The metabolic pathway of uric acid degradation to date has been elucidated only in aerobic environments and is not understood in anaerobic and microaerobic environments. In the current study, we showed that Escherichia coli, a facultative anaerobic organism, uses uric acid as a sole source of nitrogen under anaerobic and microaerobic conditions. We also showed that formate, formate dehydrogenase H, and either AegA or YgfT are involved in uric acid degradation. We propose that formate may act as an electron donor for a uric acid-degrading enzyme in this bacterium.
KEYWORDS: degradation pathway, Escherichia coli, formate, uric acid
ABSTRACT
Purine is a nitrogen-containing compound that is abundant in nature. In organisms that utilize purine as a nitrogen source, purine is converted to uric acid, which is then converted to allantoin. Allantoin is then converted to ammonia. In Escherichia coli, neither urate-degrading activity nor a gene encoding an enzyme homologous to the known urate-degrading enzymes had previously been found. Here, we demonstrate urate-degrading activity in E. coli. We first identified aegA as an E. coli gene involved in oxidative stress tolerance. An examination of gene expression revealed that both aegA and its paralog ygfT are expressed under both microaerobic and anaerobic conditions. The ygfT gene is localized within a chromosomal gene cluster presumably involved in purine catabolism. Accordingly, the expression of ygfT increased in the presence of exogenous uric acid, suggesting that ygfT is involved in urate degradation. Examination of the change of uric acid levels in the growth medium with time revealed urate-degrading activity under microaerobic and anaerobic conditions in the wild-type strain but not in the aegA ygfT double-deletion mutant. Furthermore, AegA- and YgfT-dependent urate-degrading activity was detected only in the presence of formate and formate dehydrogenase H. Collectively, these observations indicate the presence of urate-degrading activity in E. coli that is operational under microaerobic and anaerobic conditions. The activity requires formate, formate dehydrogenase H, and either aegA or ygfT. We also identified other putative genes which are involved not only in formate-dependent but also in formate-independent urate degradation and may function in the regulation or cofactor synthesis in purine catabolism.
IMPORTANCE The metabolic pathway of uric acid degradation to date has been elucidated only in aerobic environments and is not understood in anaerobic and microaerobic environments. In the current study, we showed that Escherichia coli, a facultative anaerobic organism, uses uric acid as a sole source of nitrogen under anaerobic and microaerobic conditions. We also showed that formate, formate dehydrogenase H, and either AegA or YgfT are involved in uric acid degradation. We propose that formate may act as an electron donor for a uric acid-degrading enzyme in this bacterium.
INTRODUCTION
Purine exists in all organisms, mainly as a component of adenine and guanine. Degradation of purine yields uric acid, which is used as a nitrogen source by many aerobic organisms. Many animals, plants, fungi, yeasts, and aerobic bacteria, such as Bacillus subtilis and Klebsiella pneumoniae, harbor oxygen-dependent catabolic pathways for the degradation of uric acid to allantoin and that of allantoin to ammonia (1, 2).
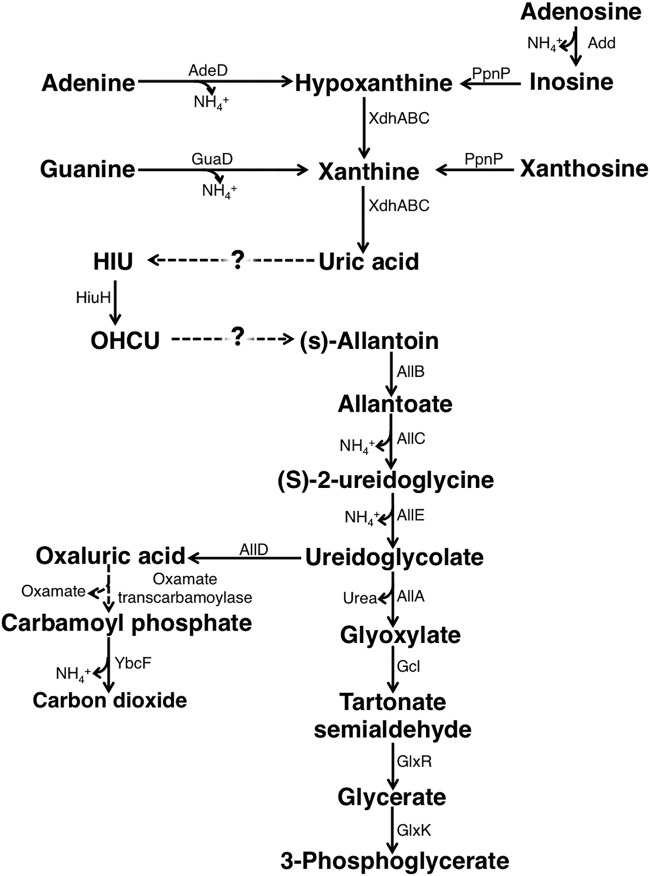
The Escherichia coli genome harbors some of the genes required for purine catabolism and allantoin degradation (Fig. 1) (3, 4). In fact, E. coli converts purine derivatives, such as adenine, guanine, xanthine, and hypoxanthine, into uric acid and can use allantoin as the sole nitrogen source (5). Therefore, although the bacterium can use adenine and adenosine as the sole nitrogen sources because it releases ammonia during deamination of adenine and adenosine to inosine and xanthosine, respectively, it has been thought that E. coli cannot use this pathway to utilize other purines (3). However, several studies have suggested that E. coli can indeed utilize other purines and that it can convert uric acid, the final product of purine metabolism, to allantoin. First, inosine or xanthosine promotes the growth of E. coli under nitrogen-limited conditions (4). Based on this observation, the bacterium may be able to catabolize inosine and xanthosine to allantoin. In addition, in an experiment involving 14C-labeled adenine as a nitrogen source, E. coli produced 14CO2, which suggested that adenine was converted to allantoin (4). Further, high-affinity urate transporter UacT, which exhibits high affinity for uric acid and not for the other purines, was identified in E. coli, suggesting the possibility of uric acid utilization (6). Based on these observations, it is possible that a new pathway for uric acid utilization exists in E. coli.
FIG 1.
Purine degradation pathway in E. coli. Add, AdeD, GuaD, XdhABC, and HiuH, the allantoin degradation enzymes, were described previously (3, 40, 47–50). E. coli genes responsible for the conversion of uric acid to HIU or for the conversion of OHSU to (S)-allantoin have not been identified to date.
The reaction from uric acid to allantoin proceeds via three steps. The first step involves the production of 5-hydroxyisouric acid (HIU) by the hydroxylation of C-5 of uric acid. The second step involves the hydrolysis of HIU to 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU). The third step involves the formation of allantoin by decarboxylation of OHCU (7).
Since the conversion of HIU to OHCU and that of OHCU to allantoin occur spontaneously, the most important step of the conversion of uric acid to allantoin is the first step of uric acid degradation (8). Three types of urate-hydroxylating enzymes have been identified to date. The first type is uric acid oxidase, found in bacteria, archaea, and eukaryotes; this enzyme does not require a cofactor (9). This type of urate oxidase requires molecular oxygen as an electron acceptor in a reaction that generates one molecule of hydrogen peroxide per one molecule of uric acid hydroxylated. The second type of urate-hydroxylating enzyme is flavin adenine dinucleotide (FAD)-dependent uric hydroxylase. It is represented by two isoenzymes that share low sequence identity. One isoenzyme is HpxO, first identified in K. pneumoniae (10), and the other is HpyO, first identified in Acinetobacter baylyi (11). These FAD-dependent uric hydroxylases incorporate hydroxyl groups generated by the reduction of dioxygen with uric acid. The third type of urate-hydroxylating enzyme is membrane cytochrome c urate oxidase, PuuD, found in Agrobacterium fabrum (12). Although enzymatic activity of a purified PuuD has not yet been confirmed, PuuD is thought to mediate the electron transfer from uric acid to oxygen, with the generated hydrogen peroxide removed by the cytochrome c domain of PuuD (12). All types of urate-hydroxylating enzymes require stoichiometric amounts of molecular oxygen to hydroxylate uric acid, and there has been no evidence of uric acid degradation outside aerobic environments (9–12).
In the current study, we identified aegA as a gene involved in menadione sensitivity of previously constructed reduced-genome strains of E. coli (13). Menadione is an artificial electron carrier, which is reduced and oxidized by electron-transferring metabolic enzymes, such as diaphorases (14). Because reduced menadione is oxidized by oxygen and produces reactive oxygen species, menadione is used to examine cell sensitivity to oxidative stress (15). We investigated the functions of AegA and its paralog YgfT and found that these proteins are involved in uric acid degradation, and that E. coli can utilize uric acid as a nitrogen source under microaerobic and aerobic conditions.
RESULTS
Identification of AegA and YgfT.
Previously, we constructed a series of reduced-genome strains of E. coli (Δ1a to Δ23a and Δ25a to Δ33a mutant strains) that lacked 38.9% of the parental chromosome by combining large chromosomal deletions, and we examined the sensitivity of those strains to menadione during stationary phase (13, 16). We found that the Δ22a mutant strain was more sensitive to menadione than the Δ21a mutant strain (13). The Δ22a mutant strain was constructed by generating a large-scale chromosomal deletion, LD3-4-1 (36,841 bp), in the Δ21a mutant strain, which suggested that a gene(s) involved in menadione sensitivity was present in the deleted region. The Δ21a mutant strain became sensitive to menadione upon deletion of the aegA gene (see Fig. S1 in the supplemental material). The menadione sensitivity was complemented by a single-copy mini-F plasmid harboring aegA, although the Δ21a and Δ21a ΔaegA mutant strains became even more sensitive after introduction of a mini-F vector plasmid (Fig. S1). These results suggested that aegA is involved in menadione sensitivity. However, deletion of aegA in the wild-type strain did not result in menadione sensitivity, suggesting that multiple mutations are required for the sensitivity (Fig. S1). We did not identify another responsible gene(s) and did not clarify the mechanism of the observed menadione sensitivity in the current study. Instead, we focused on the aegA gene.
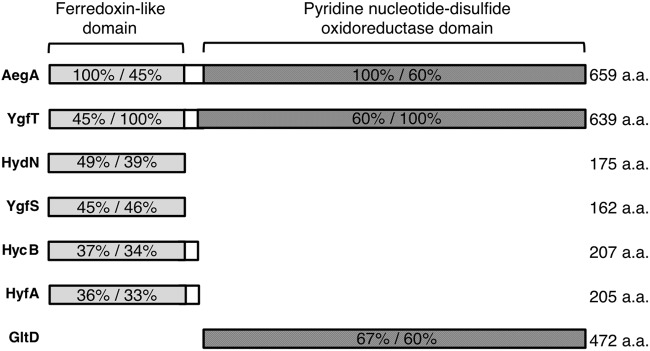
Although aegA (anaerobically expressed gene A) is reportedly expressed under anaerobic conditions (17), its biological function had not previously been elucidated. AegA is a putative oxidoreductase and has an N-terminal 4Fe-4S dicluster domain, often found within the bacterial ferredoxin, and a C-terminal pyridine nucleotide-disulfide oxidoreductase domain, which shows high similarity with the E. coli glutamate synthase β-subunit GltD (18) (Fig. 2). A BLAST search revealed that E. coli harbors an aegA paralog, ygfT (19, 20). Similar to AegA, YgfT is a putative oxidoreductase and has an N-terminal 4Fe-4S dicluster domain and a C-terminal pyridine nucleotide-disulfide oxidoreductase domain (Fig. 2). The biological function of YgfT has not been clarified. A possible σ54-dependent promoter found in promoter regions of various nitrogen-related genes (21) is located upstream of ygfT. Computational analysis suggested that ygfT and uacT (ygfU), a gene upstream of ygfT, are divergently transcribed (4).
FIG 2.
Domain organization of AegA and YgfT. The N-terminal 4Fe-4S dicluster domains of AegA and YgfT share high identity with HydN, YgfS, HycB, and HyfA. The C-terminal pyridine nucleotide-disulfide oxidoreductase domains of AegA and YgfT share high identity with the β-subunit of glutamate synthase GltD. The percent identity shared by each protein with AegA and YgfT is shown. a.a., amino acids.
Expression of aegA and ygfT.
To clarify the function(s) of aegA and ygfT, we examined the expression of these genes in E. coli strains harboring aegA-lacZ and ygfT-lacZ constructs generated by fusing the regions upstream of aegA and ygfT with lacZ, accordingly. Because these strains were constructed by inserting the upstream regions of aegA and ygfT into the chromosomal lacZ upstream region, the chromosomal aegA and ygfT loci were not disrupted and the metabolism and regulation were not affected. Using these transcriptional fusions, we examined the expression of aegA and ygfT under different conditions by measuring β-galactosidase activity, as described in Materials and Methods.
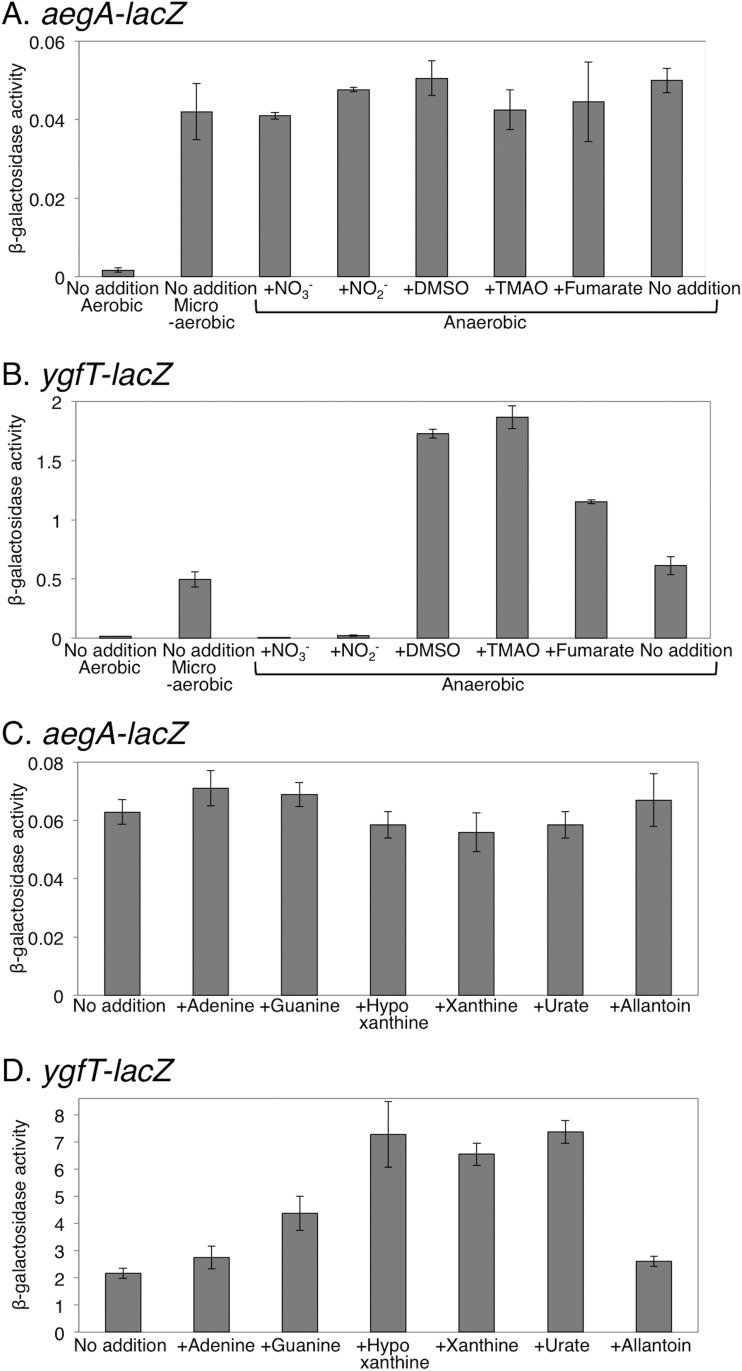
Previous work already showed that the aegA expression is induced under anaerobic conditions (17). It was also reported that the expression of some enzymes that contain ferredoxin-like domains, which are found in the N-terminal parts of AegA and YgfT, is induced in a specific environment with a specific oxidation-reduction potential (22, 23). Therefore, we first examined the effect of oxygen availability and of the other electron acceptors on the expression of aegA and ygfT. Under aerobic conditions, the expression of aegA was very low; in contrast, under microaerobic or anaerobic conditions, the expression of aegA was approximately 30 times higher than that under aerobic conditions (Fig. 3A). It was also shown that aegA expression was repressed by the addition of nitrate in medium (17). We showed that the expression of aegA was not reduced by the addition of respiratory electron acceptors, including nitrate, under anaerobic conditions (Fig. 3A). Our aegA-lacZ transcriptional fusion contained the upstream region 631 bp from the aegA start codon, while the aegA-lacZ fusion used in the previous work contained the upstream region 237 bp from the aegA start codon and the first 90 codons of aegA (17). The repression of expression in the presence of nitrate may be ascribed to the first 90 codons of aegA and may be a posttranscriptional regulation.
FIG 3.
Expression of aegA and ygfT examined under microaerobic and anaerobic conditions. (A and B) The effect of various respiratory electron acceptors on the expression of aegA (A) and ygfT (B) genes was examined. (C and D) The effect of various purine degradation intermediates on the expression of aegA (C) and ygfT (D) genes was examined under anaerobic conditions. β-Galactosidase activity is presented as units per milligram of protein.
Similarly to aegA, we examined the ygfT expression using a strain harboring ygfT-lacZ constructs. The ygfT-lacZ transcriptional fusion contained the upstream region 720 bp from the ygfT start codon, incorporating the entire intergenic region between ygfT and uacT. The expression of ygfT was low under aerobic conditions (Fig. 3B). Under microaerobic and anaerobic conditions, ygfT expression was approximately 30 and 40 times higher than that under aerobic conditions, respectively. Unlike the expression of aegA in the presence of a respiratory electron acceptor other than oxygen under anaerobic conditions, the expression of ygfT was reduced by approximately 120-fold and 28-fold in the presence of nitrates and nitrites, respectively (Fig. 3B). Conversely, the addition of dimethyl sulfoxide (DMSO), trimethylamine N-oxide (TMAO), or fumaric acid resulted in approximately 2.8-fold, 3-fold, and 1.9-fold enhancements of ygfT expression, respectively (Fig. 3B). These observations suggested that the expression of ygfT increased in an environment with a low oxidation-reduction potential and decreased in an environment with a high oxidation-reduction potential.
The ygfT gene is located in a chromosomal gene cluster presumably involved in purine catabolism (4), and uacT, encoding a urate transporter, is located upstream of ygfT, in divergent orientation. We examined the effect of purines and allantoin on the expression of aegA and ygfT. No significant change in the aegA expression was observed in the presence of purine or allantoin (Fig. 3C), while the expression of ygfT increased approximately 3.5 times in the presence of hypoxanthine, xanthine, and uric acid (Fig. 3D). However, no significant difference in ygfT expression was observed in the presence of allantoin (Fig. 3C). These observations suggested that ygfT is involved in uric acid metabolism.
Cellular uric acid degradation activity.
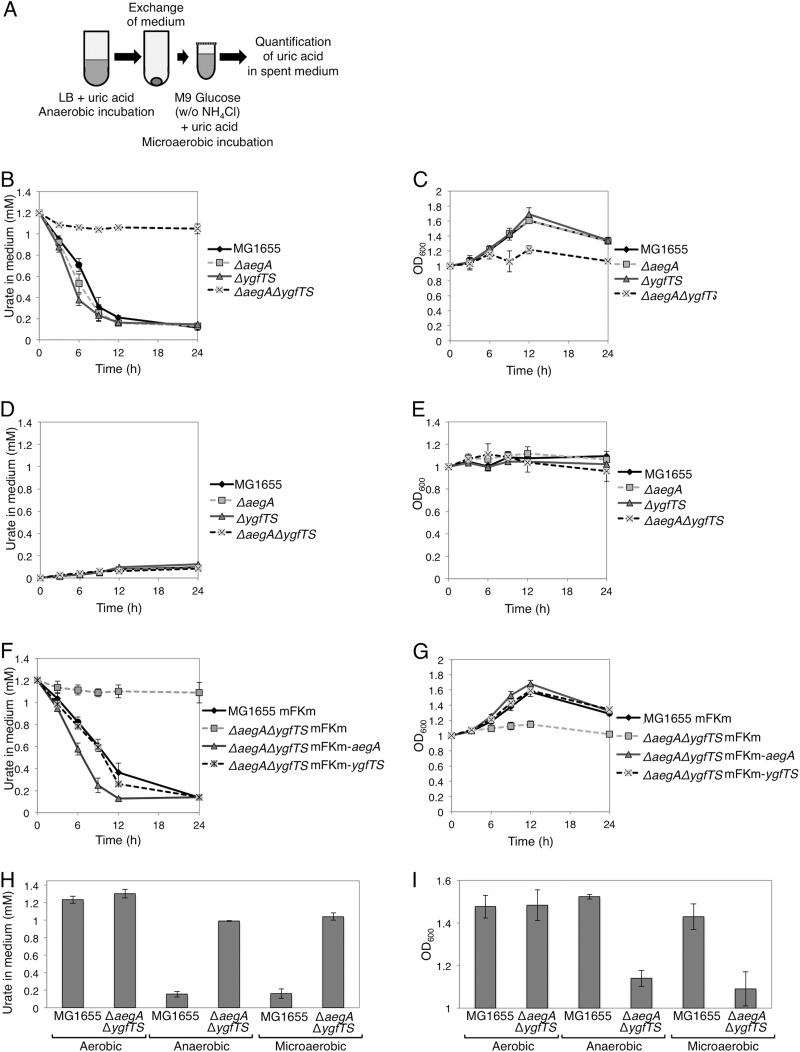
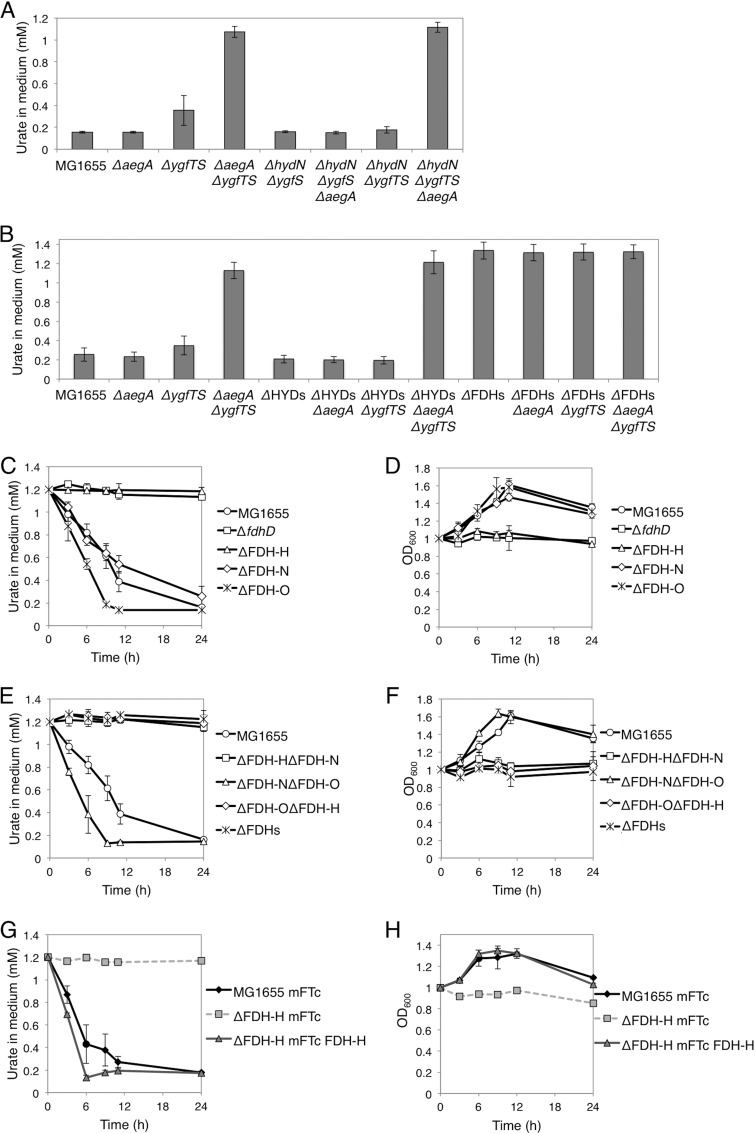
To investigate the possibility that ygfT is involved in the metabolism of uric acid, the amount of uric acid in the growth medium was determined for cells cultured in the presence of uric acid as the sole nitrogen source. A ygfT disruptant was constructed by deleting both ygfT and ygfS genes because ygfT overlaps ygfS by one nucleotide. The gene ygfS encodes a ferredoxin-like protein of unknown function. For the experiment, the wild-type strain and strains lacking aegA and/or ygfTS were anaerobically grown to the stationary phase in LB medium supplemented with uric acid. The expression of aegA and ygfT decreased when the cells were grown anaerobically in the M9 glucose medium or LB glucose medium. Hence, we used LB medium for the primary culture to increase the expression of aegA and ygfT. Then, the stationary-phase culture cells were harvested and washed. The cells were suspended (optical density at 600 nm [OD600], 1.0) in a minimal medium supplemented with glucose and uric acid as the nitrogen source and incubated under microaerobic conditions. The amount of uric acid was then determined (Fig. 4A). The uric acid degradation activity was apparent only in cells that had been grown in LB medium, washed, and incubated in a minimal medium under microaerobic conditions. Neither aegA- and ygfT-dependent uric acid degradation nor uric acid-dependent growth was observed when a small inoculum was used (OD600, 0.1 at the start of the experiment). This could be explained by a greater amount of dissolved oxygen in cultures when a small inoculum was used (OD600, 0.1) than in cultures when a large inoculum was used (OD600, 1.0). For the wild-type strain and ΔaegA and ΔygfTS mutants, the concentration of uric acid in the medium gradually decreased (Fig. 4B). No pronounced change in uric acid levels was apparent in the ΔaegA ΔygfTS mutant culture. The ΔaegA ΔygfTS mutant did not grow under these conditions, as shown in Fig. 4C, in contrast to the wild-type strain and ΔaegA and ΔygfTS mutants. Neither of the tested strains grew in a medium that had not been supplemented with uric acid, which indicated that bacterial growth was dependent on uric acid (Fig. 4D and E).
FIG 4.
Cellular uric acid degradation activity. (A) The procedure for determining uric acid degradation activity and cell growth. (B and C) Uric acid degradation activity (B) and cell growth (C) of the wild type and ΔaegA, ΔygfTS, and ΔaegA ΔygfTS mutant strains in minimal medium supplemented with uric acid as the sole nitrogen source. (D and E) Uric acid degradation activity (D) and cell growth (E) in the absence of uric acid. (F and G) Introduction of plasmid-borne aegA and ygfTS genes restores the uric acid degradation activity (F) and cell growth (G) with uric acid as the sole nitrogen source. (H and I) The amount of uric acid (H) and cell density (I) after a 24-h incubation under aerobic, microaerobic, and anaerobic conditions with uric acid as the sole nitrogen source. The cellular uric acid degradation activity was monitored by measuring the OD291 of the culture supernatant.
To confirm that the aegA and ygfTS genes were responsible for the urate-degrading activity and uric acid-dependent cell proliferation, a single-copy mini-F plasmid harboring aegA or ygfTS (mFKm-aegA or mFKm-ygfTS, respectively) was introduced into the ΔaegA ΔygfTS mutant. Urate-degrading activity and cell proliferation were observed in strains harboring the mFKm-aegA or mFKm-ygfTS plasmid but not in strains harboring the empty plasmid vector (mFKm) (Fig. 4F and G). These observations suggested that either aegA or ygfTS was required for uric acid degradation and uric acid-dependent cell proliferation under microaerobic conditions and that their functions overlap although they function independently.
To confirm that AegA is involved in urate degradation, we introduced several constructs harboring a mutated aegA gene into the ΔygfT mutant strain. Each construct harbored a missense mutation in codons corresponding to conserved amino acid residues, with three in the N-terminal ferredoxin-like domain and four in the C-terminal pyridine nucleotide-disulfide oxidoreductase domain (Fig. S2A). As shown in Fig. S2B, the seven mutants were deficient in urate-degrading activity. These observations confirmed that AegA is involved in the urate-degrading activity and indicated that the tested residues were essential for this activity.
Next, we investigated the uric acid degradation activity and growth of the wild-type and ΔaegA ΔygfTS strains cultured under aerobic, microaerobic, and anaerobic conditions. As shown in Fig. 4H and I, when either of the strains was grown under aerobic conditions, the amount of uric acid in the medium did not decrease; however, when the wild-type strain was cultured under microaerobic and anaerobic conditions, the amount of uric acid in the medium decreased, and cell proliferation was detected. These observations revealed that uric acid degradation and uric acid-dependent cell proliferation involving AegA and YgfT occur under both microaerobic and anaerobic conditions.
Involvement of formate dehydrogenases in uric acid degradation.
The N-terminal domains of AegA and YgfT are ferredoxin-like domains and share high amino acid similarity with ferredoxin-like proteins of unknown function, HydN and YgfS (Fig. 2). HydN is a ferredoxin-like protein and a putative electron transfer protein (24). YgfS, which is also a ferredoxin-like protein and a putative electron transfer protein, has not been experimentally characterized. We constructed hydN and ygfS deletion mutants and introduced the hydN ygfS double-deletion mutation into the wild-type and ΔaegA strains and the hydN deletion into the ΔygfTS and ΔaegA ΔygfTS mutant strains. The constructed strains were cultured, and the amount of uric acid in the medium was determined. As shown in Fig. 5A, double deletion of the hydN and ygfS genes did not significantly affect the strain phenotypes examined. These observations indicated that HydN and YgfS are not required for AegA- and YgfT-dependent uric acid degradation.
FIG 5.
The effect of ferredoxin-like proteins, FDH, and hydrogenases on uric acid degradation. (A) The effect of hydN and ygfS genes encoding a ferredoxin on uric acid degradation activity. (B) The amount of uric acid in the medium after a 24-h incubation. The ΔFDHs mutant strain lacks all (three) FDHs, and the ΔHYDs mutant strain lacks all (four) hydrogenases. (C and D) Urate levels in the spent medium (C) and cell growth (D) of the wild type, FDH single mutants, and fdhD mutant during a 24-h incubation. (E and F) Urate levels in the spent medium (E) and cell growth (F) of the wild type and FDH double- and triple-deletion mutants during a 24-h incubation. (G and H) The introduction of plasmid-borne fdhF restores the uric acid degradation activity (G) and cell growth (H), with uric acid as the sole nitrogen source.
The N-terminal ferredoxin-like domains of AegA and YgfT also share similarity with HycB and HyfA, which are hydrogenase electron transfer subunits (Fig. 2). Therefore, we investigated the involvement of hydrogenases in uric acid degradation. E. coli harbors four types of the hydrogenase complex, as follows: hydrogenase 1, encoded by hyaABC; hydrogenase 2, encoded by hybABOC; hydrogenase 3, encoded by hycBCDEFG; and hydrogenase 4, encoded by hyfABCDEFGHI. All these hydrogenase complexes are tightly associated with the inner membrane. Further, HycB and HyfA are constituents of hydrogenases 3 and 4, respectively, which reduce protons to hydrogen using electrons donated by formate (25). Therefore, we constructed the ΔHYDs mutant strain that lacked all genes encoding these four types of hydrogenases and ΔHYDs mutant-derived strains lacking either or both aegA and ygfTS. The constructed strains were cultured, and the amount of uric acid in the medium was determined. As shown in Fig. 5B, no significant difference was observed between the ΔHYDs mutant-derived strains and the corresponding hydrogenase-encoding strains. These observations indicated that the hydrogenases were not involved in aegA- and ygfT-dependent uric acid degradation.
N-terminal ferredoxin-like domains of AegA and YgfT also share similarity with formate dehydrogenases (FDHs), FdnH and FdoH (Fig. 2). Hence, we next examined the involvement of FDH in uric acid degradation. E. coli encodes three FDHs: FDH-H, encoded by fdhF; FDH-N, encoded by fdnGHI; and FDH-O, encoded by fdoGHI. Although FDH-N and FDH-O are membrane proteins (26, 27), FDH-H is a soluble protein (28). We constructed an ΔFDHs mutant strain that lacked all genes encoding the three FDHs and ΔFDHs mutant-derived strains that had ΔaegA and/or ΔygfTS mutations. These strains were cultured, and the amount of uric acid in the growth medium was determined. As shown in Fig. 5B, no decrease in the amount of uric acid in the medium was observed for any of the constructed ΔFDHs mutant strains. These findings indicated that at least one of the three FDH enzymes was involved in aegA- and ygfT-dependent uric acid degradation.
Next, we asked which FDH is involved in uric acid degradation. We constructed deletion strains that lacked the three formate dehydrogenases (FDH-H, FDH-N, and FDH-N). As described above, the strains were cultured in the presence of uric acid, and the amount of uric acid in the growth medium and cell growth were evaluated. As shown in Fig. 5C and D, the ΔFDH-H mutant strain lacked urate-degrading activity and did not grow under the tested conditions.
Furthermore, we constructed mutant strains that lacked two of the three FDHs (i.e., encoded only one FDH). We found that only the ΔFDH-N ΔFDH-O mutant strain, in which only FDH-H was present, was able to degrade urate and proliferate under the conditions tested (Fig. 5E and F). To confirm that FDH-H was responsible for the urate-degrading activity and uric acid-dependent cell proliferation, a single-copy mini-F plasmid (mFTc-FDH-H) harboring the fdhF gene encoding FDH-H was introduced into the ΔFDH-H mutant strain, and the phenotype was examined. Indeed, the strain exhibited urate-degrading activity and proliferated upon the introduction of mFTc-FDH-H but not after introduction of the empty plasmid vector (mFTc) (Fig. 5G and H). These observations suggested that FDH-H is required for uric acid degradation and uric acid-dependent cell proliferation.
In addition to formate dehydrogenase activity, FDH has another function, i.e., electron-transferring activity, which is different from formate dehydrogenase activity (29). To investigate whether the formate dehydrogenase activity of FDH-H was required for uric acid degradation, urate-degrading activity was examined in an ΔfdhD mutant strain lacking FdhD, which is essential for formate dehydrogenase activity but not for electron-transferring activity. FdhD is a sulfur transferase which transfers sulfur to molybdo-bis-pyranopterin guanine dinucleotide (Mo-bisPGD), an essential cofactor for formate dehydrogenase activity of FDH (30). As shown in Fig. 5C and D, the ΔfdhD mutant did not exhibit urate-degrading activity and uric acid-dependent cell proliferation. This indicated that formate dehydrogenase activity of FDH-H is indispensable for uric acid degradation.
FDH-H lacks a ferredoxin-like domain, which is found in the N-terminal regions of AegA and YgfT. FDH-H interacts with hydrogenase 3 or hydrogenase 4 to form a formate hydrogenlyase (31, 32). Further, FDH-H transfers electrons to HycB (from hydrogenase 3) and HyfA (from hydrogenase 4) (31). As described above, HycB and HyfA share similarity with the N-terminal domains of AegA and YgfT, suggesting the possibility that FDH-H donates electrons to AegA and YgfT.
Formate is required for fdhF-, aegA-, and ygfT-dependent uric acid degradation.
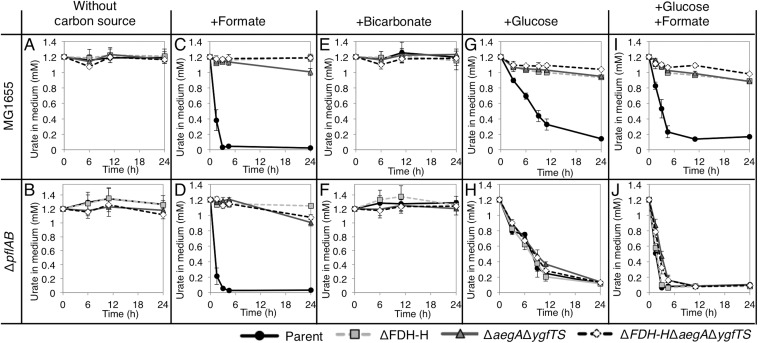
Since FDH-H was required for uric acid degradation, we next investigated whether formate was required for that activity. Under anaerobic and microaerobic conditions, pyruvate, the final product of glycolysis, is converted to formate and acetyl-coenzyme A (acetyl-CoA) by pyruvate-formate lyase, PflB, with the aid of PflA. PflAB produces a large portion of endogenous formate under anaerobic and microaerobic conditions (33). A ΔpflAB mutant, in which PflAB-dependent endogenous production of formate is impaired, was constructed, and its uric acid-degrading activity was examined. Specifically, the pflAB genes were deleted from the wild-type, ΔFDH-H mutant, ΔaegA ΔygfTS mutant, and ΔaegA ΔygfTS ΔFDH-H mutant strains. The constructed strains were cultured anaerobically until stationary phase, and the amount of uric acid in the growth medium and cell growth were examined after further incubation (Fig. 6 and S3). In the absence of a carbon source, no strain exhibited uric acid degradation (Fig. 6A and B) or growth (Fig. S3A and B). In the presence of formate as the carbon source, degradation of exogenous uric acid was observed in the ΔpflAB mutant and the wild-type strain (Fig. 6C and D) but not in the ΔFDH-H, ΔaegA ΔygfTS, or ΔaegA ΔygfTS ΔFDH-H mutant strains. These observations revealed that AegA, YgfT, and FDH-H are essential for uric acid degradation in the presence of formate.
FIG 6.
The effect of various carbon sources on uric acid degradation. (A and B) The amount of uric acid in an unsupplemented minimal medium (without any carbon source) during a 24-h incubation of wild-type derivatives (A) and ΔpflAB derivatives (B). (C and D) The effect of the supplementation of a minimal medium with formate on the amount of uric acid in the medium during a 24-h incubation of wild-type derivatives (C) and ΔpflAB derivatives (D). (E and F) The effect of the supplementation of a minimal medium with bicarbonate on the amount of uric acid in the medium during a 24-h incubation of wild-type derivatives (E) and ΔpflAB derivatives (F). (G and H) The effect of the supplementation of a minimal medium with glucose on the amount of uric acid in the medium during a 24-h incubation of wild-type derivatives (G) and ΔpflAB derivatives (H). (I and J) The effect of the supplementation of a minimal medium with glucose and formate on the amount of uric acid in the medium during a 24-h incubation of wild-type derivatives (I) and ΔpflAB derivatives (J).
Since FDH catalyzes the oxidation of formate to carbon dioxide, we next examined the possibility that the requirement for formate for urate degradation was associated with its ability to supply carbon dioxide to this process. Consequently, the amount of uric acid in the medium was determined when the tested strains were cultured in a medium supplemented with bicarbonate. However, uric acid degradation was not observed, suggesting the possibility that the requirement for formate for urate degradation was not due to its ability to supply carbon dioxide to this process (Fig. 6E and F). Because FDH forms carbon dioxide via an oxidation reaction, in which electrons are extracted from formate, formate could act as an electron donor in AegA-, YgfT-, and FDH-H-dependent uric acid degradation.
When the constructed strains were cultured in a medium supplemented with glucose as the carbon source, uric acid degradation was observed in the wild-type strain culture but not in the ΔFDH-H, ΔaegA ΔygfTS, or ΔaegA ΔygfTS ΔFDH-H mutant strain cultures (Fig. 6G). However, upon deletion of the pflAB genes, abolishing formate production by PflAB, derivatives of the ΔFDH-H, ΔaegA ΔygfTS, and ΔaegA ΔygfTS ΔFDH-H mutant strains were able to degrade exogenous uric acid (Fig. 6G and H). However, uric acid degradation by the ΔpflAB ΔFDH-H, ΔpflAB ΔaegA ΔygfTS, and ΔpflAB ΔaegA ΔygfTS ΔFDH-H mutant strains was also observed when the growth medium was supplemented with formate in addition to glucose (Fig. 6I and J). Therefore, it is unlikely that uric acid degradation results from a simple suppression of intrinsic formate production after removal of the pflAB genes. Further analysis of the AegA-, YgfT-, and FDH-H-independent uric acid degradation in ΔpflAB strains in the presence of glucose is required to clarify the mechanism(s) involved.
Identification of genes required for formate-dependent and formate-independent uric acid degradation.
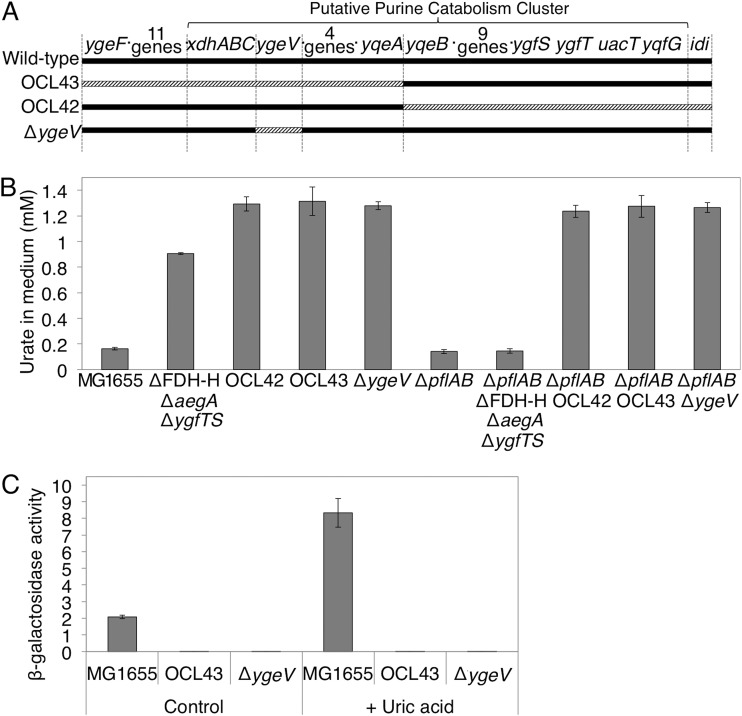
As described above, even in the absence of aegA and ygfT, or of fdhF, uric acid degradation was observed when the ΔpflAB mutant strains were incubated in the presence of glucose, suggesting that AegA, YgfT, and FDH-H were dispensable for uric acid degradation. Therefore, we attempted to identify other chromosomal regions containing genes involved in uric acid degradation. Since ygfT is located in a gene cluster presumably involved in purine catabolism (4), we investigated the possibility that other genes in this gene cluster are involved in uric acid degradation. Specifically, we generated two large-scale chromosome deletions of this gene cluster region, OCL43, which is the deletion of the chromosomal range from yqeF to yqeA, and OCL42, which is the deletion of the chromosomal range from yqeB to idi, in the ΔpflAB strains (Fig. 7A). The resultant strains were cultured in the presence of glucose, and uric acid degradation was examined. Strains with either region deleted did not exhibit urate-degrading activity, suggesting the existence of genes essential for formate-dependent exogenous uric acid degradation in each of the deleted regions (Fig. 7B). Further, ΔpflAB mutants with either of these regions deleted did not exhibit the urate-degrading activity in the presence of glucose (Fig. 7B). This suggested that genes indispensable for formate-independent uric acid degradation exist in each of the removed regions.
FIG 7.
The chromosomal regions, which are required for uric acid degradation. (A) Chromosomal regions of OCL42 and OCL43 large-scale deletion mutants. Solid lines represent the chromosome, and striped lines represent the deleted region in the indicated strain. (B) The effect of OCL43, OCL42, and ygeV deletion on the amount of uric acid in the medium after a 24-h incubation. (C) The effect of OCL43 and ygeV deletion on the expression of ygfT.
Although not all of the putatively involved genes were examined, we investigated one gene in the gene cluster, ygeV, which presumably encodes a transcriptional regulator. Consequently, ygeV deletion mutants were constructed in the wild-type and ΔpflAB backgrounds, and the uric acid degradation ability was examined in the presence of glucose. Since the ygeV gene is monocistronic, the deletion of ygeV inactivated only the ygeV gene. No urate-degrading activity was observed in either strain, indicating that ygeV is essential for both formate-dependent and formate-independent uric acid degradation (Fig. 7B).
Furthermore, the effect of OCL43 and ΔygeV on ygfT expression in the presence of uric acid was examined in the ygfT-lacZ strain. Indeed, both deletions resulted in a reduced expression of ygfT, and no induction of the gene expression by uric acid was observed (Fig. 7C). These findings demonstrated that gene(s) involved in the expression of ygfT is located in the deleted OCL43 region and that ygeV is involved. Since urate-degrading activity was not observed in the ygeV deletion mutant in the ΔpflAB background, ygeV is probably involved in the expression of other genes essential for formate-independent uric acid degradation in E. coli.
Phylogenic distribution of aegA and ygfT genes.
To investigate the conservation of the formate-dependent uric acid degradation pathway, we investigated phylogenic distribution of AegA and YgfT by using the neighbor-joining method in the MEGA 7.0 software (34). As shown in Fig. S4A, AegA and YgfT are encoded by bacteria from 19 genera, all of which belong to the Gram-negative facultative anaerobes from the Enterobacteriaceae family, including human pathogens (e.g., Shigella, Salmonella, Klebsiella, and Enterobacter spp.) and plant pathogens (e.g., Kosakonia, Brenneria, and Pectobacterium spp.). Since the aegA gene is conserved in all E. coli strains whose genomes have been sequenced to date, the function of aegA appears to be important for E. coli (35). Ten species encode both AegA and YgfT, 66 species encode only AegA, and four species encode only YgfT. Genes encoding the urate-degrading enzymes were rarely found in organisms harboring the aegA or ygfT genes, hpxO was identified in only 24 species, and hpyO was identified in only one species (Table S1).
We next examined genomic regions in the vicinity of the aegA and ygfT genes in the organisms analyzed in Fig. S4A. The ygfT, ygfS, fdhF, and uacT genes are often located close to one another (Fig. S4C). The ygfS gene encodes a ferredoxin-like protein that shares high similarity with the N-terminal domain of YgfT, fdhF encodes FDH-H, and uacT encodes a uric acid transporter. The aegA, acrD, nudK, and ypfG genes are also frequently located near each other (Fig. S4B). The acrD gene encodes a multidrug efflux pump, nudK encodes a GDP-mannose hydrolase, and ypfG encodes an uncharacterized DUF1176 (Pfam domain PF06674)-containing protein. During the degradation of colanic acid, the GDP-mannose hydrolase NudK hydrolyzes GDP-mannose to GMP and α-d-mannose-1-phosphate (36). Therefore, AegA may be involved in sweeping the nitrogen from guanine released from GDP-mannose. We noted that fdhF, which encodes FDH-H, was enriched in AegA/YgfT-encoding genomes (Fig. S4B and C). These neighborhood analyses indicated that FDH-H, and AegA or YgfT, were functionally related.
DISCUSSION
In the current study, aegA was identified as a gene involved in menadione resistance of E. coli strains with reduced genomes, and its paralog, ygfT, was identified in a gene cluster presumably involved in purine catabolism. We found that the expression of aegA and ygfT increased under anaerobic and microaerobic conditions, and that uric acid enhanced the expression of ygfT. Hence, uric acid degradation activity involving aegA and ygfT and operational under anaerobic and microaerobic conditions was identified. Although it has been suggested that E. coli might be able to degrade uric acid, the current study is the first to demonstrate such uric acid degradation. Since urate-degrading activity has thus far been only identified under aerobic conditions, and the reaction depends on molecular oxygen, identification of uric acid degradation activity under anaerobic and microaerobic conditions is highly interesting. Further, the requirement for formate and formate dehydrogenase by the AegA- and YgfT-dependent urate degradation suggests that urate degradation proceeds through a reductive pathway. Namely, the first step of the pathway is formate-dependent reduction of urate, which is followed by its hydrolysis. Further enzymatic and chemistry-based analyses of the proposed reductive pathway should reveal intermediates of urate degradation that are different from the intermediates in the known oxidative pathway (i.e., formed by urate oxidase).
Previous studies demonstrated that the N-terminal ferredoxin-like domain of AegA is similar to the HycB subunit of hydrogenase 3 (17) (Fig. 2). Further, the HycB subunit directly interacts with FDH-H and accepts electrons from FDH-H (31). However, the biological significance of the similarity remained unclear. In the current study, we found that AegA and YgfT require FDH-H and formate for uric acid degradation. These results suggest that AegA, YgfT, and HycB accept electrons from FDH-H, and that the similar domains of AegA, YgfT, and HycB are responsible for electron transfer from FDH-H. In addition, a C-terminal pyridine nucleotide-disulfide oxidoreductase domain was identified in AegA and YgfT. It shows a high level of identity with GltD, a small subunit of E. coli glutamate synthase (Fig. 2). Since GltD passes electrons from NADPH to GltB (18), the large subunit of glutamate synthase, we considered the possibility that AegA and YgfT function to reduce the enzymes necessary for uric acid degradation. Since NADPH-binding sites of the C-terminal pyridine nucleotide-disulfide oxidoreductase domain are essential for the urate-degrading activity, AegA may reduce NADP+ to NADPH using electrons from FDH-H. The structure of the C-terminal domain of AegA and YgfT is similar to that of NADP+-dependent ferredoxin oxidoreductase NfnI-L of Pyrococcus furiosus (37). NfnI-L receives electrons from NADPH and reduces two ferredoxins and one NAD+ molecule by coupling energy-producing and energy-absorbing reactions in a so-called bifurcating reaction (38). In particular, since the amino acid residues that are important for bifurcation, located in the vicinity of the FAD-binding site of NfnI-L, are also highly conserved in AegA and YgfT (see Fig. S5 in the supplemental material), it is highly likely that AegA and YgfT are bifurcating enzymes that utilize low-energy electrons from formate (E′ = –420 mV). Collectively, we propose that the electrons from formate flow as follows: first, the N-terminal ferredoxin-like domains of AegA or YgfT accept electrons from the iron-sulfur cluster of FDH-H; subsequently, the FAD cofactor of AegA or YgfT is reduced; and finally, FADH2 reduces NADP+ and the other enzyme at the same (bifurcation) or different (bifunction) time (Fig. S6).
As shown in Fig. 5C to F, of the three FDHs, AegA and YgfT required only FDH-H. While FDH-H is a soluble protein, FDH-N and FDH-O are membrane-bound complexes. AegA and YgfT, which do not have a transmembrane domain, might hence interact with FDH-H. Alternatively, FDH-H can reduce substrates with low redox potential. Previous in vitro experiments showed that FDH-H specifically reduces benzyl viologen (E′ = –359 mV), and FDH-N and FDH-O specifically reduce phenazine methosulfate (E′ = +80 mV) (28, 39). Therefore, if AegA and YgfT have a low redox potential similar to benzyl viologen, FDH-H is the only enzyme that can reduce AegA and YgfT using electrons from formate.
The mechanism by which E. coli degrades uric acid under anaerobic conditions is not known. However, since electrons from FDH-H, and AegA and YgfT, are required for formate-dependent urate degradation, uric acid might first be reduced to dihydrourate. The C-5 of dihydrourate would then be hydroxylated to hydroxyisourate, and hydroxyisourate would be hydrolyzed and decarboxylated to form allantoin. As shown in Fig. 6, the presented data suggest that a formate-independent uric acid degradation pathway exists in E. coli.
We found that the expression of aegA and ygfT was reduced in the presence of oxygen and that the expression of ygfT was reduced in the presence of nitrite and nitrate (Fig. 3A and B). Previously, it was shown that the expression of aegA is upregulated by anaerobiosis and downregulated by nitrate (17). We did not observe any inhibition of aegA expression by nitrate, although the aegA′-lacZ construct in this study contains a longer aegA promoter region than that used in the previous study. Because the aegA′-lacZ construct used in the previous study contains the first 90 codons of aegA while the aegA′-lacZ construct in this study does not, aegA expression may be downregulated by nitrate in a posttranscriptional manner.
Furthermore, we showed that YgeV is involved in uric acid degradation in E. coli. The ygeV gene presumably encodes a σ54-dependent transcriptional activator and is located in a gene cluster thought to be involved in purine catabolism. YgeV is essential for the induction of ygfT and genes involved in formate-dependent uric acid degradation (Fig. 7B and C). Since a σ54-dependent promoter is present in the intergenic region of ygfT and uacT (8), ygeV might be directly involved in the transcription of ygfT in response to hypoxanthine, xanthine, and uric acid. Interestingly, the FDH-H-encoding gene is expressed from a σ54-dependent promoter; hence, it is possible that YgeV promotes the transcription of the FDH-H gene. Genes for the utilization of allantoin as a nitrogen source are coordinately regulated by both the repressor AllR and the activator AllS (40). Considering all of the findings mentioned above, ygfT appears to be involved in the formate-dependent uric acid degradation, and ygeV appears to be required for the expression of the uric acid-inducible gene ygfT. We propose that the genes ygeV and ygfT be renamed uacR (uric acid regulator) and uacF (uric acid degradation formate-related element), respectively, reflecting their physiological roles.
FDH-H-, AegA-, and YgfT (UacF)-dependent uric acid degradation may constitute one of the important systems that enhance bacterial adaptability to the gut environment, enabling E. coli to obtain nitrogen by degrading uric acid. In fact, 30% of uric acid in a healthy human is present in the intestinal lumen (41). Further, according to a recent report, high levels of xanthine dehydrogenase are expressed by the intestinal microflora of gout patients, leading to the accumulation of large amounts of uric acid (42). Because the solubility of uric acid is very low, the accumulation of uric acid in body fluids is toxic. Such accumulation, in turn, causes gout, since humans lack uric acid-degrading enzymes. Therefore, the discovery of the urate-degrading activity of E. coli and the identification of genes required for the utilization of uric acid provide insight into purine metabolism as a function of the human intestinal microflora. Further, identification of a novel uric acid degradation pathway might lead to the development of novel therapies for gout.
We first identified aegA as a gene responsible for menadione tolerance in a reduced-genome strain, the Δ21a mutant (Fig. S1). ygfT (uacF) is in the genome of the Δ21a mutant strain. In this strain, AegA may reduce the electron flow to menadione as part of an AegA-dependent metabolic pathway, resulting in a reduced production of reactive oxygen species. Alternatively, AegA may transfer electrons from a reduced form of menadione to another electron acceptor, e.g., a uric acid-degrading enzyme, also resulting in reduced reactive oxygen species production. The deletion of the aegA gene reduced menadione tolerance of the Δ21a mutant strain but not that of the wild-type strain. In the Δ21a mutant strain, in which many metabolic pathways are deficient because many genes have been deleted, multiple metabolic pathways may be impaired, and, consequently, more electrons may flow into AegA than in the wild type.
In conclusion, in the current study, we showed for the first time that E. coli utilizes uric acid as a nitrogen source. E. coli degrades uric acid and is able to grow using uric acid as the only nitrogen source under anaerobic and microaerobic conditions. Further, AegA, YgfT (UacF), and FDH-H are required for uric acid degradation in the presence of formate.
MATERIALS AND METHODS
Bacterial strains and primers.
All E. coli strains used in the current study are derivatives of strain MG1655. Deletion mutations were constructed by using the Red recombination system (43); the mutations were transferred to the other strains by P1 transduction (44). The strain genotypes are presented in Table S2 in the supplemental material. Primers used for the construction of the deletion mutants are listed in Table S3. Primer sets for PCR are listed in Table S4. To construct the aegA′-lacZ and ygfT′-lacZ fusions, four DNA fragments (the upstream regions of lacI and chloramphenicol resistance gene [cat], the promoter region of aegA or ygfT, and lacZ) were prepared by PCR (Table S4, no. 1 to 5). The DNA fragments were then joined by second- and third-round PCR (Table S4, no. 6 to 12).
To construct the AegA point substitution strains, four DNA fragments (those encoding the C-terminal part of AegA and the N-terminal part of AegA, cat, and the downstream region of aegA) were prepared by PCR (Table S4, no. 38 to 56). Missense mutations were introduced by PCR using primers with the desired mutations. For the Cys68Ala substitution, T202 and G203 were changed to G and C, respectively. For the Cys97Ala substitution, T289, G290, and T291 were changed to G, C, and G, respectively. For the Cys121Ala substitution, T361, G362, and C363 were changed to G, C, and G, respectively. For the Cys230Ala substitution, T688, G689, and C690 were changed to G, C, and G, respectively. For the Gly334Ala G336A substitution, G1001, G1002, G1007, and T1008 were all changed to C. For the Gly447Ala substitution, G1431 was changed to C. The mutations were confirmed by sequencing.
To construct the AegA-Myc-His strain, three DNA fragments were prepared by PCR, with one encoding the C-terminal region from the termination codon of aegA (Table S4, no. 32), the Myc-His region with the cat gene (Table S4, no. 33), and the downstream region of aegA (Table S4, no. 34). The DNA fragments were joined by second- and third-round PCR (Table S4, no. 35 to 37).
To construct the His-AegA strain, four DNA fragments were prepared by PCR, as follows. The upstream region of aegA (Table S4, no. 57), pBAD-His (Table S4, no. 58), the aegA open reading frame (ORF) (Table S4, no. 59), and cat were joined with the downstream region of aegA (Table S4, no. 38). The DNA fragments were joined by second- and third-round PCR (Table S4, no. 60 and 61).
To construct the mFKm-aegA and mFKm-ygfT plasmids, three DNA fragments were prepared by PCR, aegA (Table S4, no. 16), ygfTS (Table S4, no. 17), and a kanamycin resistance (Km) gene (Table S4, no. 62). The Km DNA fragment was joined with the aegA and ygfTS fragments in a second round of PCR (Table S4, no. 18 and 19). The resultant fragments were ligated with the HpaI-mini-F fragment of plasmid pSC138 (45).
To construct the mFTc-fdhF plasmid, two DNA fragments were prepared by PCR, fdhF (Table S4, no. 27) and Tc (Table S4, no. 26). These DNA fragments were joined in a second round of PCR (Table S4, no. 28). The resultant fragment was ligated with the HpaI-mini-F fragment of plasmid pSC138 (45).
Menadione sensitivity assay.
The deletion mutants were grown on antibiotic medium 3 plates. The colonies were transferred to 2 ml of antibiotic medium 3 and incubated for 24 h at 37°C with shaking. The precultures were diluted 1/100 in 3 ml of antibiotic medium 3 and, after bubbling with N2, incubated for 24 h at 37°C with rotation. Stationary-phase cultures (0.5 ml) were transferred to sampling tubes with an O-ring and mixed with menadione solution in ethanol or with ethanol. After flashing with N2, they were incubated for 24 h at 4°C with rotation. The cultures were then diluted and plated on antibiotic medium 3 plates and incubated for 1 to 4 days at 37°C, and the colonies were counted. The concentrations of menadione used were 1.0 mM for wild-type strains and 0.1 mM for Δ21a mutant strains.
Promoter activity assay.
To test the effect of various electron acceptors on aegA and ygfT expression, the cells were grown in LB medium supplemented with 48 mM NaNO3, NaNO2, DMSO, TMAO, or fumaric acid. Anaerobic incubation was achieved by culturing (30 ml/tube) in 30-ml glass tubes topped with a butyl stopper. Microaerobic incubation was achieved by culturing (15 ml/tube) in 15-ml plastic tubes with rotation. Aerobic incubation was achieved by culturing (5 ml/flask) in 100-ml flasks. To test the effect of purine degradation intermediates on aegA and ygfT expression, the cells were grown in M9 medium (without NH4Cl) with 2% tryptone as the carbon source. The M9 tryptone medium was used to evaluate the effect of purine derivatives in more detail because the yeast extract in LB medium contains a considerable amount of purines. The medium was supplemented with 1.6 mM adenine, hypoxanthine, guanine, xanthine, uric acid, or allantoin, and the cultures were incubated under anaerobic conditions. The cultures were incubated for 24 h at 37°C with shaking (130 rpm). Next, stationary-phase cultures were chilled on ice-water slurry, the caps of containers were opened, and the cells were harvested by centrifugation. The cell pellets were washed with 50 mM Tris-HCl (pH 8.0) and resuspended in the same buffer. After disruption by sonication, the cell debris was removed by centrifugation (27,173 × g, 5 min). The β-galactosidase activity of the resultant supernatants of strains harboring the pygfT′-lacZ and paegA′-lacZ transcriptional fusions was determined by o-nitrophenyl-β-d-galactopyranoside (ONPG) hydrolysis using standard procedures (44). Protein concentrations were determined using the Bradford assay with bovine serum albumin as the standard and used to normalize the β-galactosidase-specific activity. An extinction coefficient of 4,500 M−1 cm−1 was used for calculation. Unit activity was defined as the production of 100 μmol o-nitrophenol min−1 mg protein−1. The data are the averages of the results from three independent replicates and are presented with the standard error.
Uric acid utilization assay.
The cells were grown anaerobically in LB medium supplemented with 1.6 mM uric acid. The cultures were incubated for 24 h at 37°C with shaking (130 rpm). Then, 30 ml of stationary-phase cultures was harvested by centrifugation, and the cells were washed with 0.85% NaCl and suspended in 1 ml of 0.85% NaCl. The suspensions were diluted to an OD600 of 1.0 in M9 medium (without NH4Cl), with 1.2 mM uric acid as the sole nitrogen source and 0.4% glucose as the sole carbon source. When a small inoculum was used (OD600 of 0.1 at the start of the experiment), neither aegA- and ygfT-dependent uric acid degradation nor uric acid-dependent growth was detected. When indicated, 50 mM HCOONa and 50 mM NaHCO3 were added. For the microaerobic incubation, 1.5 ml of cultures in 1.5-ml Eppendorf tubes were incubated at 37°C with rotation. Aerobic incubation was achieved by culturing (5 ml/flask) in 100-ml flasks. Anaerobic incubation was achieved by culturing (30 ml/tube) in 30-ml glass tubes topped with a butyl stopper. At the indicated times, the OD600 of the cultures was measured, and the amount of uric acid was determined by measuring the OD291 of the culture supernatant, as described previously (46). Data are the averages of the results from three independent replicates and are presented with the standard error.
Data availability.
The data that support the findings of the current study are available from the corresponding author upon request.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Keizo Shimada and Shigeki Ehira for valuable discussions.
This work was supported by a Grant-in-Aid for Scientific Research (KAKENHI) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Y.I. conducted the experiments. Y.I. and J.-I.K. designed the research and wrote the paper. J.-I.K. is responsible for the overall project design.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00573-18.
REFERENCES
- 1.Schultz AC, Nygaard P, Saxild HH. 2001. Functional analysis of 14 genes that constitute the purine catabolic pathway in Bacillus subtilis and evidence for a novel regulon controlled by the PucR transcription activator. J Bacteriol 183:3293–3302. doi: 10.1128/JB.183.11.3293-3302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong SH. 1988. The use of purine compounds as sole sources of carbon and nitrogen by Klebsiella species. Microbios 56:57–62. [PubMed] [Google Scholar]
- 3.Matsui H, Shimaoka M, Kawasaki H, Takenaka Y, Kurahashi O. 2001. Adenine deaminase activity of the yicP gene product of Escherichia coli. Biosci Biotechnol Biochem 65:1112–1118. doi: 10.1271/bbb.65.1112. [DOI] [PubMed] [Google Scholar]
- 4.Xi H, Schneider BL, Reitzer L. 2000. Purine catabolism in Escherichia coli and function of xanthine dehydrogenase in purine salvage. J Bacteriol 182:5332–5341. doi: 10.1128/JB.182.19.5332-5341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cusa E, Obradors N, Baldomà L, Badía J, Aguilar J. 1999. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J Bacteriol 181:7479–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papakostas K, Frillingos S. 2012. Substrate selectivity of YgfU, a uric acid transporter from Escherichia coli. J Biol Chem 287:15684–15695. doi: 10.1074/jbc.M112.355818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramazzina I, Folli C, Secchi A, Berni R, Percudani R. 2006. Completing the uric acid degradation pathway through phylogenetic comparison of whole genomes. Nat Chem Biol 2:144–148. doi: 10.1038/nchembio768. [DOI] [PubMed] [Google Scholar]
- 8.Gabison L, Prangé T, Colloc’h N, El Hajji M, Castro B, Chiadmi M. 2008. Structural analysis of urate oxidase in complex with its natural substrate inhibited by cyanide: Mechanistic implications. BMC Struct Biol 8:32. doi: 10.1186/1472-6807-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motojima K, Kanaya S, Goto S. 1988. Cloning and sequence analysis of cDNA for rat liver uricase. J Biol Chem 263:16677–16681. [PubMed] [Google Scholar]
- 10.de La Riva L, Badia J, Aguilar J, Bender RA, Baldoma L. 2008. The hpx genetic system for hypoxanthine assimilation as a nitrogen source in Klebsiella pneumoniae: Gene organization and transcriptional regulation. J Bacteriol 190:7892–7903. doi: 10.1128/JB.01022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michiel M, Perchat N, Perret A, Tricot S, Papeil A, Besnard M, De Berardinis V, Salanoubat M, Fischer C. 2012. Microbial urate catabolism: characterization of HpyO, a non-homologous isofunctional isoform of the flavoprotein urate hydroxylase HpxO. Environ Microbiol Rep 4:642–647. doi: 10.1111/j.1758-2229.2012.00390.x. [DOI] [PubMed] [Google Scholar]
- 12.Doniselli N, Monzeglio E, Dal Palù A, Merli A, Percudani R. 2015. The identification of an integral membrane, cytochrome c urate oxidase completes the catalytic repertoire of a therapeutic enzyme. Sci Rep 5:13798. doi: 10.1038/srep13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwadate Y, Honda H, Sato H, Hashimoto M, Kato JI. 2011. Oxidative stress sensitivity of engineered Escherichia coli cells with a reduced genome. FEMS Microbiol Lett 322:25–33. doi: 10.1111/j.1574-6968.2011.02331.x. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe N, Dickinson DA, Liu RM, Forman HJ. 2004. Quinones and glutathione metabolism. Methods Enzymol 378:319–340. doi: 10.1016/S0076-6879(04)78024-6. [DOI] [PubMed] [Google Scholar]
- 15.Hassan HM, Fridovich I. 1979. Intracellular production of superoxide radical and of hydrogen peroxide by redox active compounds. Arch Biochem Biophys 196:385–395. doi: 10.1016/0003-9861(79)90289-3. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto M, Ichimura T, Mizoguchi H, Tanaka K, Fujimitsu K, Keyamura K, Ote T, Yamakawa T, Yamazaki Y, Mori H, Katayama T, Kato J. 2005. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol Microbiol 55:137–149. doi: 10.1111/j.1365-2958.2004.04386.x. [DOI] [PubMed] [Google Scholar]
- 17.Cavicchioli R, Kolesnikow T, Chiang RC, Gunsalus RP. 1996. Characterization of the aegA locus of Escherichia coli: control of gene expression in response to anaerobiosis and nitrate. J Bacteriol 178:6968–6974. doi: 10.1128/jb.178.23.6968-6974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanoni MA, Curti B. 2005. Structure-function studies on the iron-sulfur flavoenzyme glutamate synthase: an unexpectedly complex self-regulated enzyme. Arch Biochem Biophys 433:193–211. doi: 10.1016/j.abb.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 19.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu Y-K. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J 272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reitzer L, Schneider BL. 2001. Metabolic context and possible physiological themes of sigma(54)-dependent genes in Escherichia coli. Microbiol Mol Biol Rev 65:422–444. doi: 10.1128/MMBR.65.3.422-444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Gunsalus RP. 2003. Coordinate regulation of the Escherichia coli formate dehydrogenase fdnGHI and fdhF genes in response to nitrate, nitrite, and formate: roles for NarL and NarP. J Bacteriol 185:5076–5085. doi: 10.1128/JB.185.17.5076-5085.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birkmann A, Zinoni F, Sawers G, Böck A. 1987. Factors affecting transcriptional regulation of the formate-hydrogen-lyase pathway of Escherichia coli. Arch Microbiol 148:44–51. doi: 10.1007/BF00429646. [DOI] [PubMed] [Google Scholar]
- 24.Maier T, Binder U, Böck A. 1996. Analysis of the hydA locus of Escherichia coli: two genes (hydN and hypF) involved in formate and hydrogen metabolism. Arch Microbiol 165:333–341. doi: 10.1007/s002030050335. [DOI] [PubMed] [Google Scholar]
- 25.Pinske C, Sawers RG. 2014. The importance of iron in the biosynthesis and assembly of [NiFe]-hydrogenases. Biomol Concepts 5:55–70. doi: 10.1515/bmc-2014-0001. [DOI] [PubMed] [Google Scholar]
- 26.Jormakka M, Törnroth S, Byrne B, Iwata S. 2002. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science 295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 27.Stanley NR, Sargent F, Buchanan G, Shi J, Stewart V, Palmer T, Berks BC. 2002. Behaviour of topological marker proteins targeted to the Tat protein transport pathway. Mol Microbiol 43:1005–1021. doi: 10.1046/j.1365-2958.2002.02797.x. [DOI] [PubMed] [Google Scholar]
- 28.Sawers G. 1994. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie Van Leeuwenhoek 66:57–88. doi: 10.1007/BF00871633. [DOI] [PubMed] [Google Scholar]
- 29.Iwadate Y, Funabasama N, Kato J-I. 2017. Involvement of formate dehydrogenases in stationary phase oxidative stress tolerance in Escherichia coli. FEMS Microbiol Lett 364:fnx193. doi: 10.1093/femsle/fnx193. [DOI] [PubMed] [Google Scholar]
- 30.Arnoux P, Ruppelt C, Oudouhou F, Lavergne J, Siponen MI, Toci R, Mendel RR, Bittner F, Pignol D, Magalon A, Walburger A. 2015. Sulphur shuttling across a chaperone during molybdenum cofactor maturation. Nat Commun 6:6148. doi: 10.1038/ncomms7148. [DOI] [PubMed] [Google Scholar]
- 31.McDowall JS, Murphy BJ, Haumann M, Palmer T, Armstrong FA, Sargent F. 2014. Bacterial formate hydrogenlyase complex. Proc Natl Acad Sci U S A 111:E3948–E3956. doi: 10.1073/pnas.1407927111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrews SC, Berks BC, McClay J, Ambler A, Quail MA, Golby P, Guest JR. 1997. A 12-cistron Escherichia coli operon (hyf) encoding a putative proton-translocating formate hydrogenlyase system. Microbiol 143:3633–3647. doi: 10.1099/00221287-143-11-3633. [DOI] [PubMed] [Google Scholar]
- 33.Knappe J, Blaschkowski HP, Gröbner P, Schmitt T. 1974. Pyruvate formate-lyase of Escherichia coli: the acetyl-enzyme intermediate. Eur J Biochem 50:253–263. doi: 10.1111/j.1432-1033.1974.tb03894.x. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, Rudd KE. 2012. EcoGene 3.0. Nucleic Acids Res 41:D613–D624. doi: 10.1093/nar/gks1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu W, Dunn CA, O’handley SF, Smith DL, Bessman MJ. 2006. Three new Nudix hydrolases from Escherichia coli. J Biol Chem 281:22794–22798. doi: 10.1074/jbc.M603407200. [DOI] [PubMed] [Google Scholar]
- 37.Hagen WR, Silva PJ, Amorim MA, Hagedoorn PL, Wassink H, Haaker H, Robb FT. 2000. Novel structure and redox chemistry of the prosthetic groups of the iron-sulfur flavoprotein sulfide dehydrogenase from Pyrococcus furiosus; evidence for a [2Fe-2S] cluster with Asp(Cys)3ligands. J Biol Inorg Chem 5:527–534. doi: 10.1007/PL00021452. [DOI] [PubMed] [Google Scholar]
- 38.Lubner CE, Jennings DP, Mulder DW, Schut GJ, Zadvornyy OA, Hoben JP, Tokmina-Lukaszewska M, Berry L, Nguyen DM, Lipscomb GL, Bothner B, Jones AK, Miller AF, King PW, Adams MWW, Peters JW. 2017. Mechanistic insights into energy conservation by flavin-based electron bifurcation. Nat Chem Biol 13:655–659. doi: 10.1038/nchembio.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox JC. 1989. Escherichia coli formate dehydrogenase mutants with altered selenopolymer profiles. Arch Microbiol 152:397–400. doi: 10.1007/BF00425180. [DOI] [PubMed] [Google Scholar]
- 40.Rintoul MR, Cusa E, Baldomà L, Badia J, Reitzer L, Aguilar J. 2002. Regulation of the Escherichia coli allantoin regulon: coordinated function of the repressor AllR and the activator AllS. J Mol Biol 324:599–610. doi: 10.1016/S0022-2836(02)01134-8. [DOI] [PubMed] [Google Scholar]
- 41.Sorensen LB, Levinson DJ. 1975. Origin and extrarenal elimination of uric acid in man. Nephron 14:7–20. doi: 10.1159/000180432. [DOI] [PubMed] [Google Scholar]
- 42.Guo Z, Zhang J, Wang Z, Ang KY, Huang S, Hou Q, Su X, Qiao J, Zheng Y, Wang L, Koh E, Danliang H, Xu J, Lee YK, Zhang H. 2016. Intestinal microbiota distinguish gout patients from healthy humans. Sci Rep 6:20602. doi: 10.1038/srep20602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy KC. 1998. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J Bacteriol 180:2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 45.Ogura T, Miki T, Hiraga S. 1980. Copy-number mutants of the plasmid carrying the replication origin of the Escherichia coli chromosome: evidence for a control region of replication. Proc Natl Acad Sci U S A 77:3993–3997. doi: 10.1073/pnas.77.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hibi T, Kume A, Kawamura A, Itoh T, Fukada H, Nishiya Y. 2016. Hyperstabilization of tetrameric Bacillus sp. TB-90 urate oxidase by introducing disulfide bonds through structural plasticity. Biochemistry 55:724–732. doi: 10.1021/acs.biochem.5b01119. [DOI] [PubMed] [Google Scholar]
- 47.Jochimsen B, Nygaard P, Vestergaard T. 1975. Location on the chromosome of Escherichia coli of genes governing purine metabolism. Adenosine deaminase (add), guanosine kinase (gsk) and hypoxanthine phosphoribosyltransferase (hpt). Mol Gen Genet 143:85–91. doi: 10.1007/BF00269424. [DOI] [PubMed] [Google Scholar]
- 48.Maynes JT, Yuan RG, Snyder FF. 2000. Identification, expression, and characterization of Escherichia coli guanine deaminase. J Bacteriol 182:4658–4660. doi: 10.1128/JB.182.16.4658-4660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iobbi-Nivol C, Leimkühler S. 2013. Molybdenum enzymes, their maturation and molybdenum cofactor biosynthesis in Escherichia coli. Biochim Biophys Acta 827:1086–1101. doi: 10.1016/j.bbabio.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Lee Y, Lee DH, Kho CW, Lee AY, Jang M, Cho S, Lee CH, Lee JS, Myung PK, Park BC, Park SG. 2005. Transthyretin-related proteins function to facilitate the hydrolysis of 5-hydroxyisourate, the end product of the uricase reaction. FEBS Lett 579:4769–4774. doi: 10.1016/j.febslet.2005.07.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of the current study are available from the corresponding author upon request.