Although ybeY gene belongs to the universal bacterial core genome, its biological function is incompletely understood. Here, we show that YbeY is critical for fitness and host-microbe interaction in the plant pathogen Agrobacterium tumefaciens. Consistent with the reported endoribonuclease activity of YbeY, A. tumefaciens YbeY acts as a RNase involved in maturation of 16S rRNA. This report adds a worldwide plant pathogen and natural genetic engineer of plants to the growing list of bacteria that require the conserved YbeY protein for host-microbe interaction.
KEYWORDS: Agrobacterium tumefaciens, Hfq, RNase, YbeY, iTRAQ, rRNA, ribosome, virulence
ABSTRACT
Riboregulation involving regulatory RNAs, RNA chaperones, and ribonucleases is fundamental for the rapid adaptation of gene expression to changing environmental conditions. The gene coding for the RNase YbeY belongs to the minimal prokaryotic genome set and has a profound impact on physiology in a wide range of bacteria. Here, we show that the Agrobacterium tumefaciens ybeY gene is not essential. Deletion of the gene in the plant pathogen reduced growth, motility, and stress tolerance. Most interestingly, YbeY is crucial for A. tumefaciens-mediated T-DNA transfer and tumor formation. Comparative proteomics by using isobaric tags for relative and absolute quantitation (iTRAQ) revealed dysregulation of 59 proteins, many of which have previously been found to be dependent on the RNA chaperone Hfq. YbeY and Hfq have opposing effects on production of these proteins. Accumulation of a 16S rRNA precursor in the ybeY mutant suggests that A. tumefaciens YbeY is involved in rRNA processing. RNA coimmunoprecipitation-sequencing (RIP-Seq) showed binding of YbeY to the region immediately upstream of the 16S rRNA. Purified YbeY is an oligomer with RNase activity. It does not physically interact with Hfq and thus plays a partially overlapping but distinct role in the riboregulatory network of the plant pathogen.
IMPORTANCE Although ybeY gene belongs to the universal bacterial core genome, its biological function is incompletely understood. Here, we show that YbeY is critical for fitness and host-microbe interaction in the plant pathogen Agrobacterium tumefaciens. Consistent with the reported endoribonuclease activity of YbeY, A. tumefaciens YbeY acts as a RNase involved in maturation of 16S rRNA. This report adds a worldwide plant pathogen and natural genetic engineer of plants to the growing list of bacteria that require the conserved YbeY protein for host-microbe interaction.
INTRODUCTION
The highly conserved ybeY gene belongs to the postulated minimal bacterial genome and encodes an RNase containing the UPF0054 protein motif (1, 2). The human protein C21orf57 is predicted to be a homologue of bacterial YbeY proteins, suggesting a wide phylogenetic distribution of this protein (3). The structure of YbeY partly resembles the MID domain of Argonaute proteins, which are essential for RNA-mediated regulation in eukaryotes (4–6). YbeY is an endoribonuclease involved in processing of rRNAs (especially 16S rRNA) in a broad range of bacteria, including Escherichia coli (7–9), Vibrio cholerae (10), Thermus thermophilus (11), and Yersinia enterocolitica (12), and also in the chloroplast (13). In concert with RNase R, YbeY is further involved in quality control of 70S ribosomes in E. coli (8) and, together with RNase E/G, it is involved in the maturation of the 4.5S RNA of the signal recognition particle in Corynebacterium glutamicum (14). Aside from these specific functions, YbeY plays a global role in gene regulation and influences expression of more than 1,000 genes in E. coli (9) and up to 100 genes in T. thermophilus (11), but it is presently unclear how.
Due to its role in RNA metabolism, deletion of ybeY usually impacts viability and susceptibility toward various stresses, e.g., heat stress in E. coli or certain antibiotics in V. cholerae (10, 15, 16). In some bacteria, for example V. cholerae, ybeY is an essential gene that can be deleted only in the presence of a plasmid-carried copy (10). In bacteria interacting with eukaryotes such as the plant symbiont Sinorhizobium meliloti or the human pathogens V. cholerae, Y. enterocolitica, and Brucella abortus, YbeY is critically important for successful host interaction (4, 10, 12, 17).
Another striking observation is that YbeY affects sRNA regulation and physiology in S. meliloti very much like the RNA chaperone Hfq (4). Hfq is a widespread RNA-binding protein (18). It forms a homohexamer exhibiting proximal, lateral, and distal RNA-binding surfaces (19–24). Hfq supports duplex formation of sRNAs and mRNAs by increasing their local concentration, mediating structural rearrangements, and annealing of both RNA molecules (25, 26). Hfq also influences the stability of interacting RNAs by inducing or inhibiting their degradation. Deletion of hfq usually results in a severe reduction of viability, including reduced growth, motility, and stress tolerance (27–34).
Agrobacterium tumefaciens is the causal agent of crown gall disease on mainly dicotyledonous plants. Its unique ability to transfer part of its own DNA (i.e., transfer DNA [T-DNA]) into eukaryotic hosts is responsible for Agrobacterium being the most valuable biotechnological tool for genetic manipulation of agronomically important plants like maize (35), soybean (36), and cotton (37) with an ever-increasing number of susceptible plants (38). The genome of the natural genetic engineer A. tumefaciens is divided into four replicons: a circular chromosome, a linear chromosome, the At plasmid, and the Ti (tumor-inducing) plasmid (39). Upon perception of signal molecules from plant wounds (e.g., phenolic compounds, sugars, and low pH), the VirAG two-component system initiates the infection process by activating virulence gene (vir) expression (40). The vir gene-encoded VirB/D4 type IV secretion system (T4SS) transfers a single-stranded portion of the Ti plasmid (T-DNA), along with several Vir proteins, into the plant cell. Integration of the T-DNA into the plant chromosome reprograms plant gene expression resulting in phytohormone and opine biosynthesis.
Recent RNA-sequencing studies in A. tumefaciens revealed more than 600 potential sRNAs, suggesting a large posttranscriptional network involved in the coordination of the unique physiology of this phytopathogen (41–44). Hfq plays a fundamental role in riboregulation in A. tumefaciens since it was shown by RNA coimmunoprecipitation-sequencing (RIP-Seq) analysis to bind more than 200 sRNAs and nearly one-third of all mRNAs (41). In an hfq mutant, the abundance of 136 proteins, many of them involved in ABC transport systems and motility, was altered in stationary phase. Finally, an A. tumefaciens hfq mutant exhibited severe defects in nutrient acquisition and motility and was inefficient in tumor formation (41, 45).
Here, we classify the A. tumefaciens YbeY ortholog Atu0358 as another chromosomally encoded virulence factor with a major impact on Agrobacterium physiology and tumor formation on plants. The absence of YbeY affects the biosynthesis of many Hfq-dependent proteins, albeit in an opposing direction. Cumulative evidence suggests that both proteins have different biochemical functionalities. While Hfq is an RNA chaperone, YbeY is a RNase involved in rRNA processing.
RESULTS
Atu0358 is the A. tumefaciens YbeY homologue.
The fundamental influence of riboregulation on the fitness and virulence of plant-associated alphaproteobacteria (46) motivated us to examine the impact of YbeY in the phytopathogen A. tumefaciens. The Atu0358 protein shares significant homology with other YbeY homologs (Fig. 1A). This assumption is further supported by the shared synteny of the ybeY locus in A. tumefaciens, S. meliloti, B. abortus, V. cholerae, and E. coli, where the ybeY gene is flanked upstream by the phosphate starvation-inducible gene phoH (atu0357) and downstream by a hemolysin-encoding gene (atu0359) (Fig. 1B). Transcriptome sequencing (RNA-Seq) data (42) show that ybeY is expressed under normal and virulence conditions and that it is part of a polycistronic operon with a transcription start site downstream of atu0357 (Fig. 1C).
FIG 1.
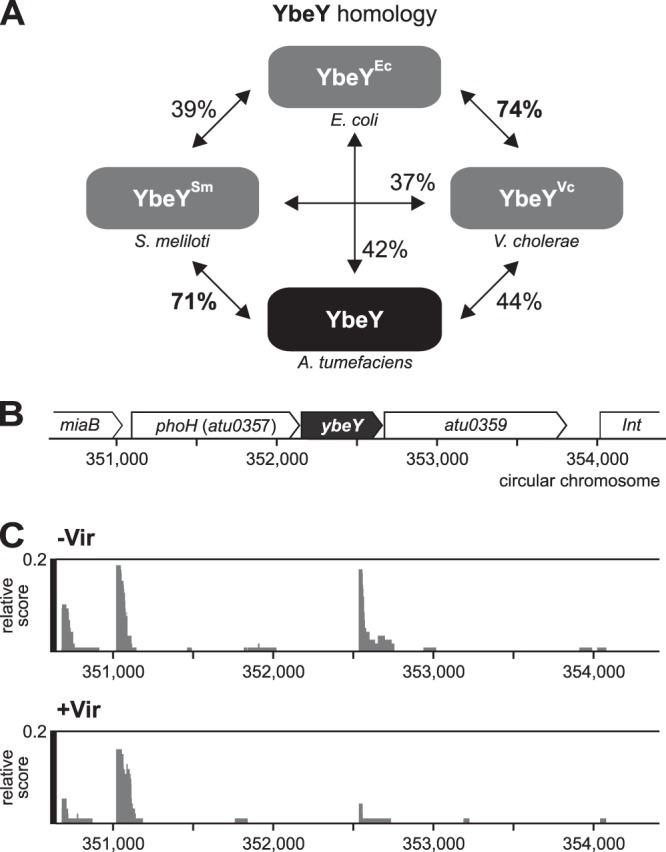
Genetic context and transcription of the A. tumefaciens ybeY. (A) Atu0358 shares high sequence identity with the YbeY orthologues from S. meliloti, V. cholerae, and E. coli. (B) The ybeY gene cluster in A. tumefaciens consists of the phosphate starvation-inducible protein gene phoH (atu0357), ybeY (atu0358), and the hemolysin gene atu0359. (C) Data from the differential RNA-Seq analysis by Wilms’ et al. (42). The transcription start site of ybeY lies upstream of phoH. cDNA libraries were generated from cultures grown without (–Vir) and with (+Vir) virulence induction by acetosyringone. Screenshots of the results after treatment with terminator 5ʹ-phosphate-dependent exonuclease (TEX) to enrich for primary 5ʹ ends are shown.
Expression of ybeY is growth phase regulated, and YbeY is less abundant than Hfq.
To assess the cellular amounts of the YbeY protein, we fused the chromosomal ybeY gene to a 3×Flag sequence at the 3ʹ end, resulting in a C-terminally tagged YbeY3×Flag protein. We compared the abundance of the YbeY3×Flag protein in complex medium from the exponential growth phase (optical density at 600 nm [OD600] = 0.5) to the stationary growth phase (OD600 = 1.5) with that of a chromosomally encoded Hfq3×Flag protein, which as previously shown (41) was present in equal amounts throughout all growth phases (Fig. 2A). In contrast, YbeY3×Flag was least abundant in the exponential phase and gradually increased throughout growth reaching a maximum in the stationary phase (Fig. 2B). Detection of the Flag-tagged proteins on one gel revealed that Hfq exceeded YbeY amounts at least 10-fold even in the stationary phase (Fig. 2C).
FIG 2.
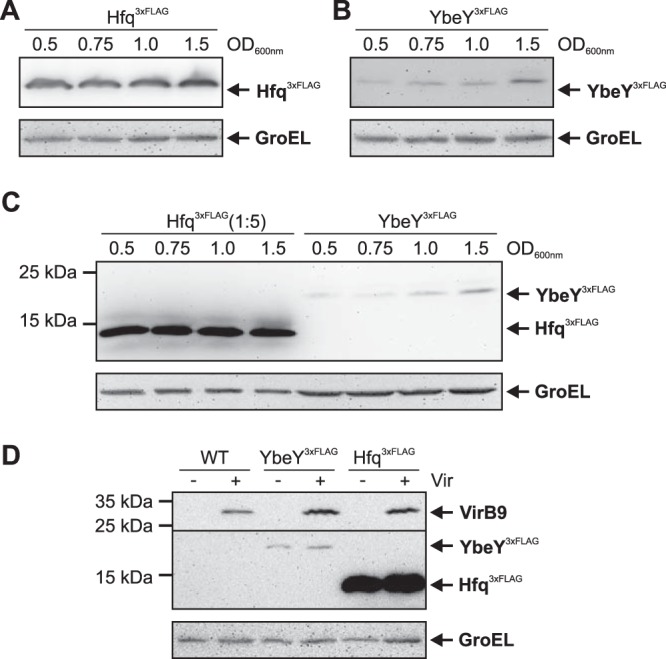
Growth-phase-dependent production of YbeY3×Flag. (A and B) The A. tumefaciens YbeY3×Flag and Hfq3×Flag strains were cultivated in YEB medium and cells were harvested from the exponential (OD600 = 0.5) to the stationary (OD600 = 1.5) growth phases. Proteins were separated by SDS-PAGE and detected via a monoclonal anti-Flag M2 antibody after Western blotting. GroEL served as a loading control. (C) Direct comparison of HfqFlag (1:5-diluted protein extracts) and YbeYFlag amounts on one Western blot. (D) WT, Hfq3×Flag, and YbeY3×Flag strains were cultivated under noninduced (–Vir) and virulence-induced (+Vir) conditions. Protein amounts were independent of the virulence inducer acetosyringone (AS). The membrane was cut (line), and the T4SS protein VirB9 was detected via a VirB9-specific antibody (upper part) to verify successful virulence induction.
Neither YbeY nor Hfq are virulence regulated because YbeY3×Flag and Hfq3×Flag amounts were unaffected in minimal medium under noninduced (–Vir) and virulence-induced (+Vir, in the presence of acetosyringone) (Fig. 2D). As in complex medium, Hfq3×Flag was much more abundant than YbeY3×Flag. Successful virulence induction was validated by immunodetection of the virulence-specific VirB9 protein, which was exclusively produced under +Vir conditions.
Growth, motility, and stress resistance depend on YbeY.
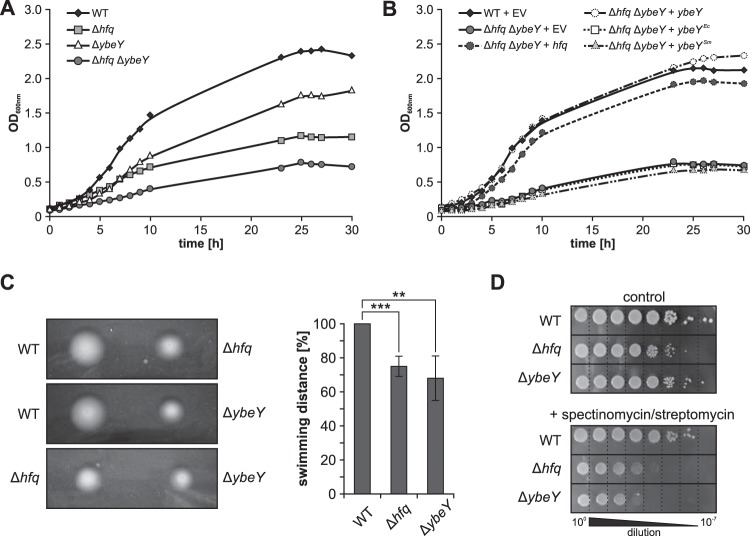
The successful construction of a ybeY mutant (ΔybeY) by deleting the ybeY open reading frame (ORF) from the chromosome demonstrated that the A. tumefaciens gene is not essential. We also obtained a Δhfq ΔybeY double mutant. However, this strain could not be propagated over longer time periods (see below). To determine the impact of YbeY on A. tumefaciens physiology, we analyzed the growth behavior of the wild type (WT) and Δhfq, ΔybeY, and a Δhfq ΔybeY deletion strains in YEB complex medium (Fig. 3A) and observed the previously described severe growth defect of the hfq mutant (45). Deletion of ybeY alone resulted in a less pronounced growth defect and the Δhfq ΔybeY double mutant exhibited the most severe growth defect, which could be rescued by ectopically expressed hfq or ybeY from A. tumefaciens (Fig. 3B). In contrast, ybeY orthologues from E. coli (YbeYEc) and S. meliloti (YbeYSm), as well as the empty vector control (EV), were unable to complement the growth defect of the Δhfq ΔybeY strain. Successful expression of both ybeYEc and ybeYSm in the double mutant was confirmed by Northern blot analysis (Fig. S1).
FIG 3.
YbeY influences A. tumefaciens growth and motility. (A) Growth experiments with WT, Δhfq, ΔybeY, and Δhfq ΔybeY strains revealed reduced growth of the ybeY mutant compared to the WT. Still, growth deficiency was less severe than in the hfq mutant. Deletion of both genes (Δhfq ΔybeY) led to a severe growth defect. (B) Growth deficiencies of the Δhfq ΔybeY mutant were rescued by ectopic expression of hfq or ybeY from A. tumefaciens. Homologues from E. coli (ybeYEc) or S. meliloti (ybeYSm) did not complement the growth defect. (C) Swimming of A. tumefaciens WT, Δhfq, and ΔybeY strains on low concentration agar. Swimming distance of Δhfq (***, P = 0.001) and ΔybeY (**, P = 0.009) strains was reduced to about 70% compared to WT. (D) Cultures of WT, Δhfq and ΔybeY strains were grown to exponential phase (OD600 = 0.5), and serial dilutions were applied to LB agar plates. WT growth and ΔybeY strain growth were comparable, whereas the growth of the Δhfq strain was reduced ∼10-fold compared to WT growth (upper panel). Addition of 10 μg/ml spectinomycin plus 3 μg/ml streptomycin had the strongest effect on the ybeY deletion strain but also inhibited growth of the hfq mutant (lower panel). EV, empty vector.
Presumably due the severe growth defect, the double mutant could not be maintained and was therefore excluded from all subsequent experiments. Like the hfq mutant, the ybeY single mutant showed reduced motility (Fig. 3C). The swimming distance of both strains on low-concentration agar plates reached only ∼70% of the WT radius.
The stress sensitivity of the WT, Δhfq and ΔybeY strains was examined on agar plates in the presence of diverse stressors. Cultures were grown to exponential phase (OD600 ∼ 0.5) and serially diluted (100 to 10−7) prior to spotting on the plates. The growth of all strains on Luria-Bertani (LB) agar (Fig. 3D, upper panel) was comparable to growth in liquid media, e.g., the growth of the hfq mutant was reduced more severely than the growth of the ybeY mutant. The commonly observed sensitivity of ybeY mutants toward heat stress was not observed in A. tumefaciens when incubated at 42°C (data not shown). A role of YbeY in translation was monitored on agar plates supplemented with the four ribosome-targeting antibiotics tetracycline (30S ribosome), gentamicin (30S), chloramphenicol (50S ribosome), and a combination of spectinomycin and streptomycin (30S). The addition of spectinomycin and streptomycin had the strongest effect on the ybeY mutant (Fig. 2C). The addition of tetracycline and chloramphenicol affected the hfq and ybeY mutants equally, whereas gentamicin impaired growth of only the Δhfq strain (see Fig. S2 in the supplemental material). The presence of the ribonucleotide reductase inhibitor hydroxyurea reduced growth of the hfq and ybeY mutants about 100- and 10-fold, respectively.
YbeY, a virulence factor of A. tumefaciens.
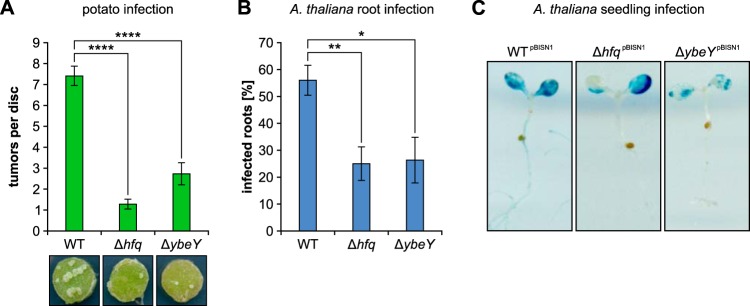
Infection assays with potato tuber discs, Arabidopsis thaliana root segments, and A. thaliana seedlings were used to monitor tumor formation as well as T-DNA transfer of A. tumefaciens. WT, Δhfq, and ΔybeY cultures were diluted, and 106 bacteria were applied to 100 potato tuber discs each. After incubation for 4 to 6 weeks, the WT induced seven to eight tumors per disc, whereas the hfq or ybeY mutant strains produced about one or maximally three tumors, respectively (Fig. 4A). Similar results were obtained in the A. thaliana root infection assay (Fig. 4B). The WT elicited tumors on more than half of all root segments, whereas the Δhfq and ΔybeY strains infected only a quarter of the roots.
FIG 4.
YbeY is crucial for A. tumefaciens virulence. (A) Infection of potato tuber discs with 106 cells of WT, Δhfq, and ΔybeY strains. After incubation for 6 weeks (16-h light/dark cycle, 23°C), tumors were counted per disc, and the average of 100 infected potato discs per strain was calculated. WT infection led to an average of seven to eight tumors per disc. The infection efficiency of Δhfq (one to two tumors/disc) and ΔybeY (two to three tumors/disc) strains was drastically reduced (****, P < 0.0001). (B) Similar results were obtained from A. thaliana root infection assays. About 25% of the 300 roots were infected by either the hfq (**, P = 0.003) or the ybeY (*, P = 0.015) mutant in contrast to the ∼55% infection efficiency of A. tumefaciens WT. (C) Qualitative measurement of T-DNA transfer was performed using WT, Δhfq, and ΔybeY strains harboring a plasmid-carried GUS-reporter gene system (pBISN1). Successful transfer of T-DNA (gus) resulted in β-glucuronidase expression in the infected plant tissue. Cleavage of the substrate X-Glc by the β‑glucuronidase stained the corresponding plant tissue (blue). Infection with WTpBISN1 led to extensive staining of the leaves, whereas seedling infection by hfqpBISN1 or ybeYpBISN1 mutants was less efficient.
To investigate T-DNA transfer efficiency and functionality of the T4SS, we infected A. thaliana seedlings with WT, Δhfq, and ΔybeY strains by the AGROBEST method (47, 48). Successful T-DNA transfer resulted in blue staining of infected plant tissue due to the transient expression of the T-DNA encoded gusA gene. Sixty plant seedlings each were infected with the three A. tumefaciens strains carrying the plasmid required for this assay. Infection with WTpBISN1 cells led to extensive staining of the leaves (Fig. 4C). Consistent with the reduced tumor formation, less plant tissue was stained upon infection with ΔhfqpBISN1 or ΔybeYpBISN1 strains.
Finally, an ectopically expressed virB-lacZ fusion (49) was used to test for successful signal transduction and vir gene expression (Fig. 5A). Expression of virB-lacZ was induced by addition of 0.1 mM acetosyringone. Compared to the WT, virB expression in the Δhfq strain was slightly increased, whereas expression was decreased about one-third in the ΔybeY strain, indicating attenuated vir gene expression in the ybeY mutant. The amounts of VirB2, VirB5, VirB8, and VirB9 were not markedly reduced in the ybeY mutant and slightly increased in the hfq mutant (Fig. 5B), suggesting that the observed defects in tumor formation are not due to reduced production of the T4SS.
FIG 5.
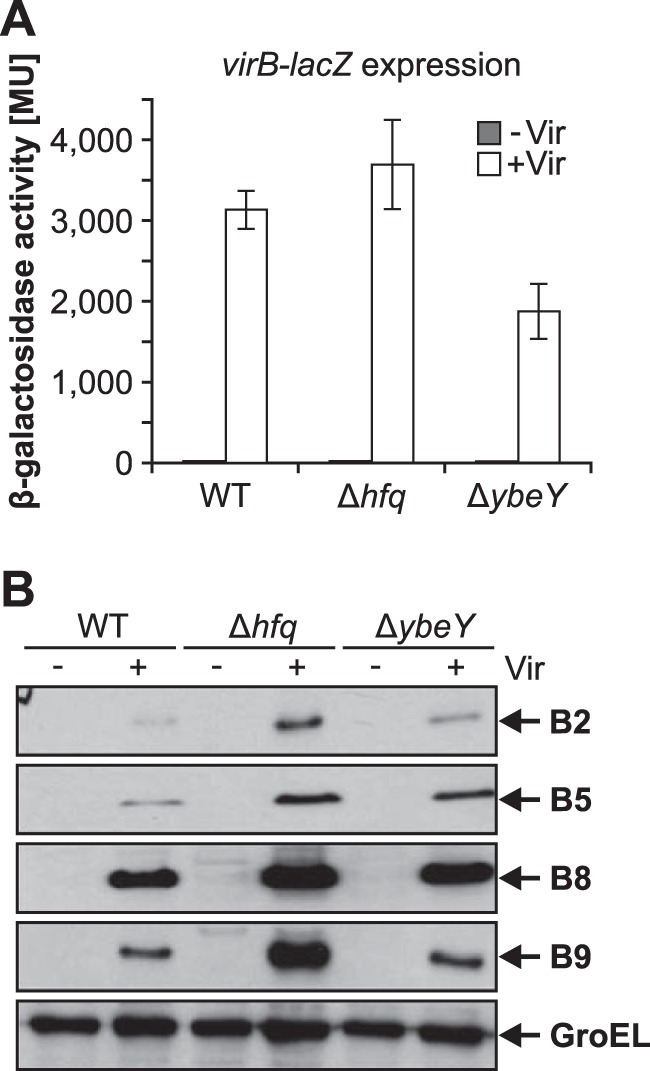
YbeY affects vir gene expression and Vir protein amounts. (A) WT, Δhfq, and ΔybeY strains harboring a plasmid-carried virB-lacZ fusion were cultured under virulence inducing (+Vir) and noninducing conditions (–Vir). Under +Vir conditions, the β-galactosidase activity was reduced in the ybeY mutant, whereas deletion of hfq did not affect virB-lacZ expression significantly compared to WT activity. (B) Protein amounts of VirB2 (12.3 kDa), VirB5 (23.3 kDa), VirB8 (26.3 kDa), and VirB9 (32.2 kDa) under the conditions described for panel A were detected via Western blotting. Vir proteins were detectable only under +Vir conditions. Vir protein amounts were slightly increased in the Δhfq strain but did not show significant differences in the ΔybeY strain. GroEL (57.4 kDa) served as a loading control.
Identification of YbeY-affected proteins by mass spectrometry.
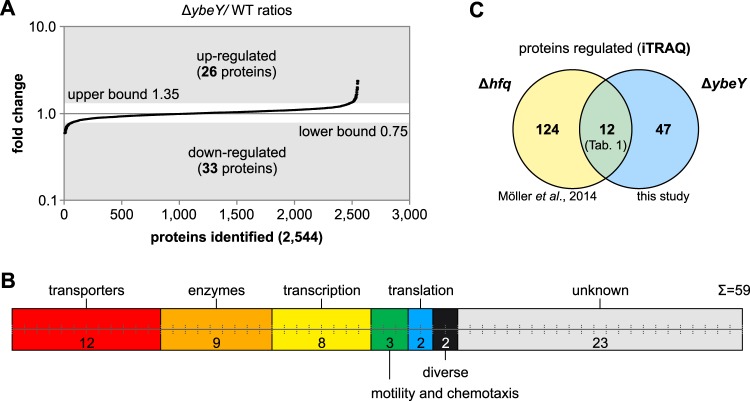
The various phenotypes of the ybeY mutant suggest a broad impact of the YbeY protein on gene expression. To identify proteins that are affected by the presence or absence of YbeY, we compared the proteomes of A. tumefaciens WT and ΔybeY strains by using isobaric tags for relative and absolute quantitation (iTRAQ) labeling prior to liquid chromatography-tandem mass spectrometry (LC-MS/MS). Total proteins were extracted from strains grown to the stationary phase (OD600 = 1.5), and previously obtained protein samples of WT and Δhfq strains (41) were used as references for quantitative comparison of the WT, Δhfq, and ΔybeY strains. In total, 2,544 proteins were uniquely identified by LC-MS/MS in three biological replicates, and the ΔybeY/WT ratios were used to calculate a confidence interval of 95% (Fig. 6A). Proteins with ΔybeY/WT ratios of >1.35 (upper bound) were considered upregulated, and proteins with a ratio of <0.75 (lower bound) were downregulated. Fifty-nine proteins were differentially expressed upon deletion of ybeY; 26 of these proteins were upregulated, and 33 were downregulated (see Table S1 in the supplemental material). Proteins were clustered according to KEGG ontology enrichment (Fig. 6B) as transporters (n = 12), enzymes (n = 9), transcription proteins (n = 8), motility and chemotaxis proteins (n = 3), translation proteins (n = 2), proteins with diverse cellular functions (n = 2), and proteins with unknown functions (n = 23).
FIG 6.
Global proteome analysis of A. tumefaciens WT and ΔybeY strains. Quantification of the WT and ΔybeY proteomes was performed using isobaric tags for relative and absolute quantitation (iTRAQ) and LC-MS/MS. (A) A total of 2,544 proteins were identified in three biological replicates. Based on a confidence interval of 95%, 26 proteins were upregulated (ΔybeY/WT ratio of >1.35) and 33 proteins were downregulated (ΔybeY/WT ratio of <0.75) in a ybeY deletion strain. (B) Proteins regulated by YbeY were clustered according to KEGG orthology enrichment into transporters, enzymes, transcription, motility and chemotaxis, translation, diverse cellular functions, and proteins with unknown function. (C) Overlap between Hfq- and YbeY-dependent proteins. Twelve YbeY-dependent proteins were previously identified to be influenced by Hfq (41).
In accordance with the overlapping phenotypes of Δhfq and ΔybeY strains, the influence of Hfq and YbeY on the A. tumefaciens proteome was partly overlapping (Fig. 6C). Twelve proteins, including nine ABC transporters, were influenced by the deletion of either hfq or ybeY. Interestingly, Hfq and YbeY had opposing effects on the coregulated proteins, e.g., proteins upregulated in Δhfq were downregulated in ΔybeY and vice versa (Table 1).
TABLE 1.
Proteins influenced by Hfq and YbeY
| GenBank accession no. | Annotation | Protein | Function | Up- or downregulationa |
|
|---|---|---|---|---|---|
| ΔybeY mutant | Δhfq mutant | ||||
| AAK88993 | Atu4447 | Sorbitol ABC transporter substrate-binding protein | Up (2.0) | Down (0.4) | |
| AAK90221 | Atu3165 | Sorbitol/mannitol ABC transporter substrate-binding protein | Up (1.5) | Down (0.4) | |
| AAK89922 | Atu3472 | BkdA2 | 2-Oxoisovalerate dehyrogenase subunit beta | Up (1.4) | Down (0.3) |
| AAK86355 | Atu0542 | Fla | Flagellin | Up (1.5) | Down (0.3) |
| AAK88868 | Atu4577 | ABC transporter substrate binding protein | Down (0.7) | Up (12.9) | |
| AAK90320 | Atu3063 | ABC transporter permease | Down (0.6) | Up (11.9) | |
| AAK88755 | Atu4695 | Oligopeptide ABC transporter substrate-binding protein | Down (0.7) | Up (10.1) | |
| AAK88128 | Atu2391 | ABC transporter, substrate binding protein (nitrate/sulfonate/taurine/bicarbonate) | Down (0.7) | Up (9.8) | |
| AAK88731 | Atu4719 | DppA | Dipeptide ABC transporter substrate-binding protein | Down (0.7) | Up (7.7) |
| AAK90716 | Atu5343 | ABC transporter substrate binding protein (oligopeptide) | Down (0.7) | Up (6.4) | |
| AAK90608 | Atu5237 | ABC transporter substrate binding protein (amino acid) | Down (0.7) | Up (5.1) | |
| AAK89127 | Atu4312 | SoxA | Sarcosine oxidase alpha subunit | Down (0.7) | Up (3.0) |
The relative abundances of proteins in the ΔybeY or Δhfq mutants in comparison to the wild-type strain is given in parentheses. For a complete list, see Table S1 in the supplemental material. Source for Δhfq mutant data: Möller et al. (41).
YbeY is involved in 16S rRNA processing in A. tumefaciens.
To assay the in vivo function of YbeY in the processing of rRNAs, we isolated total RNA from the WT plus empty vector (v), the ΔybeY mutant plus empty vector (v), and a complemented ΔybeY mutant plus ybeY strain from the exponential and stationary growth phase and separated the RNA via agarose gel electrophoresis. An additional rRNA fragment in the ΔybeY strain migrated more slowly than the 16S rRNA and was designated 17S rRNA (Fig. 7A). The additional band disappeared when the mutant was complemented with plasmid-carried ybeY.
FIG 7.
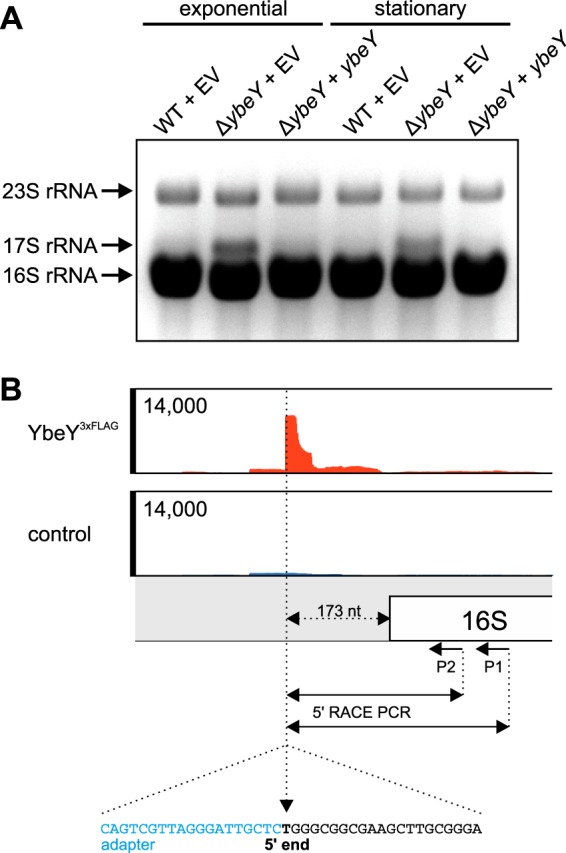
YbeY participates in rRNA processing. (A) Deletion of ybeY resulted in an additional rRNA fragment (17S rRNA). The fragment was most prominent in the exponential growth phase. Complementation (+ybeY) of the ΔybeY strain resulted in WT-like rRNA profile. (B) RIP-Seq experiments revealed binding of YbeY3×Flag to the 5ʹ upstream region of the 16S rRNA (upper panel). The 17S rRNA was isolated from an agarose gel, and the 5ʹ end was identified via 5ʹ RACE (lower panel). Sequencing of the RACE PCR fragment revealed extension of the 16S rRNA by 173 nucleotides in the ybeY mutant.
In our second global approach, we performed RIP-seq experiments with YbeY3×Flag analogous to the Hfq3×Flag RIP-Seq experiments performed previously (41). In contrast to Hfq3×Flag, YbeY3×Flag was unable to significantly enrich cellular RNAs (data not shown). However, the region immediately upstream of the 16S rRNA was enriched (Fig. 7B). When the 17S rRNA was isolated from an agarose gel and subjected to 5ʹ rapid amplification of cDNA ends (RACE), the 5ʹ end corresponded exactly to the region enriched by YbeY3×Flag, suggesting that YbeY interacts with the 173 nucleotides at the 5ʹ end of 16S rRNA during ribosome biogenesis.
Purification and coelution of YbeY and Hfq.
To characterize the YbeY protein, we constructed plasmids for heterologous production of C-terminally and N-terminally tagged YbeY6×His in E. coli and purified the proteins via an Ni-nitrilotriacetic acid (Ni-NTA) column from total protein extracts. Subsequent to affinity purification, pooled elution fractions were subjected to gel permeation chromatography (GPC; YbeY6×His in Fig. 8A and 6×HisYbeY in Fig. S3B in the supplemental material). Comparison of the elution fraction with the elution of standard proteins (Fig. S3A) suggested an oligomeric organization of two to four YbeY proteins (20.6 kDa per monomer).
FIG 8.
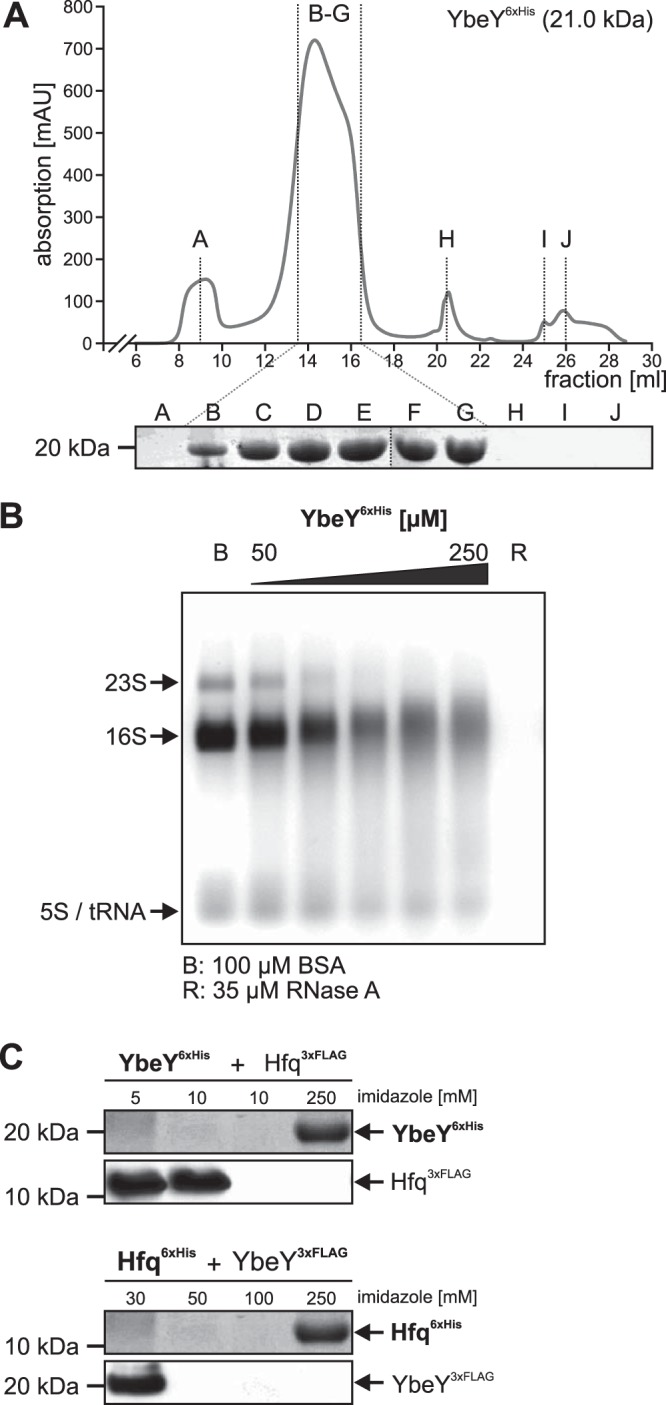
Purification and biochemical analyses of A. tumefaciens. (A) GPC with YbeY6×His (20.6 kDa, C-terminally tagged) resulted in a distinct protein peak with a maximum absorption of 719.5 mAU at 14.3 ml, indicating elution of 34- to 86-kDa large protein complexes, which corresponded to homo-oligomers of two to four YbeY6×His monomers. Protein abundance in fractions B to G was verified by SDS-PAGE. (B) RNase activity of purified YbeY. Various amounts of N-terminally tagged YbeY6×His protein (50 to 250 μM) were incubated with 5 μg total RNA from A. tumefaciens WT. Lane B, 100 μM BSA; lane R, 35 μM RNase A. (C) Pulldown experiments of YbeY6×His and Hfq3×Flag protein variants from cell extracts after incubation for 30 min on ice. YbeY6×His was bound by the Ni-NTA column, whereas Hfq3×Flag was washed off the column by low imidazole concentrations (5 to 10 mM). YbeY6×His was eluted with 250 mM imidazole. Western blot analysis with specific His- or Flag-tagged antibodies did not show coelution of Hfq3×Flag protein with YbeY6×His. Similar results were obtained when protein tags were switched between both proteins (YbeY3×Flag and Hfq6×His).
To provide evidence for RNase activity of YbeY in vitro, 50 to 250 μM GPC-purified YbeY6×His was incubated with 5 μg of total RNA from A. tumefaciens WT for 60 min at 37°C (Fig. 8B). As negative and positive controls for RNase activity, equal amounts of RNA were treated with buffer containing bovine serum albumin (BSA) or RNase A, respectively. The addition of increasing amounts of YbeY6×His (or 6×HisYbeY; data not shown) resulted in degradation of the RNA supporting an RNase activity of YbeY in vitro.
Since Hfq and YbeY influence similar phenotypes and affect the abundance of the same proteins, we sought to determine whether both proteins interact. Hfq and YbeY protein variants with either 6×His or 3×Flag tags at the C terminus were mixed and purified using Ni-NTA chromatography (Fig. 8C). As expected, the Flag-tagged proteins did not bind to Ni-NTA–Sepharose and eluted at low imidazole concentrations. Both His-tagged versions bound the Ni-NTA column and were eluted at an imidazole concentration of 250 mM. Importantly, the Flag-tagged proteins were not retained by the His-tagged proteins, suggesting that YbeY and Hfq do not interact.
DISCUSSION
The A. tumefaciens ybeY gene is differentially expressed, not essential, and a virulence factor critical for plant transformation.
As member of the prokaryotic core genome, the ybeY gene is essential in many bacteria, among them the human pathogen V. cholerae (10). The endoribonuclease activity in chloroplasts renders YbeY essential in the model plant Arabidopsis thaliana, further supporting an ancient function of the protein (13). In many bacteria, however, deletion of ybeY is tolerable, although associated with numerous physiological defects (see Table 2). This is also the case in the plant pathogen A. tumefaciens, where deletion of the gene caused pleiotropic phenotypes. Similar to the closely related plant symbiont S. meliloti (4), most defects were reminiscent of the ones in an hfq mutant but generally less severe. The defects were additive in a combined A. tumefaciens hfq ybeY double mutant that was first difficult to obtain and second unable to survive prolonged cultivation, which suggests that at least one of these riboregulators is necessary for survival. Strikingly similar phenotypes in the individual mutants do not necessarily mean that Hfq and YbeY have redundant functions, as suggested previously (4). Although both proteins play a principle role in RNA-mediated coordination of cellular processes, they do so by different activities. Hfq rather acts as RNA chaperone facilitating RNA-RNA interactions whereas YbeY—at least predominantly—acts as catalytic enzyme involved in rRNA processing (see below). Due to their global impact on RNA functionality, it will be a challenge to separate direct from indirect effects of these proteins.
TABLE 2.
Commonalities and differences between bacterial YbeY proteinsa
| Feature | Presence or absence of feature in: |
||||
|---|---|---|---|---|---|
| E. coli | V. cholerae | S. meliloti | B. abortus | A. tumefaciens | |
| Essential ybeY gene | No | Yes | No | No | No |
| Pleiotropic phenotypes of ybeY mutant | Yes | Yes | Yes | Yes | Yes |
| Virulence or symbiosis defect | ND | Yes | Yes | Yes | Yes |
| Ribonuclease activity | Yes | Yes | Yes | Yes | Yes |
| 16S rRNA maturation | Yes | Yes | No | ND | Yes |
| Oligomeric state of YbeY | ND | ND | Monomer | ND | Oligomer |
For additional details, see Discussion. ND, not determined.
Quantitative proteomics further supported a functional relationship between YbeY and Hfq and revealed Hfq-dependent and -independent proteins affected by the presence of YbeY. The abundance of 59 proteins was dysregulated in the ybeY mutant. The nature of these proteins suggests a role of YbeY in general metabolism and nutrient uptake. It is remarkable that YbeY and Hfq had opposing effects on a number of proteins, in particular on ABC transporters (Table 1). A similar response was observed in S. meliloti, where 41 genes were upregulated in the ybeY mutant and downregulated in the hfq mutant (50). The mechanistic basis of this regulatory interplay is presently unclear. Our pulldown experiments did not provide any evidence for a stable, direct interaction between Hfq and YbeY, which is consistent with previous observations (4, 10, 51). Moreover, protein levels of YbeY were not influenced by deletion of hfq and vice versa (data not shown), suggesting that both proteins act independently of each other.
In E. coli, ybeY is a heat shock gene and the ybeY mutant is temperature sensitive (15). Tracking A. tumefaciens YbeY levels throughout the growth curve showed that it was present under all conditions but accumulated toward stationary phase. This finding suggests a more important role of YbeY under starvation and stress conditions, which is fully reflected by the low cell densities of the mutant in stationary phase and its sensitivity to various noxious compounds.
YbeY is required for full virulence of Y. enterocolitica, V. cholerae, and B. abortus (10, 12, 17), which motivated us to assess virulence of the ybeY mutant in the tumor-inducing plant pathogen A. tumefaciens. Several plant infection assays looking at different stages of plant transformation placed YbeY among the virulence factors of A. tumefaciens because T-DNA transfer and concomitantly tumor formation were severely compromised in the ybeY mutant. This might be a cumulative effect of the phenotypic alterations in the ybeY mutant rather than an effect of YbeY on an individual virulence gene. Such a cumulative negative effect of the absence of ybeY on virulence was also postulated for the human pathogen B. abortus (17).
In enterohemorrhagic E. coli (EHEC), YbeY is required for normal type 3 secretion (T3SS) levels (52). This effect was not due to specific binding to or processing of the corresponding T3SS mRNA by YbeY. Instead, it was explained by an overall translation defect in the ybeY mutant caused by an accumulation of not fully matured ribosomes that leads to destabilization of untranslated mRNAs. In A. tumefaciens, the T4SS responsible for T-DNA transfer into the plant was present at normal levels in the ybeY mutant, and the signal transduction cascade responsible for plant transformation was operative. Expression of virA and virG coding for the virulence inducing two-component system was not affected (see Fig. S4 in the supplemental material), and the VirAG system was active as shown by almost normal virB-lacZ expression and WT-like VirB protein levels (Fig. 5). Although it is possible that barely noticeable changes in virulence protein levels elicit defects in T-DNA transfer, we favor the idea that translational defects and the dysregulated protein pool in the ybeY mutant are responsible for the virulence defect of the A. tumefaciens ybeY mutant. Among the suspicious proteins that might at least in part contribute to this defect are catalase CatE (underrepresented in the ybeY mutant) and the small heat shock protein HspL (overrepresented). CatE and the bifunctional peroxidase-catalase KatA protect A. tumefaciens from the plant defense molecule H2O2 during infection. Deletion of the katA gene results in avirulent bacteria (53). Since CatE supports KatA activity, downregulation of CatE might increase sensitivity toward H2O2. The small heat shock protein HspL is one of four small heat shock proteins encoded in A. tumefaciens and is a VirB8-specific chaperone promoting functionality of the T4SS (54, 55). VirB8, together with VirB7, VirB9, and VirB10, is located in the core channel across the double membrane and HspL prevents its aggregation (56, 57). Misregulated HspL levels might influence function of VirB8, thus hindering efficient T-DNA export via the T4SS. The imbalance in several nutrient acquisition systems in the ybeY mutant might be yet another reason for the attenuation in plant infection.
Different functionalities of YbeY in different organisms?
Although there is no doubt that YbeY plays a crucial role in bacterial physiology, the reported details on YbeY properties and activities are still controversial. As an isolated protein, YbeY is a nonspecific RNase. We and others showed that purified YbeY degrades bulk RNA (8, 13, 50). It is evident that such an activity must be counteracted in vivo, which led to the proposal that cellular factors restrict this indiscriminate activity and coordinate the molecular activity of YbeY. This assumption is supported by the observation that expression of the human ybeY homologue C21orf57 (huYBEY) results in uncontrolled activity and is deleterious in the yeast Saccharomyces cerevisiae, which does not have a YbeY-related protein itself (3). A recent screen for YbeY interactors in E. coli revealed the association with ribosomal protein S11 and the two GTPases Era and Der involved in ribosome biogenesis (51). Elevated levels of Era partially suppressed the growth defect of the ybeY mutant and improved 16S rRNA maturation (58), suggesting that this protein serves as “specificity factor” for YbeY.
One of the functions ascribed to E. coli YbeY is an endoribonuclease activity responsible for processing of the 3ʹ terminus of the 16S rRNA (8), a critical step in ribosome maturation that might explain why ybeY belongs to the prokaryotic core genome. A similar 3ʹ processing activity was recently reported for YqfG, the essential YbeY orthologue in Bacillus subtilis (59). Chloroplast with reduced YbeY levels displayed maturation defects in both the 5ʹ and 3ʹ ends of rRNAs, which resulted in reduced plastid translational activity (13). Structural and biochemical data conclusively support a metal-dependent RNase activity of YbeY (2, 8, 50), but the target specificities vary. E. coli YbeY was shown to be a single-strand-specific endoribonuclease (8), whereas the S. meliloti enzyme cleaved single- and double-stranded RNAs (50), an unprecedented activity among bacterial endoribonucleases.
Maturation of rRNAs is a multifactorial process involving various RNases required for trimming of the different intermediates derived from the long precursor transcript. Typically, several RNases, which can have redundant functions, cooperate in these reactions (60). For instance, RNase G in combination with RNase E cleaves the 5ʹ end of the 16S rRNA precursor in E. coli (61). On the other end of the transcript, YbeY and RNase R function together in 16S rRNA 3ʹ maturation (8). Despite the functional relationship between YbeY and RNase R, a direct interaction between the proteins was not detected, suggesting that the association with other proteins guides the processing events (51). The fact that the repertoire of cellular endo- and exoribonucleases involved in the rRNA processing varies substantially between bacteria might explain discrepancies in our current understanding of the YbeY activities in different organisms. Agrobacterium, for example, does not encode RNase G, and it is conceivable that some other RNase, which might be YbeY, fills this gap.
Surprisingly, even in closely related bacteria, the YbeY-associated activities can differ, as observed in two alphaproteobacteria associated with plants (Table 2). While YbeY is not required for 16S rRNA maturation in S. meliloti (50, 62), we found clear evidence for a 16S rRNA processing defect in the A. tumefaciens ybeY mutant (Fig. 7). Moreover, there was no indication that YbeY binds strongly to mRNAs or sRNAs in A. tumefaciens, again in contrast to reports in S. meliloti (4, 50). Instead, our RIP-Seq experiments revealed binding of YbeY to the immature 5ʹ end of 16S rRNA precursor. This is reminiscent of findings in EHEC, although different positions within the 16S rRNA were occupied (52). Another unsettled issue pertains to the oligomeric state of YbeY. While the Agrobacterium protein assembled into an oligomeric state after affinity chromatography and gel filtration, the Sinorhizobium protein behaved as a monomer in cross-linking experiments (50). Finally, distinct features of YbeY proteins from different organisms are supported by complementation experiments. Expression of the E. coli homologue complemented the symbiosis defect of the ybeY mutant in S. meliloti (7, 63), whereas the phenotypic defects of the A. tumefaciens mutant could not be rescued by E. coli or S. meliloti ybeY. It is presently unclear how these controversial findings can be reconciled, but it is evident that YbeY as a member of a limited set of universal bacterial proteins is an important protein worthy of further comparative investigations.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All bacterial strains and plasmids used in this study are listed in Table S2 in the supplemental material. Agrobacterium tumefaciens C58 strains were cultivated at 30°C in YEB complex medium or AB minimal medium (pH 5.5, supplemented with 1% [wt/vol] glucose) (64). Escherichia coli BL21 strains were cultivated in LB medium at 37°C (65). A. tumefaciens virulence was induced in AB medium inoculated to an OD600 of 0.1 and grown for 6 h at 30°C. Virulence gene expression was induced by addition of 0.1 mM acetosyringone (Sigma-Aldrich, Germany) and further cultivation (23°C, 16 h). Non-virulence-induced cultures were treated with equal volumes of dimethyl sulfoxide (DMSO). For cultivation during mutagenesis, A. tumefaciens cells were grown in LB medium (65) supplemented with either 10% (wt/vol) sucrose or 50 μg/ml kanamycin (Km).
Deletion of chromosomal ybeY.
Deletion of ybeY (atu0358) was performed by homologous recombination analogous to the deletion of hfq described by (45). A 300-bp up- and downstream region of the ybeY ORF was amplified using the primers ybeY_up_PstI_fw, ybeY_up_SmaI_rv, ybeY_down_SmaI_fw, and ybeY_down_EcoRI_rv listed in Table S3 and ligated into a pK19mobsacB suicide plasmid. The resulting ybeY_up_dwn was transformed into Agrobacterium via electroporation. Single-crossover events were selected on LB agar plates containing 50 μg/ml Km. Km-tolerant colonies were further inoculated overnight in LB medium and subsequently selected for Km sensitivity and tolerance of 10% (wt/vol) sucrose. Chromosomal deletion of ybeY was confirmed by colony PCR and Southern blot analyses.
Chromosomal integration of ybeY3×Flag.
Chromosomal integration of 3×Flag tag sequence at the ybeY 3ʹ end was performed analogous to the construction of hfq3×Flag described previously (41). The ybeY upstream region, including the ybeY ORF without the TAA stop codon was amplified using primers ybeY_up_PstI_fw, ybeY_up_SalI_rv. The downstream region was amplified using primers ybeY_down_Acc65I_fw and ybeY_down_EcoRI_rv. Both fragments were cloned into a pK19mobsacB suicide plasmid. The 3×Flag tag sequence was amplified from an A. tumefaciens hfq3×Flag strain using the primers 3×Flag_SalI_fw and 3×Flag_Acc65I_rv. The fragment was inserted between ybeY up- and downstream fragments via SalI and Acc65I restriction sites. The resulting ybeY_up_3×Flag_down plasmid was transformed into A. tumefaciens via electroporation. Integration of the ybeY3×Flag into the chromosome was performed via homologues recombination equally to ybeY mutagenesis described before.
Successful integration of the ybeY3×Flag into the A. tumefaciens chromosome was confirmed by colony PCR and Southern blot analyses.
Arabidopsis thaliana root infection.
Infection of A. thaliana Col-0 roots was performed as described previously (66). Roots of 2- to 3-week-old A. thaliana plants were cut into 0.5-cm segments and inoculated with A. tumefaciens strains for 30 min. Infected root bundles were inoculated for 2 days on MS agar plates at 23°C (16 h light/8 h dark). Roots bundles were washed in H2O plus 100 μg/ml ticarcillin and root segments (300 roots per strain) were separated on MS agar plates plus 100 μg/ml ticarcillin. After incubation for 2 to 3 weeks at 23°C (16 h light/8 h dark), infected roots were counted for tumor formation.
A. thaliana seedling infection.
To assess A. tumefaciens T-DNA transfer capability, we performed A. thaliana seedling infection assays based on the AGROBEST (Agrobacterium-mediated enhanced seedling transformation) method described previously (48), with minor modifications. The pBISN1 plasmid harboring gusA was transformed into A. tumefaciens WT, Δhfq, and ΔybeY strains via electroporation (47). A. thaliana efr-1 seeds (elongation factor Tu [EF-Tu] receptor mutant) were washed with 96% ethanol (30 s), 6% (vol/vol) NaOCl, and water (five times) and then transferred to 6-well plates with 1 ml of 1/2 MS liquid medium (1/2 MS salt, 0.5% [wt/vol] sucrose; pH 5.5). Three-day-old A. thaliana efr-1 seedlings were inoculated with 1 ml of culture grown under virulence-induced conditions (AB plus 0.1% glucose [wt/vol] plus 0.1 mM acetosyringone), followed by incubation at 23°C (16 h light/8 h dark) for 3 to 4 days. Seedlings were stained with 5-bromo-4-chloro-3-indolyl glucuronide (X-Gluc) at 37°C for 3 h. Successful T-DNA transfer resulted in blue staining of the corresponding plant tissue.
Potato infection.
Infection of potato tuber discs was performed as described earlier (54). Bacterial cells were cultivated to late exponential phase (OD600 = 0.7 to 1.0), harvested, and washed in phosphate-buffered saline (PBS) buffer. The bacterial cultures were diluted in PBS buffer to a final OD600 of 108 and 106 cells/ml. Prepared potato discs (50 per strain and dilution) were placed on water agar and inoculated with 10 μl of the bacterial solution prior to incubation for 2 days (23°C, 16 h light/8 h dark). After 2 days, potato discs were placed on water agar plates supplemented with 100 μg/ml ticarcillin to kill the bacteria. After ongoing incubation of the potato discs for 3 weeks, the tumors were scored.
β-Galactosidase assay.
A. tumefaciens cells harboring the pAC01_virB-lacZ (virB transcriptional lacZ fusion) plasmid (67) were cultivated under noninduced (+DMSO) and virulence-induced (+0.1 mM acetosyringone) conditions in AB minimal medium. The activity of the β-galactosidase was measured according to standard protocols (68).
Motility assay.
Swimming was assessed by inoculation of 5 μl of bacterial solution (OD600 = 0.5) on AB minimal medium agar (+0.1% [wt/vol] glucose) plates containing 0.5% (wt/vol) agar (Difco). Cells were incubated for 2 to 3 days at 30°C in the dark. Swimming rings were measured (diameter), and the distance was calculated relative to WT swimming.
Stress sensitivity assays.
A. tumefaciens WT, Δhfq, and ΔybeY strains were grown to an OD600 of 0.5, and cells were harvested at a final OD600 of 1.0 in distilled water. Serial 10-fold dilutions (100 to 10−7) were prepared, and 3 μl was spotted onto LB/AB agar plates supplemented with 10 μg/ml chloramphenicol, 4 μg/ml gentamicin, 10 μg/ml spectinomycin plus 3 μg/ml streptomycin, 1 μg/ml tetracycline, or 20 mM hydroxyurea prior to incubation for 2 to 3 days at 30°C.
Isolation of total RNA and Northern blot analysis.
Total RNA was isolated as described previously (45). Northern blot analysis was performed as described previously (69). For transcript analysis, 8 μg of total RNA was separated using agarose gels, blotted onto Hybond-N membranes (GE Healthcare), and hybridized with digoxigenin-labeled (Roche Applied Science, Germany) RNA probes. The oligonucleotides used for probe synthesis via in vitro transcription are listed in Table S2 in the supplemental material. Signals were detected using the Hyperfilm ECL (GE Healthcare) system.
SDS-PAGE and Western blot analysis.
For protein analysis, 1 ml of bacterial solution was harvested and resuspended in 1× SDS loading dye as described by Klüsener et al. (67). Proteins were incubated for 10 min at 95°C and separated using a 12.5% sodium dodecyl sulfate polyacrylamide gel and subsequent electrophoresis. Proteins were either stained by Coomassie blue or subjected to Western blot transfer. Identification of proteins was performed using either 1:2,000 His-conjugated (His-tagged proteins; Qiagen), monoclonal 1:3,000 VirB9, or monoclonal Flag 1:3,000 M2 (Sigma-Aldrich) antibodies. Fluorescence was detected via a Hyperfilm ECL (GE Healthcare) system.
Proteome analysis by iTRAQ.
Cultivation of A. tumefaciens Wt and ΔybeY strains, protein isolation, trypsin digestion, labeling with iTRAQ reagents, subsequent LC-MS/MS and data analysis were performed as described (41). Protein abundances significantly altered in the ybeY mutant compared to the WT were selected as previously described (70) with minor modifications. A confidence interval of 95% (Z score, 1.96) was used to determine protein ratios with a distribution outside the main distribution. The confidence interval for downregulated proteins was 0.0093 (mean ratio) – 1.96 × 1.4289 (standard deviation [SD]) and corresponded to a protein ratio of 0.76. Proteins upregulated were similarly defined (mean ratio + 1.96 × the SD) and corresponded to a protein ratio of 1.34. Proteins were considered upregulated with ΔybeY/WT ratios of >1.35 and downregulated with ratios of <0.75. Protein abundances were considered significantly altered when they reached the cutoff criteria and a variance between the three biological replicates of <40%. For protein ratios reaching the cutoff but failing the variance criterion, a combined ratio was calculated (combined ratio = ratio ± ratio × variance) as described earlier (41). Considering combined ratios includes proteins that explicitly reach the cutoff criteria for up- or downregulation.
Protein purification and GPC.
Hfq6×His and YbeY6×His fusion proteins were expressed in E. coli BL21(DE3) cells, using pET-plasmid-based expression from a T7 promoter (Novagen, Madison, WI). The proteins were purified as described previously (71). To assess the oligomeric state, we performed gel permeation chromatography. After Ni-NTA chromatography, purified His-tagged fusion proteins were separated by size via a Superdex 200 10/300 GL column (HEPES buffer [pH 7.5]) on an ÄKTA system (GE Healthcare). Additional standard proteins (Blue-Dextran [2,000 kDa], apoferritin [443 kDa], β-amylase [200 kDa], and conalbumin [67 kDa; Sigma-Aldrich]) were loaded to calculate a standard curve and estimate the molecular weights of the YbeY6×His proteins.
In vitro RNA degradation.
For RNA degradation experiments, 5 μg of total RNA was incubated with 50 to 250 μM protein in 20-μl reaction mixtures for 60 min at 37°C prior to separation on a 1.5% morpholinepropanesulfonic acid-agarose gel. In parallel, 100 μM BSA was incubated under the same conditions with 5 μg of RNA to determine the interaction of RNA with unspecific proteins. RNase A (35 μM) was added as a positive control for RNase activity. After separation the RNA was stained with EtBr and visualized under UV light.
5′ RACE.
For the RACE analyses, total RNA from the ybeY mutant grown to the stationary phase was isolated and separated via agarose gel electrophoresis. The 17S rRNA fragment was eluted from the gel using the NucleoSpin gel and a PCR Clean-Up kit (Macherey-Nagel). RACE reactions to determine the 16S* rRNA 5ʹ end were performed as described previously (72) with minor modifications (73). The adapter oligonucleotides and gene-specific primers used are listed in Table S2 in the supplemental material.
YbeY3×Flag RIP-Seq of 16S rRNA.
RNA-Seq of YbeY3×Flag-bound 16S rRNA was performed analogous to the Hfq3×Flag RIP-Seq published previously (41) and based on the procedure described earlier (74, 75). The YbeY3×Flag strain was cultivated in AB medium to stationary phase (OD600 = 0.8). YbeY3×Flag proteins were bound to monoclonal anti-3×Flag M2 antibody (Sigma-Aldrich) coupled to protein G Dynabeads (Thermo Fisher Scientific) and specifically enriched from cell lysates. RNA bound by YbeY3×Flag was isolated by using phenol-chloroform-isoamyl alcohol and precipitated by ethanol and sodium acetate. Remaining DNA was digested by DNase I (Thermo Fisher Scientific) prior to the final precipitation. cDNA libraries were prepared at Vertis Biotechnology AG (Germany) as described previously (41) and sequenced using a HiSeq 2500 (Illumina) machine in single-read mode, running 100 cycles.
Final reads (FASTQ) were trimmed by the fast_quality_trimmer program from FASTX toolkit (v0.0.13; http://hannonlab.cshl.edu/fastx_toolkit) and further processed using the READemption tool (v0.2.6) (76), as described previously (41).
The normalized reads (based on the total number of reads aligned from the respective library) from the 16S rRNA gene were enriched 9.5-fold (reads per million, 9,734; raw read count, 11,562) in the the YbeY3×Flag sample compared to YbeYWT. Enrichment was visualized using the Integrated Genome Browser (77).
Supplementary Material
ACKNOWLEDGMENT
We thank Christian Baron (Montreal) for VirB-specific antisera. We also thank Jer-Sheng Lin, Chih-Feng Wu, and the Proteomics Core Lab of the Institute of Plant and Microbial Biology for assistance in sample preparation for iTRAQ analysis, the Academia Sinica Common Mass Spectrometry Facilities for acquiring MS data on the LTQ-Orbitrap Elite, and Cynthia M. Sharma (Würzburg) for next-generation sequencing support. We thank Roman Moser and Johanna Rossmanith for critical reading of the manuscript.
This project was supported by a grant from the German Research Foundation (DFG, NA 240/11-1) to F.N. and by a joint grant from the German Academic Exchange Service (DAAD) and Taiwan National Science Council (PPP grant 0970029248P) to F.N. and E.-M.L.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00730-18.
REFERENCES
- 1.Gil R, Silva FJ, Pereto J, Moya A. 2004. Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev 68:518–537. doi: 10.1128/MMBR.68.3.518-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhan C, Fedorov EV, Shi W, Ramagopal UA, Thirumuruhan R, Manjasetty BA, Almo SC, Fiser A, Chance MR, Fedorov AA. 2005. The YbeY protein from Escherichia coli is a metalloprotein. Acta Crystallogr Sect F Struct Biol Cryst Commun 61:959–963. doi: 10.1107/S1744309105031131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosal A, Kohrer C, Babu VMP, Yamanaka K, Davies BW, Jacob AI, Ferullo DJ, Gruber CC, Vercruysse M, Walker GC. 2017. C21orf57 is a human homologue of bacterial YbeY proteins. Biochem Biophys Res Commun 484:612–617. doi: 10.1016/j.bbrc.2017.01.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandey SP, Minesinger BK, Kumar J, Walker GC. 2011. A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res 39:4691–4708. doi: 10.1093/nar/gkr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallory AC, Hinze A, Tucker MR, Bouche N, Gasciolli V, Elmayan T, Lauressergues D, Jauvion V, Vaucheret H, Laux T. 2009. Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet 5:e1000646. doi: 10.1371/journal.pgen.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. 2005. Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies BW, Kohrer C, Jacob AI, Simmons LA, Zhu J, Aleman LM, Rajbhandary UL, Walker GC. 2010. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol Microbiol 78:506–518. doi: 10.1111/j.1365-2958.2010.07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacob AI, Kohrer C, Davies BW, RajBhandary UL, Walker GC. 2013. Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation. Mol Cell 49:427–438. doi: 10.1016/j.molcel.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey SP, Winkler JA, Li H, Camacho DM, Collins JJ, Walker GC. 2014. Central role for RNase YbeY in Hfq-dependent and Hfq-independent small-RNA regulation in bacteria. BMC Genomics 15:121. doi: 10.1186/1471-2164-15-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vercruysse M, Kohrer C, Davies BW, Arnold MF, Mekalanos JJ, RajBhandary UL, Walker GC. 2014. The highly conserved bacterial RNase YbeY is essential in Vibrio cholerae, playing a critical role in virulence, stress regulation, and RNA processing. PLoS Pathog 10:e1004175. doi: 10.1371/journal.ppat.1004175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohyama H, Sakai T, Agari Y, Fukui K, Nakagawa N, Shinkai A, Masui R, Kuramitsu S. 2014. The role of ribonucleases in regulating global mRNA levels in the model organism Thermus thermophilus HB8. BMC Genomics 15:386. doi: 10.1186/1471-2164-15-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leskinen K, Varjosalo M, Skurnik M. 2015. Absence of YbeY RNase compromises the growth and enhances the virulence plasmid gene expression of Yersinia enterocolitica O:3. Microbiology 161:285–299. doi: 10.1099/mic.0.083097-0. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Zhou W, Liu G, Yang C, Sun Y, Wu W, Cao S, Wang C, Hai G, Wang Z, Bock R, Huang J, Cheng Y. 2015. The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis. Plant Physiol 168:205–221. doi: 10.1104/pp.114.255000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda T, Tanaka Y, Wachi M, Inui M. 2017. Polynucleotide phosphorylase, RNase E/G, and YbeY are Involved in the maturation of 4.5S RNA in Corynebacterium glutamicum. J Bacteriol 199:e00798-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasouly A, Schonbrun M, Shenhar Y, Ron EZ. 2009. YbeY, a heat shock protein involved in translation in Escherichia coli. J Bacteriol 191:2649–2655. doi: 10.1128/JB.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasouly A, Davidovich C, Ron EZ. 2010. The heat shock protein YbeY is required for optimal activity of the 30S ribosomal subunit. J Bacteriol 192:4592–4596. doi: 10.1128/JB.00448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budnick JA, Sheehan LM, Colquhoun JM, Dunman PM, Walker GC, Roop RM 2nd, Caswell CC. 2018. Endoribonuclease YbeY is linked to proper cellular morphology and virulence in Brucella abortus. J Bacteriol 200:e00105-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogel J, Luisi BF. 2011. Hfq and its constellation of RNA. Nat Rev Microbiol 9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sauer E. 2013. Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol 10:610–618. doi: 10.4161/rna.24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner EG. 2013. Cycling of RNAs on Hfq. RNA Biol 10:619–626. doi: 10.4161/rna.24044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olejniczak M. 2011. Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry 50:4427–4440. doi: 10.1021/bi102043f. [DOI] [PubMed] [Google Scholar]
- 22.Soper TJ, Woodson SA. 2008. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Link TM, Valentin-Hansen P, Brennan RG. 2009. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci U S A 106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson KE, Orans J, Kovach AR, Link TM, Brennan RG. 2014. Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res 42:2736–2749. doi: 10.1093/nar/gkt1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kavita K, de Mets F, Gottesman S. 2018. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr Opin Microbiol 42:53–61. doi: 10.1016/j.mib.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santiago-Frangos A, Woodson SA. 2018. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip Rev RNA 9:e1475. doi: 10.1002/wrna.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massé E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A 99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herman C, Simonsen KT, Nielsen G, Bjerrum JV, Kruse T, Kallipolitis BH, Møller-Jensen J. 2011. A role for the RNA chaperone Hfq in controlling adherent-invasive Escherichia coli colonization and virulence. PLoS One 6:e16387. doi: 10.1371/journal.pone.0016387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sittka A, Pfeiffer V, Tedin K, Vogel J. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol 63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berghoff BA, Glaeser J, Sharma CM, Zobawa M, Lottspeich F, Vogel J, Klug G. 2011. Contribution of Hfq to photooxidative stress resistance and global regulation in Rhodobacter sphaeroides. Mol Microbiol 80:1479–1495. doi: 10.1111/j.1365-2958.2011.07658.x. [DOI] [PubMed] [Google Scholar]
- 31.Arce-Rodriguez A, Calles B, Nikel PI, de Lorenzo V. 2016. The RNA chaperone Hfq enables the environmental stress tolerance super-phenotype of Pseudomonas putida. Environ Microbiol 18:3309–3326. doi: 10.1111/1462-2920.13052. [DOI] [PubMed] [Google Scholar]
- 32.Yang G, Wang L, Wang Y, Li P, Zhu J, Qiu S, Hao R, Wu Z, Li W, Song H. 2015. hfq regulates acid tolerance and virulence by responding to acid stress in Shigella flexneri. Res Microbiol 166:476–485. doi: 10.1016/j.resmic.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Barra-Bily L, Fontenelle C, Jan G, Flechard M, Trautwetter A, Pandey SP, Walker GC, Blanco C. 2010. Proteomic alterations explain phenotypic changes in Sinorhizobium meliloti lacking the RNA chaperone Hfq. J Bacteriol 192:1719–1729. doi: 10.1128/JB.01429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao M, Barnett MJ, Long SR, Teplitski M. 2010. Role of the Sinorhizobium meliloti global regulator Hfq in gene regulation and symbiosis. Mol Plant Microbe Interact 23:355–365. doi: 10.1094/MPMI-23-4-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T. 1996. High efficiency transformation of maize (Zea mays L) mediated by Agrobacterium tumefaciens. Nat Biotechnol 14:745–750. doi: 10.1038/nbt0696-745. [DOI] [PubMed] [Google Scholar]
- 36.Hinchee MAW, Connorward DV, Newell CA, Mcdonnell RE, Sato SJ, Gasser CS, Fischhoff DA, Re DB, Fraley RT, Horsch RB. 1988. Production of transgenic soybean plants using Agrobacterium-mediated DNA transfer. Nat Biotechnol 6:915–921. doi: 10.1038/nbt0888-915. [DOI] [Google Scholar]
- 37.Umbeck P, Johnson G, Barton K, Swain W. 1987. Genetically transformed cotton (Gossypium hirsutum L) plants. Nat Biotechnol 5:263–266. doi: 10.1038/nbt0387-263. [DOI] [Google Scholar]
- 38.Gelvin SB. 2003. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67:16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF Jr, Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee D Sr, Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, Nester EW. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 40.Pitzschke A, Hirt H. 2010. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J 29:1021–1032. doi: 10.1038/emboj.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Möller P, Overlöper A, Förstner KU, Wen T-N, Sharma CM, Lai E-M, Narberhaus F. 2014. Profound impact of Hfq on nutrient acquisition, metabolism and motility in the plant pathogen Agrobacterium tumefaciens. PLoS One 9:e110427. doi: 10.1371/journal.pone.0110427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilms I, Overlöper A, Nowrousian M, Sharma CM, Narberhaus F. 2012. Deep sequencing uncovers numerous small RNAs on all four replicons of the plant pathogen Agrobacterium tumefaciens. RNA Biol 9:446–457. doi: 10.4161/rna.17212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee K, Huang X, Yang C, Lee D, Ho V, Nobuta K, Fan JB, Wang K. 2013. A genome-wide survey of highly expressed noncoding RNAs and biological validation of selected candidates in Agrobacterium tumefaciens. PLoS One 8:e70720. doi: 10.1371/journal.pone.0070720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dequivre M, Diel B, Villard C, Sismeiro O, Durot M, Coppee JY, Nesme X, Vial L, Hommais F. 2015. Small RNA deep-sequencing analyses reveal a new regulator of virulence in Agrobacterium fabrum C58. Mol Plant Microbe Interact 28:580–589. doi: 10.1094/MPMI-12-14-0380-FI. [DOI] [PubMed] [Google Scholar]
- 45.Wilms I, Möller P, Stock AM, Gurski R, Lai EM, Narberhaus F. 2012. Hfq influences multiple transport systems and virulence in the plant pathogen Agrobacterium tumefaciens. J Bacteriol 194:5209–5217. doi: 10.1128/JB.00510-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becker A, Overlöper A, Schlüter JP, Reinkensmeier J, Robledo M, Giegerich R, Narberhaus F, Evguenieva-Hackenberg E. 2014. Riboregulation in plant-associated alpha-proteobacteria. RNA Biol 11:550–562. doi: 10.4161/rna.29625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Narasimhulu SB, Deng XB, Sarria R, Gelvin SB. 1996. Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell 8:873–886. doi: 10.1105/tpc.8.5.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu HY, Liu KH, Wang YC, Wu JF, Chiu WL, Chen CY, Wu SH, Sheen J, Lai EM. 2014. AGROBEST: an efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods 10:19. doi: 10.1186/1746-4811-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klüsener S, Hacker S, Tsai YL, Bandow JE, Gust R, Lai EM, Narberhaus F. 2010. Proteomic and transcriptomic characterization of a virulence-deficient phosphatidylcholine-negative Agrobacterium tumefaciens mutant. Mol Genet Genomics 283:575–589. doi: 10.1007/s00438-010-0542-7. [DOI] [PubMed] [Google Scholar]
- 50.Saramago M, Peregrina A, Robledo M, Matos RG, Hilker R, Serrania J, Becker A, Arraiano CM, Jimenez-Zurdo JI. 2017. Sinorhizobium meliloti YbeY is an endoribonuclease with unprecedented catalytic features, acting as silencing enzyme in riboregulation. Nucleic Acids Res 45:1371–1391. doi: 10.1093/nar/gkw1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vercruysse M, Kohrer C, Shen Y, Proulx S, Ghosal A, Davies BW, RajBhandary UL, Walker GC. 2016. Identification of YbeY-protein interactions involved in 16S rRNA maturation and stress regulation in Escherichia coli. mBio 7:e01785-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McAteer SP, Sy BM, Wong JL, Tollervey D, Gally DL, Tree JJ. 2018. Ribosome maturation by the endoribonuclease YbeY stabilizes a type 3 secretion system transcript required for virulence of enterohemorrhagic Escherichia coli. J Biol Chem 293:9006–9016. doi: 10.1074/jbc.RA117.000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu XQ, Li LP, Pan SQ. 2001. Feedback regulation of an Agrobacterium catalase gene katA involved in Agrobacterium-plant interaction. Mol Microbiol 42:645–657. [DOI] [PubMed] [Google Scholar]
- 54.Tsai YL, Wang MH, Gao C, Klusener S, Baron C, Narberhaus F, Lai EM. 2009. Small heat-shock protein HspL is induced by VirB protein(s) and promotes VirB/D4-mediated DNA transfer in Agrobacterium tumefaciens. Microbiology 155:3270–3280. doi: 10.1099/mic.0.030676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai YL, Chiang YR, Narberhaus F, Baron C, Lai EM. 2010. The small heat-shock protein HspL is a VirB8 chaperone promoting type IV secretion-mediated DNA transfer. J Biol Chem 285:19757–19766. doi: 10.1074/jbc.M110.110296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trokter M, Felisberto-Rodrigues C, Christie PJ, Waksman G. 2014. Recent advances in the structural and molecular biology of type IV secretion systems. Curr Opin Struct Biol 27C:16–23. doi: 10.1016/j.sbi.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsai YL, Chiang YR, Wu CF, Narberhaus F, Lai EM. 2012. One out of four: HspL but no other small heat shock protein of Agrobacterium tumefaciens acts as efficient virulence-promoting VirB8 chaperone. PLoS One 7:e49685. doi: 10.1371/journal.pone.0049685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghosal A, Babu VMP, Walker GC. 2018. Elevated levels of Era GTPase improve growth, 16S rRNA processing, and 70S ribosome assembly of Escherichia coli lacking highly conserved multifunctional YbeY endoribonuclease. J Bacteriol 200:e00278-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baumgardt K, Gilet L, Figaro S, Condon C. 2018. The essential nature of YqfG, a YbeY homologue required for 3ʹ maturation of Bacillus subtilis 16S ribosomal RNA is suppressed by deletion of RNase R. Nucleic Acids Res 46:8605–8615. doi: 10.1093/nar/gky488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith BA, Gupta N, Denny K, Culver GM. 2018. Characterization of 16S rRNA processing with pre-30S subunit assembly intermediates from Escherichia coli. J Mol Biol 430:1745–1759. doi: 10.1016/j.jmb.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 61.Li ZW, Pandit S, Deutscher MP. 1999. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J 18:2878–2885. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jimenez-Zurdo JI, Robledo M. 2017. RNA silencing in plant symbiotic bacteria: Insights from a protein-centric view. RNA Biol 14:1672–1677. doi: 10.1080/15476286.2017.1356565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davies BW, Walker GC. 2008. A highly conserved protein of unknown function is required by Sinorhizobium meliloti for symbiosis and environmental stress protection. J Bacteriol 190:1118–1123. doi: 10.1128/JB.01521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt-Eisenlohr H, Domke N, Angerer C, Wanner G, Zambryski PC, Baron C. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J Bacteriol 181:7485–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chong L. 2001. Molecular cloning: a laboratory manual, 3rd ed Science 292:446–446. doi: 10.1126/science.1060677. [DOI] [Google Scholar]
- 66.Gelvin SB. 2006. Agrobacterium transformation of Arabidopsis thaliana roots: a quantitative assay. Methods Mol Biol 343:105–113. doi: 10.1385/1-59745-130-4:105. [DOI] [PubMed] [Google Scholar]
- 67.Klüsener S, Aktas M, Thormann KM, Wessel M, Narberhaus F. 2009. Expression and physiological relevance of Agrobacterium tumefaciens phosphatidylcholine biosynthesis genes. J Bacteriol 191:365–374. doi: 10.1128/JB.01183-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 69.Wilms I, Voss B, Hess WR, Leichert LI, Narberhaus F. 2011. Small RNA-mediated control of the Agrobacterium tumefaciens GABA binding protein. Mol Microbiol 80:492–506. doi: 10.1111/j.1365-2958.2011.07589.x. [DOI] [PubMed] [Google Scholar]
- 70.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 71.Porath J. 1992. Immobilized metal ion affinity chromatography. Protein Expr Purif 3:263–281. doi: 10.1016/1046-5928(92)90001-D. [DOI] [PubMed] [Google Scholar]
- 72.Willkomm DK, Minnerup J, Huttenhofer A, Hartmann RK. 2005. Experimental RNomics in Aquifex aeolicus: identification of small noncoding RNAs and the putative 6S RNA homolog. Nucleic Acids Res 33:1949–1960. doi: 10.1093/nar/gki334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaubig LC, Waldminghaus T, Narberhaus F. 2011. Multiple layers of control govern expression of the Escherichia coli ibpAB heat shock operon. Microbiology 157:66–76. doi: 10.1099/mic.0.043802-0. [DOI] [PubMed] [Google Scholar]
- 74.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. 2008. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global posttranscriptional regulator, Hfq. PLoS Genet 4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. 2007. A small noncoding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol Microbiol 66:1174–1191. doi: 10.1111/j.1365-2958.2007.05991.x. [DOI] [PubMed] [Google Scholar]
- 76.Förstner KU, Vogel J, Sharma CM. 2014. READemption: a tool for the computational analysis of deep-sequencing-based transcriptome data. Bioinformatics 30:3421–3423. doi: 10.1093/bioinformatics/btu533. [DOI] [PubMed] [Google Scholar]
- 77.Nicol JW, Helt GA, Blanchard SG, Raja A, Loraine AE. 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.