Abstract
Ribonucleoprotein (RNP) granules are membrane-less organelles consisting of RNA-binding proteins (RBPs) and RNA. RNA granules form through liquid–liquid phase separation (LLPS), whereby weak promiscuous interactions among RBPs and/or RNAs create a dense network of interacting macromolecules and drive the phase separation. Post-translational modifications (PTMs) of RBPs have emerged as important regulators of LLPS and RNP granule dynamics, as they can directly weaken or enhance the multivalent interactions between phase-separating macromolecules or can recruit or exclude certain macromolecules into or from condensates. Here, we review recent insights into how PTMs regulate phase separation and RNP granule dynamics, in particular arginine (Arg)-methylation and phosphorylation. We discuss how these PTMs regulate the phase behavior of prototypical RBPs and how, as “friend or foe,” they might influence the assembly, disassembly, or material properties of cellular RNP granules, such as stress granules or amyloid-like condensates. We particularly highlight how PTMs control the phase separation and aggregation behavior of disease-linked RBPs. We also review how disruptions of PTMs might be involved in aberrant phase transitions and the formation of amyloid-like protein aggregates as observed in neurodegenerative diseases.
Keywords: ribonuclear protein (RNP), RNA-binding protein, post-translational modification (PTM), protein methylation, phosphorylation, liquid–liquid phase separation (LLPS), membrane-less organelle (MLO), phase transitions, protein aggregation, stress granules
Introduction
In recent years, phase separation has emerged as a novel principle of cellular organization. Numerous membrane-less organelles (MLOs)3 in the cytoplasm or nucleus, which are supramolecular assemblies of macromolecules, assemble through the process of liquid–liquid phase separation (LLPS) (1). The resulting assemblies, also called biomolecular condensates (2), concentrate certain macromolecules, while excluding others, and hence can speed up or slow down biochemical reactions or organize intracellular structures (3). Classical examples of MLOs include RNP granules, such as stress granules (SGs) and P bodies (PBs) in the cytoplasm, or nucleoli and paraspeckles in the nucleus. They consist of RNA-binding proteins (RBPs) and RNAs and often have liquid-like properties, i.e. they fuse with one another upon contact and show fast internal rearrangement and dynamic exchange of molecules with their surroundings (3–5). However, there are also cellular RNP condensates with solid-like properties, such as the Balbiani body (Bb) in oocytes, which is a large, electron-dense “aggregate” that sequesters many membranous organelles and is thought to keep them in a low-activity state during oocyte dormancy (6, 7).
In recent years, great advances have come from in vitro experiments that have reconstituted liquid-like RNP granules or other condensates with only one or a few components, e.g. purified RBPs or RNA (4, 8–13). Such in vitro studies have demonstrated that liquid-like assemblies are metastable and can harden into viscous liquids, gels, or even solid-like amyloids. Such liquid-to-solid–state transitions are thought to underlie the formation of intracellular, pathological protein aggregates in the context of neurodegenerative diseases, e.g. aggregates containing the RBPs TDP-43 or FUS in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) or Tau aggregates in Alzheimer's disease (AD) (4, 14–16). In vitro reconstitution experiments also helped to establish that biological phase separation is mainly driven by weak promiscuous interactions between multivalent protein interaction domains (8) or intrinsically disordered low complexity domains (LCDs) (17). As multivalent interaction motifs and short linear motifs in intrinsically disordered regions (IDRs) and LCDs are often post-translationally modified (18–21), it is not surprising that post-translational modifications (PTMs) are important regulators of phase separation (22, 23). PTMs change the physicochemical properties of the modified amino acids, e.g. by altering the steric properties, charge state, or bulkiness, and hence modulate interactions either positively or negatively. PTMs can alter the phase-separated state by directly weakening or enhancing the multivalent interactions between phase-separating macromolecules (Fig. 1A) or by recruiting/excluding certain macromolecules (e.g. a protein or a nucleic acid) into/from the condensate (Fig. 1B). In this fashion, PTMs can control the assembly or disassembly of condensates and MLOs and change their composition as well as their material properties (Fig. 1).
Figure 1.
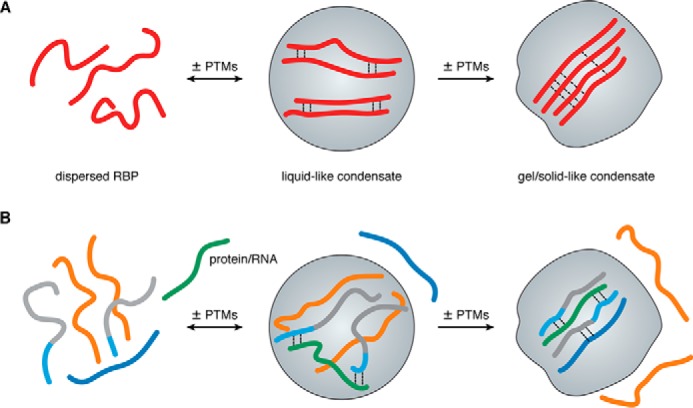
PTMs can regulate formation and material properties of condensates. A, PTMs have the potential to promote the assembly or disassembly of liquid-like condensates as well as the transition to gel- or solid-like condensates by enhancing or disrupting weak multivalent interactions between phase-separating RBPs. B, in a heterogeneous solution of macromolecules PTMs can regulate the recruitment or exclusion of specific protein or RNA molecules from or to MLOs.
In this review, we highlight recent examples that illustrate how phase separation of RBPs and intracellular RNP granules are regulated by PTMs, in particular arginine (Arg)-methylation and phosphorylation as well-studied paradigms. We discuss how these PTMs regulate the phase behavior of prototypical RBPs in vitro and how they might influence the composition, dynamics, and material properties of cellular RNP granules, such as SGs or amyloid-like condensates. We place special emphasis on disease-linked RBPs that are thought to form pathological aggregates via aberrant liquid-to-solid–state transitions and are linked to neurodegenerative diseases.
Key amino acids governing the phase separation behavior of RBPs are frequently post-translationally modified
Recently, much progress has been made toward elucidating the molecular interactions that underlie phase separation of proteins and dictate their phase behavior. In particular, recent work by Hyman and co-workers (24) has identified a sequence encoded the “molecular grammar” governing phase separation of RBPs with prion-like LCDs. This study established that interactions between aromatic and positively charged amino acids, in particular Tyr and Arg, are the key driving force for phase separation of RBPs (24). In line with these findings, several other studies have recently shown that Arg residues in RGG/RG or RG-FG repeat motifs are required for phase separation of FUS (25–27) or Ddx4, respectively (28, 29). The material properties of RBP condensates are also determined by a specific sequence code: Gly residues enhance fluidity of condensates, whereas Gln and Ser residues promote hardening (24), possibly by promoting formation of labile cross–β-sheets (30–32).
Considering that Arg, Tyr, and Ser are frequently post-translationally modified, e.g. by methyl-groups on Arg and phospho-groups on Tyr or Ser, PTMs on these residues are likely to be crucial modulators of RBP phase separation. Phosphorylation introduces a negative charge, which should tune RBP phase separation behavior either positively or negatively, depending on whether it is driven primarily by aromatic–cationic interactions or by aromatic–aromatic interactions (24). Arg-methylation does not alter the charge, but increases the bulkiness and alters the charge distribution, hydrophobicity, and hydrogen bonding properties of the guanidinium headgroup (33), all of which impacts intermolecular interactions and hence could tune the phase separation behavior.
In the following paragraphs, we will discuss arginine methylation and phosphorylation as two important control mechanisms that tune phase separation of RBPs and hence regulate the dynamic assembly and disassembly of cellular RNP granules. We contemplate whether these PTMs are “friend or foe” of RBP phase separation, i.e. whether they tend to promote or rather to suppress LLPS and RNP granule assembly/disassembly. Finally, we will briefly discuss how the interplay of Arg-methylation and phosphorylation with other PTMs may provide an even more complex biological regulation of phase separation and condensates.
Arginine methylation: a key regulator of RBP phase separation and RNP granule dynamics
Repetitive RGG- or RG-rich motifs are a prevalent sequence pattern in heterogeneous ribonucleoproteins (hnRNPs) and other nuclear RBPs (34). Arg residues located in RGG/RG motifs are the preferred sites for methylation by members of the protein arginine methyltransferase (PRMT) family (35). PRMTs catalyze the transfer of one or two methyl groups from S-adenosylmethionine to the terminal guanidino nitrogen atoms of Arg, thus generating monomethyl-arginine or symmetric or asymmetric dimethyl-arginine. In mammals, there are nine PRMTs (PRMT1–9), but the major enzyme responsible for the bulk of Arg-methylation activity is PRMT1 (36), which preferentially methylates RGG/RG motifs and generates monomethyl-arginine and asymmetric dimethyl-arginine (37).
Arg-methylation is thought to be a rather stable or “static” modification, although there is evidence for the existence of an Arg-demethylase activity (37), and JMJD6 and JmjC histone lysine demethylases have been reported to function as Arg-demethylases (38, 39). Nevertheless, Arg-methylation is considered much less dynamic than other PTMs, such as phosphorylation or acetylation (40); hence, in contrast to phosphorylation, Arg-methylation is unlikely to act as a rapid “switch” that can be readily activated and deactivated in response to changes in the cellular environment.
As RGG/RG-rich motifs are highly abundant in RBPs (34) and Arg-aromatic interactions are a key driving force of RBP phase separation (24), it is not surprising that a vast number of recent studies have provided evidence for a critical role of RGG/RG-rich motifs in RBP phase separation (25, 26, 28, 41–43). We summarize recent examples that have established Arg-methylation as a suppressor of RBP phase separation in vitro and discuss different scenarios how Arg-methylation may modulate RNP granule assembly/disassembly in cells.
Arginine methylation suppresses LLPS of RBPs in vitro
So far, three different examples (Ddx4, hnRNP-A2, and FUS) have demonstrated that Arg-methylation reduces LLPS by reducing Arg-aromatic (π) interactions that normally drive phase separation of these RBPs (Fig. 2). The first example was provided by the DEAD-box RNA helicase Ddx4, which is an essential component of germline-specific RNP granules, e.g. nuage or chromatoid bodies in mammals and P granules in Caenorhabditis elegans. Nott et al. (28) demonstrated that the disordered N-terminal RGG-rich domain of Ddx4 (DdxN1) undergoes LLPS in vitro and forms liquid-like compartments in cells. They furthermore established that formation of liquid DdxN1 droplets is driven by cation–π interactions between repeated RG and FG motifs and is suppressed by asymmetric dimethylation, introduced by co-expression of PRMT1 with Ddx4N1 in Escherichia coli (28). Subsequently, a structural study of the hnRNP-A2 prion-like LCD reported that asymmetric dimethylation of four RGG motifs (by in vitro methylation with purified PRMT1) causes a significant reduction of phase separation, by disrupting the interactions of Arg residues with aromatic residues (44). Finally, two independent studies recently reported that Arg-methylation suppresses phase separation of FUS, which shows extensive asymmetric dimethylation in its three RGG/RG repeat domains (45–47). The first study by our own laboratory compared unmethylated FUS purified from E. coli and asymmetrically dimethylated FUS obtained by in vitro methylation with PRMT1 (25). Compared with unmethylated FUS, methylated FUS exhibits significantly reduced liquid–liquid demixing and shows an enhanced droplet dynamic, as measured by fluorescence recovery after photobleaching of half-bleached FUS-enhanced GFP droplets. Moreover, when added to semi-permeabilized cells containing SGs, the methylated protein shows significantly lower SG association than unmethylated FUS (25). The other study by Qamar et al. (26) obtained hypomethylated FUS (HYPO-FUS) by purifying the protein from insect cells treated with adenosine-2′,3′-dialdehyde (AdOx), a global inhibitor of methylation. Compared with “normal” FUS purified from untreated insect cells, HYPO-FUS forms a larger number of liquid droplets and shows reduced sphericity and fewer fusion events, suggesting reduced dynamics (26). Using atomic force microscopy, they furthermore demonstrated that a portion of the HYPO-FUS assemblies have stiffer nanomechanical properties and show enhanced binding of an amyloidophyllic dye (pFTAA), suggestive of β-sheet–rich hydrogels.
Figure 2.
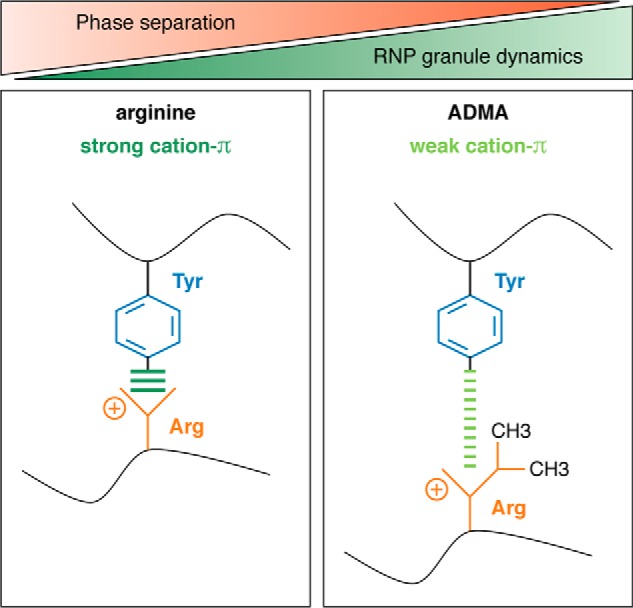
Arginine methylation weakens cation–π interactions. Arginine methylation weakens cation–π interactions of arginines with aromatic amino acids (e.g. tyrosine) and thereby weakens the intermolecular interactions that drive phase separation of RBPs, causing reduced phase separation of RBPs and increased RNP granule dynamics.
The finding that the lack of Arg-methylation promotes LLPS and more solid-like FUS assemblies is most likely of direct pathological relevance, as hypomethylated FUS has been found in insoluble protein aggregates in brains of FTD patients, whereas FUS is normally soluble and asymmetrically dimethylated in healthy brains (48, 49). Given the recent findings that Arg-methylation suppresses LLPS of FUS and keeps the protein in a dynamic, liquid-like state (25, 26), it can be speculated that loss of FUS Arg-methylation is a pathogenic event, promoting LLPS and liquid-to-solid–state transition of FUS, eventually leading to pathological FUS aggregates. In this respect, it will be interesting to determine how loss of FUS Arg-methylation arises and whether other Arg-methylated RBPs that aggregate in neurodegenerative diseases, e.g. hnRNP-A family proteins, EWS or TAF15 (50), are also hypomethylated in patients and show enhanced LLPS and solidification upon loss of Arg-methylation.
So far, three different examples (Ddx4N1, hnRNP-A2 LCD, and FUS) have shown a suppression of LLPS by Arg-methylation (25, 26, 28, 44), and there are currently no examples where Arg-methylation had the opposite effect. This could indicate that Arg-methylation in general is a PTM that reduces Arg-aromatic (π) interactions (Fig. 2) and thus reduces phase separation of RBPs, in particular of RBPs with numerous Arg and Tyr residues, e.g. hnRNP-UL1, hnRNP-R, Drosha, or Syncrip (24). Further experimental tests will eventually prove or disprove this hypothesis.
Arginine methylation regulates RNP granule assembly through diverse mechanisms
Methylation of Arg residues was not only shown to influence phase separation of RBPs in vitro, but was also found to regulate RNP granule dynamics and function in cells, although the underlying molecular mechanisms are in most cases still unknown. Studying Arg-methylation in cells is challenging, as “unmethyl-Arg” and “methyl-Arg” cannot be faithfully mimicked by amino acid mutations, and inhibition or overexpression of PRMTs affects a multitude of Arg-methylated proteins, making it impossible to attribute a certain phenotypic change to one particular Arg-methylated protein. Nevertheless, from the reported examples, two general principles can be extracted regarding how Arg-methylation of RBPs may regulate RNP granules: (a) by altering protein–protein or protein–RNA interactions, and (b) by altering nucleocytoplasmic transport of RBPs and hence altering their concentration in the nucleus or cytoplasm (Fig. 3). Interestingly, Arg-methylation does not always have a suppressive effect on RNP granule formation, as might be expected from its suppressive effect on RBP LLPS in vitro (see above and Fig. 2). Instead, it can also promote RNP granule assembly, e.g. by recruiting a certain factor (protein or RNA) that in turn promotes phase separation and RNP granule formation, or by increasing the concentration of a phase-separating RBP in a particular compartment, hence promoting RBP condensation in this compartment. Thus, even though Arg-methylation generally tends to reduce phase separation of RBPs (25, 26, 28, 44), it has a more complex regulatory role in the in vivo context and can affect the dynamics and functional properties of RNP granules in numerous different ways.
Figure 3.
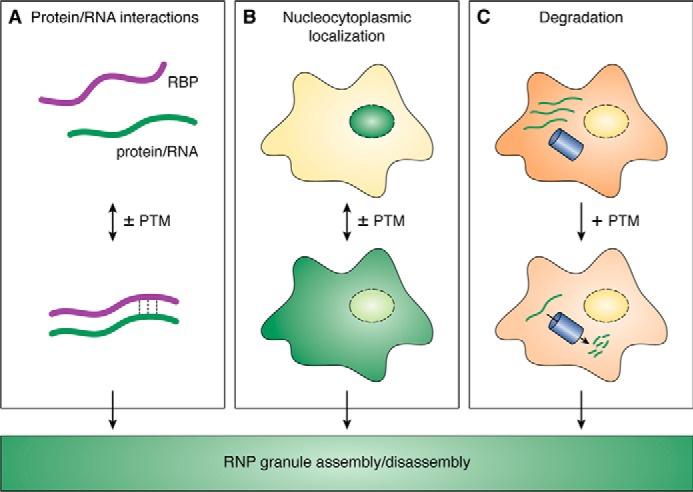
Different mechanisms how PTMs can regulate RNP granule assembly or disassembly in cells. A, PTMs of RBPs can regulate protein–protein or protein–RNA interactions and thereby modulate RNP granules. B, nucleocytoplasmic localization of RBPs can be regulated by PTMs. Altered RBP concentrations in the nucleus or cytoplasm may affect the assembly/disassembly of RNP granules in the respective cellular compartment. C, PTMs can regulate RNP granules by promoting degradation by autophagy or the proteasome. Reduced cellular concentrations of a specific RBP may affect RNP granules.
First evidence for a “suppression” of RNP granules by Arg-methylation was provided in 2006 by Dolzhanskaya et al. (51), who reported that global inhibition of methylation using AdOx increases the number of cells with FMRP-containing cytoplasmic granules, which are negative for SG marker proteins. More recently, Arg-methylation has been more directly linked to the suppression of SGs, as it was shown that hypermethylation of the SG-nucleating protein G3BP1 by PRMT overexpression suppresses SG formation (52). Conversely, pharmacological inhibition of PRMT1 or PRTM5 or AdOx treatment reduces Arg-methylation levels of G3BP1 and elevates the number of arsenite-induced SGs. They furthermore demonstrated that arsenite stress quickly and reversibly decreases G3BP1 methylation on three Arg residues, suggesting that G3BP1 demethylation might be involved in SG formation (52). A follow-up study has implicated the JmjC domain-containing histone arginine demethylase JMJD6 as G3BP1-demethylating enzyme: JmJD6 partially localizes to SGs, and its overexpression promotes SG formation and demethylation of G3BP1 (53). Whether Arg-methylation directly affects LLPS of FMRP or G3BP1, e.g. suppressing it through reduced cation–π interactions (Fig. 2), remains to be demonstrated using in vitro methylation and in vitro phase separation experiments.
As noted above, there is also ample evidence for a “promotion” of RNP granules by Arg-methylation. In 2016, it was shown that the RGG domain of the symmetrically dimethylated Lsm4 protein is required for PB formation, and PRMT5 depletion strongly reduces Lsm4 methylation and PB formation (54). Similarly, RAP55A, a member of the Scd6 or Lsm14 family, is an asymmetrically dimethylated PB protein that fails to localize to PBs upon PRMT1 depletion, indicating that Arg-methylation stimulates PB localization of RAP55A (55). This appears to be functionally relevant, as Arg-methylation of the RAP55A yeast orthologue Scd6 promotes its SG localization and translation repression activity by enhancing eIF4G1 binding (56). Interestingly, a number of other translational repressors, e.g. Sbp1, Npl3, and Ded1, are Arg-methylated and repress translation through binding to eIF4G1 (57). Moreover, the disease-linked RBPs FMRP, ATXN-2, and hnRNP-A1 are Arg-methylated and play a role in translational control (58–60). Thus, it will be interesting to determine whether Arg-methylation is a general regulator of translational repressors, e.g. by regulating their interaction with other proteins and/or mRNAs, thus affecting their recruitment into RNP granules and controlling their activity as translational repressors.
Arg-methylation cannot only promote the assembly of liquid-like condensates, but recently was also reported to promote the solid-like Bb in germ cells. The Bb is a large, electron-dense “aggregate” in oocytes, enriched in germline-specific mRNAs and a single protein with a prion-like LCD, called Xvelo in C. elegans or Bucky ball (Buc) in zebrafish (6, 7). The C terminus of Buc contains a tri-RG motif that is symmetrically dimethylated and is required for Bb formation: Arg-methylated Buc recruits the Tudor-domain containing protein Tdrd6, which regulates Buc aggregation and promotes Bb assembly and hence is essential for germ cell development (61). Other examples where Arg-methylation promotes RNP assembly through recruitment of the Tudor-domain containing protein SMN include the symmetrically dimethylated snRNP proteins SmD1 and SmD3 and the asymmetrically dimethylated protein FUS. Methylated SmD1 and SmD3 recruit SMN and in turn promote assembly snRNPs and spliceosomes (62), whereas FUS recruits SMN via its methylated RGG domains to promote assembly of Cajal body–associated nuclear structures called Gems (63, 64). Together, these example demonstrate that Arg-methylation frequently promotes RNP granule assembly by positively affecting protein–protein interactions, e.g. with Tudor domain-containing proteins, which in turn trigger RNP granule assembly (Fig. 3A).
Another mechanism how Arg-methylation may affect RNP granule assembly or disassembly is through the regulation of nucleocytoplasmic transport, leading to an altered concentration of certain factors in the cytoplasm versus the nucleus (Fig. 3B). Evidence for this is so far limited, but was proposed to underlie the reduced SG localization of cold-inducible RNA-binding protein (CIRP) and of SERPINE1 mRNA-binding protein 1 (SERBP1) upon AdOx treatment (65, 66). SG recruitment of CIRP upon oxidative stress or heat shock is abolished in AdOx-treated HeLa cells (65), potentially because nucleocytoplasmic localization of CIRP is regulated by Arg-methylation (67); hence, unmethylated CIRP might be unable to exit the nucleus and fail to localize to cytoplasmic SGs. Similarly, SERBP1 shows reduced cytoplasmic localization upon loss of Arg-methylation (66) and hence may not partition into SGs upon AdOx treatment (68). How exactly nucleocytoplasmic transport of CIRP and SERBP1 is regulated by Arg-methylation remains to be established, but there are several other examples where Arg-methylation has been clearly shown to regulate Transportin-dependent nuclear import of RBPs, e.g. of PABPN1 or FUS (48, 49, 69). This additional level of regulation should be kept in mind as a further possibility of how Arg-methylation may affect condensation of certain RBPs in the nucleus or cytoplasm and hence may control the assembly or disassembly of nuclear or cytosolic RNP granules.
In summary, numerous examples illustrate that Arg-methylation is a key regulatory PTM controlling RNP granules' dynamics and functions. The above-mentioned examples illustrate that this regulation can entail different mechanisms, e.g. modulation of protein–protein interactions, or altered nucleocytoplasmic transport and hence altered protein concentrations in the nucleus or cytoplasm. The different scenarios most likely lead to altered phase separation behavior of individual RNP granule components, although this still remains to be demonstrated through in vitro phase separation experiments. A future challenge will be to combine in vitro studies (such as those described above) with cellular or in vivo experiments to reveal how Arg-methylation regulates phase separation of specific RBPs and in turn modulates RNP granule assembly and dynamics.
Other PTMs on arginine residues and their role in phase separation and RNP granule dynamics
Arginine residues in RGG/RG motifs are also subject to other types of PTMs, most notably citrullination. The conversion of Arg to citrulline is catalyzed by the peptidyl arginine deiminase (PAD) protein family, which converts the positively charged guanidino group of Arg to a neutral urea group; hence citrulline has different chemical properties than unmethylated Arg. RGG/RG-rich motifs are consensus sequences for PAD4-mediated citrullination, and interestingly, PAD4 overexpression was recently found to competitively inhibit Arg-methylation and to increase the solubility of different RBPs (e.g. FUS, TAF15, EWS, and hnRNP-A1) in cells (70). In line with these findings, PAD-mediated citrullination of FUS was shown to abolish LLPS of FUS in vitro (26). This suggests that citrullination reduces the multivalent Arg–Tyr interactions that drive phase separation of FUS (24, 26) and hence has a similar “suppressive” effect on RBP phase separation as Arg-methylation. Whether citrullination of disease-linked RBPs is altered in neurodegenerative diseases with RBP aggregates remains to be examined. Interestingly, a single nucleotide polymorphism in the PAD4 gene, associated with low PAD4 expression, is linked with a higher ALS risk and earlier disease onset (70). This could indicate that PAD4-mediated RBP citrullination is decreased in ALS patients and may contribute to aberrant phase transitions of disease-linked RBPs, e.g. FUS, TAF15, EWS, and hnRNP-A1. Under which conditions PAD4-mediated citrullination occurs and what determines the relative ratios of PRMT-mediated Arg-methylation and PAD-mediated citrullination of RGG/RG motifs is poorly understood. Further studies are necessary to dissect the interplay between Arg-methylation and citrullination and to clarify their roles in phase separation and RNP granule dynamics.
Besides methylation and citrullination, several other enzymatic and nonenzymatic PTMs of Arg residues are known, including phosphorylation, ADP-ribosylation (71), and methylglyoxal adducts, the latter being abundant on histones (72). Given the important role of Arg residues in phase separation of RBPs (24), it will be interesting to determine whether these other PTMs arise on RBPs under physiological or disease conditions and how they affect phase separation and RNP granule dynamics.
Phosphorylation: an important regulator of LLPS and membrane-less organelles
Phosphorylation is the most frequent PTM (73), and phosphorylation by kinases and dephosphorylation by phosphatases provide a major control mechanism to many fundamental processes in eukaryotic cells. In contrast to Arg-methylation, phosphorylation can rapidly and reversibly modify proteins and act as a rapid switch to respond to signals and quickly modulate protein function. The most frequently phosphorylated residue is serine (∼90%), followed by threonine and tyrosine (∼10%). Ser residues were found to affect the hardening behavior of RBPs (24), and Ser and Tyr residues are often highly enriched in prion-like LCDs that drive LLPS of RBPs (24, 74). Unlike Arg-methylation, phosphorylation alters the charge of the modified amino acid side chains, i.e. it introduces two negative charges (PO42−). This drastically alters their steric and chemical properties and provides novel possibilities for intra- and intermolecular electrostatic interactions (18). According to the recently established molecular grammar of RBP phase separation, this is expected to tune RBP phase separation behavior either positively or negatively, depending on whether LLPS is driven primarily by aromatic–cationic interactions or by aromatic–aromatic interactions, respectively (24). Accordingly, several recent studies have provided evidence for a critical role of phosphorylation in regulating the phase separation behavior of RBPs in vitro and the assembly or disassembly of RNP granules in cells.
Phosphorylation regulates LLPS and aggregation of disease-linked RBPs in vitro
In contrast to Arg-methylation, phosphorylation does not always have a suppressive effect on LLPS of RBPs in vitro, but there are examples for both negative and positive regulation of LLPS by phosphorylation.
One example is the neurodegeneration-linked protein FUS, which is phosphorylated at multiple Ser/Thr residues by DNA-dependent protein kinase (DNA-PK) (75–77). Phosphorylation with DNA-PK interferes with phase separation of the FUS LCD (30, 76, 78), reduces binding to FUS-LCD hydrogels (79), and abrogates phase separation of multivalent interacting proteins tethered to FUS-LCD (80). DNA-PK–mediated phosphorylation or phosphomimetic substitutions of 12 Ser/Thr residues to acidic glutamate in full-length FUS also reduce phase separation and prevent its subsequent liquid-to-solid–state transition and fibril or aggregate formation (76). NMR experiments demonstrated that this was due to less frequent transient intermolecular contacts (76) and interference with hydrogen bonding and cross–β-interactions (30, 78). In line with these in vitro findings, phosphomimetic FUS was less aggregation-prone when overexpressed in yeast or mammalian cells (76). Under which conditions FUS is phosphorylated in vivo is still poorly understood; so far, only the treatment of cells with DNA damage–inducing drugs, e.g. calicheamicin or calyculin-A, was found to transiently induce FUS phosphorylation on multiple Ser/Thr residues (75, 77). It has been proposed that transient phosphorylation of FUS after DNA damage may occur in FTD patients with FUS aggregates, as post-mortem brains of these patients show elevated levels of γ-H2AX (75), a marker for DNA double–strand breaks. Whether phosphorylation of FUS (and the other members of the FET protein family, EWS and TAF15) indeed occurs in disease conditions or as a physiological stress response, and how it affects other functional properties, such as protein interactions or subcellular localization, are still matters of debate (22, 75, 77) and remain to be clarified by future studies.
Another example is TDP-43 (TAR DNA–binding protein of 43 kDa), which is found in cytoplasmic and nuclear aggregates in the degenerating brain regions in most ALS and FTD patients (81, 82) and up to 30–50% of AD patients (83, 84). A phosphomimetic S48E substitution in the N-terminal domain (NTD) of TDP-43, a site that is constitutively phosphorylated in different cell lines (85–88), was found to reduce LLPS of TDP-43 in vitro and to form less viscous nuclear assemblies than WT TDP-43, indicating reduced intermolecular interactions (87). In line with previous studies reporting that oligomerization of TDP-43 via the NTD is essential for proper splicing activity (89, 90), the S48E mutation impairs the splicing regulatory activity of TDP-43 (87), indicating that phase separation of TDP-43 may be important for the protein's physiological function in splicing regulation. Interestingly, the data on S48E also demonstrate that regions outside prion-like LCDs of RBPs can regulate phase separation via PTMs, in this case phosphorylation in the globular NTD of TDP-43 (87).
Surprisingly, it has so far not been examined how phosphorylation and dephosphorylation of TDP-43 in the C-terminal LCD affect its phase separation behavior in vitro. TDP-43 is known to be abnormally phosphorylated on multiple Ser residues in the LCD (e.g. Ser-379, Ser-403, Ser-404, Ser-409, and Ser-410) in ALS and FTD patients (91, 92), which was shown to be mediated by casein kinase 1δ (CK1δ) (93–95), GADD34-bound casein kinase 1ϵ (CK1ϵ) (96), or Tau tubulin kinases TTBK1/2 (97, 98). Dephosphorylation of TDP-43 was shown to be mediated by protein phosphatases PP1α and PP1β (99) or the phosphatase calcineurin (100). In cells and model organisms, some studies found that CK1δ- or TTBK1/2-mediated phosphorylation or inhibition of calcineurin-mediated dephosphorylation causes aggregation of phosphorylated TDP-43 and promotes toxicity and neurodegeneration (97, 98, 100–102). In contrast, Li et al. (103) reported that phospho-mimicking substitutions of the C-terminal serines decrease TDP-43 aggregation and alleviate cytotoxicity in a neuronal cell line, suggesting that phosphorylation may rather serve as a compensatory mechanism antagonizing TDP aggregation. In vitro LLPS experiments will hopefully be able to resolve whether C-terminal TDP-43 phosphorylation suppresses or enhances phase separation and aggregation of TDP-43.
In contrast to FUS and TDP-43, where phosphorylation has a suppressive effect on LLPS, the microtubule-associated protein Tau exemplifies a protein whose phase separation is promoted by phosphorylation. Tau is a neuron-specific protein that stabilizes microtubules, but forms insoluble neurofibrillary tangles in AD and other neurodegenerative disorders (104). Although Tau is not a classical RBP, it has long been known that Tau binds RNA and that RNA induces fibrillization of Tau in vitro (105, 106). Additionally, Tau was shown to associate with SGs by binding to RBPs that are typically found in SGs, e.g. TIA-1 (107–109). More recently, Tau or its isolated microtubule binding domain was shown to undergo LLPS in vitro (15, 16, 110, 111), and GFP-tagged Tau forms dynamic, droplet-like Tau condensates in neurons (16, 108). Tau condensation appears to have an important physiological function, as Tau droplets were shown to concentrate tubulin and to nucleate microtubule bundles, suggesting that intracellular Tau condensates may locally promote microtubule polymerization, e.g. in the neuronal growth cone, at branching points or after axon injury (111).
Nonetheless, physiological Tau condensation can become pathological and give rise to aberrant Tau aggregates as seen in neurodegenerative disorders; Tau droplets rapidly transition from a liquid to a gel-like state in vitro and even mature into thioflavin S–positive aggregates (16). Interestingly, Ser phosphorylation in the microtubule binding domain of Tau by MARK2 kinase promotes LLPS of Tau, indicating that the additional negative charges promote electrostatic interactions that drive Tau phase separation (15). Similar observations were made when hyper-phosphorylated (insect cell-derived) Tau was compared with partially dephosphorylated (alkaline phosphatase-treated) Tau and E. coli-derived unphosphorylated Tau, demonstrating that phosphorylation promotes phase separation of full-length Tau (16). This is most likely pathologically relevant, as hyperphosphorylated Tau is found in Tau aggregates of AD and FTD patients (112–114), and the AD-risk factor ApoE4 elevates Tau phosphorylation in neurons (115). Thus, phosphorylation of Tau (e.g. by MARK2 or other kinases (116)) may drive pathological Tau aggregation by promoting LLPS and a liquid-to-solid–state transition of Tau. Interestingly, Tau was shown to associate with a number of RBPs and to promote SG formation by interacting with TIA-1 (107–109). In this respect, it would be interesting to examine how phosphorylation and altered Tau LLPS behavior influence phase separation of TIA-1 and other SG-associated RBPs, e.g. using in vitro reconstitution experiments. This could reveal an interesting connection between a microtubule-nucleating protein and RBP phase separation as well as RNP granule dynamics.
Phosphorylation regulates the dynamics of RNP granules, e.g. SGs, in cells
Evidence for an important role of phosphorylation in the regulation of RNP granule dynamics mainly comes from the observation that inhibition or overexpression of certain kinases leads to altered RNP granule assembly or disassembly (117–121). In many cases, the involved substrate(s) and underlying molecular mechanisms are still unclear, but there are a few examples from which we can learn about possible mechanisms regarding how phosphorylation may modulate RNP granules: (a) by modulating LLPS of certain RBPs, thus promoting their condensation or decondensation and hence RNP granule assembly or disassembly, and (b) by promoting degradation of certain RBPs (by the proteasome or autophagy), hence reducing their cellular concentration and altering RNP granule assembly or disassembly (Fig. 3C). Just like Arg-methylation, phosphorylation can have either a suppressive or a promoting effect on RNP granule formation, although in most paradigms examined so far, phosphorylation was found to promote RNP granule disassembly, i.e. to “suppress” certain RNP compartments.
The first prominent kinase that shows a suppressive effect on certain MLOs is dual specificity tyrosine phosphorylation–regulated kinase 3 (DYRK3), a kinase with broad specificity that phosphorylates multiple Ser and Thr residues in unstructured domains (117, 122). In 2013, DYRK3 was shown to localize to SGs via its N-terminal LCD and to regulate their dissolution via its kinase activity (117). More recently, the same group examined the role of DYRK3 during mitosis, as it has long been known that numerous cytosolic and nuclear MLOs disappear during mitosis. They found that DYRK3 acts as a “dissolvase” of multiple MLOs during mitosis (118). Stable isotope labeling with amino acids in cell culture-based proteomics showed that DYRK3 interacts with multiple RBPs that are known components of splicing speckles, SGs, and the centrosome. During mitosis, as the nuclear envelope dissolves, the DYRK3-to-substrate ratio suddenly increases for certain nuclear or cytosolic DYRK3 substrates, which drives them from a condensed state into a dispersed state and causes dissolution of the corresponding MLO (118).
Besides DYRK3, casein kinase 2 (CK2) was recently found to cause SG disassembly, most likely via phosphorylation of the SG-nucleating protein G3BP1 (119). It has long been known that phosphorylation of G3BP1 on Ser-149 regulates the protein's oligomerization, SG recruitment, and RNase activity (123, 124), but only recently was CK2 identified as the kinase that phosphorylates G3BP1 on Ser-149 and thus promotes SG disassembly (119). Whether CK2 also acts on other key SG regulators, such as other SG nucleating RBPs or translation inhibition factors, and hence affects SG dynamics through multiple different mechanisms remains to be examined. Future research will also have to address how different kinases coordinately regulate SG dynamics. It seems likely that at least some of them directly phosphorylate key SG nucleators (e.g. G3BP1) and regulate their phase separation behavior.
A third kinase that shows a “suppressive” effect on SGs is spleen tyrosine kinase (Syk), although this mechanism appears to involve SG degradation rather than SG disassembly. Syk is recruited to G3BP1-positive SGs, where it actively phosphorylates SG-localized proteins on Tyr residues and promotes SG clearance by autophagy (120). Degradation of entire RNP granules or individual RBPs could be a common theme of how phosphorylation down-regulates RNP granules and their cellular functions (Fig. 3C). Recently, Ime2 kinase was shown to phosphorylate the amyloid-like translational repressor Rim4 in budding yeast, causing Rim4 disassembly and rapid degradation by the proteasome (121). This mechanism is important for progression of yeast cells through meiosis, as yeast employs amyloid-like structures to mediate translational repression during pre-meiotic G1 phase but have to release translational repression to progress through meiosis (125). This is achieved by phosphorylation in the LCD of Rim4, causing its decondensation and subsequent proteasomal degradation. It is tempting to speculate that clearance of amyloid-like assemblies by phosphorylation might be a general mechanism that is also used to clear disease-associated protein aggregates, e.g. Tau or TDP-43. Indeed, Tau and TDP-43 aggregates are known to be hyperphosphorylated in post-mortem brains of AD, FTD, or ALS patients (82, 95, 112, 113, 126), suggesting that this could be a typical cellular response to the presence of amyloid-like aggregates. Perhaps deficiencies in certain chaperones or in proteasomal degradation in the aging brain prevent efficient disassembly or clearance of hyperphosphorylated Tau or TDP-43 aggregates, leading to their persistence.
As mentioned above, phosphorylation cannot only have a negative, suppressive effect on RNP granules, but also can promote their formation (Fig. 4). Kinases that were reported to have a “promoting” effect on SGs include 5′-AMP-activated protein kinase-α2 (AMPK-α2), which localizes to SGs upon stress treatment of HeLa cells. Pharmacological inhibition or knockdown of AMPK-α interferes with SG formation, whereas pharmacological activation of the enzyme stimulates SG assembly (127, 128). Other examples are the mTOR (mechanistic target of rapamycin) effector kinases S6 kinase 1 and 2 (S6K1 and S6K2) that were shown to localize to SGs and to be required for SG assembly and maintenance after mild oxidative stress (129).
Figure 4.
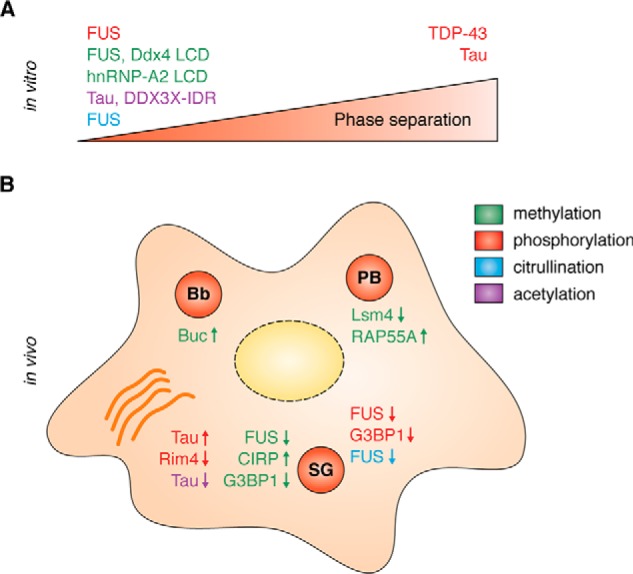
PTMs can have either a suppressive or a promoting effect on phase separation and RNP granule formation. Color code in legend indicates the respective PTM. For details and references see text. A, in vitro: PTMs were shown to reduce or enhance phase separation of the indicated RBP or LCD or IDR. B, in cells: PTMs can regulate the assembly, disassembly, and composition of different RNP granules, e.g. SG, PB, and Bb, and of amyloid-like aggregates (orange). An arrow pointing up indicates a promotion of condensate formation or RBP recruitment by the respective PTM, and an arrow pointing down indicates condensate disassembly or RBP exclusion from the condensate by the respective PTM.
Taken together, phosphorylation appears to be a crucial control mechanism governing RNP granule assembly, disassembly, and/or degradation (Figs. 3 and 4). For the most part, it is still unknown which exact molecular interactions are modulated by the involved kinases and whether phosphatases are also involved in the regulation. Another interesting question is how a kinase can access its phosphorylation sites within condensates, especially if phosphorylation sites are partially buried within solid-like assemblies, e.g. Rim4 aggregates. Further study of the molecules and interactions involved will hopefully uncover how phosphorylation regulates the dynamics of various RNP granules and whether it can be exploited as a promising drug target to disassemble or prevent formation of pathological RNP aggregates.
Arginine methylation and phosphorylation as regulators of LLPS and RNP granules: friend or foe?
As discussed above, there are numerous paradigms indicating an important role of Arg-methylation and phosphorylation in the regulation of LLPS of RGG/RG-containing RBPs, both from in vitro studies and from studies of RNP granules in cells or model organisms (Fig. 4). Depending on the modified proteins, both PTMs can have either a suppressive or a promoting effect on LLPS and RNP granule assembly, i.e. they cannot be clearly categorized as “friend” or “foe” of phase separation. However, so far there are more examples where Arg-methylation and phosphorylation suppress LLPS of RBPs and trigger the disassembly of RNP granules, so for the most part, they tend to negatively regulate biological phase separation.
Suppression of LLPS by Arg-methylation or phosphorylation may be especially relevant for aggregation-prone RBPs that get highly concentrated in cytoplasmic RNP granules and therefore are at risk of undergoing aberrant liquid-to-solid transitions and engaging in amyloid-like cross–β-sheet interactions, e.g. the highly abundant Arg/Ser/Tyr-rich proteins FUS, EWS, and TAF15 (24). In young and healthy cells, numerous quality control mechanisms are in place to control LLPS and aberrant phase transitions of these proteins, e.g. chaperones and factors that promote degradation of misfolded proteins, such as the ubiquitin–proteasome system and autophagy (130). Arg-methylation could be an additional “protein quality control” mechanism that declines during aging, as several PRMTs and global Arg-methylation levels are reduced in aged cells or rats (131). Arg-methylation appears to be especially relevant in terminally differentiated and long-lived neurons, e.g. motor neurons, which show tissue-specific expression of PRMT8 and require Arg-methylation for a proper stress response and for maintaining motor neuron health during aging (132). Suppression of aberrant RBP phase separation by Arg-methylation may be part of the protective mechanism, and hence it would be interesting to examine whether loss of PRMT8 promotes pathological RBP phase transitions and alters RNP granule dynamics and function in neurons. Taken together, Arg-methylation may be a “friend” of protein homeostasis and quality control of liquid phase separation and hence a “friendly” and protective PTM in neurons.
Whether phosphorylation of disease-linked aggregating proteins is indeed a protective “friend” or rather a threatening “foe” still remains to be clarified, and it is likely that no general rule applies to all aggregating proteins. On the one hand, phosphorylation of amyloid-like aggregates may be an important quality control mechanism, favoring decondensation of solid assemblies and subsequent clearance by the proteasome or autophagy, as recently shown for Rim4 aggregates in yeast (121). On the other hand, phosphorylation of Tau and TDP-43 promotes their aggregation, and both proteins are known to be hyperphosphorylated in post-mortem brains of AD, FTD, or ALS patients (82, 95, 112, 113, 126). Interestingly, both Tau and TDP-43 can be phosphorylated by Tau tubulin kinases 1 and 2 (TTBK1/2) (97, 133), and elevated levels of TTBK1/2 have been identified in brains of AD (134) as well as FTD-Tau and FTD-TDP patients (98), where they co-localize with pTDP-43 (97). A phosphatase that was found to be elevated in the aging brain and cortices of ALS patients is the nuclear PP1 isoform PP1γ (158), furthermore indicating that misregulated phosphorylation and dephosphorylation events may contribute to pathological protein aggregation and neurodegeneration.
Interplay of arginine methylation and phosphorylation with other PTMs
Besides post-translational Arg-methylation and Ser/Tyr-phosphorylation, many other PTMs are found on RBPs, in particular in intrinsically disordered regions and short linear motifs (21). RBPs can carry single or multiple PTMs, or a combination of different PTMs on the same or different amino acids, which can lead to additive, cooperative, or competitive responses, providing many layers of complex biological regulation (18). How the different types of PTMs coordinately regulate phase separation of RBPs and RNP granule dynamics, is still largely undetermined. RBPs are frequently both methylated and phosphorylated, suggesting that they could cooperate or interfere with one another. For instance, for the SG nucleating protein G3BP1, it has been proposed that demethylation may cooperate with dephosphorylation to promote SG assembly and translational inhibition (52, 119). Other examples could include FUS or hnRNP-A1, which are constitutively Arg-methylated and become phosphorylated during DNA damage, oxidative stress, or heat stress, respectively (22, 135).
Other PTMs that appear to cross-talk with phosphorylation are lysine acetylation and O-linked GlcNAc (O-GlcNAc) modification. Lys acetylation was shown to decrease Tau LLPS and aggregation (136, 137) as well as LLPS of the N-terminal IDR of DDX3X (DDX3X-IDR1) (138), and the deacetylase HDAC6 was shown to be required for SG formation (139). Interestingly, acetylation on Lys-321 of Tau prevents phosphorylation of the downstream Ser-324 residue, which inhibits Tau function and promotes its aggregation (136). Thus, acetylation and phosphorylation coordinately regulate Tau assembly and function. Similar mechanisms may exist for other acetylated and phosphorylated RBPs, e.g. TDP-43 (95, 140). The second example is O-GlcNAcylation on Ser/Thr residues, which reversibly occurs on numerous components of the translational machinery in response to stress and promotes SG and PB formation (141) as well as selective translation of stress mRNAs during the heat-stress response (142). O-GlcNAcylation appears to be tightly interlinked with phosphorylation, as both PTMs occur on Ser and Thr residues. Mass spectrometry studies have shown that O-GlcNAcylation and phosphorylation are often found on the same proteins, and protein kinases are more extensively O-GlcNAcylated than proteins in general, demonstrating cross-talk between the two PTMs (143, 144). An interesting target protein that is both phosphorylated and O-GlcNAcylated is Tau, and curiously, loss of O-GlcNAcylation in forebrain neurons induces progressive neurodegeneration, an increased production of hyperphosphorylated Tau and Tau aggregation (145). Another example is hnRNP-A1, whose nucleocytoplasmic localization is regulated by phosphorylation and O-GlcNAcylation (135, 146). It will be interesting to study the role of O-GlcNAcylation on Tau and hnRNP-A1 phase separation and SG association, and how both types of PTM (and possibly other PTMs) coordinately regulate the phase separation and aggregation behavior of these disease-linked proteins.
So far, most PTMs have been studied in isolation, either through in vitro modification approaches or cellular studies exploiting chemical inhibition or silencing/overexpression of a certain modifying enzyme. Recently, advances in semi-synthetic strategies have allowed for the site-specific introduction of single or multiple different PTMs into recombinant proteins, as successfully demonstrated for Lys acetylation and Tyr and Ser phosphorylation in the microtubule-binding domain of Tau (147). Such strategies provide new opportunities for investigating other types of PTMs that occur within Tau or other heavily post-translationally modified proteins. This may eventually allow us to decipher the “PTM code” of key disease-linked proteins, such as Tau, FUS, or TDP-43.
Perspective
Arg-methylation and phosphorylation have been recognized as important regulators of LLPS and RNP granule dynamics; however, they are by far not the only PTMs that have been shown to modulate phase separation and RNP granules. Several deubiquitylases as well as the small ubiquitin-like proteins NEDD8 and SUMO were shown to regulate SG assembly and disassembly in yeast and mammalian cells (148–151). Ubiquitin and other small ubiquitin-like proteins are covalently attached to target proteins and often provide an altered binding surface and altered protein–protein interactions. Furthermore, ubiquitin and poly(ADP-ribose) (PAR) were recently shown to influence phase separation in trans, i.e. ubiquitin binds to the UBA domain of Ubiquilin 2 (UBQLN2) and thus disperses UBQLN2 condensates (152), whereas PAR binds to PAR-binding motifs in TDP-43 and thus promotes phase separation and SG localization of TDP-43, but prevents pathological TDP-43 phosphorylation (153). This suggests a highly complex regulation of phase separation by a multitude of different PTMs, and we are only beginning to unravel this complex puzzle.
Beyond RBPs and RNP granules, PTMs also control phase separation of key chromatin and transcription regulators (154–156) as well as signaling proteins in various signaling cascades (19, 157). Hence, the control of phase separation by PTMs is not only an important principle for RBPs and RNP granules, but most likely for all phase-separating proteins and biomolecular condensates.
Acknowledgment
We thank Dr. Saskia Hutten for critical comments on the manuscript and figures.
This work was supported in part by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) within the Emmy Noether Grant DO 1804/1-1 (to D. D.) and Munich Cluster for Systems Neurology Grant EXC 1010, SyNergy (to D. D.). This article is part of the thematic series, Phase separation of RNA-binding proteins in physiology and disease. The authors declare that they have no conflicts of interest with the contents of this article.
- MLO
- membrane-less organelle
- RBP
- RNA-binding protein
- RNP
- ribonucleoprotein
- PTM
- post-translational modification
- LLPS
- liquid–liquid phase separation
- FUS
- fused in sarcoma
- FTD
- frontotemporal dementia
- LCD
- low complexity domain
- ALS
- amyotrophic lateral sclerosis
- NTD
- N-terminal domain
- AD
- Alzheimer's disease
- IDR
- intrinsically disordered region
- SG
- stress granule
- PB
- P body
- Bb
- Balbiani body
- snRNP
- small nuclear ribonucleoprotein
- PAR
- poly(ADP-ribose)
- PRMT
- protein arginine methyltransferase
- HYPO-FUS
- hypomethylated FUS
- DNA-PK
- DNA-dependent protein kinase
- AMPK-α2
- 5′-AMP-activated protein kinase-α2
- PAD
- peptidyl arginine deiminase
- AdOx
- adenosine-2′,3′-dialdehyde.
References
- 1. Alberti S. (2017) Phase separation in biology. Curr. Biol. 27, R1097–R1102 10.1016/j.cub.2017.08.069 [DOI] [PubMed] [Google Scholar]
- 2. Banani S. F., Rice A. M., Peeples W. B., Lin Y., Jain S., Parker R., and Rosen M. K. (2016) Compositional control of phase-separated cellular bodies. Cell 166, 651–663 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shin Y., and Brangwynne C. P. (2017) Liquid phase condensation in cell physiology and disease. Science 357, eaaf4332 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- 4. Patel A., Lee H. O., Jawerth L., Maharana S., Jahnel M., Hein M. Y., Stoynov S., Mahamid J., Saha S., Franzmann T. M., Pozniakovski A., Poser I., Maghelli N., Royer L. A., Weigert M., et al. (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- 5. Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., and Hyman A. A. (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- 6. Boke E., Ruer M., Wühr M., Coughlin M., Lemaitre R., Gygi S. P., Alberti S., Drechsel D., Hyman A. A., and Mitchison T. J. (2016) Amyloid-like self-assembly of a cellular compartment. Cell 166, 637–650 10.1016/j.cell.2016.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Woodruff J. B., Hyman A. A., and Boke E. (2018) Organization and function of non-dynamic biomolecular condensates. Trends Biochem. Sci. 43, 81–94 [DOI] [PubMed] [Google Scholar]
- 8. Li P., Banjade S., Cheng H. C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J. V., King D. S., Banani S. F., Russo P. S., Jiang Q. X., Nixon B. T., and Rosen M. K. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J., Grishin N. V., Frantz D. E., Schneider J. W., Chen S., Li L., et al. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A. P., Kim H. J., Mittag T., and Taylor J. P. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burke K. A., Janke A. M., Rhine C. L., and Fawzi N. L. (2015) Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell 60, 231–241 10.1016/j.molcel.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saha S., and Hyman A. A. (2017) RNA gets in phase. J. Cell Biol. 216, 2235–2237 10.1083/jcb.201706034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jain A., and Vale R. D. (2017) RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramaswami M., Taylor J. P., and Parker R. (2013) Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154, 727–736 10.1016/j.cell.2013.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ambadipudi S., Biernat J., Riedel D., Mandelkow E., and Zweckstetter M. (2017) Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun. 8, 275 10.1038/s41467-017-00480-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wegmann S., Eftekharzadeh B., Tepper K., Zoltowska K. M., Bennett R. E., Dujardin S., Laskowski P. R., MacKenzie D., Kamath T., Commins C., Vanderburg C., Roe A. D., Fan Z., Molliex A. M., Hernandez-Vega A., et al. (2018) Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 37, e98049 10.15252/embj.201798049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uversky V. N. (2015) The multifaceted roles of intrinsic disorder in protein complexes. FEBS Lett. 589, 2498–2506 10.1016/j.febslet.2015.06.004 [DOI] [PubMed] [Google Scholar]
- 18. Bah A., and Forman-Kay J. D. (2016) Modulation of intrinsically disordered protein function by post-translational modifications. J. Biol. Chem. 291, 6696–6705 10.1074/jbc.R115.695056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chong P. A., and Forman-Kay J. D. (2016) Liquid-liquid phase separation in cellular signaling systems. Curr. Opin. Struct. Biol. 41, 180–186 10.1016/j.sbi.2016.08.001 [DOI] [PubMed] [Google Scholar]
- 20. Xie H., Vucetic S., Iakoucheva L. M., Oldfield C. J., Dunker A. K., Obradovic Z., and Uversky V. N. (2007) Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J. Proteome Res. 6, 1917–1932 10.1021/pr060394e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tompa P., Davey N. E., Gibson T. J., and Babu M. M. (2014) A million peptide motifs for the molecular biologist. Mol. Cell 55, 161–169 10.1016/j.molcel.2014.05.032 [DOI] [PubMed] [Google Scholar]
- 22. Rhoads S. N., Monahan Z. T., Yee D. S., and Shewmaker F. P. (2018) The role of post-translational modifications on prion-like aggregation and liquid-phase separation of FUS. Int. J. Mol. Sci. 19, E886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Itakura A. K., Futia R. A., and Jarosz D. F. (2018) It pays to be in phase. Biochemistry 57, 2520–2529 10.1021/acs.biochem.8b00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang J., Choi J. M., Holehouse A. S., Lee H. O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D., Poser I., Pappu R. V., Alberti S., and Hyman A. A. (2018) A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 10.1016/j.cell.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hofweber M., Hutten S., Bourgeois B., Spreitzer E., Niedner-Boblenz A., Schifferer M., Ruepp M.-D., Simons M., Niessing D., Madl T., and Dormann D. (2018) Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e13 10.1016/j.cell.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 26. Qamar S., Wang G., Randle S. J., Ruggeri F. S., Varela J. A., Lin J. Q., Phillips E. C., Miyashita A., Williams D., Ströhl F., Meadows W., Ferry R., Dardov V. J., Tartaglia G. G., Farrer L. A., et al. (2018) FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173, 720–734.e15 10.1016/j.cell.2018.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bogaert E., Boeynaems S., Kato M., Guo L., Caulfield T. R., Steyaert J., Scheveneels W., Wilmans N., Haeck W., Hersmus N., Schymkowitz J., Rousseau F., Shorter J., Callaerts P., Robberecht W., et al. (2018) Molecular dissection of FUS points at synergistic effect of low-complexity domains in toxicity. Cell Rep. 24, 529–537.e4 10.1016/j.celrep.2018.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nott T. J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T. D., Bazett-Jones D. P., Pawson T., Forman-Kay J. D., and Baldwin A. J. (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chong P. A., Vernon R. M., and Forman-Kay J. D. (2018) RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 430, 4650–4665 10.1016/j.jmb.2018.06.014 [DOI] [PubMed] [Google Scholar]
- 30. Murray D. T., Kato M., Lin Y., Thurber K. R., Hung I., McKnight S. L., and Tycko R. (2017) Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171, 615–627.e16 10.1016/j.cell.2017.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hughes M. P., Sawaya M. R., Boyer D. R., Goldschmidt L., Rodriguez J. A., Cascio D., Chong L., Gonen T., and Eisenberg D. S. (2018) Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science 359, 698–701 10.1126/science.aan6398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guenther E. L., Cao Q., Trinh H., Lu J., Sawaya M. R., Cascio D., Boyer D. R., Rodriguez J. A., Hughes M. P., and Eisenberg D. S. (2018) Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation. Nat. Struct. Mol. Biol. 25, 463–471 10.1038/s41594-018-0064-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Evich M., Stroeva E., Zheng Y. G., and Germann M. W. (2016) Effect of methylation on the side-chain pKa value of arginine. Protein Sci. 25, 479–486 10.1002/pro.2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thandapani P., O'Connor T. R., Bailey T. L., and Richard S. (2013) Defining the RGG/RG motif. Mol. Cell 50, 613–623 10.1016/j.molcel.2013.05.021 [DOI] [PubMed] [Google Scholar]
- 35. Bedford M. T., and Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 10.1016/j.molcel.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tang J., Frankel A., Cook R. J., Kim S., Paik W. K., Williams K. R., Clarke S., and Herschman H. R. (2000) PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 275, 7723–7730 10.1074/jbc.275.11.7723 [DOI] [PubMed] [Google Scholar]
- 37. Yang Y., and Bedford M. T. (2013) Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 10.1038/nrc3409 [DOI] [PubMed] [Google Scholar]
- 38. Chang B., Chen Y., Zhao Y., and Bruick R. K. (2007) JMJD6 is a histone arginine demethylase. Science 318, 444–447 10.1126/science.1145801 [DOI] [PubMed] [Google Scholar]
- 39. Walport L. J., Hopkinson R. J., Chowdhury R., Schiller R., Ge W., Kawamura A., and Schofield C. J. (2016) Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 7, 11974 10.1038/ncomms11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fackelmayer F. O. (2005) Protein arginine methyltransferases: guardians of the Arg? Trends Biochem. Sci. 30, 666–671 10.1016/j.tibs.2005.10.002 [DOI] [PubMed] [Google Scholar]
- 41. Boeynaems S., Bogaert E., Kovacs D., Konijnenberg A., Timmerman E., Volkov A., Guharoy M., De Decker M., Jaspers T., Ryan V. H., Janke A. M., Baatsen P., Vercruysse T., Kolaitis R. M., Daelemans D., et al. (2017) Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e5 10.1016/j.molcel.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C. C., Eckmann C. R., Myong S., and Brangwynne C. P. (2015) The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194 10.1073/pnas.1504822112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saha S., Weber C. A., Nousch M., Adame-Arana O., Hoege C., Hein M. Y., Osborne-Nishimura E., Mahamid J., Jahnel M., Jawerth L., Pozniakovski A., Eckmann C. R., Jülicher F., and Hyman A. A. (2016) Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell 166, 1572–1584.e16 10.1016/j.cell.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ryan V. H., Dignon G. L., Zerze G. H., Chabata C. V., Silva R., Conicella A. E., Amaya J., Burke K. A., Mittal J., and Fawzi N. L. (2018) Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol. Cell 69, 465–479.e7 10.1016/j.molcel.2017.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rappsilber J., Friesen W. J., Paushkin S., Dreyfuss G., and Mann M. (2003) Detection of arginine dimethylated peptides by parallel precursor ion scanning mass spectrometry in positive ion mode. Anal. Chem. 75, 3107–3114 10.1021/ac026283q [DOI] [PubMed] [Google Scholar]
- 46. Ong S. E., Mittler G., and Mann M. (2004) Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods 1, 119–126 10.1038/nmeth715 [DOI] [PubMed] [Google Scholar]
- 47. Scaramuzzino C., Monaghan J., Milioto C., Lanson N. A. Jr., Maltare A., Aggarwal T., Casci I., Fackelmayer F. O., Pennuto M., and Pandey U. B. (2013) Protein arginine methyltransferase 1 and 8 interact with FUS to modify its sub-cellular distribution and toxicity in vitro and in vivo. PLoS ONE 8, e61576 10.1371/journal.pone.0061576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dormann D., Madl T., Valori C. F., Bentmann E., Tahirovic S., Abou-Ajram C., Kremmer E., Ansorge O., Mackenzie I. R., Neumann M., and Haass C. (2012) Arginine methylation next to the PY-NLS modulates transportin binding and nuclear import of FUS. EMBO J. 31, 4258–4275 10.1038/emboj.2012.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Suárez-Calvet M., Neumann M., Arzberger T., Abou-Ajram C., Funk E., Hartmann H., Edbauer D., Kremmer E., Göbl C., Resch M., Bourgeois B., Madl T., Reber S., Jutzi D., Ruepp M. D., et al. (2016) Monomethylated and unmethylated FUS exhibit increased binding to transportin and distinguish FTLD-FUS from ALS-FUS. Acta Neuropathol. 131, 587–604 10.1007/s00401-016-1544-2 [DOI] [PubMed] [Google Scholar]
- 50. Taylor J. P., Brown R. H. Jr, and Cleveland D. W. (2016) Decoding ALS: from genes to mechanism. Nature 539, 197–206 10.1038/nature20413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dolzhanskaya N., Merz G., Aletta J. M., and Denman R. B. (2006) Methylation regulates the intracellular protein-protein and protein-RNA interactions of FMRP. J. Cell Sci. 119, 1933–1946 10.1242/jcs.02882 [DOI] [PubMed] [Google Scholar]
- 52. Tsai W. C., Gayatri S., Reineke L. C., Sbardella G., Bedford M. T., and Lloyd R. E. (2016) Arginine demethylation of G3BP1 promotes stress granule assembly. J. Biol. Chem. 291, 22671–22685 10.1074/jbc.M116.739573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tsai W. C., Reineke L. C., Jain A., Jung S. Y., and Lloyd R. E. (2017) Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule nucleating protein G3BP1. J. Biol. Chem. 292, 18886–18896 10.1074/jbc.M117.800706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Arribas-Layton M., Dennis J., Bennett E. J., Damgaard C. K., and Lykke-Andersen J. (2016) The C-terminal RGG domain of human Lsm4 promotes processing body formation stimulated by arginine dimethylation. Mol. Cell. Biol. 36, 2226–2235 10.1128/MCB.01102-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Matsumoto K., Nakayama H., Yoshimura M., Masuda A., Dohmae N., Matsumoto S., and Tsujimoto M. (2012) PRMT1 is required for RAP55 to localize to processing bodies. RNA Biol. 9, 610–623 10.4161/rna.19527 [DOI] [PubMed] [Google Scholar]
- 56. Poornima G., Shah S., Vignesh V., Parker R., and Rajyaguru P. I. (2016) Arginine methylation promotes translation repression activity of eIF4G-binding protein, Scd6. Nucleic Acids Res. 44, 9358–9368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rajyaguru P., She M., and Parker R. (2012) Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol. Cell 45, 244–254 10.1016/j.molcel.2011.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wall M. L., and Lewis S. M. (2017) Methylarginines within the RGG-motif region of hnRNP A1 affect its IRES trans-acting factor activity and are required for hnRNP A1 stress granule localization and formation. J. Mol. Biol. 429, 295–307 10.1016/j.jmb.2016.12.011 [DOI] [PubMed] [Google Scholar]
- 59. Kaehler C., Guenther A., Uhlich A., and Krobitsch S. (2015) PRMT1-mediated arginine methylation controls ATXN2L localization. Exp. Cell Res. 334, 114–125 10.1016/j.yexcr.2015.02.022 [DOI] [PubMed] [Google Scholar]
- 60. Stetler A., Winograd C., Sayegh J., Cheever A., Patton E., Zhang X., Clarke S., and Ceman S. (2006) Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum. Mol. Genet. 15, 87–96 10.1093/hmg/ddi429 [DOI] [PubMed] [Google Scholar]
- 61. Roovers E. F., Kaaij L. J. T., Redl S., Bronkhorst A. W., Wiebrands K., de Jesus Domingues A. M., Huang H. Y., Han C. T., Riemer S., Dosch R., Salvenmoser W., Grün D., Butter F., van Oudenaarden A., and Ketting R. F. (2018) Tdrd6a regulates the aggregation of Buc into functional subcellular compartments that drive germ cell specification. Dev. Cell 46, 285–301.e9 10.1016/j.devcel.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Friesen W. J., Massenet S., Paushkin S., Wyce A., and Dreyfuss G. (2001) SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell 7, 1111–1117 10.1016/S1097-2765(01)00244-1 [DOI] [PubMed] [Google Scholar]
- 63. Yamazaki T., Chen S., Yu Y., Yan B., Haertlein T. C., Carrasco M. A., Tapia J. C., Zhai B., Das R., Lalancette-Hebert M., Sharma A., Chandran S., Sullivan G., Nishimura A. L., Shaw C. E., et al. (2012) FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2, 799–806 10.1016/j.celrep.2012.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sun S., Ling S. C., Qiu J., Albuquerque C. P., Zhou Y., Tokunaga S., Li H., Qiu H., Bui A., Yeo G. W., Huang E. J., Eggan K., Zhou H., Fu X. D., Lagier-Tourenne C., and Cleveland D. W. (2015) ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat. Commun. 6, 6171 10.1038/ncomms7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. De Leeuw F., Zhang T., Wauquier C., Huez G., Kruys V., and Gueydan C. (2007) The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp. Cell Res. 313, 4130–4144 10.1016/j.yexcr.2007.09.017 [DOI] [PubMed] [Google Scholar]
- 66. Lee Y. J., Hsieh W. Y., Chen L. Y., and Li C. (2012) Protein arginine methylation of SERBP1 by protein arginine methyltransferase 1 affects cytoplasmic/nuclear distribution. J. Cell Biochem. 113, 2721–2728 10.1002/jcb.24151 [DOI] [PubMed] [Google Scholar]
- 67. Aoki K., Ishii Y., Matsumoto K., and Tsujimoto M. (2002) Methylation of Xenopus CIRP2 regulates its arginine- and glycine-rich region-mediated nucleocytoplasmic distribution. Nucleic Acids Res. 30, 5182–5192 10.1093/nar/gkf638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee Y. J., Wei H. M., Chen L. Y., and Li C. (2014) Localization of SERBP1 in stress granules and nucleoli. FEBS J. 281, 352–364 10.1111/febs.12606 [DOI] [PubMed] [Google Scholar]
- 69. Fronz K., Güttinger S., Burkert K., Kühn U., Stöhr N., Schierhorn A., and Wahle E. (2011) Arginine methylation of the nuclear poly(A) binding protein weakens the interaction with its nuclear import receptor, transportin. J. Biol. Chem. 286, 32986–32994 10.1074/jbc.M111.273912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tanikawa C., Ueda K., Suzuki A., Iida A., Nakamura R., Atsuta N., Tohnai G., Sobue G., Saichi N., Momozawa Y., Kamatani Y., Kubo M., Yamamoto K., Nakamura Y., and Matsuda K. (2018) Citrullination of RGG motifs in FET proteins by PAD4 regulates protein aggregation and ALS susceptibility. Cell Rep. 22, 1473–1483 10.1016/j.celrep.2018.01.031 [DOI] [PubMed] [Google Scholar]
- 71. Slade D. J., Subramanian V., Fuhrmann J., and Thompson P. R. (2014) Chemical and biological methods to detect post-translational modifications of arginine. Biopolymers 101, 133–143 10.1002/bip.22256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Galligan J. J., Wepy J. A., Streeter M. D., Kingsley P. J., Mitchener M. M., Wauchope O. R., Beavers W. N., Rose K. L., Wang T., Spiegel D. A., and Marnett L. J. (2018) Methylglyoxal-derived posttranslational arginine modifications are abundant histone marks. Proc. Natl. Acad. Sci. U.S.A. 115, 9228–9233 10.1073/pnas.1802901115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Khoury G. A., Baliban R. C., and Floudas C. A. (2011) Proteome-wide post-translational modification statistics: frequency analysis and curation of the Swiss-Prot database. Sci. Rep. 1, srep00090 10.1038/srep00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Alberti S., Halfmann R., King O., Kapila A., and Lindquist S. (2009) A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146–158 10.1016/j.cell.2009.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Deng Q., Holler C. J., Taylor G., Hudson K. F., Watkins W., Gearing M., Ito D., Murray M. E., Dickson D. W., Seyfried N. T., and Kukar T. (2014) FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J. Neurosci. 34, 7802–7813 10.1523/JNEUROSCI.0172-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Monahan Z., Ryan V. H., Janke A. M., Burke K. A., Rhoads S. N., Zerze G. H., O'Meally R., Dignon G. L., Conicella A. E., Zheng W., Best R. B., Cole R. N., Mittal J., Shewmaker F., and Fawzi N. L. (2017) Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36, 2951–2967 10.15252/embj.201696394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rhoads S. N., Monahan Z. T., Yee D. S., Leung A. Y., Newcombe C. G., O'Meally R. N., Cole R. N., and Shewmaker F. P. (2018) The prion-like domain of FUS is multiphosphorylated following DNA damage without altering nuclear localization. Mol. Biol. Cell 29, 1786–1797 10.1091/mbc.E17-12-0735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Luo F., Gui X., Zhou H., Gu J., Li Y., Liu X., Zhao M., Li D., Li X., and Liu C. (2018) Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation. Nat. Struct. Mol. Biol. 25, 341–346 10.1038/s41594-018-0050-8 [DOI] [PubMed] [Google Scholar]
- 79. Han T. W., Kato M., Xie S., Wu L. C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., and McKnight S. L. (2012) Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- 80. Lin Y., Currie S. L., and Rosen M. K. (2017) Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 292, 19110–19120 10.1074/jbc.M117.800466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 10.1126/science.1134108 [DOI] [PubMed] [Google Scholar]
- 82. Neumann M., Kwong L. K., Lee E. B., Kremmer E., Flatley A., Xu Y., Forman M. S., Troost D., Kretzschmar H. A., Trojanowski J. Q., and Lee V. M. (2009) Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 117, 137–149 10.1007/s00401-008-0477-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Arai T., Mackenzie I. R., Hasegawa M., Nonoka T., Niizato K., Tsuchiya K., Iritani S., Onaya M., and Akiyama H. (2009) Phosphorylated TDP-43 in Alzheimer's disease and dementia with Lewy bodies. Acta Neuropathol. 117, 125–136 10.1007/s00401-008-0480-1 [DOI] [PubMed] [Google Scholar]
- 84. Josephs K. A., Whitwell J. L., Weigand S. D., Murray M. E., Tosakulwong N., Liesinger A. M., Petrucelli L., Senjem M. L., Knopman D. S., Boeve B. F., Ivnik R. J., Smith G. E., Jack C. R. Jr., Parisi J. E., Petersen R. C., and Dickson D. W. (2014) TDP-43 is a key player in the clinical features associated with Alzheimer's disease. Acta Neuropathol. 127, 811–824 10.1007/s00401-014-1269-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., and Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 10.1093/nar/gkr1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang A., Conicella A. E., Schmidt H. B., Martin E. W., Rhoads S. N., Reeb A. N., Nourse A., Ramirez Montero D., Ryan V. H., Rohatgi R., Shewmaker F., Naik M. T., Mittag T., Ayala Y. M., and Fawzi N. L. (2018) A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 37, e97452 10.15252/embj.201797452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rigbolt K. T., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., and Blagoev B. (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4, rs3 [DOI] [PubMed] [Google Scholar]
- 89. Afroz T., Hock E. M., Ernst P., Foglieni C., Jambeau M., Gilhespy L. A. B., Laferriere F., Maniecka Z., Plückthun A., Mittl P., Paganetti P., Allain F. H. T., and Polymenidou M. (2017) Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat. Commun. 8, 45 10.1038/s41467-017-00062-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhang Y. J., Caulfield T., Xu Y. F., Gendron T. F., Hubbard J., Stetler C., Sasaguri H., Whitelaw E. C., Cai S., Lee W. C., and Petrucelli L. (2013) The dual functions of the extreme N terminus of TDP-43 in regulating its biological activity and inclusion formation. Hum. Mol. Genet. 22, 3112–3122 10.1093/hmg/ddt166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hasegawa M., Arai T., Nonaka T., Kametani F., Yoshida M., Hashizume Y., Beach T. G., Buratti E., Baralle F., Morita M., Nakano I., Oda T., Tsuchiya K., and Akiyama H. (2008) Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 64, 60–70 10.1002/ana.21425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Inukai Y., Nonaka T., Arai T., Yoshida M., Hashizume Y., Beach T. G., Buratti E., Baralle F. E., Akiyama H., Hisanaga S., and Hasegawa M. (2008) Abnormal phosphorylation of Ser409/410 of TDP-43 in FTLD-U and ALS. FEBS Lett. 582, 2899–2904 10.1016/j.febslet.2008.07.027 [DOI] [PubMed] [Google Scholar]
- 93. Salado I. G., Redondo M., Bello M. L., Perez C., Liachko N. F., Kraemer B. C., Miguel L., Lecourtois M., Gil C., Martinez A., and Perez D. I. (2014) Protein kinase CK-1 inhibitors as new potential drugs for amyotrophic lateral sclerosis. J. Med. Chem. 57, 2755–2772 10.1021/jm500065f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kametani F., Nonaka T., Suzuki T., Arai T., Dohmae N., Akiyama H., and Hasegawa M. (2009) Identification of casein kinase-1 phosphorylation sites on TDP-43. Biochem. Biophys. Res. Commun. 382, 405–409 10.1016/j.bbrc.2009.03.038 [DOI] [PubMed] [Google Scholar]
- 95. Kametani F., Obi T., Shishido T., Akatsu H., Murayama S., Saito Y., Yoshida M., and Hasegawa M. (2016) Mass spectrometric analysis of accumulated TDP-43 in amyotrophic lateral sclerosis brains. Sci. Rep. 6, 23281 10.1038/srep23281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Goh C. W., Lee I. C., Sundaram J. R., George S. E., Yusoff P., Brush M. H., Sze N. S. K., and Shenolikar S. (2018) Chronic oxidative stress promotes GADD34-mediated phosphorylation of the TAR DNA-binding protein TDP-43, a modification linked to neurodegeneration. J. Biol. Chem. 293, 163–176 10.1074/jbc.M117.814111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liachko N. F., McMillan P. J., Strovas T. J., Loomis E., Greenup L., Murrell J. R., Ghetti B., Raskind M. A., Montine T. J., Bird T. D., Leverenz J. B., and Kraemer B. C. (2014) The tau tubulin kinases TTBK1/2 promote accumulation of pathological TDP-43. PLoS Genet. 10, e1004803 10.1371/journal.pgen.1004803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Taylor L. M., McMillan P. J., Liachko N. F., Strovas T. J., Ghetti B., Bird T. D., Keene C. D., and Kraemer B. C. (2018) Pathological phosphorylation of tau and TDP-43 by TTBK1 and TTBK2 drives neurodegeneration. Mol. Neurodegener. 13, 7 10.1186/s13024-018-0237-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gu J., Wang W., Miao S., Chen F., Wu F., Hu W., Iqbal K., Gong C. X., and Liu F. (2018) Protein phosphatase 1 dephosphorylates TDP-43 and suppresses its function in tau exon 10 inclusion. FEBS Lett. 592, 402–410 10.1002/1873-3468.12976 [DOI] [PubMed] [Google Scholar]
- 100. Liachko N. F., Saxton A. D., McMillan P. J., Strovas T. J., Currey H. N., Taylor L. M., Wheeler J. M., Oblak A. L., Ghetti B., Montine T. J., Keene C. D., Raskind M. A., Bird T. D., and Kraemer B. C. (2016) The phosphatase calcineurin regulates pathological TDP-43 phosphorylation. Acta Neuropathol. 132, 545–561 10.1007/s00401-016-1600-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Nonaka T., Suzuki G., Tanaka Y., Kametani F., Hirai S., Okado H., Miyashita T., Saitoe M., Akiyama H., Masai H., and Hasegawa M. (2016) Phosphorylation of TAR DNA-binding protein of 43 kDa (TDP-43) by truncated casein kinase 1δ triggers mislocalization and accumulation of TDP-43. J. Biol. Chem. 291, 5473–5483 10.1074/jbc.M115.695379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Choksi D. K., Roy B., Chatterjee S., Yusuff T., Bakhoum M. F., Sengupta U., Ambegaokar S., Kayed R., and Jackson G. R. (2014) TDP-43 phosphorylation by casein kinase Iϵ promotes oligomerization and enhances toxicity in vivo. Hum. Mol. Genet. 23, 1025–1035 10.1093/hmg/ddt498 [DOI] [PubMed] [Google Scholar]
- 103. Li H. Y., Yeh P. A., Chiu H. C., Tang C. Y., and Tu B. P. (2011) Hyperphosphorylation as a defense mechanism to reduce TDP-43 aggregation. PLoS ONE 6, e23075 10.1371/journal.pone.0023075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wang Y., and Mandelkow E. (2016) Tau in physiology and pathology. Nat. Rev. Neurosci. 17, 5–21 10.1038/nrg.2015.11 [DOI] [PubMed] [Google Scholar]
- 105. Kampers T., Friedhoff P., Biernat J., Mandelkow E. M., and Mandelkow E. (1996) RNA stimulates aggregation of microtubule-associated protein tau into Alzheimer-like paired helical filaments. FEBS Lett. 399, 344–349 10.1016/S0014-5793(96)01386-5 [DOI] [PubMed] [Google Scholar]
- 106. Wang X., Wang D., Zhao J., Qu M., Zhou X., He H., and He R. (2006) The proline-rich domain and the microtubule binding domain of protein tau acting as RNA binding domains. Protein Pept. Lett. 13, 679–685 10.2174/092986606777790566 [DOI] [PubMed] [Google Scholar]
- 107. Vanderweyde T., Yu H., Varnum M., Liu-Yesucevitz L., Citro A., Ikezu T., Duff K., and Wolozin B. (2012) Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J. Neurosci. 32, 8270–8283 10.1523/JNEUROSCI.1592-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Vanderweyde T., Apicco D. J., Youmans-Kidder K., Ash P. E. A., Cook C., Lummertz da Rocha E., Jansen-West K., Frame A. A., Citro A., Leszyk J. D., Ivanov P., Abisambra J. F., Steffen M., Li H., Petrucelli L., and Wolozin B. (2016) Interaction of tau with the RNA-binding protein TIA1 regulates tau pathophysiology and toxicity. Cell Rep. 15, 1455–1466 10.1016/j.celrep.2016.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Maziuk B. F., Apicco D. J., Cruz A. L., Jiang L., Ash P. E. A., da Rocha E. L., Zhang C., Yu W. H., Leszyk J., Abisambra J. F., Li H., and Wolozin B. (2018) RNA binding proteins co-localize with small tau inclusions in tauopathy. Acta Neuropathol. Commun. 6, 71 10.1186/s40478-018-0574-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zhang X., Lin Y., Eschmann N. A., Zhou H., Rauch J. N., Hernandez I., Guzman E., Kosik K. S., and Han S. (2017) RNA stores tau reversibly in complex coacervates. PLoS Biol. 15, e2002183 10.1371/journal.pbio.2002183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Hernández-Vega A., Braun M., Scharrel L., Jahnel M., Wegmann S., Hyman B. T., Alberti S., Diez S., and Hyman A. A. (2017) Local nucleation of microtubule bundles through tubulin concentration into a condensed tau phase. Cell Rep. 20, 2304–2312 10.1016/j.celrep.2017.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gong C. X., Liu F., Grundke-Iqbal I., and Iqbal K. (2005) Post-translational modifications of tau protein in Alzheimer's disease. J. Neural Transm. 112, 813–838 10.1007/s00702-004-0221-0 [DOI] [PubMed] [Google Scholar]
- 113. Köpke E., Tung Y. C., Shaikh S., Alonso A. C., Iqbal K., and Grundke-Iqbal I. (1993) Microtubule-associated protein tau. Abnormal phosphorylation of a non-paired helical filament pool in Alzheimer disease. J. Biol. Chem. 268, 24374–24384 [PubMed] [Google Scholar]
- 114. Braak H., and Braak E. (1995) Staging of Alzheimer's disease-related neurofibrillary changes. Neurobiol. Aging 16, 271–278 10.1016/0197-4580(95)00021-6 [DOI] [PubMed] [Google Scholar]
- 115. Wang C., Najm R., Xu Q., Jeong D. E., Walker D., Balestra M. E., Yoon S. Y., Yuan H., Li G., Miller Z. A., Miller B. L., Malloy M. J., and Huang Y. (2018) Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector. Nat. Med. 24, 647–657 10.1038/s41591-018-0004-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Martin L., Latypova X., Wilson C. M., Magnaudeix A., Perrin M. L., Yardin C., and Terro F. (2013) Tau protein kinases: involvement in Alzheimer's disease. Ageing Res. Rev. 12, 289–309 10.1016/j.arr.2012.06.003 [DOI] [PubMed] [Google Scholar]
- 117. Wippich F., Bodenmiller B., Trajkovska M. G., Wanka S., Aebersold R., and Pelkmans L. (2013) Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152, 791–805 10.1016/j.cell.2013.01.033 [DOI] [PubMed] [Google Scholar]
- 118. Rai A. K., Chen J. X., Selbach M., and Pelkmans L. (2018) Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216 10.1038/s41586-018-0279-8 [DOI] [PubMed] [Google Scholar]
- 119. Reineke L. C., Cheema S. A., Dubrulle J., and Neilson J. R. (2018) Chronic starvation induces noncanonical pro-death stress granules. J. Cell Sci. 131, pii: jcs220244 10.1242/jcs.220244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Krisenko M. O., Higgins R. L., Ghosh S., Zhou Q., Trybula J. S., Wang W. H., and Geahlen R. L. (2015) Syk is recruited to stress granules and promotes their clearance through autophagy. J. Biol. Chem. 290, 27803–27815 10.1074/jbc.M115.642900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Carpenter K., Bell R. B., Yunus J., Amon A., and Berchowitz L. E. (2018) Phosphorylation-mediated clearance of amyloid-like assemblies in meiosis. Dev. Cell 45, 392–405.e6 10.1016/j.devcel.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Aranda S., Laguna A., and de la Luna S. (2011) DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB J. 25, 449–462 10.1096/fj.10-165837 [DOI] [PubMed] [Google Scholar]
- 123. Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J. M., Bertrand E., and Tazi J. (2003) The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 124. Tourrière H., Gallouzi I. E., Chebli K., Capony J. P., Mouaikel J., van der Geer P., and Tazi J. (2001) RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol. Cell. Biol. 21, 7747–7760 10.1128/MCB.21.22.7747-7760.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Berchowitz L. E., Kabachinski G., Walker M. R., Carlile T. M., Gilbert W. V., Schwartz T. U., and Amon A. (2015) Regulated formation of an amyloid-like translational repressor governs gametogenesis. Cell 163, 406–418 10.1016/j.cell.2015.08.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Goedert M., and Spillantini M. G. (2011) Pathogenesis of the tauopathies. J. Mol. Neurosci. 45, 425–431 10.1007/s12031-011-9593-4 [DOI] [PubMed] [Google Scholar]
- 127. Mahboubi H., Koromilas A. E., and Stochaj U. (2016) AMP kinase activation alters oxidant-induced stress granule assembly by modulating cell signaling and microtubule organization. Mol. Pharmacol. 90, 460–468 10.1124/mol.116.105494 [DOI] [PubMed] [Google Scholar]
- 128. Mahboubi H., Barisé R., and Stochaj U. (2015) 5′-AMP-activated protein kinase α regulates stress granule biogenesis. Biochim. Biophys. Acta 1853, 1725–1737 10.1016/j.bbamcr.2015.03.015 [DOI] [PubMed] [Google Scholar]
- 129. Sfakianos A. P., Mellor L. E., Pang Y. F., Kritsiligkou P., Needs H., Abou-Hamdan H., Désaubry L., Poulin G. B., Ashe M. P., and Whitmarsh A. J. (2018) The mTOR-S6 kinase pathway promotes stress granule assembly. Cell Death Differ. 25, 1766–1780 10.1038/s41418-018-0076-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Alberti S., Mateju D., Mediani L., and Carra S. (2017) Granulostasis: protein quality control of RNP granules. Front. Mol. Neurosci. 10, 84 10.3389/fnmol.2017.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Hong E., Lim Y., Lee E., Oh M., and Kwon D. (2012) Tissue-specific and age-dependent expression of protein arginine methyltransferases (PRMTs) in male rat tissues. Biogerontology 13, 329–336 10.1007/s10522-012-9379-2 [DOI] [PubMed] [Google Scholar]
- 132. Simandi Z., Pajer K., Karolyi K., Sieler T., Jiang L. L., Kolostyak Z., Sari Z., Fekecs Z., Pap A., Patsalos A., Contreras G. A., Reho B., Papp Z., Guo X., Horvath A., Kiss G., Keresztessy Z., et al. (2018) Arginine methyltransferase PRMT8 provides cellular stress tolerance in aging motoneurons. J. Neurosci. 38, 7683–7700 10.1523/JNEUROSCI.3389-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Sergeant N., Bretteville A., Hamdane M., Caillet-Boudin M. L., Grognet P., Bombois S., Blum D., Delacourte A., Pasquier F., Vanmechelen E., Schraen-Maschke S., and Buée L. (2008) Biochemistry of Tau in Alzheimer's disease and related neurological disorders. Expert Rev. Proteomics 5, 207–224 10.1586/14789450.5.2.207 [DOI] [PubMed] [Google Scholar]
- 134. Sato S., Cerny R. L., Buescher J. L., and Ikezu T. (2006) Tau-tubulin kinase 1 (TTBK1), a neuron-specific tau kinase candidate, is involved in tau phosphorylation and aggregation. J. Neurochem. 98, 1573–1584 10.1111/j.1471-4159.2006.04059.x [DOI] [PubMed] [Google Scholar]
- 135. Guil S., Long J. C., and Cáceres J. F. (2006) hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol. Cell. Biol. 26, 5744–5758 10.1128/MCB.00224-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Carlomagno Y., Chung D. C., Yue M., Castanedes-Casey M., Madden B. J., Dunmore J., Tong J., DeTure M., Dickson D. W., Petrucelli L., and Cook C. (2017) An acetylation-phosphorylation switch that regulates tau aggregation propensity and function. J. Biol. Chem. 292, 15277–15286 10.1074/jbc.M117.794602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Ferreon J. C., Jain A., Choi K. J., Tsoi P. S., MacKenzie K. R., Jung S. Y., and Ferreon A. C. (2018) Acetylation disfavors tau phase separation. Int. J. Mol. Sci. 19, E1360 10.3390/ijms19051360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Saito M., Hess D., Eglinger J., Fritsch A. W., Kreysing M., Weinert B. T., Choudhary C., and Matthias P. (2019) Acetylation of intrinsically disordered regions regulates phase separation. Nat. Chem. Biol. 15, 51–61 10.1038/s41589-018-0180-7 [DOI] [PubMed] [Google Scholar]
- 139. Kwon S., Zhang Y., and Matthias P. (2007) The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 21, 3381–3394 10.1101/gad.461107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Cohen T. J., Hwang A. W., Restrepo C. R., Yuan C. X., Trojanowski J. Q., and Lee V. M. (2015) An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun. 6, 5845 10.1038/ncomms6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ohn T., Kedersha N., Hickman T., Tisdale S., and Anderson P. (2008) A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10, 1224–1231 10.1038/ncb1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhang X., Shu X. E., and Qian S. B. (2018) O-GlcNAc modification of eIF4GI acts as a translational switch in heat shock response. Nat. Chem. Biol. 14, 909–916 10.1038/s41589-018-0120-6 [DOI] [PubMed] [Google Scholar]
- 143. Trinidad J. C., Barkan D. T., Gulledge B. F., Thalhammer A., Sali A., Schoepfer R., and Burlingame A. L. (2012) Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics 11, 215–229 10.1074/mcp.O112.018366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Hart G. W., Slawson C., Ramirez-Correa G., and Lagerlof O. (2011) Cross-talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 10.1146/annurev-biochem-060608-102511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Wang A. C., Jensen E. H., Rexach J. E., Vinters H. V., and Hsieh-Wilson L. C. (2016) Loss of O-GlcNAc glycosylation in forebrain excitatory neurons induces neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 113, 15120–15125 10.1073/pnas.1606899113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Roth S., and Khalaila I. (2017) The effect of O-GlcNAcylation on hnRNP A1 translocation and interaction with transportin1. Exp. Cell Res. 350, 210–217 10.1016/j.yexcr.2016.11.023 [DOI] [PubMed] [Google Scholar]
- 147. Haj-Yahya M., and Lashuel H. A. (2018) Protein semisynthesis provides access to tau disease-associated post-translational modifications (PTMs) and paves the way to deciphering the tau PTM code in health and diseased states. J. Am. Chem. Soc. 140, 6611–6621 10.1021/jacs.8b02668 [DOI] [PubMed] [Google Scholar]
- 148. Xie X., Matsumoto S., Endo A., Fukushima T., Kawahara H., Saeki Y., and Komada M. (2018) Deubiquitylases USP5 and USP13 are recruited to and regulate heat-induced stress granules through their deubiquitylating activities. J. Cell Sci. 131, jcs10856 10.1242/jcs.210856 [DOI] [PubMed] [Google Scholar]
- 149. Jayabalan A. K., Sanchez A., Park R. Y., Yoon S. P., Kang G. Y., Baek J. H., Anderson P., Kee Y., and Ohn T. (2016) NEDDylation promotes stress granule assembly. Nat. Commun. 7, 12125 10.1038/ncomms12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Jongjitwimol J., Baldock R. A., Morley S. J., and Watts F. Z. (2016) Sumoylation of eIF4A2 affects stress granule formation. J. Cell Sci. 129, 2407–2415 10.1242/jcs.184614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Nostramo R., Varia S. N., Zhang B., Emerson M. M., and Herman P. K. (2016) The catalytic activity of the Ubp3 deubiquitinating protease is required for efficient stress granule assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 36, 173–183 10.1128/MCB.00609-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Dao T. P., Kolaitis R. M., Kim H. J., O'Donovan K., Martyniak B., Colicino E., Hehnly H., Taylor J. P., and Castañeda C. A. (2018) Ubiquitin modulates liquid–liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell 69, 965–978.e6 10.1016/j.molcel.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. McGurk L., Gomes E., Guo L., Mojsilovic-Petrovic J., Tran V., Kalb R. G., Shorter J., and Bonini N. M. (2018) Poly(ADP-ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 71, 703–717.e9 10.1016/j.molcel.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Boehning M., Dugast-Darzacq C., Rankovic M., Hansen A. S., Yu T., Marie-Nelly H., McSwiggen D. T., Kokic G., Dailey G. M., Cramer P., Darzacq X., and Zweckstetter M. (2018) RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 25, 833–840 10.1038/s41594-018-0112-y [DOI] [PubMed] [Google Scholar]
- 155. Larson A. G., Elnatan D., Keenen M. M., Trnka M. J., Johnston J. B., Burlingame A. L., Agard D. A., Redding S., and Narlikar G. J. (2017) Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Lu H., Yu D., Hansen A. S., Ganguly S., Liu R., Heckert A., Darzacq X., and Zhou Q. (2018) Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323 10.1038/s41586-018-0174-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Su X., Ditlev J. A., Hui E., Xing W., Banjade S., Okrut J., King D. S., Taunton J., Rosen M. K., and Vale R. D. (2016) Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 10.1126/science.aad9964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Jawaid A., Woldemichael B. T., Kremer E. A., Laferriere F., Gaur N., Afroz T., Polymenidou M., and Mansuy I. M. (2018) Memory decline and its reversal in aging and neurodegeneration involve miR-183/96/182 biogenesis. Mol. Neurobiol. 10.1007/s12035-018-1314-3 [DOI] [PubMed] [Google Scholar]


