Fire blight disease has relatively few management options, with antibiotic application at bloom time being chief among them. As modification to elongation factor P (EF-P) is vital to virulence in several species, both EF-P and its modifying enzymes make attractive targets for novel antibiotics. However, it will be useful to understand how bacteria might overcome the hindrance of EF-P function so that we may be better prepared to anticipate bacterial adaptation to such antibiotics. The present study indicates that the mutation of hrpA3 could provide a partial offset for the loss of EF-P activity. In addition, little is known about EF-P functional interactions or the HrpA3 predicted RNA helicase, and our genetic approach allowed us to discern a novel gene associated with EF-P function.
KEYWORDS: DEAH-box RNA helicase, elongation factor P, Erwinia amylovora, apple, fire blight, ribosome
ABSTRACT
Elongation factor P (EF-P) facilitates the translation of certain peptide motifs, including those with multiple proline residues. EF-P must be posttranslationally modified for full functionality; in enterobacteria, this is accomplished by two enzymes, namely, EpmA and EpmB, which catalyze the β-lysylation of EF-P at a conserved lysine position. Mutations to efp or its modifying enzymes produce pleiotropic phenotypes, including decreases in virulence, swimming motility, and extracellular polysaccharide production, as well as proteomic perturbations. Here, we generated targeted deletion mutants of the efp, epmA, and epmB genes in the Gram-negative bacterium Erwinia amylovora, which causes fire blight, an economically important disease of apples and pears. As expected, the Δefp, ΔepmA, and ΔepmB mutants were all defective in virulence on apples, and all three mutants were complemented in trans with plasmids bearing wild-type copies of the corresponding genes. By analyzing spontaneous suppressor mutants, we found that mutations in the hrpA3 gene partially or completely suppressed the colony size, extracellular polysaccharide production, and virulence phenotypes in apple fruits and apple tree shoots but not the swimming motility phenotypes of the Δefp, ΔepmA, and ΔepmB mutants. The deletion of hrpA3 alone did not produce any alterations in any characteristics measured, indicating that the HrpA3 protein is not essential for any of the processes examined. The hrpA3 gene encodes a putative DEAH-box ATP-dependent RNA helicase. These results suggest that the loss of the HrpA3 protein at least partially compensates for the lack of the EF-P protein or β-lysylated EF-P.
IMPORTANCE Fire blight disease has relatively few management options, with antibiotic application at bloom time being chief among them. As modification to elongation factor P (EF-P) is vital to virulence in several species, both EF-P and its modifying enzymes make attractive targets for novel antibiotics. However, it will be useful to understand how bacteria might overcome the hindrance of EF-P function so that we may be better prepared to anticipate bacterial adaptation to such antibiotics. The present study indicates that the mutation of hrpA3 could provide a partial offset for the loss of EF-P activity. In addition, little is known about EF-P functional interactions or the HrpA3 predicted RNA helicase, and our genetic approach allowed us to discern a novel gene associated with EF-P function.
INTRODUCTION
The translation of mRNA into protein by ribosomes is an essential function of all cells. The ribosome may pause during this translation as a function of steric hindrance of amino acids or uncharged tRNA, which reduces the rate of protein synthesis (1). These pauses may either require intervention for cell function or serve as a regulatory step in protein synthesis, such as pauses regulating the transcription of downstream genes (2).
Elongation factor P (EF-P), a protein discovered in Escherichia coli, is associated with the 70S ribosome and stimulates peptide bond formation (3, 4). EF-P is a ubiquitous prokaryotic homologue of eukaryotic eIF5A, a protein that promotes translation of proline-rich motifs (5). The association of EF-P with the ribosome is transient and dynamic (6). EF-P can diminish ribosomal stalling at proline-rich regions of polypeptides by stimulating peptide bond formation in the ribosome (7–9). In addition, other motifs, both with and without proline, have been shown to require EF-P for efficient translation (10–12). EF-P-mediated alleviation of translational stalling is thought to play a role in protein abundance only when translation initiation is not the rate-limiting step in the synthesis of a protein (13).
EF-P must be posttranslationally modified for full functionality. In E. coli and Salmonella spp., EF-P is β-lysylated at a conserved lysine position by two proteins, namely, EpmA (YjeA and PoxA) and EpmB (YjeK) (14). EpmB is a lysine-2,3-aminomutase that converts l-lysine into (R)-β-lysine, which is ligated to the conserved Lys34 residue of EF-P by EpmA. While EF-P β-lysylation was originally described in enterobacteria, it is now clear that several other posttranslational modification pathways exist in more divergent bacteria. A lysine residue is posttranslationally modified in Bacillus subtilis by a range of enzymes (15). In Pseudomonas aeruginosa, EF-P is instead rhamnosylated at a conserved arginine residue by EarP (16). Unlike most species in which mutations to modifying enzymes produce a similar phenotype to efp mutants, B. subtilis-modifying enzyme mutants show distinct phenotypes, including altered proline translation rate and sensitivity to certain chemicals (15).
EF-P is required for full virulence in several species, including Salmonella enterica, Shigella flexneri, and Agrobacterium tumefaciens (17–19). The mutants also exhibit a wide range of other phenotypes, including increased chemical sensitivity, decreased swimming motility, and decreased extracellular polysaccharide production (17, 20, 21).
Fire blight is a serious bacterial disease of rosaceous plants, including apple and pear, which is caused by the enterobacterium Erwinia amylovora. Bacteria typically enter the plant through flowers (22) and are able to spread systemically within the plant, causing severe tissue necrosis, yield losses, and potentially tree mortality. Antibiotics, such as streptomycin, are heavily used for fire blight disease management, which not only increases environmental levels of antibiotics but also produces high pressure for antibiotic resistance (23). Like many other enterobacteria, E. amylovora requires both a type III secretion system and the production of extracellular polysaccharides for pathogenicity (24, 25).
Recently, we described an E. amylovora epmB (yjeK) mutant that displays environmentally dependent changes to its proteome, as well as a decrease in virulence and the production of the extracellular polysaccharide amylovoran (21). Amylovoran is an essential virulence factor for E. amylovora, and mutations in the amylovoran biosynthesis operon (ams) result in a total loss of pathogenicity (25), and reduced amylovoran production in the epmB mutant likely contributes to its reduced ability to cause fire blight. In the present study, we identified extragenic suppressor mutants of an efp deletion mutant that were able to restore multiple phenotypes to wild type. These suppressors all contained mutations in hrpA3 (DEAH family RNA helicase-like protein A [26]), which encodes a predicted ATP-dependent RNA helicase. These data help elucidate the precise role of EF-P in protein translation.
RESULTS AND DISCUSSION
Δefp, ΔepmA, and ΔepmB mutants display similar virulence defects.
Previously, we analyzed the virulence defect of an E. amylovora epmB (yjeK) Tn5 transposon mutant (21). The epmB Tn5 mutant was defective in virulence in apple fruits and tree shoots. In addition, the epmB Tn5 mutant had defects in extracellular polysaccharide production and swimming motility, increased chemical sensitivity, and major alterations to the proteome (21). In several systems, the phenotypes of efp, epmA, and epmB mutants have been shown to be nearly identical, which is consistent with their roles related to EF-P production and function (20).
Here, we created individual, targeted deletion mutants of the efp, epmA, and epmB genes in E. amylovora (Δefp, ΔepmA, and ΔepmB) (see Table S1 in the supplemental material). These mutants were inoculated onto apple fruitlets, and all produced severely attenuated disease development (see Fig. S1 in the supplemental material). The virulence defect phenotypes of all three mutants were complemented in trans with plasmid-borne copies of the respective genes (Fig. S1). These results indicate that efp, epmA, and epmB all have similar mutant phenotypes in E. amylovora, and genetic complementation confirms the specific role of each gene in the phenotype.
Spontaneous suppressors of the Δefp, ΔepmA, and ΔepmB small-colony phenotype.
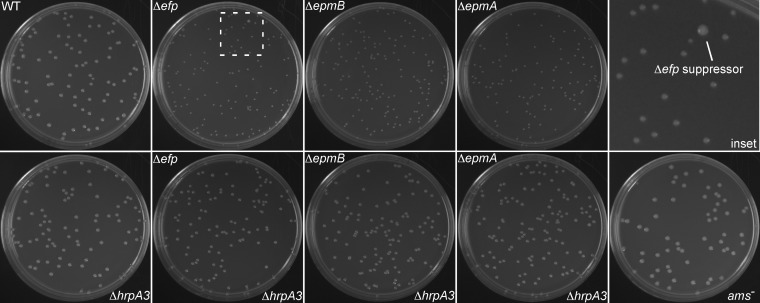
While growing the Δefp, ΔepmA, and ΔepmB mutants on rich-medium LB agar plates, it was noted that all of them produced small colonies compared to the wild type (Fig. 1 and 2). When plated for single colonies, Δefp, ΔepmA, and ΔepmB strains periodically displayed occasional larger colonies with sizes similar to wild-type E. amylovora colonies (Fig. 1, inset). When these wild-type-sized colony strains were isolated, they consistently produced only large colonies, suggesting that the phenotype was stable. PCR tests verified the Δefp, ΔepmA, and ΔepmB genotypes of the wild-type-sized colony strains, indicating that these represented potential suppressor mutants of the small-colony phenotype of the Δefp, ΔepmA, and ΔepmB mutants. Six independent wild-type-sized colony isolates from independent cultures of the Δefp mutant (ΔefpL1 to ΔefpL6) and one wild-type-sized colony isolate in the ΔepmB genetic background (ΔepmBL1) were obtained in pure culture (Table S1). All suppressors were isolated from unique parent strains to ensure that suppressors were not simply clones.
FIG 1.
Deletion of hrpA3 restores colony size to that of the wild type in Δefp, ΔepmA, and ΔepmB genetic backgrounds. LB plates are shown after 72 hours of growth at 28°C. Genotypes are indicated on the figure. “Inset” represents a magnification of the boxed area in the Δefp panel and contains an example spontaneous suppressor colony. Plates are 100- by 15-mm petri dishes.
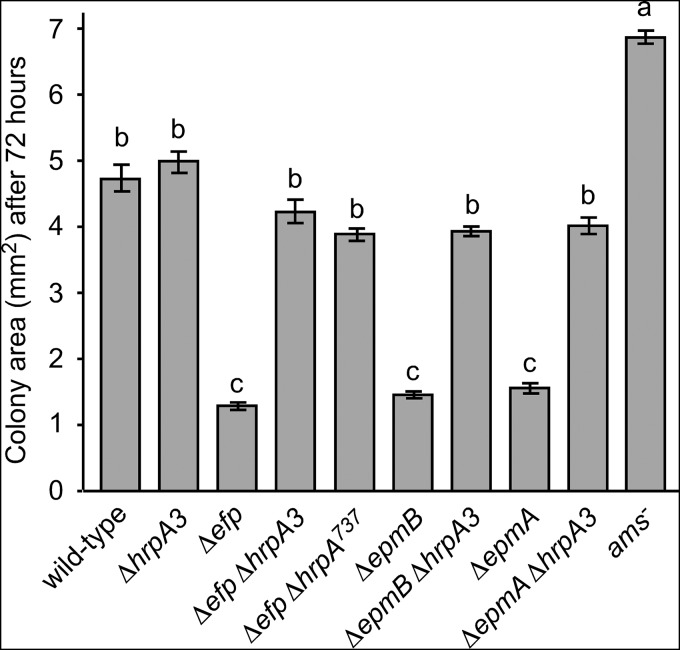
FIG 2.
Quantification of colony sizes shown in Fig. 1. Colony sizes for the indicated strains were measured after 72 hours of growth on LB plates at 28°C. Colony size was quantified using NIH ImageJ to measure the area of each colony. Bars represent the average areas of 10 colonies for each genotype from a single representative experiment, and error bars indicate standard error (SE). Strains sharing a letter have no statistically significant difference based on a Tukey test. The entire experiment was performed at least four times with similar results each time.
All sequenced suppressors had mutations in hrpA3.
The complete genome sequences of seven wild-type-sized colony isolates (ΔefpL1 to ΔefpL6 and ΔepmBL1) were determined, assembled, and compared with the sequence of the parental wild-type HKN06P1 E. amylovora lab strain. Only one gene contained sequence polymorphisms in all seven of the suppressors. This gene, hrpA3, encodes a putative RNA helicase protein (coding DNA sequence [CDS] GenBank accession no. CBA20795 in E. amylovora strain CFBP1430).
Suppressors ΔefpL1 to ΔefpL6 all contained frameshift mutations predicted to disrupt the C-terminal half of the HrpA3 protein, while suppressor strain ΔepmBL1 contained a large deletion of the 5′ region of the hrpA3 gene (Fig. 3; see File S1 in the supplemental material). Despite being isolated from independent parental strains, suppressors ΔefpL2, ΔefpL3, and ΔefpL5 all contained the same hrpA3 mutant allele, suggesting that they were actually independent mutation events resulting in the same hrpA3 allele. These results imply that the mutation of hrpA3 can suppress the small-colony phenotype of efp and epmB mutants and that the loss of a functional HrpA3 protein can compensate in some way for the loss of a functional EF-P protein, at least in terms of colony size. All of the mutations resulted in major predicted alterations to the HrpA3 protein structure, and no amino acid substitution mutations were obtained. This finding suggests that a major disruption of the HrpA3 protein structure is required in order to suppress the effects of the loss of EF-P.
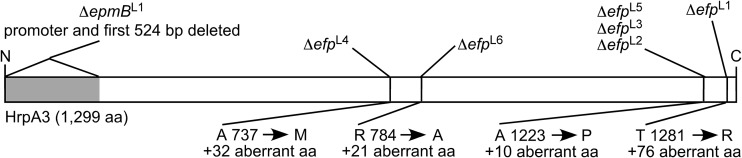
FIG 3.
Polymorphisms in the hrpA3 gene detected in seven independent suppressors of the Δefp and ΔepmB small-colony phenotype. ΔefpL1 to ΔefpL6 are from six independent Δefp suppressors; ΔepmBL1 is from a ΔempB suppressor. For the frameshift mutations, alterations to the predicted protein product of the hrpA3 gene are indicated as the position of the first altered amino acid, along with the length of additional, aberrant amino acids produced by the frameshift. N and C indicate the amino and carboxy termini, respectively, of the HrpA3 protein.
hrpA3 codes for a predicted DEAH-box RNA helicase. While these proteins are referred to as “helicases,” their double-stranded unwinding activity is unclear (27). Characterized mostly in eukaryotes, DEXD/H box RNA helicases are thought to be involved in multiple cellular processes, including translational initiation and termination, ribosome biogenesis, and RNA degradation (28). In Borrelia burgdorferi, hrpA3 is necessary for infectivity in mice (29), while in E. coli it is necessary for the processing of the daa fimbrial operon (30).
Deletion of hrpA3 suppresses the small-colony phenotype of Δefp, ΔepmA, and ΔepmB mutants.
To confirm that mutations to hrpA3 were responsible for the wild-type-sized colony phenotype of the Δefp suppressor strains, the hrpA3 gene was deleted in wild-type, Δefp, ΔepmA, and ΔepmB genetic backgrounds. In addition, a partial genomic deletion of hrpA3 (ΔhrpA3737) was constructed in the Δefp genetic background. This deletion terminated the predicted protein product at amino acid position 737, which corresponded to the earliest change seen among the frameshift mutations (Δefp737) (Fig. 3). The Δefp ΔhrpA3, Δefp ΔhrpA3737, ΔepmA ΔhrpA3, and ΔepmB ΔhrpA3 double mutants all had colony sizes similar to those of the wild type on LB agar plates (Fig. 1 and 2). The ΔhrpA3 single mutant had colonies similar in size to those of the wild type (Fig. 1 and 2). In addition, a mutant with a Tn5 insertion in the promoter region of the ams operon (ams−) (21) was analyzed for colony size, and the colonies were larger than the wild type (Fig. 1 and 2).
These results confirm that a mutation of hrpA3 can suppress the small-colony phenotype of Δefp, ΔepmA, and ΔepmB mutants, as implicated by the suppressor genome sequencing results. In addition, a partial deletion of hrpA3 (ΔhrpA3737) was sufficient to suppress the small-colony phenotype of the Δefp mutant. However, disruption of hrpA3 alone did not affect colony size. The normal colony size of the ΔhrpA3 single mutant indicated that the hrpA3 gene is dispensable for growth on LB agar.
Deletion of hrpA3 suppresses the amylovoran production defect of Δefp, ΔepmA, and ΔepmB mutants.
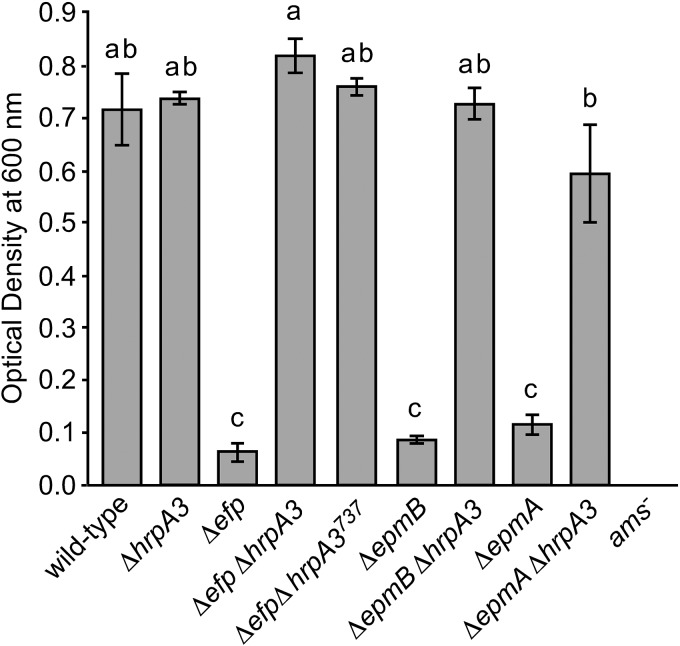
Previously, we reported that a Tn5 mutation in epmB (yjeK) resulted in greatly diminished amylovoran production (21). Consistent with the previous findings, the Δefp, ΔepmA, and ΔepmB mutants produced a minimal amount of the extracellular polysaccharide amylovoran (Fig. 4). In contrast, the Δefp ΔhrpA3, Δefp ΔhrpA3737, ΔepmA ΔhrpA3, and ΔepmB ΔhrpA3 double mutants had amylovoran production levels similar to those of the wild type (Fig. 4). As expected, the ams− mutant did not produce detectable amylovoran; however, the ΔhrpA3 single mutant had amylovoran production levels similar to those of the wild type. These results indicate that the mutation of hrpA3 can suppress the amylovoran production defect of the Δefp, ΔepmA, and ΔepmB mutants. However, the mutation of hrpA3 alone does not affect amylovoran production, indicating that hrpA3 is dispensable for amylovoran production by E. amylovora.
FIG 4.
Deletion of hrpA3 restores the amylovoran extracellular polysaccharide production level to that of the wild type in Δefp, ΔepmA, and ΔepmB genetic backgrounds. Amylovoran production in the indicated strains was assayed via a cetylpyridinium chloride precipitation assay, where the precipitate OD at 600 nm is measured using a spectrophotometer. The experiment was repeated three times, with similar results each time. Error bars represent SE. Strains sharing the same letter have no statistically significant difference based on a Tukey test.
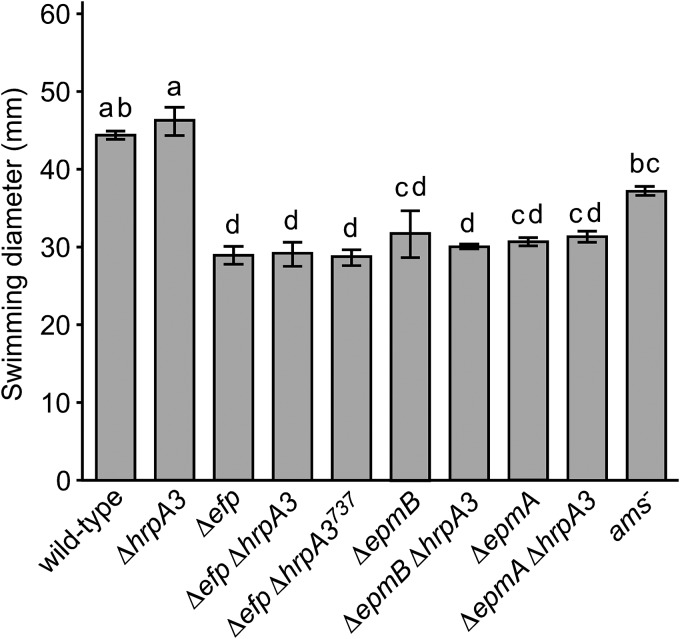
Deletion of hrpA3 does not suppress swimming motility defects of the Δefp, ΔepmA, and ΔepmB mutants.
Previously, we reported that a Tn5 mutation in epmB (yjeK) resulted in greatly diminished swimming motility in low-density agar medium, and this correlated with reduced abundance of particular motility-associated proteins (21). Similarly, the Δefp, ΔepmA, and ΔepmB mutants all had reduced swimming motility (Fig. 5). In addition, the swimming motility of the Δefp ΔhrpA3, Δefp ΔhrpA3737, ΔepmA ΔhrpA3, and ΔepmB ΔhrpA3 double mutants was similar to the cognate Δefp, ΔepmA, and ΔepmB single mutants, respectively (Fig. 5). The ΔhrpA3 mutant had a similar swimming motility as that of the wild type. These results indicate that disruption of hrpA3 cannot suppress the swimming motility defect of the Δefp, ΔepmA, and ΔepmB mutants and that hrpA3 by itself is not required for E. amylovora swimming motility. This finding indicates that not all phenotypes of the Δefp, ΔepmA, and ΔepmB mutants are suppressed by the mutation of hrpA3, suggesting that the loss of the HrpA3 protein cannot compensate for all the defects caused by a lack of the EF-P protein.
FIG 5.
Deletion of hrpA3 does not increase swimming motility in Δefp, ΔepmA, and ΔepmB genetic backgrounds. Bars represent average diameters of visible bacteria for the indicated strains on M9 minimal medium plates after 48 hours at 28°C. The experiment was repeated four times with similar results each time. Error bars represent SE. Strains sharing the same letter have no statistically significant difference based on a Tukey test.
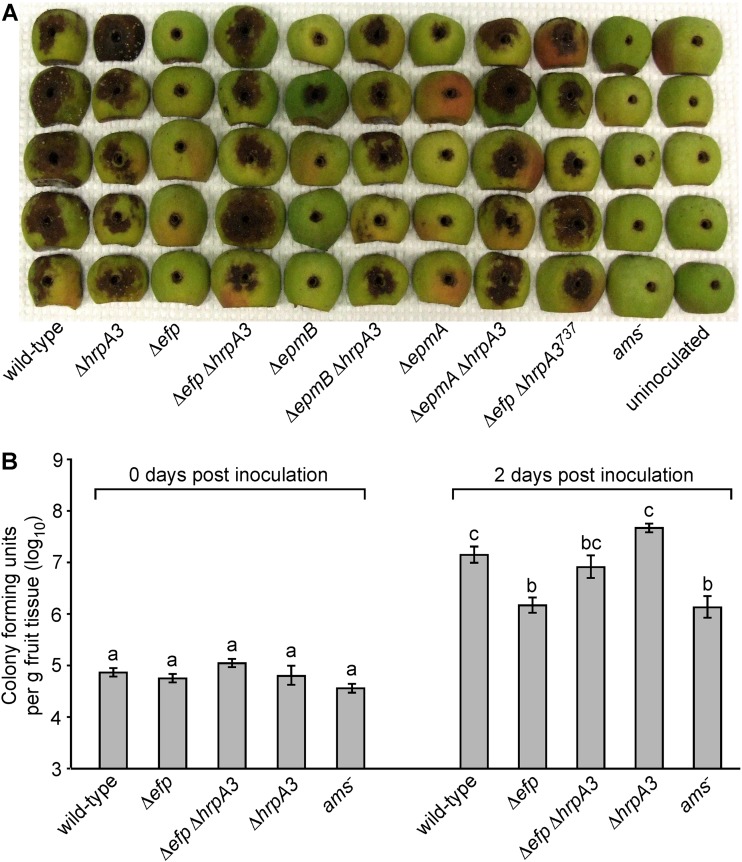
Deletion of hrpA3 partially suppresses virulence defects of the Δefp, ΔepmA, and ΔepmB mutants.
Previously, we reported that a Tn5 mutation in epmB (yjeK) resulted in greatly diminished virulence and growth on apple fruitlets and virulence in apple tree shoots (21). Consistent with this, the Δefp, ΔepmA, and ΔepmB mutants were all minimally virulent on apple fruit, producing little of the necrosis and bacterial ooze indicative of fire blight disease development, while the wild-type strain produced severe necrosis and ooze on all fruits inoculated (Fig. 6A). In contrast, the Δefp ΔhrpA3, Δefp ΔhrpA3737, ΔepmA ΔhrpA3, and ΔepmB ΔhrpA3 double mutants produced symptoms on apple fruitlets similar to those produced by the wild type, and the ΔhrpA3 single mutant also produced similar disease symptoms on apple fruitlets as those of the wild type (Fig. 6A). As expected, the ams− mutant produced no symptoms on the fruit, which is consistent with amylovoran being essential for E. amylovora virulence (25). These results indicate that deletion of hrpA3 partially suppresses the virulence defect of the Δefp, ΔepmA, and ΔepmB mutants.
FIG 6.
Deletion of hrpA3 partially restores virulence in the Δefp, ΔepmA, and ΔepmB genetic backgrounds. (A) Symptom development in apple fruitlets at 7 days after inoculation with 2 × 106 cells of the indicated E. amylovora strains. (B) Growth of the indicated bacterial strains in apple fruitlets inoculated as in panel A, over the first 2 days of infection, expressed as CFU per gram of apple tissue. The experiment was repeated three times, with similar results each time. Within a given time point, bars sharing a letter have no statistically significant difference according to a Tukey test, and error bars represent SE.
E. amylovora growth in apple fruit tissues typically correlates with severity of disease development (31). Growth of the wild-type, Δefp, Δefp ΔhrpA3, ΔhrpA3, and ams− E. amylovora strains in the apple fruitlets was quantified 2 days after inoculation, when the majority of population increase is typically observed (Fig. 6B). Populations of the Δefp ΔhrpA3 double mutant were numerically higher in inoculated fruitlet tissue than populations of the Δefp mutant, and Δefp ΔhrpA3 populations were not statistically separable from either the wild-type or the Δefp mutant populations. This indicates that deletion of hrpA3 partially corrected the growth defect of the Δefp mutant in immature apple fruit tissues. The growth of the Δefp mutant was similar to that of the ams− mutant, indicating that the Δefp mutant has a severe defect in growth similar to that of the avirulent ams− mutant.
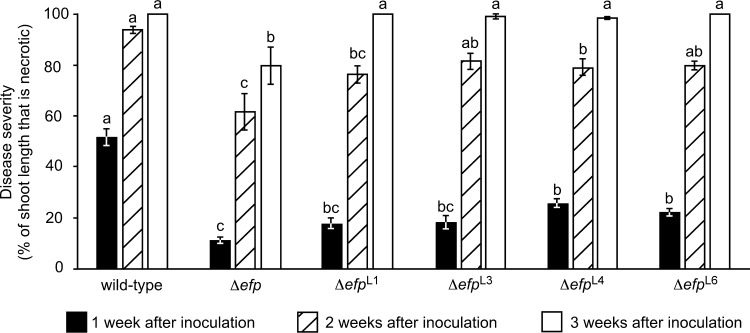
Shoot blight is one of the most common and devastating manifestations of fire blight disease. While there is a good correlation between virulence in apple fruitlets and shoot blight virulence in whole trees (31), the shoot tissue host environment poses different challenges to E. amylovora. Therefore, the virulence of four representative spontaneous suppressor strains (ΔefpL1, ΔefpL3, ΔefpL4, and ΔefpL6) was tested on second-leaf ‘Gala’ apple tree shoots by shoot tip wound inoculation. The selected suppressor strains represented the four different hrpA3 truncation alleles (Fig. 3). Each mutant was inoculated onto actively growing shoot tips, and the length of symptomatic branches as a percentage of total branch length was measured at 1, 2, and 3 weeks postinoculation.
While not as virulent as the wild type, ΔefpL1, ΔefpL3, ΔefpL4, and ΔefpL6 produced numerically higher disease severity values than the Δefp mutant at all time points (Fig. 7). At 1 week after inoculation, disease severity produced by ΔefpL4 and ΔefpL6 had a statistically significant difference from the Δefp mutant. After 2 weeks, the disease severity produced by ΔefpL3 and ΔefpL6 was not statistically separable from the disease severity caused by the wild type, and ΔefpL3, ΔefpL4, and ΔefpL6 produced significantly more severe symptoms than the Δefp mutant. After 3 weeks, the disease severity caused by all four suppressor strains had a statistically significant difference from the Δefp mutant, and most tree shoots inoculated with the suppressor strains and the wild type were completely necrotic. Together, the results indicate that the loss of HrpA3 results in significantly increased virulence of the Δefp mutant.
FIG 7.
Δefp suppressor mutants have increased virulence in apple tree shoots compared with that of the Δefp parent strain. Apple trees were inoculated through shoot tip wounds with the indicated E. amylovora strains, and the extent of resulting necrosis in the inoculated shoots was measured at 1-week intervals. Bars represent the extent of necrosis in 25 shoots per strain; error bars indicate SE. Within a given time point, bars sharing a letter have no statistically significant difference according to a Tukey test. This experiment was performed twice with similar results each time; results of one representative experiment are shown.
CONCLUSIONS
Suppressor mutants have long been used to uncover novel partners in biological pathways, in this case allowing us to identify an otherwise unknown gene related to EF-P function, hrpA3. The present study has revealed that mutations disrupting the predicted RNA helicase HrpA3 can partially compensate for the loss of functional EF-P. Since hrpA3 deletions and truncations display nearly identical phenotypes in Δefp, ΔepmA, and ΔepmB mutants, it follows that the loss of HrpA3 appears to compensate specifically for the loss of a functional EF-P and that this compensation does not involve a restoration of function to the EF-P protein.
The restoration of amylovoran production in the Δefp ΔhrpA3, Δefp ΔhrpA3737, ΔepmA ΔhrpA3, and ΔepmB ΔhrpA3 double mutants to wild-type levels could at least partly account for their increased virulence compared to that of the Δefp, ΔepmA and ΔepmB single mutants. However, increased amylovoran production is unlikely to account for the restoration of colony size to that of the wild type in the Δefp ΔhrpA3, Δefp ΔhrpA3737, ΔepmA ΔhrpA3, and ΔepmB ΔhrpA3 double mutants, as the ams− mutant displayed colonies that were larger than the wild type. This effect is possibly due to a decrease in the colony dome shape sometimes seen in extracellular polysaccharide mutants (32, 33). Thus, at least two unique measurable phenotypes (colony size and amylovoran production) are restored by the mutation of hrpA3 in Δefp, ΔepmA, and ΔepmB strains.
Deleting hrpA3 in a wild-type E. amylovora background produced no measurable phenotypic change in any assay performed in this study, indicating that hrpA3 is not an essential gene in E. amylovora. Several other putative ATP-dependent RNA helicase genes are found in the E. amylovora genome, including hrpB, deaD, and dbpA. It is possible that aspects of HrpA3 function are duplicated by one or more of these other putative RNA helicases; however, our results indicate that HrpA3 has some unique functions related specifically to EF-P.
It is possible that HrpA3 directly or indirectly interacts with or structurally changes the ribosome complex in a way that partially alleviates ribosomal stalling, either universally or under certain circumstances. In an interactome study in E. coli, HrpA3 was found to interact with both 50S and 30S subunits of the ribosome, as well as elongation factor thermounstable (EF-Tu), uncharacterized tRNA/rRNA methyltransferase YfiF, and several other proteins (34). It is not immediately clear why disruption of a DEAH-box RNA helicase would ameliorate the effects of the loss of EF-P. However, DEAH family RNA helicases have a number of functions related to mRNA processing and translation that could be related to this effect, including ribosomal biogenesis, translation initiation, and the bacterial degradosome (35).
Given the role of HrpA3 orthologues in the degradation of particular RNAs (29, 30, 36), it is possible that deletion of hrpA3 in E. amylovora efp mutants simply alters the abundance of certain transcripts in a manner which happens to suppress specific Δefp phenotypes. For example, mutation of the DEAH-box RNA helicase gene hrpB of Xanthomonas citri pv. citri reduced biofilm formation, increased sliding motility, and reduced symptom development in sweet orange (37). Furthermore, mutation of DEAH-box RNA helicase Z5898 of E. coli O157:H7 resulted in the absence of flagella (38).
Mechanistically, ΔhrpA3 suppression of Δefp phenotypes could occur through altered processing of translational stall events. mRNAs in stalled ribosomes may be cleaved and both the mRNA and defective protein targeted for degradation (39, 40). HrpA3 has been found to be associated with the degradosome of E. coli (41). It is possible that HrpA3 helps enable degradation of certain mRNAs in stalled ribosomes. Thus, the Δefp ΔhrpA3 mutant may have more time than the Δefp mutant to resolve ribosomal stalls (42) without aborting translation, leading to a partial alleviation of Δefp mutant phenotypes in the Δefp ΔhrpA3 mutant. Long transcripts, such as the 16-kb ams transcript (43), might actually be translated more efficiently in the Δefp ΔhrpA3 mutant than in the Δefp mutant, resulting in an energetic benefit to the cell.
MATERIALS AND METHODS
Growth media and conditions.
A nalidixic acid-resistant derivative of E. amylovora strain HKN06P1 (31) (referred to as wild type) was used throughout. E. amylovora strains were routinely grown at 28°C in lysogeny broth (LB) or M9 minimal medium (6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl, 1.0 g NH4Cl, and 0.24 g MgSO4 [all per liter], supplemented with 0.4% sorbitol and 0.2 g/liter of nicotinic acid). E. coli strains were grown in LB at 37°C. When appropriate, antibiotics were used at the following concentrations: nalidixic acid, 20 μg ml−1; kanamycin, 50 μg ml−1; carbenicillin, 100 μg ml−1; and chloramphenicol, 25 μg ml−1. Bacterial suspensions of approximately 108 CFU/ml were prepared by resuspending the indicated strains in 10 mM MgCl2 at a specific optical density (OD) at 600 nm. At this wavelength, an OD of 0.1 for wild-type and hrpA3 deletion strains or 0.09 for Δefp, ΔepmB, and ΔepmA strains corresponded to approximately 108 CFU/ml.
General DNA manipulations.
Standard molecular biology techniques were used throughout (44). All PCRs were performed using Q5 polymerase (New England Biolabs) and products were initially cloned using the TOPO-TA kit (Invitrogen). Plasmids and oligonucleotides used in this study are listed in Tables S2 and S3 in the supplemental material, respectively.
Creation of deletion and complementation strains.
Deletions of indicated genes were generated using overlap extension PCR to generate segments of DNA homologous of DNA upstream and downstream of the target gene, without the open reading frame present. The resulting products were cloned into the suicide plasmid pDS132 (45). These plasmids were transferred into the nalidixic acid-resistant wild-type strain through biparental mating with E. coli SM10λpir. Initial recombination events were selected by plating onto chloramphenicol and nalidixic acid, and secondary recombination events were selected by plating onto 5% sucrose. Successful deletions were selected for the loss of the gene by PCR amplification of the cloned area. Complementation plasmids were produced as described previously (21) and are listed in Table S2.
Isolation of spontaneous suppressor strains.
Suppressor strains were selected based on colony size. To ensure that these suppressor strains were not simply clones, multiple independent efp deletions were created and used as separate parent strains. The background genotypes of all suppressor mutations were confirmed by PCR.
Quantification of colony size.
Images of 100-mm-diameter plates were taken 72 hours after plating approximately 50 CFU of each strain. Ten colonies from each strain were selected at random and their areas were quantified using ImageJ (46). This experiment was repeated at least four times.
Genome sequencing and SNP mapping.
Full genomes of the wild-type HKN06P1, Δefp, six independently derived Δefp suppressor lines, and one ΔepmB suppressor line were sequenced on an Illumina MiSeq instrument using 250 × 250 paired-end sequencing at the Penn State Genomics Core Facility at University Park. The DNA libraries were constructed at the Penn State Genomics Core Facilities using the TruSeq DNA PCR-free kit. Results were analyzed using Geneious 11.0.2 (https://www.geneious.com). The genome of Δefp was aligned to the wild type, and suppressor genomes were subsequently aligned to Δefp using the Geneious mapper. Variations were identified both inside and outside coding sequences, with a minimum variant frequency of 0.25 and a maximum variant P value of 10−6 and a minimum strand bias P value of 10−5 when exceeding 65% bias. Only putative single nucleotide polymorphisms (SNPs), insertions, and deletions with coverage of over 50×, variant frequency over 95%, and a variant P value of less than 0.001 were considered for further analysis.
Amylovoran production and motility assays.
Amylovoran was quantified using cetylpyridinium chloride as described in reference 32. Motility assays were conducted as described previously (21) on M9 minimal medium plates with 0.3% agar. Motility was quantified as the visible diameter of bacterial spread after 48 hours using ImageJ (21). All experiments were performed at least three separate times.
Disease assays.
Growth in immature ‘Gala’ apple fruitlets was quantified as described in (31). Briefly, bacterial suspensions of 2 × 106 CFU/ml in MgCl2 were added into apples, and CFU/g was determined by serial dilution and plating of macerated apples on the indicated days. This experiment was performed at least 3 times. To determine strain virulence on apple trees, greenhouse-grown second-leaf ‘Gala’ apple trees on Malling.9 rootstocks were inoculated as previously described (31). The first emerging leaves were cut with scissors dipped in bacterial suspensions of 1 × 108 CFU/ml in MgCl2. Twenty shoots were inoculated per strain, on approximately five separate trees. The length of the shoot showing visible symptoms was recorded every week and was expressed as a percentage of the total length of the shoot. The entire experiment was performed twice.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by a Penn State College of Agricultural Sciences student competitive grant to S.M.K.; a grant from the State Horticultural Association of Pennsylvania to T.W.M. and K.A.P.; a United States Department of Agriculture National Institute of Food and Agriculture (USDA NIFA) predoctoral fellowship grant to S.M.K. (grant no. 2017-67011-26030); a USDA NIFA Research and Extension Experiences for Undergraduates grant to T.W.M. (grant no. 2016-67032-25007), under which A.C.H. was awarded a fellowship; and a USDA NIFA Hatch Appropriations grant under project no. PEN04649 and accession no. 1016093.
The funders played no role in the study design, data collection, data interpretation, or the decision to submit the study for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00722-18.
REFERENCES
- 1.Buskirk AR, Green R. 2017. Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos Trans R Soc B 372:20160183. doi: 10.1098/rstb.2016.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yanofsky C. 1981. Attenuation in the control of expression of bacterial operons. Nature 289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- 3.Glick BR, Ganoza MC. 1975. Identification of a soluble protein that stimulates peptide bond synthesis. Proc Natl Acad Sci U S A 72:4257–4260. doi: 10.1073/pnas.72.11.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glick BR, Green RM, Ganoza MC. 1979. Purification of factor EF-P, a protein that stimulates peptide-bond synthesis with certain aminoacyl-tRNA analogues. Can J Biochem 57:749–757. doi: 10.1139/o79-093.383236 [DOI] [Google Scholar]
- 5.Saini P, Eyler DE, Green R, Dever TE. 2009. Hypusine-containing protein eIF5A promotes translation elongation. Nature 459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tollerson R II, Witzky A, Ibba M. 2017. Elongation factor P interactions with the ribosome are independent of pausing. mBio 8:e01056-17. doi: 10.1128/mBio.01056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. 2013. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 8.Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. 2013. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 9.Woolstenhulme CJ, Guydosh NR, Green R, Buskirk AR. 2015. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep 11:13–21. doi: 10.1016/j.celrep.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peil L, Starosta AL, Lassak J, Atkinson GC, Virumäe K, Spitzer M, Tenson T, Jung K, Remme J, Wilson DN. 2013. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc Natl Acad Sci U S A 110:15265–15270. doi: 10.1073/pnas.1310642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hersch SJ, Wang M, Zou SB, Moon K-M, Foster LJ, Ibba M, Navarre WW. 2013. Divergent protein motifs direct elongation factor P-mediated translational regulation in Salmonella enterica and Escherichia coli. mBio 4:e00180-13. doi: 10.1128/mBio.00180-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starosta AL, Lassak J, Peil L, Atkinson GC, Woolstenhulme CJ, Virumäe K, Buskirk A, Tenson T, Remme J, Jung K, Wilson DN. 2014. A conserved proline triplet in val-tRNA synthetase and the origin of elongation factor P. Cell Rep 9:476–483. doi: 10.1016/j.celrep.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hersch SJ, Elgamal S, Katz A, Ibba M, Navarre WW. 2014. Translation initiation rate determines the impact of ribosome stalling on bacterial protein synthesis. J Biol Chem 289:28160–28171. doi: 10.1074/jbc.M114.593277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park J-H, Johansson HE, Aoki H, Huang BX, Kim H-Y, Ganoza MC, Park MH. 2012. Post-translational modification by β-lysylation is required for activity of Escherichia coli elongation factor P (EF-P). J Biol Chem 287:2579–2590. doi: 10.1074/jbc.M111.309633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witzky A, Hummels KR, Tollerson R II, Rajkovic A, Jones LA, Kearns DB, Ibba M. 2018. EF-P posttranslational modification has variable impact on polyproline translation in Bacillus subtilis. mBio 9:e00306-18. doi: 10.1128/mBio.00306-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lassak J, Keilhauer EC, Fürst M, Wuichet K, Gödeke J, Starosta AL, Chen J-M, Søgaard-Andersen L, Rohr J, Wilson DN, Häussler S, Mann M, Jung K. 2015. Arginine-rhamnosylation as new strategy to activate translation elongation factor P. Nat Chem Biol 11:266–270. doi: 10.1038/nchembio.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navarre WW, Zou SB, Roy H, Xie JL, Savchenko A, Singer A, Edvokimova E, Prost LR, Kumar R, Ibba M, Fang FC. 2010. PoxA, YjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol Cell 39:209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marman HE, Mey AR, Payne SM. 2014. Elongation factor P and modifying enzyme PoxA are necessary for virulence of Shigella flexneri. Infect Immun 82:3612–3621. doi: 10.1128/IAI.01532-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng WT, Banta LM, Charles TC, Nester EW. 2001. The chvH locus of Agrobacterium encodes a homologue of an elongation factor involved in protein synthesis. J Bacteriol 183:36–45. doi: 10.1128/JB.183.1.36-45.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou SB, Hersch SJ, Roy H, Wiggers JB, Leung AS, Buranyi S, Xie JL, Dare K, Ibba M, Navarre WW. 2012. Loss of elongation factor P disrupts bacterial outer membrane integrity. J Bacteriol 194:413–425. doi: 10.1128/JB.05864-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klee SM, Mostafa I, Chen S, Dufresne C, Lehman BL, Sinn JP, Peter KA, McNellis TW. 2018. An Erwinia amylovora yjeK mutant exhibits reduced virulence, increased chemical sensitivity and numerous environmentally dependent proteomic alterations. Mol Plant Pathol 19:1667–1678. doi: 10.1111/mpp.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson SV. 1986. The role of the stigma in fire blight infections. Phytopathology 76:476–482. doi: 10.1094/Phyto-76-476. [DOI] [Google Scholar]
- 23.Russo NL, Burr TJ, Breth DI, Aldwinckle HS. 2008. Isolation of streptomycin-resistant isolates of Erwinia amylovora in New York. Plant Dis 92:714–718. doi: 10.1094/PDIS-92-5-0714. [DOI] [PubMed] [Google Scholar]
- 24.Bauer DW, Beer SV. 1991. Further characterization of an hrp gene cluster of Erwinia amylovora. Mol Plant Microbe Interact 4:493–499. doi: 10.1094/MPMI-4-493. [DOI] [Google Scholar]
- 25.Bellemann P, Geider K. 1992. Localization of transposon insertions in pathogenicity mutants of Erwinia amylovora and their biochemical characterization. J Gen Microbiol 138:931–940. doi: 10.1099/00221287-138-5-931. [DOI] [PubMed] [Google Scholar]
- 26.Moriya H, Kasai H, Isono K. 1995. Cloning and characterization of the hrpA gene in the terC region of Escherichia coli that is highly similar to the DEAH family RNA helicase genes of Saccharomyces cerevisiae. Nucleic Acids Res 23:595–598. doi: 10.1093/nar/23.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cordin O, Banroques J, Tanner NK, Linder P. 2006. The DEAD-box protein family of RNA helicases. Gene 367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 28.Tanner NK, Linder P. 2001. DExDH box RNA helicases: from generic motors to specific dissociation functions. Mol Cell 8:251–262. doi: 10.1016/S1097-2765(01)00329-X. [DOI] [PubMed] [Google Scholar]
- 29.Salman-Dilgimen A, Hardy P-O, Dresser AR, Chaconas G. 2011. HrpA, a DEAH-Box RNA helicase, is involved in global gene regulation in the Lyme disease spirochete. PLoS One 6:e22168. doi: 10.1371/journal.pone.0022168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koo JT, Choe J, Moseley SL. 2004. HrpA, a DEAH-box RNA helicase, is involved in mRNA processing of a fimbrial operon in Escherichia coli. Mol Microbiol 52:1813–1826. doi: 10.1111/j.1365-2958.2004.04099.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee SA, Ngugi HK, Halbrendt NO, O'Keefe G, Lehman B, Travis JW, Sinn JP, McNellis TW. 2010. Virulence characteristics accounting for fire blight disease severity in apple trees and seedlings. Phytopathology 100:539–550. doi: 10.1094/PHYTO-100-6-0539. [DOI] [PubMed] [Google Scholar]
- 32.Halupecki E, Bazzi C, Jock S, Geider K, Đermić D, Cvjetković B. 2006. Characterization of Erwinia amylovora strains from Croatia. Eur J Plant Pathol 114:435–440. doi: 10.1007/s10658-006-0003-7. [DOI] [Google Scholar]
- 33.Bellemann P, Bereswill S, Berger S, Geider K. 1994. Visualization of capsule formation by Erwinia amylovora and assays to determine amylovoran synthesis. Int J Biol Macromol 16:290–296. doi: 10.1016/0141-8130(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 34.Butland G, Peregrín-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 35.Redder P, Hausmann S, Khemici V, Yasrebi H, Linder P. 2015. Bacterial versatility requires DEAD-box RNA helicases. FEMS Microbiol Rev 39:392–412. doi: 10.1093/femsre/fuv011. [DOI] [PubMed] [Google Scholar]
- 36.Salman-Dilgimen A, Hardy P-O, Radolf JD, Caimano MJ, Chaconas G. 2013. HrpA, an RNA helicase involved in RNA processing, is required for mouse infectivity and tick transmission of the Lyme disease spirochete. PLoS Pathog 9:e1003841. doi: 10.1371/journal.ppat.1003841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Granato LM, Picchi CS, Andrade MO, Takita MA, de Souza AA, Wang N, Machado MA. 2016. The ATP-dependent RNA helicase HrpB plays an important role in motility and biofilm formation in Xanthomonas citri subsp. citri. BMC Microbiol 16:55. doi: 10.1186/s12866-016-0655-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Xu X, Lan R, Xiong Y, Ye C, Ren Z, Liu L, Zhao A, Wu L-F, Xu J. 2013. An O island 172 encoded RNA helicase regulates the motility of Escherichia coli O157:H7. PLoS One 8:e64211. doi: 10.1371/journal.pone.0064211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sunohara T, Jojima K, Yamamoto Y, Inada T, Aiba H. 2004. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA 10:378–386. doi: 10.1261/rna.5169404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sunohara T, Jojima K, Tagami H, Inada T, Aiba H. 2004. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J Biol Chem 279:15368–15375. doi: 10.1074/jbc.M312805200. [DOI] [PubMed] [Google Scholar]
- 41.Carabetta VJ, Silhavy TJ, Cristea IM. 2010. The response regulator SprE (RssB) is required for maintaining poly(A) polymerase I-degradosome association during stationary phase. J Bacteriol 192:3713–3721. doi: 10.1128/JB.00300-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elgamal S, Artsimovitch I, Ibba M. 2016. Maintenance of transcription-translation coupling by elongation factor P. mBio 7:e01373-16. doi: 10.1128/mBio.01373-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bugert P, Geider K. 1995. Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora. Mol Microbiol 15:917–933. doi: 10.1111/j.1365-2958.1995.tb02361.x. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 45.Philippe N, Alcaraz J-P, Coursange E, Geiselmann J, Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255. doi: 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 46.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.