Our findings provide new insights into the mechanism of E. coli phototaxis, showing that all five chemoreceptor types respond to light and their interactions play an important role in cell behavior. Our results also open up new avenues for examining and manipulating E. coli taxis. Since light is a universal stimulus, it may provide a way to quantify interactions among different types of receptors. Because light is easier to control spatially and temporally than chemicals, it may be used to study swimming behavior in complex environments. Since phototaxis can cause migration of E. coli bacteria in light gradients, light may be used to control bacterial density for studying density-dependent processes in bacteria.
KEYWORDS: chemotaxis, Escherichia coli, phototaxis
ABSTRACT
Blue light has been shown to elicit a tumbling response in Escherichia coli, a nonphototrophic bacterium. The exact mechanism of this phototactic response is still unknown. Here, we quantify phototaxis in E. coli by analyzing single-cell trajectories in populations of free-swimming bacteria before and after light exposure. Bacterial strains expressing only one type of chemoreceptor reveal that all five E. coli receptors (Aer, Tar, Tsr, Tap, and Trg) are capable of mediating responses to light. In particular, light exposure elicits a running response in the Tap-only strain, the opposite of the tumbling responses observed for all other strains. Therefore, light emerges as a universal stimulus for all E. coli chemoreceptors. We also show that blue light exposure causes a reversible decrease in swimming velocity, a proxy for proton motive force. This result is consistent with a previously proposed hypothesis that, rather than sensing light directly, chemoreceptors sense light-induced perturbations in proton motive force, although other factors are also likely to contribute.
IMPORTANCE Our findings provide new insights into the mechanism of E. coli phototaxis, showing that all five chemoreceptor types respond to light and their interactions play an important role in cell behavior. Our results also open up new avenues for examining and manipulating E. coli taxis. Since light is a universal stimulus, it may provide a way to quantify interactions among different types of receptors. Because light is easier to control spatially and temporally than chemicals, it may be used to study swimming behavior in complex environments. Since phototaxis can cause migration of E. coli bacteria in light gradients, light may be used to control bacterial density for studying density-dependent processes in bacteria.
INTRODUCTION
Phototaxis, the light-dependent movement of microorganisms, was observed in certain species of purple bacteria as early as the 19th century (1). Along with halobacteria and cyanobacteria, purple bacteria are phototrophic, i.e., they use light as a source of energy. Phototaxis confers an obvious advantage to phototrophic bacteria, as it allows them to migrate to optimal illumination conditions (2, 3). Escherichia coli, on the other hand, is a surprising example of a nonphototrophic bacterium for which exposure to blue light results in changes in motile behavior (4–7).
E. coli motility is governed by a few simple principles that allow it to find the most favorable environment efficiently. E. coli is propelled by a bundle of rotating helical flagella and swims by alternating between two types of motion, namely, runs, during which cells swim in one direction along an approximately straight path, and tumbles, during which cells randomly reorient. Runs correspond to counterclockwise (CCW) rotation of all flagellar motors, which results in a tight bundle of flagella propelling the cell forward. During tumbles, one or more flagella rotate clockwise (CW), breaking from the bundle and causing random reorientation of the cell before the next run (8). Therefore, the fraction of time spent tumbling (the tumble bias) depends on the fraction of time each flagellar motor rotates CW. Tumble bias changes in response to intracellular cues, such as the proton motive force (PMF) (9), or extracellular chemical cues (10), resulting in a behavior known as taxis.
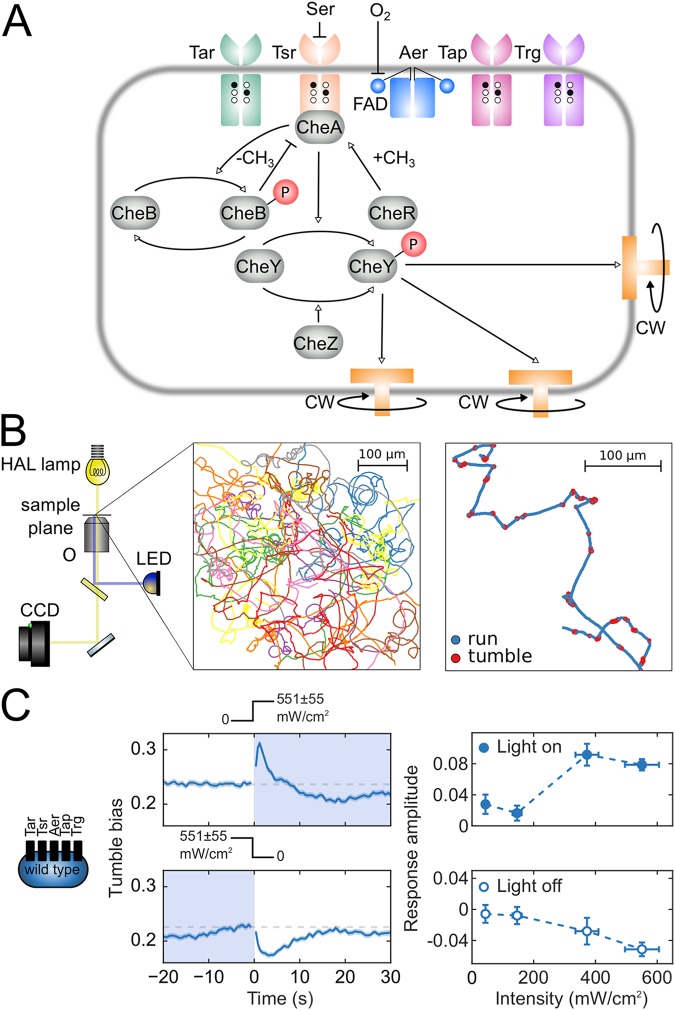
This behavior is controlled by a simple signaling network (Fig. 1A) (10). The intracellular signaling molecule CheY, in its active phosphorylated form (CheY-P), binds to the flagellar motors, causing a switch in the rotational direction from CCW to CW. CheY is phosphorylated by the kinase CheA and dephosphorylated by the phosphatase CheZ. E. coli has five types of transmembrane receptors (i.e., Tar, Tsr, Aer, Tap, and Trg) (Fig. 1A), of varying abundance, that sense a range of environmental signals (11, 12). These receptors form complexes with CheA, coupling its kinase activity to environmental conditions. For example, binding of chemical repellents to the receptors’ ligand-binding domains results in increased CheA activity, a higher concentration of CheY-P, a higher probability of CW motor rotation, and therefore higher tumble bias. Conversely, binding of chemoattractants deactivates CheA, resulting in lower tumble bias. Finally, CheA activity is also regulated by receptor methylation, which is controlled by the methyltransferase CheR and the methylesterase CheB, the latter of which is active only when phosphorylated by active CheA (10) (Fig. 1A). CheR and CheB regulation of CheA activity forms a negative feedback loop that allows bacteria to adapt to new conditions. The net result is that E. coli run lengths increase as conditions become more favorable, cells migrate toward a better environment, and the cells eventually adapt as the tumble bias returns to its basal value (10).
FIG 1.
Phototaxis in E. coli. (A) Schematic of the chemotaxis network. Five types of chemoreceptors (Tar, Tsr, Aer, Trg, and Tap), sensitive to different extracellular and intracellular cues, modulate the activity of the kinase CheA, which phosphorylates the signaling molecule CheY. Phosphorylated CheY binds to flagellar motors, causing them to switch to CW rotation. Chemotactic adaptation is mediated by the methyltransferase CheR and the methylesterase CheB. Methylation of receptor sites (black circles) by CheR increases kinase activity. CheB, when phosphorylated by active CheA, demethylates receptors (white circles) and decreases kinase activity. (B) Experimental and data analysis framework for studying phototaxis in E. coli. An inverted light microscope with a 20× objective (O) and a wide-range halogen (HAL) lamp (yellow light path) images swimming cells, and a blue LED (blue light path) stimulates cells. Movies of swimming bacteria are captured by a CCD camera. Bacteria are detected in each movie frame and their coordinates are linked into trajectories, which are filtered and analyzed to assign runs and tumbles. (C, left and center) Response of the wild-type strain RP437 (schematic indicates that all receptor types are present) to a turn on and turn off of blue light with an intensity of 551 ± 55 mW/cm2. Light exposure is indicated by shading and by the light intensity profiles above the plots. Each point on the time trace is the average of the tumble biases of ∼6,000 to 7,000 trajectories. The shading around the time trace represents the standard error of the mean tumble bias. (C, right) Amplitude of the responses to turn on (top) and turn off (bottom) as a function of light intensity.
The taxis signaling network is characterized by (i) its extreme sensitivity (an E. coli cell can respond to concentration changes as small as ∼3 nM, corresponding to just a few molecules per cell volume [13]), (ii) its wide dynamic range (a cell is sensitive to changes in concentration of up to 5 orders of magnitude [10]), and (iii) its ability to integrate diverse extracellular cues, including not only concentrations of various chemicals (chemotaxis) but also temperature (thermotaxis), pH (pH taxis), and light (phototaxis) (12, 14, 15). Chemotaxis in E. coli has been studied extensively and serves as a paradigm for the way living cells modulate their behavior in response to environmental signals (10, 16–19). However, there have been only a few studies of E. coli phototactic responses (4–7), and the adaptive value of phototaxis remains unclear.
Here, we study phototaxis by analyzing single-cell trajectories in populations of E. coli bacteria swimming freely in two dimensions, before and after exposure to blue light. Our results show that light is a universal tactic signal and elicits responses mediated by all five types of receptors. Single-receptor mutant measurements confirm that Tar and Aer receptors mediate increased tumbling in bacteria exposed to light, in agreement with prior studies (7). The roles of the other three receptors in phototaxis were previously unknown. We find that Tsr and Trg also mediate tumbling in response to light, whereas Tap mediates a running response. Although Tap is a low-abundance receptor, we observe that several multireceptor strains containing Tap exhibit running responses to light, proving the ability of Tap to control motility. A reversible decrease in bacterial swimming velocity that we observe upon light exposure suggests that light perturbs electron transport and/or PMF. Based on these results, we propose a mechanism for a universal tactic response to light in which E. coli receptors sense light-induced perturbations of PMF or parameters coupled to PMF.
RESULTS
Responses to blue light require at least one receptor, as well as functional CheY, CheR, and CheB.
We analyzed swimming trajectories from thousands of cells stimulated by turning on or turning off blue light (Fig. 1B; see Fig. S1 and S4 in the supplemental material; also see Materials and Methods). Like Wright et al. (7), we observed that wild-type E. coli cells tumbled more in response to a turn on and then returned to the prestimulus tumble bias, exhibiting the opposite response to a turn off (Fig. 1C). The light intensities needed to elicit a detectable response were similar to those reported by Taylor and Koshland (4) and by Taylor et al. (5) but were significantly larger than those reported by Wright and coworkers, i.e., more than 300 mW/cm2, compared to 7 mW/cm2 (7). We attribute the discrepancy to different growth conditions in the latter study (see the supplemental material). When we used the same substrates in growth and motility media as Wright et al. (7) (M9 minimal medium supplemented with 5 mg/ml glycerol and motility buffer with 5 mM lactate), we reproduced similar responses at a lower light intensity (Fig. S5). Under our conditions, using the nonglycolytic substrate succinate for both growth and motility media (M9 minimal medium supplemented with 4 mg/ml succinate and motility buffer with 4 mg/ml succinate), the response amplitude increased with light intensity and saturated at ∼400 mW/cm2 (Fig. 1C).
We confirmed that this response is mediated by the chemotactic network by performing control experiments with mutants missing different components of the network (Fig. S6). We observed no response to light in a receptorless strain (UU1250), a strain lacking functional CheY (CR20), or a strain expressing a constitutively active mutant of CheY, CheYD13K (PS2001::pMS164) (Table 1). Therefore, similar to other types of tactic stimuli, blue light modulates the activity of receptor signaling complexes, and this signal is communicated to the flagellar motors through the signaling molecule CheY-P. Blue light does not appear to affect the switching of flagellar motor rotation directly.
TABLE 1.
Strains used in this work
| Strain | Genotypea | Comments | Reference or source |
|---|---|---|---|
| RP437 | Wild-type chemotaxis strain | 55 | |
| ΔR | ΔcheR | Christopher Rao | |
| ΔB | ΔcheB | Christopher Rao | |
| CR20 | CheY::FRT | Runner | Christopher Rao (56) |
| UU1250 | Δ(tar-tap) Δtsr Δaer Δtrg | Receptorless strain | John S. Parkinson (25) |
| PS2001::pMS164 | ΔcheB ΔcheY ΔcheZ | Lacks CheB, CheY, and CheZ and contains pMS164 plasmid | Philippe Cluzel (52) |
| UU1615 | Δaer Δtar Δtap Δtrg | Tsr-only strain | John S. Parkinson (38) |
| UU1624 | Δaer Δtsr Δtap Δtrg | Tar-only strain | John S. Parkinson (38) |
| UU1250::pSB20 | Δ(tar-tap) Δtsr Δaer Δtrg | Aer-only strain; contains pSB20 plasmid | John S. Parkinson (7) |
| UU1250::pPA705 | Δ(tar-tap) Δtsr Δaer Δtrg | Trg-only strain; contains pPA705 plasmid | This work |
| TP6 | Δ(tar-tap) Δtsr Δaer Δtrg | Tap-only strain; contains pTP1 plasmid | This work |
| RP8604 | Δtsr Δtrg | Tar-Aer-Tap strain | John S. Parkinson |
| RP1131 | trg::Tn10 | ΔTrg strain | John S. Parkinson |
| UU1117 | Δaer | ΔAer strain | John S. Parkinson (57) |
| UU1623 | Δtsr Δtrg Δtap | Tar-Aer strain | John S. Parkinson |
FRT, flippase recombination target.
We also confirmed that the observed adaptation to blue light is mediated by the chemotactic network proteins CheR and CheB. A strain lacking the receptor demethylase CheB (ΔB) (Table 1) had a very high tumble bias, which did not increase measurably upon light exposure (Fig. S6). The lack of response indicates that fully methylated receptors cannot be further activated by light. A strain lacking the receptor methyltransferase CheR (ΔR) (Table 1) exhibited an initial sharp increase in tumble bias, similar to the wild-type strain, but the tumble bias then failed to return to a lower value (Fig. S6). The lack of adaptation is expected; in the absence of CheR, receptor methylation levels are low and adaptation through demethylation is thus impossible. In summary, the lack of response in the ΔCheB strain indicates that a tumbling response to light is caused by receptor activation in the wild-type strain. The postresponse kinetics in the ΔCheR strain indicate that adaptation in the wild-type strain is mediated by CheR through the negative feedback loop of the chemotaxis network. (Note that, rather than simply leveling off, the tumble bias of ΔCheR continued to increase slowly, which we speculate is due to a slow secondary effect such as the recently reported motor remodeling [20].)
All five E. coli chemoreceptors mediate blue-light-induced changes in tumble bias.
(i) Turn-on responses. The results described above demonstrate that the blue light response in wild-type E. coli requires functioning receptor complexes. To determine the independent contributions of individual receptor types, we measured the responses of mutants expressing only a single receptor type (see Materials and Methods). Abundances (i.e., copy numbers) of different receptor types can span 2 orders of magnitude in wild-type strains (21). Therefore, the low-copy-number receptors Aer, Tap, and Trg were expressed from a plasmid, with the concentration of inducer adjusted to ensure that the tumble bias in the mutant strain in the absence of stimulus was similar to that of the wild-type strain. We confirmed the results of Wright et al. (7) by observing tumbling responses in Tar- and Aer-only strains (Fig. 2A). In addition, we detected responses in Tsr-, Trg-, and Tap-only strains. The Tsr-only strain exhibited a weak but consistent tumbling response, and the Trg-only strain showed a tumbling response with delayed onset (Fig. 2A).
FIG 2.
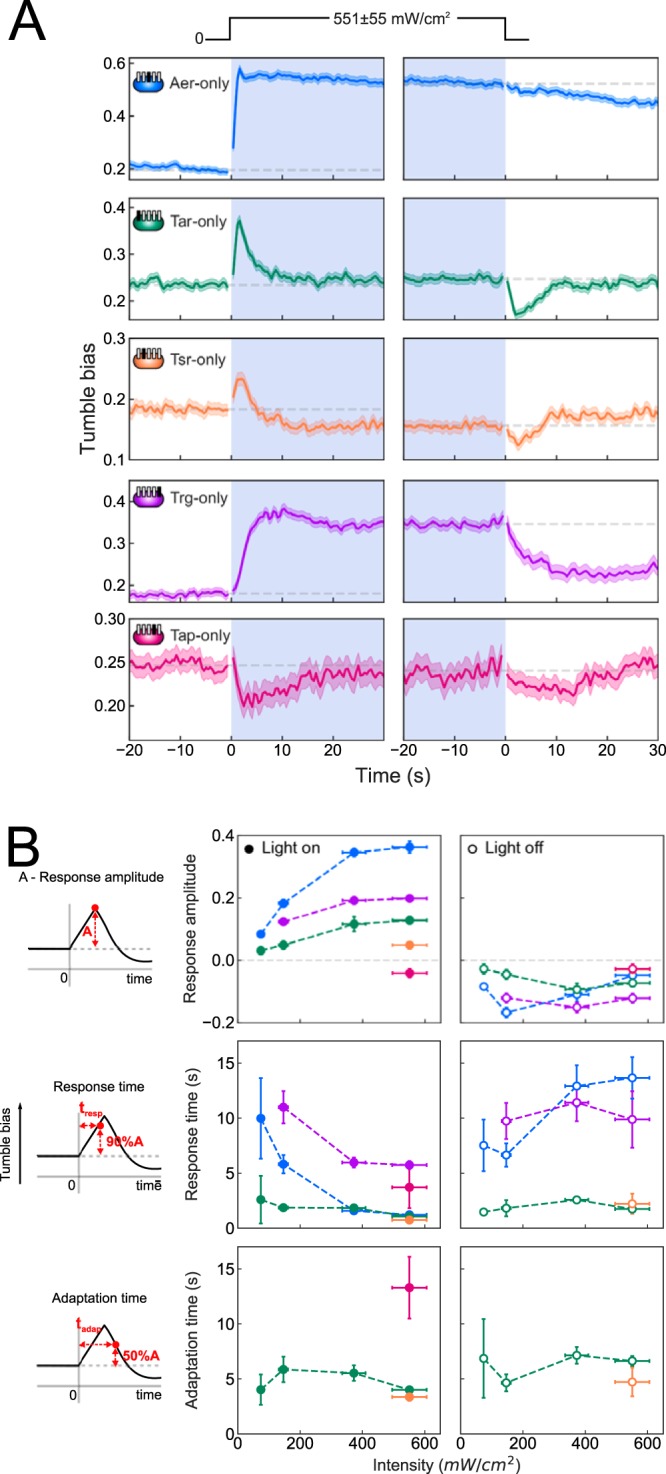
Blue light responses for single-receptor E. coli strains. (A) Tumble bias traces for single-receptor E. coli strains (Table 1), i.e., Aer-only (blue), Tar-only (green), Tsr-only (orange), Trg-only (purple), and Tap-only (magenta) strains. Approximately 200 to 2,000 trajectories were used to calculate the average tumble bias at each time point. Schematics indicate which receptors are present (filled rectangles) or absent (empty rectangles) in each strain. All responses were measured at a blue light intensity of 551 ± 55 mW/cm2, as indicated by the light intensity profile. Gray dashed lines show the prestimulus tumble bias for turn-on and turn-off responses. (B) Blue light intensity dependence of parameters for the responses to turn on (filled circles) and turn off (empty circles) for all strains. Response amplitude, response time, and adaptation time were calculated as illustrated by the schematics.
The Tap-only strain showed a unique response, compared to the other strains, which is a key result of this study. Tap, like Trg, can be methylated, but it lacks the NWETF motif that recruits the methyltransferase CheR (22). Therefore, we expected Tap to have a low methylation level and hence low activity in the absence of other receptors. Indeed, we observed that the Tap-only strain had a significantly lower tumble bias than did the wild-type strain and did not respond to blue light exposure (Fig. S7). The lack of response suggests that, unlike Trg, the Tap receptor is not activated by light exposure; however, it is possible that Tap can be deactivated by light exposure and thus mediate a running response. To test this hypothesis, we performed light response measurements of the Tap-only mutant in the presence of phenol. Phenol is a repellent for Tap and therefore activates Tap receptors, increasing the tumble bias to a higher baseline level (23). Indeed, the Tap-only strain had a higher tumble bias in the presence of 2.5 mM phenol and exhibited running responses (deactivation) in blue light (Fig. 2A; also see Fig. S7). Phenol does not absorb in the blue region of the spectrum (24), which rules out the possibility that Tap responds to excited-state phenol rather than to light itself. In further controls with the Tar-only strain in the presence of 2.5 mM phenol (Fig. S7), we observed qualitatively the same responses to light turn on and turn off as in the absence of phenol, although the time scales of the responses and adaptation were affected. Taken together, these results indicate that, unlike the Tar, Tsr, Aer, and Trg receptors, the Tap receptor is deactivated by light exposure and mediates a running response in E. coli.
The observed single-receptor responses exhibited adaptation behavior consistent with what is known about the mechanisms of adaptation for different receptors (Fig. 2A). Both Tar- and Tsr-only strains adapted to a steady-state tumble bias <10 s after the light turn on, which is consistent with methylation-dependent adaptation (Fig. 2B). In contrast, Tap- and Trg-only strains did not adapt appreciably to light turn on, consistent with the fact that they cannot recruit CheR (22). We did observe partial slow adaptation kinetics in both mutants, which may be due to the recently reported mechanism of motor remodeling (20). The Aer receptor adapts through an unknown methylation-independent pathway that also tends to be slower (25); consequently, we did not observe significant adaptation for the Aer-only strain within the duration of our measurements.
(ii) Turn-off responses. For most of the single-receptor strains, responses to light turn off were symmetrical to the responses to light turn on (Fig. 2A). For example, Tsr- and Tar-only strains exhibited running responses to turn off with adaptation kinetics similar to those of turn-on responses, further confirming that adaptation is mediated by the negative feedback loop of the chemotaxis network (Fig. 2A). In Trg- and Tap-only strains, responses to light turn off were also opposite in sign to those to light turn on, but with little to no adaptation (Fig. 2A). In contrast, the Aer-only strain exhibited markedly slower response kinetics when high-intensity light was turned off, compared to when light was turned on (Fig. 2A). This result suggests that the effect of light goes beyond receptor activation and Aer may be sensing a secondary effect from which it takes some time to recover, rather than light itself (Fig. 2A). At low light intensity, however, Aer responses to light turn off and turn on became symmetric (Fig. S8). This intensity dependence suggests that Aer has a sensing mechanism that is qualitatively different at low and high light intensities.
(iii) Response time analysis. We also analyzed the response times of the various strains, defined as the time from the application of the stimulus to the response peak. We switched light on and off in less than one movie frame (0.08 s), and conformational changes of receptors in response to stimuli are known to occur on a subsecond time scale (26). Responses to light exposure were immediate within the time resolution of the experiment for Tar-, Tsr-, and Aer-only strains at high light intensity, while both Tap- and Trg-only strains demonstrated gradual responses (Fig. 2A). Slower response times may indicate that receptors are responding to the light-induced perturbations of cellular processes on longer time scales, rather than responding directly to light. Our measurements cannot determine whether the observed gradual response kinetics result from a uniformly slower response across the cell population or from immediate single-cell-level responses with different time delays (27). Single-cell taxis assays that provide long (∼100-s) tumble bias traces for individual bacteria may allow differentiation between these two explanations (27).
Figure 2B summarizes the dependence of the response amplitudes, response times, and adaptation times on light intensity for representative strains. There are several trends worth noting. The amplitude of the responses to light exposure increased with intensity and saturated at ∼400 mW/cm2, similar to the wild-type response (Fig. 2B, left). This is in contrast to the results of Wright et al., who reported saturation above ∼10 mW/cm2 (7). However, as discussed above, growth conditions affect the sensitivity of the responses (see Fig. S5 and the information in the supplemental material). Although the peak tumble biases at saturating light intensity for single-receptor mutants were greater than that for the wild-type strain, they were still lower than that for continuously tumbling bacteria (Fig. 2A). Response times to light turn on in Aer- and Trg-only strains decreased with light intensity (Fig. 2B, middle). No dependence of response and adaptation times on light intensity was observed for the Tar-only strain (Fig. 2B, middle and right).
Effects of Tap on light responses in multireceptor strains.
Our results indicate that light is a universal tactic stimulus that affects all five E. coli chemotactic receptors. When all receptors respond to the same stimulus, it is far from obvious how the signals from different receptors are integrated to produce a response in wild-type strains. Chemotaxis signaling units, formed by trimers of receptor homodimers and CheA dimers, are organized into a hexagonal lattice that serves as a structural platform for interactions between receptors (28–30). Signals from low-abundance receptors are thus amplified by interactions with high-abundance receptors and could still contribute to the chemotactic response (31). Therefore, the contributions of individual receptor types to the overall response could be nonadditive. As shown in Fig. 1, in a wild-type strain in which four of five receptors (all except Tap) mediate a tumbling response to light, the integrated result of the individual receptor contributions is a tumbling response with amplitude comparable to that of the Tar-only strain. This suggests that, despite its low abundance, the running receptor Tap may contribute to the response of the wild-type strain and decrease the amplitude of the tumbling response.
Since Tap is the only receptor type to mediate a running response to light exposure, we investigated whether it could switch the sign of the response in multireceptor mutants, i.e., strains expressing a subset of the five chemoreceptors (Table 1). Indeed, we found that adding the Tap receptor to the ΔTsr ΔTrg ΔTap mutant, generating a ΔTsr ΔTrg strain, caused a switch from a tumbling response to a running response (Fig. 3A). This result provides independent validation of the running response observed in a Tap-only strain. By examining a number of multireceptor strains, we found that the presence of Tap was necessary but not alone sufficient to produce a running response. Thus, we observed a running response to light in a ΔAer strain but not in a ΔTrg strain (Fig. 3B).
FIG 3.
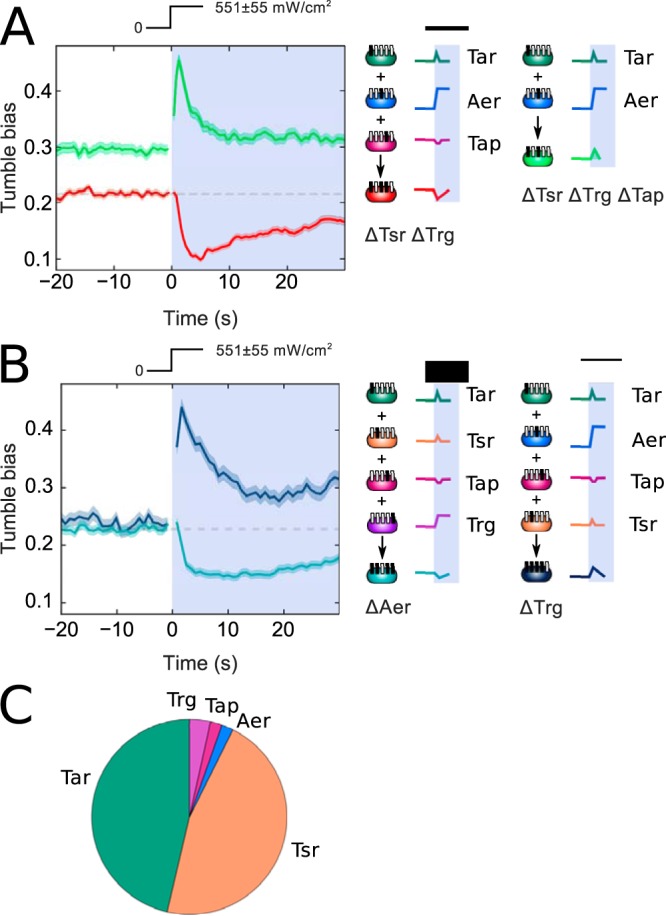
Effects of receptor composition on the blue light responses in multireceptor strains. (A) Tumble bias traces for ΔTsr ΔTrg (red), and ΔTsr ΔTrg ΔTap (green) strains. (B) Tumble bias traces for ΔTrg (dark blue) and ΔAer (cyan) strains. Schematics show responses observed for individual receptors present in these multireceptor strains. All responses were measured at a blue light intensity of 551 ± 55 mW/cm2, as indicated by the light intensity profile. Approximately 900 to 4,000 trajectories were used to calculate the average tumble bias at each time point. (C) Pie chart of the relative abundances of different receptor types in the wild-type strain, based on data from reference 21.
We expect receptor abundance in multireceptor strains to be similar to that in the wild-type strain, where the abundance of Tap is much lower than that of Tar or Tsr (Fig. 3C). Therefore, the running responses we observe must result from amplification of Tap-mediated signaling by other receptors. The importance of interactions between receptors in determining the overall response is further underlined by nonlinear effects of other receptors. For example, replacing the low-abundance Trg receptor with the similarly low-abundance Aer receptor resulted in a switch from a running response to a tumbling response, as illustrated by the ΔAer and ΔTrg strains (Fig. 3B).
Blue light exposure causes a reversible decrease in swimming velocity.
Despite primarily sensing different environmental signals, the five chemoreceptors in E. coli universally respond to the same blue light stimulus, one in the opposite direction from the other four. According to current understanding, Aer is the only receptor that binds a blue-light-absorbing chromophore (flavin adenine dinucleotide [FAD]) as a cofactor and therefore could be directly photosensitive (32). The mechanism of photosensitivity is unclear for the remaining four receptors. It has been speculated that E. coli monitors some intracellular parameter perturbed by the absorption of blue photons. Wright and coauthors suggested, for example, that the Tar receptor might be sensing perturbations in electron transport induced by blue light, which would affect the PMF (7). We explored whether the PMF hypothesis is a plausible explanation for the light sensitivity of chemoreceptors.
PMF is generated during respiration by electron and proton translocation across the membrane (33). It is used to synthesize ATP and to energize other processes in the cell, such as ion transport and flagellar rotation (33). It was shown previously that the flagellar rotation rate is linearly proportional to PMF under both high and low viscous loads, with the latter corresponding to the load on flagella in free-swimming bacteria (34, 35). Therefore, with all else being equal, the bacterial swimming velocity can serve as a proxy measure of PMF. If light does affect PMF, then we can expect to see corresponding trends in swimming velocity.
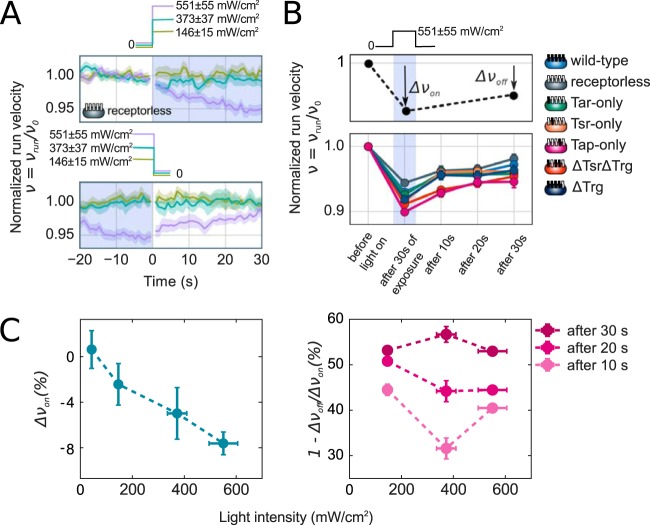
Velocity traces for the receptorless strain UU1250 at different light intensities are shown in Fig. 4A. Light exposure caused a gradual decrease in the swimming velocity. Since the receptorless strain does not exhibit a change in tumble bias in response to light (Fig. S6), trends in swimming velocity cannot be attributed to an imperfect run-tumble assignment (see Materials and Methods). Similar decreases in velocity were observed for all other strains assayed, including the wild-type strain (Fig. 4B). The magnitude of the decrease in normalized swimming velocity after 30 s of light exposure, Δνon, depended on the light intensity and varied from 0% at ≤44 mW/cm2 to ∼8% at 550 mW/cm2, on average (Fig. 4B). Moreover, the velocity decrease appeared to be reversible. About 50% of the velocity decrease was recovered within 30 s after the light was turned off, at all intensity levels. It is likely that full recovery could be achieved with sufficient waiting time, but our assay precluded measurements beyond ∼30 s. Under our experimental conditions, we could not distinguish between a velocity recovery in previously exposed bacteria and a velocity increase due to unexposed bacteria swimming into the field of view after 30 s (see Materials and Methods). Nevertheless, our results strongly suggest a reversible decrease in velocity, which is consistent with reversible perturbation of PMF by blue light exposure, as opposed to irreversible motor photodamage.
FIG 4.
Effects of light on running velocity. (A) Light exposure causing reversible decreases in swimming velocity. Normalized velocity traces are shown for the receptorless strain UU1250. Approximately 2,000 trajectories were used to calculate velocity at each time point. Light intensity is indicated by the trace color. The velocity is normalized to its prestimulus value (calculated in a 20-s window before light exposure [see Materials and Methods]). (B) Plot of the normalized swimming velocity for different E. coli strains. The normalized velocity was calculated in a 4-s window after 30 s of exposure to light and then 10, 20, and 30 s after the light was turned off. Light exposure and intensity are indicated by the intensity profile above the plot. (C) Dependence of velocity decrease and recovery on light intensity. Velocity decrease (Δνon) and velocity recovery (1 − Δνon/Δνoff) were calculated as illustrated by the schematic in panel B. Velocity recovery is shown 10, 20, and 30 s after light turn off, as indicated by the color.
DISCUSSION
A tumbling response to blue light in E. coli was first demonstrated in 1975 (4). Later, Wright et al. (7) identified two types of receptors essential for the tumbling response to blue light, namely, Tar and Aer. Our work shows that the response to light in E. coli is mediated by all of the receptor types, including Tsr, Trg, and Tap. Downstream from the receptors, the effect of light on the chemotaxis network is consistent with that of other tactic stimuli. Light thus emerges as a universal tactic stimulus that affects all five E. coli chemoreceptors. While four of five receptors mediate tumbling, or a repellent response to light, the Tap receptor mediates running, or an attractant response. We find that, despite being a low-abundance receptor, Tap is capable of determining the direction of the light response in multireceptor mutants, likely aided by amplification of Tap-mediated signaling by other receptors.
Receptor-receptor interactions that lead to signal amplification are mediated by a hexagonal lattice formed by the core functional units of chemotaxis, i.e., two heterotrimers of receptor homodimers, which bind one CheA homodimer, and two molecules of the coupling protein CheW (29). Formation of hexagonal lattice clusters was shown for two major receptors, Tsr and Tar, based on cryo-electron microscopic data (36) and on the cooperativity of dose-response curves (37), respectively. Moreover, cross-linking studies have demonstrated that both Aer and Trg, in the absence of other receptors, can form trimers of dimers, the structural units of a hexagonal lattice (29, 38, 39), although to our knowledge formation of higher-order structures has not been shown directly. While there have been no experimental studies of cluster formation by Tap to date, the data strongly suggest that most, if not all, receptor types participate in receptor-receptor interactions and can amplify each other’s signaling responses.
Our results on the effect of light on swimming velocity are consistent with the hypothesis that blue light perturbs electron transport or affects PMF, as well as with the previous observation that phototaxis in E. coli requires a functioning electron transport chain (5). Perturbation of electron transport (and therefore of cellular metabolism in general) by blue light may help explain the observation by Abana et al. that E. coli growth is slowed under similar light intensities (520 mW/cm2) (40). The mechanism of PMF perturbation is not clear, and our experiments do not provide clues to help elucidate it. We propose that light may cause photoreduction of electron carriers such as FAD in the cytoplasmic pool, thereby disrupting electron transport. FAD is a good candidate carrier, because it absorbs in the blue spectral region and is known to undergo photoconversion between its different redox and protonation states as a cofactor of light oxygen voltage (LOV) and blue light sensor using FAD (BLUF) flavoprotein light sensors (41–43). The alternative explanation is that PMF is affected because blue light disrupts the integrity of the membrane. There is evidence that prolonged exposure to UV light can cause membrane damage (44). However, any mechanism must be largely reversible, as our results demonstrate.
The different response kinetics that we observed for single-receptor mutants may reflect different blue-light-sensing mechanisms of individual receptors (Fig. 2; also see Fig. S8 in the supplemental material). Response times appear to fall into two categories, namely, fast (e.g., Tar), with the response being essentially immediate within the time resolution of our measurements and likely reflecting a direct light absorption process, and slow (e.g., Trg), with light-induced changes being presumably more indirect and taking longer to have an effect. In the Aer-only strain, the response is essentially immediate at higher intensities, which is consistent with direct sensing of light by Aer through the photoreduction of its cofactor FAD (45). However, as a receptor for “energy taxis,” Aer can also sense perturbations in electron transport, which have been proposed to occur through changes in the redox state of respiratory enzymes (46). We speculate that this sensing mechanism is reflected in the slower response times observed at lower light intensities (Fig. S8). In contrast, Tsr has been shown to respond directly to changes in PMF and may display fast response kinetics due to the low threshold needed to elicit a response (46). How the remaining receptors sense light remains unclear. Based on the response kinetics, we suspect that Tar responds to light or parameters that change immediately with light intensity, while Tap and Trg sense more indirect light-induced effects (Fig. 2).
To place our results in perspective, we compared the intensity values that we used with those that bacteria may actually encounter in nature. E. coli bacteria live a biphasic life cycle, with their primary habitat in the mammalian gut. Between being excreted from one host and finding the next one, E. coli bacteria inhabit nutrient-sparse water or soil environments (47). During the environmental phase of its lifestyle, E. coli can be exposed to the light from the sun. The intensity of the solar illumination at the surface of Earth is ∼140 mW/cm2 across the visible spectrum, with ∼60 mW/cm2 falling within the 60-nm band of the blue light response spectrum, as measured by Wright et al. (7). This value is comparable to the lowest intensity at which we observed significant responses for some of the strains in this study (i.e., 74 mW/cm2) (Fig. 2), although not for the wild-type strain (Fig. 1). Under different growth conditions, however, the wild-type strain exhibits a stronger response at these low light intensities (Fig. S5), consistent with observations by Wright and coworkers (7). Therefore, responses may be more significant for certain free-living E. coli strains. While the biological significance of the blue light response remains to be seen, our results indicate that migration of E. coli bacteria due to exposure to sunlight is plausible.
We propose that blue light may be a useful tool in investigations of chemotactic behavior. Because light is a universal stimulus for all E. coli receptors, it may provide a way to quantify interactions among different types of receptors. In addition, light is much easier to control both in time and in space, compared with chemicals. Therefore, using light as a stimulus may allow the study of tactic behavior of bacteria in a heterogeneous environment with multiple light gradients, thereby bridging the gap between the types of gradients bacteria likely encounter in nature and those that can be generated experimentally.
MATERIALS AND METHODS
Microbiology.
(i) Cell growth and media. Bacteria were grown for 20 to 24 h at 30°C from a single colony, in 1 ml of M9 minimal medium supplemented with 4 mg/ml succinate (1× M9 salts [12.8 g/liter Na2HPO4·7H2O, 3 g/liter KH2PO4, 0.5 g/liter NaCl, 1 g of NH4Cl]; 2 μM MgSO4; 0.1 mM CaCl2; 0.5 mM each Met, Leu, Thr, and His; 100 μg/ml thiamine; 4 mg/ml succinate) unless otherwise noted, with shaking at 265 rpm and with appropriate antibiotics if necessary (34 μg/liter chloramphenicol or 100 μg/liter ampicillin). The overnight culture was diluted 50-fold in 1 ml of the same medium and grown at 30°C for 8 to 12 h (to an optical density at 600 nm [OD600] of ∼0.25 to 0.3), with shaking at 265 rpm and with appropriate inducers if necessary. The following concentrations of inducers were used for strains with plasmids: 50 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for UU1250::pSB20, 1.5 μM sodium salicylate (NaSal) for UU1250::pTP1, 0.8 μM NaSal for UU1250::pA705, and 40 or 50 μM IPTG for PS2001::pMS164, as indicated.
The overday culture was harvested after 8 to 12 h, during the early exponential phase, by centrifugation (1,300 × g for 10 min) and was gently resuspended in the appropriate volume of motility buffer (27) (70 mM NaCl, 100 mM Tris-HCl [pH 7.5], 4 mg/ml succinate, 100 μM methionine) to reach a final OD600 of 0.15. Bacteria were placed back in the shaker to oxygenate the medium. Methionine was added to a final concentration of 100 μM prior to sample chamber assembly. Phenol was added to a final concentration of 2.5 mM where noted.
We note that different combinations of growth and motility media have been used in previous work on phototaxis in E. coli (7). Here, we chose a nonglycolytic substrate (succinate) consistent across both growth and motility media, but we report comparisons of different substrates used in the literature in the supplemental material.
(ii) Strains. Bacterial strains and plasmids used in this study are listed in Tables 1 and 2. Plasmid pTP1 was constructed from plasmid pKG117 by subcloning the wild-type tap gene between NdeI and BamHI restriction sites (Table 2). Synthesis and subcloning were performed by GenScript.
TABLE 2.
Plasmids used in this work
| Plasmid | Genotypea | Comments | Reference or source |
|---|---|---|---|
| pSB20 | Aer Ampr | IPTG-inducible wild-type Aer | 7 |
| pPA705 | Trg Cmr | NaSal-inducible wild-type Trg | 28 |
| pKG117 | Aer Cmr | NaSal-inducible wild-type Aer | 7 |
| pTP1 | Tap Cmr | NaSal-inducible wild-type Tap | This work |
| pMS164 | CheYD13K Cmr | IPTG-inducible CheYD13K | 52 |
Ampr, ampicillin resistance; Cmr, chloramphenicol resistance.
Two-dimensional swimming assay.
Slides (3 inches by 1 inch, product no. 3010; Thermo-Fisher) and coverslips (22 mm by 22 mm, no. 1; VWR) were sonicated in acetone for ∼15 min, rinsed, sonicated in KOH for 15 min, rinsed, and dried by centrifugation (1,000 rpm for 3 min). Cleaning was performed on the day of each experiment, because we found that storing cleaned slides in distilled water for even 1 day resulted in the accumulation of defects on the glass surface. Prior to each experiment, slides and coverslips were passivated with bovine serum albumin (BSA) (product no. B9000S; New England BioLabs) to prevent sticking of bacteria. Slides and coverslip were incubated with 2 mg/ml BSA for ∼20 min, rinsed with a copious amount of water, and dried with nitrogen. To assemble the chamber, a drop of motility buffer (5 μl) containing E. coli cells was placed on a slide and gently covered with a coverslip. Care was taken to prevent the formation of air bubbles. To prevent buffer evaporation, open sides were sealed with fast curing epoxy (Devcon 5 min epoxy). The distance between the slide and coverslip was determined by the thickness of the liquid layer of bacterial medium and was ∼10 μm, which approximately corresponds to the depth of field of our 20× objective.
Microscopy.
We used an inverted optical microscope (Axio Observer A1; Zeiss) with a 20× objective (A-Plan 20×/0.45 M27; Zeiss), in phase-contrast mode, to image swimming bacteria. For observation, bacteria were illuminated from the top with a halogen lamp (HAL 12 V/100 W). Light from the lamp was heat filtered and passed through a 500-nm long-pass filter (product no. ET500lp; Chroma) to exclude the possibility of excitation of bacteria by the observation light (Fig. 1B).
Excitation light from a blue light-emitting diode (LED) was introduced through the back port of the microscope. A blue LED (product no. M455L3; Thorlabs) with a collimation assembly (retaining ring, lens tube, and aspheric condenser lens [product no. ACL2520-A; Thorlabs]) was mounted using a Zeiss Axioskop microscope lamphouse port adapter (product no. SM1A23; Thorlabs). Excitation light was passed through a 440 ± 5-nm bandpass filter (product no. CT440/10bp; Chroma) and was directed toward the field of view by a 500-nm dichroic mirror (product no. 500dcxr; Chroma). To achieve even illumination of the field of view, we followed the standard procedure for Koehler illumination. The microscope field stop was opened to match the field of view (∼1.2 mm in diameter for the 20× objective).
The output light intensity of the LED was determined by the current from the LED driver (product no. LEDD1B; Thorlabs) controlled by a data acquisition (DAQ) card (product no. NI PCI-6221; National Instruments) and defined using a LabView interface. Neutral density (ND) filters of ND 1 and ND 0.5 (product no. NE10B ND and NE05B ND, respectively; Thorlabs) were installed in the filter slider to gain finer control over the resulting light intensity. Light intensity in the sample plane at different supply voltages was measured with a power meter (power meter 1916C equipped with photodiode sensor 918D-SL-OD3; Newport) placed on the microscope stage such that the illumination fitted the area of the sensor. To calculate light power density, the total power was divided by the illumination area. The unevenness of the illumination was estimated from the distribution of pixel brightness values in an image taken by the camera below its saturation and was found to be ∼10% of the mean. We used this value as an estimate for the error in light power density.
Movies of swimming bacteria were captured using a charge-coupled device (CCD) camera (Grasshopper 3; Point Grey Research) mounted at the microscope side port. Camera calibration with USAF target 1951 (product no. R3L3S1P; Thorlabs) gave us an estimate of the pixel size of ∼0.26 μm. The size of the area captured by the camera in the sample plane was 532 μm by 532 μm. Movies were recorded at a frame rate of 12 frames per second.
Data analysis.
(i) Trajectory preparation and filtering. Analysis of bacterial trajectories was performed using an automated workflow implemented in Python. First, we detected bacteria in each frame of a recorded movie using the OpenCV computer vision library (48). Then, coordinates were linked into trajectories using the trackPy package (Fig. 1B) (49). At that point, we removed all trajectories shorter than 1 s, or 12 frames, from further consideration. Then we calculated instantaneous linear velocities and accelerations and angular velocities and accelerations using 1-frame windows, following the procedure described by Dufour et al. (50).
Next, we used the following approach to filter out spurious trajectories that belonged to bacteria tethered to the surface, drifting, or swimming too slowly. For every trajectory, we calculated the average angular velocity and the 95th percentile of the linear velocity. The two-dimensional distribution of all trajectories in these coordinates contains two clusters, with one cluster corresponding to normally swimming bacteria and the less populated cluster containing trajectories of very slow or surface-tethered bacteria (see Fig. S3 in the supplemental material). For each bacterial strain, we found the two coordinates of the maximum of the swimming cluster, i.e., the most probable values of angular velocity and 95th percentile of velocity, and kept only the trajectories that lay within a certain radius R from the maximum. With the exception of the few strains that exhibited very strong responses to light, we defined R as R = 4<MAD>, where <MAD> is the median absolute deviation (MAD) from the maximum of the distribution, averaged across all strains with a functional chemotaxis network. Filtering was performed separately for different strains, to avoid bias due to variations in the swimming behavior. We found that filtering did not affect disproportionately the trajectories of the bacteria that had been exposed to light; the fractions of trajectories within 4<MAD> for bacteria before, during, and after light exposure were approximately the same.
The area accessible to bacteria in a slide-coverslip chamber is larger than the illuminated area. Therefore, unexposed bacteria swimming from outside the area illuminated by blue light could subsequently swim into the field of view and affect the observed kinetics. However, the illuminated area is still larger than the observation area captured by the camera, and such bacteria would have to swim 0.2 to 0.3 mm to reach the field of view. To estimate on what timescale these bacteria could contribute to the observed kinetics, we calculated the mean square displacement (MSD) for trajectories of the wild-type E. coli strain (strain RP437) as a function of time. From a power law fit of the MSD, we estimated that it would take unexposed bacteria at least 30 s to reach the field of view, although the time would vary for different strains, depending on the swimming velocity and tumbling frequency. Based on this estimate, we considered only the portions of tumble bias and velocity time traces within 30 s after the light was turned off or on.
(ii) Run-tumble assignment. To assign run and tumble states, we used a hidden Markov model with gaussian-distributed emissions, similar to that described by Dufour et al. (50). In this type of model, the state of the system (e.g., run or tumble) is not directly observed (hidden) but its outputs (emissions), such as velocity, acceleration, and angular acceleration, can be observed. The hidden states and known outputs are related by an emission probability, while the transition between states is given by a transition matrix.
We implemented the hidden Markov model using the Python package hmmlearn to infer the sequence of hidden swimming states from time traces of the observable parameters (51). Parameters of the model (the transition probability matrix and the emission probabilities of the observables) are estimated from a reference data set consisting of >20,000 prestimulus trajectories of wild-type bacteria. Training is performed iteratively. At each iteration, velocities and accelerations are normalized to the average swimming velocity (the 95th percentile of the velocity is used over the first iteration), model parameters are estimated from a resulting sequence of observables, and an optimal sequence of states is inferred. The process is repeated until the change in normalized velocity between two consecutive iterations is <2%. To account for the variation in swimming velocity within one trajectory, we used a model with three states, namely, fast run, slow run, and tumble. The running velocity was calculated from states corresponding to a fast run. Although we performed run-tumble assignments for all of the collected trajectories, only those longer than 100 frames were used for the plots shown here, because shorter trajectories do not contain enough points for accurate run-tumble assignments, as shown by Dufour et al. (50).
A hidden Markov model trained on the data for the wild-type strain learned accurate definitions of motility states in terms of swimming parameters (velocity, acceleration, and angular acceleration). However, transition probabilities learned from wild-type data may not be applicable to strains with a tumble bias significantly different from that of the wild-type strain. For this reason, we used a gaussian mixture model trained on wild-type data to detect run and tumble states in the ΔCheB strain, for which we expect a significantly greater tumble bias than in the wild-type strain. The gaussian mixture model was implemented using the Python package scikit-learn (51).
We validated our assignment by comparing tumble bias time traces obtained with the aforementioned analysis to those obtained with alternative run-tumble assignment criteria introduced by Alon et al. (52). Under these alternate criteria, bacteria were considered to be tumbling if the velocity was below the 90th percentile of velocities divided by 2 and if the angular velocity was above 6 rad/s. We compared tumble bias traces obtained with each assignment method. Although the absolute values of tumble bias were offset depending on the specific method used to assign runs and tumbles, the trends (responses and adaptation to light exposure) did not depend on the analysis method. In addition, following Wright et al., we used angular velocity or rate of change in direction (RCD) (7), which is proportional to tumble bias over a range of values (53), as a population measure of the E. coli chemotactic response. We found similar trends in the RCD traces as well. Finally, we also calculated the tumble frequency as an alternative to tumble bias. As shown in Fig. S4 in the supplemental material for one of the strains under study, the change in tumble frequency upon light exposure followed the same trend as tumble bias. The prestimulus value of tumble frequency, ∼1.2 s−1, is in agreement with the literature (54). Therefore, we conclude that our results are robust with respect to the method of analysis. The Python library that we developed for detection and analysis of trajectories is available at the GitHub repository (https://github.com/tatyana-perlova/py-taxis).
For each movie, the average tumble bias across all trajectories was calculated in each movie frame. Tumble bias traces from movies taken under the same conditions were pooled together and averaged, yielding a smooth tumble bias trace as a function of time for each strain or condition. Each tumble bias trace in the figures is the result of combining at least two biological replicas. We used the standard error between the tumble biases of individual bacterial trajectories as a measure of uncertainty of the average tumble bias (Fig. 1C, shaded areas). The amplitude A was defined as the maximum change in tumble bias upon light exposure, compared to the prestimulus value, and was calculated over a 0.5-s window to minimize the contribution of noise (Fig. 2B). For extraction of response and adaptation times, tumble bias (TB) response traces during light exposure were fitted to the following equation:
The logarithmic term was added to account for the slow adaptation kinetics for some of the receptor mutants. The response time (tresp) was defined as the time it takes for the resulting fitted function to reach 90% ± 1% of the amplitude (Fig. 2B) or
Similarly, the adaptation time (tadap) was defined as the time it takes for the difference between the prestimulus value of tumble bias and the fitting function to reach 50% ± 1% of the amplitude (Fig. 2B). The errors in determining responses and adaptation times were calculated as the standard error of the tumble bias at tresp or tadap (width of the shaded area around the tumble bias trace) (Fig. 2) divided by the slope of the linear fit to the trace at that point. For plotting, tumble bias time traces were averaged using a 10-frame moving window.
(iii) Running velocity analysis. The running velocity was defined as the velocity during frames assigned to runs. For the nontumbling receptorless strain UU1250, we used velocity in every frame. Time traces of the normalized run velocity (Fig. 4A) were calculated as follows: each individual velocity trace was divided by the prestimulus velocity calculated in a 20-s window prior to light exposure. Traces were then shifted to align light-on or light-off frames and were averaged using a 10-frame nonoverlapping rolling window. Because the tumble assignment procedure is not perfect, not all tumbles are detected, which is reflected in a sudden decrease in swimming velocity immediately after light exposure for strains with a tumbling response to light (data not shown). To measure the effect of light on the running velocity while minimizing the effect of false-negative results, we used the following procedure: for each individual movie, velocities were calculated in 4-s windows after 30 s of light exposure and then 10, 20, and 30 s after the light was turned off (Fig. 4B). Therefore, we do not take into account velocity immediately after the light is turned on. For Aer-only and Trg-only strains, tumble bias does not return to prestimulus values even after 30 s of exposure. Therefore, these strains were excluded from this analysis. Observation was limited to 30 s to avoid contributions of unexposed bacteria swimming into the field of view. Velocities calculated in this way for each trace were then normalized to the prestimulus velocity calculated in a 20-s window and averaged across all of the traces for a particular strain. Decreases and recovery of normalized run velocity were calculated as illustrated in Fig. 4B, by averaging data points for different strains. The velocity decrease (Δνon) was an average decrease of normalized run velocity after 30 s of light exposure. The velocity recovery characterizes the fraction of Δνon recovered 30 s after light turn off and was calculated as (Δνon − Δνoff)/Δνon (Fig. 4B).
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Gennis, Christopher Rao, George Ordal, and Yann Dufour for advice and discussion. We thank all members of the Chemla laboratory and the Gruebele group, especially Roshni Bano, for help and suggestions throughout the course of this work. We thank Anton Goloborodko for help with data analysis. Mutant strains used in this study were kindly provided by John Parkinson.
Funding was provided by National Science Foundation grant PHY-1430124 (Physics Frontiers Center [PFC]: Center for the Physics of Living Cells [CPLC]).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00762-18.
REFERENCES
- 1.Häder DP. 1987. Photosensory behavior in procaryotes. Microbiol Rev 51:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armitage JP, Hellingwerf KJ. 2003. Light-induced behavioral responses (‘phototaxis’) in prokaryotes. Photosynth Res 76:145–155. doi: 10.1023/A:1024974111818. [DOI] [PubMed] [Google Scholar]
- 3.Purcell EB, Crosson S. 2008. Photoregulation in prokaryotes. Curr Opin Microbiol 11:168–178. doi: 10.1016/j.mib.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Taylor BL, Koshland DE. 1975. Intrinsic and extrinsic light responses of Salmonella typhimurium and Escherichia coli. J Bacteriol 123:557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor BL, Miller JB, Warrick HM, Koshland DE. 1979. Electron acceptor taxis and blue light effect on bacterial chemotaxis. J Bacteriol 140:567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H, Inokuchi H, Adler J. 1995. Phototaxis away from blue light by an Escherichia coli mutant accumulating protoporphyrin IX. Proc Natl Acad Sci U S A 92:7332–7336. doi: 10.1073/pnas.92.16.7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright S, Walia B, Parkinson JS, Khan S. 2006. Differential activation of Escherichia coli chemoreceptors by blue-light stimuli. J Bacteriol 188:3962–3971. doi: 10.1128/JB.00149-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mears PJ, Koirala S, Rao CV, Golding I, Chemla YR. 2014. Escherichia coli swimming is robust against variations in flagellar number. eLife 3:e01916. doi: 10.7554/eLife.01916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebbapragada A, Johnson MS, Harding GP, Zuccarelli AJ, Fletcher HM, Zhulin IB, Taylor BL. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci U S A 94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sourjik V, Wingreen NS. 2012. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol 24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grebe TW, Stock J. 1998. Bacterial chemotaxis: the five sensors of a bacterium. Curr Biol 8:R154–R157. doi: 10.1016/S0960-9822(98)00098-0. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, Sourjik V. 2012. Opposite responses by different chemoreceptors set a tunable preference point in Escherichia coli pH taxis. Mol Microbiol 86:1482–1489. doi: 10.1111/mmi.12070. [DOI] [PubMed] [Google Scholar]
- 13.Mao H, Cremer PS, Manson MD. 2003. A sensitive, versatile microfluidic assay for bacterial chemotaxis. Proc Natl Acad Sci U S A 100:5449–5454. doi: 10.1073/pnas.0931258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paster E, Ryu WS. 2008. The thermal impulse response of Escherichia coli. Proc Natl Acad Sci U S A 105:5373–5377. doi: 10.1073/pnas.0709903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor BL, Zhulin IB, Johnson MS. 1999. Aerotaxis and other energy-sensing behavior in bacteria. Annu Rev Microbiol 53:103–128. doi: 10.1146/annurev.micro.53.1.103. [DOI] [PubMed] [Google Scholar]
- 16.Baker MD, Wolanin PM, Stock JB. 2006. Signal transduction in bacterial chemotaxis. Bioessays 28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 17.Berg HC. 2000. Motile behavior of bacteria. Phys Today 53:24. doi: 10.1063/1.882934. [DOI] [Google Scholar]
- 18.Hazelbauer GL, Falke JJ, Parkinson JS. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci 33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vladimirov N, Sourjik V. 2009. Chemotaxis: how bacteria use memory. Biol Chem 390:1097–1104. doi: 10.1515/BC.2009.130. [DOI] [PubMed] [Google Scholar]
- 20.Yuan J, Branch RW, Hosu BG, Berg HC. 2012. Adaptation at the output of the chemotaxis signalling pathway. Nature 484:233–236. doi: 10.1038/nature10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Hazelbauer GL. 2004. Cellular stoichiometry of the components of the chemotaxis signaling complex. J Bacteriol 186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barnakov AN, Barnakova LA, Hazelbauer GL. 1998. Comparison in vitro of a high- and a low-abundance chemoreceptor of Escherichia coli: similar kinase activation but different methyl-accepting activities. J Bacteriol 180:6713–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto K, Macnab RM, Imae Y. 1990. Repellent response functions of the Trg and Tap chemoreceptors of Escherichia coli. J Bacteriol 172:383–388. doi: 10.1128/jb.172.1.383-388.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berlman IB. 1971. Handbook of fluorescence spectra of aromatic molecules, 2nd ed Academic Press, New York, NY. [Google Scholar]
- 25.Bibikov S, Miller A, Gosink KK, Parkinson JS. 2004. Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J Bacteriol 186:3730–3737. doi: 10.1128/JB.186.12.3730-3737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vladimirov N, Løvdok L, Lebiedz D, Sourjik V. 2008. Dependence of bacterial chemotaxis on gradient shape and adaptation rate. PLoS Comput Biol 4:e1000242. doi: 10.1371/journal.pcbi.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min TL, Mears PJ, Golding I, Chemla YR. 2012. Chemotactic adaptation kinetics of individual Escherichia coli cells. Proc Natl Acad Sci U S A 109:9869–9874. doi: 10.1073/pnas.1120218109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studdert CA, Parkinson JS. 2004. Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci U S A 101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. 2012. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci U S A 109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkinson JS, Hazelbauer GL, Falke JJ. 2015. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol 23:257–266. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sourjik V, Berg HC. 2002. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci U S A 99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor BL. 2007. Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol Microbiol 65:1415–1424. doi: 10.1111/j.1365-2958.2007.05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White D, Drummond J, Fuqua C. 2011. The physiology and biochemistry of prokaryotes, 4th ed, chapter 4, Membrane bioenergetics: the proton potential. Oxford University Press, Oxford, UK. [Google Scholar]
- 34.Fung DC, Berg HC. 1995. Powering the flagellar motor of Escherichia coli with an external voltage source. Nature 375:809–812. doi: 10.1038/375809a0. [DOI] [PubMed] [Google Scholar]
- 35.Gabel CV, Berg HC. 2003. The speed of the flagellar rotary motor of Escherichia coli varies linearly with protonmotive force. Proc Natl Acad Sci U S A 100:8748–8751. doi: 10.1073/pnas.1533395100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. 2007. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci U S A 104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Endres RG, Oleksiuk O, Hansen CH, Meir Y, Sourjik V, Wingreen NS. 2008. Variable sizes of Escherichia coli chemoreceptor signaling teams. Mol Syst Biol 4:211. doi: 10.1038/msb.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gosink KK, del Carmen Burón-Barral M, Parkinson JS. 2006. Signaling interactions between the aerotaxis transducer Aer and heterologous chemoreceptors in Escherichia coli. J Bacteriol 188:3487–3493. doi: 10.1128/JB.188.10.3487-3493.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ames P, Studdert CA, Reiser RH, Parkinson JS. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci U S A 99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abana CM, Brannon JR, Ebbott RA, Dunigan TL, Guckes KR, Fuseini H, Powers J, Rogers BR, Hadjifrangiskou M. 2017. Characterization of blue light irradiation effects on pathogenic and nonpathogenic Escherichia coli. MicrobiologyOpen doi: 10.1002/mbo3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura S, Nakamura T, Ogura Y. 1963. Absorption spectrum of flavin mononucleotide semiquinone. J Biochem 53:143–146. doi: 10.1093/oxfordjournals.jbchem.a127669. [DOI] [PubMed] [Google Scholar]
- 42.Losi A, Gärtner W. 2011. Old chromophores, new photoactivation paradigms, trendy applications: flavins in blue light-sensing photoreceptors. Photochem Photobiol 87:491–510. doi: 10.1111/j.1751-1097.2011.00913.x. [DOI] [PubMed] [Google Scholar]
- 43.Conrad KS, Manahan CC, Crane BR. 2014. Photochemistry of flavoprotein light sensors. Nat Chem Biol 10:801–809. doi: 10.1038/nchembio.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenzie K, Maclean M, Grant MH, Ramakrishnan P, MacGregor SJ, Anderson JG. 2016. The effects of 405 nm light on bacterial membrane integrity determined by salt and bile tolerance assays, leakage of UV-absorbing material and SYTOX green labelling. Microbiology 162:1680–1688. doi: 10.1099/mic.0.000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kao Y-T, Saxena C, He T-F, Guo L, Wang L, Sancar A, Zhong D. 2008. Ultrafast dynamics of flavins in five redox states. J Am Chem Soc 130:13132–13139. doi: 10.1021/ja8045469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edwards JC, Johnson MS, Taylor BL. 2006. Differentiation between electron transport sensing and proton motive force sensing by the Aer and Tsr receptors for aerotaxis. Mol Microbiol 62:823–837. doi: 10.1111/j.1365-2958.2006.05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blount ZD. 2015. The unexhausted potential of E. coli. Elife 4:e05826. doi: 10.7554/eLife.05826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bradski G. 2000. The OpenCV Library. Dr Dobb’s http://www.drdobbs.com/open-source/the-opencv-library/184404319. [Google Scholar]
- 49.Allan D, Caswell TA, Keim N, Boulogne F, Perry RW, Uieda L. 2014. Trackpy v030 http://soft-matter.github.io/trackpy/v0.3.0/index.html. [Google Scholar]
- 50.Dufour YS, Gillet S, Frankel NW, Weibel DB, Emonet T. 2016. Direct correlation between motile behavior and protein abundance in single cells. PLoS Comput Biol 12:e1005041. doi: 10.1371/journal.pcbi.1005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830. http://www.jmlr.org/papers/volume12/pedregosa11a/pedregosa11a.pdf. [Google Scholar]
- 52.Alon U, Camarena L, Surette MG, Aguera y Arcas B, Liu Y, Leibler S, Stock JB. 1998. Response regulator output in bacterial chemotaxis. EMBO J 17:4238–4248. doi: 10.1093/emboj/17.15.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khan S, Castellano F, Spudich JL, McCray JA, Goody RS, Reid GP, Trentham DR. 1993. Excitatory signaling in bacteria probed by caged chemoeffectors. Biophys J 65:2368–2382. doi: 10.1016/S0006-3495(93)81317-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mittal N, Budrene EO, Brenner MP, Van Oudenaarden A. 2003. Motility of Escherichia coli cells in clusters formed by chemotactic aggregation. Proc Natl Acad Sci U S A 100:13259–13263. doi: 10.1073/pnas.2233626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parkinson JS, Houts SE. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol 151:106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Min TL, Mears PJ, Chubiz LM, Rao CV, Golding I, Chemla YR. 2009. High-resolution, long-term characterization of bacterial motility using optical tweezers. Nat Methods 6:831–835. doi: 10.1038/nmeth.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bibikov SI, Biran R, Rudd KE, Parkinson JS. 1997. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol 179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.