometric amounts with rat Nopp140-associated protein of 57 kDa (NAP57) (Meier and Blobel, 1994), a protein component of box H/ACA snoRNPs. Interestingly, rat NAP57 shares conserved regions with the S. cerevisiae Cbf5 gene product that may play several roles in the transcription, processing, and pseudouridylation of yeast pre-rRNA (Cadwell et al., 1997; Lafontaine et al., 1998). The human homolog of NAP57, called dyskerin, is a product of the DKC1 gene. Mutations in DKC1 lead to dyskeratosis congenita, a rare X-linked (Xq28) recessive disease in which progressive bone marrow failure is the primary cause of mortality. The Drosophila homolog of NAP57 is Nop60B (Phillips et al., 1998), the product of the minifly (mfl) gene that maps to the right arm of chromosome 2 within region 60B-C (thus its designation Nop60B) (Giordano et al., 1999). Mutations in mfl lead to reduced body size, abnormal eggs, and reduced fertility. Interestingly, Cbf5, NAP57, dyskerin, and Nop60B are all related to TruB, a pseudouridine synthase for tRNAs in Escherichia coli. The implication herein is that Nopp140 may be involved with pseudouridine conversion by the box H/ACA snoRNP particles. The yeast homolog Srp40 also associates with box H/ACA snoRNP particles (Yang et al., 2000). Besides box H/ACA snoRNPs, Nopp140 coprecipitates with components of the box C/D snoRNPs. Specifically, Nopp140 associates with fibrillarin and the newly characterized mammalian NAP65 (Nop5/58p in yeast), both of which are components of box C/D snoRNPs (Yang et al., 2000).
In addition to these associations, vertebrate Nopp140 shuttles between the nucleus and the cytoplasm (Meier and Blobel, 1992). This observation is consistent with a chaperone function of Nopp140 in the transport of karyophilic proteins into the nucleus, or in the export of nuclear products to the cytoplasm. Phosphorylation may regulate this chaperone function. That is, Nopp140 binds nuclear localization signal (NLS)-containing proteins in vitro when its multiple serines are phosphorylated by CKII, whereas the dephosphorylated version of Nopp140 fails to do so (Meier and Blobel, 1990).
One of the most intriguing interactions exists between Nopp140 and p80 coilin (Isaac et al., 1998). Coilin is a marker protein for nuclear organelles that were traditionally called coiled bodies (Andrade et al., 1991), but recently renamed Cajal bodies (CBs) (Gall et al., 1999) in honor of S.R. Ramón y Cajal who first described the structures as accessory organelles to the nucleoli (Ramón y Cajal, 1903). CBs are often found in the vicinity of nucleoli, adjoining nucleoli, or within the nucleoli of plant and animal cells (reviewed by Matera, 1999; Gall, 2000). The molecular composition of CBs continues to unfold (Bohmann et al., 1995b; Gall et al., 1999; Matera, 1999; Gall, 2000). Besides the aforementioned p80 coilin, somatic cell CBs contain pre-mRNA splicing components such as snRNAs U1, U2, U4, U5, U6, the trimethylguanosine cap epitope, and Sm proteins (Matera, 1999; Gall, 2000). CBs also contain nucleolar proteins Nopp140, NAP57, fibrillarin, Gar1p, topoisomerase I, and the ribosomal protein, S6. CBs contain snoRNAs U3 and U8 (Narayanan et al., 1999; Speckman et al., 1999), but somatic cell CBs do not contain nucleolin, B23, or any rRNA. CBs, therefore, are not directly involved in ribosome biogenesis. Interactions between CBs and nucleoli are well established (Bohmann et al., 1995a; Isaac et al., 1998; Sleeman et al., 1998; Platani et al., 2000). Deletion mutants of p80 coilin altered CBs and nucleoli (Bohmann et al., 1995a; Sleeman et al., 1998). Conversely, deletion mutants of Nopp140 caused dominant negative effects on the normal distribution of nucleolar proteins within nucleoli and CBs (Isaac et al., 1998). Physical interactions were observed by Platani et al. (2000) who used time-lapse fluorescence microscopy to show that CBs move to and from nucleoli. Association of Nopp140 with both nucleoli and CBs supports the hypothesis that Nopp140 shuttles RNA processing complexes (snoRNPs) to and from nucleoli.
The conclusion from these introductory comments is that Nopp140 appears to have multiple and diverse functions. Herein, we introduce two splice variants of Nopp140 in Drosophila melanogaster that differ in their carboxy ends. DmNopp140 appears to be the sequence homolog of vertebrate Nopp140 in overall peptide domain composition and arrangement. DmNopp140-RGG is identical to DmNopp140 throughout most of its primary sequence (residues 1–551), but its carboxy terminal tail contains an RGG domain that is highly reminiscent of the carboxy RGG domain in vertebrate nucleolin (Lapeyre et al., 1987). As far as we know, this is the first example of a Nopp140-like protein that contains a peptide domain typically reserved for RNA-binding proteins (Burd and Dreyfuss, 1994). Comparative molecular and genetic analyses of the two Drosophila Nopp140 variants should provide valuable insights to Nopp140's diverse functions, while at the same time expanding our knowledge of nucleolar functions, both traditional and novel.
MATERIALS AND METHODS
Recovery, Sequencing, and Cloning of Drosophila Nopp140 cDNAs
We used standard molecular biology techniques (Ausubel et al., 1987–1995) to screen aliquots of a D. melanogaster stage 10 egg chamber cDNA lambda phage library. The probe was a random primed, 32P-labeled subclone of our Xenopus nucleolin cDNA (Rankin et al., 1993; accession number X63091). Specifically, the subclone was a 444-base pair fragment that spanned the NcoI site at the translation start site to a downstream PstI site. It encodes most of the alternating acidic and basic regions within the amino terminal third of the smaller of two Xenopus nucleolin proteins (Meβmer and Dreyer, 1993, for a comparison of the two nucleolin proteins in X. laevis). We used low stringency washes (2× SSC without SDS at room temperature) to detect related Drosophila cDNA sequences. Four strongly positive plaques were picked and rescreened, again under low stringency to establish clonal purity. Individual plaques were amplified, and phage DNA was prepared and digested with EcoRI to liberate the Drosophila cDNAs from the lambda genome. The Drosophila inserts were ligated into pBluescript KS(+) (Strategene, La Jolla, CA) and sequenced in both directions by using Sanger's dideoxy method for DNA sequencing. We used Sequenase (USB, Cleveland, OH) according to the manufacturer's recommendations.
One of the Drosophila inserts that displayed a strong hybridization signal with the Xenopus probe was only 787 base pairs in length (B72A). Its deduced translation product contained alternating acidic and basic regions, and thus it was highly reminiscent of the alternating acidic and basic regions within vertebrate nucleolin and Nopp140. We used this insert to rescreen the Drosophila cDNA library, this time using higher stringency washes (0.5× SSC, 0.1× SDS at 60°C). Rescreening identified several larger inserts that we sequenced. One of the inserts provided a nearly full-length cDNA that encoded a Nopp140-like protein. The deduced protein sequence, however, contained a RGG carboxy terminus, and we refer to the protein as DmNopp140-RGG. To provide the missing 5′ end of the cDNA, we obtained an expressed sequence tag (LD10913) from Genome Systems (St. Louis, MO) that proved to be a complete cDNA encoding DmNopp140-RGG (our accession number AF162774).
While sequencing the library's cDNA that encodes DmNopp140-RGG, the Berkeley Drosophila Genome Project (BDGP) published the Drosophila genome (FlyBase, 1999). We used the cDNA sequence for DmNopp140-RGG in a BLAST search of the genome and found the Nopp140 gene in polytene region 79A5 on the proximal left arm of chromosome 3. The BDGP predicted two conceptual transcripts from this gene. They are splice variants that encode DmNopp140-RGG (CT22845) and DmNopp140 (CT22833). We next obtained several additional expressed sequence tags (ESTs) from Research Genetics (Huntsville, AL) and used restriction enzyme digestion patterns to determine which of the several clones expressed the two proteins (Figure 1B). Full-length cDNAs encoding DmNopp140 (EST SD10348) and DmNopp140-RGG (EST LD10913) were ligated into pEGFP-C3 (CLONTECH, Palo Alto, CA). This allowed us to express the red-shifted version of the green fluorescent protein (EGFP) fused in frame to the amino terminal ends of DmNopp140 and DmNopp140-RGG. To engineer a carboxy terminal truncation of DmNopp140-RGG (referred to as DmNopp140ΔRGG), two polymerase chain reaction (PCR) primers were designed to amplify the DmNopp140-RGG cDNA (LD10913) except for the sequences that encode the RGG tail. The upstream primer was complementary to the noncoding strand, and it contained an EcoRI site (underlined) just upstream of the ATG start codon (in bold) (5′-GCGAATTCTCATGACAGACCTGCTAAAGATAGCC-3′). The downstream primer (5′-AAGGATCCTTATCCGTTGTTGTGCTTCTTAAAGTC-G-3′) was complementary to the coding strand, and it contained a stop codon (in bold) that would have normally encoded amino acid residue 562 of DmNopp140-RGG. The underlined BamHI site was included for cloning purposes. The resulting PCR product was digested with EcoRI and BamHI and then ligated into the pEGFP-C3 vector at the respective restriction sites. The EcoRI site in the upstream PCR primer was positioned such that the ATG start codon was in frame with the sequences that encode the EGFP. The region of DmNopp140-RGG that is deleted in DmNopp140ΔRGG is shown as italicized letters in Figure 1C. All recombinant plasmids were purified twice by cesium banding before their use in transfection assays (see below). The cytomegalovirus immediate early promoter within pEGFP-C3 controls expression of the fusion proteins even in Drosophila cells (Echalier, 1997).
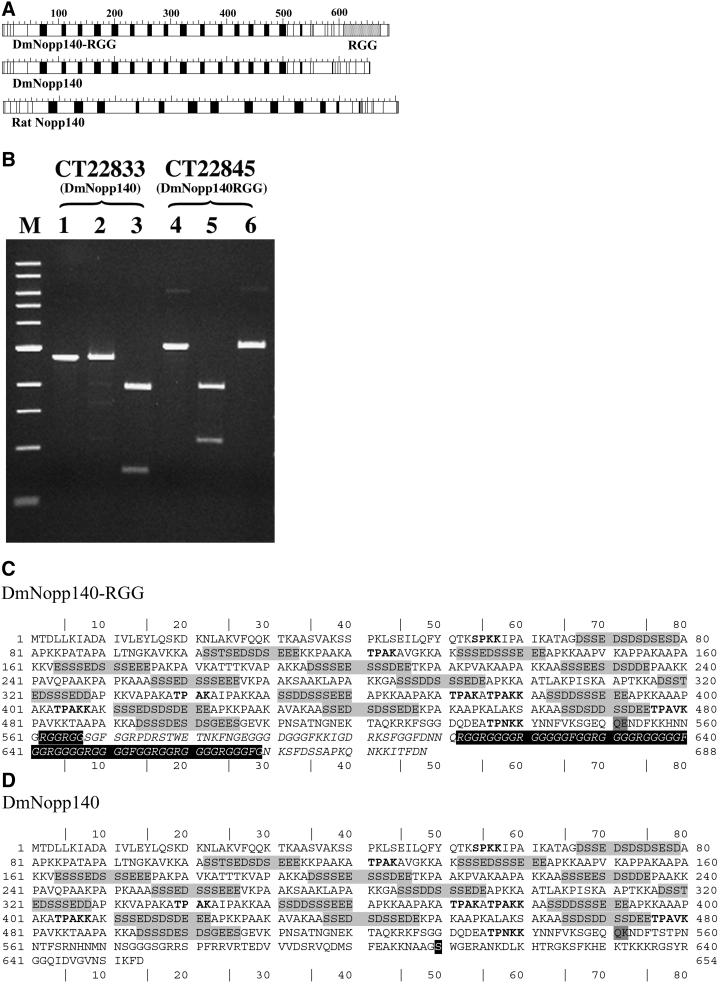
Figure 1.
Sequence comparisons of DmNopp140-RGG and DmNopp140. (A) Acidic (black boxes) and basic (white boxes) regions alternate within the central domains of both Drosophila variants. This alternating pattern is similar to that within rat Nopp140. DmNopp140 is the Drosophila homolog of mammalian Nopp140, whereas DmNopp140-RGG is unique due to its carboxyl-terminal RGG domain. (B) Expressed sequence tags exist for both Drosophila variants. Restriction maps allowed us to predict digestion patterns and thus to verify the clones. The purified cDNA insert that encodes conceptual transcript 22833 (DmNopp140) was left undigested (lane 1), incubated with PvuI that failed to cut as predicted (lane 2), or digested with AvaI that generated two bands of predicted size (lane 3). The purified cDNA insert that encodes conceptual transcript 22845 (DmNopp140-RGG) was left undigested (lane 4), incubated with PvuI that generated two predicted fragments (lane 5), or incubated with AvaI that failed to cut as expected (lane 6). The digestion patterns confirmed the existence of two separate cDNA clones. (C) The deduced amino acid sequence of DmNopp140-RGG. CT22845 consists of three exons. The first encodes amino acids 1–34, the second encodes 35–551, and the third encodes 552–688. The serine-rich acidic regions are highlighted in light gray. MPF phosphorylation motifs are in bold. The RGG domain spans residues 612–669. Two additional upstream RGG motifs reside at 562–567. (D) The deduced amino acid sequence of DmNopp140. CT22833 consists of four exons. The first two exons are identical to those in CT22845. Thus, the two proteins are identical up to residue 551 (dark gray box), after which their sequences diverge. The third exon in CT22833 encodes residues 552–604 and the fourth exon encodes residues 605–654. The carboxy terminus of DmNopp140 is 64% identical to the carboxy terminus of rat Nopp140. A highly conserved serine residue within this terminus (residue 610 in the black box) is a putative substrate site for cAMP-dependent protein kinase.
Bacterial Expression and In Vitro Phosphorylation
The full-length cDNA encoding DmNopp140-RGG was removed from EST LD10913 by using a BspHI site that is located at the translation start site and a BamHI site within the 3′ untranslated region. The cDNA was ligated into pET30 (Novagen, Madison, WI) at the NcoI (compatible with BspHI) and BamHI sites such that the DmNopp140-RGG coding sequence was positioned in frame with sequences within pET30 that encode the His tag. We transformed E. coli strain JM109 DE3 pLysS with the recombinant pET30 plasmid. To express DmNopp140-RGG, 5 ml of an overnight culture was transferred to 1 liter of LB broth. The culture was allowed to grow to an OD600 of 0.6 at which point isopropyl β-d-thiogalactoside was added to a final concentration of 1 mM. Cells were incubated at 37°C for an additional 3 h and then harvested by centrifugation at ∼6000 × g. Cells were resuspended in 30 ml of His binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl pH 7.9) supplemented with 0.1 mM phenylmethylsulfonyl fluoride, 1.0 μg/ml pepstatin, and 1.0 μg/ml leupeptin. Cells were lysed by sonication for 2 min on ice. The cell lysate was cleared by centrifugation at 25,000 × g for 20 min at 4°C. The supernatant was passed through a 0.4-μm syringe filter directly onto a HIS-tag column by using Novagen's recommendations. Eluate was collected and dialyzed overnight at 4°C in 75 mM KCl, 10 mM Tris-HCl, pH 7.2, 1 mM EDTA, and 0.1 mM phenylmethylsulfonyl fluoride. EDTA was included in the dialysis buffer to chelate nickel that leaches off the column. We concentrated the protein by placing the dialysis bag (molecular weight cut-off of 13,000–14,000) into solid polyvinylpyrrolidone (average molecular weight = 360,000). This resulted in rapid dehydration without adversely changing ionic strength.
The enriched protein was phosphorylated in vitro by using γ-labeled [32P]ATP at 800 Ci/mmol (ICN, Costa Mesa, CA) and either casein kinase II or Cdk1/cyclin B protein kinase (mitosis-promoting factor [MPF]). Both enzymes were purchased from New England Biolabs (Beverly, MA). Phosphorylations were performed according to the manufacturer's recommendations.
Cell Culture and Transient Transfection
All media and antibiotics were from Invitrogen (Carlsbad, CA). Drosophila Schneider II cells were grown in Schneider's Drosophila medium supplemented with 10% fetal calf serum (FCS) and 50 μg/ml penicillin-streptomycin-glutamine in a 25°C ambient air incubator. HeLa cell stocks were maintained at 37°C in 5% CO2 by using DMEM that was supplemented with 10% FCS and 50 μg/ml Gentamicin.
For transfection of HeLa cells, ∼1 × 105 cells were grown on 22- × 22-mm coverslips in six-well culture plates (Falcon 3046; Falcon Plastics, Oxnard, CA) at 37°C in a 5% CO2 atmosphere. Transfection was by DNA-calcium phosphate precipitation by using the N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid method of Chen and Okayama (1988). After adding the DNA precipitate, cells were incubated at 35°C in 3% CO2 for 12–18 h (overnight). Cells were then washed twice with 1× phosphate-buffered saline (PBS) and either fixed with 2.0% paraformaldehyde in 0.6× PBS for 1.5 h at room temperature, or recultured in DMEM with 10% FCS at 37°C in a 5% CO2 atmosphere for an additional 24 h before 1× PBS washing and 2% paraformaldehyde fixation. Transfection methods used for mammalian cells were used for the Schneider II cells, except that the Drosophila cells were maintained in Schneider's Drosophila medium and in ambient air at 25°C throughout all procedures.
Antibodies and Antibody Staining
After formaldehyde fixation, cells (still attached to coverslips) were washed several times with 1× PBS and then treated with 0.1% Triton X-100 in 1× PBS for 5 min. The cells were again washed with 1× PBS and blocked with 10% horse serum or 3% bovine serum albumin in 1× PBS. Anti-human nucleolin was a gift from Dr. Benigno Valdez (Baylor College of Medicine, Houston, TX). The S4 anti-human fibrillarin (Lischwe et al., 1985; Ochs et al., 1985) was a gift from Dr. Robert Ochs (Scripts Research Institute, La Jolla, CA). The anti-human p80 coilin rabbit serum (R288) was developed by Andrade et al. (1993), but provided to us by Dr. Joe Gall (Carnegie Institution of Washington, Baltimore, MD). Primary antibodies were diluted appropriately in 10% horse serum or 3% bovine serum albumin and placed onto the coverslips for 1–2 h at room temperature. Respective affinity-purified secondary antibodies (ICN, Costa Mesa, CA) were tagged either with fluorescein or rhodamine.
In Vitro mRNA Production and Oocyte Injection
We prepared synthetic mRNAs encoding either the green fluorescent protein (GFP)-tagged DmNopp140 or the GFP-tagged DmNopp140-RGG by using a T7 Message Machine kit from Ambion (Austin, TX). DNA inserts encoding the GFP tag along with either DmNopp140-RGG or DmNopp140 were ligated into pBluescript (Stratagene) to make use of its T7 promoter for in vitro transcription as described by Ambion. We injected the transcripts into Xenopus oocytes by using procedures described by Heine et al. (1993). After injection the oocytes were cultured overnight at 18°C in OR2 medium. The next day, oocyte nuclear contents were prepared for light microscopy as described by Gall (1998).
RESULTS
Recovery and Sequence of DmNopp140-RGG
In an effort to recover Drosophila cDNAs that encode either nucleolin or Nopp140, we originally used a 5′ subclone of our Xenopus nucleolin cDNA (Rankin et al., 1993) to screen a stage 10 egg chamber cDNA library under low stringency. The Xenopus subclone encodes the majority of the alternating acidic and basic regions within the amino terminal third of Xenopus nucleolin. We recovered and sequenced several overlapping Drosophila inserts. Our full-length cDNA (accession number AF162774) encodes what we refer to as DmNopp140-RGG. The Drosophila protein resembles vertebrate Nopp140 (Meier, 1996) in that it contains a long central series of alternating acidic and basic regions (Figure 1A).
We used the DmNopp140-RGG cDNA sequence in a BLAST search of the BDGP. Conceptual gene CG7421 maps within polytene region 79A5 on the left arm of polytene chromosome 3 (see the various links at http://hedgehog.lbl.gov:8000/cgi-bin/annot/gene?CG7421). The pre-mRNA is 3247 nucleotides with the translation start codon positioned at residues 88–90. The BDGP lists two splice variants for the gene, conceptual transcripts CT22833 and CT22845. CT22845 encodes DmNopp140-RGG, and its sequence is nearly identical to our cDNA sequence. Three exons constitute CT22845; they include nucleotides 88–186, 389-1942, and 2580–2993 of the pre-mRNA. These exons encode amino acids 1–33, 34–551, and 552–688, respectively (Figure 1C).
Conversely, CT22833 encodes DmNopp140 (Figure 1, A and D) with a carboxyl-terminal tail that is highly conserved among previously identified Nopp140 proteins (Meier, 1996; Table 1). Four exons constitute CT22833. The first two exons include nucleotides 88–186 and 389-1942, and again they encode amino acid residues 1–33 and 34–551 as in CT22845. The next two exons include nucleotides 2184–2342 and 3096–3248, and they, respectively, encode amino acids 552–604 and 605–654 within DmNopp140 (Figure 1D). Thus, the two splice variant mRNAs CT22845 and CT22833 are identical in their first two exons that encode amino acid residues 1–551. Due to alternative splicing, however, the two transcripts contain mutually exclusive exons that encode totally different carboxy termini. The two Drosophila proteins differ in sequence beginning at residue 552. The dark gray boxes in Figure 1, C and D, contain residue 551 that is common to both proteins and residue 552 that differs between the two proteins.
Table 1.
FASTA results relative to the carboxy terminus of DmNopp140
| Species/protein | % Identity | Compared overlap |
|---|---|---|
| Human Nopp140 | 65 | 97 amino acids |
| Rat Nopp140 | 64 | 97 amino acids |
| Xenopus Nopp180 | 62 | 77 amino acids |
| Yeast Srp40 | 40 | 103 amino acids |
Two lines of evidence from the BDGP indicate that both mRNAs are expressed. First, Genscan or Genie programs used by the BDGP suggest that CG7421 encodes both transcripts. Second, ESTs exist for both transcripts. We purchased several ESTs from Genome Systems and Research Genetics and used the predicted restriction enzyme digestion patterns (CT22833 vs. CT22845) to verify that at least one of the purchased clones encoded DmNopp140 versus DmNopp140-RGG (Figure 1B).
DmNopp140-RGG contains 688 amino acid residues, whereas DmNopp140 is 654 residues in length (Figure 1, C and D). The prototypical rat Nopp140 is longer with 704 residues. Both DmNopp140-RGG and DmNopp140 contain alternating acidic (light gray shading) and basic regions that constitute a large central domain in both proteins. The acidic regions within the two Drosophila proteins are similar to those within rat Nopp140; they are rich in glutamic acid, aspartic acid, and serine. Also like rat Nopp140, the basic regions in the two Drosophila variants are rich in alanine, proline, and lysine.
Two readily apparent differences exist between the rat protein and the two Drosophila proteins. The first is the number of alternating acidic and basic regions is greater in the Drosophila proteins (15 acidic regions) versus the rat protein (12 acidic regions). We note, however, that Xenopus xNopp180 has 18 acidic regions (Cairns and McStay, 1995). The second difference is that the overall length of the central acidic-basic domain in the two Drosophila variants is shorter compared with the central domains in the rat and Xenopus proteins. Although the differences may seem minor, they could explain some unexpected results pertaining to the failure of these proteins to localize to CBs in Xenopus oocytes and HeLa cells (see below).
The carboxy terminus of DmNopp140 is fairly well conserved across the eukaryotes (Meier, 1996; Isaac et al., 1998). FASTA results display 65 and 64% identity over 94 amino acid stretches when the carboxy tail of DmNopp140 (beginning at residue 552) is compared with that of human and rat Nopp140 proteins, respectively (Table 1). Furthermore, Meier (1996) and Isaac et al. (1998) described two separate subdomains that comprise the carboxy terminus of Nopp140 homologs. The first half of the carboxy terminus, referred to as Ca (Isaac et al., 1998), shows good homology among vertebrates (e.g., 54% identity between rat and Xenopus). The latter half of the carboxy domain, referred to as Cb, has good homology throughout the eukaryotes in general (e.g., 59% identity between rat and yeast) with even higher degrees of homology between the metazoans (e.g., 81% identity between rat and Xenopus) (Meier, 1996). Interestingly, the breakpoint between the peptide encoded by the third exon (residues 552–604 = DmNopp140-Ca) and the peptide encoded by the fourth exon (residues 605–654 = DmNopp140-Cb) correlates well with the first and second halves of rat Nopp140. That is, DmNopp140-Ca has 50% identity with the comparable sequence in both human and rat Nopp140, whereas DmNopp140-Cb has 78 and 76% identities with the comparable sequences in human and rat Nopp140s, respectively. These homology values for DmNopp140-Ca (50%) and DmNopp140-Cb (78%) are in good agreement with the reported metazoan homologies of 54 and 81% identity, respectively.
DmNopp140-RGG, on the other hand, contains an RGG domain near its carboxy terminus. Although two adjacent RGG motifs reside at position 562–567, most of the RGG motifs in DmNopp140-RGG lie within a defined domain spanning residues 612–669. We call this latter region the RGG domain. The RGG domain in DmNopp140-RGG contains 58 residues: 44 glycines, 10 arginines, and 4 phenylalanines. All 10 arginines within the Drosophila RGG domain are followed by at least two glycines. This domain of DmNopp140-RGG is very similar to the one near the carboxy terminus of vertebrate (Chinese hamster ovary; CHO) nucleolin (Lapeyre et al., 1987). The RGG domain in CHO nucleolin is 52 residues in length; it consists of 37 glycines, 10 arginines, and 5 phenylalanines (Lapeyre et al., 1987). Nine of the 10 arginines in the CHO RGG domain are followed by at least two glycines. Interestingly, the RGG domain in both DmNopp140 and vertebrate nucleolin does not constitute the very ends of the proteins; 19 residues follow the RGG domain in DmNopp140-RGG, whereas 12 residues follow the RGG domain in CHO nucleolin. The significance of this terminal carboxy tail remains unknown. We conclude that DmNopp140-RGG is novel; it appears to be a composite of Nopp140 throughout most of its length and a carboxy terminus rich in RGG motifs that are usually reserved for RNA-binding proteins such as nucleolin (Burd and Dreyfuss, 1994).
MPF and CKII In Vitro Phosphorylations
The deduced peptide sequence of DmNopp140-RGG indicated eight putative MPF (cdk1/cyclin B) phosphorylation sites within the repeating basic regions (Figure 1C, bold SPKK or TPAK sites). Similar sites have been mapped within the basic regions of vertebrate nucleolin (Belenguer et al., 1990; Peter et al., 1990; Zhu et al., 1999). To show that DmNopp140-RGG is an in vitro substrate for MPF, we expressed DmNopp140-RGG as an amino terminal His-tagged fusion protein in E. coli and then purified the protein by using a nickel affinity column. The purified protein was labeled in vitro by MPF in the presence of [ γ-32P]ATP (Figure 2, lanes A and A′). If it occurs in vivo, MPF phosphorylation of Nopp140 may regulate nucleolar disassembly during prophase, whereas its dephosphorylation may regulate nucleologenesis during telophase (Pai et al., 1995).
Figure 2.
In vitro MPF and CKII phosphorylation. Left, Coomassie-stained gel showing His-tagged DmNopp140-RGG that was expressed in E. coli and purified using nickel affinity columns. Proteolysis was unavoidable. Right, corresponding autoradiogram. Lanes A and A′, protein incubated with MPF and [γ-32P]-labeled ATP. Lanes B and B′, protein incubated with CKII and γ-labeled ATP. Lanes C and C′, purified protein incubated with γ-labeled ATP, but no enzyme. The same amount of protein was used in each assay. CKII phosphorylation greatly exceeds MPF phosphorylation, probably due to the greater number of CKII sites versus MPF sites. Excess phosphorylation by CKII resulted in a detectable shift in molecular weight (arrow).
Rat Nopp140 is a substrate for CKII enzymes (Meier, 1996). CKII specifically phosphorylates the multiple serine residues within the central acidic regions. CKII readily phosphorylated the His-tagged DmNopp140-RGG in vitro (Figure 2, lanes B and B′). The extent of phosphorylation was much greater with CKII than with MPF (Figure 2, compare lanes A′ and B′). Although we cannot rule out differences in enzyme activities, the enhanced phosphorylation by CKII versus MPF is consistent with the large number of CKII sites (∼82) versus the eight putative MPF sites. Phosphorylation by CKII retarded the mobility of the protein in the SDS-polyacrylamide gel (Figure 2, arrow in lanes B and B′) as previously described for the CKII phosphorylation of rat Nopp140 (Meier, 1996). Both Drosophila variants are identical at these MPF and CKII phosphorylation sites, and we fully expect that DmNopp140 will show the same phosphorylation profile as shown herein for DmNopp140-RGG. As with nucleolin (Csermely et al., 1993; Bonnet et al., 1996), CKII phosphorylation of Nopp140 in vivo probably occurs during interphase in response to growth signals.
Exogenous Expression in Drosophila Schneider II Cells
The respective cDNAs encoding either DmNopp140-RGG or DmNopp140 were ligated into expression vectors downstream and in frame with sequences encoding the GFP. We purified the plasmids by cesium chloride gradient ultracentrifugation and then used them to cotransfect Drosophila Schneider II cells (Figure 3). Strong upstream cytomegalovirus promoters directed constitutive transcription for all constructs.
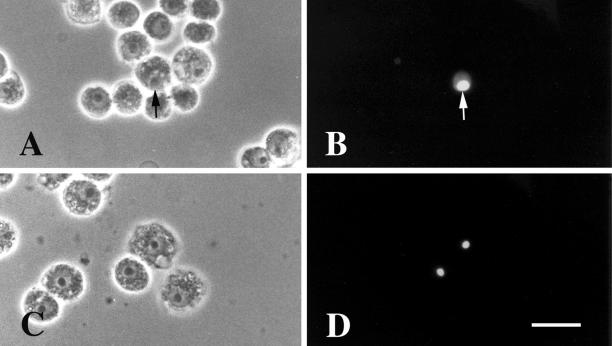
Figure 3.
Drosophila Schneider II cells transfected to express GFP-DmNopp140-RGG and GFP-DmNopp140. GFP-DmNopp140-RGG localized to nucleoli (arrows in A and B). No other nuclear bodies were apparent (B). GFP-DmNopp140 also localized to nucleoli (C and D). No other nuclear bodies were apparent (D). Bar (A–D), 40 μm.
Drosophila Schneider II cells contain one prominent nucleolus. GFP-DmNopp140-RGG (Figure 3, A and B) and GFP-DmNopp140 (Figure 3, C and D) localized to the single nucleolus in the relatively few cells that were transfected. The observation confirmed our expectations that both variants are nucleolar proteins in Drosophila cells. Although the number of transfected Schneider II cells was limiting, we observed no other nuclear structures (e.g., Cajal bodies) in cells exogenously expressing GFP-DmNopp140-RGG or GFP-DmNopp140.
Exogenous Expression in Xenopus Oocytes
Xenopus oocytes contain large, multiple extrachromosomal nucleoli that allow for unencumbered localization of nucleolar components (Shah et al., 1996). Toward this end, we injected Xenopus oocytes with synthetic transcripts encoding either GFP-DmNopp140 or GFP-DmNopp140-RGG. After mRNA injection, the oocytes were incubated at 18°C overnight to allow for protein synthesis and cytoplasmic to nuclear translocation, after which the nuclear contents were prepared for light microscopy according to Gall (1998). The nuclear preparations were stained with 4,6-diamidino-2-phenylindole (DAPI) to localize the rDNA within the FCs of the multiple nucleoli (Figure 4, C and H). Clearly, GFP-DmNopp140-RGG (Figure 4B) and GFP-DmNopp140 (Figure 4G) localized to the DFC regions immediately surrounding the FCs. This is the subcompartment of the DFC that is enriched for endogenous fibrillarin (Shah et al., 1996). We know that mammalian Nopp140 associates with fibrillarin in box C/D snRNPs (Yang et al., 2000). Thus, the localization of both Drosophila variants to this specific site within the DFCs of Xenopus nucleoli is consistent with the possibility that both Drosophila variants are intimately involved in snoRNP transport and/or in pre-rRNA processing.
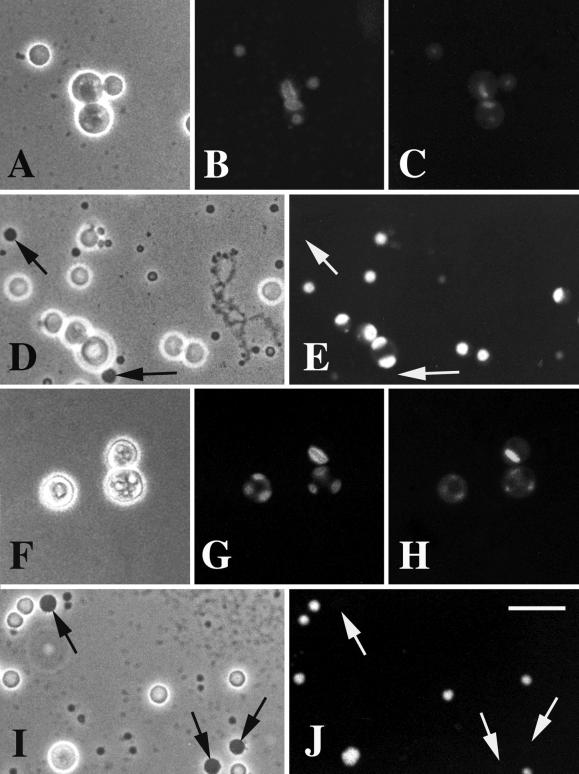
Figure 4.
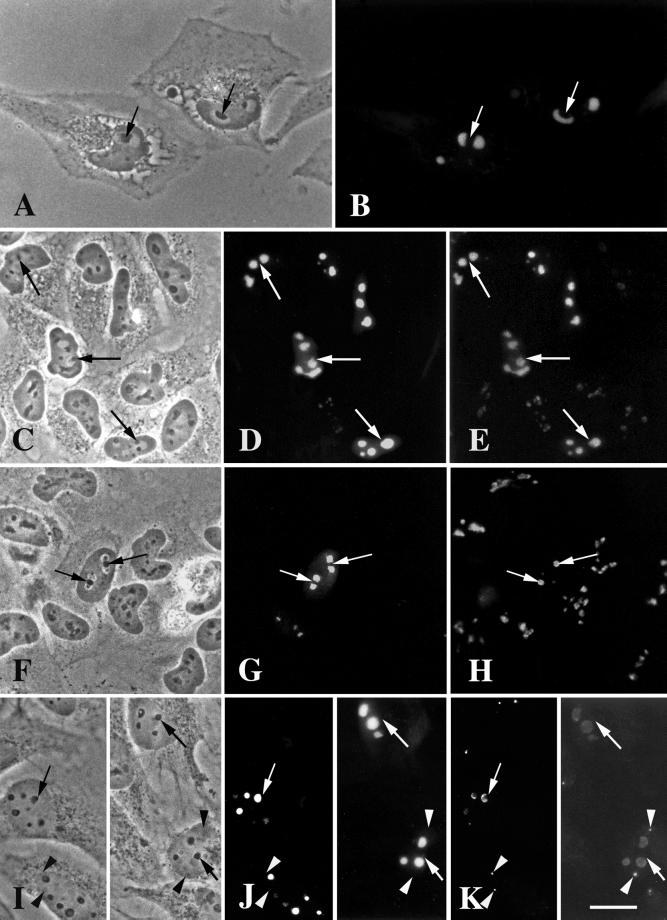
DmNopp140-RGG and DmNopp140 localize to the DFCs of Xenopus oocyte nucleoli, but they fail to localize to the sphere organelles. Xenopus oocytes were injected with synthetic transcripts that encoded either GFP-DmNopp140-RGG (A–E) or GFP-DmNopp140 (F–J). (A, D, F, and I) Phase contrast images showing a few of the ∼1000 nucleoli in each oocyte nucleus. GFP-DmNopp140-RGG (B) and GFP-DmNopp140 (G) both localized within the DFC regions of the nucleoli, specifically within a subregion immediately surrounding the FCs. These FCs contained detectable amounts of the nucleolar DNA as demonstrated by DAPI staining (C and H). Some nucleoli typically contained multiple FCs and DFCs (G and H). Both GFP-DmNopp140-RGG (D and E) and DmNopp140 (I and J) failed to localize to the large sphere organelles (arrows) that are homologous to Cajal bodies. Bar (A–J), 20 μm.
In three separate trials, GFP-Nopp140-RGG and GFP-DmNopp140 labeled only nucleoli after 18–24 h post injection (Figure 4, E and J, respectively). No other germinal vesicle bodies contained either variant. The oocyte CBs (formerly called spheres or C snurposomes; Gall, 2000) were completely devoid of any GFP labeling (Figure 4, E and J, arrows). This was surprising because the large Cajal bodies in amphibian oocyte nuclei are known to contain Nopp140 (Gall et al., 1999). Isaac et al. (1998) demonstrated a lag time of between 12 and 24 h before Nopp140 appeared in the CBs of transfected COS-1 cells, and this might explain our failure to see labeling in the oocyte CBs. On the other hand, Wu et al. (1994) showed a very rapid (1–4 h) localization of coilin to Xenopus oocyte CBs after injecting mRNAs. The length of time between our oocyte injections and nuclear preparations (∼18 h) should have been sufficient time for at least some of the Drosophila proteins to accumulate within the CBs. We know that the central domain of alternating acidic and basic regions of Nopp140 is required for Cajal body localization (Isaac et al., 1998). Perhaps sufficient sequence differences exist between xNopp180 and the two Drosophila variants (see above) such that the Drosophila proteins fail to localize to CBs in the Xenopus oocyte nucleus.
Exogenous Expression of DmNopp140-RGG in HeLa Cells
We used the same expression construct used for the Schneider II cells to express and localize DmNopp140-RGG in HeLa cells. As expected, GFP-DmNopp140-RGG localized to the phase-dark nucleoli (Figure 5, A and B). In transfected cells that expressed a moderate amount of GFP-DmNopp140-RGG, we saw a punctate staining pattern over the nucleoli (Figure 5B), suggesting that GFP-DmNopp140-RGG localized to subdomains of the nucleoli. These subdomains may be analogous to the subdivision within the DFCs that we observed in the Xenopus oocyte nucleoli (Figure 4B). Endogenous fibrillarin colocalized well with DmNopp140-RGG in nucleoli of HeLa cells expressing moderate amounts of DmNopp140-RGG (Figure 5, D and E, arrows). In HeLa cells expressing greater quantities of GFP-DmNopp140-RGG (Figure 5, D and E, arrowheads), the punctate staining was not readily apparent due to greater accumulations of GFP-DmNopp140-RGG within the nucleoli. Nevertheless, fibrillarin continued to colocalize.
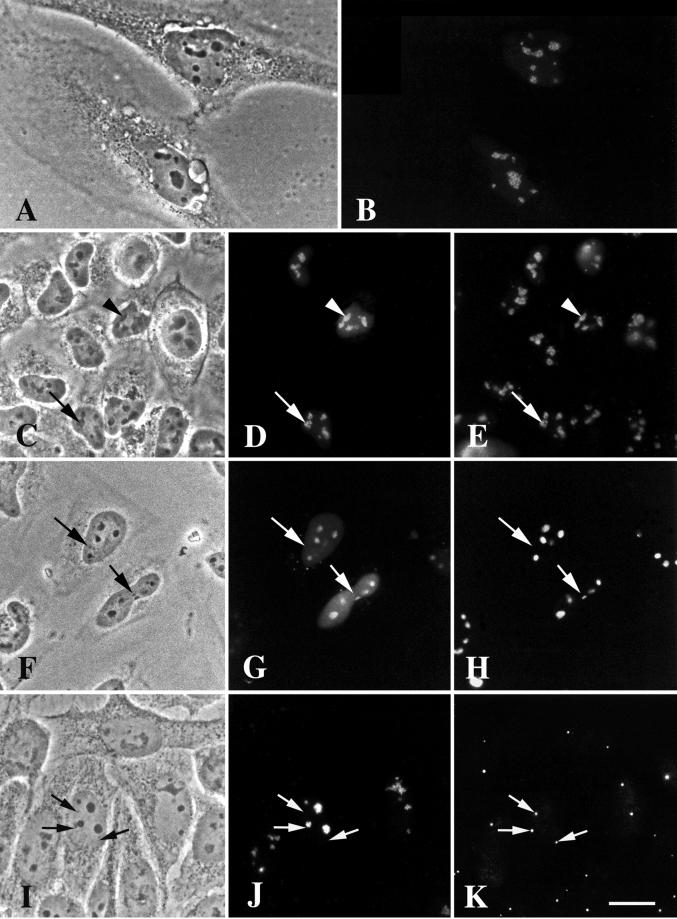
Figure 5.
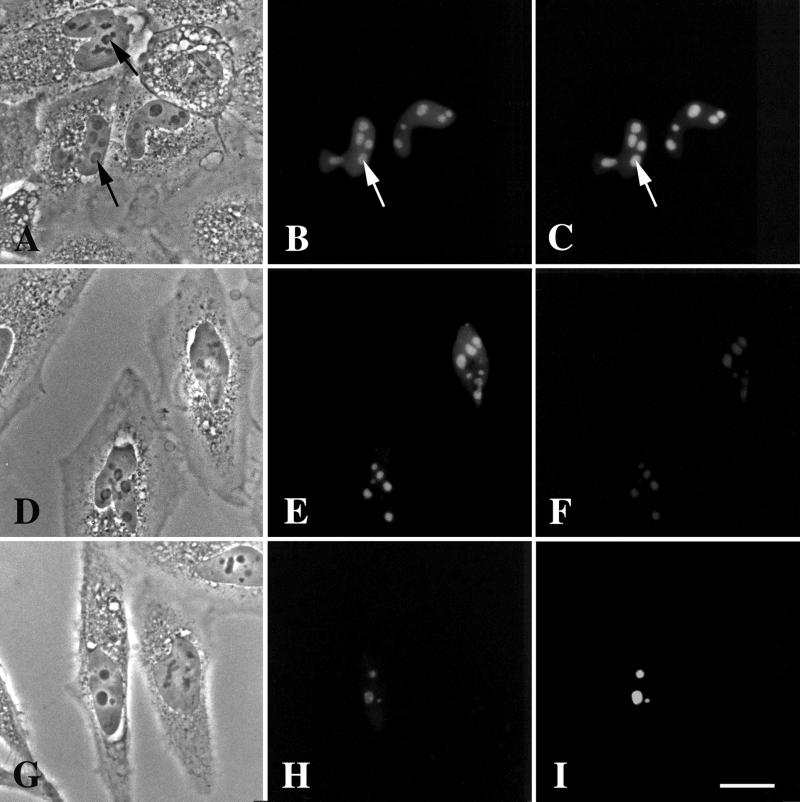
DmNopp140-RGG localizes to intact, phase-dark nucleoli but fails to localize to endogenous Cajal bodies in transfected HeLa cells. (A and B) GFP-DmNopp140-RGG (B) localized to nucleoli in a punctate staining pattern. (C–E) Exogenous GFP-DmNopp140-RGG (D) colocalized with endogenous fibrillarin (E) that was detected with the S4 anti-human fibrillarin antibody and a rhodamine-conjugated secondary antibody. Arrowheads in C–E point to nucleoli that contain relatively large amounts of DmNopp140-RGG, whereas arrows in C–E point to nucleoli that maintain a punctate staining pattern with moderate amounts of DmNopp140-RGG. (F–H) Exogenous GFP-DmNopp140-RGG (G) colocalized with endogenous nucleolin (H) that was detected with an anti-nucleolin antibody and a rhodamine-conjugated secondary antibody. Arrows in F–H point to nuclear bodies that fail to stain with DmNopp140-RGG or anti-nucleolin. Conversely, all labeled structures evident in B, D, and G corresponded well to phase-dark nucleoli in A, C, and F, respectively. (I–K) Exogenous GFP-DmNopp140-RGG (J) again localized to intact phase-dark nucleoli, but it failed to colocalize with coilin in endogenous CBs as detected by the anti-human coilin antibody R288 (K). Precisely aligned arrows point to endogenous CBs. Bar (A–K), 20 μm.
We counterstained other transfected HeLa cells with anti-human nucleolin (Figure 5H). As expected, GFP-DmNopp140-RGG and endogenous nucleolin colocalized to the relatively large, phase-dark nucleoli. Smaller nuclear organelles were evident by phase contrast microscopy (Figure 5F, arrows), but they did not contain GFP-DmNopp140-RGG (Figure 5G) nor did they stain with anti-nucleolin (Figure 5H). This observation is significant because the nucleoplasm in HeLa cells that overexpressed GFP-DmNopp140-RGG was lightly but uniformly labeled (Figure 5G), and we observed no other nuclear organelle (e.g., CBs) that contained GFP-DmNopp140-RGG other than the large nucleoli that also contained endogenous nucleolin. Because nucleolin is not a component of somatic cell CBs, we conclude that DmNopp140-RGG localized only to nucleoli when exogenously expressed in HeLa cells.
Because DmNopp140-RGG failed to localize to the large CBs in Xenopus oocyte nuclei, we critically tested whether GFP-DmNopp140-RGG could localize to endogenous CBs in HeLa cells. After transfection, the cells were counterstained with R288, a rabbit antiserum directed against human p80 coilin (Andrade et al., 1993). Transfected cells again contained nucleoli that were well labeled by GFP-DmNopp140-RGG (Figure 5J), but the endogenous CBs shown in Figure 5K were completely devoid of GFP-DmNopp140-RGG. The arrows in Figure 5, I–K, are well matched, denoting the position of the endogenous CBs. The cells in Figure 5, I–K, were washed free of the DNA-Ca2+ 24 h after its addition and then incubated under normal conditions for an additional 24 h before fixation. This should have been ample time for GFP-DmNopp140-RGG to transit the nucleoli and accumulate within CBs (Isaac et al. 1998). We observed identical results with transfected cells that were fixed 24 h after the DNA-Ca2+ was initially added. The failure of GFP-DmNopp140-RGG to localize to endogenous CBs in HeLa cells was consistent with its failure to localize to CBs in Xenopus oocytes.
Exogenous Expression of DmNopp140 in HeLa Cells
As with GFP-DmNopp140-RGG, we used the same expression construct used for the Schneider II cells to express and localize GFP-DmNopp140 in HeLa cells. Unlike GFP-DmNopp140-RGG, however, GFP-DmNopp140 caused HeLa cell nucleoli to partition into phase-light and phase-dark regions (Figure 6, A and B). GFP-DmNopp140 localized exclusively to the phase-light regions (Figure 6B). In some cells, the phase-light regions extended well into the nucleoplasm, but they remained attached to the phase-dark regions of the nucleoli (Figure 6, A, C, F, and I, arrows).
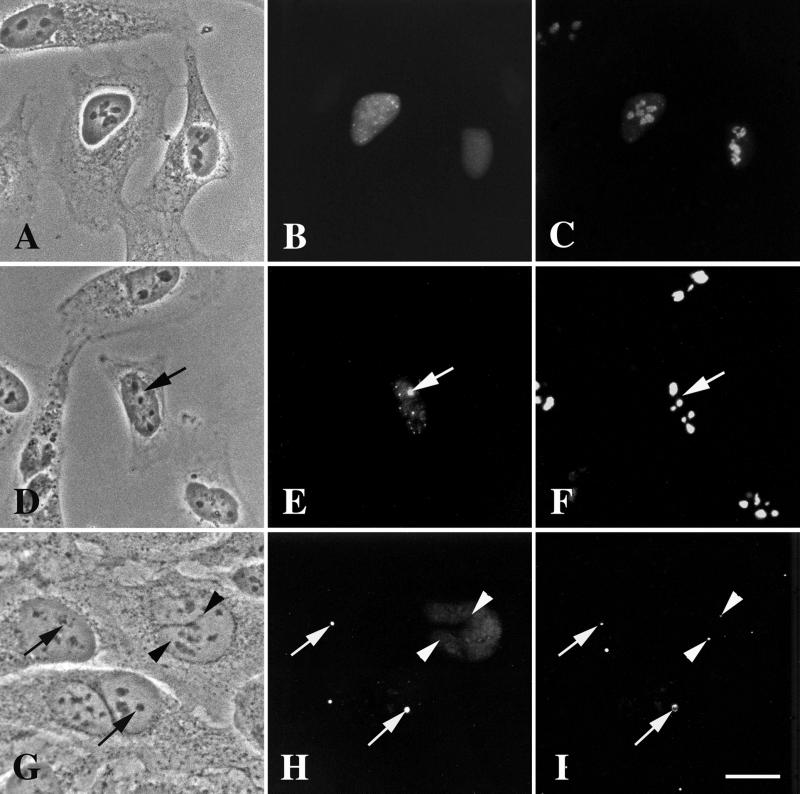
Figure 6.
DmNopp140 partitions HeLa nucleoli into phase-light and phase-dark regions, and it also fails to localize to endogenous CBs. (A and B) GFP-DmNopp140 localized exclusively to the phase-light regions of the nucleoli (B). The phase-dark regions (A and B) were completely devoid on GFP-DmNopp140. (C–E) GFP-DmNopp140 (D) colocalized with endogenous fibrillarin (E) in the phase-light regions of the partitioned nucleoli. The S4 anti-human fibrillarin antibody and a rhodamine-conjugated secondary antibody detected the endogenous fibrillarin (E). Precisely aligned arrows point to phase-dark regions. (F–H) GFP-DmNopp140 (G) again localized to the phase-light regions, whereas endogenous nucleolin (H) localized to the phase-dark regions (F–H, arrows). An anti-human nucleolin antibody and a rhodamine-conjugated secondary antibody detected the endogenous nucleolin (H). (I–K) GFP-DmNopp140 localized to the phase-light regions of partitioned nucleoli, and precisely matched arrows show the phase-dark regions. The anti-human coilin antiserum R288 detected endogenous CBs (K, arrowheads). GFP-DmNopp140 failed to localize to endogenous CBs, but endogenous coilin appeared to accumulate on the periphery of or within the phase-light regions (K). Bar (A–K), 20 μm.
We counterstained the partitioned nucleoli with anti-fibrillarin (Figure 6E) and anti-nucleolin (Figure 6H) to initially define the molecular composition of the phase-light and phase-dark regions. GFP-DmNopp140 colocalized with fibrillarin in the phase-light regions (compare the matched arrows in Figure 6, D and E). This colocalization is in accord with coimmunoprecipitation results that showed an association between mammalian Nopp140 and fibrillarin within C/D box snoRNP particles (Yang et al., 2000). Whereas GFP-DmNopp140 accumulated within the phase-light regions (Figure 6G), endogenous nucleolin localized within the phase-dark regions (Figure 6, F–H, compare the matched arrows). The partitioning of endogenous fibrillarin (phase-light region) from endogenous nucleolin (phase-dark region) in HeLa cells expressing GFP-DmNopp140 suggests that nucleolin and fibrillarin do not form tight associations in vivo.
We also tested the ability of GFP-DmNopp140 to localize to endogenous CBs in transfected HeLa cells. Counterstaining the cells with anti-human coilin showed that GFP-DmNopp140, like GFP-DmNopp140-RGG, failed to localize to endogenous CBs (Figure 6, J and K, arrowheads). As with DmNopp140-RGG, its failure to associate with endogenous CBs is consistent with the results obtained with GFP-DmNopp140 expression in Xenopus oocytes.
Interestingly, in about half of the transfected cells expressing GFP-DmNopp140, endogenous coilin redistributed to the phase-light regions. There was a mixture of observed coilin redistributions, and we present these diverse redistributions in right- and left-hand panels for Figure 6, I–K. First, endogenous coilin was observed to accumulate on the very periphery of some phase-light regions. This is evident in the transfected cell in the center of the left-hand panels of Figure 6, I–K. In this particular cell, the nucleolus is partitioned into a phase-dark spot (arrow) and a phase-light(er) region. Anti-coilin labeled only the left side of this phase-light region (same cell in the left-hand panel of Figure 6K). Conversely, we observed no redistribution of endogenous coilin in the transfected cell in lower portion of the left-hand panels. In this particular cell, the nucleoli were again partitioned, but endogenous coilin remained within small CBs (Figure 6K, arrowheads point to 0.5–1-μm spheres in the left-hand panel); the anti-coilin completely failed to stain the phase-light regions containing GFP-DmNopp140.
In the right-hand panels of Figure 6, I–K, two transfected cells expressed GFP-DmNopp140, but the partitioning of their nucleoli was more dramatic (arrows point to the phase-dark regions, whereas the phase-light regions spill out into the nucleoplasm). Anti-coilin again stained the periphery of these phase-light regions, but there was now internal staining as well. Occasionally, we observed what appeared to be a Cajal body within the periphery of a phase-light region as seen in the uppermost phase-light region in the right-hand panel of Figure 6K. In addition to anti-coilin staining the phase-light regions, separate CBs were also evident in cells expressing GFP-DmNopp140 (arrowheads in the lower nucleus of the right-hand panels). Again, GFP-DmNopp140 failed to localize to these CBs.
In summary, GFP-DmNopp140 causes nucleoli to partition into phase-light and phase-dark regions. GFP-DmNopp140 and fibrillarin colocalize to the phase-light regions, whereas nucleolin localizes to the phase-dark regions. GFP-DmNopp140 fails to localize to endogenous CBs, but endogenous coilin appears to associate with the phase-light regions, preferentially on the periphery of these regions in many cases.
Coexpression of DmNopp140-RGG and DmNopp140 in HeLa Cells
We coexpressed GFP-DmNopp140 (Figure 7, B, E, and H) and RFP-DmNopp140-RGG (Figure 7, C, F, and I) in HeLa cells to determine whether they would colocalize together within partitioned or intact nucleoli. Both proteins colocalized to fairly intact nucleoli (Figure 7A) when expressed in approximately equal amounts based on fluorescence intensities (Figure 7, B and C). There were no prominent phase-light nucleolar regions protruding into the nucleoplasm as observed in Figure 6A. On closer examination, however, some nucleoli appeared slightly partitioned, but not much more than what we occasionally observed in nontransfected cells (Figure 7A, compare the two arrows). Cells that expressed greater amounts of GFP-DmNopp140 (Figure 7E) relative to RFP-DmNopp140-RGG (Figure 7F) again contained partitioned nucleoli with prominent phase-light and phase-dark regions (Figure 7D). Interestingly, DmNopp140-RGG localized with DmNopp140 within these phase-light regions, suggesting an association may occur between these two proteins. Finally, nucleoli appeared morphologically normal in cells that expressed less DmNopp140 (Figure 7H) compared with DmNopp140-RGG (Figure 7I). Both proteins again colocalized to the intact nucleoli in these cells. The observations in Figure 7 collectively indicate that the extent to which nucleoli partition into phase-light and phase-dark regions is proportional to the amount of DmNopp140 expressed within the cell, but that an equal coexpression of DmNopp140-RGG may dampen this nucleolar partitioning.
Figure 7.
Coexpression of GFP-DmNopp140 and RFP-DmNopp140-RGG in transfected HeLa cells. (B, E, and H) Expression of GFP-DmNopp140. (C, F, and I) Expression of RFP-DmNopp140-RGG. (A–C) GFP-DmNopp140 and RFP-DmNopp140-RGG were expressed in approximately equal amounts based on fluorescence signals. Both proteins colocalized to the nucleoli. One nucleolus in a transfected cell appeared partially segregated by phase contrast microscopy (A, lower arrow), but no more so than a nucleolus in a nontransfected cell (A, upper arrow). (D–F) GFP-DmNopp140 (E) was overexpressed with respect to RFP-DmNopp140-RGG (F). Nucleoli appeared segregated by phase contrast microscopy (D), yet both proteins colocalized to the phase-light regions. (G–I) GFP-DmNopp140 (H) was underexpressed relative to RFP-DmNopp140-RGG (I). Nucleoli appeared morphologically normal by phase contrast microscopy (G), yet both proteins colocalized. Bar (A–I), 20 μm.
Localization Patterns of DmNopp140ΔRGG in HeLa Cells
The carboxy RGG domain of DmNopp140-RGG is very similar to the carboxy RGG domain of vertebrate nucleolin. When the RGG domain of nucleolin is deleted, the resulting truncation translocates to the nucleus (the bipartite NLS is further upstream), but it fails to associate with nucleoli (Créancier et al., 1993; Heine et al., 1993; Meßmer and Dreyer, 1993; Schmidt-Zachmann and Nigg, 1993). The RGG domain on its own, however, is not a nucleolar localization signal; nonnucleolar proteins fused to an NLS and to the nucleolin RGG domain also fail to localize to nucleoli (Meβmer and Dreyer, 1993; Schmidt-Zachmann and Nigg, 1993).
To test the localization properties of the RGG domain in DmNopp140-RGG, we used the PCR to amplify only the cDNA sequence that encodes amino acids 1–561 of DmNopp140-RGG. The deleted segment extended upstream of the actual RGG domain to include two additional tandem RGG motifs at residues 562–567. The deleted residues (562–688) are italicized in Figure 1C. We refer to the expressed truncation as DmNopp140ΔRGG. In more than half of the transfected cells, GFP-DmNopp140ΔRGG failed to localize to nucleoli (Figure 8B). In a few these cells, we could see nucleoli that were barely labeled above background. Instead of localizing to the nucleoli, GFP-DmNopp140ΔRGG localized to the nucleoplasm (Figure 8B). The nucleoli in these cells appeared morphologically normal (Figure 8A), and fibrillarin remained localized to these nucleoli (Figure 8C).
Figure 8.
Expression of the carboxyl-terminal truncation, DmNopp140ΔRGG, in transfected HeLa cells. (A–C) GFP-DmNopp140ΔRGG failed to localize to intact phase-dark nucleoli. The cells were counterstained with the S4 anti-human fibrillarin antibody and a rhodamine-conjugated secondary antibody (C). Most of the endogenous fibrillarin remained associated with the nucleoli. (D–F) In approximately half the transfected cells GFP-DmNopp140ΔRGG caused nucleoli to partition into phase-light (precisely matched arrows D–F) and phase-dark regions, reminiscent of the effects caused by GFP-DmNopp140. The cells were counterstained with anti-human nucleolin and a rhodamine-conjugated secondary antibody (F). Nucleolin localized only within the phase-dark regions that were completely devoid of GFP-DmNopp140ΔRGG. (G–I) Transfected HeLa cells again expressed GFP-DmNopp140ΔRGG (H). Some cells displayed a diffuse distribution of GFP-DmNopp140ΔRGG, whereas other cells displayed partitioned nucleoli with DmNopp140ΔRGG localizing to the phase-light regions (precisely aligned arrows in G and H). The cells were counterstained with the R288 antiserum directed against human coilin (I). Arrowheads show endogenous CBs that did not contain GFP-DmNopp140ΔRGG. Conversely, arrows show phase-light regions that appear to contain endogenous coilin. Bar (A–I), 20 μm.
In many of the other transfected cells (approaching half), we observed accumulations of GFP-DmNopp140ΔRGG in large phase-light regions of partitioned nucleoli (Figure 8, D and E, precisely aligned arrows). Endogenous nucleolin again maintained its localization within the phase-dark regions (Figure 8F), whereas endogenous fibrillarin colocalized with DmNopp140ΔRGG in the phase-light regions (our unpublished data). We conclude from these observations that DmNopp140ΔRGG generally fails to associate with morphologically normal phase-dark nucleoli when expressed in low-to-moderate levels. At higher expression levels, however, GFP-DmNopp140-ΔRGG mimics full-length DmNopp140 in causing nucleoli to partition into phase-light and phase-dark regions. Again, fibrillarin and nucleoli partition to phase-light and phase-dark regions, respectively.
Cells expressing GFP-DmNopp140-ΔRGG were counterstained with anti-human p80 coilin (Figure 8, G–I). In many cells DmNopp140-ΔRGG again distributed throughout the nucleoplasm, but it failed to localize to endogenous CBs that were detected with the anti-coilin (Figure 8, H and I, precisely aligned arrowheads). In cells that contained partitioned nucleoli, however, endogenous coilin again accumulated on the periphery of or within phase-light regions (Figure 8, G–I, precisely aligned arrowheads). A good example of peripheral localization of endogenous coilin is shown just below the center of Figure 8I. The top part of this peripheral region appears to contain a spherical CB.
From the observations presented in Figure 8, we conclude that DmNopp140-ΔRGG behaves much like GFP-DmNopp140 in causing nucleoli to partition into phase-light and phase-dark regions. Because DmNopp140-ΔRGG partitions nucleoli in a manner similar to that observed for DmNopp140, the amino terminus and/or the large central domain of DmNopp140 must be responsible for this observed partitioning. The distinctive RGG domain in DmNopp140-RGG may prevent or dampen any propensity of the amino terminus or the central domain within DmNopp140-RGG to partition nucleoli, because partitioning does not occur in cells expressing GFP-DmNopp140-RGG. Finally, DmNopp140-ΔRGG fails to localize to endogenous CBs, but endogenous coilin appears to localize to the phase-light regions containing DmNopp140-ΔRGG. We can conclude that the RGG domain does not prevent DmNopp140-RGG from localizing to CBs. The reason that the two intact Drosophila variants fail to localize to endogenous CBs must lie in sequence or structural differences within the amino terminus or central region compared with the rat protein that we know localizes to CBs.
DISCUSSION
Two Drosophila Nopp140 Variants
The Nopp140 gene in D. melanogaster maps within polytene segment 79A5. This region is proximal to the centromere (80F) on the left arm of chromosome 3. Transcription is in the direction of the centromere. Genes upstream of Nopp140 are eagle (a steroid hormone receptor/transcription factor) in 79A4; the gene encoding cyclin H (for cell cycle regulation); and conceptual genes CG7407, CG7414, and CG7148, all of unknown function. CG7145 maps downstream of Nopp140; it encodes a 1-pyrroline-5-carboxylate dehydrogenase-like enzyme. An enhancer-promoter type P-element transposon [EP(3)3138] maps within the promoter of CG7145, ∼5.5 kbp downstream of the 3′ end of Nopp140 (FlyBase, 1999). In the future, this P-element should allow us to create deficiencies that eliminate the Nopp140 gene in our efforts to understand Nopp140 function in metazoans.
The Nopp140 gene encodes a predicted pre-mRNA of 3247 nucleotides that is differentially spliced to produce two transcripts, each encoding a Nopp140 variant. DmNopp140 is the sequence homolog of rat Nopp140. Its overall domain composition and organization is very similar to the prototypical rat Nopp140, and this is particularly true for its central and carboxyl-terminal domains. The overall length of the central domain within DmNopp140 (and DmNopp140-RGG), however, is shorter than that in rat Nopp140. Despite its central domain being shorter, DmNopp140 has 14 acidic regions instead of 10 as in rat Nopp140. Xenopus xNopp180, on the other hand, has 18 acidic regions (Cairns and McStay, 1995).
The carboxy termini of various Nopp140 proteins serve to best define homology (Meier, 1996; Table 1). The entire carboxy terminus of DmNopp140 is 64% identical over a 97-amino acid comparison with the carboxy terminus of the prototypical rat Nopp140. The individual carboxy subdomains of DmNopp140 (Ca and Cb) are as close in homology to the respective sequences in rat and human Nopp140 as the two Xenopus subdomains are to the same respective domains in human and rat Nopp140. That individual exons encode these two subdomains in Drosophila suggests evolutionary constraints on distinct subdomain function. These functions have yet to be determined precisely, but the properties of these two subdomains have been described (Isaac et al., 1998). Finally, a consensus cAMP-dependent protein kinase phosphorylation site is present in the conserved carboxy terminus of all Nopp140 homologs (Meier, 1996). The presence of the putative site suggests that Nopp140 is a direct substrate for signal transduction-mediated phosphorylation cascades that may regulate molecular interactions of Nopp140 within nucleoli or Cajal bodies. DmNopp140 contains a similar site (serine 638), but whether this site is used in vivo is not yet known.
DmNopp140 Partitions HeLa Cell Nucleoli
Despite sequence similarities to the mammalian Nopp140 proteins, DmNopp140 causes HeLa cell nucleoli to partition into clearly discernible phase-light and phase-dark regions (Figure 6A). Partitioning was so severe in some cells (Figure 6, A and B) that we were often able to identify transfected cells expressing exogenous DmNopp140 simply by using phase contrast microscopy. Endogenous fibrillarin colocalized with DmNopp140 to the phase-light regions (Figure 6, C–E), whereas endogenous nucleolin localized to the phase-dark regions (Figure 6, F–H). DmNopp140 completely failed to associate with the phase-dark regions. The separation of DmNopp140 from nucleolin is reminiscent of dominant negative effects caused by the transient expression of the carboxy terminal domain of rat Nopp140, the hemagglutinin-tagged NoppC described by Isaac et al. (1998). In their study, exogenous expression of NoppC clearly partitioned fibrillarin and nucleolin in a manner similar, but not identical to our exogenous expression of full-length DmNopp140 in HeLa cells. NoppC chased endogenous Nopp140, fibrillarin, and NAP57 out of the nucleoli and into the nucleoplasm. The three proteins did not localize to any nucleolar cap or phase-light regions of the partitioned nucleoli. Nucleolin and UBF (upstream binding factor), on the other hand, were not affected by the expression of NoppC; they remained within the phase-dark nucleoli. In our studies, endogenous fibrillarin was “chased” into the phase-light regions of the partitioned nucleoli, whereas nucleolin remained behind in the phase-dark regions.
Chen et al. (1999) described similar nucleolar partitioning in HeLa cells when they overexpressed a carboxy truncation of human Nopp140 (hNopp140N382, containing the first 382 amino acid residues out of the 699 total). In their study, the truncation colocalized with fibrillarin and the large subunit of polymerase I within partitioned nucleolar caps. Chen et al. (1999) proposed that hNopp140N382 caused cap formation by interacting with the large subunit of RNA polymerase I in a dominant negative manner to block rRNA transcription. The authors cited that similar nucleolar caps form when cells are treated with actinomycin D at concentrations known to block rRNA transcription (see the descriptions of actinomycin D-segregated nucleoli by Busch and Smetana, 1970).
It will be interesting to determine whether the partitioned nucleoli that we see due to the exogenous expression of DmNopp140 (rather than by actinomycin D) are still functional. Barring any dominant negative effects on transcription caused by DmNopp140, we may be able to determine the active sites for transcription and processing with respect to the phase-light and phase-dark regions. The partitioned nucleoli may allow us to explore possible molecular associations between nucleolar components in vivo. For example, the colocalization of DmNopp140 and endogenous fibrillarin within the phase-light regions is consistent with the observation that Nopp140 associates with fibrillarin in box C/D small nuclear RNA (snRNA) complexes (Yang et al., 2000). Fibrillarin associates with U3 and many of the other box C/D snoRNPs. Conversely, nucleolin also interacts with the U3 snoRNP (a box C/D snoRNP) for cleavage within the 5′ external transcribed spacer of pre-rRNA (Ginisty et al., 1998). It remains uncertain whether nucleolin interacts with the many other box C/D snoRNPs that selectively methylate pre-rRNA. The clear segregation of endogenous fibrillarin (phase-light region) from endogenous nucleolin (phase-dark region) in HeLa cells that express DmNopp140 suggests that nucleolin and fibrillarin do not associate directly in vivo, despite that they normally colocalize within DFCs. We predict, therefore, that box C/D snoRNPs (with fibrillarin as antigen) and box H/ACA snoRNPs (with NAP65 as antigen) will colocalize within the partitioned phase-light regions, whereas nucleolin and B23 remain within the phase-dark regions of partitioned nucleoli in cells expressing DmNopp140. Questions remain: what other nucleolar components reside within the phase-light versus the phase-dark regions? Does U3 localize to both the phase-light and the phase-dark regions?
DmNopp140-RGG Is a Novel Splice Variant
With its RGG carboxy terminus, DmNopp140-RGG is a novel and striking variant of DmNopp140. Naturally occurring variants of other nucleolar proteins have been described. For instance, X. laevis expresses two versions of nucleolin that are encoded by separate genes (Meβmer and Dreyer, 1993). Xenopus is pseudotetraploid (Kobel and DuPasquier, 1986), and the two modern nucleolin genes may have descended from a common ancestral gene by duplication and divergence. The two nucleolin proteins differ primarily in the number of amino terminal acidic and basic regions. Furthermore, two splice variants of B23 exist in rat (Chang and Olson, 1989, 1990; Wang et al., 1994). B23.1 is the prominent nucleolar protein expressed in all tissues, whereas B23.2 is a shorter variant that localizes to the cytoplasm and perhaps the nucleoplasm, but not the nucleolus (Wang et al., 1993). Alternative splicing results in the deletion of the carboxyl-terminal 35 residues and the substitution of two additional upstream residues to convert B23.1 to B23.2 (Chang and Olson, 1989, 1990). Interestingly, B23.1, but not B23.2, binds nucleic acids (Wang et al., 1994). As a third example, two isoforms of human Nopp140 have been reported (Pai and Yeh, 1996). The α form predominates, but the novel β form contains a 10-amino acid insert in the fourth proline-rich basic region of the central domain. No functional differences are known to exist between the α and β forms of human Nopp140. Compared with these relatively minor differences in the number of alternating acidic and basic regions or the even the deletion variant of the B23, the discovery of DmNopp140-RGG indicates that novel proteins related to the prototypical rat Nopp140 but with strikingly different domains may perform related but nonoverlapping functions.
RGG Domain of DmNopp140-RGG
The extensive RGG domain in DmNopp140-RGG is very similar to the RGG domain found in vertebrate nucleolin. Nucleolin's RGG domain is necessary but not sufficient for proper nucleolar localization (Créancier et al., 1993; Heine et al., 1993; Meβmer and Dreyer, 1993; Schmidt-Zachmann and Nigg, 1993). Likewise, DmNopp140-ΔRGG generally fails to associate with nucleoli in roughly half the transfected HeLa cells (Figure 8B). In these cells, DmNopp140-ΔRGG distributes to the nucleoplasm. Thus, the RGG domain of DmNopp140-RGG appears to be necessary for nucleolar localization at least in mammalian cells. We speculate that the RGG domain in DmNopp140-RGG may share the same interactions with nucleolar proteins (Cartegni et al., 1996; Bouvet et al., 1998) or RNAs (Ghisolfi et al., 1992a,b; Hanakahi et al., 2000) that have been attributed to nucleolin by way of its RGG domain.
Besides nucleolin, other nucleolar proteins that contain RGG motifs include (but are not limited to) fibrillarin (Lischwe et al., 1985; Ochs et al., 1985) Gar1; Nopp44/46 in Trypanosoma brucei (Das et al., 1998); and the three yeast nucleolar proteins gar2p, Ssb1p, and Nop3p. Ribosomal protein S2 also contains RGG motifs (Suzuki et al., 1991). The arginine residues within RGG motifs of nucleolin and fibrillarin are asymmetrically dimethylated (NG,NG-dimethylarginine), and we strongly suspect that the arginines within the RGG domain of DmNopp140-RGG will be similarly dimethylated. This posttranslational modification does not change the charge of the arginine side chains, but it certainly makes the arginine side chains bulkier and more hydrophobic, in all likelihood to modulate molecular interaction. Arginine methylation within RNA-binding proteins is believed to regulate their protein–RNA (Tao and Frankel, 1992), protein–protein interactions (Liu and Dreyfuss, 1995; Friesen et al., 2001), or perhaps their nucleocytoplasmic shuttling (Shen et al., 1998). The functional significance of RGG methylation in regulating protein–protein interactions is just now coming to light (Friesen et al., 2001).
DmNopp140-ΔRGG
The distribution of DmNopp140-ΔRGG to the nucleoplasm or nucleolar phase-light regions in HeLa cells is in sharp contrast to the nucleolar localization observed for rat Nopp140ΔC in monkey COS-1 cells (Isaac et al., 1998). Rat Nopp140ΔC accumulated in the cytoplasm, intact nucleoli, CBs, and in phase-dense nuclear rings referred to as R-rings. Rat Nopp140ΔC did not accumulate in the nucleoplasm (Isaac et al., 1998) as did DmNopp140-ΔRGG. In many other transfected HeLa cells, DmNopp140-ΔRGG localized to the phase-light regions of partitioned nucleoli (Figure 8H). Rat Nopp140ΔC did not appear to partition the nucleoli. One possible explanation for these differences between rat Nopp140ΔC and DmNopp140-ΔRGG is that the two deletions may not be directly comparable. The rat Nopp140ΔC was truncated at residue 586, thus deleting the last 118 residues (587–704). This deletion stretched into the back end of the central domain (Figure 1A). We deleted residues 561–688 from DmNopp140-RGG, thus leaving behind 10 residues that are unique to DmNopp140-RGG versus DmNopp140 (Figure 1, C and D). Further work is necessary to determine whether this 10-residue segment, now the very carboxy end, is critical for the distribution of DmNopp140ΔRGG to the nucleoplasm compared with the rat Nopp140ΔC. Alternatively, sequence and structural differences between the amino termini and the central domains of rat Nopp140ΔC and DmNopp140-ΔRGG may account for these differences.
Cajal Bodies Versus Phase-Light Regions
Both DmNopp140-RGG and DmNopp140 fail to localize to CBs in the nuclei of Xenopus oocytes and HeLa cells. The best explanation for this discrepancy between the two Drosophila variants and rat Nopp140 may lie in their respective tertiary structures. Direct associations between Nopp140 and coilin have been established using the two-hybrid system and coimmunoprecipitations (Isaac et al., 1998). Specifically, full-length rat Nopp140 associates well with the amino-terminal region (the first 161 amino acids) of coilin. Although deletion of the conserved carboxy terminus of rat Nopp140 (both subregions Ca and Cb) greatly diminishes the interaction with coilin, coilin fails to interact with the individual amino terminal domain of Nopp140, the individual central domain, and the individual carboxy domain (Isaac et al., 1998). From these results, it appears that the entire Nopp140 sequence is necessary to achieve an association with coilin, perhaps by folding into a particular tertiary structure. Any interaction between Nopp140 and coilin must be reserved for the CBs because p80 coilin does not normally localize to nucleoli (but see below). A particular tertiary structure of Nopp140 that is necessary for coilin interaction must shift when Nopp140 gains access to nucleoli. Such a structural shift in Nopp140 may then permit Nopp140 to interact with other nucleolar components instead of coilin. Phosphorylation of Nopp140 (Meier, 1996) or of coilin (Hebert and Matera, 2000) by CKII may affect their respective structures to thus allow or prevent their intermolecular associations. Perhaps the two Drosophila variants are sufficiently different in primary sequence and thus tertiary structure that they fail to interact with endogenous coilin in the CBs of Xenopus oocytes and HeLa cells.
Clues as to why the two Drosophila variants fail to localize to CBs may be found in the studies describing treacle. Treacle is a human protein related to Nopp140 in that it contains a homologous central repeat domain with 10 repeating acidic and basic regions (Dixon et al., 1997; Wise et al., 1997). Like Nopp140, treacle is highly phosphorylated by CKII, and it localizes to the DFCs of nucleoli. Treacle, however, is distinct from Nopp140 in that it fails to associate with CBs (Isaac et al., 2000). Treacle is the product of the human TCOF1 gene (Dixon et al., 1997), mutations in which give rise to the Treacher Collins syndrome, an autosomal dominant disorder that affects craniofacial development (Dixon, 1996). Sequence differences between treacle and Nopp140 reside primarily in their amino and carboxy termini, and these differences may account for treacle's failure to localize to CBs (Isaac et al., 2000). Analogous differences between the amino and carboxy termini of rat Nopp140 and the two Drosophila variants may therefore account for the failure of the two Drosophila variants to localize to CBs.
Despite that the Drosophila Nopp140 variants fail to localize to endogenous CBs, endogenous coilin appears to associate with the phase-light regions of partitioned nucleoli in about half the cells expressing DmNopp140 or DmNopp140ΔRGG. In these cells, coilin accumulates on it periphery of the phase-light regions, but some phase-light regions also appear to contain small amounts of p80 coilin within their interiors. Previous observations established that coilin can localize to intranucleolar structures under certain conditions. For example, coilin localizes to nucleoli within liver cells and brown adipocytes of hibernating hazel dormice (Malatesta et al., 1994) and within certain human breast cancer cells (Ochs et al., 1994). Simple overexpression of GFP-tagged full-length coilin causes its accumulation in spherical structures within nucleoli, whereas two GFP-coilin truncations, GFP-coilin(1–248) GFP-coilin(1–315), colocalize with fibrillarin and Nopp140 within the DFCs of HeLa nucleoli (Hebert and Matera, 2000). Several other results suggest that phosphorylation plays an important role in coilin's localization to nucleoli. First, endogenous coilin and U2 snRNPs can redistribute to intranucleolar structures within HeLa cells when Ser/Thr dephosphorylation is inhibited by 10 nM okadaic acid (Lyon et al., 1997; Sleeman et al., 1998). Second, a dominant mutant of coilin (S202D) that mimics constitutive phosphorylation localizes to intranucleolar structures and causes the disassembly of all other extranucleolar CBs (Lyon et al., 1997; Sleeman et al., 1998). Conversely, the S184A mutation in human coilin mimics the dephosphorylated state, and the mutant colocalized with fibrillarin in 30% of the cells expressing the mutation (Hebert and Matera, 2000). Finally, a cryptic nucleolar localization signal within coilin may be differentially exposed depending on the state of coilin phosphorylation (Hebert and Matera, 2000). In many of these cases, coilin associated with a defined spherical structure within the nucleoli, structures that are reminiscent of the CBs. We refrain from identifying the phase-light regions as swollen FCs because the phase-light regions contain exogenous DmNopp140 and endogenous fibrillarin, both of which are considered to be DFC markers. It is interesting to note that fibrillarin is a component of both CBs and the phase-light regions of the partitioned nucleoli. Nucleolin, on the other hand, is not a component of either the somatic cell CBs or the phase-light regions. Future work with antibodies directed against known FC and DFC markers should better define the molecular composition of the phase-light regions and how these phase-light regions relate to CBs.
Finally, we also predict that molecular interactions (and functions) of DmNopp140-RGG versus DmNopp140 will prove different within nucleoli. The molecular cytology and molecular genetics available in Drosophila should give us ample opportunities to explore these differences. For example, we fully expect DmNopp140 to interact with the minifly gene product Nop60B (Phillips et al., 1998) in Drosophila. But will the same interaction occur with DmNopp140-RGG? Drosophila also allows us to explore developmental and tissue-specific expression patterns of the two variants. What we learn about the molecular interactions of DmNopp140 versus DmNopp140-RGG, and the expression patterns of these two splice variants, should advance our knowledge of novel and traditional nucleolar functions.
ACKNOWLEDGMENTS
We thank Drs. John Tower and Allan Spradling (Department of Embryology, Carnegie Institution of Washington, Baltimore, MD) for an aliquot of their stage 10 egg chamber cDNA library. We also thank Dianne Stewart and Allan Spradling for the Schneider II cells. We thank Drs. Zheng'an Wu and Joe Gall (Department of Embryology, Carnegie Institution of Washington) for the R288 rabbit serum directed against human coilin and for insightful discussions regarding CBs. We thank Dr. Benigno (Ben) Valdez (Baylor College of Medicine, Houston, TX) for antibodies directed against human nucleolin and Dr. Robert Ochs (Scripts Research Institute, La Jolla, CA) for the anti-human S4 anti-fibrillarin antibody. This work was supported by National Science Foundation grant MCB-9727917.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–04-0162. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–04-0162.
REFERENCES
- Andrade LEC, Chan EKL, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LEC, Tan EM, Chan EKL. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci USA. 1993;90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, More DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1995. [Google Scholar]
- Bachellerie J-P, Cavaillé J. Guiding ribose methylation of rRNA. Trends Biochem Sci. 1997;22:257–261. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- Belenguer P, Caizergues-Ferrer M, Labbe J-C, Dorée M, Amalric F. Mitosis-specific phosphorylation of nucleolin by p34cdc2protein kinase. Mol Cell Biol. 1990;10:3607–3618. doi: 10.1128/mcb.10.7.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K, Ferreira JA, Lamond AI. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol. 1995a;131:817–831. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K, Ferreira J, Santama N, Weis K, Lamond AI. Molecular analysis of the coiled body. J Cell Sci. 1995b;19:107–113. doi: 10.1242/jcs.1995.supplement_19.16. [DOI] [PubMed] [Google Scholar]
- Bonnet H, Filhol O, Truchet I, Brethenou P, Cochet C, Amalric F, Bouche G. Fibroblast growth factor-2 binds to the regulatory B subunit of CH2 and directly stimulates CK2 activity toward nucleolin. J Biol Chem. 1996;271:24781–24787. doi: 10.1074/jbc.271.40.24781. [DOI] [PubMed] [Google Scholar]
- Bouvet P, Diaz JJ, Kleinbeiter K, Madjar JJ, Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Busch H, Smetana K. The Nucleolus. New York: Academic Press; 1970. [Google Scholar]
- Cadwell C, Yoon H-J, Zebarjadian Y, Carbon J. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol Cell Biol. 1997;17:6175–6183. doi: 10.1128/mcb.17.10.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns C, McStay B. Identification and cDNA cloning of a Xenopusnucleolar phosphoprotein, xNopp180, that is the homolog of the rat nucleolar protein Nopp140. J Cell Sci. 1995;108:3339–3347. doi: 10.1242/jcs.108.10.3339. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Maconi M, Morandi E, Cobianchi F, Riva S, Biamonti G. hnRNP A1 selectively interacts through its gly-rich domain with different RNA-binding proteins. J Mol Biol. 1996;259:337–348. doi: 10.1006/jmbi.1996.0324. [DOI] [PubMed] [Google Scholar]
- Chang J-H, Olson MOJ. A single gene codes for two forms of rat nucleolar protein B23 mRNA. J Biol Chem. 1989;264:11732–11737. [PubMed] [Google Scholar]
- Chang J-H, Olson MOJ. Structure of the gene for rat nucleolar protein B23. J Biol Chem. 1990;265:18227–18233. [PubMed] [Google Scholar]
- Chen C, Okayama H. Calcium phosphate-mediated gene transfer: a highly efficient system for stably transforming cells with plasmid DNA. Biotechniques. 1988;6:632–638. [PubMed] [Google Scholar]
- Chen H-K, Pai C-Y, Huang J-Y, Yeh N-H. Human Nopp140, which interacts with RNA polymerase I: implications for rRNA gene transcription and nucleolar structural organization. Mol Cell Biol. 1999;19:8536–8546. doi: 10.1128/mcb.19.12.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Créancier L, Prats H, Zanibellato C, Amalric F, Bugler B. Determination of the functional domains involved in nucleolar targeting of nucleolin. Mol Biol Cell. 1993;4:1239–1250. doi: 10.1091/mbc.4.12.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, Schnaider T, Cheatham B, Olson MOJ, Kahn CR. Insulin induces the phosphorylation of nucleolin. A possible mechanism of insulin-induced RNA efflux from nuclei. J Biol Chem. 1993;268:9747–9752. [PubMed] [Google Scholar]
- Das A, Park J-H, Hagen CB, Parsons M. Distinct domains of a nucleolar protein mediate protein kinase binding, interaction with nucleic acids and nucleolar localization. J Cell Sci. 1998;111:2615–2623. doi: 10.1242/jcs.111.17.2615. [DOI] [PubMed] [Google Scholar]
- Dixon MJ. Treacher Collins syndrome. Hum Mol Genet. 1996;5:1391–1396. doi: 10.1093/hmg/5.supplement_1.1391. [DOI] [PubMed] [Google Scholar]
- Dixon J, Edwards SJ, Anderson I, Scambler PJ, Dixon MJ. Identification of the complete coding sequence and genomic organization of the Treacher Collins syndrome gene. Genome Res. 1997;7:223–234. doi: 10.1101/gr.7.3.223. [DOI] [PubMed] [Google Scholar]
- Dumbar TS, Gentry GA, Olson MOJ. Interaction of nucleolar phosphoprotein B23 with nucleic acids. Biochemistry. 1989;28:9495–9501. doi: 10.1021/bi00450a037. [DOI] [PubMed] [Google Scholar]
- Dundr M, Raska I. Nonisotopic ultrastructural mapping of transcription sites within the nucleolus. Exp Cell Res. 1993;208:275–281. doi: 10.1006/excr.1993.1247. [DOI] [PubMed] [Google Scholar]
- Echalier G. Drosophila Cells in Culture. San Diego: Academic Press; 1997. [Google Scholar]
- Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- FlyBase. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. http://flybase.bio.indiana.edu/ ( http://flybase.bio.indiana.edu/). ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Spread preparation of Xenopusgerminal vesicle contents. In: Spector D, Goldman R, Leinwand L, editors. Cells: A Laboratory Manual. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. pp. 52.1–52.4. [Google Scholar]
- Gall JG. Cajal bodies: the first 100 years. Annu Rev Dev Biol. 2000;16:272–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia SN, Pillus L. Net results of nucleolar dynamics. Cell. 1999;97:825–828. doi: 10.1016/s0092-8674(00)80794-1. [DOI] [PubMed] [Google Scholar]
- Ghisolfi L, Gerard J, Amalric F, Erard M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J Biol Chem. 1992a;267:2955–2959. [PubMed] [Google Scholar]
- Ghisolfi L, Kharrat A, Joseph G, Amalric F, Erard M. Concerted activities of the RNA recognition and the glycine-rich C-terminal domains of nucleolin are required for efficient complex formation with pre-ribosomal RNA. Eur J Biochem. 1992b;209:541–548. doi: 10.1111/j.1432-1033.1992.tb17318.x. [DOI] [PubMed] [Google Scholar]
- Ghisolfi-Nieto L, Joseph G, Puvion-Dutilleul F, Amalric F, Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J Mol Biol. 1996;260:34–53. doi: 10.1006/jmbi.1996.0380. [DOI] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Hélène S, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Giordano E, Peluso I, Senger S, Furia M. minifly, a Drosophilagene required for ribosome biogenesis. J Cell Biol. 1999;144:1123–1133. doi: 10.1083/jcb.144.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolov AA. Cell Biology Monographs. New York: Springer Verlag; 1985. The nucleolus and ribosome biogenesis; pp. 1–263. [Google Scholar]
- Hanakahi LA, Bu Z, Maizels N. The C-terminal domain of nucleolin accelerates nucleic acid annealing. Biochemistry. 2000;39:15493–15499. doi: 10.1021/bi001683y. [DOI] [PubMed] [Google Scholar]
- Heine MA, Rankin ML, DiMario PJ. The gly/arg-rich (GAR) domain of Xenopusnucleolin facilitates in vitro nucleic acid binding and in vivo nucleolar localization. Mol Biol Cell. 1993;4:1189–1204. doi: 10.1091/mbc.4.11.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell. 2000;11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera AH, Olson MOJ. Association of protein C23 with rapidly labeled nucleolar RNA. Biochemistry. 1986;25:6258–6264. doi: 10.1021/bi00368a063. [DOI] [PubMed] [Google Scholar]
- Herrera JE, Savkur R, Olson MOJ. The ribonuclease activity of nucleolar protein B23. Nucleic Acids Res. 1995;23:3974–3979. doi: 10.1093/nar/23.19.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozák P, Cook PR, Schöfer C, Mosgöller W, Wachtler F. Site of transcription of ribosomal RNA and intranucleolar structure in HeLa cells. J Cell Sci. 1994;107:639–648. doi: 10.1242/jcs.107.2.639. [DOI] [PubMed] [Google Scholar]
- Isaac C, Yang Y, Meier UT. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac C, Marsh KL, Paznekas WA, Dixon J, Dixon MJ, Wang Jabs E, Meier UT. Characterization of the nucleolar gene product, treacle, in Treacher Collins syndrome. Mol Biol Cell. 2000;11:3061–3071. doi: 10.1091/mbc.11.9.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson FB, Sinclair DA, Guarente L. Molecular biology of aging. Cell. 1999;96:291–302. doi: 10.1016/s0092-8674(00)80567-x. [DOI] [PubMed] [Google Scholar]
- Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Kobel HR, DuPasquier L. Genetics of polyploid. Xenopus. Trends Genet. 1986;2:310–315. [Google Scholar]
- Lafontaine DLJ, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B, Bourbon H, Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc Natl Acad Sci USA. 1987;84:1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Meier UT, Dobrowolska G, Krebs EG. Specific interaction between casein kinase 2 and the nucleolar protein Nopp140. J Biol Chem. 1997;272:3773–3779. doi: 10.1074/jbc.272.6.3773. [DOI] [PubMed] [Google Scholar]
- Lischwe MA, Ochs RL, Reddy R, Cook RG, Yeoman LC, Tan EM, Reichlin M, Busch H. Purification and partial characterization of nucleolar scleroderma antigen (Mr 43,000; pI 8.5) rich in NG,NG-dimethylarginine. J Biol Chem. 1985;260:14304–14310. [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon CE, Bohmann K, Sleeman J, Lamond A. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp Cell Res. 1997;230:84–93. doi: 10.1006/excr.1996.3380. [DOI] [PubMed] [Google Scholar]
- Malatesta M, Zancanaro C, Martin TE, Chan EKL, Amalric F, Lührmann R, Vogel P, Fakan S. Is the coiled body involved in nucleolar functions? Exp Cell Res. 1994;211:415–419. doi: 10.1006/excr.1994.1106. [DOI] [PubMed] [Google Scholar]
- Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Meier UT. Comparison of the rat nucleolar protein Nopp140 with its yeast homolog SRP40. Differential phosphorylation in vertebrates and yeast. J Biol Chem. 1996;271:19376–19384. [PubMed] [Google Scholar]
- Meier UT, Blobel G. A nuclear localization signal binding protein in the nucleolus. J Cell Biol. 1990;111:2235–2245. doi: 10.1083/jcb.111.6.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meßmer B, Dreyer C. Requirements for nuclear translocation and nucleolar accumulation of nucleolin of Xenopus laevis. Eur J Cell Biol. 1993;61:369–382. [PubMed] [Google Scholar]
- Miau L-H, Chang C-J, Tsai W-H, Lee S-C. Identification and characterization of a nucleolar phosphoprotein, Nopp140, as a transcription factor. Mol Cell Biol. 1997;17:230–239. doi: 10.1128/mcb.17.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougey EB, Pape LK, Sollner-Webb B. A U3 small nuclear ribonucleoprotein-requiring processing event in the 5′ external transcribed spacer of Xenopusprecursor rRNA. Mol Cell Biol. 1993;13:5990–5998. doi: 10.1128/mcb.13.10.5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Seckman W, Terns R, Terns MP. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol Cell. 1999;10:2131–2147. doi: 10.1091/mbc.10.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs RL, Lischwe MA, Spohn WH, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell. 1985;54:123–134. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Ochs RL, Stein TW, Jr, Tan EM. Coiled bodies in the nucleolus of breast cancer cells. J Cell Sci. 1994;107:385–399. doi: 10.1242/jcs.107.2.385. [DOI] [PubMed] [Google Scholar]
- Okuda M, et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Olson MOJ. The role of proteins in nucleolar structure and function. In: Straus P, Wilson S, editors. The Eukaryotic Nucleus: Molecular Biochemistry and Macromolecular Assemblies. Vol. 2. Caldwell, NJ: The Telford Press; 1990. pp. 519–559. [Google Scholar]
- Olson MO, Dundr M, Szebeni A. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- Orrick LR, Olson MOJ, Busch H. Comparison of nucleolar proteins of normal rat liver and Novikoff hepatoma ascites cells by two-dimensional polyacrylamide gel electrophoresis. Proc Natl Acad Sci. 1973;70:1316–1320. doi: 10.1073/pnas.70.5.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai C-Y, Chen H-K, Sheu H-L, Yeh N-H. Cell-cycle-dependent alterations of a highly phosphorylated nucleolar protein p130 are associated with nucleologenesis. J Cell Sci. 1995;108:1911–1920. doi: 10.1242/jcs.108.5.1911. [DOI] [PubMed] [Google Scholar]
- Pai C-Y, Yeh N-H. Cell proliferation-dependent expression of two isoforms of the nucleolar phosphoprotein p130. Biochem Biophys Res Commun. 1996;221:581–587. doi: 10.1006/bbrc.1996.0639. [DOI] [PubMed] [Google Scholar]
- Peculis BA, Steitz JA. Distribution of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopusoocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T, Politz JC. The nucleolus and the four ribonucleoproteins of translation. J Cell Biol. 2000;148:1091–1095. doi: 10.1083/jcb.148.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Nakagawa J, Dorée M, Labbé JC, Nigg EA. Identification of major nucleolar proteins as candidate mitotic substances of cdc2 kinase. Cell. 1990;60:791–801. doi: 10.1016/0092-8674(90)90093-t. [DOI] [PubMed] [Google Scholar]
- Pfeifle J, Anderer FA. Isolation and localization of phosphoprotein pp 135 in the nucleoli of various cell lines. Eur J Biochem. 1984;139:417–424. doi: 10.1111/j.1432-1033.1984.tb08021.x. [DOI] [PubMed] [Google Scholar]
- Pfeifle J, Boller K, Anderer FA. Phosphoprotein pp135 is an essential component of the nucleolus organizer region (NOR) Exp Cell Res. 1986;162:11–22. doi: 10.1016/0014-4827(86)90422-2. [DOI] [PubMed] [Google Scholar]
- Phillips B, Billin AN, Cadwell C, Buchholz R, Erickson C, Merriam JR, Carbon J, Poole SJ. The Nop60B gene of Drosophilaencodes an essential nucleolar protein that functions in yeast. Mol Gen Genet. 1998;260:20–29. doi: 10.1007/s004380050866. [DOI] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol. 2000;151:1561–1574. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón y Cajal SR. Un sencillo metodo de coloacion seletiva del reticulo protoplasmico y sus efectos en los diversos organos nerviosos de vertebrados y invertebrados. Trab Lab Invest Biol. 1903;2:129–221. [Google Scholar]
- Rankin ML, Heine MA, Xiao S, LeBlanc MD, Nelson JW, DiMario PJ. A complete nucleolin cDNA sequence from Xenopus laevis. Nucleic Acids Res. 1993;21:169. doi: 10.1093/nar/21.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur R, Olson MOJ. Preferential cleavage in pre-ribosomal RNA by protein B23 endoribonuclease. Nucleic Acids Res. 1998;26:4508–4515. doi: 10.1093/nar/26.19.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Zachmann M, Nigg EA. Protein localization to the nucleolus: a search for targeting domains in nucleolin. J Cell Sci. 1993;105:799–806. doi: 10.1242/jcs.105.3.799. [DOI] [PubMed] [Google Scholar]
- Shah SB, Terry CD, Wells DA, DiMario PJ. Structural changes in oocyte nucleoli of Xenopus laevisduring oogenesis and meiotic maturation. Chromosoma. 1996;105:111–121. doi: 10.1007/BF02509521. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Jordan EG. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Shen EC, Henry MF, Weiss VH, Valentini SR, Silver PA, Lee MS. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J, Lyon CE, Platani M, Kreivi J-P, Lamond A. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp Cell Res. 1998;243:290–304. doi: 10.1006/excr.1998.4135. [DOI] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Speckman W, Narayanan A, Terns R, Terns M. Nuclear retention elements of U3 small nucleolar RNA. Mol Cell Biol. 1999;19:8412–8421. doi: 10.1128/mcb.19.12.8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Olvera J, Woll IG. Primary structure of rat ribosomal protein S2. J Biol Chem. 1991;266:20007–20010. [PubMed] [Google Scholar]
- Szebeni A, Olson MOJ. Nucleolar protein B23 has molecular chaperone activities. Protein Sci. 1999;8:905–912. doi: 10.1110/ps.8.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Frankel AD. Specific binding of arginine to TAR RNA. Proc Natl Acad Sci USA. 1992;89:2723–2726. doi: 10.1073/pnas.89.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandelaer M, Thiry M. The phosphoprotein pp135 is an essential constituent of the fibrillar components of nucleoli and of coiled bodies. Histochem Cell Biol. 1998;110:169–177. doi: 10.1007/s004180050278. [DOI] [PubMed] [Google Scholar]
- Visintin R, Amon A. The nucleolus: the magician's hat for cell cycle tricks. Curr Opin Cell Biol. 2000;12:372–377. doi: 10.1016/s0955-0674(00)00102-2. [DOI] [PubMed] [Google Scholar]
- Wang D, Baumann A, Szebeni A, Olson MOJ. The nucleic acid binding activity of nucleolar protein B23.1 resides in its carboxyl-terminal end. J Biol Chem. 1994;269:30994–30998. [PubMed] [Google Scholar]
- Wang H, Boisvert D, Kim KK, Kim R, Kim SH. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution. EMBO J. 2000;19:317–323. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Umekawa H, Olson MOJ. Expression and subcellular locations of two forms of nucleolar protein B23 in rat tissues and cells. Cell Mol Biol Res. 1993;39:33–42. [PubMed] [Google Scholar]
- Weinstein LB, Steitz JA. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- Wise CA, Chiang LC, Paznekas WA, Sharma M, Musy MM, Ashley JA, Lovett M, Jabs EW. TCOF1gene encodes a putative nucleolar phosphoprotein that exhibits mutations in Treacher Collins Syndrome throughout its coding region. Proc Natl Acad Sci USA. 1997;94:3110–3115. doi: 10.1073/pnas.94.7.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Gall JG. Human p80-coilin is targeted to sphere organelles in the amphibian germinal vesicle. Mol Biol Cell. 1994;5:1119–1127. doi: 10.1091/mbc.5.10.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Isaac C, Wang C, Dragon F, Pogacic V, Meier UT. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol Biol Cell. 2000;11:567–577. doi: 10.1091/mbc.11.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatsepina OV, Rousselet A, Chan PK, Olson MOJ, Jordan EG, Bornens M. The nucleolar phosphoprotein B23 redistributes in part to the spindle poles during mitosis. J Cell Sci. 1999;112:455–466. doi: 10.1242/jcs.112.4.455. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Lu D, DiMario P. Nucleolin, defective for MPF phosphorylation, localizes normally during mitosis and nucleologenesis. Histochem Cell Biol. 1999;111:477–487. doi: 10.1007/s004180050384. [DOI] [PubMed] [Google Scholar]