Publisher's Note: There is a Blood Commentary on this article in this issue.
Key Points
Complex karyotype defined by the presence of ≥3 chromosomal abnormalities should not be axiomatically considered unfavorable in CLL.
High cytogenetic complexity with ≥5 chromosomal aberrations emerges as prognostically adverse, independently of other biomarkers.
Abstract
Recent evidence suggests that complex karyotype (CK) defined by the presence of ≥3 chromosomal aberrations (structural and/or numerical) identified by using chromosome-banding analysis (CBA) may be relevant for treatment decision-making in chronic lymphocytic leukemia (CLL). However, many challenges toward the routine clinical application of CBA remain. In a retrospective study of 5290 patients with available CBA data, we explored both clinicobiological associations and the clinical impact of CK in CLL. We found that patients with ≥5 abnormalities, defined as high-CK, exhibit uniformly dismal clinical outcomes, independently of clinical stage, TP53 aberrations (deletion of chromosome 17p and/or TP53 mutations [TP53abs]), and the expression of somatically hypermutated (M-CLL) or unmutated immunoglobulin heavy variable genes. Thus, they contrasted with CK cases with 3 or 4 aberrations (low-CK and intermediate-CK, respectively) who followed aggressive disease courses only in the presence of TP53abs. At the other end of the spectrum, patients with CK and +12,+19 displayed an exceptionally indolent profile. Building upon CK, TP53abs, and immunoglobulin heavy variable gene somatic hypermutation status, we propose a novel hierarchical model in which patients with high-CK exhibit the worst prognosis, whereas those with mutated CLL lacking CK or TP53abs, as well as CK with +12,+19, show the longest overall survival. Thus, CK should not be axiomatically considered unfavorable in CLL, representing a heterogeneous group with variable clinical behavior. High-CK with ≥5 chromosomal aberrations emerges as prognostically adverse, independent of other biomarkers. Prospective clinical validation is warranted before ultimately incorporating high-CK in risk stratification of CLL.
Visual Abstract
Introduction
Chronic lymphocytic leukemia (CLL) is a malignancy of mature clonal B cells that mainly affects the elderly population and displays exceptional clinical and biological heterogeneity.1-3 Many host- and tumor-related features with prognostic and/or predictive value have been identified over the years, assisting in the stratification of patients into subgroups with distinct clinical course and response to treatment.4-23 Among tumor-related biomarkers, those recommended by the International Workshop on Chronic Lymphocytic Leukemia (iwCLL) for prognostic assessment before treatment initiation in both general practice and clinical trials pertain to the genomic background of the malignant clone, more particularly the TP53 gene, and the somatic hypermutation status (SHM) of the rearranged immunoglobulin heavy variable (IGHV) gene expressed by the clonotypic B-cell receptor immunoglobulin.24
The genomic landscape of CLL is heterogeneous, lacking a specific cytogenetic abnormality.25 Historically, the first evidence for the genetic heterogeneity of CLL emerged from chromosome banding analyses (CBAs) from the early 1990s revealing various numerical and structural abnormalities.26-28 These studies also indicated that the presence of an increased number of cytogenetic abnormalities was associated with more aggressive clinical outcomes, highlighting the prognostic significance of complex karyotype (CK) defined by the presence of at least 3 numerical and/or structural abnormalities.28
However, CBA analysis was never widely incorporated into the routine diagnostic algorithm of CLL, mainly due to technical considerations, particularly concerning the relative difficulty in obtaining sufficient metaphases of the CLL clone; this difficulty translated into a low detection rate of chromosome abnormalities, at least until relatively recently.29,30 This scenario, combined with the finding that fluorescence in situ hybridization (FISH) could detect at least 1 of only 4 recurrent aberrations with prognostic relevance [namely deletions of chromosome 11q (del(11q)), 13q (del(13q)), and 17p (del(17p)); and trisomy of chromosome 12 (+12)] in ∼80% of patients,12 rendered CBA a less popular approach for assessing the CLL genetic background.
According to the recently updated iwCLL recommendations, thorough genetic risk stratification in CLL requires FISH analysis complemented by mutational screening for the TP53 gene.24 However, arguably, FISH offers only a partial view of the cytogenetic landscape of CLL, whereas CBA presents the opportunity to globally assess the karyotype of the malignant clone, thus potentially offering valuable complementary information and, eventually, refinement of risk stratification.6,8,26,31-35 From a practical perspective, it is relevant to mention that, thanks to the introduction of modern cell stimulation protocols, the methodologic limitations of older protocols have been overcome, allowing for robust CBA.29,32,36
Recently, CBA in CLL has attracted great interest given the reports suggesting that in addition to representing an independent prognostic marker,6,13,37-41 CK may also constitute a novel predictive marker for refractoriness to not only chemotherapy-based treatment regimens42-45 but also to novel agents; these novel agents include B-cell signaling kinase inhibitors and the Bcl-2 inhibitor venetoclax, independently of the presence of TP53 aberrations [TP53abs; deletion of chromosome 17p (del(17p)); and/or TP53 mutation].46-50 However, the available evidence derives from small cohorts of patients in various disease phases and with markedly different treatment exposures. This situation precludes definitive conclusions from being drawn regarding the precise predictive value of CK and the optimal management of CK patients.
Responding to these developments, the recently updated iwCLL guidelines state that CBA before treatment initiation is “desirable” in the context of clinical trials and useful also in general practice, provided that an established methodology is available.24 However, many challenges toward routine clinical application of CBA must still be overcome, thereby indicating the need for rigorous definitions as well as systematic investigation in a large series, which is the aim of the present study of the European Research Initiative on CLL (ERIC).
Patients and methods
Patients
The present multicenter retrospective study included 5479 individuals with CLL (n = 5082 [93%]) and high-count (clinical) monoclonal B-cell lymphocytosis51 (n = 397 [7%]) from 17 European institutions (Table 1) in whom cytogenetic data from CBA were available; 2198 of 5479 cases have been reported previously.6,34,37,40,41 A total of 189 cases (3%) were excluded from further analysis due to having fewer metaphases than required for reliable assessment (definitions given in the following section).
Table 1.
Main clinicobiological features of the patients included in the study
| Feature | Entire cohort (n = 5290) | Non-CK (0-2 abs; n = 4496) | CK ≥3 abs (n = 794) | Low-CK/intermediate-CK (3-4 abs; n = 523) | High-CK (≥5 abs; n = 271) | P, non-CK vs CK | P, low-CK/intermediate-CK vs high-CK |
|---|---|---|---|---|---|---|---|
| Male | 3302/5290, 62% | 2790/4496, 62% | 522/794, 66% | 351/523, 67% | 171/271, 63% | .047 | .56 |
| Median age (diagnosis) | 64.6 y | 64.3 y | 64.7 y | 64.2 y | 66.1 y | .58 | .02 |
| MBL | 383/4454, 9% | 353/3813, 9% | 30/641, 5% | 27/412, 7% | 3/229, 1% | .0001 | .004 |
| Binet A | 3030/4454, 68% | 2643/3813, 69% | 387/641, 60% | 263/412, 64% | 124/229, 54% | <.0001 | .017 |
| Binet B/C | 1041/4454, 23% | 817/3813, 22% | 224/641, 35% | 122/412, 29% | 102/229, 45% | <.0001 | .0002 |
| U-CLL | 1514/3453, 44% | 1187/2939, 40% | 327/514, 64% | 201/351, 57% | 126/163, 77% | <.0001 | <.0001 |
| TP53abs | 657/4968, 13% | 337/4204, 8% | 320/764, 42% | 151/501, 30% | 169/263, 64% | <.0001 | <.0001 |
| del(11q) | 487/4500, 11% | 353/3714, 9% | 165/622, 26% | 119/413, 29% | 46/209, 22% | <.0001 | .07 |
| Trisomy 12 | 685/4500, 15% | 557/3714, 15% | 150/622, 24% | 117/413, 28% | 33/209, 16% | <.0001 | .0005 |
| idel(13q) | 1734/4500, 38% | 1621/3714, 44% | 113/622, 18% | 86/413, 21% | 27/209, 13% | <.0001 | <.0001 |
The statistically significant level was defined as .008 following the Bonferroni correction for multiple testing. abs, aberrations; CK, ≥3 abs; low-CK, 3 abs; intermediate-CK, 4 abs; high-CK, ≥5 abs; MBL, monoclonal B-cell lymphocytosis; TP53abs, deletion of chromosoe 17p and/or TP53 mutation; del(11q), deletion of chromosome 11q; idel(13q), isolated deletion of chromosome 13q.
CBA was performed within the first year from diagnosis and before the administration of any treatment in 4402 (85%) of 5179 patients and in 4499 (92%) of 4868 patients, respectively. No significant differences in obtaining an adequate number of metaphases were observed across the various institutions. The study was conducted under all recommended national and international ethical and legal recommendations after approval by the local ethics review committee of each participating institution. Demographic, clinical, and biological data for the patient cohort are summarized in Table 1.
Cytogenetic analysis
Stimulation protocols used for metaphase induction were based on either phorbol-12-myristate-13-acetate (TPA) (n = 2631 [50%]) or immunostimulatory cytosine guanine dinucleotide (CpG)-oligonucleotide DSP30 plus interleukin 2 (IL-2) (n = 2659 [50%]) following standard procedures.6,29,37,38,52 No differences regarding the number of obtained metaphases were observed between the 2 protocols. Details regarding the actual protocols are provided in the supplemental Methods, available on the Blood Web site.
Karyotypes were classified according to the 2016 International System for Human Cytogenetic Nomenclature.53 For a karyotype to be deemed normal, a minimum of 15 metaphases had to be examined; 10 metaphases were the minimum in the case of abnormal findings. Single-cell abnormalities were taken into consideration only if verified according to FISH analysis.
A karyotype was defined as complex if ≥3 clonal aberrations (numerical and/or structural; unbalanced and balanced aberrations were considered as a single event) were present in CBA54,55; aberrations detected only according to FISH were not taken into consideration regarding the definition of CK. Interphase FISH analysis was performed in 4766 (90%) cases using the probes for the 13q14, 11q22 (ATM), and 17p13 (TP53) regions and trisomy 12 (CEP 12).
Other biomarkers
Immunogenetic analysis
Amplification of IGHV–immunoglobulin heavy diversity–immunoglobulin heavy joining rearrangements was performed in 3453 (65%) patients as previously described.56 Based on the SHM status, namely the germline identity of the clonotypic rearranged IGHV genes, patients were classified as having unmutated CLL (U-CLL) (≥98% identity) or mutated CLL (M-CLL) (<98% identity).
Analysis of TP53 gene mutations
Mutational screening for the TP53 gene included exons 4-8 but also exons 9-10 for some centers and was performed in 2861 (54%) of 5290 cases, mainly those negative for del(17p) per FISH analysis (n = 2482). Most cases (70%) were analyzed by using Sanger sequencing. The remaining patients were analyzed with next-generation sequencing (NGS); only clones with variant allele frequency >10% were considered.57 The detection rate of TP53 mutations was similar independently of the applied methodologies for cases carrying CK (21% and 26% for cases analyzed with NGS and Sanger sequencing, respectively; P = .15).
Statistical analysis
Descriptive statistics for discrete parameters included counts and frequency distributions; for quantitative variables, statistical measures included medians, standard deviations, and minimum–maximum values. Overall survival (OS), the end point of the present study, was measured from the date of CBA until last follow-up or death. The impact of CK on time-to-first-treatment among patients with early-stage disease has been reported elsewhere.6 Survival curves were constructed with the Kaplan-Meier method, and the log-rank test was used to determine differences between survival proportions. Univariable Cox regression was applied to assess the prognostic significance of CK and other prognostic factors on survival outcome. Multivariable Cox regression models were implemented to test the simultaneous effect of factors on outcomes, taking into account the relative effect of the remaining parameters. For the multivariable analysis, we considered only cases with available data for all the factors included in the model (n = 2376) because imputing the values of the biomarkers could introduce substantial bias. However, no major differences were observed between the entire cohort and the proportion of cases included in the multivariable analysis (supplemental Table 1). Survival analysis was performed with a significance level of 5%; for descriptive statistics, the statistically significant level was defined as .008 following the Bonferroni correction for multiple testing. All analyses were performed with Statistica Software version 10.0 (StatSoft, Inc.).
Results
CK in CLL: main features and associations
Following the current definition for CK (ie, ≥3 structural and/or numerical aberrations), CK was detected in 794 (15%) of 5290 cases (supplemental Figure 1), in accordance with previous reports in cohorts analyzed close to diagnosis.6 CK was significantly associated with advanced clinical stage, TP53abs, U-CLL, del(11q), and +12, as well as lower prevalence of isolated del(13q) [idel(13q)] detected according to FISH analysis (P < .008 for all comparisons vs non-CK cases) (Table 1). Interestingly, CK was detected even among cases with clinical monoclonal B-cell lymphocytosis (30 of 383 [8%]).
CK was detected more often in cases analyzed with the CpG/IL-2 protocol compared with the TPA protocol (508 of 2659 [19%] vs 286 of 2630 [11%]; P < .001) (supplemental Tables 2 and 3). This difference may be attributed, at least in part, to the reported higher effectiveness of the CpG/IL-2 stimulation protocol.29,58 In total, abnormal karyotypes carrying other than idel(13q) were detected in 55% and 43% (P < .001) of the cases analyzed with the CpG/IL-2 and the TPA protocols, respectively (supplemental Figure 2). Cases analyzed with the CpG/IL-2 methodology were enriched for TP53abs (16% vs 10% [P = .0001] vs the TPA stimulation protocol).
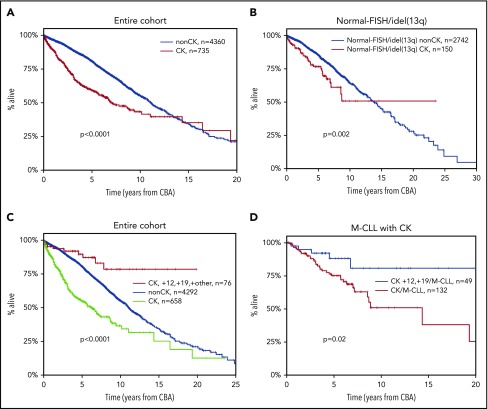
Regarding the clinical impact, CK was associated with shorter OS (median OS, 6.9 years; lower quartile–upper quartile [LQ-UQ], 2.5-18.2 years; P < .0001) (Figure 1A). This finding retained independent significance even in the multivariable analysis (hazard ratio [HR], 1.578; 95% confidence interval [CI], 1.267-1.966; P < .001) (supplemental Table 4) along with advanced clinical stage, TP53abs, and U-CLL.
Figure 1.
Kaplan-Meier curves for OS. (A) Patients with CK (≥3 aberrations, red line) vs non-CK cases (0-2 aberrations, blue line) in the entire cohort. The observed crossover can be explained by the few “events” at the tail of the CK curve, where mostly censored cases are included. (B) Patients with FISH-normal/idel(13q) detected by FISH who carry CK (≥3 aberrations, red line) vs non-CK FISH-normal/idel(13q) cases (blue line). (C) Patients with CK and +12,+19 (red line) vs CLL with CK (green line) and the remaining non-CK CLL (blue line). (D) Patients with CK and +12,+19 carrying mutated IGHV genes (M-CLL, blue line) vs the remaining M-CLL with CK (red line).
We and others have reported that CK can be present even among cases with idel(13q) or normal FISH [FISH-normal/idel(13q)], identifying cases with dismal clinical outcome within this otherwise “FISH-favorable” group.6,32,33 In the present cohort, 159 (5%) of 2963 cases with FISH-normal/idel(13q) carried CK, with a significantly higher incidence of CK among the idel(13q) subgroup [idel(13q), 113 of 1746 (6.4%); FISH-normal, 46 of 1229 (3.7%); P = .001]. These cases exhibited significantly shorter OS compared with the FISH-normal/idel(13q) cases lacking cytogenetic complexity (0-2 aberrations on CBA) (median OS of 7.88 years [LQ-UQ, 3.5-12.74 years] vs a median OS of 13.7 years [LQ-UQ, 7.5-20.1 years], respectively; P = .002) (Figure 1B). Interestingly, the great majority of FISH-normal/idel(13q) cases with CK were negative for TP53 gene mutations (100 [87%] of 115 cases with available data).
CK is not always adverse: the case of +12,+19 CLL
We and others have previously reported that the coexistence of +12 and +19 defines a CLL subgroup with a constellation of distinctive clinicobiological features, including ubiquitous expression of immunoglobulin G–switched heavy chains, biased expression of lambda light chains, high prevalence of CD38 positivity and monoclonal paraproteinemia, low prevalence of TP53abs, and almost exclusive usage of mutated IGHV genes.8,59,60 Moreover, these cases carry extra trisomies, mostly +18, as well as structural abnormalities, hence fulfilling the criteria for CK; however, they exhibit a significantly more indolent course compared with cases with sole +12.
In the present cohort, a total of 81 CK cases with +12,+19 (10% of all CK CLL) were identified who carried either extra trisomies (n = 43) or extra structural abnormalities (n = 38). No differences were observed between the 2 subgroups regarding either demographic/biological features or clinical outcome (supplemental Table 5; supplemental Figure 3). However, it should be mentioned that the number of patients in each +12,+19 subgroup was limited, and hence some caution is warranted. Interestingly, CK cases belonging to the +12,+19 variant exhibited significantly longer OS (median OS, not reached [NR]) compared with not only the remaining CK CLL (median OS, 6.2 years; LQ-UQ, 2.2-14.4 years; P < .001) but also the non-CK CLL (median OS, 11.1 years; LQ-UQ, 6.1-17.3 years; P < .0001) (Figure 1C). Similar to previous reports,8 CK cases with +12,+19 exhibited longer OS compared with cases with sole +12 detected according to CBA (data not shown). Thus, this profile identifies a subgroup that, despite formally considered as CK, exhibits an extremely indolent course with only 7 deaths in 81 cases and only 57% having received treatment at a median follow-up of 7.2 years. This survival advantage of +12,+19 CK cases compared with the remaining CK CLL was retained even when the analysis was restricted to M-CLL (median OS +12,+19 CK, NR; median OS CK/M-CLL, 9.24 years [LQ-UQ, 4.9-NR], respectively; P = .02) (Figure 1D).
Not all CKs are equivalent: high vs low complexity
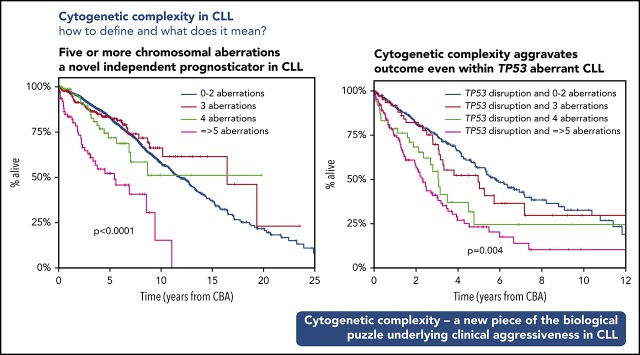
Published evidence suggests that among cases with CK, those carrying ≥5 abnormalities may exhibit a worse clinical outcome compared with those with 3 or 4 aberrations.6,39 However, the relevant series were rather small, hindering definitive conclusions. To address this issue and also capitalizing on the large cohort size of the present study, CK cases were subdivided into 3 subgroups based on whether they were carrying 3 (n = 355 [45%]), 4 (n = 168 [21%]), or ≥5 (n = 271 [34%]) abnormalities. These subgroups were defined as low, intermediate, and high CKs (low-CK, intermediate-CK, and high-CK), respectively.
High-CK cases were significantly enriched for TP53abs as well as U-CLL, reaching up to 65% and 76%, respectively (P < .001 compared with low-CK and intermediate-CK) (Figure 2A), whereas low-CK and intermediate-CK cases exhibited rather similar demographic and biological profiles (supplemental Table 6). Prompted by this finding and also considering recent independent reports61,62 alluding to the significance of small TP53 clones detectable only by using NGS, we investigated whether non-TP53 aberrant high-CK cases might also carry minor TP53-mutant subclones using NGS with a sensitivity of 2%. Interestingly, among 25 analyzed cases, none was found positive for low-frequent TP53 mutations.
Figure 2.
Different biological profiles and clinical outcome among patients with CK (≥3 aberrations [abs]) depending on the number of chromosomal abnormalities. (A) Frequency of U-CLL (unmutated IGHV genes), TP53abs (deletion of chromosome 17p and/or TP53 mutations), del(11q) (deletion of chromosome 11q), and normal-FISH/idel(13q) (normal FISH or isolated deletion of chromosome 13q according to Döhner hierarchical model). Patients with CK and ≥5 aberrations (high-CK) are enriched for U-CLL and TP53abs compared with CK patients with 3 aberrations (low-CK) and those with 4 aberrations (intermediate-CK). Patients with normal-FISH(idel(13q) are detected within all CK groups. (B-D) Kaplan-Meier curves for OS. (B) All patients with CK in the entire cohort. Low-CK, intermediate-CK, and high-CK cases are represented with the blue, red, and green lines, respectively. (C) Patients without TP53abs. High-CK patients exhibit the shortest OS (purple line), whereas there is no difference between low-CK (red line), intermediate-CK (green line), and the remaining non-CK CLL (blue line). (D) Patients with TP53abs. The number of aberrations aggravates the clinical outcome, with high-CK (purple line) exhibiting the shortest OS.
Moreover, cases with low-CK and intermediate-CK exhibited a rather similar distribution of aberrant chromosomal regions, with “CLL-recurrent” aberrations predominating; in contrast, high-CK cases showed a broader spectrum of aberrations affecting almost all chromosomes (Figure 3), independently of TP53 status (supplemental Figure 4). In line with our previous observation regarding the existence of FISH-normal/idel(13q) cases harboring CK,6,32 we also found such cases in all CK subgroups defined here, accounting for 30%, 28%, and 21% of low-CK, intermediate-CK, and high-CK cases, respectively (Figure 2A).
Figure 3.
Distribution of chromosome gains and losses as well as chromosomal breakpoints in the CKs of the present series within 3 aberrations (low-CK), 4 aberrations (intermediate-CK), and ≥5 aberrations (high-CK). (A) 3 aberrations (low-CK); (B) 4 aberrations (intermediate-CK); (C) ≥5 aberrations (high-CK). Gains, right green bars; losses, left red bars; translocation breakpoints, right blue bars adjacent to chromosomes. Ideograms were prepared with the CYDAS software package, freely available at www.cydas.org.
High-CK exhibited significantly shorter OS (median OS, 3.1 years; LQ-UQ, 1.3-8.3 years; P < .001) compared with either low-CK or intermediate-CK cases (median OS for low-CK and intermediate-CK, 12.3 and 7.25 years; LQ-UQ, 5.1-18.1 and 3.75-NR, respectively; P = .04 between low-CK and intermediate-CK) (Figure 2B); these cases resembled the remaining, non-CK CLL (median OS, 11.1 years; LQ-UQ, 6.1-17.3 years) (supplemental Figure 5A). The same results were obtained when cases with CK and +12,+19 were excluded from the analysis (supplemental Figure 5B).
The dismal impact of high-CK compared with the intermediate-CK and low-CK cases was even more striking among cases lacking TP53abs (median OS, 5.1 years; LQ-UQ, 1.8-8.9 years; P < .0001) (Figure 2C). In contrast, no difference was found between low-CK or intermediate-CK vs the remaining, non-CK CLL lacking TP53abs (median OS, 14.8 years, NR, and 11.8 years; LQ-UQ, 6.7-19.1 years, 4.3 years-NR, and 6.7-17.9 years, respectively; P = .27).
When the analysis was restricted to cases carrying TP53abs, high-CK exhibited the shortest OS compared with intermediate- and low-CK (median OS, 2.5, 3.1, and 5 years, respectively; P = .004) (Figure 2D), suggesting that a complex genetic background aggravates the already dismal clinical outcome of cases with TP53abs. The remaining non-CK cases harboring TP53abs exhibited a median OS of 6.6 years (LQ-UQ, 3.2 years-NR).
Turning to immunogenetic categories, within U-CLL, high-CK cases exhibited the shortest OS (median OS, 2.33 years; LQ-UQ, 1.2-7.9 years; P < .001) compared with either low-CK or intermediate-CK cases (median OS, 10.1 and 4.4 years; LQ-UQ, 5 years-NR and 2.7 years-NR, respectively [P = .003] between low-CK and intermediate-CK) (supplemental Figure 6A). In M-CLL, high-CK was associated with the worst clinical outcome (median OS, 6.1 years; LQ-UQ, 3.2-8.4 years; P < .001), whereas low-CK and intermediate-CK exhibited similar OS (supplemental Figure 6B).
Based on these findings, we considered high-CK cases as an independent subgroup, distinct from either low-CK or intermediate-CK cases that we merged into 1 subgroup. Interestingly, when evaluated as a single parameter, low-CK/intermediate-CK were borderline significant in the univariable analysis for OS (HR, 1.216; 95% CI, 1.007-1.470; P = .042), whereas they failed to reach significance in the multivariate analysis (HR, 1.214; 95% CI, 0.918-1.606; P = .17). In contrast, high-CK emerged as an independent adverse prognosticator on multivariable analysis (HR, 2.320; 95% CI, 1.603-3.092; P < .001), along with advanced clinical stage, TP53, and SHM status (Table 2).
Table 2.
Univariable and multivariable analysis for OS
| Parameter | Univariable analysis (n = 5095) | Multivariable analysis (n = 2376) | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Male | 1.202 | 1.070-1.350 | .001 | 1.159 | 0.974-1.379 | .09 |
| Low-CK/intermediate-CK | 1.216 | 1.007-1.470 | .042 | 1.214 | 0.918-1.606 | .17 |
| High-CK | 2.059 | 1.789-2.370 | <.001 | 2.226 | 1.603-3.092 | <.001 |
| idel(13q) | 0.894 | 0.792-1.010 | .07 | — | — | — |
| Trisomy 12 | 1.310 | 1.125-1.525 | <.001 | 1.206 | 0.979-1.487 | .08 |
| del(11q) | 1.942 | 1.659-2.273 | <.001 | 1.152 | 0.914-1.451 | .23 |
| TP53abs | 2.904 | 2.517-3.350 | <.001 | 1.960 | 1.558-2.465 | <.001 |
| U-CLL | 2.851 | 2.467-3.295 | <.001 | 2.320 | 1.927-2.793 | <.001 |
| Binet B/C | 2.036 | 1.793-2.312 | <.001 | 1.575 | 1.313-1.888 | <.001 |
High-CK (≥5 aberrations) is an independent predictor for shorter OS contrasting low-CK/intermediate-CK (3 and 4 aberrations, respectively), which failed to retain significance in the multivariable analysis. Abbreviations are explained in Table 1.
CK, TP53 aberrations and B-cell receptor immunoglobulin SHM: an integrated model
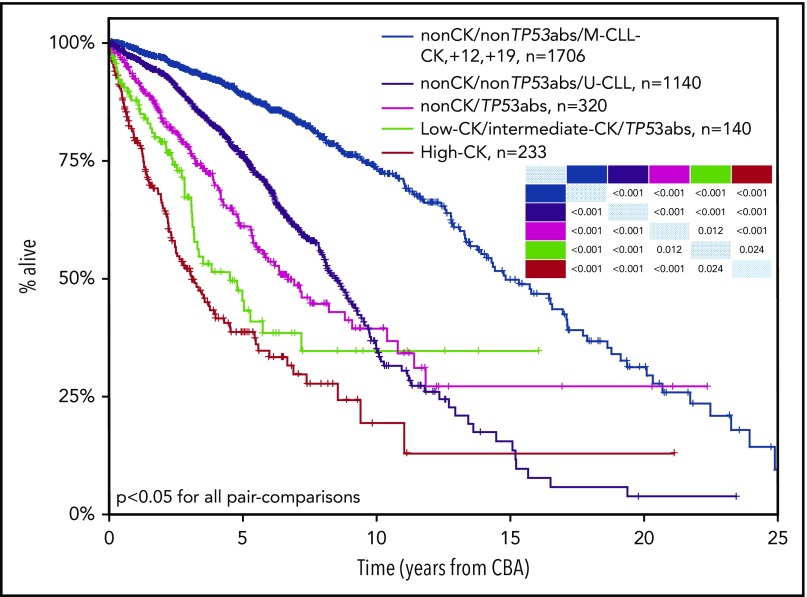
Integrating CK, TP53abs, and IGHV gene SHM, we developed a hierarchical model leading to the identification of 5 groups ranked from the shortest to the longest OS, as follows: (1) high-CK (median OS, 3.1 years; LQ-UQ, 1.3-8.3 years); (2) low-CK and intermediate-CK with TP53abs (median OS, 4.3 years; LQ-UQ, 2.3 years-NR); (3) non- CK/TP53abs (median OS, 6.6 years; LQ-UQ, 3.2 years-NR); (4) non-CK/non-TP53abs/U-CLL (median OS, 8.4 years; LQ-UQ, 5.1 years-NR); and (5) non-CK/non-TP53abs/M-CLL and CK with+12,+19 (median OS, 14.7 years; LQ-UQ, 9.4-21.5 years) (P < .05 for all pair comparisons) (Figure 4). In this proposed hierarchical model, all cases with high-CK were considered as 1 group independently of the presence of TP53abs. This decision was based on the fact that when cases with CK and coexisting TP53abs were placed in the model as a separate subgroup, no significant difference was observed compared with high-CK cases without TP53abs (P = .06).
Figure 4.
Kaplan-Meier curves based on a hierarchical model for OS incorporating CK, TP53abs (deletion of chromosome 17p and/or TP53 mutations), and the expression of somatically hypermutated (M-CLL) or unmutated (U-CLL) immunoglobulin heavy variable genes (IGHV). High-CK (≥5 aberrations, red line) exhibits the shortest OS followed by cases with TP53abs and 3 or 4 aberrations (low-CK and intermediate-CK, respectively; low-CK/intermediate-CK/TP53abs, green line), non-CK cases with TP53abs (non-CK/TP53abs, purple line), and non-CK/non-TP53abs cases with unmutated IGHV genes (non-CK/non-TP53abs/U-CLL, black line). Patients with the longest OS are those with non-CK/TP53abs and mutated IGHV genes (M-CLL), as well as patients with CK and +12,+19 (non-CK/non-TP53abs/M-CLL–CK,+12,+19, blue line). P values for all pair comparisons are provided with an inset table in which the colored cells indicate the respective subgroups based on the color of each Kaplan-Meier curve.
Discussion
In the largest study thus far conducted, we conclude that CK defined according to the presence of ≥3 numerical and/or structural abnormalities should not be axiomatically considered unfavorable in CLL, representing a heterogeneous group with variable clinical behavior ranging from remarkably indolent to extremely aggressive. High-CK, defined as the presence of at least 5 abnormalities, was associated with dismal clinical outcome, independently of the SHM and TP53 status. In contrast, low-CK and intermediate-CK defined according to the presence of 3 or 4 aberrations, respectively, seem to be clinically relevant only in the presence of TP53abs.
This differential impact on clinical outcome between high-CK vs low-CK/intermediate-CK subgroups could be partially attributed to the enrichment of U-CLL and TP53abs within the former, reaching up to 76% and 65%, respectively. Interestingly, among 25 cases analyzed by using NGS, none was found positive for low-frequent TP53 mutations, indicating that TP53abs do not constitute a sole explanation for high-CK. Moreover, when considering the distribution of aberrations along different chromosomes, high-CK cases displayed a distinct profile from the low-CK/intermediate-CK ones. In particular, the spectrum of affected chromosome regions was significantly broader, indicating that high-CK is reflecting increased genomic instability (Figure 3). Contrasting myeloid neoplasms with high karyotypic complexity that are characterized by a distinctive pattern of abnormalities (deletions of chromosomes 5q, 7q, and 17p),63 high-CK CLL exhibits accumulation of diverse chromosomal abnormalities in addition to the “CLL-typical” ones (deletion of 13q, 11q, 17p, and trisomy 12). On these grounds, the possibility that high-CK may in fact represent merely a surrogate for genomic instability in CLL cannot be excluded; however, this possibility should be tested experimentally before any conclusions can be drawn.
Conceivably, cases with 3 or 4 abnormalities (ie, the low-CK and intermediate-CK subgroups, respectively) might be prone to clonal evolution, acquire additional genomic aberrations, and upgrade to high-CK. Clonal evolution in CLL has been associated with U-CLL and TP53abs and, when present, is linked to resistance to treatment and shorter OS.64-66 In the current cohort, the great majority of the analyzed samples were obtained upon or near diagnosis, meaning that high-CK can also be an early event. However, to address the issue of clonal evolution, large cohorts with longitudinal samples are needed, which is beyond the scope of the present study.
Further highlighting that not all CK are equivalent, CK cases harboring +12,+19 were found to display an extremely indolent course even when the analysis was restricted to M-CLL. This outcome further supports previous reports that +12,+19 CLL represents a unique subgroup with a distinctive biological background and clinical behavior.8 Currently, the ontogenetic trajectory and mechanisms leading to the emergence of such clones remain unknown.
The relative significance of CK, particularly in relation to TP53 status, in patients with CLL treated with novel agents remains to be determined conclusively, given that the available evidence is derived from retrospective studies in small series with rather discrepant results. These discrepancies can be partially explained by the differences between cohorts46,47,49,50,67; however, it should also be noted that in all published studies, CK has been considered as a homogeneous group with no further differentiation according to the number of abnormalities, which, as we show herein, is crucial independent of the TP53 status.
Regarding the optimal methodology for detecting CK in CLL, our results mirror previous reports by us and others that CpG/IL-2 is superior to TPA stimulation because it was capable of identifying more cases with CK.29 Interestingly, in subgroup analysis, the distinction among low-CK/intermediate-CK and high-CK was clearly demarcated among cases analyzed with the CpG/IL-2 protocol (supplemental Figure 7A), whereas intermediate-CK and high-CK cases detected by using TPA methodologies found similar OS (supplemental Figure 7B). This scenario suggests that the TPA protocol may have failed to reveal the full spectrum of chromosomal aberrations within the CLL clone, thus leading to potential underestimation of CK. In our experience, this outcome is not mainly due to an insufficient number of obtained metaphases but rather to the difference in the quality of the obtained clonal metaphases, which is higher with the CpG/IL-2 protocol, thus facilitating the detection of the respective chromosomal aberrations. However, the large number of CK cases identified in the present cohort allowed robust subgroup analysis, hence reaching solid conclusions.
The retrospective nature of our study hinders robust correlations between CK and the response to particular treatment regimens. However, the great majority of the analyzed patients were treated with chemotherapy-based regimens upon treatment indication. Moreover, our study included “general practice” patients mostly recruited before 2015, and therefore only a small minority were treated with novel agents. Therefore, whether different CK subgroups may be associated with differential responses to such agents remains to be elucidated in future studies, ideally concerning prospective cohorts. It should be further highlighted that CBA represents the traditional and well-established methodology to define cytogenetic complexity not only in CLL but in almost all hematologic malignancies. Nonetheless, novel molecular techniques such as microarrays may be useful for the characterization of the clonal genomic background.68-72 However, the lack of harmonization, as well as the paucity of solid evidence regarding the clinical impact of cytogenetic complexity detected by microarrays in CLL, raises concerns about the unconditional use of microarrays in the everyday clinical setting. Lately, whole-genome sequencing has been reported as an alternative option for providing global genomic information.73-75 Whole-genome sequencing is for the time being, however, mostly used in the research field with further validation being needed before even suggesting integration of such an approach into the clinical routine.
In summary, we report that CK defined according to the presence of ≥3 numerical and/or structural abnormalities detected by using CBA should not be axiomatically considered unfavorable in CLL because it represents a heterogeneous group with variable clinical behavior. High-CK defined according to the presence of ≥5 chromosomal aberrations emerges as prognostically adverse, independently of clinical stage, SHM, and TP53 status, whereas low-CK and intermediate-CK are clinically relevant only if coexisting with TP53abs. Remarkably, cases carrying a CK with +12,+19 represent a unique subgroup with excellent prognosis. CK along with SHM and TP53 status enabled construction of a hierarchical model capable of identifying subgroups of patients with markedly distinct clinical outcomes. However, prospective clinical validation is clearly warranted before ultimately incorporating high-CK into risk stratification in CLL in everyday practice.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors thank the members of the Spanish Cooperative Group for Hematological Cytogenetics and the Spanish CLL Group for providing clinical and biological data.
This research was supported in part by Greek Precision Medicine Network in Oncology; H2020 “AEGLE, An analytics framework for integrated and personalized healthcare services in Europe” by the European Union; “ODYSSEAS,” implemented under the “Action for the Strategic Development on the Research and Technological Sector,” funded by the Operational Programme “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014-2020) and cofinanced by Greece and the European Union (European Regional Development Fund); the Swedish Research Council, the Lion’s Cancer Research Foundation, Uppsala; the Slovenian Research Agency; Associazione Italiana per la Ricerca sul Cancro AIRC (Investigator Grant #20246 to P.G. and 5 per mille Research Program #21198), Milano, Italy, Ricerca; ERA-NET TRANSCAN-2 JTC 2016, GCH-CLL; MIUR-PRIN 2015ZMRFEA, Rome, Italy; support by the Fondo di Ateneo per la Ricerca 2013, 2014, 2016, 2018 of the University of Ferrara (G.M.R., A.C.), Ricerca Finalizzata Ministero della Sanità (A.C., project RF-2011-02349712), Ministero dell’Istruzione, dell’Università e della Ricerca PRIN 2015 (A.C., project 2015ZMRFEA), BEAT Leukemia Foundation Milan Italy, and AIL Ferrara, Italy; and AZV 15-30015A and 15-31834A by the Ministry of Health, Czech Republic.
Footnotes
Contact panagiotis.baliakas@igp.uu.se or kostas.stamatopoulos@certh.gr for original data.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: P.B. designed the study, performed research, and wrote the paper; S.J., M.I., A.P., K.P., F.N.-K., Z.D., G.M.R., A.V., A.X., J.D., F.B.-M., E.S., P.A., K.D., G.P., V.E., M.D., T.I., R.C., M.D., M.J.C., N.R.-X., C.M., M.J., A.C.L., P.P., H.P., F.C., A.A., L.T., N.S., F.D., P.G., A.P.K., A.C., S.P., B.E., A.A., D.O., and C.H. provided data and performed research; and K.S. designed the study and wrote the paper, which was approved by all the authors.
Conflict-of-interest disclosure: K.S. received research support from Janssen Pharmaceuticals, Gilead Sciences, and Novartis SA. P.G. received research support from AbbVie, Janssen Pharmaceuticals, Gilead Sciences, and Novartis and honoraria from AbbVie, Acerta, BeiGene, Janssen, Gilead, and Sunesis. S.J. is employed by MLL Munich Leukemia Laboratory. H.P. received personal fees from Novartis Pharmaceuticals Corporation, Janssen, Takeda GmbH, and Celgene International unrelated to the present study. L.T. received research support from Janssen and Gilead. The remaining authors declare no competing financial interests.
A complete list of the members of the European Research Initiative on CLL (ERIC) can be found on the ERIC Web site at www.ericll.org/our-members.
Correspondence: Kostas Stamatopoulos, Institute of Applied Biosciences, Center for Research and Technology Hellas, 57001 Thermi, Thessaloniki, Greece; e-mail: kostas.stamatopoulos@gmail.com.
REFERENCES
- 1.Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer. 2016;16(3):145-162. [DOI] [PubMed] [Google Scholar]
- 2.Hallek M, Pflug N. Chronic lymphocytic leukemia. Ann Oncol. 2010;21(suppl 7):vii154-vii164. [DOI] [PubMed] [Google Scholar]
- 3.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804-815. [DOI] [PubMed] [Google Scholar]
- 4.Baliakas P, Hadzidimitriou A, Sutton LA, et al. Clinical effect of stereotyped B-cell receptor immunoglobulins in chronic lymphocytic leukaemia: a retrospective multicentre study. Lancet Haematol. 2014;1(2):e74-e84. [DOI] [PubMed] [Google Scholar]
- 5.Baliakas P, Hadzidimitriou A, Agathangelidis A, et al. Prognostic relevance of MYD88 mutations in CLL: the jury is still out. Blood. 2015;126(8):1043-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baliakas P, Iskas M, Gardiner A, et al. Chromosomal translocations and karyotype complexity in chronic lymphocytic leukemia: a systematic reappraisal of classic cytogenetic data. Am J Hematol. 2014;89(3):249-255. [DOI] [PubMed] [Google Scholar]
- 7.Baliakas P, Mattsson M, Stamatopoulos K, Rosenquist R. Prognostic indices in chronic lymphocytic leukaemia: where do we stand how do we proceed? J Intern Med. 2016;279(4):347-357. [DOI] [PubMed] [Google Scholar]
- 8.Baliakas P, Puiggros A, Xochelli A, et al. Additional trisomies amongst patients with chronic lymphocytic leukemia carrying trisomy 12: the accompanying chromosome makes a difference. Haematologica. 2016;101(7):e299-e302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catovsky D, Wade R, Else M. The clinical significance of patients’ sex in chronic lymphocytic leukemia. Haematologica. 2014;99(6):1088-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cortese D, Sutton LA, Cahill N, et al. On the way towards a ‘CLL prognostic index’: focus on TP53, BIRC3, SF3B1, NOTCH1 and MYD88 in a population-based cohort. Leukemia. 2014;28(3):710-713. [DOI] [PubMed] [Google Scholar]
- 11.Del Giudice I, Rossi D, Chiaretti S, et al. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica. 2012;97(3):437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910-1916. [DOI] [PubMed] [Google Scholar]
- 13.Eichhorst B, Hallek M. Prognostication of chronic lymphocytic leukemia in the era of new agents. Hematology Am Soc Hematol Educ Program. 2016;2016:149-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848-1854. [PubMed] [Google Scholar]
- 15.Mansouri L, Cahill N, Gunnarsson R, et al. NOTCH1 and SF3B1 mutations can be added to the hierarchical prognostic classification in chronic lymphocytic leukemia. Leukemia. 2013;27(2):512-514. [DOI] [PubMed] [Google Scholar]
- 16.Parikh SA, Strati P, Tsang M, West CP, Shanafelt TD. Should IGHV status and FISH testing be performed in all CLL patients at diagnosis? A systematic review and meta-analysis. Blood. 2016;127(14):1752-1760. [DOI] [PubMed] [Google Scholar]
- 17.Rai KR, Jain P. Chronic lymphocytic leukemia (CLL)—then and now. Am J Hematol. 2016;91(3):330-340. [DOI] [PubMed] [Google Scholar]
- 18.Rossi D, Gerber B, Stüssi G. Predictive and prognostic biomarkers in the era of new targeted therapies for chronic lymphocytic leukemia. Leuk Lymphoma. 2017;58(7):1548-1560. [DOI] [PubMed] [Google Scholar]
- 19.Wierda WG, O’Brien S, Wang X, et al. Prognostic nomogram and index for overall survival in previously untreated patients with chronic lymphocytic leukemia. Blood. 2007;109(11):4679-4685. [DOI] [PubMed] [Google Scholar]
- 20.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840-1847. [PubMed] [Google Scholar]
- 21.International CLL-IPI working group An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. Lancet Oncol. 2016;17(6):779-790. [DOI] [PubMed] [Google Scholar]
- 22.Baliakas P, Mattsson M, Hadzidimitriou A, et al. No improvement in long-term survival over time for chronic lymphocytic leukemia patients in stereotyped subsets #1 and #2 treated with chemo(immuno)therapy. Haematologica. 2018;103(4):e158-e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baliakas P, Moysiadis T, Hadzidimitriou A, et al. Tailored approaches grounded on immunogenetic features for refined prognostication in chronic lymphocytic leukemia [published online ahead of print 27 September 2018]. Haematologica. doi: 10.3324/haematol.2018.195032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallek M, Cheson BD, Catovsky D, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood. 2018;131(25):2745-2760. [DOI] [PubMed] [Google Scholar]
- 25.Puiggros A, Blanco G, Espinet B. Genetic abnormalities in chronic lymphocytic leukemia: where we are and where we go. BioMed Res Int. 2014;2014:435983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dierlamm J, Michaux L, Criel A, Wlodarska I, Van den Berghe H, Hossfeld DK. Genetic abnormalities in chronic lymphocytic leukemia and their clinical and prognostic implications. Cancer Genet Cytogenet. 1997;94(1):27-35. [DOI] [PubMed] [Google Scholar]
- 27.Juliusson G, Gahrton G. Chromosome aberrations in B-cell chronic lymphocytic leukemia. Pathogenetic and clinical implications. Cancer Genet Cytogenet. 1990;45(2):143-160. [DOI] [PubMed] [Google Scholar]
- 28.Juliusson G, Oscier DG, Fitchett M, et al. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323(11):720-724. [DOI] [PubMed] [Google Scholar]
- 29.Haferlach C, Bacher U. Cytogenetic methods in chronic lymphocytic leukemia. Methods Mol Biol. 2011;730:119-130. [DOI] [PubMed] [Google Scholar]
- 30.Gahrton G, Robèrt KH, Friberg K, Zech L, Bird AG. Nonrandom chromosomal aberrations in chronic lymphocytic leukemia revealed by polyclonal B-cell-mitogen stimulation. Blood. 1980;56(4):640-647. [PubMed] [Google Scholar]
- 31.Dubuc AM, Davids MS, Pulluqi M, et al. FISHing in the dark: how the combination of FISH and conventional karyotyping improves the diagnostic yield in CpG-stimulated chronic lymphocytic leukemia. Am J Hematol. 2016;91(10):978-983. [DOI] [PubMed] [Google Scholar]
- 32.Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgV(H) status and immunophenotyping. Leukemia. 2007;21(12):2442-2451. [DOI] [PubMed] [Google Scholar]
- 33.Haferlach C, Dicker F, Weiss T, et al. Toward a comprehensive prognostic scoring system in chronic lymphocytic leukemia based on a combination of genetic parameters. Genes Chromosomes Cancer. 2010;49(9):851-859. [DOI] [PubMed] [Google Scholar]
- 34.Puiggros A, Collado R, Calasanz MJ, et al. Patients with chronic lymphocytic leukemia and complex karyotype show an adverse outcome even in absence of TP53/ATM FISH deletions. Oncotarget. 2017;8(33):54297-54303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rigolin GM, Cibien F, Martinelli S, et al. Chromosome aberrations detected by conventional karyotyping using novel mitogens in chronic lymphocytic leukemia with “normal” FISH: correlations with clinicobiologic parameters. Blood. 2012;119(10):2310-2313. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen-Khac F, Borie C, Callet-Bauchu E, Eclache V, Struski S. Cytogenetics in the management of chronic lymphocytic leukemia: an update by the Groupe francophone de cytogénétique hématologique (GFCH). Ann Biol Clin (Paris). 2016;74(5):561-567. [DOI] [PubMed] [Google Scholar]
- 37.Blanco G, Puiggros A, Baliakas P, et al. Karyotypic complexity rather than chromosome 8 abnormalities aggravates the outcome of chronic lymphocytic leukemia patients with TP53 aberrations. Oncotarget. 2016;7(49):80916-80924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dicker F, Schnittger S, Haferlach T, Kern W, Schoch C. Immunostimulatory oligonucleotide-induced metaphase cytogenetics detect chromosomal aberrations in 80% of CLL patients: a study of 132 CLL cases with correlation to FISH, IgVH status, and CD38 expression. Blood. 2006;108(9):3152-3160. [DOI] [PubMed] [Google Scholar]
- 39.Jaglowski SM, Ruppert AS, Heerema NA, et al. Complex karyotype predicts for inferior outcomes following reduced-intensity conditioning allogeneic transplant for chronic lymphocytic leukaemia. Br J Haematol. 2012;159(1):82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rigolin GM, Cavallari M, Quaglia FM, et al. In CLL, comorbidities and the complex karyotype are associated with an inferior outcome independently of CLL-IPI. Blood. 2017;129(26):3495-3498. [DOI] [PubMed] [Google Scholar]
- 41.Rigolin GM, Saccenti E, Guardalben E, et al. In chronic lymphocytic leukaemia with complex karyotype, major structural abnormalities identify a subset of patients with inferior outcome and distinct biological characteristics. Br J Haematol. 2018;181(2):229-233. [DOI] [PubMed] [Google Scholar]
- 42.Badoux XC, Keating MJ, Wang X, et al. Fludarabine, cyclophosphamide, and rituximab chemoimmunotherapy is highly effective treatment for relapsed patients with CLL. Blood. 2011;117(11):3016-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herling CD, Klaumünzer M, Rocha CK, et al. Complex karyotypes and KRAS and POT1 mutations impact outcome in CLL after chlorambucil-based chemotherapy or chemoimmunotherapy. Blood. 2016;128(3):395-404. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi K, Hu B, Wang F, et al. Clinical implications of cancer gene mutations in patients with chronic lymphocytic leukemia treated with lenalidomide. Blood. 2018;131(16):1820-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Bris Y, Struski S, Guièze R, et al. Major prognostic value of complex karyotype in addition to TP53 and IGHV mutational status in first-line chronic lymphocytic leukemia. Hematol Oncol. 2017;35(4):664-670. [DOI] [PubMed] [Google Scholar]
- 46.Mato AR, Hill BT, Lamanna N, et al. Optimal sequencing of ibrutinib, idelalisib, and venetoclax in chronic lymphocytic leukemia: results from a multicenter study of 683 patients. Ann Oncol. 2017;28(5):1050-1056. [DOI] [PubMed] [Google Scholar]
- 47.Mato AR, Thompson M, Allan JN, et al. Real-world outcomes and management strategies for venetoclax-treated chronic lymphocytic leukemia patients in the United States. Haematologica. 2018;103(9):1511-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts AW, Davids MS, Pagel JM, et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med. 2016;374(4):311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson PA, O’Brien SM, Wierda WG, et al. Complex karyotype is a stronger predictor than del(17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer. 2015;121(20):3612-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson MA, Tam C, Lew TE, et al. Clinicopathological features and outcomes of progression of CLL on the BCL2 inhibitor venetoclax. Blood. 2017;129(25):3362-3370. [DOI] [PubMed] [Google Scholar]
- 51.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muthusamy N, Breidenbach H, Andritsos L, et al. Enhanced detection of chromosomal abnormalities in chronic lymphocytic leukemia by conventional cytogenetics using CpG oligonucleotide in combination with pokeweed mitogen and phorbol myristate acetate. Cancer Genet. 2011;204(2):77-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.International Standing Committee on Human Cytogenetic Nomenclature ISCN: An International System for Human Cytogenomic Nomenclature. Basel, NY: Karger; 2016. [Google Scholar]
- 54.Peterson JF. The complexities of defining a complex karyotype in hematological malignancies: a need for standardization? Acta Haematol. 2017;138(1):65-66. [DOI] [PubMed] [Google Scholar]
- 55.Slovak ML, Kopecky KJ, Cassileth PA, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96(13):4075-4083. [PubMed] [Google Scholar]
- 56.Agathangelidis A, Darzentas N, Hadzidimitriou A, et al. Stereotyped B-cell receptors in one-third of chronic lymphocytic leukemia: a molecular classification with implications for targeted therapies. Blood. 2012;119(19):4467-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malcikova J, Tausch E, Rossi D, et al. ; European Research Initiative on Chronic Lymphocytic Leukemia (ERIC)–TP53 network . ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia—update on methodological approaches and results interpretation. Leukemia. 2018;32(5):1070-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Put N, Konings P, Rack K, et al. ; Belgian Cytogenetic Group for Hemato-Oncology (BCGHO) . Improved detection of chromosomal abnormalities in chronic lymphocytic leukemia by conventional cytogenetics using CpG oligonucleotide and interleukin-2 stimulation: a Belgian multicentric study. Genes Chromosomes Cancer. 2009;48(10):843-853. [DOI] [PubMed] [Google Scholar]
- 59.Ibbotson R, Athanasiadou A, Sutton LA, et al. Coexistence of trisomies of chromosomes 12 and 19 in chronic lymphocytic leukemia occurs exclusively in the rare IgG-positive variant. Leukemia. 2012;26(1):170-172. [DOI] [PubMed] [Google Scholar]
- 60.Sellmann L, Gesk S, Walter C, et al. Trisomy 19 is associated with trisomy 12 and mutated IGHV genes in B-chronic lymphocytic leukaemia. Br J Haematol. 2007;138(2):217-220. [DOI] [PubMed] [Google Scholar]
- 61.Rossi D, Khiabanian H, Spina V, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123(14):2139-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nadeu F, Delgado J, Royo C, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127(17):2122-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391-2405. [DOI] [PubMed] [Google Scholar]
- 64.Huang SJ, Bergin K, Smith AC, et al. Clonal evolution as detected by interphase fluorescence in situ hybridization is associated with worse overall survival in a population-based analysis of patients with chronic lymphocytic leukemia in British Columbia, Canada. Cancer Genet. 2017;210:1-8. [DOI] [PubMed] [Google Scholar]
- 65.Stilgenbauer S, Sander S, Bullinger L, et al. Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica. 2007;92(9):1242-1245. [DOI] [PubMed] [Google Scholar]
- 66.Wawrzyniak E, Kotkowska A, Blonski JZ, et al. Clonal evolution in CLL patients as detected by FISH versus chromosome banding analysis, and its clinical significance. Eur J Haematol. 2014;92(2):91-101. [DOI] [PubMed] [Google Scholar]
- 67.Lazarian G, Tausch E, Eclache V, et al. TP53 mutations are early events in chronic lymphocytic leukemia disease progression and precede evolution to complex karyotypes. Int J Cancer. 2016;139(8):1759-1763. [DOI] [PubMed] [Google Scholar]
- 68.Gunnarsson R, Isaksson A, Mansouri M, et al. Large but not small copy-number alterations correlate to high-risk genomic aberrations and survival in chronic lymphocytic leukemia: a high-resolution genomic screening of newly diagnosed patients. Leukemia. 2010;24(1):211-215. [DOI] [PubMed] [Google Scholar]
- 69.Kay NE, Eckel-Passow JE, Braggio E, et al. Progressive but previously untreated CLL patients with greater array CGH complexity exhibit a less durable response to chemoimmunotherapy. Cancer Genet Cytogenet. 2010;203(2):161-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ouillette P, Collins R, Shakhan S, et al. Acquired genomic copy number aberrations and survival in chronic lymphocytic leukemia. Blood. 2011;118(11):3051-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Puiggros A, Puigdecanet E, Salido M, et al. Genomic arrays in chronic lymphocytic leukemia routine clinical practice: are we ready to substitute conventional cytogenetics and fluorescence in situ hybridization techniques? Leuk Lymphoma. 2013;54(5):986-995. [DOI] [PubMed] [Google Scholar]
- 72.Schoumans J, Suela J, Hastings R, et al. Guidelines for genomic array analysis in acquired haematological neoplastic disorders. Genes Chromosomes Cancer. 2016;55(5):480-491. [DOI] [PubMed] [Google Scholar]
- 73.Klintman J, Barmpouti K, Knight SJL, et al. Clinical-grade validation of whole genome sequencing reveals robust detection of low-frequency variants and copy number alterations in CLL. Br J Haematol. 2018;182(3):412-417. [DOI] [PubMed] [Google Scholar]
- 74.Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Puente XS, Beà S, Valdés-Mas R, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519-524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.