Abstract
The aquaglyceroporins are a subfamily of aquaporins that conduct both water and glycerol. Aquaporin-3 (AQP3) has an important physiological function in renal water reabsorption, and AQP3-mediated hydrogen peroxide (H2O2) permeability can enhance cytokine signaling in several cell types. The related aquaglyceroporin AQP7 is required for dendritic cell chemokine responses and antigen uptake. Selective small-molecule inhibitors are desirable tools for investigating the biological and pathological roles of these and other AQP isoforms. Here, using a calcein fluorescence quenching assay, we screened a library of 7360 drug-like small molecules for inhibition of mouse AQP3 water permeability. Hit confirmation and expansion with commercially available substances identified the ortho-chloride–containing compound DFP00173, which inhibited mouse and human AQP3 with an IC50 of ∼0.1–0.4 μm but had low efficacy toward mouse AQP7 and AQP9. Surprisingly, inhibitor specificity testing revealed that the methylurea-linked compound Z433927330, a partial AQP3 inhibitor (IC50, ∼0.7–0.9 μm), is a potent and efficacious inhibitor of mouse AQP7 water permeability (IC50, ∼0.2 μm). Stopped-flow light scattering measurements confirmed that DFP00173 and Z433927330 inhibit AQP3 glycerol permeability in human erythrocytes. Moreover, DFP00173, Z433927330, and the previously identified AQP9 inhibitor RF03176 blocked aquaglyceroporin H2O2 permeability. Molecular docking to AQP3, AQP7, and AQP9 homology models suggested interactions between these inhibitors and aquaglyceroporins at similar binding sites. DFP00173 and Z433927330 constitute selective and potent AQP3 and AQP7 inhibitors, respectively, and contribute to a set of isoform-specific aquaglyceroporin inhibitors that will facilitate the evaluation of these AQP isoforms as drug targets.
Keywords: aquaporin, molecular docking, glycerol, inhibitor, hydrogen peroxide, aquaporin inhibitor, calcein, CHO cell assay, HyPer, residual pore
Introduction
Aquaporin-3 (AQP3)2 is a member of the major intrinsic protein family of transmembrane channels that are known to conduct water, glycerol, as well as hydrogen peroxide (H2O2) (1, 2). Two other AQP isoforms of the so-called aquaglyceroporin subfamily, AQP7 and AQP9, are expressed in the mouse. AQP3 expression sites in the body include the kidneys (3, 4), gastrointestinal tract (5, 6), airway epithelia (7), conjunctival epithelium of the eye (5), urinary bladder (8), skin keratinocytes (9), and human erythrocytes (10). AQP3 gene deletion in mice results in extreme polyuria (11). Pharmacological inhibition of AQP3 has thus been suggested as an approach to induce aquaresis (12), selective salt-sparing removal of excess body water that may be desirable for some clinical conditions such as heart failure and hyponatremia. Surprisingly, the extremely rare AQP3-deficient humans do not show any clinical manifestations, including no polyuria (13). However, it cannot be excluded that this observation is due to compensatory adaptations in AQP3-deficient individuals. Consequently, AQP3 inhibitors could still have potential for treating human disorders of water retention.
Beside uses for aquaporin inhibitors in water imbalance disorders, several newer AQP3−/− knockout and knockdown animal studies suggest a number of applications for potent, selective, and nontoxic AQP3 inhibitors. This includes mouse models of multistage skin tumorigenesis, non-small-cell lung cancer, and breast cancer (14–17). Moreover, AQP3-mediated H2O2 conductance confers sensitivity to cytokines in T cells, alveolar macrophages, and keratinocytes. Thus, AQP3−/− knockout mice are largely protected from hapten-induced contact hypersensitivity, psoriasis, and allergic airway inflammation (18–20).
A second murine aquaglyceroporin, AQP7, is expressed in adipose tissue, renal proximal tubules, muscle, and pancreatic β-cells (21). Knockout mouse phenotypes suggest associated functions in adipocyte glycerol release during lipolysis, glycerol reabsorption in proximal tubules, energy supply to heart muscle, as well as regulation of β-cell glycerol content (21–26). More recently, AQP7 was also detected in skin dendritic cells. Similar to Aqp3−/− knockout mice, Aqp7−/− knockout mice were protected from hapten-induced contact hypersensitivity (27). Up until now, it has not been clarified whether AQP7 conducts H2O2 and has a role in cytokine signaling. However, it is conceivable that combined inhibition of AQP3 and AQP7 could be useful in the treatment of atopic dermatitis.
At present, only few studies have described AQP3 inhibitors (28). Cu2+ as well as Ni2+ ions bind to extracellular loop sites of human AQP3 (29, 30). Inhibitor potency was not fully clarified in these studies because it varied with the utilized buffer system. When bicarbonate buffer was used, copper (IC50, ∼5 μm) appeared to potently and selectively inhibit the water and glycerol permeability of AQP3 but not of the homologous AQP7. On the other hand, in the presence of chelators such as Tricine and HEPES buffers, the apparent Cu2+ potency was in the millimolar range (29, 30). It should be noted that normal human plasma Cu2+ levels range from 1 to 15 μm (http://www.hmdb.ca/ (31)).3 More recently, the gold(III) bipyridyl compound Auphen was described as an AQP3 inhibitor. Auphen inhibition of AQP3 conductance to glycerol was potent, with an IC50 of ∼0.8 μm (32) in human erythrocytes. Besides AQP3, Auphen also blocked AQP7 (IC50, ∼6.5 μm) in an adipocyte cell line (33). Because of the apparent clinical potential of AQP3 inhibitors as well as to obtain new tools for experimental research, we decided to identify novel AQP3 inhibitors. We previously identified inhibitors of the third murine aquaglyceroporin, AQP9, by screening a commercially available library of small molecules (34). The library contains chemically stable, diverse, drug-like molecules with beneficial properties for further development. Here we describe the identification of a potent and selective mouse (m) and human (h) AQP3 inhibitor. Structure–activity relationship analysis and AQP isoform specificity analysis have additionally resulted in the serendipitous discovery of a potent AQP7 inhibitor.
Results
AQP3 inhibitor screen
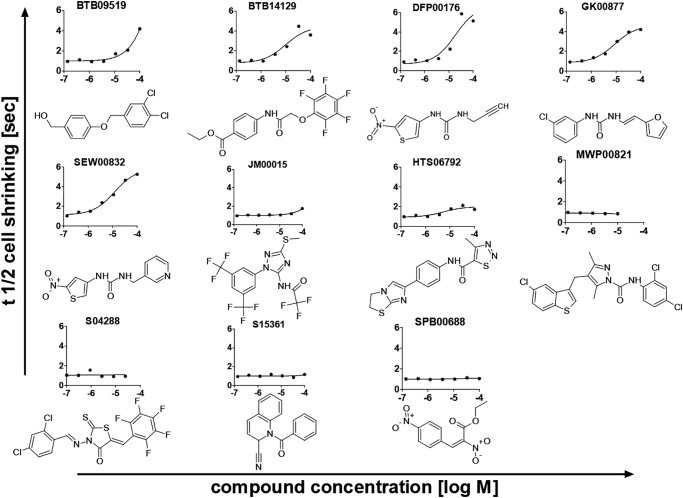
Chinese hamster ovary (CHO) cells expressing mAQP3 under the control of a tetracycline promoter were grown in 96-well plates for identifying AQP3 inhibitors in a calcein quenching–based assay of cell water permeability (34). Plates 1–23 of the Maybridge Hitfinder Library version 8, comprising a total of 7360 different molecules, were screened at a final concentration of 100 μm in assay buffer containing 1% DMSO. HTS06792 was identified previously as a weak mAQP9 inhibitor (34). Further analyses suggested partial inhibition of mAQP3. HTS06792 was thus used as a positive control in each screening plate, whereas DMSO-containing wells were used as negative controls. Hits were arbitrarily defined as substances that induced an increase in t½ of cell shrinking time of more than 2.5 s (compared with ∼1 s for HTS06792). Ten substances fitting this criterion were subsequently repurchased and subjected to dose–response analyses for hit confirmation. Inhibition of mAQP3 could be confirmed for five compounds. In addition, JM00015 showed mAQP3 inhibition at the highest tested concentration but at a lower efficacy than expected (Fig. 1).
Figure 1.
Dose–response curves for the ten most potent screening hits as well as for the positive control HTS06792. Shown is t½ of cell shrinking, indicating that water permeability was measured in calcein-loaded, mAQP3-expressing CHO cells. Screening hits were repurchased before analysis to confirm hit identity. Supplier product codes are indicated above each substance. Compounds MWP00821 and S04288 were not sufficiently soluble in DMSO and were thus tested at lower concentrations than the other compounds. AQP3 inhibition could be confirmed for five hit compounds. In addition, JM00015 showed AQP3 inhibition at the highest tested concentration and at lower efficacy than expected.
All confirmed active compounds appeared to be of relatively linear structure, with exception of the weak active JM00015. Besides the positive control, hits could broadly be categorized into the single hit BTB09519 and a group of substances described by a urea-containing linker (DFP00176, GK00877, or SEW00832) or acetamide linker (BTB14129); a left-hand 2-nitrothiophene, ethyl-benzoate, or chlorobenzene group; and a more variable right-hand side, as intuitively depicted (Fig. 1). These substances shared structural elements that are similar to the previously identified benzothiadiazole urea–containing AQP9 inhibitors RF03176 and HTS13772 (34, 35). Indeed, AQP3 inhibition by RF03176 was identified previously (34), and this substance was the seventh best hit in the current screen. We did initially not pursue RF03176 further because of its previously described low potency against AQP3 (34).
Structure–activity relationship
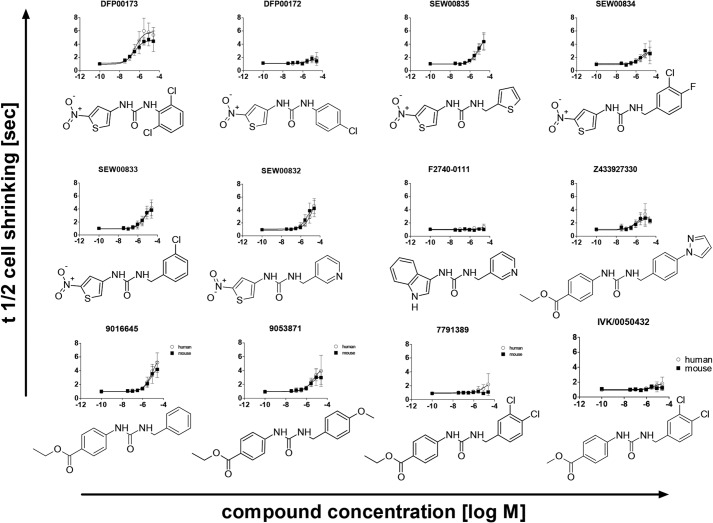
To explore this putative hit series further, a set of 12 commercially available, structurally similar compounds was purchased. These comprised six 1-(5-nitrothiophene-3-yl)urea compounds, a 1-(1H-indole-3-yl)urea compound, four ethyl 4-(carbamoylamino)benzoate compounds, and a methyl 4-(carbamoylamino)benzoate compound (Fig. 2, top left to bottom right). Potency and efficacy were tested on both mouse and human AQP3 isoforms, respectively. Two of these compounds contained a urea linker, and ten compounds comprised a methylurea linker. The most potent AQP3 inhibitor, DFP00173, was identified among the two compounds comprising a urea linker. Between these compounds, a 2,6-dichlorophenyl right-hand side in DFP00173 was clearly preferred to 4-chlorophenyl in DFP00172. Comparison of several methylurea linker compounds (SEW00835, SEW00833, and SEW00832 versus 9016645) suggests that 2-nitrothiophene and ethyl benzoate left-hand sides result in similar potency. However, direct comparisons are not possible because of the lack of commercially available molecules with identical right-hand sides. Considering right-hand sides, 3,4-chlorophenyl (7791389) appears to decrease efficacy and potency compared with phenyl (9016645) and methoxyphenyl groups (9053871), respectively. Interestingly, a right-hand phenylpyrazole (Z433927330) group resulted in a potent but moderately efficacious compound. Overall, no clear potency differences were observed between the inhibition of mAQP3 and hAQP3.
Figure 2.
Potency and structure–activity relationship analysis of mAQP3 and hAQP3 inhibition, respectively, by commercially available hit analogues. Shown is t½ of cell shrinking, indicating that water permeability was measured in calcein-loaded, m/hAQP3-expressing CHO cells. No clear potency differences between mAQP3 and hAQP3 inhibition were observed. Error bars indicate standard deviation.
AQP inhibitor specificity
We selected the two most potent compounds, DFP00173 and Z433927330, as well as compound 9016645 for further characterization of inhibitor specificity toward mouse aquaglyceroporins. Of these, 9016645 showed moderate selectivity for AQP3, whereas DFP00173 selectively inhibited AQP3, with only minor inhibition of AQP7 and AQP9. Z433927330 inhibited AQP7 potently and with good efficacy, whereas inhibition of AQP3 and AQP9 was lower and seemed to be less potent (Fig. 3, A–C). Comparison of 9016645 and Z433927330 inhibition profiles suggested that a shift in isoform specificity and potency was caused by the additional right-hand pyrazole.
Figure 3.
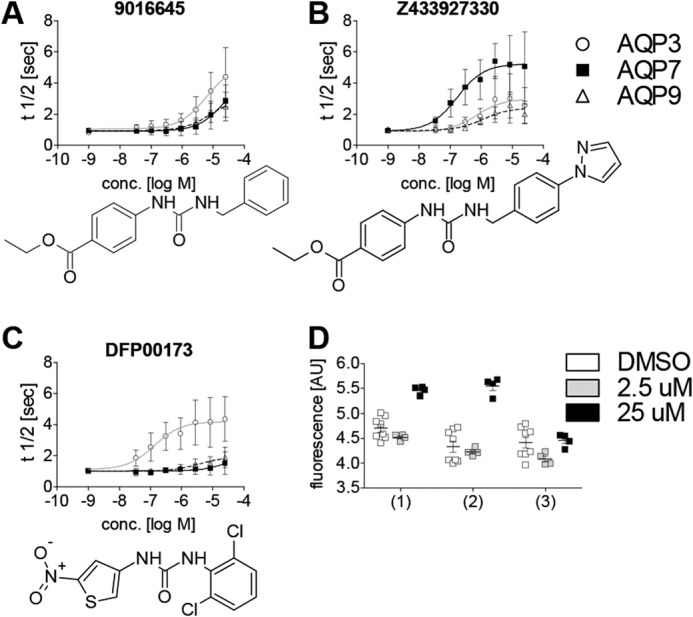
AQP isoform specificity of three selected compounds. A–C, cell water permeability was measured in calcein-loaded recombinant CHO cell lines. The structurally similar compounds 9016645 (A) and Z433927330 (B) show reversed isoform specificity. DFP00173 (C) specifically inhibits AQP3 over the homologous AQP isoforms AQP7 and AQP9. 9016645: IC50, ∼6 μm (mAQP3); IC50, N.D. (mAQP7); IC50, N.D. (mAQP9). Z433927330: IC50, ∼0.2 μm (mAQP7); IC50, ∼0.7 μm (mAQP3); IC50, ∼1.1 μm (mAQP9). DFP00173: IC50, ∼0.1 μm (mAQP3); IC50, N.D. (mAQP7); IC50, N.D. (mAQP9). The AQP isoform was identified as a significant source for variation in two-way ANOVA for all three substances at p < 0.0001; n = 3. D, cell proliferation and viability of CHO cells are not reduced after 48 h of incubation with AQP inhibitors compared with solvent (DMSO). 1, DFP00173; 2, Z433927330; 3, RF03176. Calcein retention was not affected at 2.5 μm. Exposure to 25 μm of either DFP00173 or Z433927330 resulted in increased fluorescence (p < 0.001; n = 4 for inhibitors and 8 for DMSO). Error bars indicate standard deviation. AU, arbitrary units.
To investigate potential cytotoxic or anti-proliferative effects, we incubated proliferating CHO cells for 48 h in the presence of the inhibitors DFP00173 and Z433927330 and the previously described AQP9 inhibitor RF03176 (34) before loading cells with Calcein-AM. A 5.8-fold increase in cell number was expected during this incubation period. Furthermore, the fluorophore can only be retained in viable cells (36). We found no negative effect of either substance on calcein fluorescence, indicating no apparent negative effects of inhibitors on CHO cell proliferation or vitality in the tested concentration range up to 25 μm (Fig. 3D). However, incubation with 25 μm DFP00173 and Z433927330 but not 25 μm RF03176 resulted in higher fluorescence, indicating increased proliferation or dye retention or a combination thereof.
Hydrogen peroxide permeability
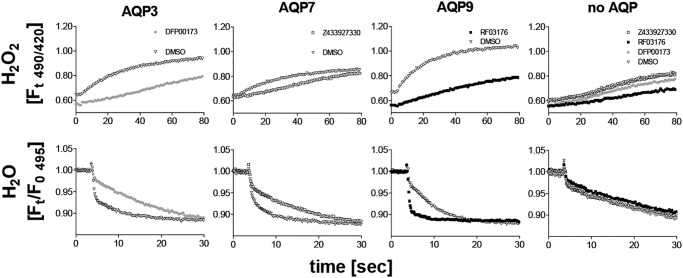
AQP3 and AQP9 are known to function as hydrogen peroxide channels in chemokine signaling in T cells and neutrophils (18, 37, 38). H2O2 permeability of AQP7 has currently not been demonstrated. However, AQP7 is required for chemokine responses in skin dendritic cells (27). Consequently, we tested whether AQP inhibitors are suitable to block AQP-mediated H2O2 permeability. AQP3, AQP7, and AQP9 were stably expressed in CHO cells along with the H2O2 sensor HyPer-3 (39). Addition of extracellular H2O2 resulted in an increase in 490/420-nm fluorescence intensity ratio in all HyPer-3–expressing cell lines (Fig. 4). Faster changes were observed in AQP3-, AQP7-, and AQP9-expressing cell lines compared with HyPer-3–expressing cells without ectopic AQP expression. H2O2 permeability in AQP-expressing cell lines was reduced by isoform cognate inhibitors. The inhibitors did not show clear effects on H2O2 permeability in CHO–HyPer-3 cells without ectopic AQP expression.
Figure 4.
Shown are H2O2 permeability (top row) and water permeability (bottom row) in H2O2 sensor (HyPer-3)–expressing CHO cell lines. Expression of mAQP3, mAPQ7, and mAPQ9 conferred a faster fluorescence increase in response to H2O2 addition (150 μm), compared with control cells (no AQP). Addition of cognate inhibitors resulted in a slower fluorescence increase in AQP-expressing cell lines after H2O2 addition, whereas no clear effects on control cells (no AQP) were observed. Similarly, AQP inhibitors reduced water permeability in AQP-expressing cell lines only (cell shrinking in response to sucrose addition 3.6 s into each read). The same HyPer-3–expressing cell lines were loaded with calcein before water permeability measurements. We note that the calcein fluorescence intensity is about 10-fold higher than HyPer-3 fluorescence under the conditions used. HyPer-3 did not seem to interfere with water permeability measurements. Furthermore, we noted a small baseline fluorescence decrease induced by DFP00173 and RF03176 treatment that was present before H2O2 addition. The reason for this effect is unknown. Means of four recordings are shown.
Glycerol permeability of human erythrocytes
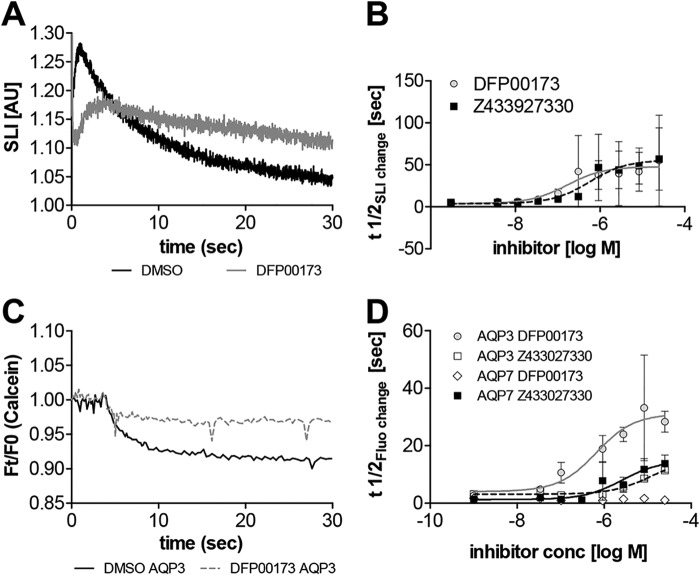
Human erythrocytes express water-permeable AQP1 as well as glycerol and water-permeable AQP3 (10). To confirm AQP3 inhibition in a second assay as well as to test inhibition of AQP3 glycerol permeability, we isolated whole human erythrocytes that were exposed to inward-directed glycerol gradients (Fig. 5, A and B). Changes in scattered light intensity were recorded over time in a stopped-flow apparatus. Treatment with DFP00173 and Z433927330, respectively, resulted in inhibition of erythrocyte glycerol permeability as well as inhibition of initial water permeability (Fig. 5A). The observed potencies for DFP00173 inhibition of glycerol permeability in erythrocytes (IC50, ∼ 0.2 μm) and Z433927330 (IC50, ∼ 0.6 μm) agreed well with the observed inhibition of water permeability in CHO cells. However, inhibition of glycerol permeability by Z433927330 appeared to be more efficacious than inhibition of CHO cell water permeability, causing similar levels of reduced permeability as DFP00173 (Fig. 5B). Furthermore, we tested the inhibition of AQP3 and AQP7 glycerol permeability by these two compounds in CHO cells (Fig. 5, C and D). We found highly specific inhibition of AQP3 glycerol permeability by DFP00173, which did not apparently inhibit AQP7 glycerol permeability. On the other hand, we found moderate selectivity of Z433927330 for inhibition of AQP7 glycerol permeability compared with inhibition of AQP3 permeability. We note that inhibition efficacy has to be compared cautiously between cell lines because we observed significantly higher glycerol permeability in AQP7-expressing cells compared with AQP3-expressing cells.
Figure 5.
Inhibition of glycerol permeability. A, addition of extracellular glycerol causes a rapid scattered light intensity (SLI) increase, indicative of water exit, mainly through hAQP1 and hAPQ3, followed by a slower scattered light intensity decrease, indicative of swelling induced by the osmotic influx of water, following glycerol entry mainly through AQP3. Representative example traces show clear inhibition of erythrocyte water and glycerol permeability by addition of 25 μm DFP00173 compared with solvent (1% DMSO). B, relative potency of erythrocyte glycerol permeability inhibition, measured as t½ of the scattered light intensity decrease. Apparent IC50 values were ∼0.2 μm (DFP00173) and ∼0.6 μm (Z433927330). Substance was identified as a significant source for variation in two-way ANOVA at p < 0.0001; n = 6. C, example traces of glycerol permeability in calcein-loaded CHO-mAQP3 cells in an isotonic shrinking assay. DFP00173-treated (25 μm) and solvent control (1% DMSO) cell traces of relative fluorescence intensity are shown. Cells were preincubated in 500 mm glycerol buffer before addition of 500 mm membrane-impermeable sucrose. Fluorescence quenching indicates cellular glycerol efflux along its gradient. D, dose–response curves of glycerol permeability inhibition in CHO cells expressing mAQP3 and mAQP7, respectively. Fluorescence quenching kinetics expressed as t½ were measured. Apparent IC50 values were ∼0.7 μm for DFP00173 (mAQP3), N.D. for DFP00173 (mAQP7), ∼11.5 μm for Z433927330 (mAQP3), and ∼2.6 μm for Z433927330 (mAQP7). n = 3. AU, arbitrary units. Error bars in B and D indicate standard deviation.
Homology modeling and molecular docking
We previously identified a putative binding site for RF03176 and similar compounds on hAQP9 at the cytoplasmic side of the pore. This binding site was examined by site-specific mutations in hAQP9 (35). Based on calculated logP values (RF03176, 2.80; DFP00173, 3.97; Z433927330, 3.72), we reasoned that inhibitors could diffuse across the lipid bilayer to reach the cytoplasmic side of the pore. This assumption was tested in epithelial barrier assays using mouse cortical collecting duct mpkCCD cells (40), which express very little AQP2 under basal culture conditions (41). Moreover, AQP2 is not expected to reach the apical membrane in the absence of vasopressin. Indeed, we found that all three substances crossed an electrically tight mpkCCD cell monolayer at a rate comparable with DMSO from the basal to the apical side, whereas cell-impermeable TO-PRO-3 remained undetectable at the apical side (Fig. S2).
Because of the chemical similarity between RF03176 and the currently identified hit series, we hypothesized similar binding of the newly identified compounds to mouse aquaglyceroporins at the cytoplasmic pore side. To investigate this, we generated homology models of the three mouse aquaglyceroporins mAQP3, mAQP7, and mAQP9. Initial calculations of the pore dimensions suggested that the pore appears to be too narrow for inhibitor binding at the so-called aromatic/arginine region and too shallow for strong inhibitor binding in its remaining extracellular segment (Fig. S1). Subsequent molecular docking, utilizing the entire pore of these models as a potential interaction surface, confirmed this hypothesis. Analysis of the top 50 poses for the newly identified ligands DFP00173, Z433927330 and 9016645, respectively, displayed a predicted binding site similar to RF03176 at the cytoplasmic side of the pore of all three aquaglyceroporins. Hydrogen bonds formed between the inhibitor urea linker and backbone carbonyls of loop B are a common motif (Fig. 6). Importantly, this binding prevents carbonyl–water or carbonyl–solute interactions, which are crucial for channel permeability. Further hydrogen bonds with one or both of the NPA box asparagines are formed in some AQP–inhibitor combinations. These asparagines of the two NPA boxes are well conserved among aquaporins and appear to be critical for selective channel permeability (42).
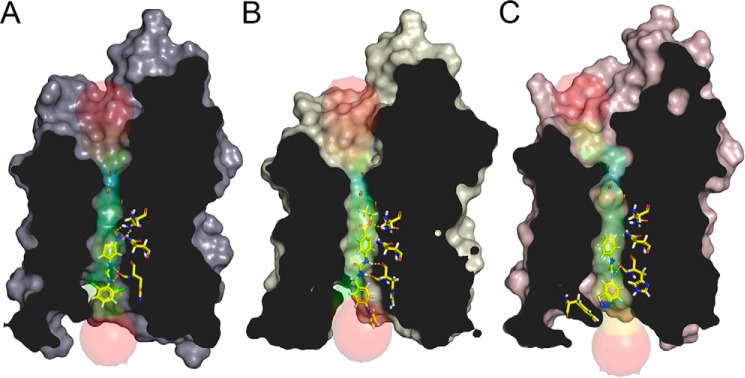
Figure 6.
Representative AQP-inhibitor binding sites. A–C, the calculated diameter of the AQP pore is shown in transparent surface view, colored from blue (narrowest) to red (widest). Docked ligands and select amino acids are shown in stick representation. Hydrogen bonds form between the inhibitor urea linker and backbone carbonyls of each AQP isoform. Asparagines of the NPA boxes form interactions between AQP3 and DFP00173 (A) and between AQP7 and Z433927330 (B). Phe-180 is unique to AQP9 and may form a positive edge interaction with an RF03176 nitrogen lone pair, thereby stabilizing a conformation that constricts the entrance to the pore (C).
To exemplify inhibitor–AQP interactions, DFP00173 interacts in a similar fashion with all three mouse aquaporins, with the dichlorophenyl group at the cytoplasmic opening of the pore and hydrogen bonding between the urea backbone of the ligand and loop B of the aquaporins, as well as hydrogen bonding between asparagines and the nitro group (Fig. 6A).
Compound Z433927330 differs from 9016645 by a pyrazole group (Figs. 2 and 3). Molecular docking suggests that the pyrazole does not grossly influence the position of these molecules in aquaglyceroporins (Fig. 6B). Besides the general interaction of loop B carbonyls with the urea linker, oxygens from the ester functional group act as hydrogen bond acceptors for asparagine hydrogens here.
Docking calculations suggest that the hydrophobic phenyl group in RF03176 is located close to the loop B asparagine, creating a tight constriction at this site. A phenylalanine (Phe-180) at the cytosolic entrance in m/hAQP9 is located proximal to the RF03176 benzothiadiazole. Interactions between nitrogen lone pairs of the thiadiazole group and the δ-positive edge of Phe-180 are conceivable (Fig. 6C). An identical interaction is seen in hAQP9 (35).
The homology models suggest interesting differences between mouse aquaglyceroporins. In line with similar channel substrate permeability, pore-lining residues are highly conserved. Differences that may explain inhibitor specificity are, however, observed in residues at the cytoplasmic pore entrance. Specifically, a Phe-180 in mAQP9 is replaced by Val-179 in mAQP3 and Thr-159 in mAQP7. Furthermore, the chemical environment surrounding the conserved histidine in loop B (His-81 in mAQP3) is noticeably different. In mAQP3, a glutamate (Glu-96) is involved in a hydrogen bond to the ϵ nitrogen of the histidine, which reduces its rotameric freedom. In mAQP7 the unprotonated δ nitrogen of the corresponding histidine (His-61) constrains freedom of rotation as a hydrogen bond acceptor to an asparagine (Asn-70). The latter is in a position accessible for pore solutes. In contrast, the residues at these two positions are both methionines in mAQP9 (Met-91 and Met-97, respectively). This allows the histidine (His-82) to adopt a different configuration that makes the pore narrower.
Discussion
Here we describe the identification of novel mAQP3 and mAQP7 inhibitors. Together with previously identified specific mAQP9 inhibitors, this provides a complete set of potent mouse aquaglyceroporin inhibitors, in part with high isoform specificity suitable for dissecting the function of these channels in cell-based assays.
These are not the first aquaporin inhibitors described in the literature. However, as raised by Verkman et al. (43), several previously described AQP inhibitors were not sufficiently validated upon discovery. Consequently, the effect of some AQP inhibitors could not be reproduced by independent laboratories (44–46). Here as well as in a previous study, we tested inhibitor specificity among related and less related AQP isoforms (34). We consider the observed inhibitor specificity as a strong argument for fidelity of the inhibitors described in these studies.
In further support for this class of inhibitors, we previously described the effect of hAQP9 point mutations on inhibition by RF03176 and structurally similar molecules (35). DFP00173 and Z433927330 share structural similarities with RF03176. Moreover, we confirmed inhibition of hAQP3 glycerol permeability by some of the described inhibitors in human erythrocytes, utilizing stopped-flow scattered light intensity recordings, a method considered a gold standard in inhibitor validation (43). Relative inhibitor potency measurements suggested good agreement of inhibitor potency between CHO cell water permeability and erythrocyte glycerol permeability assays.
A number of aquaporins, including AQP3, AQP8, and AQP9, have recently been described to function as H2O2 channels in cytokine and growth factor signaling, primarily in leukocytes (18, 19, 37, 38, 47–51). Similarly, altered cytokine responses have been described for AQP7-deficient dendritic cells (27). However, altered water and/or glycerol permeability of AQP7 deficient cells was proposed as an explanation. To our knowledge, this is the first study that describes AQP7-mediated H2O2 permeability, providing an alternative explanation for reduced cytokine responses in AQP7-deficient dendritic cells.
Verkman et al. (43) have previously branded aquaporins as “important but elusive drug targets.” The elusive part of the assessment was based in part on a low hit discovery rate in an unpublished screen for AQP1 inhibitors conducted by the authors. Only a few weak AQP1 inhibitors could be discovered in a screen of 100,000 small molecules (43). As an explanation, narrow AQP pore geometry as well as the necessity to find molecules binding deeply in this pore to inhibit water (and solute) transport has been given. In agreement with this view, we found that the pore diameter at the aromatic/arginine region seems to be too narrow to contain the AQP inhibitors discovered in this study. This results in a very limited surface that remains available for potential inhibitor binding at the extracellular orifice of mouse aquaglyceroporins. In contrast, we found that the aquaglycerol channel pore on the cytoplasmic side of the aromatic/arginine constriction can ideally fit linear aromatic molecules. It may well be confirmed that the discovery of inhibitors for pure water channels such as AQP1 is more challenging. This study identified at least one hit series containing potent as well as specific AQP3 and AQP7 inhibitors, respectively, in a medium-throughput screen of 7360 molecules. Moreover, in a previous screen, we identified two hit series containing potent and specific AQP9 inhibitors in a screen of only 1920 molecules (34). These outcomes suggest that the pore geometry of aquaglyceroporins permits inhibitor discovery.
We have previously characterized the binding of several hAQP9 inhibitors in a pore region facing the cytoplasm using computational and experimental methods (35). Our analyses in this study suggest that inhibition cooperatively results from steric hindrance of the pore by bulky functional groups and disruption of the pore–water hydrogen-bonding network. The role of steric hindrance is exemplified by the differences between DFP00173, comprising two ortho-chlorides, and the closely related but less potent DFP00172 (para-chloride) and SEW00833 (meta-chloride), respectively. Aquaporin water and solute permeability require backbone carbonyls in loops B and E as hydrogen bond acceptors and NPA asparagines as hydrogen bond donors. All of our described inhibitors form hydrogen bonds to backbone carbonyls, thus mimicking channel–substrate interactions. Indeed, AQP7 and APQ9 are permeable for urea (34, 52), which is a commonly observed linker in the aquaglyceroporin inhibitors we identified.
In light of this common inhibitor binding, the experimentally observed inhibitor specificity requires further consideration. Molecular docking analyses did not provide clear explanations for this selectivity. Interestingly, although occupying carbonyls of the channel polypeptide backbone, the inhibitor urea linker offers its own carbonyl as a potential substitute. At physiologically relevant temperatures, inhibitors and channels are dynamic. It is thus conceivable that alternative hydrogen bonding involving the urea carbonyl facilitates permeability through a residual pore. Thus, overall channel diameter as well as local inhibitor-bound isoform-specific pore constrictions may explain the most striking experimentally observed inhibitor specificities.
Notably, among our homology models, mAQP3 is narrowest at the NPA region. The nitro group in DFP00173 is predicted to form a very strong interaction with the asparagine of the first NPA box. Furthermore, Ile-71 restricts alternative backbone inhibitor interactions, locking DFP00173 in a position that does not allow residual pore permeability. The analogous Val-51 provides more room for alternative positioning in mAQP7. Similarly, favorable positioning seems to be hindered in mAQP9 by Phe-180, as ligands can only take up discrete positions along the carbonyl backbone.
Residual pore permeability may also provide an explanation for the previously observed RF03176 specificity to m/hAQP9 (34). AQP9-specific Met-91 and Met-97 are positioned in such a way at the cytoplasmic pore entrance that access to the underlying loop B backbone carbonyl His-82 may be obscured when RF03176 is bound. In support of this mechanism, we have previously mutated Met-91 and His-82 of hAQP9 (35). A M91N mutation could conceivably have provided better access to His-82 and did result in almost complete loss of inhibition. A H82A mutation may also have provided better access to the backbone carbonyl at this position and did result in lower inhibitor potency. For mAQP3 and mAQP7, similar shielding of the corresponding histidine carbonyl does not seem to be the case. For Z433927330, molecular docking interactions are almost identical to those of 9016645, whereas the additional pyrazole in Z433927330 resulted in higher experimentally observed potency. A simple explanation is that the additional pyrazole group in Z433927330 offers additional hydrogen bonding possibilities. However, docking calculations did not correlate well with the experimental observations. Further analyses will be required to find explanations for these effects.
In conclusion, we identified a complete set of aquaglyceroporin inhibitors. All described compounds are commercially available for independent verification. We confirmed the identity and purity of the three currently most useful substances, DFP00173, Z433927330, as well as the previously described HTS13286 (34), of one batch from the described sources by MS. The substances should be useful in future cell-based experiments, e.g. investigation of AQPs in cytokine signaling. All three substances provide useful starting points for further development of substances that are suited for in vivo experiments and potentially for drug development.
Experimental procedures
CHO cell experiments
Generation of the utilized CHO cells with tetracycline-inducible ectopic AQP expression has been described previously (34, 35). For H2O2 permeability assays, CHO cells with stably integrated tetracycline repressor were stably transfected with VspI-linearized pC1–Hyper-3 (39) and pC1-SypHer (53) plasmids, respectively, before selection of transfected cells with G418 (800 μg/ml, Thermo Fisher Scientific) for 2 weeks and subsequent sorting of a pool of YFP fluorescent cells on a BD FACSAria II flow cytometer (BD Biosciences). Human AQP3 was ligated from IMAGE clone ID 3877096 (Source BioScience) into pcDNA5/FRT/TO (Thermo Fisher Scientific) following KpnI/NotI digestion of the parent vectors. These DNA recombinations were designed with the help of AiO software (54).
The use of a plate reader to measure water permeability has been described previously (55) and was used with modifications. CHO cells were cultured in DMEM/F12/10% donor bovine serum, 250 μg/ml hygromycin, and 10 μg/ml blasticidin (all from Thermo Fisher Scientific) in a humidified incubator at 37 °C and 5% CO2.
Three days before water permeability or H2O2 permeability assays, 6000 cells/well were seeded in black, clear-bottom, poly-lysine–coated 96-well plates (Greiner, 655 946). Twenty-four hours after seeding, AQP expression was induced with tetracycline; in water permeability assays, tetracycline was titrated for each cell line to reach uninhibited cell shrinking of t½ of ∼1 s. For H2O2 permeability assays, all cell lines were induced with 5 ng/ml tetracycline, which leads to a maximal increase in water permeability in the tested cell lines. Cells were grown for an additional 48 h. Before water permeability assays, cells were incubated in fresh medium containing 5 mm probenecid, 2.5 μm calcein O,O′-diacetate tetrakis(acetoxymethyl) ester (Calcein-AM, BD Biosciences) for 45 min. The plates were transferred to a Fluostar Optima plate reader, and fluorescence intensity was recorded for 30 s (water permeability) and 79 s (H2O2 permeability). Optical filters were 485BP12/520 for calcein and 420BP10/490BP10 excitation and 520LP emission for HyPer-3 recordings. Osmotic cell shrinkage was induced 3.6 s into reads by addition of 1 volume of 500 mm sucrose in assay buffer. The other assay buffer components were as follows: MgSO4, 0.8 mm; KCl, 5.0 mm; CaCl2, 1.8 mm; NaHEPES, 25 mm; NaCl, 111.5 mm; and probenecid, 5.0 mm (pH 7.4). Similarly, 1 volume of 150 μm H2O2 was added 3.6 s into HyPer-3 recordings. All assay buffers contained 1% DMSO to keep DMSO concentrations of inhibitor-treated wells constant during volume additions (44). For cytotoxicity assays, CHO cells were grown in the presence of the indicated inhibitor concentrations for 48 h, followed by cell loading with Calcein-AM as for water permeability assays and similar fluorescence recording at a single time point.
Inhibitor compounds
Maybridge Hitfinder Library version 8 was purchased from Thermo Fisher Scientific. Follow-up compounds were collectively purchased via MolPort (Riga, Latvia) from various vendors: S04288, MWP00821, SEW00832, BTB14129, GK00877, BTB09519, S15361, DFP00176, SPB00688, JM00015, HTS06792, DFP00173, DFP00172, SEW00835, SEW00834, and SEW00833 (all from Maybridge/Thermo Fisher Scientific); F2740-0111 (Life Chemicals Inc.); Z433927330 (ENAMINE); 9016645, 9053871, and 7791389, (ChemBridge Corp.); and IVK/0050432 (Alinda Chemical, Ltd.). All compounds were dissolved in DMSO and applied in assay buffer containing a final concentration of 1% DMSO.
Erythrocyte isolations and stopped-flow light scattering
Freshly collected human whole blood was washed three times in PBS (spinning at 800 × g) to remove serum and the cellular buffy coat. Stopped-flow light scattering experiments were carried out on a BioLogic MPS-200 stopped-flow reaction analyzer (BioLogic, Claix, France) as described previously (56). For measurement of glycerol permeability, suspensions of erythrocytes (∼1% hematocrit) in PBS were subjected to a 100 mm inwardly directed gradient of glycerol. The erythrocyte volume changes were recorded as the kinetics of the scattered light intensity at 20 °C at a wavelength of 530 nm and with a dead time of 1.6 ms and a mixing efficiency of 99% in less than 1 ms. Data were fit to a single exponential function, and the related half-life time (t½) of the cell swelling phase during entry of glycerol and water into the erythrocytes was measured. During this phase, the t½ values represent an index of the glycerol permeability of the analyzed erythrocytes. A series of stopped-flow light scattering experiments were done for each inhibitor compound, testing the effects of concentrations obtained by serial dilution. All compounds were dissolved in DMSO and applied in 1% DMSO for 10 min at 20 °C prior the light scattering measurement. Erythrocyte suspensions containing 1% DMSO (10 min, 20 °C) represented the control condition.
Homology modeling
To rationalize AQP inhibitor isoform selectivity, we generated homology models of murine AQP3, AQP7, and AQP9. The sequences were aligned to the Escherichia coli glycerol transporter GlpF using ClustalW version 2.1 (57, 58) and inspected manually. The 2.2-Å X-ray structure of GlpF (PDB code 1FX8) (59) was used as a template for homology modeling, and all three input files were submitted to the I-TASSER server (60). The geometry of the resulting homology models was optimized by fragment guided molecular dynamics simulation using the FG-MD server (61), followed by manual correction of a few pore residues using the Coot software (62). Noteworthy is phenylalanine 64 in AQP9, which was further into the pore than expected, probably because of the lack of pore waters in the generation of the models. Pore radii were calculated utilizing the program HOLE (63).
Molecular docking
The ligands DFP00172, RF03176, Z4339927330, and 9016645 were docked into the pore of the AQP3, AQP7, and AQP9 using the LeadIT Molecular Docking software. To avoid bias, the pore in its entirety was defined as a binding site. A library containing the four ligands of interest was created, and docking was performed using standard settings. The results were optimized using Hyde, a program in the LeadIT software package (version 2.3.2, BioSolveIT GmbH, Sankt Augustin, Germany).
Statistical analysis
Dose–response curves were compared between AQP isoforms and substances by two-way ANOVA in GraphPad Prism 5.0.
Author contributions
Y. S., P. G., G. C., and M. R. conceptualization; Y. S., P. G., A. M., U. J., G. C., and M. R. data curation; Y. S., P. G., A. M., T. S., I. A., M. K. O., U. J., P. K., J. D. N., S. N., G. C., and M. R. formal analysis; Y. S., P. G., A. M., U. J., G. C., and M. R. validation; Y. S., P. G., A. M., G. C., and M. R. investigation; Y. S. and M. R. visualization; Y. S., P. G., A. M., T. S., U. J., and M. R. methodology; Y. S., P. G., A. M., U. J., G. C., and M. R. writing-original draft; Y. S., P. G., A. M., T. S., I. A., M. K. O., U. J., P. K., J. D. N., S. N., G. C., and M. R. writing-review and editing; I. A., U. J., G. C., and M. R. supervision; I. A., U. J., P. K., J. D. N., S. N., G. C., and M. R. funding acquisition; M. R. resources.
Supplementary Material
This work was supported by funds from Aalborg University, the Danish Research Council, and Stiftelsen Olle Engkvist Byggmästare. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.
- AQP
- aquaporin
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- CHO
- Chinese hamster ovary
- m
- mouse
- h
- human
- Calcein-AM
- calcein O,O′-diacetate tetrakis(acetoxymethyl) ester
- ANOVA
- analysis of variance
- N.D.
- not determined
- NPA
- conserved asparagine-proline-alanine motive.
References
- 1. Ishibashi K., Sasaki S., Fushimi K., Uchida S., Kuwahara M., Saito H., Furukawa T., Nakajima K., Yamaguchi Y., and Gojobori T. (1994) Molecular cloning and expression of a member of the aquaporin family with permeability to glycerol and urea in addition to water expressed at the basolateral membrane of kidney collecting duct cells. Proc. Natl. Acad. Sci. U.S.A. 91, 6269–6273 10.1073/pnas.91.14.6269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Miller E. W., Dickinson B. C., and Chang C. J. (2010) Aquaporin-3 mediates hydrogen peroxide uptake to regulate downstream intracellular signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 15681–15686 10.1073/pnas.1005776107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Echevarria M., Windhager E. E., Tate S. S., and Frindt G. (1994) Cloning and expression of AQP3, a water channel from the medullary collecting duct of rat kidney. Proc. Natl. Acad. Sci. U.S.A. 91, 10997–11001 10.1073/pnas.91.23.10997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ecelbarger C. A., Terris J., Frindt G., Echevarria M., Marples D., Nielsen S., and Knepper M. A. (1995) Aquaporin-3 water channel localization and regulation in rat kidney. Am. J. Physiol. 269, F663–F672 [DOI] [PubMed] [Google Scholar]
- 5. Frigeri A., Gropper M. A., Turck C. W., and Verkman A. S. (1995) Immunolocalization of the mercurial-insensitive water channel and glycerol intrinsic protein in epithelial cell plasma membranes. Proc. Natl. Acad. Sci. U.S.A. 92, 4328–4331 10.1073/pnas.92.10.4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramírez-Lorca R., Vizuete M. L., Venero J. L., Revuelta M., Cano J., Ilundáin A. A., and Echevarría M. (1999) Localization of aquaporin-3 mRNA and protein along the gastrointestinal tract of Wistar rats. Pflugers Arch. 438, 94–100 10.1007/s004240050884 [DOI] [PubMed] [Google Scholar]
- 7. Nielsen S., King L. S., Christensen B. M., and Agre P. (1997) Aquaporins in complex tissues: II: subcellular distribution in respiratory and glandular tissues of rat. Am. J. Physiol. 273, C1549–C1561 10.1152/ajpcell.1997.273.5.C1549 [DOI] [PubMed] [Google Scholar]
- 8. Frigeri A., Gropper M. A., Umenishi F., Kawashima M., Brown D., and Verkman A. S. (1995) Localization of MIWC and GLIP water channel homologs in neuromuscular, epithelial and glandular tissues. J. Cell Sci. 108, 2993–3002 [DOI] [PubMed] [Google Scholar]
- 9. Umenishi F., Verkman A. S., and Gropper M. A. (1996) Quantitative analysis of aquaporin mRNA expression in rat tissues by RNase protection assay. DNA Cell Biol. 15, 475–480 10.1089/dna.1996.15.475 [DOI] [PubMed] [Google Scholar]
- 10. Roudier N., Verbavatz J. M., Maurel C., Ripoche P., and Tacnet F. (1998) Evidence for the presence of aquaporin-3 in human red blood cells. J. Biol. Chem. 273, 8407–8412 10.1074/jbc.273.14.8407 [DOI] [PubMed] [Google Scholar]
- 11. Ma T., Song Y., Yang B., Gillespie A., Carlson E. J., Epstein C. J., and Verkman A. S. (2000) Nephrogenic diabetes insipidus in mice lacking aquaporin-3 water channels. Proc. Natl. Acad. Sci. U.S.A. 97, 4386–4391 10.1073/pnas.080499597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rojek A., Praetorius J., Frøkiaer J., Nielsen S., and Fenton R. A. (2008) A current view of the mammalian aquaglyceroporins. Annu. Rev. Physiol. 70, 301–327 10.1146/annurev.physiol.70.113006.100452 [DOI] [PubMed] [Google Scholar]
- 13. Roudier N., Ripoche P., Gane P., Le Pennec P. Y., Daniels G., Cartron J.-P., and Bailly P. (2002) AQP3 deficiency in humans and the molecular basis of a novel blood group system, GIL. J. Biol. Chem. 277, 45854–45859 10.1074/jbc.M208999200 [DOI] [PubMed] [Google Scholar]
- 14. Hara-Chikuma M., and Verkman A. S. (2008) Prevention of skin tumorigenesis and impairment of epidermal cell proliferation by targeted aquaporin-3 gene disruption. Mol. Cell Biol. 28, 326–332 10.1128/MCB.01482-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xia H., Ma Y.-F., Yu C.-H., Li Y.-J., Tang J., Li J.-B., Zhao Y.-N., and Liu Y. (2014) Aquaporin 3 knockdown suppresses tumour growth and angiogenesis in experimental non-small cell lung cancer. Exp. Physiol. 99, 974–984 10.1113/expphysiol.2014.078527 [DOI] [PubMed] [Google Scholar]
- 16. Xiong G., Chen X., Zhang Q., Fang Y., Chen W., Li C., and Zhang J. (2017) RNA interference influenced the proliferation and invasion of XWLC-05 lung cancer cells through inhibiting aquaporin 3. Biochem. Biophys. Res. Commun. 485, 627–634 10.1016/j.bbrc.2017.02.013 [DOI] [PubMed] [Google Scholar]
- 17. Satooka H., and Hara-Chikuma M. (2016) Aquaporin-3 controls breast cancer cell migration by regulating hydrogen peroxide transport and its downstream cell signaling. Mol. Cell Biol. 36, 1206–1218 10.1128/MCB.00971-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hara-Chikuma M., Chikuma S., Sugiyama Y., Kabashima K., Verkman A. S., Inoue S., and Miyachi Y. (2012) Chemokine-dependent T cell migration requires aquaporin-3-mediated hydrogen peroxide uptake. J. Exp. Med. 209, 1743–1752 10.1084/jem.20112398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hara-Chikuma M., Satooka H., Watanabe S., Honda T., Miyachi Y., Watanabe T., and Verkman A. S. (2015) Aquaporin-3-mediated hydrogen peroxide transport is required for NF-κB signalling in keratinocytes and development of psoriasis. Nat. Commun. 6, 7454 10.1038/ncomms8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ikezoe K., Oga T., Honda T., Hara-Chikuma M., Ma X., Tsuruyama T., Uno K., Fuchikami J., Tanizawa K., Handa T., Taguchi Y., Verkman A. S., Narumiya S., Mishima M., and Chin K. (2016) Aquaporin-3 potentiates allergic airway inflammation in ovalbumin-induced murine asthma. Sci. Rep. 6, 25781 10.1038/srep25781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Skowronski M. T., Lebeck J., Rojek A., Praetorius J., Füchtbauer E. M., Frøkiaer J., and Nielsen S. (2007) AQP7 is localized in capillaries of adipose tissue, cardiac and striated muscle: implications in glycerol metabolism. Am. J. Physiol. Renal Physiol. 292, F956–F965 10.1152/ajprenal.00314.2006 [DOI] [PubMed] [Google Scholar]
- 22. Hibuse T., Maeda N., Funahashi T., Yamamoto K., Nagasawa A., Mizunoya W., Kishida K., Inoue K., Kuriyama H., Nakamura T., Fushiki T., Kihara S., and Shimomura I. (2005) Aquaporin 7 deficiency is associated with development of obesity through activation of adipose glycerol kinase. Proc. Natl. Acad. Sci. U.S.A. 102, 10993–10998 10.1073/pnas.0503291102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hibuse T., Maeda N., Nakatsuji H., Tochino Y., Fujita K., Kihara S., Funahashi T., and Shimomura I. (2009) The heart requires glycerol as an energy substrate through aquaporin 7, a glycerol facilitator. Cardiovasc. Res. 83, 34–41 10.1093/cvr/cvp095 [DOI] [PubMed] [Google Scholar]
- 24. Maeda N., Funahashi T., Hibuse T., Nagasawa A., Kishida K., Kuriyama H., Nakamura T., Kihara S., Shimomura I., and Matsuzawa Y. (2004) Adaptation to fasting by glycerol transport through aquaporin 7 in adipose tissue. Proc. Natl. Acad. Sci. U.S.A. 101, 17801–17806 10.1073/pnas.0406230101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sohara E., Rai T., Miyazaki J., Verkman A. S., Sasaki S., and Uchida S. (2005) Defective water and glycerol transport in the proximal tubules of AQP7 knockout mice. Am. J. Physiol. Renal Physiol. 289, F1195–F1200 10.1152/ajprenal.00133.2005 [DOI] [PubMed] [Google Scholar]
- 26. Matsumura K., Chang B. H., Fujimiya M., Chen W., Kulkarni R. N., Eguchi Y., Kimura H., Kojima H., and Chan L. (2007) Aquaporin 7 is a β-cell protein and regulator of intraislet glycerol content and glycerol kinase activity, β-cell mass, and insulin production and secretion. Mol. Cell Biol. 27, 6026–6037 10.1128/MCB.00384-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hara-Chikuma M., Sugiyama Y., Kabashima K., Sohara E., Uchida S., Sasaki S., Inoue S., and Miyachi Y. (2012) Involvement of aquaporin-7 in the cutaneous primary immune response through modulation of antigen uptake and migration in dendritic cells. FASEB J. 26, 211–218 10.1096/fj.11-186627 [DOI] [PubMed] [Google Scholar]
- 28. Calamita G., Perret J., and Delporte C. (2018) Aquaglyceroporins: drug targets for metabolic diseases? Front. Physiol. 9, 851 10.3389/fphys.2018.00851,10.3389/fpls.2018.00851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zelenina M., Bondar A. A., Zelenin S., and Aperia A. (2003) Nickel and extracellular acidification inhibit the water permeability of human aquaporin-3 in lung epithelial cells. J. Biol. Chem. 278, 30037–30043 10.1074/jbc.M302206200 [DOI] [PubMed] [Google Scholar]
- 30. Zelenina M., Tritto S., Bondar A. A., Zelenin S., and Aperia A. (2004) Copper inhibits the water and glycerol permeability of aquaporin-3. J. Biol. Chem. 279, 51939–51943 10.1074/jbc.M407645200 [DOI] [PubMed] [Google Scholar]
- 31. Wishart D. S., Feunang Y. D., Marcu A., Guo A. C., Liang K., Vázquez-Fresno R., Sajed T., Johnson D., Li C., Karu N., Sayeeda Z., Lo E., Assempour N., Berjanskii M., Singhal S., et al. (2018) HMDB 4.0: The Human Metabolome Database for 2018. Nucleic Acids Res. 46, D608–D617 10.1093/nar/gkx1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martins A. P., Marrone A., Ciancetta A., Galán Cobo A., Echevarría M., Moura T. F., Re N., Casini A., and Soveral G. (2012) Targeting aquaporin function: potent inhibition of aquaglyceroporin-3 by a gold-based compound. PLoS ONE 7, e37435 10.1371/journal.pone.0037435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Madeira A., de Almeida A., de Graaf C., Camps M., Zorzano A., Moura T. F., Casini A., and Soveral G. (2014) A gold coordination compound as a chemical probe to unravel aquaporin-7 function. ChemBioChem 15, 1487–1494 10.1002/cbic.201402103 [DOI] [PubMed] [Google Scholar]
- 34. Jelen S., Wacker S., Aponte-Santamaría C., Skott M., Rojek A., Johanson U., Kjellbom P., Nielsen S., de Groot B. L., and Rützler M. (2011) Aquaporin-9 protein is the primary route of hepatocyte glycerol uptake for glycerol gluconeogenesis in mice. J. Biol. Chem. 286, 44319–44325 10.1074/jbc.M111.297002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wacker S. J., Aponte-Santamaría C., Kjellbom P., Nielsen S., de Groot B. L., and Rützler M. (2013) The identification of novel, high affinity AQP9 inhibitors in an intracellular binding site. Mol. Membr Biol. 30, 246–260 10.3109/09687688.2013.773095 [DOI] [PubMed] [Google Scholar]
- 36. Lichtenfels R., Biddison W. E., Schulz H., Vogt A. B., and Martin R. (1994) CARE-LASS (calcein-release-assay), an improved fluorescence-based test system to measure cytotoxic T lymphocyte activity. J. Immunol. Methods 172, 227–239 10.1016/0022-1759(94)90110-4 [DOI] [PubMed] [Google Scholar]
- 37. Moniaga C. S., Watanabe S., Honda T., Nielsen S., and Hara-Chikuma M. (2015) Aquaporin-9-expressing neutrophils are required for the establishment of contact hypersensitivity. Sci. Rep. 5, 15319 10.1038/srep15319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Watanabe S., Moniaga C. S., Nielsen S., and Hara-Chikuma M. (2016) Aquaporin-9 facilitates membrane transport of hydrogen peroxide in mammalian cells. Biochem. Biophys. Res. Commun. 471, 191–197 10.1016/j.bbrc.2016.01.153 [DOI] [PubMed] [Google Scholar]
- 39. Bilan D. S., Pase L., Joosen L., Gorokhovatsky A. Y., Ermakova Y. G., Gadella T. W., Grabher C., Schultz C., Lukyanov S., and Belousov V. V. (2013) HyPer-3: a genetically encoded H2O2 probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chem. Biol. 8, 535–542 10.1021/cb300625g [DOI] [PubMed] [Google Scholar]
- 40. Duong Van Huyen J., Bens M., and Vandewalle A. (1998) Differential effects of aldosterone and vasopressin on chloride fluxes in transimmortalized mouse cortical collecting duct cells. J. Membr. Biol. 164, 79–90 10.1007/s002329900395 [DOI] [PubMed] [Google Scholar]
- 41. Hasler U., Mordasini D., Bens M., Bianchi M., Cluzeaud F., Rousselot M., Vandewalle A., Feraille E., and Martin P. Y. (2002) Long term regulation of aquaporin-2 expression in vasopressin-responsive renal collecting duct principal cells. J. Biol. Chem. 277, 10379–10386 10.1074/jbc.M111880200 [DOI] [PubMed] [Google Scholar]
- 42. de Groot B. L., and Grubmüller H. (2005) The dynamics and energetics of water permeation and proton exclusion in aquaporins. Curr. Opin. Struct. Biol. 15, 176–183 10.1016/j.sbi.2005.02.003 [DOI] [PubMed] [Google Scholar]
- 43. Verkman A. S., Anderson M. O., and Papadopoulos M. C. (2014) Aquaporins: important but elusive drug targets. Nat. Rev. Drug Discov. 13, 259–277 10.1038/nrd4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang B., Kim J. K., and Verkman A. S. (2006) Comparative efficacy of HgCl2 with candidate aquaporin-1 inhibitors DMSO, gold, TEA+ and acetazolamide. FEBS Lett. 580, 6679–6684 10.1016/j.febslet.2006.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Søgaard R., and Zeuthen T. (2008) Test of blockers of AQP1 water permeability by a high-resolution method: no effects of tetraethylammonium ions or acetazolamide. Pflugers Arch. 456, 285–292 10.1007/s00424-007-0392-2 [DOI] [PubMed] [Google Scholar]
- 46. Esteva-Font C., Jin B. J., Lee S., Phuan P. W., Anderson M. O., and Verkman A. S. (2016) Experimental evaluation of proposed small-molecule inhibitors of water channel aquaporin-1. Mol. Pharmacol. 89, 686–693 10.1124/mol.116.103929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rhee S. G. (2006) Cell signaling: H2O2, a necessary evil for cell signaling. Science 312, 1882–1883 10.1126/science.1130481 [DOI] [PubMed] [Google Scholar]
- 48. Hara-Chikuma M., Watanabe S., and Satooka H. (2016) Involvement of aquaporin-3 in epidermal growth factor receptor signaling via hydrogen peroxide transport in cancer cells. Biochem. Biophys. Res. Commun. 471, 603–609 10.1016/j.bbrc.2016.02.010 [DOI] [PubMed] [Google Scholar]
- 49. Bertolotti M., Bestetti S., García-Manteiga J. M., Medraño-Fernandez I., Dal Mas A., Malosio M. L., and Sitia R. (2013) Tyrosine kinase signal modulation: a matter of H2O2 membrane permeability? Antioxid. Redox Signal. 19, 1447–1451 10.1089/ars.2013.5330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bertolotti M., Farinelli G., Galli M., Aiuti A., and Sitia R. (2016) AQP8 transports NOX2-generated H2O2 across the plasma membrane to promote signaling in B cells. J. Leukocyte Biol. 100, 1071–1079 10.1189/jlb.2AB0116-045R [DOI] [PubMed] [Google Scholar]
- 51. Medraño-Fernandez I., Bestetti S., Bertolotti M., Bienert G. P., Bottino C., Laforenza U., Rubartelli A., and Sitia R. (2016) Stress regulates aquaporin-8 permeability to impact cell growth and survival. Antioxid. Redox Signal. 24, 1031–1044 10.1089/ars.2016.6636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tsukaguchi H., Shayakul C., Berger U. V., Mackenzie B., Devidas S., Guggino W. B., van Hoek A. N., and Hediger M. A. (1998) Molecular characterization of a broad selectivity neutral solute channel. J. Biol. Chem. 273, 24737–24743 10.1074/jbc.273.38.24737 [DOI] [PubMed] [Google Scholar]
- 53. Poburko D., Santo-Domingo J., and Demaurex N. (2011) Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J. Biol. Chem. 286, 11672–11684 10.1074/jbc.M110.159962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Karreman C. (2002) AiO, combining DNA/protein programs and oligo-management. Bioinformatics 18, 884–885 10.1093/bioinformatics/18.6.884 [DOI] [PubMed] [Google Scholar]
- 55. Fenton R. A., Moeller H. B., Nielsen S., de Groot B. L., and Rützler M. (2010) A plate reader-based method for cell water permeability measurement. Am. J. Physiol. Renal Physiol. 298, F224–F230 10.1152/ajprenal.00463.2009 [DOI] [PubMed] [Google Scholar]
- 56. Calamita G., Ferri D., Gena P., Carreras F. I., Liquori G. E., Portincasa P., Marinelli R. A., and Svelto M. (2008) Altered expression and distribution of aquaporin-9 in the liver of rat with obstructive extrahepatic cholestasis. Am. J. Physiol. Gastrointest. Liver Physiol. 295, G682–G690 10.1152/ajpgi.90226.2008 [DOI] [PubMed] [Google Scholar]
- 57. Thompson J. D., Higgins D. G., and Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 59. Fu D., Libson A., Miercke L. J., Weitzman C., Nollert P., Krucinski J., and Stroud R. M. (2000) Structure of a glycerol-conducting channel and the basis for its selectivity. Science 290, 481–486 10.1126/science.290.5491.481 [DOI] [PubMed] [Google Scholar]
- 60. Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y. (2015) The I-TASSER suite: protein structure and function prediction. Nat. Methods 12, 7–8 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang J., Liang Y., and Zhang Y. (2011) Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure 19, 1784–1795 10.1016/j.str.2011.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., and Sansom M. S. (1996) J. Mol. Graph. 14, 354–360, 376 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.